ABSTRACT
Inflammation has a pronounced impact on the intestinal ecosystem by driving an expansion of facultative anaerobic bacteria at the cost of obligate anaerobic microbiota. This pathogen “blooming” is also a hallmark of enteric Salmonella enterica serovar Typhimurium (S. Tm) infection. Here, we analyzed the contribution of bacterial and host factors to S. Tm “blooming” in a gnotobiotic mouse model for S. Tm-induced enterocolitis. Mice colonized with the Oligo-Mouse-Microbiota (OMM12), a minimal bacterial community, develop fulminant colitis by day 4 after oral infection with wild-type S. Tm but not with an avirulent mutant. Inflammation leads to a pronounced reduction in overall intestinal bacterial loads, distinct microbial community shifts, and pathogen blooming (relative abundance >50%). S. Tm mutants attenuated in inducing gut inflammation generally elicit less pronounced microbiota shifts and reduction in total bacterial loads. In contrast, S. Tm mutants in nitrate respiration, salmochelin production, and ethanolamine utilization induced strong inflammation and S. Tm “blooming.” Therefore, individual Salmonella-specific inflammation-fitness factors seem to be of minor importance for competition against this minimal microbiota in the inflamed gut. Finally, we show that antibody-mediated neutrophil depletion normalized gut microbiota loads but not intestinal inflammation or microbiota shifts. This suggests that neutrophils equally reduce pathogen and commensal bacterial loads in the inflamed gut.
KEYWORDS: Salmonella colitis, microbiota, mouse model, nutritional immunity
INTRODUCTION
Salmonella enterica serovar Typhimurium (S. Tm) is a frequent cause of Salmonellosis in humans with millions of infections worldwide (1). The infection is usually self-limiting; however, in very young, old, or immunocompromised patients, S. Tm can induce life-threatening systemic disease with several thousands of deaths per year (2). The intestinal microbiota efficiently lowers the risk of infection by mediating colonization resistance (CR). When CR is reduced such as upon antibiotic therapy (3), a high-fat meal (4), or an infant or low-complex microbiota (5), S. Tm can invade the gut ecosystem and cause disease. In mice, the infection proceeds in several phases (6): First, S. Tm expands to high numbers in the gut by consuming hydrogen (7), diet- and microbiota-liberated mucosal carbohydrates (8, 9), oxygen (10), anaerobic electron acceptors, and fumarate (11, 12). Only after reaching a certain density (>105 CFU/g), S. Tm invades the gut mucosa at sufficient numbers and triggers intestinal inflammation (13, 14).
Gut inflammation dramatically alters the intestinal environment, leading to drastic changes in microbial community composition and favoring the S. Tm overgrowth (Salmonella “blooming”) over obligate anaerobic commensal bacteria (15, 16). Evidently, Salmonella “blooming” is caused by two contrasting effects of inflammation (17, 18): (i) selective fostering of pathogen growth by nutritional changes and (ii) differential killing of the mostly anaerobic commensal microbiota by innate immune defense mechanisms against which Salmonella is more resistant. In this study, we analyzed the contribution of both bacterial and host factors to Salmonella “blooming” in a gnotobiotic mouse model for S. Tm-induced enterocolitis.
An increase in the concentration of luminal oxygen levels, as well as anaerobic electron acceptors generated by the inflamed mucosa (10, 19), favors S. Tm growth over exclusively fermenting anaerobes (11, 20). This enables the pathogen to consume host-, diet-, and microbiota-derived metabolites, including mucin-derived sugars, ethanolamine, 1,2-propanediol, fructose-asparagine, succinate, and lactate (21–26), and gain a competitive advantage over competing commensals. The production of the siderophore salmochelin, a glycosylated variant of enterochelin that is not bound by the antimicrobial protein lipocalin 2 (LCN-2), also boosts S. Tm in the inflamed gut (27). On the other hand, acute S. Tm inflammation also exerts significant “collateral damage” to the beneficial microbiota (28). The microbiota is inhibited by antimicrobial molecules (27, 29) and bile salts (30), against which Salmonella exhibits a higher degree of resistance.
Moreover, neutrophils that are a hallmark of acute Salmonella-induced inflammation (31, 32) infiltrate the mucosa where they effectively reduce pathogen tissue loads by an NADPH-oxidase-mediated defense (13, 33). Transmigrated live neutrophils, which engulf luminal S. Tm, are present in the gut lumen early after infection (34). Neutrophils also form intraluminal structures (“casts” or pseudomembranes), which encapsulate commensals and thus prevent their epithelial contact and translocation (35). In S. Tm infection, luminal neutrophils impose a drastic bottleneck upon the pathogen population (36) and are likely to also kill commensals. In particular, the neutrophil effector molecules LCN-2, calprotectin, and elastase have been associated with microbiota alterations in response to pathogen-induced inflammation in mice (37, 38).
Overall, selective nutritional use and differential killing sustain long-term S. Tm “blooming” and the establishment of a “supershedder” state, that is, individuals that show high pathogen loads in stool (39). However, the relative importance of these contributing effects to S. Tm “blooming” remains elusive as previous studies focused on individual mechanisms and used different experimental protocols and S. Tm strains. Moreover, it is challenging to disentangle inflammation- and antibiotic-induced changes in the microbiome in models that rely on antibiotic pretreatment. To overcome this limitation, we analyzed the course of S. Tm-induced inflammation in a gnotobiotic infection model that does not rely on antibiotic pretreatment. Mice stably colonized with the Oligo-Mouse-Microbiota (OMM12; 12 mouse-derived commensals) exhibit partial colonization resistance and develop S. Tm gut inflammation after oral infection (40). Members of the OMM12 are well studied with regard to their ecology and metabolic potential and can also be studied in in vitro community culture models (41, 42). Moreover, metabolic and immune phenotypes of OMM12 mice have been extensively characterized in comparison to germ-free and specific pathogen free (SPF) mice (43, 44). Therefore, this model is widely used and steadily gaining ground in research of colonization resistance against pathogens (4, 9, 45) or the functional study of other members of the gut microbial community, including bacteria, phages, and fungi (46–49). Here, we provide a detailed characterization of enteric S. Tm infection and its effect on the gut microbial community in the OMM12 model. We study the course of infection of different S. Tm mutants and assess their ability to cause inflammation-induced S. Tm “blooming.” Finally, we employ a neutrophil depletion protocol to address the role of neutrophils in S. Tm “blooming,” luminal bacterial killing, and microbiota alterations.
RESULTS
Course of oral S. Tm infection in OMM12 mice
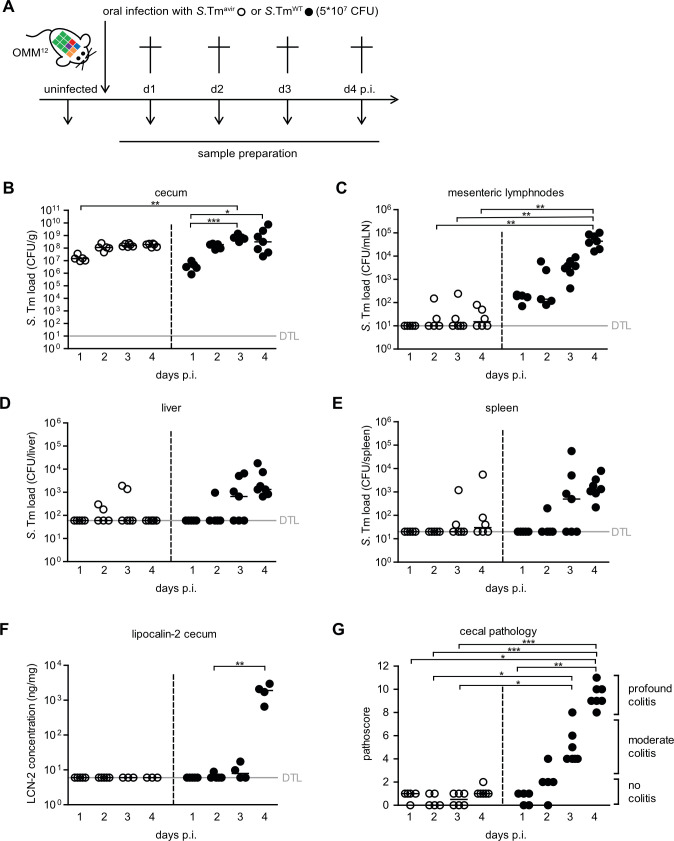
Mice stably colonized with the OMM12 community (OMM12 mice) were infected intragastrically with 5 × 107 CFUs of either wild-type S. Tm (S. TmWT) or an avirulent S. Tm mutant (S. Tmavir; ΔinvG; sseD::aphT) lacking functional Salmonella pathogenicity island (SPI)-1- and SPI-2-encoded type III secretion systems (T3SS). At days 1–4 post infection (p.i.), groups of mice were sacrificed, and intestinal and organ samples were taken for analyses (Fig. 1A). Cecum loads of both S. Tm strains increased steadily from day 1 to day 4 p.i. No difference was observed between S. Tmavir and S. TmWT loads at all tested days (P > 0.05, Kruskal–Wallis test with Dunn’s multiple comparison test; Fig. 1B). Compared to S. Tmavir, S. TmWT loads were higher at day 4 p.i. in the mesenteric lymph nodes (mLNs; P < 0.01, Fig. 1C). No significant difference could be detected in the spleen (P > 0.05; Fig. 1E). Starting at day 3 p.i., S. TmWT caused strong gut inflammation as determined by vastly increased cecal LCN-2 levels (Fig. 1F) and cecal histopathology (Fig. 1G). Mice infected with avirulent S. Tm did not exhibit overt pathological changes in the cecum or increased LCN-2 (Fig. 1F and G).
Fig 1.
Course of enteric S. Tm infection and systemic dissemination. (A) Experimental scheme. OMM12 mice were orally infected with either S. Tmavir or S. TmWT. Mice were sacrificed at days 1, 2, 3, and 4 p.i. (infection with S. Tmavir: day 1 p.i. n = 5, day 2 p.i. n = 5, day 3 p.i. n = 6, and day 4 p.i. n = 6; infection with S. TmWT: day 1 p.i. n = 5, day 2 p.i. n = 5, day 3 p.i. n = 7, and day 4 p.i. n = 7). S. Tm loads in (B) cecal content, (C) mLNs, (D) liver, and (E) spleen were determined by plating. (F) LCN-2 amount in cecal content d4 was determined by enzyme-linked immunosorbent assay (ELISA). (G) Histopathological analysis of cecal tissue. Cecal tissue sections of the mice were stained with hematoxylin/eosin to determine the degree of submucosal edema, neutrophil infiltration, and epithelial damage (1–3: no pathological changes; 4–6: moderate inflammation; and above 7: severe inflammation). Statistical analysis was performed using Kruskal–Wallis test with Dunn’s multiple comparison test (*P < 0.05, **P < 0.01, and ***P < 0.001). Each dot represents one mouse, black lines indicate median, and gray lines indicate detection limit (DTL).
S. Tm WT infection leads to pathogen blooming, a decrease in gut luminal bacterial load, and community shifts
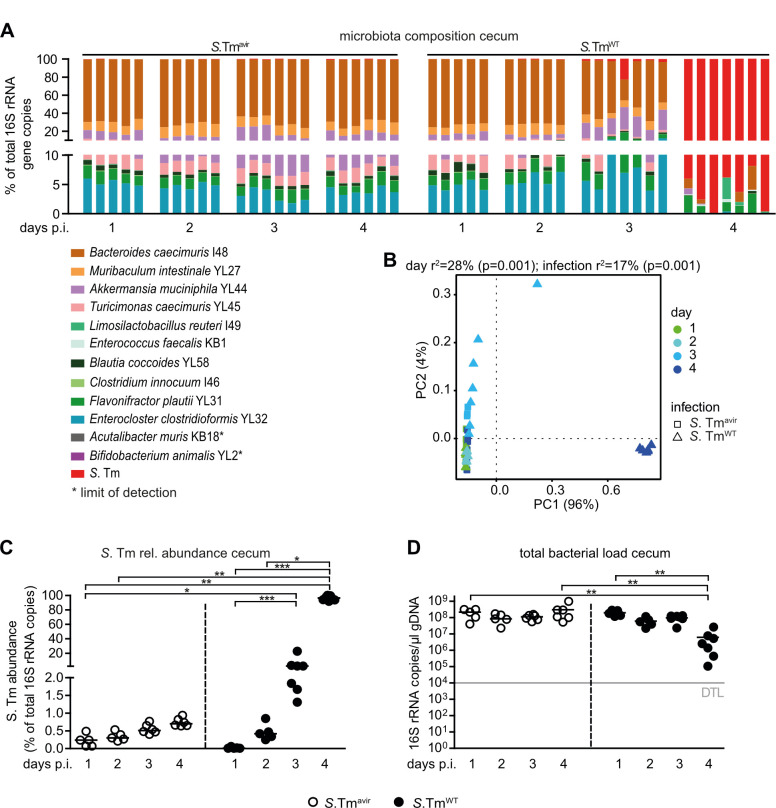
Next, we determined the OMM12 community composition including S. Tm loads in cecum content (Fig. 2; Fig. S1) and feces (Fig. S2) by strain-specific quantitative PCR (qPCR). Calculations of relative abundance revealed that S. TmWT dominated (rel. abundance >50%) the bacterial community in feces and cecum content by day 4 p.i. (Fig. 2A through C; Fig. S2A through C), coinciding with developing gut inflammation (Fig. 1F and G). In contrast, no significant increase in relative abundance over time was observed for S. Tmavir (Fig. 2A through C; Fig. S2A through C). ADONIS permutational multivariate analysis of variance (PERMANOVA) analysis on the Bray–Curtis distance metric revealed significant differences of microbial community composition by day and between S. TmWT- and S. Tmavir-infected mice in feces and cecal content (Fig. 2AB; Fig. S2A and B; Tables S1 and S2). Strikingly, S. TmWT infection and resulting inflammation led to a drastic decrease in total gut bacterial loads determined by 16S rRNA copies (Fig. 2D; Fig. S2D). This was the case for the majority of OMM12 strains but Enterococcus faecalis KB1 (Fig. S1).
Fig 2.
S. TmWT causes pronounced shifts in microbiota composition at day 4 p.i. in OMM12 mice. (A) Analysis of microbiota composition in cecal content. Microbiota composition was determined by strain-specific qPCR assay and is shown as relative abundances of the individual strains (% of cumulated 16S rRNA gene copy numbers). (B) Principal coordinates analysis (PCoA) based on the distance matrix of Bray–Curtis dissimilarity of relative OMM12 abundance profiles shows the effect of time after infection. Points are colored by time (days) after infection. (C) Relative abundance of S. Tm in cecal contents at different time points. (D) Absolute amount of 16S rRNA gene copies (determined by a universal primer/probe combination). Statistical analysis was performed using Kruskal–Wallis test with Dunn’s multiple comparison test (*P<0.05, **P < 0.01, and ***P < 0.001). Each dot represents one mouse, black lines indicate median, and gray lines indicate DTL.
We also infected OMM12 mice with S. Tm mutants in either the SPI-1 (S. TmΔSPI-1)- or the SPI-2 (S.TmΔSPI-2)-encoded T3SS for 4 days. We compared S. Tm loads and microbiota composition to S. TmWT and S. Tmavir data shown in Fig. 1. S. TmΔSPI-1 and S. TmΔSPI-2 colonized the intestine at largely similar levels (Fig. S3A). Interestingly, systemic S. Tm loads were similar between the mutants (Fig. S4A through C). S. TmΔSPI-2 (triggering gut inflammation via the SPI-1 T3SS) induced more cecal pathology and higher S. Tm abundance compared to the SPI-1-deficient mutant although this difference was not statistically significant (Fig. S3B and C). Microbiota composition in the cecum content was significantly different between all groups, as determined by ADONIS PERMANOVA analysis on the Bray–Curtis distance metric (Fig. S3E; Table S3). Blooms were only evident in S.TmΔSPI-2- and S. TmWT-infected mice (Fig. S3F). The total bacterial load in S.T mΔSPI-1- and S. TmΔSPI-2-infected mice was altered when compared to S. Tmavir but not significantly changed when compared to the S. TmWT-infected group (Fig. S3G). This applied also for most of the individual OMM12 strains (Fig S4D through O). Taken together, S. TmWT induced strong gut inflammation in OMM12 mice, beginning at day 3 p.i. and depending on the activity of both Salmonella T3SS. Severe inflammation at day 4 p.i. was paralleled with S. TmWT blooming, drastic reduction of total gut luminal bacterial loads, and overall changes in microbiota composition.
Anaerobic respiration and ethanolamine utilization fuel S. Tm blooms and dysbiosis
To analyze the relevance of anaerobic and aerobic respiration for S. Tm blooming, we generated S. Tm mutants deficient in nitrate respiration (S. TmNi.; ΔnarZ; narG::cat; napA::aphT) and in nitrate and tetrathionate respiration (S. TmNi. + Te.; ΔnarZ; narG::cat; napA::aphT; ttrS::tet). Moreover, to probe the role of siderophore-mediated iron acquisition and ethanolamine utilization, we generated S. Tm entA::cat (S. TmEntA) and eutC::aphT mutant (S. TmEA) strains. In addition, we created an S. Tm cyxA mutant, attenuated in aerobic respiration by the deletion of the cytochrome bd-II oxidase gene (S. Tmcyx; ΔcyxA).
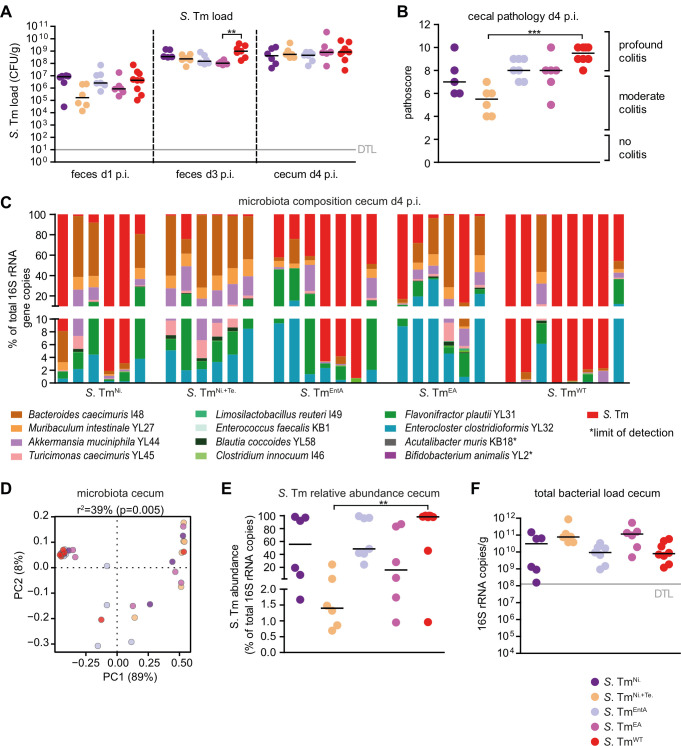
We infected groups of OMM12 mice with 5 × 107 CFUs of the mutant strains or S. TmWT. Fecal samples were collected on different days after infection in order to monitor the course of intestinal S. Tm colonization. Mice were sacrificed at day 4 p.i., cecal content was harvested for microbiota analysis, inflammation was quantified, and systemic Salmonella loads were determined (Fig. 3). S. Tm strains colonized the gut of OMM12 mice equally well (Fig. 3A). Only S. TmEA showed significantly reduced loads at day 3 p.i. compared to S. TmWT (P < 0.01, Kruskal–Wallis test with Dunn’s multiple comparison test; Fig. 3A). There was no difference in systemic Salmonella loads at day 4 p.i. between groups (Fig. S5). S. TmWT, S. TmEA, and S. TmEntA induced profound colitis by day 4 p.i., while S. TmNi. + Te induced significantly less colitis symptoms (P < 0.001, Fig. 3B). For S. Tmcyx, no significant differences could be detected in pathogen loads in intestine or organs in comparison to S. TmWT (Fig. S6). PERMANOVA analysis with ADONIS on the Bray–Curtis distance metric revealed significant differences between the cecal community structures of S. TmNi. + Te and S. TmWT or S. TmEntA-infected mice (Fig. 3D; Table S4) and abundance of most OMM12 bacteria was increased in S. TmNi. + Te-infected mice (Fig. S7). For S. Tmcyx, no significant difference was found with respect to microbiota composition in comparison to S. TmWT (Fig. S6 and S7; Table S5). S. TmNi. + Te mutant did not establish “blooms,” while relative abundances of the other S. Tm mutants were similar to S. TmWT (Fig. 3E), and total bacterial loads were increased in mice infected with S. TmNi. + Te compared to S. TmWT-infected mice (Fig. 3F).
Fig 3.
Course of infection of S. Tm mutants in anaerobic respiration, salmochelin production, and ethanolamine utilization in OMM12 mice. OMM12 mice were orally infected with different S. Tm mutant strains: S. TmNi. (n = 6), S. TmNi. + Te. (n = 6), S. TmEntA (n = 7), S. TmEA (n = 6), or S. TmWT (n = 8). Mice were sacrificed at day 4 p.i. (A) S. Tm loads in feces and cecal content at days 1, 3, and 4 p.i. were determined by plating. (B) Histopathological analysis of cecal tissue at day 4 p.i. Cecal tissue sections of the mice were stained with hematoxylin/eosin to determine the degree of submucosal edema, neutrophil infiltration, and epithelial damage (1–3: no pathological changes; 4–6: moderate inflammation; and above 7: severe inflammation). (C) Analysis of microbiota composition in cecal content. Microbiota composition was determined by strain-specific qPCR assay and is shown as relative abundances of the individual strains (% of cumulated 16S rRNA gene copy numbers). (D) PCoA based on the distance matrix of Bray–Curtis dissimilarity of relative OMM12 abundance profiles shows the effect of the different S. Tm mutant strains. Points are colored by time (days) after infection. (E) Relative abundance of S. Tm and (F) absolute amount of 16S rRNA gene copies (determined by a universal primer/probe combination) in cecal contents 4 days p.i. Statistical analysis was performed using Kruskal–Wallis test with Dunn’s multiple comparison test (*P < 0.05, **P < 0.01, and ***P < 0.001). Each dot represents one mouse, black lines indicate median, and gray lines indicate DTL.
Neutrophils reduce overall luminal bacterial loads but do not promote Salmonella “blooms”
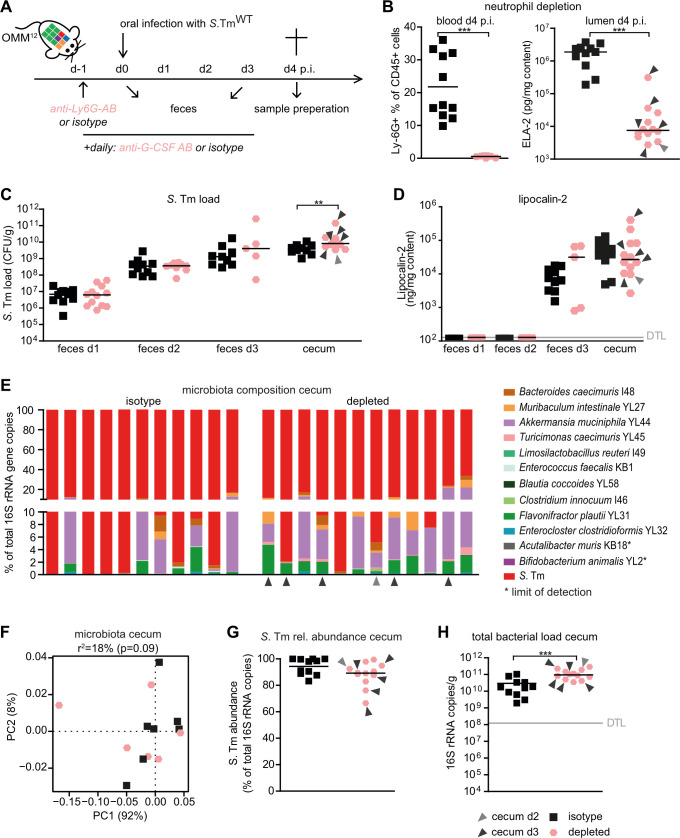
Next, we set out to test the hypothesis that neutrophils are the cause of decreased luminal bacterial loads in S. TmWT-infected OMM12 mice at day 4 p.i. Therefore, we employed a neutrophil depletion protocol using α-Ly6G and α-G-CSF antibodies or isotype control antibodies in OMM12 mice that were additionally infected with S. TmWT (Fig. 4A). Antibody-mediated depletion was confirmed by fluorescence activated cell sorting (FACS) analysis of blood samples gating on CD45+, SYTOX-, CD3-, CD11b+, Ly-6G+, and Ly-6Cintermediate cells and by quantifying neutrophil-elastase in cecal content (Fig. 4B). Only mice with confirmed neutrophil depletion (<4% Ly6G+/CD45+ cells) were included in the analysis.
Fig 4.
Neutrophils reduce the total gut luminal bacterial loads but are dispensable for the induction of Salmonella “blooms” and dysbiosis. (A) Experimental scheme. OMM12 were treated with one dose of α-Ly6G antibody or isotype control 1 day before infection with S. TmWT. In addition, α-G-CSF antibody or isotype control was daily administered starting from day −1 until day 3 p.i. All antibodies were given via the intraperitoneal route. (B) Monitoring of neutrophil depletion at day 4 p.i. in blood by FACS gating on CD45+, SYTOX-, CD3-, CD11b+, Ly-6G+, and Ly-6C-intermediate cells or in cecal content by neutrophil elastase-specific ELISA. FACS data are indicated as the mean. Statistical analysis was performed using the Mann–Whitney U test. (C) S. Tm loads in feces at days 1, 2, and 3 and cecal content at day 4 p.i. were determined by plating. (D) LCN-2 levels in feces and cecal content (ng/mg content). LCN-2, elastase levels, and CFUs are shown as median, and data were analyzed using the Mann–Whitney U test. (E) Analysis of cecal microbiota composition at day 4 p.i. with different S. TmWT and neutrophil depletion or isotype control treatments. Microbiota composition is shown as relative abundance and expressed as % of cumulated 16S rRNA gene copy numbers. (F) PCoA based on the distance matrix of Bray–Curtis dissimilarity of relative OMM12 abundance profiles shows the effect of the different treatments. (G) Relative abundance of S. Tm and (H) absolute amount of 16S rRNA gene copies (determined by a universal primer/probe combination) in cecal contents 4 days p.i. Light and dark gray arrows indicate samples analyzed at day 2 or 3 p.i., respectively. Statistical analysis was performed using the Mann–Whitney U test (*P < 0.05, **P < 0.01, and ***P < 0.001). Each dot represents one mouse, black lines indicate median, and gray lines indicate DTL.
Some neutrophil-depleted mice developed severe infection and had to be euthanized at day 2 or 3 p.i. (indicated with gray and black arrows). Cecal S. TmWT loads were similar in both groups of mice at days 1–3 p.i. but significantly higher in neutrophil-depleted mice at day 4 p.i. (Fig. 4C). At the point of sacrifice, all mice developed pronounced gut inflammation as determined by LCN-2 levels (Fig. 4D). Systemic loads of S. TmWT were higher in the mLN, spleen, and liver of neutrophil-depleted mice (Fig. S8A through C).
There was no apparent difference in the overall cecal microbiota composition between the groups at day 4 p.i. (Fig. 4E and F; Table S6). Moreover, no difference in rel. S. TmWT abundance was observed between the groups (Fig. 4G). However, a lower overall amount of total gut luminal bacteria was observed in the isotype group (Fig. 4H copies/mg content isotype vs depleted P < 0.001, Mann–Whitney U test), indicating that neutrophils are causal for inflammation-associated reduction of gut luminal bacterial loads in S. Tm-infected OMM12 mice. Reduction was most pronounced for Bacteroides caecimuris I48, Muribaculum intestinale YL27, Akkermansia muciniphila YL44, Turicimonas muris YL45, Blautia coccoides YL58, Flavonifractor plautii YL31, and Enterocloster clostridioformis YL32—other bacteria were less affected (Fig. S8D through O).
DISCUSSION
Together, the host and its intestinal symbionts establish microbiota-nourishing immunity that forms an efficient barrier against S. Tm infection (50). Epithelial hypoxia, short-chain fatty acid production, and depletion of growth substrates are cornerstones of this barrier. Using its virulence factors, S. Tm can overcome colonization resistance, trigger inflammation, and alter the gut luminal environment to benefit from it—at the expense of intestinal commensal bacteria. Inflammation-driven pathogen “blooming” has a major impact on S. Tm dissemination and evolution (51). Overgrowth of related Enterobacteriaceae promotes the exchange of conjugative plasmids (52) and phages (53). This increases the chance of spreading antibiotic resistances and fosters evolution of highly virulent strains (54), emphasizing the broad implications of S. Tm adaptation of the inflammatory milieu in the mammalian gut. Here, we analyze the extent to which S. Tm-induced intestinal inflammation, virulence, and gut luminal neutrophils contribute to alterations in the gut microbiota loads and community profiles in OMM12 mice, a widely used infection model (4, 9, 55) based on a well-characterized minimal bacterial community (56).
Due to the partial colonization resistance phenotype of OMM12 mice, wild-type S. Tm induced severe gut inflammation within 4 days after oral infection without antibiotic pretreatment. Mice were characterized by an overall reduction of luminal bacterial loads and S. Tm blooming (i.e., S. Tm being the dominant species). This overgrowth was not observed with an avirulent mutant (ΔinvG ΔssaV; S. Tmavir), which can colonize the gut but is defective in inducing inflammation. This confirms the results from previous studies in the streptomycin-treated mouse colitis model and highlights the pronounced negative impact of intestinal inflammation on the majority of intestinal commensals while fostering pathogen overgrowth (15).
Comparing the course of infection in S. Tm TTSS mutants, we found that S. TmΔSPI-1 was more strongly attenuated in inducing gut inflammation than S. TmΔSPI-2 at day 4 p.i. This suggests that in OMM12 mice, the SPI-1 TTSS contributes more to overall inflammation than the SPI-2 TTSS. Previous work in the streptomycin model showed that SPI-1 triggers inflammation within hours after infection. This response is due to effector proteins that initiate a complex immunological signaling cascade, culminating in the production of IFNγ (57–61). In contrast, in OMM12 mice, overt inflammation starts only by day 3 p.i. (Fig. 1F and G), which is mainly due to initially lower S. Tm loads (<107 CFU/g) compared to streptomycin-treated mice (>108 CFU/g). Mutants lacking a functional SPI-1 or a SPI-2 TTSS caused equally attenuated colitis at day 4 p.i. However, at early time points postinfection (days 1 and 2 p.i.), the S. TmΔSPI-1 mutant is strongly attenuated and inflammation triggered via the SPI-2 TTSS only developed at day 3 p.i. (62).
S. Tm mutants in anaerobic respiration as well as iron uptake via salmochelin and ethanolamine utilization were previously shown to be attenuated in growth in the inflamed gut in direct competition with S. TmWT (11, 20, 27, 63). In this study, we aimed to identify mechanisms that promote S. Tm blooming in competition against the OMM12 microbiota. We show that a double mutant in anaerobic nitrate and tetrathionate respiration (S. TmNi+Te) has a reduced capacity to bloom and induce microbial community shifts. As gut inflammation in S. TmNi+Te-infected mice was also significantly reduced at day 4 p.i., we cannot conclude that anaerobic respiration creates an advantage in competition against OMM12 in the inflamed gut. In contrast, mutants in O2 and nitrate respiration, ethanolamine utilization, and salmochelin production (S. Tmcyx, S. TmNi, S. TmEA, and S. TmEntA) induced strong inflammation, bloomed, and altered the microbiota composition to a similar extent as S. TmWT. Therefore, these mechanisms are, at least individually, not required for competition against OMM12 bacteria in the inflamed gut. In the previous work using the streptomycin-treated colitis model, S. Tmcyx was strongly attenuated in intestinal colonization (10). While the phenotype of partial colonization resistance of OMM12 mice was key for conducting the experiments without previous microbiota perturbation, it also has its limitations. The OMM12 microbial community lacks key functional members, such as Escherichia coli or other facultative anaerobic bacteria that compete with S. Tm for nutrients and O2 in the inflamed gut upon recovery from streptomycin treatment. To this end, a more complex microbial model community such as the recently established human-derived defined complex communities might be used (64).
Our data indicate that neutrophils protect the host from systemic Salmonella spread and reduce total gut luminal bacterial loads but do not contribute to Salmonella “blooms” and concomitant dysbiosis. Notably, antibody-mediated depletion of neutrophils resulted in the reduction of LCN-2 and elastase-2 levels and concomitantly resulted in an overall increase in total gut luminal bacterial loads. This confirms the pronounced antibacterial function of transmigrated neutrophils in the gut lumen reported previously (34, 35, 37). In contrast, neutrophil depletion did not change the relative abundance of gut luminal S. Tm. This suggests that neutrophil-derived antimicrobial effectors equally reduce numbers of S. Tm and members of the OMM12 and are not causally involved in S. Tm blooming in this model. Rather, “blooming” is mainly mediated by the pathogens’ diverse capabilities of taking advantage of the altered substrate range in the inflamed intestine.
Gnotobiotic OMM12 mice are increasingly used as a standardized infection model for S. Tm and other pathogenic Enterobacteriaceae (4, 9, 45). Here, we present a detailed characterization of the course of S. TmWT infection and mutants in major virulence factors. Our study reveals that OMM12 mice infected with S. TmWT develop fulminant colitis by day 4 p.i. characterized by pathogen blooming and shifts in microbiota composition. We conclude that OMM12 mice are a valuable model to dissect the impact of S. TmWT-induced inflammation on the gut ecosystem, including all its members, without the need for previous microbiota perturbation.
MATERIALS AND METHODS
Bacterial strains
For detailed information about individual Oligo-MM12 strains refer to reference (40). All Oligo-MM12 strains are part of the mouse intestinal bacteria collection [miBC (65)] and are also available via the German Collection of Microorganisms and Cell Cultures (www.dsmz.de/miBC). Salmonella strains used and created in this study are based on S. Tm strain SL1344 (Table 1).
TABLE 1.
Bacterial strains used in this study
| Strain | Strain ID | Genotype | Reference |
|---|---|---|---|
| S. TmWT | SB300 | S. Tm strain SL1344 | (66) |
| S. TmAvir | M557 | ΔinvG; sseD::aphT | (67) |
| S. TmSPI-1 | SB161 | ΔinvG | (68) |
| S .TmSPI-2 | M556 | sseD::aphT | (67) |
| MBE1 | MBE1 | narZ::cat | This study |
| MBE2 | MBE2 | narG::cat | This study |
| MBE3 | MBE3 | napA::aphT | This study |
| MBE4 | MBE4 | narZ::cat | This study |
| MBE5 | MBE5 | ΔnarZ | This study |
| MBE6 | MBE6 | ΔnarZ; narG::cat | This study |
| S. TmNi. | MBE7 | ΔnarZ; narG::cat; napA::aphT | This study |
| S. TmNi. + Te. | MBE8 | ΔnarZ; narG::cat; napA::aphT; ttrS::tet | This study |
| MBE9 | MBE9 | entA::cat | This study |
| S. TmEntA | MBE10 | entA::cat | This study |
| MBE11 | MBE11 | eutC::aphT | This study |
| S. TmEA | MBE12 | eutC::aphT | This study |
Construction of S. Tm strains and plasmids
All bacterial strains used in this study are listed in Table 1. S. TmNi. (MBE7: ΔnarZ, narG::cat, napA::aphT) was generated in a stepwise manner. First, single S. Tm mutant strains were generated (MBE1: narZ::cat, MBE2: narG::cat and MBE3: napA::aphT) using λ red recombination as described previously (69). Briefly, antibiotic resistance genes as well as flippase recognition target (FRT) sites were PCR-amplified using specific knock out primers (ko-primers, Table 2) and the plasmids pKD3 (cat) or pKD4 (aphT) listed in Table 3. PCR products were subsequently electroporated into S. TmWT harboring pKD46 (Table 3). The narZ::cat allele from MBE1 was transduced to S. TmWT by P22-transduction (70) in order to create MBE4. The narZ::cat was then deleted using flippase (FLP)-recombinase encoded on pCP20 (Table 3). The resulting strain MBE5 (ΔnarZ) was transduced with P22-phage lysate of MBE2 in order to create MBE6 (ΔnarZ, narG::cat). MBE6 was finally transduced with P22-phage lysate of MBE3 to generate MBE7 (ΔnarZ, narG::cat, napA::aphT). In order to construct S. TmNi.Ni. + Te. (MBE8: ΔnarZ, narG::cat, napA::aphT, ttrS::tet), the ttrS::tet allele from S. Tm stain M961 [ΔsodCI, ΔsodCII, BCB4::tet (71)] was P22-transduced into MBE7. S. TmEntA (MBE10: entA::cat) was created by P22-transduction of the entA::cat allele from MBE9 to S. TmWT. MBE9 was previously generated by λ red recombination. S. TmEA. (MBE12: eutC::aphT) was constructed by P22-transduction of the eutC::aphT allele from MBE11 to S. TmWT. MBE11 was generated before using λ red recombination. In all cases, genetic modifications were verified by PCR using gene-specific check-up primers (Table 2). The construction and sequences of plasmids harboring full-length 16S rRNA sequences used as standard for qPCR reactions are detailed in reference (40).
TABLE 2.
Primers used for construction of Salmonella mutantsa
| Designation | Sequence 5′–3′ | Construction of | Amplicon size (bp) |
|---|---|---|---|
| narZ-fwd-ko | AATGGTTTAACGCCAAATCGACAGGATGGCGGAAAATTTATCGAAGCAGGAGAAATGTC atatgaatatcctccttagtt | ||
| narZ-rev-ko | GGTGTGACAGCCAATACATTTATCGAGATTCAGTACCATCCCAACCTGTGAGCGTATTT tgtgtaggctggagctgcttc | MBE1 (narZ::cat) | |
| narZ fwd-check up | GCGTCGTAAGCCTAAACA | ||
| narZ rev-check up | CCGGTCCAGACGTTTTTA | 3,948 | |
| narG-fwd-ko | CTTAGTTAAGCAATGTCGATTTATCAGAGAGCCGTAAGGTTCCACACAGGAGAAACCCG atatgaatatcctccttagtt | ||
| narG-rev-ko | GGTATGACAGCCGATGCACTTATCGAGATTCAGCACCATGCCGACTTGTGAACGAATTT tgtgtaggctggagctgcttc | MBE2 (narG::cat) | |
| narG fwd-check up | TTCTCACGCCCATTCAGC | ||
| narG rev-check up | CCAGACGTTTTTACAGGT | 3,921 | |
| napA-fwd-ko | GGCGTACTGGCGGTGTCGCTGGTTTATCACCAGCAGGATGAGCAAGGTGAGGAAACACCatatgaatatcctccttagtt | ||
| napA-rev-ko | ACATCACGCAGGAAGCGGCGGCGGCCATTTTGGGGTTTCGCTGTACGGGACATAACGCG tgtgtaggctggagctgcttc | MBE3 (napA::aphT) | |
| napA fwd-check up | ACAATTGAGTCAGTACGC | ||
| napA rev-check up | GCTGGCGAAGACATTTTC | 2,756 | |
| ttrS fwd-check up | CGGCTTGTTGTTGATCTAA | ||
| ttrS rev-check up | CCCAGACTTTCCAGTAAAA | 1,873 | |
| entA-fwd-ko | CTGGCGAAAAACCCGACCATTGACGCCTGGTGGGCGCTGCTTTCTCGCGGGGTAGAGTA atatgaatatcctccttagtt | ||
| entA-rev-ko | AGTGTTCTGACTGGTAGCGTTCAATTCATCCAGCGTTAAATGCCGTTTCCAGATCATCG tgtgtaggctggagctgcttc | MBE9 (entA::cat) | |
| entA fwd-check up | GTAAAGTACACGGCGATAT | ||
| entA rev-check up | TAAACAATGCCCAGATGCG | 932 | |
| eutC-fwd-ko | GTCTGACCAAACGGGCGGGCGATCCGTCACTGTTCTTCTGATGACGCGGGGATAACACC atatgaatatcctccttagtt | ||
| eutC-rev-ko | ATCACGCGCATGGCAGTCACTGAAGGTCGAATTAAATCTAATGCAGGCATGATGTCTCC tgtgtaggctggagctgcttc | MBE11 (eutC::aphT) | |
| eutC fwd-check up | CTGGAAACGATGGGCATTAT | ||
| eutC rev-check up | TAATTTAAGTTCCCGCGCAA | 1,086 |
Genes of interest were replaced by either chloramphenicol (cat) or kanamycin (aphT) resistance genes using the λ red recombination system (69). PCR products were generated with primers harboring a homology region adjacent to the gene of interest (capital letters) and a sequence targeting the plasmids pKD3 (cat) and pKD4 (aphT, small letters). Check-up primers were used to verify the correct insertion of the antibiotic cassette.
TABLE 3.
Plasmids used for λ red and FLP recombination
| Plasmid | Function | Genotype | Reference |
|---|---|---|---|
| pKD3 | Harbors a chloramphenicol resistance gene (cat) | cat with FRT sites, AmpR | (69) |
| pKD4 | Harbors a kanamycin resistance gene (aphT) | aphT with FRT sites, AmpR | (69) |
| pKD46 | Harbors 2,154 nt (31088–33241) of phage λ, enzymes for recombination | araC, ParaB γ, β and exo, repA101ts, AmpR | (69) |
| pCP20 | Harbors FLP-recombinase | FLP+, λ cI857+, λ pR Repts, AmpR, CmR | (72) |
Culture conditions and bacterial in vitro assays
Culture conditions as well as cryopreservation protocols of individual Oligo-MM12 strains are described in reference (40). For mouse infection, a single colony of an S. Tm strain was inoculated in 3 mL of lysogeny broth (LB) medium containing 0.3 M NaCl (LB0.3) and grown on a wheel rotor for 12 h at 37°C. The starter culture was diluted in a 1:20 ratio in fresh LB0.3 and incubated for another 4 h at 37°C under constant rotation. The subculture was washed in ice-cold phosphate-buffered saline (PBS), and the pellet was subsequently re-suspended in PBS. Mice were finally gavaged with 5 × 107 CFUs of S. Tm strains. For investigating phenotypes conferred by gene knock outs, individual colonies of S. Tm strains were aerobically inoculated with 5 mL of M9 medium (Na2HPO4 · 2H2O 40 mM, KH2PO4 20 mM, NaCl 9 mM, NH4Cl 37.39 mM, D-glucose 1.1 mM, MgSO4 1 mM, CaCl2 100 µM, Thiamine 10 mg/mL, and Histidine 500 mg/L) without antibiotics on a wheel rotor for 12 h at 37°C. Aerobic as well as anaerobic (7% H2, 10% CO2, and rest N2) subcultures with a starting OD600 of 0.02 were subsequently carried out in 10 mL of M9 medium with and without nitrate (20 mM). For testing S. Tm strains deficient in ethanolamine utilization, the M9 medium was additionally supplemented with ethanolamine (5 mM) and tetrathionate (40 mM). Aerobic subcultures were performed in glass Erlenmeyer flasks sealed with aluminum foil, whereas anaerobic subcultures were conducted in 100 mL Wheaton glass serum bottles with pre-reduced medium (pre-reduction for at least 2 days under anoxic conditions: 3% H2 and rest N2). Subcultures were incubated at 37°C and shaken at 180 rpm, and samples were taken for OD600 measurement and determination of nitrate concentrations (73). In vitro competition assays were performed as described in reference (74). Briefly, starter cultures of the competing strains in 5 mL of LB or M9 medium without antibiotics incubated on a wheel rotor for 12 h at 37°C were mixed in a 1:1 ratio with a starting OD600 of 0.05 of each strain and used to inoculate 14 mL cultures with either plain mucin broth (Type II porcine mucin 2.5 g/L, Morpholino propanesulfonic acid sodium 9.25 g/L, MgSO4 2.2 mM, trace elements 1,000×: CaCl2 · 2H2O 0.3 mM, ZnSO4 · 7H2O 0.1 mM, FeSO4 · 7H2O 0.045 mM, Na2SeO3 0.2 mM, Na2MoO4 · 2H2O 0.2 mM, MnSO4 · H2O 2 mM, CuSO4 · 5H2O 0.1 mM, CoCl2 · 6H2O 3 mM, and NiSO4 · 6H2O 0.1 mM) or mucin broth containing nitrate (40 mM). For competition assays underlining the importance of tetrathionate respiration, the mucin broth was supplemented with tetrathionate (40 mM) and histidine (500 mg/L) to enhance growth. Cultures were statically incubated in 15 mL plastic tubes for 24 h at 37°C. CFUs of S. Tm strains were determined by plating on LB agar plates containing selective antibiotics. The deficiency of acquiring iron via siderophores was investigated using Chromazurol S (CAS) agar. Next, 1 mL of starter cultures in 5 mL of LB medium without antibiotics incubated on a wheel rotor for 12 h at 37°C was centrifuged, and the pellet was re-suspended in sterile ice-cold PBS. Then, 5 µL were adjusted to an OD600 of 0.1, spotted on CAS agar plates, and incubated at 37°C for 48 h. The diameters of the colony as well as the diameter of the colony + the orange CAS halo were measured, and the CAS-reactive ring was calculated in mm according to the formula: 0.5× [(diameter of colony + CAS halo) – diameter of colony].
DNA extraction from intestinal contents
Fecal and cecal DNA were extracted using the QIAamp DNA Stool Mini Kit (Qiagen) according to the manufacturer’s protocol with modifications. The protocol was extended with an initial bead-beating step using glass beads (0.5–0.75 mm) and zirconia beads (<100 µm). Lysozyme (20 mg/mL) was additionally added to the lysis buffer.
qPCR of 16S rRNA genes
16S rRNA-specific primers and hydrolysis probes were designed and validated according to the Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines as detailed previously (40, 75).
qPCR reactions were performed in a LightCycler96 thermo cycler (Roche) using white LightCycler 480 Multiwell Plate 96 plates (Roche). One 20 µL reaction mixture contained 250 nM of hydrolysis probe, 300 nM of each corresponding primer, 5 ng of template genomic DNA (gDNA), water, and FastStart Essential DNA Probes Master (Roche). The sequences of primers and hydrolysis probes, the qPCR efficiencies, as well as the specificity/detection limits of qPCR assays are depicted in reference (40). All qPCR reactions were performed in duplicate applying the following conditions: 95°C for 10 min, followed by 45 cycles of 95°C for 15 s and 60°C for 1 min. Fluorescence for each cycle was recorded after the step at 60°C. Quantification cycle as well as the baseline were automatically determined by the software LightCycler 96 version 1.1 (Roche).
Animal experiments
Oligo-MM12 mice were generated as previously described (40) and sterilely bred in the animal housing facility of the Max von Pettenkofer-Institute. Sex- and age-matched mice were orally infected with 5 × 107 CFUs of S. Tm strains under germ-free conditions and maintained in gnotocages (Han, Bioscape). The protocol for neutrophil depletion was adapted from reference (36). Briefly, neutrophils were depleted by one intraperitoneal (i.p.) injection with 150 µg of α- m Ly-6G (clone: 1A8, BioXCell) prior to infection with S. TmWT followed by daily i.p. injections with 10 µg of α-mouse G-CSF (clone: 67604, R&D Systems). Control mice received 150 µg of Rat IgG2a (clone: 2A3, BioXCell) once and daily injections with 10 µg of Rat IgG1 (clone: 43414, R&D Systems). Antibodies were dissolved in sterile PBS. Mice were euthanized by cervical dislocation, and organs were aseptically removed. Salmonella loads in mesenteric lymph nodes, liver, spleen, as well as in cecal content and feces were determined by plating on MacConkey agar plates (Roth). The mouse LCN-2/NGAL detection kit (R&D Systems), ELISA kit, and horseradish peroxidase (HRP) Streptavidin (BioLegend) were used for measuring LCN-2 levels. Neutrophil-specific elastase was quantified using the Mouse Neutrophil Elastase/ELA2 DuoSet ELISA kit and the DuoSet Ancillary Reagent Kit2 (both from R&D Systems). Cecal pathology was scored at necropsy. Optimal cutting temperature compound (Sakura, Torrance) was used to embed cecal tissue. Tissues were flash frozen and cut into 5 µm cryosections. Cryosections were hematoxylin and eosin (H&E)-stained and scored as detailed in reference (76). Scoring parameters include submucosal edema, infiltration, loss of goblet cells, as well as epithelial damage. Scoring was performed in a blinded manner and yielded a total score of 0–13 points according to the severity of inflammatory symptoms (scores: 0–3: no pathological change, 4–8: mild inflammation, and 9–13: severe inflammation). All animal experiments were approved by the local authorities (Regierung von Oberbayern) and were performed according to the legal requirements.
FACS
Efficient depletion of neutrophil in the blood was monitored by FACS. Three to four drops of blood taken from the tail vein were collected in 1 mL of pre-cooled FACS buffer (PBS and 1% heat-inactivated FACS). For subsequent erythrolysis, samples were centrifuged and FACS buffer was exchanged with 1 mL of BD FACS Lysing Solution (BD). After 10 min of incubation in the dark at RT, samples were centrifuged again, Lysing Solution was discarded, and cells were resuspended in 100 µL of FACS buffer. Cells were subsequently stained with α-CD45-PerCP (clone: 30-F11, BioLegend, 1:100), α-CD11b-APC-Cy7 (clone: M1/70, BioLegend, 1:200), α-Ly-6G-Pacific Blue (clone: 1A8, BioLegend, 1:200), α-Ly-6C-FITC (clone: AL-21, BD, 1:400), α-CD3-PE (clone: 17A2, BioLegend, 1:200), and α-CD16/CD32 (93, FC-Block, eBioscience, 1:100). Following 30 min of incubation at 4°C, cells were washed and analyzed with the FACSCanto II (BD). SYTOX red (5 nM) was used to discriminate dead and living cells. Data were recorded using the BD FACSDiva software (BD) and analyzed using the FlowJo software. Neutrophils were identified as CD45+, SYTOX-, CD3-, CD11b+, Ly-6G+, and Ly-6C-intermediate cells.
Statistical analysis
CFU data, LCN-2 levels, and pathoscores were expressed as median. More than two different groups were compared to each other using Kruskal–Wallis test, with Dunn’s multiple comparison test. Two groups were compared using the Mann–Whitney U test (Prism 5; GraphPad Software, San Diego, CA, USA). The percentage of individual bacteria was expressed as mean ± SD. Differences between individual bacteria were compared using a two-way ANOVA, with Bonferroni posttest (Prism 5; GraphPad Software, San Diego, CA, USA). FACS data were expressed as the mean and analyzed using unpaired t test (Prism 5; GraphPad Software, San Diego, CA, USA).
To analyze clustering of qPCR data, Pearson distance matrix was used containing dissimilarity values for each pairwise comparison. Strength and statistical significance of sample grouping were determined by applying the nonparametric Adonis method based on PERMANOVA, together with the parametric significance test PERMDISP, which analyzes multivariate homogeneity of group dispersions. The used scripts are available in QIIME (77).
In all cases, P values < 0.05 were considered as statistically significant.
Fold changes in absolute abundance were calculated with absolute values that were normalized to a million gene copies determined by universal probe.
ACKNOWLEDGMENTS
The work was supported by grants from the BMBF (Medizinische Infektionsgenomik), DFG Priority program SPP1656, research grants DFG STE 1971/2-1, and the German Center for Infection Research (DZIF).
M.B. and B.S. conceived and designed the study. M.B., C.E., and B.S. wrote the paper. M.B., C.E., S.H., D.R., T.D., S.B., P.S., S.H., and M.D. performed the experiments. M.B., C.E., D.G., P.M., and T.D. analyzed the data. M.B. and A.B. contributed important material. All authors approved the definitive version of the manuscript.
Contributor Information
Bärbel Stecher, Email: stecher@mvp.lmu.de.
Manuela Raffatellu, University of California San Diego School of Medicine, La Jolla, California, USA.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/iai.00318-23.
Fig. S1 to S8.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. GBD 2017 Disease and Injury Incidence and Prevalence Collaborators . 2018. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990-2017: a systematic analysis for the global burden of disease study 2017. Lancet 392:1789–1858. doi: 10.1016/S0140-6736(18)32279-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. GBD 2017 Non-Typhoidal Salmonella Invasive Disease Collaborators . 2019. The global burden of non-typhoidal Salmonella invasive disease: a systematic analysis for the global burden of disease study 2017. Lancet Infect Dis 19:1312–1324. doi: 10.1016/S1473-3099(19)30418-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barthel M, Hapfelmeier S, Quintanilla-Martínez L, Kremer M, Rohde M, Hogardt M, Pfeffer K, Rüssmann H, Hardt W-D. 2003. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun 71:2839–2858. doi: 10.1128/IAI.71.5.2839-2858.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wotzka SY, Kreuzer M, Maier L, Arnoldini M, Nguyen BD, Brachmann AO, Berthold DL, Zünd M, Hausmann A, Bakkeren E, et al. 2019. Escherichia coli limits Salmonella Typhimurium infections after diet shifts and fat-mediated microbiota perturbation in mice. Nat Microbiol 4:2164–2174. doi: 10.1038/s41564-019-0568-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kim Y-G, Sakamoto K, Seo S-U, Pickard JM, Gillilland MG, Pudlo NA, Hoostal M, Li X, Wang TD, Feehley T, Stefka AT, Schmidt TM, Martens EC, Fukuda S, Inohara N, Nagler CR, Núñez G. 2017. Neonatal acquisition of clostridia species protects against colonization by bacterial pathogens. Science 356:315–319. doi: 10.1126/science.aag2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stecher B. 2021. Establishing causality in Salmonella-microbiota-host interaction: the use of gnotobiotic mouse models and synthetic microbial communities. Int J Med Microbiol 311:151484. doi: 10.1016/j.ijmm.2021.151484 [DOI] [PubMed] [Google Scholar]
- 7. Maier L, Vyas R, Cordova CD, Lindsay H, Schmidt TSB, Brugiroux S, Periaswamy B, Bauer R, Sturm A, Schreiber F, von Mering C, Robinson MD, Stecher B, Hardt W-D. 2013. Microbiota-derived hydrogen fuels Salmonella Typhimurium invasion of the gut ecosystem. Cell Host Microbe 14:641–651. doi: 10.1016/j.chom.2013.11.002 [DOI] [PubMed] [Google Scholar]
- 8. Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, Naidu N, Choudhury B, Weimer BC, Monack DM, Sonnenburg JL. 2013. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 502:96–99. doi: 10.1038/nature12503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Eberl C, Weiss AS, Jochum LM, Durai Raj AC, Ring D, Hussain S, Herp S, Meng C, Kleigrewe K, Gigl M, Basic M, Stecher B. 2021. E. coli enhance colonization resistance against Salmonella Typhimurium by competing for galactitol, a context-dependent limiting carbon source. Cell Host Microbe 29:1680–1692. doi: 10.1016/j.chom.2021.09.004 [DOI] [PubMed] [Google Scholar]
- 10. Rivera-Chávez F, Zhang LF, Faber F, Lopez CA, Byndloss MX, Olsan EE, Xu G, Velazquez EM, Lebrilla CB, Winter SE, Bäumler AJ. 2016. Depletion of butyrate-producing Clostridia from the gut microbiota drives an aerobic luminal expansion of Salmonella. Cell Host Microbe 19:443–454. doi: 10.1016/j.chom.2016.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lopez CA, Winter SE, Rivera-Chávez F, Xavier MN, Poon V, Nuccio S-P, Tsolis RM, Bäumler AJ. 2012. Phage-mediated acquisition of a type III secreted effector protein boosts growth of Salmonella by nitrate respiration. mBio 3:e00143-12. doi: 10.1128/mBio.00143-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nguyen BD, Cuenca VM, Hartl J, Gül E, Bauer R, Meile S, Rüthi J, Margot C, Heeb L, Besser F, Escriva PP, Fetz C, Furter M, Laganenka L, Keller P, Fuchs L, Christen M, Porwollik S, McClelland M, Vorholt JA, Sauer U, Sunagawa S, Christen B, Hardt W-D. 2020. Import of aspartate and malate by DcuABC drives H2/fumarate respiration to promote initial Salmonella gut-lumen colonization in mice. Cell Host Microbe 27:922–936. doi: 10.1016/j.chom.2020.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ackermann M, Stecher B, Freed NE, Songhet P, Hardt WD, Doebeli M. 2008. Self-destructive cooperation mediated by phenotypic noise. Nature 454:987–990. doi: 10.1038/nature07067 [DOI] [PubMed] [Google Scholar]
- 14. Stecher B, Chaffron S, Käppeli R, Hapfelmeier S, Freedrich S, Weber TC, Kirundi J, Suar M, McCoy KD, von Mering C, Macpherson AJ, Hardt W-D. 2010. Like will to like: abundances of closely related species can predict susceptibility to intestinal colonization by pathogenic and commensal bacteria. PLoS Pathog 6:e1000711. doi: 10.1371/journal.ppat.1000711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, Chaffron S, Macpherson AJ, Buer J, Parkhill J, Dougan G, von Mering C, Hardt W-D. 2007. Salmonella enterica serovar Typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol 5:2177–2189. doi: 10.1371/journal.pbio.0050244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Barman M, Unold D, Shifley K, Amir E, Hung K, Bos N, Salzman N. 2008. Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infect Immun 76:907–915. doi: 10.1128/IAI.01432-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stecher B, Hardt W-D. 2011. Mechanisms controlling pathogen colonization of the gut. Curr Opin Microbiol 14:82–91. doi: 10.1016/j.mib.2010.10.003 [DOI] [PubMed] [Google Scholar]
- 18. Zhang APP, Pigli YZ, Rice PA. 2010. Structure of the LexA-DNA complex and implications for SOS box measurement. Nature 466:883–886. doi: 10.1038/nature09200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Byndloss MX, Olsan EE, Rivera-Chávez F, Tiffany CR, Cevallos SA, Lokken KL, Torres TP, Byndloss AJ, Faber F, Gao Y, Litvak Y, Lopez CA, Xu G, Napoli E, Giulivi C, Tsolis RM, Revzin A, Lebrilla CB, Bäumler AJ. 2017. Microbiota-activated PPAR-gamma signaling inhibits dysbiotic enterobacteriaceae expansion. Science 357:570–575. doi: 10.1126/science.aam9949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, Roth JR, Bäumler AJ. 2010. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467:426–429. doi: 10.1038/nature09415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stecher B, Barthel M, Schlumberger MC, Haberli L, Rabsch W, Kremer M, Hardt WD. 2008. Motility allows S. Typhimurium to benefit from the mucosal defence. Cell Microbiol 10:1166–1180. doi: 10.1111/j.1462-5822.2008.01118.x [DOI] [PubMed] [Google Scholar]
- 22. Thiennimitr P, Winter SE, Winter MG, Xavier MN, Tolstikov V, Huseby DL, Sterzenbach T, Tsolis RM, Roth JR, Bäumler AJ. 2011. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci U S A 108:17480–17485. doi: 10.1073/pnas.1107857108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ali MM, Newsom DL, González JF, Sabag-Daigle A, Stahl C, Steidley B, Dubena J, Dyszel JL, Smith JN, Dieye Y, Arsenescu R, Boyaka PN, Krakowka S, Romeo T, Behrman EJ, White P, Ahmer BMM. 2014. Fructose-asparagine is a primary nutrient during growth of Salmonella in the inflamed intestine. PLoS Pathog 10:e1004209. doi: 10.1371/journal.ppat.1004209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Faber F, Thiennimitr P, Spiga L, Byndloss MX, Litvak Y, Lawhon S, Andrews-Polymenis HL, Winter SE, Bäumler AJ, Sperandio V. 2017. Respiration of microbiota-derived 1,2-propanediol drives Salmonella expansion during colitis. PLoS Pathog 13:e1006129. doi: 10.1371/journal.ppat.1006129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Spiga L, Winter MG, Furtado de Carvalho T, Zhu W, Hughes ER, Gillis CC, Behrendt CL, Kim J, Chessa D, Andrews-Polymenis HL, Beiting DP, Santos RL, Hooper LV, Winter SE. 2017. An oxidative central metabolism enables Salmonella to utilize microbiota-derived succinate. Cell Host Microbe 22:291–301. doi: 10.1016/j.chom.2017.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gillis CC, Hughes ER, Spiga L, Winter MG, Zhu W, Furtado de Carvalho T, Chanin RB, Behrendt CL, Hooper LV, Santos RL, Winter SE. 2018. Dysbiosis-associated change in host metabolism generates lactate to support Salmonella growth. Cell Host Microbe 23:570. doi: 10.1016/j.chom.2018.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Raffatellu M, George MD, Akiyama Y, Hornsby MJ, Nuccio S-P, Paixao TA, Butler BP, Chu H, Santos RL, Berger T, Mak TW, Tsolis RM, Bevins CL, Solnick JV, Dandekar S, Bäumler AJ. 2009. Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe 5:476–486. doi: 10.1016/j.chom.2009.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stecher B, Hardt WD. 2008. The role of microbiota in infectious disease. Trends Microbiol 16:107–114. doi: 10.1016/j.tim.2007.12.008 [DOI] [PubMed] [Google Scholar]
- 29. Miki T, Goto R, Fujimoto M, Okada N, Hardt WD. 2017. The bactericidal lectin regiiibeta prolongs gut colonization and enteropathy in the streptomycin mouse model for salmonella diarrhea. Cell Host Microbe 21:195–207. doi: 10.1016/j.chom.2016.12.008 [DOI] [PubMed] [Google Scholar]
- 30. Crawford RW, Keestra AM, Winter SE, Xavier MN, Tsolis RM, Tolstikov V, Bäumler AJ, Nassif X. 2012. Very long o-antigen chains enhance fitness during Salmonella-induced colitis by increasing bile resistance. PLoS Pathog 8:e1002918. doi: 10.1371/journal.ppat.1002918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Day DW, Mandal BK, Morson BC. 1978. The rectal biopsy appearances in Salmonella colitis. Histopathology 2:117–131. doi: 10.1111/j.1365-2559.1978.tb01700.x [DOI] [PubMed] [Google Scholar]
- 32. Boyd JF. 1985. Pathology of the alimentary tract in Salmonella Typhimurium food poisoning. Gut 26:935–944. doi: 10.1136/gut.26.9.935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Felmy B, Songhet P, Slack EMC, Müller AJ, Kremer M, Van Maele L, Cayet D, Heikenwalder M, Sirard J-C, Hardt W-D. 2013. NADPH oxidase deficient mice develop colitis and bacteremia upon infection with normally avirulent, TTSS-1- and TTSS-2-deficient Salmonella Typhimurium. PLoS One 8:e77204. doi: 10.1371/journal.pone.0077204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Loetscher Y, Wieser A, Lengefeld J, Kaiser P, Schubert S, Heikenwalder M, Hardt W-D, Stecher B. 2012. Salmonella transiently reside in luminal neutrophils in the inflamed gut. PLoS One 7:e34812. doi: 10.1371/journal.pone.0034812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Molloy MJ, Grainger JR, Bouladoux N, Hand TW, Koo LY, Naik S, Quinones M, Dzutsev AK, Gao J-L, Trinchieri G, Murphy PM, Belkaid Y. 2013. Intraluminal containment of commensal outgrowth in the gut during infection-induced dysbiosis. Cell Host & Microbe 14:318–328. doi: 10.1016/j.chom.2013.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Maier L, Diard M, Sellin ME, Chouffane E-S, Trautwein-Weidner K, Periaswamy B, Slack E, Dolowschiak T, Stecher B, Loverdo C, Regoes RR, Hardt W-D. 2014. Granulocytes impose a tight bottleneck upon the gut luminal pathogen population during Salmonella Typhimurium colitis. PLoS Pathog 10:e1004557. doi: 10.1371/journal.ppat.1004557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gill N, Ferreira RBR, Antunes LCM, Willing BP, Sekirov I, Al-Zahrani F, Hartmann M, Finlay BB. 2012. Neutrophil elastase alters the murine gut microbiota resulting in enhanced Salmonella colonization. PLoS One 7:e49646. doi: 10.1371/journal.pone.0049646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu JZ, Jellbauer S, Poe AJ, Ton V, Pesciaroli M, Kehl-Fie TE, Restrepo NA, Hosking MP, Edwards RA, Battistoni A, Pasquali P, Lane TE, Chazin WJ, Vogl T, Roth J, Skaar EP, Raffatellu M. 2012. Zinc sequestration by the neutrophil protein calprotectin enhances Salmonella growth in the inflamed gut. Cell Host Microbe 11:227–239. doi: 10.1016/j.chom.2012.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lawley TD, Bouley DM, Hoy YE, Gerke C, Relman DA, Monack DM. 2008. Host transmission of Salmonella enterica serovar Typhimurium is controlled by virulence factors and indigenous intestinal Microbiota. Infect Immun 76:403–416. doi: 10.1128/IAI.01189-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Brugiroux S, Beutler M, Pfann C, Garzetti D, Ruscheweyh H-J, Ring D, Diehl M, Herp S, Lötscher Y, Hussain S, Bunk B, Pukall R, Huson DH, Münch PC, McHardy AC, McCoy KD, Macpherson AJ, Loy A, Clavel T, Berry D, Stecher B. 2016. Genome-guided design of a defined mouse microbiota that confers colonization resistance against Salmonella enterica serovar Typhimurium. Nat Microbiol 2:16215. doi: 10.1038/nmicrobiol.2016.215 [DOI] [PubMed] [Google Scholar]
- 41. Weiss AS, Burrichter AG, Durai Raj AC, von Strempel A, Meng C, Kleigrewe K, Münch PC, Rössler L, Huber C, Eisenreich W, Jochum LM, Göing S, Jung K, Lincetto C, Hübner J, Marinos G, Zimmermann J, Kaleta C, Sanchez A, Stecher B. 2022. In vitro interaction network of a synthetic gut bacterial community. ISME J 16:1095–1109. doi: 10.1038/s41396-021-01153-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Weiss AS, Niedermeier LS, von Strempel A, Burrichter AG, Ring D, Meng C, Kleigrewe K, Lincetto C, Hübner J, Stecher B. 2023. Nutritional and host environments determine community ecology and keystone species in a synthetic gut bacterial community. Nat Commun 14:4780. doi: 10.1038/s41467-023-40372-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hoces D, Lan J, Sun W, Geiser T, Stäubli ML, Cappio Barazzone E, Arnoldini M, Challa TD, Klug M, Kellenberger A, Nowok S, Faccin E, Macpherson AJ, Stecher B, Sunagawa S, Zenobi R, Hardt W-D, Wolfrum C, Slack E. 2022. Metabolic reconstitution of germ-free mice by a gnotobiotic Microbiota varies over the circadian cycle. PLoS Biol 20:e3001743. doi: 10.1371/journal.pbio.3001743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Afrizal A, Jennings SAV, Hitch TCA, Riedel T, Basic M, Panyot A, Treichel N, Hager FT, Wong E-Y, Wolter B, et al. 2022. Enhanced cultured diversity of the mouse gut microbiota enables custom-made synthetic communities. Cell Host Microbe 30:1630–1645. doi: 10.1016/j.chom.2022.09.011 [DOI] [PubMed] [Google Scholar]
- 45. Osbelt L, Wende M, Almási É, Derksen E, Muthukumarasamy U, Lesker TR, Galvez EJC, Pils MC, Schalk E, Chhatwal P, Färber J, Neumann-Schaal M, Fischer T, Schlüter D, Strowig T. 2021. Klebsiella oxytoca causes colonization resistance against multidrug-resistant K. pneumoniae in the gut via cooperative carbohydrate competition. Cell Host Microbe 29:1663–1679. doi: 10.1016/j.chom.2021.09.003 [DOI] [PubMed] [Google Scholar]
- 46. Studer N, Desharnais L, Beutler M, Brugiroux S, Terrazos MA, Menin L, Schürch CM, McCoy KD, Kuehne SA, Minton NP, Stecher B, Bernier-Latmani R, Hapfelmeier S. 2016. Functional intestinal bile acid 7α-dehydroxylation by Clostridium scindens associated with protection from Clostridium difficile infection in a gnotobiotic mouse model. Front Cell Infect Microbiol 6:191. doi: 10.3389/fcimb.2016.00191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. van Tilburg Bernardes E, Pettersen VK, Gutierrez MW, Laforest-Lapointe I, Jendzjowsky NG, Cavin J-B, Vicentini FA, Keenan CM, Ramay HR, Samara J, MacNaughton WK, Wilson RJA, Kelly MM, McCoy KD, Sharkey KA, Arrieta M-C. 2020. Intestinal fungi are causally implicated in microbiome assembly and immune development in mice. Nat Commun 11:2577. doi: 10.1038/s41467-020-16431-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lourenço M, Chaffringeon L, Lamy-Besnier Q, Pédron T, Campagne P, Eberl C, Bérard M, Stecher B, Debarbieux L, De Sordi L. 2020. The spatial heterogeneity of the gut limits predation and fosters coexistence of bacteria and bacteriophages. Cell Host Microbe 28:390–401. doi: 10.1016/j.chom.2020.06.002 [DOI] [PubMed] [Google Scholar]
- 49. Ye H, Borusak S, Eberl C, Krasenbrink J, Weiss AS, Chen S-C, Hanson BT, Hausmann B, Herbold CW, Pristner M, Zwirzitz B, Warth B, Pjevac P, Schleheck D, Stecher B, Loy A. 2023. Ecophysiology and interactions of a taurine-respiring bacterium in the mouse gut. Nat Commun 14:5533. doi: 10.1038/s41467-023-41008-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Miller BM, Bäumler AJ. 2021. The habitat filters of microbiota-nourishing immunity. Annu Rev Immunol 39:1–18. doi: 10.1146/annurev-immunol-101819-024945 [DOI] [PubMed] [Google Scholar]
- 51. Stecher B, Maier L, Hardt WD. 2013. Blooming' in the gut: how dysbiosis might contribute to pathogen evolution. Nat Rev Microbiol 11:277–284. doi: 10.1038/nrmicro2989 [DOI] [PubMed] [Google Scholar]
- 52. Stecher B, Denzler R, Maier L, Bernet F, Sanders MJ, Pickard DJ, Barthel M, Westendorf AM, Krogfelt KA, Walker AW, Ackermann M, Dobrindt U, Thomson NR, Hardt WD. 2012. Gut inflammation can boost horizontal gene transfer between pathogenic and commensal enterobacteriaceae. Proc Natl Acad Sci U S A 109:1269–1274. doi: 10.1073/pnas.1113246109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Diard M, Bakkeren E, Cornuault JK, Moor K, Hausmann A, Sellin ME, Loverdo C, Aertsen A, Ackermann M, De Paepe M, Slack E, Hardt W-D. 2017. Inflammation boosts bacteriophage transfer between Salmonella spp. Science 355:1211–1215. doi: 10.1126/science.aaf8451 [DOI] [PubMed] [Google Scholar]
- 54. Wotzka SY, Nguyen BD, Hardt WD. 2017. Salmonella Typhimurium diarrhea reveals basic principles of enteropathogen infection and disease-promoted DNA exchange. Cell Host Microbe 21:443–454. doi: 10.1016/j.chom.2017.03.009 [DOI] [PubMed] [Google Scholar]
- 55. Lamy-Besnier Q, Chaffringeon L, Lourenço M, Payne RB, Trinh JT, Schwartz JA, Sulakvelidze A, Debarbieux L. 2021. Prophylactic administration of a bacteriophage cocktail is safe and effective in reducing Salmonella enterica serovar Typhimurium burden in vivo . Microbiol Spectr 9:e0049721. doi: 10.1128/spectrum.00497-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Weiss AS, Burrichter AG, Durai Raj AC, von Strempel A, Meng C, Kleigrewe K, Münch PC, Rössler L, Huber C, Eisenreich W, Jochum LM, Göing S, Jung K, Lincetto C, Hübner J, Marinos G, Zimmermann J, Kaleta C, Sanchez A, Stecher B. 2022. In vitro interaction network of a synthetic gut bacterial community. ISME J 16:1095–1109. doi: 10.1038/s41396-021-01153-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rhee SJ, Walker WA, Cherayil BJ. 2005. Developmentally regulated intestinal expression of IFN-gamma and its target genes and the age-specific response to enteric Salmonella infection. J Immunol 175:1127–1136. doi: 10.4049/jimmunol.175.2.1127 [DOI] [PubMed] [Google Scholar]
- 58. Godinez I, Haneda T, Raffatellu M, George MD, Paixão TA, Rolán HG, Santos RL, Dandekar S, Tsolis RM, Bäumler AJ. 2008. T cells help to amplify inflammatory responses induced by Salmonella enterica serotype Typhimurium in the intestinal mucosa. Infect Immun 76:2008–2017. doi: 10.1128/IAI.01691-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Knodler LA, Vallance BA, Celli J, Winfree S, Hansen B, Montero M, Steele-Mortimer O. 2010. Dissemination of invasive Salmonella via bacterial-induced extrusion of mucosal epithelia. Proc Natl Acad Sci U S A 107:17733–17738. doi: 10.1073/pnas.1006098107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sellin ME, Müller AA, Felmy B, Dolowschiak T, Diard M, Tardivel A, Maslowski KM, Hardt W-D. 2014. Epithelium-intrinsic NAIP/Nlrc4 Inflammasome drives infected enterocyte expulsion to restrict Salmonella replication in the intestinal mucosa. Cell Host Microbe 16:237–248. doi: 10.1016/j.chom.2014.07.001 [DOI] [PubMed] [Google Scholar]
- 61. Songhet P, Barthel M, Stecher B, Müller AJ, Kremer M, Hansson GC, Hardt W-D, Bereswill S. 2011. Stromal IFN-γR-signaling modulates goblet cell function during Salmonella Typhimurium infection. PLoS ONE 6:e22459. doi: 10.1371/journal.pone.0022459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hapfelmeier S, Stecher B, Barthel M, Kremer M, Müller AJ, Heikenwalder M, Stallmach T, Hensel M, Pfeffer K, Akira S, Hardt W-D. 2005. The Salmonella pathogenicity island (SPI)-1 and SPI-2 type III secretion systems allow Salmonella serovar Typhimurium to trigger colitis via myd88-dependent and myd88-independent mechanisms. J Immunol 174:1675–1685. doi: 10.4049/jimmunol.174.3.1675 [DOI] [PubMed] [Google Scholar]
- 63. Thiennimitr P, Winter SE, Bäumler AJ. 2012. Salmonella, the host and its Microbiota. Curr Opin Microbiol 15:108–114. doi: 10.1016/j.mib.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Cheng AG, Ho P-Y, Aranda-Díaz A, Jain S, Yu FB, Meng X, Wang M, Iakiviak M, Nagashima K, Zhao A, Murugkar P, Patil A, Atabakhsh K, Weakley A, Yan J, Brumbaugh AR, Higginbottom S, Dimas A, Shiver AL, Deutschbauer A, Neff N, Sonnenburg JL, Huang KC, Fischbach MA. 2022. Design, construction, and in vivo augmentation of a complex gut microbiome. Cell 185:3617–3636. doi: 10.1016/j.cell.2022.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Clavel T, Lagkouvardos I, Blaut M, Stecher B. 2016. The mouse gut microbiome revisited: from complex diversity to model ecosystems. Int J Med Microbiol 306:316–327. doi: 10.1016/j.ijmm.2016.03.002 [DOI] [PubMed] [Google Scholar]
- 66. Hoiseth SK, Stocker BA. 1981. Aromatic-dependent Salmonella Typhimurium are non-virulent and effective as live vaccines. Nature 291:238–239. doi: 10.1038/291238a0 [DOI] [PubMed] [Google Scholar]
- 67. Ehrbar K, Hapfelmeier S, Stecher B, Hardt WD. 2004. InvB is required for type III-dependent secretion of SopA in Salmonella enterica serovar Typhimurium. J Bacteriol 186:1215–1219. doi: 10.1128/JB.186.4.1215-1219.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kaniga K, Bossio JC, Galán JE. 1994. The Salmonella Typhimurium invasion genes invF and invG encode homologues of the AraC and PulD family of proteins. Mol Microbiol 13:555–568. doi: 10.1111/j.1365-2958.1994.tb00450.x [DOI] [PubMed] [Google Scholar]
- 69. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Schmieger H. 1972. Phage P22-mutants with increased or decreased transduction abilities. Mol Gen Genet 119:75–88. doi: 10.1007/BF00270447 [DOI] [PubMed] [Google Scholar]
- 71. Deiwick J, Hensel M. 1999. Regulation of virulence genes by environmental signals in Salmonella Typhimurium. Electrophoresis 20:813–817. doi: [DOI] [PubMed] [Google Scholar]
- 72. Cherepanov PP, Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14. doi: 10.1016/0378-1119(95)00193-a [DOI] [PubMed] [Google Scholar]
- 73. Miranda KM, Espey MG, Wink DA. 2001. A rapid, simple spectrophotometric method for simultaneous detection of nitrate and nitrite. Nitric Oxide 5:62–71. doi: 10.1006/niox.2000.0319 [DOI] [PubMed] [Google Scholar]
- 74. Bapteste E, Bicep C, Lopez P. 2012. Evolution of genetic diversity using networks: the human gut Microbiome as a case study. Clin Microbiol Infect 18:40–43. doi: 10.1111/j.1469-0691.2012.03856.x [DOI] [PubMed] [Google Scholar]
- 75. Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622. doi: 10.1373/clinchem.2008.112797 [DOI] [PubMed] [Google Scholar]
- 76. Gerlach RG, Jäckel D, Stecher B, Wagner C, Lupas A, Hardt W-D, Hensel M. 2007. Salmonella pathogenicity island 4 encodes a giant non-fimbrial adhesin and the cognate type 1 secretion system. Cell Microbiol 9:1834–1850. doi: 10.1111/j.1462-5822.2007.00919.x [DOI] [PubMed] [Google Scholar]
- 77. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S8.