Abstract
Cholesterol is the precursor of all steroids, but how cholesterol flux is controlled in steroidogenic tissues is poorly understood. The cholesterol exporter ABCG1 is an essential component of the reverse cholesterol pathway and its global inactivation results in neutral lipid redistribution to tissue macrophages. The function of ABCG1 in steroidogenic tissues, however, has not been explored. To model this, we inactivated Abcg1 in the mouse adrenal cortex, which led to an adrenal-specific increase in transcripts involved in cholesterol uptake and de novo synthesis. Abcg1 inactivation did not affect adrenal cholesterol content, zonation, or serum lipid profile. Instead, we observed a moderate increase in corticosterone production that was not recapitulated by the inactivation of the functionally similar cholesterol exporter Abca1. Altogether, our data imply that Abcg1 controls cholesterol uptake and biosynthesis and regulates glucocorticoid production in the adrenal cortex, introducing the possibility that ABCG1 variants may account for physiological or subclinical variation in stress response.
Keywords: Abcg1, cholesterol, glucocorticoids, steroids, adrenal cortex
Steroid hormones mediate a myriad of physiological responses, from the control of blood pressure (mineralocorticoids) and sexual maturation (sex hormones) to the regulation of glucose homeostasis and stress response (glucocorticoids) (1). The extensive impact of steroid hormones on human physiology demands a fine regulation of steroid production. Alterations of this fine balance may result in pathological phenotypes, including adrenocortical insufficiency or steroid hypersecretion, for which many monogenic or polygenic determinants still need to be identified (2, 3).
As the obligatory precursor of all steroids, cholesterol is a key modulator of steroidogenesis, both in a quantitative and qualitative fashion. Disruption of cholesterol homeostasis results in adrenal insufficiency (eg, in Smith-Lemli-Opitz disease) (4), while the dysregulated accumulation of cholesterol leads to increased cholesterol storage and physical and biochemical cellular distress (eg, in lipoid congenital adrenal hyperplasia) (5-7). Besides, fine tuning of cholesterol homeostasis is critical for regulation of steroidogenesis within a physiological range. For instance, interfering with cholesterol content in plasma membranes directly impacts the synthesis of pregnenolone, which is a common precursor to all steroids (8). In addition, the master transcriptional activator of steroidogenesis, Steroidogenic Factor 1 (NR5A1), not only induces the expression of critical steroidogenic enzymes but also triggers the expression of cholesterogenic genes to provide more substrate for steroidogenesis (9). Furthermore, our group previously showed that intracellular cholesterol deprivation reroutes steroidogenesis to a more androgenic profile, implicating cholesterol in the prioritization of steroidogenic pathways (10).
Levels of intracellular cholesterol are therefore finely balanced between cholesterol acquisition (contributed by uptake from the circulation, de novo biosynthesis, and hydrolysis of cholesteryl esters) and disposal (mediated by excretion of cholesterol to the circulation, cholesterol esterification for long-term storage, and cholesterol deployment for biosynthesis of downstream products) (11). Intracellular cholesterol homeostasis in adrenocortical cells is thought to rely on the sterol regulatory element-binding factor 2, which acts as a master transcriptional activator of the cholesterol biosynthetic pathway and the cholesterol import machinery upon conditions of sterol depletion (7, 11-13). However, the molecular programs that control cholesterol availability in steroidogenic cells are not fully characterized.
Abcg1 is an adenosine triphosphate-dependent transporter involved in the maintenance of tissue and cellular cholesterol homeostasis. In mice and humans, it is expressed in a variety of cell types including adrenocortical cells (14-22). Its subcellular localization is still a matter of debate: Abcg1 has been found in both endosomes and in the plasma membrane, and in association with actin filaments (23-27). It is thought to mobilize cholesterol from the endoplasmic reticulum and to redistribute it to the plasma membrane, favoring cholesterol efflux to a variety of extracellular acceptors (15, 28-30). The role of Abcg1 in steroidogenic tissues, however, is unknown.
Here we study the adrenal cortex to determine the role of Abcg1 in a steroidogenic tissue. Abcg1 inactivation in mouse adrenals results in increased transcripts for cholesterol biosynthesis and uptake, leading to increased corticosterone production. Our data suggest that Abcg1 is a key regulator of cholesterol flux and glucocorticoid production.
Methods
Experimental Animals
All animal procedures were approved by the Veterinary Office of the Canton Bern in Switzerland. Generation of the aldosterone synthase-Cre strain (Cyp11b2tm1.1(cre)Brlt), and the compound conditional Abcg1 and Abca1 strain (B6.Cg-Abca1tm1Jp Abcg1tm1Tall/J), was previously described (31, 32). To generate the bigenic mice carrying 1 Cre allele and 2 conditional alleles either within the Abca1 or the Abcg1 locus (referred to as Abca1 cKO and Abcg1 cKO, respectively), males of the Cre-bearing strain were crossed with compound heterozygous females for the conditional Abca1 and Abcg1 alleles. Pups expressing the Cre recombinase and either the Abca1tm1Jp or the Abcg1tm1Tall/J allele were selected and crossed with isogenic littermates. Littermates carrying the Cre allele alone, or 1 of the 2 conditional alleles, were used as controls. All mice were kept on a mixed sv129-C57BL/6 genetic background, with free access to chow and water, under a 12-hour light/12-hour dark cycle. Unless otherwise specified, all mice used for this work were 2-month-old females. Adrenal weight was measured on an analytical balance on freshly dissected adrenal glands following clearance of the surrounding fat tissue.
Gene Expression Analysis
RNA was isolated from whole adrenals cleaned of the adherent fat or from livers and homogenized in TRI Reagent (Sigma) using the Direct-zol miniprep RNA kit (Zymo Research), following the manufacturer's protocol. RNA was reverse transcribed using the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). Gene expression analysis was performed by real-time quantitative PCR using the PowerUp SYBR Master Mix and the QuantStudio 1 thermocycler (Thermo Fisher Scientific). Technical duplicates were used to minimize variability. For mouse studies, the following primers were used: Abcg1, Fw: ACATCGAATTCAAGGACCTT, Rv: CCCAGAGATCCCTTTCAAAA; Abca1, Fw: AACTTTCAAGATGCTGACTG, Rv: AAAGAACTCCACATGCTCTC; Ldlr, Fw: GTTGCAGCAGAAGACTCAT, Rv: CACCCACTTGCTAGCGAT; Scarb1, Fw: CAGGTGCTCAAGAATGTCC, Rv: TAGAAAGGGACGGGGATC; Hmgcr, Fw: AATGCCTTGTGATTGGAGTT, Rv: CCGGGAAGAATGTCATGAA; Sqle, Fw: AAAGAAAGAACAGCTGGAGT, Rv: TAGCTGCTCCTGTTAATGTC; Insig1, Fw: ATAGCCACCATCTTCTCCTC, Rv: TCTCTCTTGAACTTGTGTGG; Gck, Fw: TGTAAGGCACGAAGACATAG, Rv: GTTGTTCCCTTCTGCTCC; Pck1, Fw: GTGGAAGGTCGAATGTGTG, Rv: TTGATAGCCCTTAAGTTGCC; G6pc, Fw: GTTCAACCTCGTCTTCAAGT Rv: CTGTTGCTGTAGTAGTCGG; Nr3c1, Fw: CTATGAACTTCGCAGGCC Rv: GAGAACTCACATCTGGTCTC; Gapdh, Fw: ATCAACGACCCCTTCATTG, Rv: TTGATGACAAGCTTCCCATT; Actb, Fw: GACCTGACAGACTACCTCAT, Rv: CTCGAAGTCTAGAGCAACAT. Transcripts encoding glyceraldehyde 3-phosphate dehydrogenase or actin beta were used as internal control, and data were expressed using the 2−ddCt method.
Transcriptome Profiling
Preparation of whole-adrenal RNA isolates was conducted as indicated in the Gene Expression Analysis section. RNA samples were quantified using the RiboGreen assay (Thermo Fisher Scientific). Sample quality was analyzed on a Fragment Analyzer 5200 (Agilent) using the Fragment Analyzer HS RNA kit(15NT) (Agilent, DNF-472-1000). Illumina Stranded mRNA Prep Preparation including polyA enrichment was used according to the manufacturer's recommendations to construct libraries from total RNA. Subsequently, the Illumina NovaSeq and NextSeq platforms with a NovaSeq 200cy Kit (v1.5) and a NextSeq 300cy Kit (v2), respectively, were used to sequence the libraries. The produced paired-end reads that passed Illumina's chastity filter were demultiplexed using Illumina's bcl2fastq software version 2.20.0.422 (no further refinement or selection). Illumina adapter residuals were trimmed using cutadapt (v4.0 with Python 3.9.16). Quality of the reads in fastq format was checked with the software FastQC (version 0.11.9). Raw reads shorter than 10 bp, having average Q-values below 24, or incorporating uncalled “N” bases were filtered out using the BBTools software suite (version 38.86). The splice-aware RNA mapping software STAR (version 2.7.10a) was used to map the remaining reads to the mm10 reference genome provided by IGenomes (archive-2015-07-17-14-33-26). To count the uniquely mapped reads to annotated genes, the software htseq-count (HTSeq version 0.13.5) was used. Normalization of the raw counts and differential gene expression analysis was carried out with the R software package DESeq2 (version 1.38.3). Combined evidence from previous works suggest that only about 50% to 60% of cells contributing to whole-adrenal transcriptome profiling efforts are recombined steroidogenic cells of interest (31, 33). Therefore, we expected differentially expressed genes to be less abundant in our whole-adrenal extracts with respect to more enriched cell populations (eg, sorted cortical cells) and used a relaxed fold change threshold (1.2) to capture these genes. Library construction, sequencing, and data analysis described in this section were performed by Microsynth AG (Balgach, Switzerland). Profiling results are stored within the Gene Expression Omnibus repository under the accession number GSE242081.
Histology, Immunofluorescence, and Microscopy
Adrenals were dissected, cleared of the fat tissue, and fixed overnight in 4% paraformaldehyde. Four-µm paraffin sections were processed for protein immunodetection as previously described (10). Briefly, antigen retrieval was performed in 10 mM Sodium Citrate pH 6, and incubation was conducted overnight using a mouse monoclonal anti-Disabled-2/p96 (Dab2; BD Transduction Laboratories, cat. no. 610464; RRID: AB_397837) and a rabbit polyclonal anti-Akr1b7 [kindly provided by Dr Pierre Val and Dr Antoine Martinez; RRID: AB_3075891 (34)]. Indirect staining was performed using the goat anti-rabbit IgG (H + L) highly cross-adsorbed secondary antibody conjugated with Alexa Fluor™ 488 (from Thermo Fisher Scientific, cat. no. A11008; RRID: AB_143165), and a goat anti-mouse IgG (H + L) cross-adsorbed secondary antibody conjugated with Alexa Fluor™ 647 (from Thermo Fisher Scientific, cat. no. A21235; RRID: AB_2535804). 4′,6-diamidino-2-phenylindole was used for counterstaining. Images were captured with a Nikon Eclipse Ti-E microscope. Hematoxylin and eosin staining was carried out on neighboring sections compared to the immunofluorescence experiment. For Oil Red O staining, mouse adrenals were snap frozen and 5-µm sections were processed in a mixed solution of Oil Red O and dextrin, followed by nuclear counterstaining with Mayer's hemalum (all products from Merck).
Steroid Profiling and Blood Tests
Mouse serum was obtained using cardiac puncture of mice euthanized by intraperitoneal injection of pentobarbital. This terminal procedure was chosen because it allowed the collection of paired blood and adrenal tissue samples while causing a significantly lower stress response in mice compared to other euthanasia methods (35). Twenty-five µL of serum was used for further liquid chromatography-mass spectrometry (LC-MS) analysis using an established in house LC-MS method (36). Briefly, samples were collected and stored at −20 °C. Following thawing, 38 µL of internal standard was added to 25 µL of sample and extracted with ZnSO4 and methanol. After centrifugation, the organic phase was purified using a solid-phase extraction on an OasisPrime HLB 96-well plate using a positive pressure 96-well processor (both Waters, UK). For LC-MS analysis, a Vanquish UHPLC (equipped with an ACQUITY UPLC HSS T3 Column, 100 Å, 1.8 µm, 1 mm × 100 mm column; Waters, Switzerland) was coupled to a Q Exactive Plus Orbitrap (both Thermo Fisher Scientific, Reinach, Switzerland). Separation was achieved using gradient elution over 17 minutes using water and methanol both supplemented with 0.1% formic acid (all Sigma-Aldrich, Buchs, Switzerland) as mobile phases. Data analysis was performed using TraceFinder 4.1 (Thermo Fisher Scientific). The method was validated according to international standards. Steroid hormone concentrations were calculated in nmol/L. Values detected below the lower limit of accurate quantification were not used for statistics. Adrenocorticotropin hormone (ACTH) in serum was measured using an enzyme-linked immunosorbent assay kit (Abcam, cat. no. ab263880; RRID: AB_2910221), following the manufacturer's protocol. While ACTH is routinely assayed in plasma, we preferred quantification in serum as suggested by the kit based on the equivalence of serum and plasma for ACTH measurement in humans (37). Total cholesterol, high-density lipoproteins (HDL), and low-density lipoproteins/very-low density lipoproteins particles were measured using a cholesterol assay kit (Abcam, cat. no. ab65390), while triglycerides were assayed with a triglyceride assay kit (Abcam, cat. no. ab65336), following the manufacturer's instructions.
In Situ Hybridization
For double enzymatic in situ hybridization, mice adrenal glands were fixed in 4% paraformaldehyde at 4 °C for 24 hours and 5-μm-thick sections from Formalin-Fixed Paraffin-Embedded blocks were cut. In situ hybridization was performed following the manufacturer's recommendation of the BaseScope Duplex Reagent Kit Intro Pack-Mm (Advanced Cell Diagnostics, cat. no. 323871). Standard conditions were used: 15 minutes incubation for the Antigen retrieval step and 30 minutes for Protease III treatment. In situ hybridization staining was performed manually with the following combinations of RNAscope® probes (all from Bio-Techne). BA-Mm-Abca1-3ZZ-st-C2 probe, recognizing Abca1 (cat no. 1218611-C2) detected with the Fast Red signal; BA-Mm-Abcg-E3-1ZZ-st-C1 probe, recognizing Abcg1, (cat no. 1218601-C1) detected with the green signal. Basescope Duplex Positive Control Probe-Mouse(Mm)-C1-Ppib-1ZZ/C2-Polr2a-3ZZ and Basescope Duplex Negative Control Probe-C1-DapB-3ZZ/C2-DapB-3ZZ (cat no. 322982) were used respectively as positive and negative controls. Nuclei were visualized using hematoxylin, and slides were mounted with Vectamount mounting medium (Vector Labs, cat. no. H5000). Images were acquired on a NanoZoomer S60 digital slide scanner at 40× (Hamamatsu).
Cholesterol Quantification
Adrenal glands were dissected, clear of the surrounding fat pad, and homogenized in radioimmunoprecipitation assay buffer (Pierce, cat. no. 89900) supplemented with a protease and phosphatase inhibitor by Thermo Scientific (cat. no. A32961) at 4 °C, using lysing matrix tubes (MP Biomedicals, cat. no. 6913100) and a Bead Mill Homogenizer by Omni International. Tissue lysates were incubated for 1 hour in ice and spun down on a bench centrifuge for 20 minutes at 4 °C to get rid of unprocessed debris. Quantification of cholesterol was carried out using a Cholesterol/Cholesterol Ester-GloTM Assay by Promega (cat. no. J3190) following the manufacturer's instructions, with the exception that adrenal lysates were diluted from 1:10 to 1:40 in the lysis buffer provided by the kit to fit the calibration curve. The assay was performed either with or without cholesterol esterase, to allow for quantification of both total and free cholesterol, respectively. Values for esterified cholesterol were obtained by subtraction of free from total cholesterol. All cholesterol values were normalized by protein concentration assayed using a DC protein assay (Bio-Rad, cat. no. 5000112).
Statistical Analysis
Two-tailed Student's t-test was used for comparisons between any 2 groups. For every comparison, the F-test was used to assess inequality of variances. In case of inequality of variances, the Welch correction was adopted. One-way ANOVA and Dunnett’s multiple comparison test were used for comparisons between groups of 3 or more, unless otherwise specified. Prism 10 software (GraphPad) was used for statistical analysis. All data were included; no exclusion method was applied. Data are presented as mean ± SEM.
Results
Loss of Adrenocortical Abcg1 Increases Transcripts Involved in Cholesterol Metabolism
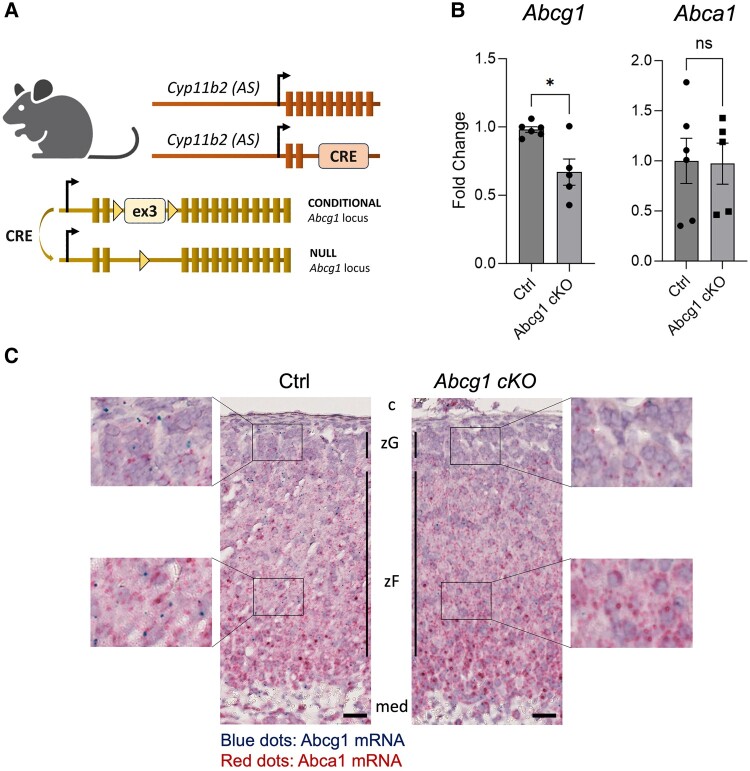
To investigate the role of Abcg1 in the adrenal cortex, we generated a mouse model where both Abcg1 alleles were conditionally inactivated using an aldosterone synthase (Cyp11b2)-specific Cre recombinase (Fig. 1A). The efficiency and extent of recombination were determined by quantifying Abcg1 transcripts within control and conditional knockout adrenals (henceforth referred to as Abcg1 cKO), and by in situ visualization of Abcg1 mRNAs. Specifically, Abcg1 transcripts were reduced by about 40% in recombined whole adrenals (Fig. 1B), and recombination occurred throughout the entire cortex (Fig. 1C). Importantly, transcripts encoding Abca1, a functionally similar adenosine triphosphate-dependent cholesterol exporter (11), were not affected by Abcg1 knockout (Fig. 1B).
Figure 1.
Effective gene recombination in Abcg1 cKO mice. (A) Schematic representation of the mouse model used to inactivate Abcg1 in adrenocortical cells using Cre-mediated recombination of Abcg1's third exon. (B) Quantitation of transcripts encoding Abcg1 and Abca1 in control and Abcg1 cKO adrenal glands. (C) In situ depiction of Abcg1 (blue dots) and Abca1 (red dots) transcripts in control (left) and Abcg1 cKO adrenal sections (right), including insets’ virtual magnifications on each side. All mice used for this figure were 2-month-old females. Scale bar = 25μm. *P ≤ .05.
Abbreviations: AS, aldosterone synthase; c, capsule; med, medulla; ns, not significant; zF, zona Fasciculata; zG, zona Glomerulosa.
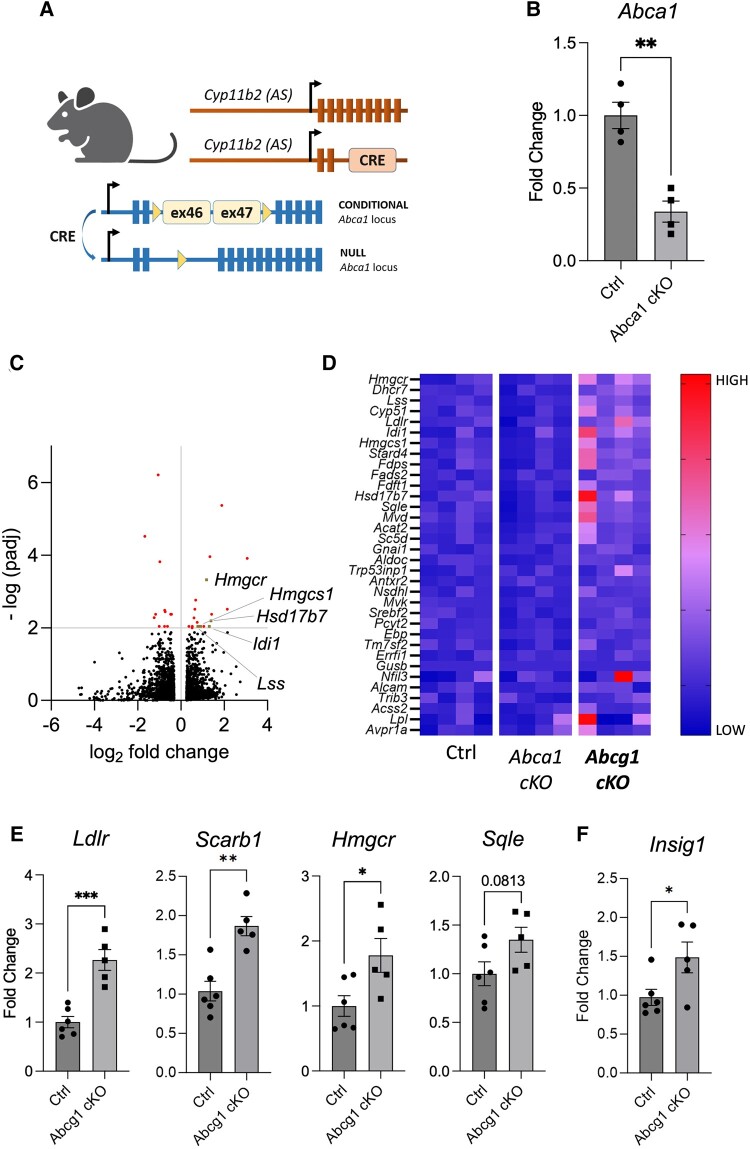
To assess the impact of Abcg1 on adrenal physiology, we profiled the transcriptome of Abcg1 cKO adrenals and compared it with the transcriptome of control and Abca1 cKO counterparts, which were also used as controls (Fig. 2A and 2B). Using a cutoff of 1.2 for fold change and 0.01 for adjusted P-value, we found 19 upregulated and 12 downregulated genes specifically in Abcg1 knockout adrenal glands (Fig. 2C). Gene set enrichment analysis revealed that cholesterol metabolism was the most affected pathway, with 34 genes contributing to the cholesterol set enrichment (HALLMARK_CHOLESTEROL_HOMEOSTASIS dataset) (Fig. 2D). Using quantitative PCR, we validated 3 of these upregulated genes, either implicated in cholesterol uptake (Ldlr) or biosynthesis (Hmgcr, Sqle) (Fig. 2E). The gene encoding the HDL receptor (Scarb1), which is the main route for cholesterol delivery to steroidogenic pathways (38), resulted upregulated using quantitative PCR (Fig. 2E), despite not contributing to the enrichment of the gene set enrichment analysis dataset (Fig. 2C and 2D). To determine the adrenal perception of cholesterol load, we also quantified Insig1, which is normally reduced upon accumulation of sterols (39, 40). Surprisingly, we found that Insig1 was upregulated in Abcg1 cKO adrenals (Fig. 2F), suggesting that cholesterol metabolism in Abcg1-deficient glands is dysregulated.
Figure 2.
Inactivation of adrenocortical Abcg1 results in increased transcripts for cholesterol uptake and synthesis. (A) Schematic representation of the mouse model used to inactivate Abca1 in adrenocortical cells using Cre-mediated recombination of exons 46 and 47. (B) Quantification of Abca1 transcripts in control and Abca1 cKO adrenal glands. (C) Volcano plot depicting 12 downregulated and 19 upregulated genes (red or beige dots) in Abcg1 cKO adrenals compared to the combined (summed) datasets of controls and Abca1 cKO counterparts, using cutoffs of 1.2 for fold change and 0.01 for adjusted P-value. Each beige dot is associated with a gene name as indicated in the plot. (D) Heat map depicting color-coded expression levels of 34 transcripts responsible for the enrichment of the HALLMARK_CHOLESTEROL_HOMEOSTASIS dataset in Gene Set Enrichment Analysis. (E and F) Quantitation of transcripts involved in cholesterol regulation, uptake, and de novo synthesis in control and Abcg1 cKO adrenal glands. All mice used for this figure were 2-month-old females. *P ≤ .05; **P ≤ .01; ***P ≤ .001.
Abbreviation: AS, aldosterone synthase.
Altogether, our data indicate that Abcg1 deficiency in the adrenal cortex disrupts intracellular cholesterol homeostasis by driving the expression of transcripts that normally promote increased cholesterol production and uptake.
Loss of Abcg1 Results in Increased Corticosterone
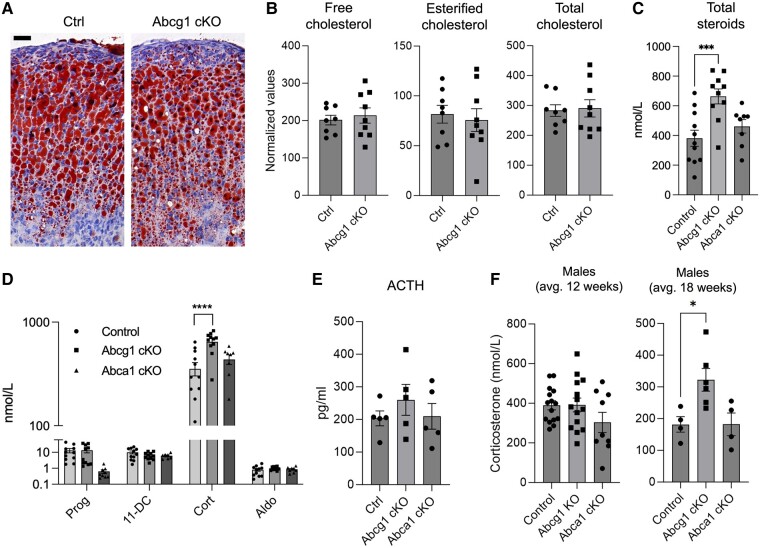
To determine whether increased cholesterol-related transcripts driven by Abcg1 inactivation results in increased cholesterol storage, we performed an Oil Red O staining of adrenal sections and observed no difference between Abcg1 cKO and control tissues (Fig. 3A). Direct quantification of total, free, and esterified cholesterol in the adrenals confirmed that cellular cholesterol compartments are not impacted by inactivation of Abcg1 (Fig. 3B).
Figure 3.
Inactivation of adrenocortical Abcg1 results in increased corticosterone synthesis. (A) Oil Red O staining (red) of control and Abcg1 cKO adrenocortical sections. Mayer's hemalum was used to counterstain nuclei (blue). Images are representative of 4 animals per genotype. Scale bar = 25 µm. (B) Free, esterified, and total cholesterol in whole adrenal glands from control and Abcg1 cKO animals. (C) Aggregated quantification of adrenal steroids detected in mouse sera using liquid chromatography-mass spectrometry, ie, pregnenolone, Prog, 11-DC, Cort, and Aldo. (D) Steroid concentrations in sera of control and Abcg1 cKO mice. Most pregnenolone values were below the threshold of accurate quantification, likely because of intense processivity into downstream products, and are not reported in this graph. (E) Levels in sera from control and Abcg1 cKO mice. (F) Levels of corticosterone in male control and Abcg1 cKO mice at different ages. Except for panel F, all mice used for this figure were 2-month-old females. *P ≤ .05, ***P ≤ .001, ****P ≤ .0001.
Abbreviations: 11-DC, 11-deoxycorticosterone; ACTH, adrenocorticotropin hormone; Aldo, aldosterone; avg, average; Cort, corticosterone; Prog, progesterone.
We then investigated whether the increase in cholesterol-related transcripts might lead to an increase of steroid biosynthesis. The adrenal steroid output (ie, the sum of pregnenolone, progesterone, 11-deoxycorticosterone, corticosterone, and aldosterone) showed a 74% increase in Abcg1 cKO mice compared to control animals. Instead, Abca1 cKO mice did not display any change in adrenal steroid metabolites (Fig. 3C). Most of the variation in Abcg1 cKO steroid profile was explained by increased corticosterone, the main glucocorticoid in mice, whereas the other steroids were not affected (Fig. 3D). The increase in corticosterone, although significant, was not sufficient to suppress the level of its main secretagogue, ACTH (Fig. 3E). While these results are based on female mice, male counterparts displayed a comparable increase in corticosterone but at an older age (average 18 weeks for males, compared with 12 weeks for females) (Fig. 3D and 3F).
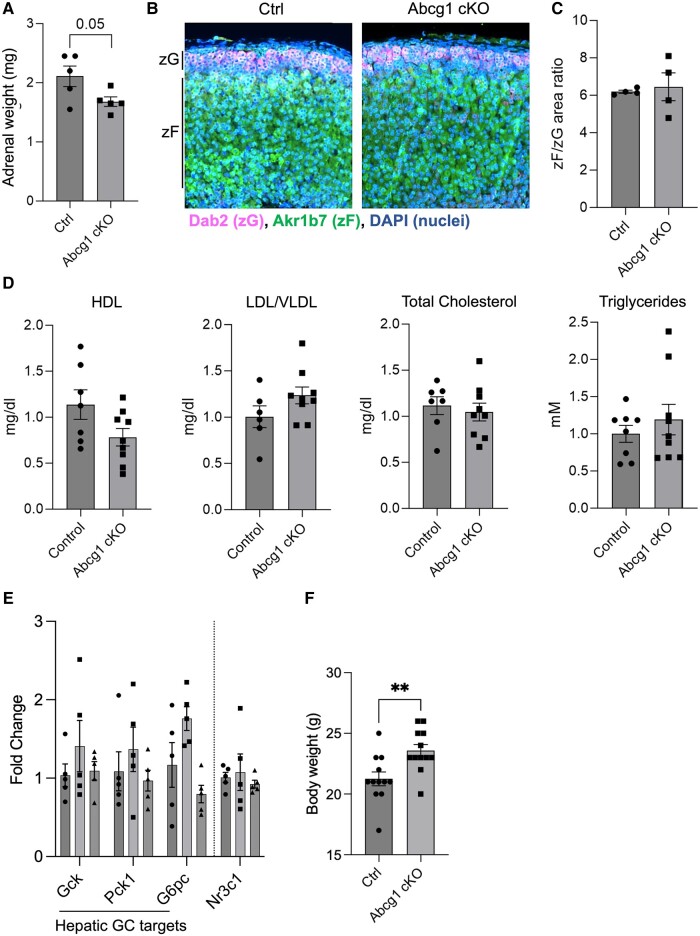
We then evaluated whether the increase in corticosterone was associated with increased adrenal size or altered zonation. First, we assessed adrenal weight, which revealed Abcg1 cKO adrenals mice were unchanged, compared to controls, with a paradoxical trend toward a decrease in adrenal weight (Fig. 4A). Next, we stained for the zone-specific markers Dab2 [identifying the zona Glomerulosa (zG)] and Akr1b7 [identifying the zona Fasciculata (zF)], which showed no difference between control and Abcg1 cKO mice in the zF-to-zG area ratio (Fig. 4B and 4C). These results indicate that neither increased adrenal mass nor expansion of the zF explains the increased corticosterone production in Abcg1 cKO mice.
Figure 4.
Abcg1 cKO mice display increased body weight but unaltered adrenal mass, zonation, and serum lipid profile. (A) Adrenal weight measured on freshly dissected whole adrenals in control and Abcg1 cKO mice. (B) Representative depiction of immunofluorescence assay on adrenocortical sections from control and Abcg1 cKO mice. Images are representative of 4 animals per genotype. Scale bar = 50 µm. (C) Ratio of the zF area (measured as the area stained by Akr1b7) and the zG area (measured as the area stained by Dab2). (D) Lipid profile in control and Abcg1 cKO mouse sera. (E). Quantification of glucocorticoid-sensitive (ie, Gck, Pck1, G6pc) and insensitive (Nr3c1) genes in livers from control, Abcg1 cKO, and Abca1 cKO animals. (F) Quantification of live animal weight. All mice used for this figure were 2-month-old females. **P ≤ .01.
Abbreviations: GC, glucocorticoids; zF, zona Fasciculata; zG, zona Glomerulosa
Furthermore, to exclude that corticosterone production was influenced by a change in systemic lipid metabolism in Abcg1 cKO mice, we performed serum lipid profiling, which revealed no differences in HDL, low-density lipoproteins, total cholesterol, or triglycerides between Abcg1 cKO and control mice (Fig. 4D).
Finally, the systemic response to increased glucocorticoid was estimated in Abcg1 cKO mice by quantifying 3 glucocorticoid target genes in the liver, ie, Gck, Pck1, and G6pc (41-44), which showed a nonsignificant trend of increase compared to control and Abca1 cKO animals (Fig. 4E). Instead, no such trend was observed for Nr3c1, whose expression levels are not sensitive to circulating glucocorticoids (Fig. 4E) (44). In addition, Abcg1 cKO mice displayed a mild increase in body weight compared to control animals (Fig. 4F), compatible with a moderate but prolonged exposure to increased corticosterone (45).
Altogether, our data suggest that loss of Abcg1 results in increased intracellular cholesterol uptake and biosynthesis, leading to higher glucocorticoid production.
Discussion
We show that inactivation of Abcg1 in the adrenal cortex leads to increased expression of genes that promote cholesterol availability (from uptake and biosynthesis), as well as an increase in glucocorticoid production. The increase in glucocorticoid production was observed in both female and male mice, albeit at an older age in male mice, possibly due to a slower rate of recombination and/or tissue turnover in these mice (46, 47).
Although our work does not provide an integrated analysis of 24-hour urine corticosterone metabolites, the absence of ACTH suppression and the analysis of corticosterone-responsive liver genes suggest that Abcg1 cKO mice show only a mild increase of daily corticosterone output, most likely within physiological range. Consistent with this conclusion, we expect only a minor (if any) impact on glucose metabolism, which was not directly investigated in this work. The increase in body weight in Abcg1 cKO mice is compatible with a protracted exposure to moderately increased corticosterone levels (45). In addition, the ACTH values averaging 200 pg/mL throughout all our animal groups possibly reflect a mild stress stimulation, compatible with reported values in rats upon pentobarbital-mediated terminal anesthesia (48).
Surprisingly, our data are in contrast with the mild glucocorticoid insufficiency and decreased cortical cholesteryl esters found by Hoekstra and colleagues in mice following global deletion of Abcg1 (20). This discrepancy could be explained by a possible decrease in corticotropin releasing hormone and/or ACTH in mice with global Abcg1 deletion, which were not assayed in the study. Alternatively, global loss of Abcg1 could lead to functional impairment or dysgenesis of the adrenal cortex, underlying a not yet described role of Abcg1 during intrauterine development. This latter hypothesis is less plausible, though, because of the low level of ABCG1 expression reported in human fetal tissues (21). In our work, we use a conditional mouse model that leads to inactivation of Abcg1 specifically in the steroidogenic cells of the adrenal cortex during the first weeks of postnatal development (31), which allows us to rule out prenatal or systemic effects of Abcg1 deletion on the phenotype. However, an accurate quantification of the extent of recombination in Abcg1 cKO adrenals is technically challenging. Therefore, we cannot exclude the possibility that the differences between Hoekstra and olleagues’ work (20) and ours are due to a different degree of Abcg1 recombination in adrenocortical cells.
Our finding that adrenal Abcg1 inactivation results in upregulation of transcripts important for cholesterol biosynthesis and uptake is in line with the increases seen in Hmgcr, Farnesyl pyrophosphate (Fpp), and Ldlr in the liver from global Abcg1 KO mice (15). This similarity suggests that the genetic network regulated by Abcg1 is conserved among different tissues.
Abcg1 inactivation, however, did not affect transcripts encoding genes directly implicated in steroidogenic conversions, raising the hypothesis that increased adrenal steroidogenesis might be due to excess cholesterol in Abcg1 cKO mice flowing directly into the steroidogenic machinery and fueling the production of the end-product corticosterone. This hypothesis implies that the amounts of cholesterol entering the steroidogenic pathway are loosely controlled, and exposure to functional cholesterol sources (eg, lipoproteins) may directly trigger increased steroidogenesis. While, to our knowledge, this has not been formally tested in vivo, steroidogenesis is directly stimulated by exposure to lipoproteins in primary adrenal cells and in the established NCI-H295R adrenal cell line (49) (and our data, not shown).
It is interesting to note that aldosterone, despite being an adrenal functional end-product, is not affected by Abcg1 inactivation. This is surprising in consideration of the fact that exposure to cholesterol results in increased aldosterone production in vitro (49, 50). We suspect this difference is because aldosterone synthase (Cyp11b2) expression, unlike the expression of 11-beta-hydroxylase (Cyp11b1—the last step in corticosterone biosynthesis) is finely tuned in mice by a range of physiological stimuli. In fact, the expression of Cyp11b2 in mice and rats, unlike in cells, is regulated in such a way that only a subset of zG cells express the enzyme at a given time (51). Excess sodium can suppress Cyp11b2 expression almost completely, while poor dietary sodium intake produces a marked increase in Cyp11b2 (52). Instead, Cyp11b1 is constitutively expressed in zF cells and converts any available substrate into corticosterone (52), including any excess cholesterol that can be present in Abcg1 cKO adrenals. Therefore, we expect that the local concentration of cholesterol and steroid precursors may not affect aldosterone production.
Finally, although the extent to which our findings in mice are relevant to human pathophysiology remains to be explored, our data introduce the possibility that Abcg1 variants may account for physiological or subclinical variation in stress response among healthy subjects. The Human Gene Mutation Database lists 25 different mutations or polymorphisms that have been described in ABCG1 having a possible or probable pathological outcome (53-60). The individuals carrying these variants present with a series of phenotypes or risk associations predominantly linked to cardiovascular disorders, including impaired HDL homeostasis and increased risk for coronary heart disease. However, steroidogenic capacity in these individuals has not been assessed. Given the association between higher serum cortisol concentrations and cardiovascular risk profile (61), it would be of interest to assess basal and stimulated glucocorticoid levels in individuals carrying these alleles, which might explain interindividual variability in basal cortisol or physiological cortisol responses, and excess cortisol levels in individuals carrying risk alleles.
Acknowledgments
We thank Dr. Pierre Val and Dr. Antoine Martinez for sharing the antibodies used for immunofluorescence and Dr. Idoia Martinez de Lapiscina for valuable discussion. We also thank the Translational Research Unit Platform at the University of Bern for supporting the histological procedures. BioRender.com was used to generate schematics.
Contributor Information
Jani Liimatta, Division of Pediatric Endocrinology, Diabetology and Metabolism, Department of Pediatrics, Inselspital, Bern University Hospital, Bern 3010, Switzerland; Department for BioMedical Research, University Hospital Inselspital, University of Bern, Bern 3010, Switzerland; Kuopio Pediatric Research Unit (KuPRU), University of Eastern Finland and Kuopio University Hospital, Kuopio 70200, Finland.
Evelyn Curschellas, Department of Chemistry, Biochemistry and Pharmacy, Medical Faculty, University of Bern, Bern 3010, Switzerland.
Emre Murat Altinkilic, Division of Pediatric Endocrinology, Diabetology and Metabolism, Department of Pediatrics, Inselspital, Bern University Hospital, Bern 3010, Switzerland; Department for BioMedical Research, University Hospital Inselspital, University of Bern, Bern 3010, Switzerland.
Rawda Naamneh Elzenaty, Department for BioMedical Research, University Hospital Inselspital, University of Bern, Bern 3010, Switzerland.
Philipp Augsburger, Department for BioMedical Research, University Hospital Inselspital, University of Bern, Bern 3010, Switzerland.
Therina du Toit, Division of Pediatric Endocrinology, Diabetology and Metabolism, Department of Pediatrics, Inselspital, Bern University Hospital, Bern 3010, Switzerland; Department for BioMedical Research, University Hospital Inselspital, University of Bern, Bern 3010, Switzerland; Department of Nephrology and Hypertension, Inselspital, Bern University Hospital, University of Bern, Bern 3010, Switzerland.
Clarissa D Voegel, Department for BioMedical Research, University Hospital Inselspital, University of Bern, Bern 3010, Switzerland; Department of Nephrology and Hypertension, Inselspital, Bern University Hospital, University of Bern, Bern 3010, Switzerland.
David T Breault, Department of Pediatrics, Harvard Medical School, Boston Children's Hospital, Boston, MA 02115, USA; Harvard Stem Cell Institute, Cambridge, MA 02138, USA.
Christa E Flück, Division of Pediatric Endocrinology, Diabetology and Metabolism, Department of Pediatrics, Inselspital, Bern University Hospital, Bern 3010, Switzerland; Department for BioMedical Research, University Hospital Inselspital, University of Bern, Bern 3010, Switzerland.
Emanuele Pignatti, Division of Pediatric Endocrinology, Diabetology and Metabolism, Department of Pediatrics, Inselspital, Bern University Hospital, Bern 3010, Switzerland; Department for BioMedical Research, University Hospital Inselspital, University of Bern, Bern 3010, Switzerland.
Funding
This work was funded by the Novartis Foundation for Medical-Biological Research (E.P., 22B088), the NCCR RNA&Disease Translational Fellowship Grant (E.P.), the International Fund Congenital Adrenal Hyperplasia (E.P.), the University of Bern via the Initiator grant (E.P.), the Uniscientia Foundation Zürich/Vaduz (C.e.F.), and the research fellowship grants from the Sigrid Jusélius Foundation and the Foundation for Pediatric Research (both from Helsinki, Finland) (J.L.).
Author Contributions
J.L., E.C., M.A., R.N.E., and P.A. assisted with the experiments. T.d.T and C.V. performed the liquid chromatography and mass spectrometry LC-MS analysis. D.T.B. contributed the Cyp11b2tm1.1(cre)Brlt mouse model and edited the manuscript. C.e.F. supervised the project and contributed the laboratory infrastructure. E.P. designed and supervised the project, carried out the experiments, and wrote the manuscript.
Disclosures
The authors have nothing to disclose.
Data Availability
Some or all datasets generated during and/or analyzed during the current study are not publicly available but are available from the corresponding author on reasonable request.
References
- 1. Miller WL, Auchus RJ. The molecular biology, biochemistry, and physiology of human steroidogenesis and its disorders. Endocr Rev. 2011;32(1):81‐151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vaduva P, Bonnet F, Bertherat J. Molecular basis of primary aldosteronism and adrenal cushing syndrome. J Endocr Soc. 2020;4(9):bvaa075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pignatti E, Flück CE. Adrenal cortex development and related disorders leading to adrenal insufficiency. Mol Cell Endocrinol. 2021;527:111206. [DOI] [PubMed] [Google Scholar]
- 4. Donoghue SE, Pitt JJ, Boneh A, White SM. Smith-Lemli-Opitz syndrome: clinical and biochemical correlates. J Pediatr Endocrinol Metab. 2018;31(4):451‐459. [DOI] [PubMed] [Google Scholar]
- 5. Bose HS, Sugawara T, Strauss JF III, Miller WL; International Congenital Lipoid Adrenal Hyperplasia Consortium . The pathophysiology and genetics of congenital lipoid adrenal hyperplasia. N Engl J Med. 1996;335(25):1870‐1878. [DOI] [PubMed] [Google Scholar]
- 6. Mizuno Y, Ishii T, Hasegawa T. In vivo verification of the pathophysiology of lipoid congenital adrenal hyperplasia in the adrenal cortex. Endocrinology. 2019;160(2):331‐338. [DOI] [PubMed] [Google Scholar]
- 7. Miller WL. Disorders in the initial steps of steroid hormone synthesis. J Steroid Biochem Mol Biol. 2017;165(Pt A):18‐37. [DOI] [PubMed] [Google Scholar]
- 8. Deng B, Shen W-J, Dong D, Azhar S, Kraemer FB. Plasma membrane cholesterol trafficking in steroidogenesis. FASEB J. 2019;33(1):1389‐1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Baba T, Otake H, Inoue M, et al. Ad4BP/SF-1 regulates cholesterol synthesis to boost the production of steroids. Commun Biol. 2018;1(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pignatti E, Altinkilic EM, Bräutigam K, et al. Cholesterol deprivation drives DHEA biosynthesis in human adrenals. Endocrinology. 2022;163(7):bqac076. [DOI] [PubMed] [Google Scholar]
- 11. Luo J, Yang H, Song B-L. Mechanisms and regulation of cholesterol homeostasis. Nat Rev Mol Cell Biol. 2020;21(4):225‐245. [DOI] [PubMed] [Google Scholar]
- 12. Shimomura I, Shimano H, Horton JD, Goldstein JL, Brown MS. Differential expression of exons 1a and 1c in mRNAs for sterol regulatory element binding protein-1 in human and mouse organs and cultured cells. J Clin Invest. 1997;99(5):838‐845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shimizu-Albergine M, Van Yserloo B, Golkowski MG, Ong S-E, Beavo JA, Bornfeldt KE. SCAP/SREBP pathway is required for the full steroidogenic response to cyclic AMP. Proc Natl Acad Sci U S A. 2016;113(38):E5685‐E5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bojanic DD, Tarr PT, Gale GD, et al. Differential expression and function of ABCG1 and ABCG4 during development and aging. J Lipid Res. 2010;51(1):169‐181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kennedy MA, Barrera GC, Nakamura K, et al. ABCG1 has a critical role in mediating cholesterol efflux to HDL and preventing cellular lipid accumulation. Cell Metab. 2005;1(2):121‐131. [DOI] [PubMed] [Google Scholar]
- 16. Sturek JM, Castle JD, Trace AP, et al. An intracellular role for ABCG1-mediated cholesterol transport in the regulated secretory pathway of mouse pancreatic β cells. J Clin Invest. 2010;120(7):2575‐2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yvan-Charvet L, Ranalletta M, Wang N, et al. Combined deficiency of ABCA1 and ABCG1 promotes foam cell accumulation and accelerates atherosclerosis in mice. J Clin Invest. 2007;117(12):3900‐3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bensinger SJ, Bradley MN, Joseph SB, et al. LXR signaling couples sterol metabolism to proliferation in the acquired immune response. Cell. 2008;134(1):97‐111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tarr PT, Edwards PA. ABCG1 and ABCG4 are coexpressed in neurons and astrocytes of the CNS and regulate cholesterol homeostasis through SREBP-2. J Lipid Res. 2008;49(1):169‐182. [DOI] [PubMed] [Google Scholar]
- 20. Hoekstra M, Ouweneel AB, Nahon JE, et al. ATP-binding cassette transporter G1 deficiency is associated with mild glucocorticoid insufficiency in mice. Biochim Biophys Acta Mol Cell Biol Lipids. 2019;1864(4):443‐451. [DOI] [PubMed] [Google Scholar]
- 21. Klucken J, Büchler C, Orsó E, et al. ABCG1 (ABC8), the human homolog of the Drosophila white gene, is a regulator of macrophage cholesterol and phospholipid transport. Proc Natl Acad Sci U S A. 2000;97(2):817‐822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sjöstedt E, Zhong W, Fagerberg L, et al. An atlas of the protein-coding genes in the human, pig, and mouse brain. Science. 2020;367(6482):eaay5947. [DOI] [PubMed] [Google Scholar]
- 23. Tarling EJ, Edwards PA. ATP binding cassette transporter G1 (ABCG1) is an intracellular sterol transporter. Proc Natl Acad Sci U S A. 2011;108(49):19719‐19724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang N, Ranalletta M, Matsuura F, Peng F, Tall AR. LXR-induced redistribution of ABCG1 to plasma membrane in macrophages enhances cholesterol mass efflux to HDL. Arterioscler Thromb Vasc Biol. 2006;26(6):1310‐1316. [DOI] [PubMed] [Google Scholar]
- 25. Sano O, Ito S, Kato R, et al. ABCA1, ABCG1, and ABCG4 are distributed to distinct membrane meso-domains and disturb detergent-resistant domains on the plasma membrane. PLoS One. 2014;9(10):e109886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tarling EJ, Edwards PA. Intracellular localization of endogenous mouse ABCG1 IS MIMICKED BY BOTh ABCG1-L550 and ABCG1-P550-brief report. Arterioscler Thromb Vasc Biol. 2016;36(7):1323‐1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pandzic E, Gelissen IC, Whan R, et al. The ATP binding cassette transporter, ABCG1, localizes to cortical actin filaments. Sci Rep. 2017;7(1):42025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gelissen IC, Harris M, Rye K-A, et al. ABCA1 and ABCG1 synergize to mediate cholesterol export to apoA-I. Arterioscler Thromb Vasc Biol. 2006;26(3):534‐540. [DOI] [PubMed] [Google Scholar]
- 29. Wang N, Lan D, Chen W, Matsuura F, Tall AR. ATP-binding cassette transporters G1 and G4 mediate cellular cholesterol efflux to high-density lipoproteins. Proc Natl Acad Sci U S A. 2004;101(26):9774‐9779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kobayashi A, Takanezawa Y, Hirata T, et al. Efflux of sphingomyelin, cholesterol, and phosphatidylcholine by ABCG1. J Lipid Res. 2006;47(8):1791‐1802. [DOI] [PubMed] [Google Scholar]
- 31. Freedman BD, Kempna PB, Carlone DL, et al. Adrenocortical zonation results from lineage conversion of differentiated zona glomerulosa cells. Dev Cell. 2013;26(6):666‐673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Westerterp M, Gourion-Arsiquaud S, Murphy AJ, et al. Regulation of hematopoietic stem and progenitor cell mobilization by cholesterol efflux pathways. Cell Stem Cell. 2012;11(2):195‐206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lopez JP, Brivio E, Santambrogio A, et al. Single-cell molecular profiling of all three components of the HPA axis reveals adrenal ABCB1 as a regulator of stress adaptation. Sci Adv. 2021;7(5):eabe4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Aigueperse C, Martinez A, Lefrançois-Martinez AM, Veyssière G, Jean CI. Cyclic AMP regulates expression of the gene coding for a mouse vas deferens protein related to the aldo-keto reductase superfamily in human and murine adrenocortical cells. J Endocrinol. 1999;160(1):147‐154. [DOI] [PubMed] [Google Scholar]
- 35. Boivin GP, Bottomley MA, Schiml PA, Goss L, Grobe N. Physiologic, behavioral, and histologic responses to various euthanasia methods in C57BL/6NTac male mice. J Am Assoc Lab Anim Sci. 2017;56(1):69‐78. [PMC free article] [PubMed] [Google Scholar]
- 36. Andrieu T, du Toit T, Vogt B, Mueller MD, Groessl M. Parallel targeted and non-targeted quantitative analysis of steroids in human serum and peritoneal fluid by liquid chromatography high-resolution mass spectrometry. Anal Bioanal Chem. 2022;414(25):7461‐7472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chakera AJ, McDonald TJ, Knight BA, Vaidya B, Jones AG. Current laboratory requirements for adrenocorticotropic hormone and renin/aldosterone sample handling are unnecessarily restrictive. Clin Med (Lond). 2017;17(1):18‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Connelly MA, Williams DL. SR-BI and cholesterol uptake into steroidogenic cells. Trends Endocrinol Metab. 2003;14(10):467‐472. [DOI] [PubMed] [Google Scholar]
- 39. Sever N, Lee PCW, Song B-L, Rawson RB, DeBose-Boyd RA. Isolation of mutant cells lacking insig-1 through selection with SR-12813, an agent that stimulates degradation of 3-hydroxy-3-methylglutaryl-coenzyme a reductase. J Biol Chem. 2004;279(41):43136‐43147. [DOI] [PubMed] [Google Scholar]
- 40. Lee PCW, Sever N, DeBose-Boyd RA. Isolation of sterol-resistant Chinese hamster ovary cells with genetic deficiencies in both insig-1 and insig-2. J Biol Chem. 2005;280(26):25242‐25249. [DOI] [PubMed] [Google Scholar]
- 41. Bose SK, Hutson I, Harris CA. Hepatic glucocorticoid receptor plays a greater role than adipose GR in metabolic syndrome despite renal compensation. Endocrinology. 2016;157(12):4943‐4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Imai E, Stromstedt PE, Quinn PG, Carlstedt-Duke J, Gustafsson JA, Granner DK. Characterization of a complex glucocorticoid response unit in the phosphoenolpyruvate carboxykinase gene. Mol Cell Biol. 1990;10(9):4712‐4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kooi BT V, Onuma H, Oeser JK, et al. The glucose-6-phosphatase catalytic subunit gene promoter contains both positive and negative glucocorticoid response elements. Mol Endocrinol. 2005;19(12):3001‐3022. [DOI] [PubMed] [Google Scholar]
- 44. Præstholm SM, Correia CM, Goitea VE, et al. Impaired glucocorticoid receptor expression in liver disrupts feeding-induced gene expression, glucose uptake, and glycogen storage. Cell Rep. 2021;37(5):109938. [DOI] [PubMed] [Google Scholar]
- 45. Palmowski A, Nielsen SM, Boyadzhieva Z, et al. The effect of low-dose glucocorticoids over two years on weight and blood pressure in rheumatoid arthritis: individual patient data from five randomized trials. Ann Intern Med. 2023;176(9):1181‐1189. [DOI] [PubMed] [Google Scholar]
- 46. Grabek A, Dolfi B, Klein B, Jian-Motamedi F, Chaboissier M-C, Schedl A. The adult adrenal cortex undergoes rapid tissue renewal in a sex-specific manner. Cell Stem Cell. 2019;25(2):290‐296.e2. [DOI] [PubMed] [Google Scholar]
- 47. Lyraki R, Schedl A. The sexually dimorphic adrenal cortex: implications for adrenal disease. Int J Mol Sci. 2021;22(9):4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vahl TP, Ulrich-Lai YM, Ostrander MM, et al. Comparative analysis of ACTH and corticosterone sampling methods in rats. Am J Physiol Endocrinol Metab. 2005;289(5):E823‐E828. [DOI] [PubMed] [Google Scholar]
- 49. Cherradi N, Bideau M, Arnaudeau S, et al. Angiotensin II promotes selective uptake of high density lipoprotein cholesterol esters in bovine adrenal glomerulosa and human adrenocortical carcinoma cells through induction of scavenger receptor class B type I. Endocrinology. 2001;142(10):4540‐4549. [DOI] [PubMed] [Google Scholar]
- 50. Xing Y, Cohen A, Rothblat G, et al. Aldosterone production in human adrenocortical cells is stimulated by high-density lipoprotein 2 (HDL2) through increased expression of aldosterone synthase (CYP11B2). Endocrinology. 2011;152(3):751‐763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pignatti E, Leng S, Carlone DL, Breault DT. Regulation of zonation and homeostasis in the adrenal cortex. Mol Cell Endocrinol. 2017;441:146‐155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nishimoto K, Harris RBS, Rainey WE, Seki T. Sodium deficiency regulates rat adrenal zona glomerulosa gene expression. Endocrinology. 2014;155(4):1363‐1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Liu X, Lai H, Xin S, et al. Whole-exome sequencing identifies novel mutations in ABC transporter genes associated with intrahepatic cholestasis of pregnancy disease: a case-control study. BMC Pregnancy Childbirth. 2021;21(1):110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Toma C, Shaw AD, Overs BJ, et al. De novo gene variants and familial bipolar disorder. JAMA Netw Open. 2020;3(5):e203382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dron JS, Wang J, McIntyre AD, et al. Six years’ experience with LipidSeq: clinical and research learnings from a hybrid, targeted sequencing panel for dyslipidemias. BMC Med Genomics. 2020;13(1):23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jin SC, Homsy J, Zaidi S, et al. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat Genet. 2017;49(11):1593‐1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Iossifov I, O’Roak BJ, Sanders SJ, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515(7526):216‐221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Motazacker MM, Peter J, Treskes M, Shoulders CC, Kuivenhoven JA, Hovingh GK. Evidence of a polygenic origin of extreme high-density lipoprotein cholesterol levels. Arterioscler Thromb Vasc Biol. 2013;33(7):1521‐1528. [DOI] [PubMed] [Google Scholar]
- 59. Xu Y, Wang W, Zhang L, et al. A polymorphism in the ABCG1 promoter is functionally associated with coronary artery disease in a Chinese Han population. Atherosclerosis. 2011;219(2):648‐654. [DOI] [PubMed] [Google Scholar]
- 60. Furuyama S, Uehara Y, Zhang B, et al. Genotypic effect of ABCG1 gene promoter -257T > G polymorphism on coronary artery disease severity in Japanese men. J Atheroscler Thromb. 2009;16(3):194‐200. [DOI] [PubMed] [Google Scholar]
- 61. Pilz S, Theiler-Schwetz V, Trummer C, et al. Associations of serum cortisol with cardiovascular risk and mortality in patients referred to coronary angiography. J Endocr Soc. 2021;5(5):bvab017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Some or all datasets generated during and/or analyzed during the current study are not publicly available but are available from the corresponding author on reasonable request.