Abstract
Breast cancer is now the most common cancer globally, accounting for 12% of all new annual cancer cases worldwide. Despite epidemiologic studies having established a number of risk factors, knowledge of chemical exposure risks is limited to a relatively small number of chemicals. In this exposome research study, we used non-targeted, high-resolution mass spectrometry of pregnancy cohort biospecimens in the Child Health and Development Studies to test for associations with breast cancer identified via the California Cancer Registry. Second and third trimester archival samples were analyzed from 182 women who subsequently developed breast cancer and 384 randomly selected women who did not develop breast cancer. Environmental chemicals were annotated with the Toxin and Toxin-Target Database for chemical signals that were higher in breast cancer cases and used with an exposome epidemiology analytic framework to identify suspect chemicals and associated metabolic networks. Network and pathway enrichment analyses showed consistent linkage in both second and third trimesters to inflammation pathways, including linoleate, arachidonic acid and prostaglandins, and identified new suspect environmental chemicals associated with breast cancer, i.e., an N-substituted piperidine insecticide and a common commercial product, 2,4-dinitrophenol, linked to variations in amino acid and nucleotide pathways in second trimester and benzo[a]carbazole and a benzoate derivative linked to glycan and amino sugar metabolism in third trimester. The results identify new suspect environmental chemical risk factors for breast cancer and provide an exposome epidemiology framework for discovery of suspect environmental chemicals and potential mechanistic associations with breast cancer.
Keywords: Cancer, Exposome, Metabolomics, Pregnancy
1. Introduction
Environmental impacts on human health are commonly studied using epidemiologic methods, which support tests for association of individual exposures with health outcome. Humans experience many environmental exposures, however, and these occur along with chemicals derived from diet, intestinal microbiome, dietary supplements, pharmaceuticals, and personal use products. The cumulative measure of environmental influences and associated biological responses throughout the lifespan, which complements the genome in determination of health outcomes, is termed the exposome (Miller and Jones 2014; Vermeulen et al. 2020; Wild 2005). Methods for comprehensive measurement of a human exposome are not available, but progress is ongoing in development of improved surveillance of external exposures as well as biomonitoring of internal exposures. The availability of these methods creates an opportunity to begin to understand the interaction of multiple exposures as causal factors in complex human diseases.
Metabolomics is the study of small molecules in biologic systems and often focused on a relatively small number of precursors and biologic intermediates that are essential to biologic structures and functions. High-resolution metabolomics (HRM) uses advanced mass spectrometry and data science to obtain a broader coverage of chemicals in biological samples, especially to obtain insight into an individual’s exposures and associated biological responses (Jones et al. 2012). The metabolome as a functional readout of the interactions of a person’s genes with exposures from diet and environment (Johnson et al. 2017; Jones et al. 2016; Niedzwiecki et al. 2019) has been used extensively in combination with exposure measurements, such as air pollution (Liang et al. 2018; Ritz et al. 2022) and targeted environmental chemicals, such as 4,4′-dichlorodiphenyl-trichloroethane (DDT), polychlorinated biphenyls (PCB) and polybrominated biphenyls (PBB) (Hu et al. 2020; Walker et al. 2019), to gain understanding of functional associations with exposures.
High-resolution metabolomics with non-targeted mass spectrometry methods for biomonitoring delivers tens of thousands of data points per biologic samples, well beyond the number of chemicals measured using targeted methods (Uppal et al. 2016). Early studies showed that these mass spectrometry methods detect chemicals in human plasma derived from diet, intestinal microbiome, dietary supplements, pharmaceuticals, personal use products and environmental exposures (Johnson et al. 2010; Soltow et al. 2013; Walker et al. 2016). Although the total number of chemicals detected remains uncertain, available evidence indicate that an omics scale detection is achieved, i.e., detection of tens of thousands of chemical signals is of a magnitude similar to the number of genes in the human genome. To date, there has been limited use of the information-rich, high-resolution mass spectrometry data in a non-targeted manner for discovery of new chemical associations with disease.
Use of omics scale biomonitoring to discover associations with disease has been termed “exposome epidemiology” (Jones and Cohn 2020). The current study was undertaken to develop exposome epidemiology concepts for discovery of potential new environmental factors contributing to breast cancer. The long-term goal for complex diseases such as breast cancer, which involves multiple genetic, behavioral and environmental factors, is to integrate omics scale biomonitoring with other factors to ultimately improve prediction and intervention strategies to decrease disease burden. The results obtained in the present study provide evidence for new candidate environmental breast carcinogens and information concerning possible mechanisms by which these agents contribute to breast cancer.
In this study, archival blood samples from the Child Health and Development Studies (CHDS) cohort collected during pregnancy were used to perform a non-targeted exposome-wide association study of breast cancer. The CHDS cohort includes samples collected from 1959 to 1967 from about 20,000 pregnancies in Oakland, California, USA. Breast cancer diagnoses were identified by linkage to the California Cancer Registry through 1997, and the current study used archival second (T2) and third (T3) trimester samples of 182 women who subsequently developed breast cancer for comparison to samples from 384 women who did not develop breast cancer.
2. Materials and methods
2.1. Analytical workflow for detection of suspect chemicals associated with subsequent breast cancer outcome
We developed an exposome epidemiology approach (codes for analysis are available in Supplemental Table S1) in which we first performed non-targeted statistical analysis to select HRM features that were higher in cases than in non-cases. The decision to only look at features that were higher in association with subsequent breast cancer was based upon 1) the assumption that causative features would be higher while protective features would likely be lower in association breast cancer outcome and 2) preliminary analyses showed that more features were negatively associated and these were enriched in features annotated as diet-derived phytochemicals, which would require different databases than those for toxic substances. Additionally, an untargeted analysis of possible breast cancer-preventive exposures in this cohort was considered worthy of independent investigation.
In the selection of features positively associated with breast cancer outcome (higher abundance in breast cancer than non-breast cancer), we used raw p <0.05 as a cutoff for selection based upon the expectation that multiple environmental factors with small effect size may contribute to breast cancer. Furthermore, we considered this more liberal selection criteria, compared to more rigorous False Discovery Rate (FDR) criteria, as more appropriate for a small-population discovery study because it provides a better balance between Type 1 and Type 2 statistical error in detection of exposures with small effect size (Uppal et al. 2016). We selected HRM features that were higher in cases than non-cases to search for potentially causative factors by matching to a xenobiotic database, Toxin and Toxin-Target Database (T3DB) (Lim et al. 2010; Wishart et al. 2015), recognizing that protective factors that are decreased in association with breast cancer would prioritize use of HRM features that were lower in cases and an alternative database, such as Food Database (FoodDB) (Scalbert et al. 2011). HRM features that were increased with breast cancer and annotated as environmental chemicals were then used for network analysis with the data-dependent community detection tool, xMWAS (Uppal et al. 2018) to select most highly associated mass spectral features. Pathway enrichment analysis of the most central communities of mass spectral features was then performed with mummichog (Li et al. 2013), and targeted mass spectrometry was used to improve understanding of the annotated chemicals associated with breast cancer outcome.
2.2. Samples and assays
The CHDS recruited women residing in the Oakland and East Bay, California area who were members of the Kaiser Foundation Health Plan and received obstetric care for pregnancies between 1959 and 1966 with deliveries extending into 1967 (van den Berg et al. 1988). >98% of all eligible women enrolled.
Blood samples were collected from these mothers during pregnancy in each trimester and in the early post-partum period by CHDS research staff without request for fasting, processed to isolate serum, and stored since then at − 20° Celsius. Second and third trimester archival samples available were analyzed for HRM and environmental chemicals. The samples used for the present study include second (T2, n = 182)) and third (T3, n = 172)) trimester archival samples of 201 women who subsequently developed breast cancer, compared to second (T2, n = 384) and third (T3, n = 351) trimester archival samples from 413 women who did not develop breast cancer. Breast cancer cases were identified by linkage to the California Cancer Registry for cases diagnosed through 1997. Record abstraction for cancer diagnoses to the California Cancer Registry is based primarily on pathology reports, and case identification is considered to be >99% complete after a 2-year lag (Perkins et al.). Cases were defined as mothers with incident invasive or noninvasive breast cancer diagnosed at a median age at diagnosis of 54 years (interquartile range, 13 years) with available prenatal serum and a standardized gross placental exam. Non-cases were an 8% sample of women not known to have breast cancer randomly selected among mothers with available prenatal serum and a standardized gross placental exam (Cohn et al. 2017). Breast cancer rates for included and excluded subsets in the CHDS cohort were highly comparable suggesting selection did not impose significant bias: 1.89 per 1,000 person-years (95% Confidence Interval ([95% CI] = 1.60, 2.23) for included vs. 1.88 per 1,000 (95% CI = 1.59, 2.20) for excluded. It is possible, however, that we missed some cases of breast cancer, including among women who are identified in this study as “non-cases”. In this case, failure to identify cases among the non-case group would result in underestimating differences between cases and non-cases in these analyses and would not be expected to impact findings. The CHDS founding mothers voluntarily participated in an in-person interview and gave permission to access their own medical records and those of their children to researchers. The institutional review board of the Public Health Institute approved the present study, and we complied with all federal guidelines governing the use of human participants. Forty-six percent of cases and forty-five percent of the non-cases had available data and serum.
2.3. Chemicals
HPLC grade acetonitrile and methanol, LC-MS water and 98% formic acid were obtained from Sigma-Aldrich (St. Louis, MO). A mixture of 14 stable isotopic chemicals were used as an internal standard (Go et al. 2015) included [13C6]-D-glucose, [15N]-indole, [2-15N]-L-lysine dihydrochloride, [13C5]-L-glutamic acid, [13C7]-benzoic acid, [3,4-13C2]-cholesterol, [15N]-L-tyrosine, [trimethyl-13C3]-caffeine, [15N2]-uracil, [3,3-13C2]-cystine, [1,2-13C2]-palmitic acid, [15N,13C5]-L-methionine, [15N]-choline chloride and 2′-deoxyguanosine-15N132, C10-5′-monophosphate from Cambridge Isotope Laboratories, Inc (Andover, PA).
2.4. High-resolution mass spectrometry
Serum samples were analyzed with liquid chromatography-high resolution mass spectrometry (LC-HRMS) as described previously (Jarrell et al. 2021; Jarrell et al. 2020). Briefly, 50 μL of serum was treated 2:1 (v/v) with acetonitrile, and 2.5 μL of the stable isotope standard mixture was added. Proteins were precipitated by incubation at 4 °C for 30 min and removed by centrifugation for 10 min at 21,000×g at 4 °C. Supernatants were placed in autosampler vials and maintained at 4 °C in an autosampler until analysis. Two pooled human reference samples including NIST SRM1950 and Qstd [pooled plasma from 2 separate lots from Equitech-Bio, Inc (Kerrville, Texas)] were included. NIST SRM1950 was run at the beginning and end of the full sample set, and Qstd was included at the beginning and end of each batch of 20 samples.
Samples and reference materials were analyzed with three technical replicates using a High-Field Q-Exactive mass spectrometer (Thermo Fisher) with C18 chromatography and electrospray ionization (ESI) in negative mode. Data collection occurred continuously throughout 5 min of chromatographic separation from 85 to 1,275 mass-to-charge ratio (m/z). Data extraction was performed using apLCMS and xMSanalyzer, generating mass spectral features consisting of m/z, retention time (RT) and peak intensity. Feature and sample filtering retained features with a median CV of 50% or less, a minimum mean Pearson correlation coefficient of 0.7 between technical replicates of each sample, and presence in at least 30% of samples.
2.5. Selection of features associated with subsequent breast cancer
Age and pregnancy estrogens interacted and together were associated with breast cancer outcomes in this sample (Cohn et al. 2017). We reasoned that controlling for these covariates had the potential to mask associations with environmental chemicals because environmental chemicals and biologic responses to exposures can accumulate with age. Hence, analyses were performed without adjustments for covariates. Metabolome-Wide Association Study (MWAS) was performed with LIMMA [Linear Models for Microarray and RNA-Seq Data (Ritchie et al. 2015)] to select m/z features that were positively associated with breast cancer outcome. LIMMA uses linear models to assess differential expression simultaneously for many mass spectral signals in a manner that borrows information across mass spectral signals to improve stability for individual signals. The method was developed and validated for microarray studies and previously applied successfully to untargeted mass spectrometry analyses. In LIMMA, a moderated t-statistic is used for significance analysis and provides p-values computed for each mass spectral signal and for each comparison. This differs from an ordinary t-statistic by having standard errors moderated across signals using a simple Bayesian model. Intensity values were log2 transformed prior to analysis with LIMMA to reduce heteroscedasticity, and LIMMA was performed using the R package, xmsPANDA (https://github.com/kuppal2/xmsPANDA), with retention of features at raw p < 0.05.
2.6. Annotation of mass spectral features and metabolite identification
To select mass spectral features of environmental interest, features positively associated with subsequent breast cancer diagnosis were subjected to a multistage clustering algorithm and annotated with xMSannotator (Uppal et al. 2017) using T3DB (Lim et al. 2010; Wishart 2015) at 5 ppm tolerance. For selected chemicals, identities were confirmed when possible by accurate m/z match, co-elution with authentic standards and ion dissociation mass spectrometry (MS2); Level 1 identification by criteria of Schymanski et al (Schymanski et al. 2014)].
2.7. Targeted mass spectrometry analysis in identification of features
Annotated chemicals at the center of the xMWAS network structures underwent additional investigation by ion dissociation, structure analysis, and spectral matching to publicly available spectral libraries. Plasma samples with the highest intensities of the chemicals of interest were selected and analyzed with C18 liquid chromatography (Dionex Ultimate 3000) and MS2 ion dissociation (Thermo Scientific Fusion) with negative ESI. The mass spectrometer was set to scan a minimal range of m/z at 60,000 resolution for MS1. Then, an inclusion list isolated the m/z of interest. MS2 spectra were acquired in HCD mode with a normalized collision energy of 30% and analyzed at 30,000 resolution using the dual-pressure linear ion trap. Raw MS1 and MS2 spectra were analyzed in xCalibur QualBrowser (Thermo). Fragmentation patterns were analyzed with ChemDraw (PerkinElmer Informatics) and compared against experimental spectra in the MassBank of North America (MoNA) library. MoNA MS2 spectra were selected for negative ESI and similar collision energy. When MoNA spectra were not available, experimental spectra were searched against the NIST Tandem Mass Spectral Library, which provides a spectral match percentage but no information on collision energy or ionization polarity. To align with the goal of high-throughput analyses, chromatographic gradients and fragmentation settings were maintained to be consistent with the original analysis and not extensively optimized for each chemical. Under these conditions, the complexity of the plasma sample matrix and the low abundance of environmental chemicals does not allow for the isolation of one precursor and its fragments. Therefore, in some spectra, the precursor mass is easily visible within 10 ppm error, but the fragments are not clearly visible due to interference from other ions. Spectral peaks corresponding to product ions were identified by their theoretical structures in “ChemDraw” with unit resolution in the dual-pressure linear ion trap detector.
2.8. Network and pathway analyses
Annotated environmental chemicals were tested for associations with m/z features using xMWAS based on partial least-squares regression (Uppal et al. 2018). This data-dependent approach supports agnostic detection of top communities of mass spectral features associated with outcome (breast cancer diagnosis). In this analysis, selection of statistical thresholds for visualization of communities is arbitrary, i.e., the purpose is to identify the top communities associated with subsequent breast cancer diagnosis. In these analyses, retention of features with p < 0.05 maximizes opportunity to detect associations with small effect size. The top communities of associated features can be visualized with different |r| correlation thresholds, in which a higher threshold yields more sparse community structures compared to lower thresholds. Higher |r| thresholds limit the communities to the most central drivers of association while lower |r| thresholds are valuable for subsequent pathway enrichment analyses to identify metabolic pathway associations useful for mechanistic insight. In the present analyses, thresholds for inclusion in the networks were |r| > 0.30 and p < 0.05. Features associated with network structures were used for pathway enrichment analysis using mummichog (v3) (Li et al. 2013). Enriched pathways were filtered for those that included at least 3 significantly associated metabolites at p < 0.05.
3. Results
3.1. Study population demographics
T2 and T3 samples were available from 201 individuals who subsequently developed breast cancer. These individuals had a median age of 31 y at time of blood collection, and the individuals without breast cancer had a median age of 36 (p < 0.0001). Serum collection occurred between 1960 and 1964, with T2 blood collection occurring at a median gestational age of 161 and 160 days in cases and non-cases, respectively. For T3, blood collections were at a median gestational age of 249 and 250 days, respectively. Characteristics of cases and non-cases are provided in Table 1.
Table 1.
Characteristics of Study Population.
| Non-Cases (N = 413) |
Cases (N = 201) |
|||
|---|---|---|---|---|
| Characteristic | Median | (IQR) | Median | (IQR) |
| Year of mother’s birth | 1936 | (9) | 1931 | (11) |
| Year of blood draw | 1962 | (2) | 1962 | (2) |
| Gestational day of T2 blood draw | 160 | (33) | 161 | (29) |
| Gestational day of T3 blood draw | 250 | (17) | 249 | (16) |
| BMI at first prenatal visit (kg/m2)a | 22.1 | (4.3) | 22.6 | (4.2) |
| Characteristic | Percent | Percent | ||
|
| ||||
| Race | ||||
| non-Hispanic Caucasian | 70 | 69 | ||
| African American | 20 | 19 | ||
| Hispanic | 2 | 1 | ||
| Asian | 6 | 7 | ||
| Mixed | 2 | 3 | ||
3.2. Metabolic feature profiling on T2 and T3 serum
To select m/z features which were positively associated with subsequent breast cancer diagnosis, we retained 9042 m/z features present in at least 80% of samples for statistical analyses and selection of features for annotation. For T2 and T3 measurements, no features had FDR < 0.05 and therefore none are likely to be useful biomarkers for breast cancer risk. For statistical analyses, T2 and T3 analyses were done independently because of the dynamic, changing, maternal and fetal interactions that occur between T2 and T3 and because of the potential for extensive pregnancy-related metabolic changes to mask more subtle associations with subsequent breast cancer diagnosis. Any of the m/z features selected with raw p < 0.05 could contribute to breast cancer risk, and 521 features had p < 0.05 for T2, and 557 features had p <0.05 for T3. Of these m/z features, 188 were higher (333 were lower) in breast cancer than non-cases in T2 samples while 151 were higher (406 were lower) in breast cancer than non-cases in T3 samples (Table 2). These positively associated m/z features were selected to search for possible matches to environmental chemicals.
Table 2.
HRM profiling of T2 and T3 serum samples comparing breast case (BC) and non-case groups (p < 0.05).
| T2 (182 BC, 384 non-cases) | T3 (172 BC, 351 non-cases) | |
|---|---|---|
| Total number of metabolic features | 9042 | 9042 |
| Features differing at. p < 0.05 | 521 | 557 |
| Differing features with BC/non-cases ratio > 1.0 | 188 | 151 |
3.3. Environmental chemical annotation of suspect chemicals in breast cancer group
Most of the selected m/z features are likely to represent endogenous metabolites, but environmental chemicals contributing to subsequent breast cancer could also be present. To search for suspect environmental chemicals associated with subsequent breast cancer diagnosis, we used xMSannotator, a network-based computational tool, with the Toxin and Toxin-Target Database, T3DB (Wishart 2015; Wishart et al. 2007), to further characterize the mass spectral features which were higher in their intensities in women who went on to develop breast cancer. xMSannotator uses a multistage clustering algorithm in which intensity profiles, retention time characteristics, mass defect, and isotope/adduct patterns are used to assign confidence levels to annotation results relative to publicly available databases, such as T3DB. The annotated environmental chemicals for T2 and T3 are shown in Table 3 and Table 4, respectively. More environmental chemicals were annotated in T2 than T3 serum, yielding 17 and 7, respectively, and these included matches to chemicals widely used for anti-inflammatory, anticonvulsant and antipsychotic drugs, quaternary ammonium salt, pesticides, herbicides, fungicides, plasticizers, preservatives, cleaning materials, and flavorings. Importantly, at this level of investigation, these annotations are only suspect chemicals and require further investigation and verification (see below). The average abundance of these chemicals based upon relative intensities of the mass spectral signals was between 10% and 30% higher in women who went on to develop breast cancer compared to those who did not (p ≤ 0.05, Tables 3 and 4).
Table 3.
T3DB-annotated m/z features higher in T2 serum of women who subsequently developed breast cancer (BC) compared with non-cases (p < 0.05).
| Feature | Name | m/z | RT (sec) | % Higher in BC | Category |
|---|---|---|---|---|---|
| F1* | 4-Hydroxyretinoic acid | 315.1957 | 177 | 20 | Vitamin A metabolite |
| F2* | Unidentified | 334.2090 | 125 | 20 | Unidentified |
| F3* | 2-Nitromethylene-piperidine | 141.0670 | 278 | 10 | Insecticide |
| F4* | 2,4-Dinitrophenol | 183.0046 | 283 | 10 | Multiple commercial uses |
| F5* | Oxalic Acid | 88.9883 | 129 | 10 | Cleaning agent |
| F6* | N-acetyl-proline | 156.0666 | 283 | 10 | Endogenous metabolite |
| F7* | Unidentified | 345.0777 | 51 | 20 | Unidentified |
| F8 | Unidentified | 225.0712 | 139 | 30 | Unidentified |
| F9 | Quaternium-52 | 610.4093 | 195 | 20 | Quaternary ammonium salt |
| F10 | Ethyl cyanoacrylate | 124.0404 | 14 | 10 | Cyanoacrylate glue component |
| F11 | 2-Chloro-4,5-xylyl N-hydroxy-N-methylcarbamate | 228.0427 | 284 | 10 | Cholinesterase inhibitor |
| F12 | Psoralen, Angelicin | 167.0135 | 20 | 20 | Skin nodule treatment, |
| F13 | Hexazinone | 273.1344 | 32 | 10 | Herbicide |
| F14 | MHP (Methyl hydrogen phthalate) | 179.0350 | 138 | 10 | Organic compound |
| F15 | 2-Hydroxyphenyl methylcarbamate | 166.0508 | 212 | 10 | Pesticide |
| F16 | Levodopa (also exists as natural product) | 178.0510 | 23 | 20 | Dopamine precursor drug |
| F17 | Phenylmercuric acetate | 337.0157 | 22 | 20 | Preservative |
Targeted mass spectrometry analysis was performed to aid in identification of features F1 to F7 which were found to occur in central communities of Fig. 1, indicated by asterisk.
T3DB annotations are provided for others (F8-F17).
Table 4.
T3DB-annotated m/z features higher in T3 serum of women who subsequently developed breast cancer (BC) compared with those in the non-case women (p < 0.05).
| Feature | Name | m/z | RT (sec) | % Higher in BC | Category |
|---|---|---|---|---|---|
| F18* | Ergosterol | 271.1195 | 25 | 10 | Vitamin D2 precursor |
| F19* | Benzoate derivative | 214.9728 | 22.8 | 10 | Food preservative |
| F20* | Benzo[a]carbazole | 217.0866 | 21 | 10 | Heterocyclic amine |
| F21* | Glucose | 215.0327 | 23 | 10 | Endogenous metabolite |
| F22 | Trimethadione | 142.0509 | 277 | 10 | Anticonvulsant |
| F23 | Heptabromobiphenyl | 734.4246 | 195 | 20 | Aromatic compound |
| F24 | Chlorpromazine | 317.0876 | 135 | 10 | Antipsychotics |
Targeted mass spectrometry analysis was performed to aid in identification of features F18-F21 which were found to occur in central communities of Fig. 3, indicated by asterisk.
T3DB annotation is provided for others (F22-F24).
3.4. Metabolome-Wide association study (MWAS) of T2 environmental chemicals
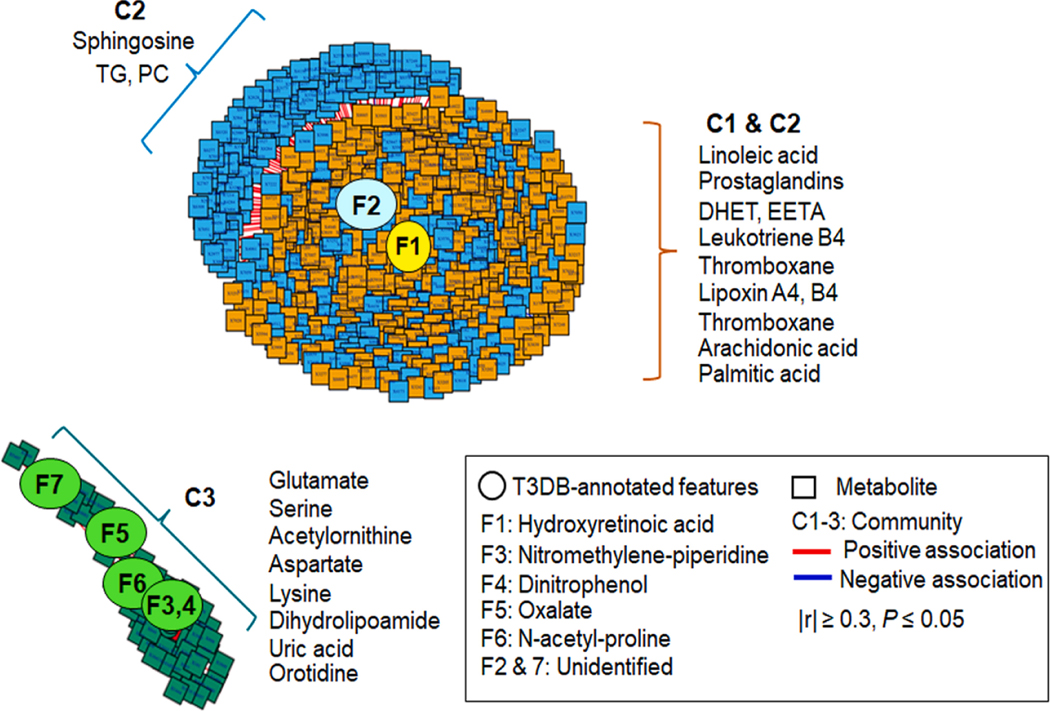
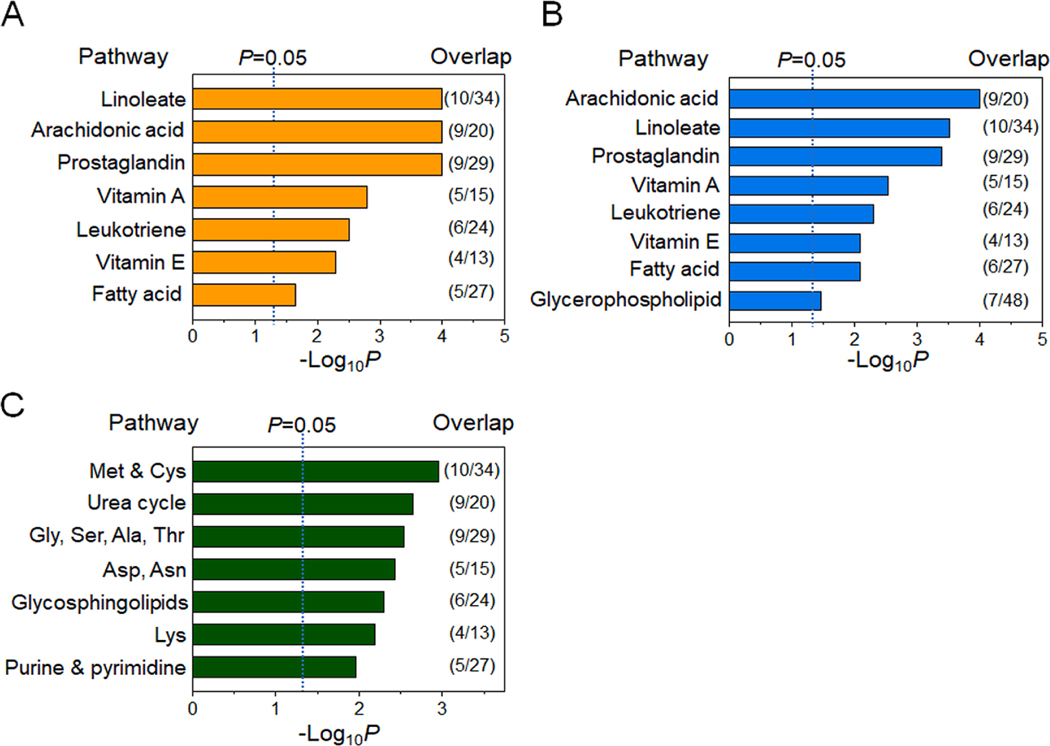
Prior studies have identified inflammatory lipid pathways (linoleate, arachidonate, prostaglandin), lipid and energy metabolism pathways (fatty acids, TCA cycle), oxidative stress pathways (methionine and cysteine), and nitrogen metabolism pathways (urea cycle, pyrimidine, purine) as top metabolic pathway associations with environmental chemicals and breast cancer (Hu et al. 2019; Li et al. 2020; Walker et al. 2019). Data-driven analysis of features annotated as environmental chemicals (Table 3) showed three metabolic communities labeled in Fig. 1 as Community 1 (C1, orange), Community 2 (C2, blue) and Community 3 (C3, green). C1 was positively associated with m/z 315.1957 (F1, Fig. 1, Table 3). Pathway enrichment analysis of the 716 metabolic features associated with C1 showed that the associated pathways included linoleate, arachidonate and prostaglandins (Fig. 2A), which are closely related to inflammation and oxidative stress. Database searches showed that m/z 315.1957 matched multiple lipids (e.g., prostaglandins, resolvins), consistent with the strong associations with related lipid species having activities in inflammation. The vitamin A pathway was also associated with C1 (Fig. 2A), and targeted mass spectrometry with MS/MS analysis showed a likelihood that m/z 315.1957 was 4-hydroxyretinoic acid (F1, Supplemental Fig. S1).
Fig. 1.
Metabolome-Wide Association Study (MWAS) of T3DB-annotated chemicals higher in second trimester (T2) serum. Association of seventeen T3DB-annotated features (Table 3) that are higher in breast cancer than non-cases with the metabolome (9,042 metabolic features) from T2 serum (n = 384 for non-cases, n = 182 for breast cancer) are examined using xMWAS. The outcome of xMWAS analysis is visualized with two separate networks at |r| ≥ 0.3. The networks include three communities (C1, C2, C3) and show central seven chemicals (F1-F7) with tight connections [C1 (orange). C2 (blue), C3 (green)]. Representative metabolites of pathways associated with community are shown next to each community. DHET: dihydroxyeicosatrienoic acid, EETA: epoxyeicosatrienoic acid, TG: triglyceride, PC: phosphatidylcholine.
Fig. 2.
Metabolic pathway associated with second trimester (T2) chemicals. Pathway enrichment analysis with metabolites of three network communities in Fig. 1 was conducted using mummichog. A) A total of seven pathways were found altered with m/z 315.1957 (F1) in C1, B) Eight pathways were altered with m/z 334.2090 (F2) in C2, and C) Seven pathways were altered with environmental chemicals and drug metabolites (F3-F7) (p < 0.05). The ratio of selected metabolites mapped to the listed pathway over the number of total pathway metabolites detected is provided to the right of each bar.
C1 extensively overlapped with Community 2 (C2, blue), which was positively associated with m/z 334.2090 (F2, Fig. 1, Table 3). In addition to pathway associations with prostaglandins, linoleic acid and arachidonic acid, a subset of C2 included phospholipids, e.g., sphingosine, phosphatidyl choline (PC), and triglyceride (blue, Fig. 1), associated with glycerophospholipid metabolism. Thus, this chemical defines a separate cluster of metabolites associated with breast cancer than those in C1, but this feature remains unidentified because subsequent mass spectrometry analysis failed to provide support for identification. Together, results from C1 and C2 support prior findings that inflammatory lipid and other lipid pathways associated with DDT, PCB and PBB are associated with breast cancer, but do not reveal any new suspect environmental chemicals.
A third community, C3, had metabolomic associations (green) which were separated from C1 and C2 and positively associated with m/z features annotated as environmental chemicals, including an insecticide (F3, nitromethylene-piperidine), a wood preservation and dye production chemical (F4, 2,4-dinitrophenol), oxalate (F5), and F6, m/z 156.0666 and F7, m/z 345.0777 (Fig. 1, Table 3). Of these, oxalate was previously confirmed, and supportive targeted mass spectrometry data was obtained for 2-nitromethylene-piperidine and 2,4-dinitrophenol (F3 and F5, Supplemental Fig. S1). The m/z feature 156.0666 (F6) was present at high concentrations in most samples and appeared likely to be an endogenous metabolite, N-acetylproline (F6, Supplemental Fig. S1). No supportive mass spectrometry data could be obtained for the m/z feature 345.0777 (F7). The 56 metabolic features in C3 that were positively associated with these annotated environmental chemicals were related to pathways for antioxidant methionine (Met) and cysteine (Cys) regulation, urea cycle, amino acids (glycine, serine, alanine, threonine, aspartate, asparagine, lysine), glycerophospholipids, and purine and pyrimidine nucleotide metabolism (Fig. 2C). Representative metabolites associated with these pathways of each community are indicated next to each community (Fig. 1) and included amino acids (glutamate, serine, lysine, aspartate), a pyrimidine (orotidine), a purine (uric acid), and a coenzyme for mitochondrial electron transfer and the TCA cycle (dihydrolipoamide). Collectively, the results for T2 serum of women who subsequently developed breast cancer show that two suspect environmental chemicals, nitromethylene-piperidine and 2,4-dinitrophenol, along with oxalate, are closely associated with metabolic perturbations related to amino acid and nucleotide metabolism. Other communities included lipids functioning in inflammation, sphingolipids and lipids, but no suspect environmental chemicals were identified for these communities.
3.5. MWAS of T3 environmental chemicals and alterations in metabolic pathway
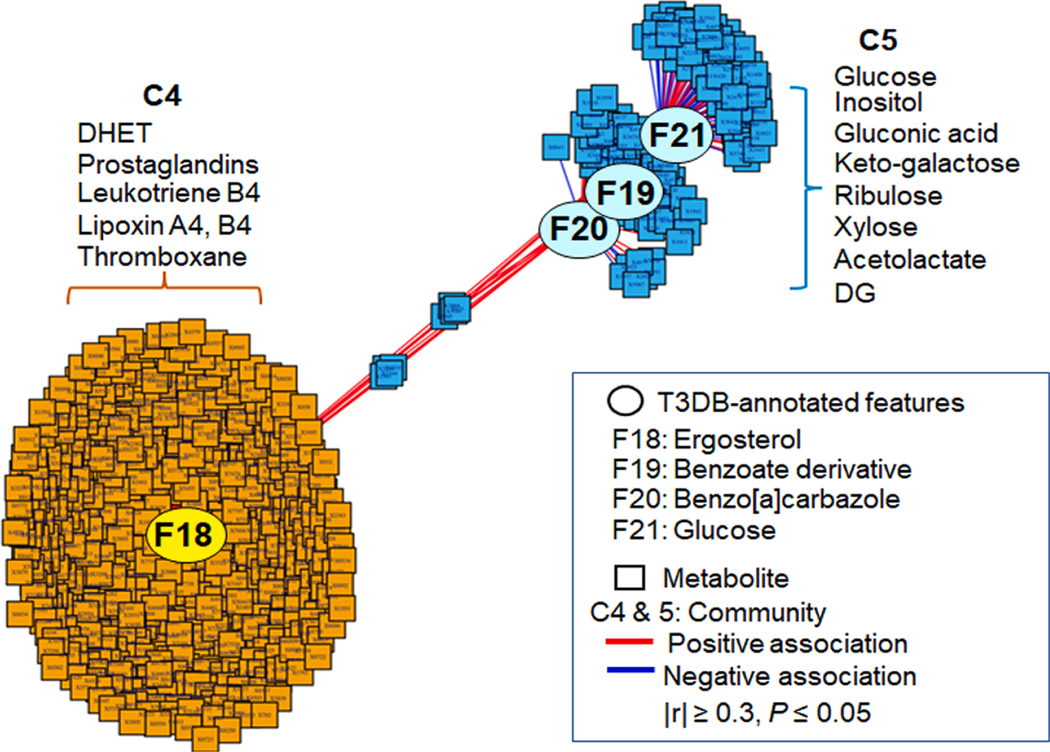
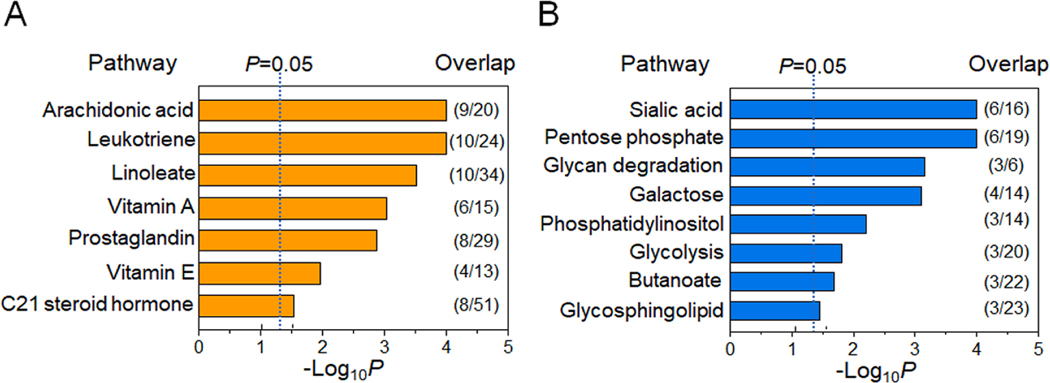
Following the same experimental approach shown above, we examined the relationship of annotated matches of T3 suspect environmental chemicals (Table 4) and changes in metabolites using xMWAS (Uppal et al. 2018). Two major metabolic communities were identified (Fig. 3), with the first community (labeled C4, orange) including metabolites associated exclusively with m/z 271.1195 (F18) and second community (C5, blue) including metabolites associated with m/z 214.9728 (F19), m/z 217.0866 (F20) and m/z 215.0327 (F21) (Fig. 3). Targeted mass spectrometry analysis of m/z 271.1195 (F18) showed that this feature was likely ergosterol or related derivative (F18, Supplemental Fig. S2). Pathway enrichment analysis of the 650 metabolic features of C4 showed associated pathways for arachidonate, leukotriene, linoleate, vitamin A, prostaglandins, vitamin E and steroid hormones (Fig. 4A). Most of these pathways are closely related to inflammation and oxidative stress, as also found in T2 associations in C1 and C2 (Fig. 2A, 2B). Targeted mass spectrometry analyses showed that m/z 214.9713 (F19) was likely a bromine adduct of a benzoic acid metabolite (F19, Supplemental Fig. S2); m/z 217.0866 (F20) had an MS2 spectrum with 83% match to benzo[a]carbazole (F20, Supplemental Fig. S2); m/z 215.0327 (F21) was a high abundance signal from glucose (F21, Supplemental Fig. S2). Pathway enrichment analysis of the 155 metabolic features in C5 showed associations for sialic acid, pentose phosphate, N-glycan degradation, galactose, phosphatidylinositol, glycolysis and gluconeogenesis, butanoate, and glycosphingolipid pathways (Fig. 4B). Representative metabolites associated with these pathways are indicated next to each community (Fig. 3). Collectively, suspect environmental chemicals in Communities 4 and 5 include a benzoate derivative and benzo[a]carbazole, and metabolic pathway associations suggesting that breast cancer effects could be mediated through inflammatory lipids, glucose-related metabolism and estrogenic signaling.
Fig. 3.
Metabolome-Wide Association Study (MWAS) of T3DB-annotated chemicals higher in third trimester (T3) serum. Association of seven T3DB-annotated chemicals (Table 4) that are higher in breast cancer than non-cases with the metabolome (9,042 metabolic features) from T3 serum (n = 351 for non-cases, n = 172 for breast cancer) are examined using xMWAS. The network is visualized at |r| ≥ 0.3 and includes two communities (C4 and C5) showing central four chemicals with tight connections [C4 (orange)] than C5 (blue)]. Representative metabolites of pathways associated with community are shown next to each community.
Fig. 4.
Metabolic pathway associated with T3 chemicals. Pathway enrichment analysis with metabolites of two network communities in Fig. 3 was conducted using mummichog. A) Seven pathways were found altered with F18 chemical in C1and B) eight pathways were altered with chemicals (F19-F21) in C2 (p < 0.05). The ratio of selected metabolites mapped to the listed pathway over the number of total pathway metabolites detected is provided to the right of each bar.
3.6. Comparisons of T2 and T3 suspect chemical-metabolic pathway associations
In comparison of findings for T3 and T2, metabolic pathways for C4 in T3 showed considerable overlap with the pathways associated with C1 and C2 in T2. In contrast, the remaining community, C5, in T3 differed substantially from the remaining community, C3, in T2. Specifically, in T3, we identified a suspect benzoate and benzo[a]carbazole in breast cancer cases that were associated with pathways of carbohydrate metabolism functioning in extracellular matrix turnover and complex carbohydrate metabolism. In T2, we identified a suspect insecticide, 2-nitromethylpiperidine, a widely used commercial chemical, 2,4-dinitrophenol, and oxalic acid, in breast cancer cases that were associated with central pathways functioning in amino acid homeostasis, urea cycle, pyrimidine and purine metabolism, and defense against oxidative stress.
4. Discussion
The present study applies a new exposome epidemiology approach to discover suspect environmental chemicals that may be implicated in breast cancer development. New suspect chemicals from this analysis include an insecticide, nitromethylene-piperidine; a common commercial chemical, 2,4-dinitrophenol; a heterocyclic amine with diverse commercial applications, benzo[a]carbazole; a benzoate derivative which could be derived from natural products or environmental chemicals; and oxalate, an endogenous metabolite that is also used as a cleaning agent. These chemicals were at higher abundance in serum of women decades before breast cancer diagnosis. Importantly, the network analysis is data-dependent, raising the possibility that individual chemicals could have a causal role in breast cancer or that breast cancer could occur through interaction of multiple chemicals in causal mechanisms. In the latter case, individual variations in exposure could have important impact on risk.
New approaches to identify suspect carcinogens are critical because research over the last 50 years has not translated to a strategy for individual breast cancer prevention. The suspect chemical signal F18 (m/z 271.1195) in Community 4, was consistent with identification as ergosterol, which is derived from yeast and precursor of vitamin D2. Pathway mapping of this community showed overlap with inflammatory lipid pathways found in Communities 1 and 2, and also included vitamin A metabolism, which is consistent with feature F1 in Community 1, 4-hydroxyretinoic acid, being a suspect chemical. Many diet-derived carotenoids can be converted to isobaric species to 4-hydroxyretinoic acid, and these are not distinguished by the HRM methods used. Thus, future research will be needed to address the potential for diet-environment interactions, such as implied by the overlap of estrogen-, vitamin D- and vitamin A-linked metabolites. Pregnancy impacts initiation, progression, and susceptibility to breast cancer (Troisi et al. 2018), and therefore pregnancy provides an appropriate time frame for study. Later age at pregnancy is a long-established breast cancer risk factor (Albrektsen et al. 2005; MacMahon et al. 1970) which has become increasingly common and is still unexplained. Because first birth rates have increased 6-fold for women ages 35–39 from 1973 to 2006 (Martin et al. 2015), more detailed understanding of the respective contributions and interactions must be a priority to learn how to mitigate risk for this population group of higher risk women.
In T2, an additional chemical cluster containing an insecticide (nitromethylene-piperidine), a cleaning agent (oxalate), a chemical used in wood preservation and dye production (dinitrophenol), and two unidentified chemical features (F2, F7), were associated with multiple amino acid pathways and nitrogen metabolism (urea cycle, pyrimidine, purine). The amino acid pathways overlap with pathways previously found to vary with persistent organic pollutants, p,p’-DDT (Hu et al. 2020) and PBBs (Walker et al. 2019). In a multigenerational study, p,p’-DDT exposure in women before puberty was found to be associated with breast cancer in mothers (Cohn et al. 2007) and in utero o’,p’-DDT exposure was associated with breast cancer in daughters (Cohn et al. 2015). Polybrominated biphenyls (PBB) are also persistent organic pollutants that cause breast cancer (IARC 2015). Potential mechanistic connections between dintrophenol, nitromethylene-piperidine, oxalate, and these POPs are not apparent, and mechanisms by which POPs disrupt central amino acid and nitrogen metabolism pathways are not known. In particular, amino acids are essential for growth and development, and disruption of pyrimidine and purine metabolism can be expected to have long-term consequences on metabolic programming which could contribute to cancer susceptibility. Thus, these findings warrant further investigation into underlying mechanisms related to breast cancer development.
In T3, the large metabolic community (C4, Fig. 3) containing inflammatory lipids was linked to a sparse community (C5, Fig. 3) containing glucose, benzo[a]carbazole and a suspect benzoate derivative. Of potential importance in this network, the glucose signal was negatively associated with metabolites connected to sialic acid, N-glycan and other pathways associated with extracellular matrix and turnover of connective tissue. Benzoate is conjugated with glycine for elimination, and the role of glycine in folate-dependent metabolism through the vitamin B6-dependent hydroxymethyltransferase raises the possibility that benzoate derivatives could have unrecognized pathogenic effects. Similarly, the heterocyclic amine, benzo[a]carbazole, could be bioactivated by cytochrome P450 enzymes to generate mutagenic species which have not been characterized. Future hypothesis-driven research will be needed to address these possibilities.
The exposome epidemiology approach used in the present study has important assumptions, i.e., that chemical exposures which increase cancer risk can occur decades before breast cancer occurrence, that these exposures are detectable and at higher abundance in serum decades prior to breast cancer detection, and that network analyses of HRM data are sufficient to detect these exposures and link them to biologic responses. With these assumptions, all HRM features from archival blood serum of women collected decades before breast cancer diagnosis can be used in a non-targeted manner to select ones that are increased (p < 0.05) in association with breast cancer occurrence. This selection criterion is not suitable for biomarker development because of multiple testing; however, this cutoff is suitable for discovery of potential chemical risk factors because any of the features at this cutoff could be correct. The selected HRM features are then used with a toxic exposome database, T3DB, to obtain accurate mass matches to known toxic chemicals, and these are subjected to non-targeted network and pathway enrichment analyses to identify environmental chemical-metabolic network associations linked to breast cancer outcome.
Exposome epidemiology will benefit from populations of tens or hundreds of thousands of individuals by enabling detection of exposures impacting small numbers of women and/or having only small contributions individually to breast cancer risk. The present study with only hundreds rather than thousands of individuals has limited capability to detect carcinogens with small effect size. Additionally, selection of HRM features for statistical selection in the current study required that features be present in 80% of the samples. Environmental chemicals and personal use products such as hair dyes cannot be expected to be present in 80% of the women and unlikely to be detectable. Application of untargeted gas chromatography-high-resolution mass spectrometry, which provides capabilities to measure other hydrophobic and volatile environmental chemicals (Hu et al. 2021), can be used to enhance coverage of environmental chemicals. Thus, future studies will benefit from larger population sizes, inclusion of complementary chemical analyses, and use of statistical methods which can accommodate sparse environmental chemical detection.
5. Conclusion
Recent technological and statistical advances in high-resolution metabolomics (HRM) provide capabilities for omics scale biomonitoring of chemicals derived from the environment along with endogenous metabolism and chemicals from the diet, intestinal microbiome, dietary supplements, pharmaceuticals, and personal use products. Because the metabolome is a functional readout of the interactions of a person’s genes with exposures, HRM of biologic samples provides one of the most accessible ways to connect environmental exposures with biologic status to anticipate breast cancer. As shown in the present study, these can be used with network and pathway analysis to identify suspect environmental chemicals and functional communities linked to breast cancer outcome. Such network approaches can be broadly applied to discover how life-long exposures impact personal cancer risks. For breast cancer, we believe that this approach will yield critically needed protocols to enhance protection or mediate pregnancy-associated risk.
Supplementary Material
Funding
This study was supported by Department of Defense Breast Cancer Research Program Grants DAMD17-99-1-9358 and W81XWH-20-1-0102, National Institute of Environmental Health and Science grant R21 ES031824, R01 ES031980 and P30 ES019776, and National Institute of Diabetes and Digestive and Kidney Diseases RC 2 DK118619. The collection of cancer incidence data used in our study was supported by the California Department of Public Health as part of the statewide cancer reporting program mandated by California Health and Safety Code Section 103885; the National Cancer Institute Surveillance, Epidemiology, and End Results Program under Contract HHSN261201000140C awarded to the Cancer Prevention Institute of California, Contract HHSN261201000035C awarded to the University of Southern California, and Contract HHSN261201000034C awarded to the Public Health Institute; and the Centers for Disease Control and Prevention National Program of Cancer Registries, under Agreement U58DP003862-01 awarded to the California Department of Public Health. The ideas and opinions expressed herein are those of the authors and endorsement by the State of California, Department of Public Health, National Institutes of Health, and Centers for Disease Control and Prevention or their contractors and subcontractors, or any of the funders of this research is not intended nor should be inferred.
Abbreviations:
- BC
breast cancer
- FDR
false discovery rate
- HRM
high-resolution metabolomics
- HRMS
high-resolution mass spectrometry
- LC-HRMS
liquid chromatography-high resolution mass spectrometry
- MWAS
metabolome-wide association study
- m/z
mass to charge
- RT
retention time
- T2
second trimester
- T3
third trimester
- T3DB
the toxin and toxin-targeted database
Footnotes
Author statement
All authors take public responsibility for the content of the work submitted for review. The opinions expressed are those of the authors and do not necessarily reflect the official positions of Departments of Defense, National Institute of Health, Public Health Institute or Emory University.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.envint.2023.108112.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data availability
Data will be made available on request.
References
- Albrektsen G, Heuch I, Hansen S, Kvåle G, 2005. Breast cancer risk by age at birth, time since birth and time intervals between births: exploring interaction effects. Br. J. Cancer 92 (1), 167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg BJ, Christianson RE, Oechsli FW, 1988. The California Child Health and Development Studies of the School of Public Health, University of California at Berkeley. Paediatr. Perinat. Epidemiol 2 (3), 265–282. [DOI] [PubMed] [Google Scholar]
- Cohn BA, Wolff MS, Cirillo PM, Sholtz RI, 2007. DDT and breast cancer in young women: new data on the significance of age at exposure. Environ. Health Perspect 115 (10), 1406–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn BA, La Merrill M, Krigbaum NY, Yeh G, Park J-S, Zimmermann L, Cirillo PM, 2015. DDT Exposure in Utero and Breast Cancer. J. Clin. Endocrinol. Metab 100 (8), 2865–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn BA, Cirillo PM, Hopper BR, Siiteri PK, 2017. Third Trimester Estrogens and Maternal Breast Cancer: Prospective Evidence. J. Clin. Endocrinol. Metab 102, 3739–3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go Y-M, Kim CW, Walker DI, Kang DW, Kumar S, Orr M, Uppal K, Quyyumi AA, Jo H, Jones DP, 2015. Disturbed flow induces systemic changes in metabolites in mouse plasma: a metabolomics study using ApoE(−)/(−) mice with partial carotid ligation. Am. J. Physiol. Regul. Integr. Comp. Physiol 308 (1), R62–R72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Li S, Cirillo PM, Krigbaum NY, Tran V, Jones DP, Cohn BA, 2019. Metabolome Wide Association Study of Serum Poly and Perfluoroalkyl Substances (PFASs) in Pregnancy and Early Postpartum . Reprod. Toxicol 87, 70–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Li S, Cirillo P, Krigbaum N, Tran V, Ishikawa T, La Merrill MA, Jones DP, Cohn B, 2020. Metabolome Wide Association Study of serum DDT and DDE in Pregnancy and Early Postpartum. Reprod. Toxicol. 92, 129–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Walker DI, Liang Y, Smith MR, Orr ML, Juran BD, Ma C, Uppal K, Koval M, Martin GS, Neujahr DC, Marsit CJ, Go YM, Pennell KD, Miller GW, Lazaridis KN, Jones DP, 2021. A scalable workflow to characterize the human exposome. Nat. Commun 12, 5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IARC. Polychlorinated Biphenyls and Polybrominated Biphenyls ed^eds; 2015.
- Jarrell ZR, Smith MR, Hu X, Orr M, Liu KH, Quyyumi AA, Jones DP, Go YM, 2020. Plasma acylcarnitine levels increase with healthy aging. Aging (Albany NY) 12 (13), 13555–13570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrell ZR, Smith MR, He X, Orr M, Jones DP, Go YM, 2021. Firsthand and Secondhand Exposure Levels of Maltol-Flavored Electronic Nicotine Delivery System Vapors Disrupt Amino Acid Metabolism. Toxicol. Sci 182, 70–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JM, Yu T, Strobel FH, Jones DP, 2010. Nov. A practical approach to detect unique metabolic patterns for personalized medicine. Analyst 135 (11), 2864–2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CH, Athersuch TJ, Collman GW, Dhungana S, Grant DF, Jones DP, Patel CJ, Vasiliou V, 2017. Yale school of public health symposium on lifetime exposures and human health: the exposome; summary and future reflections. Hum. Genomics 11, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DP, Cohn BA, 2020. A vision for exposome epidemiology: The pregnancy exposome in relation to breast cancer in the Child Health and Development Studies. Reprod. Toxicol 92, 4–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DP, Park Y, Ziegler TR, 2012. Nutritional metabolomics: progress in addressing complexity in diet and health. Annu. Rev. Nutr 32 (1), 183–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DP, Walker DI, Uppal K, Rohrbeck P, Mallon CT, Go YM, 2016. Metabolic Pathways and Networks Associated With Tobacco Use in Military Personnel. J. Occup. Environ. Med 58, S111–S116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Park Y, Duraisingham S, Strobel FH, Khan N, Soltow QA, Jones DP, Pulendran B, Ouzounis CA, 2013. Predicting network activity from high throughput metabolomics. PLoS Comput. Biol 9 (7), e1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Cirillo P, Hu X, Tran V, Krigbaum N, Yu S, Jones DP, Cohn B, 2020. Understanding mixed environmental exposures using metabolomics via a hierarchical community network model in a cohort of California women in 1960’s. Reprod. Toxicol 92, 57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D, Moutinho JL, Golan R, Yu T, Ladva CN, Niedzwiecki M, Walker DI, Sarnat SE, Chang HH, Greenwald R, Jones DP, Russell AG, Sarnat JA, 2018. Use of high-resolution metabolomics for the identification of metabolic signals associated with traffic-related air pollution. Environ. Int 120, 145–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim E, Pon A, Djoumbou Y, Knox C, Shrivastava S, Guo AC, Neveu V, Wishart DS, 2010. T3DB: a comprehensively annotated database of common toxins and their targets. Nucleic Acids Res. 38, D781–D786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMahon B, Cole P, Lin TM, Lowe CR, Mirra AP, Ravnihar B, Salber EJ, Valaoras VG, Yuasa S, 1970. Age at first birth and breast cancer risk. Bull. World Health Organ 43, 209–221. [PMC free article] [PubMed] [Google Scholar]
- Martin J, Hamilton B, Osterman M, Curtin S, Matthews T, 2015. Births: final data for 2013. National vital statistics reports: from the Centers for Disease Control and Prevention, National Center for Health Statistics, National Vital Statistics System 64, 1. [PubMed] [Google Scholar]
- Miller GW, Jones DP, 2014. The nature of nurture: refining the definition of the exposome. Toxicol. Sci 137, 1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedzwiecki MM, Walker DI, Vermeulen R, Chadeau-Hyam M, Jones DP, Miller GW, 2019. The Exposome: Molecules to Populations. Annu. Rev. Pharmacol. Toxicol 59 (1), 107–127. [DOI] [PubMed] [Google Scholar]
- Perkins C, Wright W, Schlag R. Cancer Incidence and Mortality in California, 1988–1994, CA: California Department of Health Servicies, Cancer Surveilance Section. [Google Scholar]
- Ritchie ME, Phipson B, Wu D.i., Hu Y, Law CW, Shi W, Smyth GK, 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43 (7), e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritz B, Yan Q.i., He D.i., Wu J, Walker DI, Uppal K, Jones DP, Heck JE, 2022. Child serum metabolome and traffic-related air pollution exposure in pregnancy. Environ. Res 203, 111907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scalbert A, Andres-Lacueva C, Arita M, Kroon P, Manach C, Urpi-Sarda M, Wishart D, 2011. Databases on food phytochemicals and their health-promoting effects. J. Agric. Food Chem 59 (9), 4331–4348. [DOI] [PubMed] [Google Scholar]
- Schymanski EL, Jeon J, Gulde R, Fenner K, Ruff M, Singer HP, Hollender J, 2014. Identifying small molecules via high resolution mass spectrometry: communicating confidence. Environ. Sci. Tech 48 (4), 2097–2098. [DOI] [PubMed] [Google Scholar]
- Soltow QA, Strobel FH, Mansfield KG, Wachtman L, Park Y, Jones DP, 2013. High-performance metabolic profiling with dual chromatography-Fourier-transform mass spectrometry (DC-FTMS) for study of the exposome. Metabolomics 9 (S1), 132–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troisi R, Bjørge T, Gissler M, Grotmol T, Kitahara CM, Myrtveit Sæther SM, Ording AG, Sköld C, Sørensen HT, Trabert B, Glimelius I, 2018. The role of pregnancy, perinatal factors and hormones in maternal cancer risk: a review of the evidence. J. Intern. Med 283 (5), 430–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppal K, Walker DI, Liu K, Li S, Go Y-M, Jones DP, 2016. Computational Metabolomics: A Framework for the Million Metabolome. Chem. Res. Toxicol 29 (12), 1956–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppal K, Walker DI, Jones DP, 2017. xMSannotator: An R Package for Network-Based Annotation of High-Resolution Metabolomics Data. Anal. Chem 89 (2), 1063–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppal K, Ma C, Go Y-M, Jones DP, Wren J, 2018. xMWAS: a data-driven integration and differential network analysis tool. Bioinformatics 34 (4), 701–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen R, Schymanski EL, Barabasi A-L, Miller GW, 2020. The exposome and health: Where chemistry meets biology. Science 367 (6476), 392–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker DI, Uppal K, Zhang L, Vermeulen R, Smith M, Hu W, Purdue MP, Tang X, Reiss B, Kim S, Li L, Huang H, Pennell KD, Jones DP, Rothman N, Lan Q, 2016. High-resolution metabolomics of occupational exposure to trichloroethylene. Int. J. Epidemiol 45 (5), 1517–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker DI, Marder ME, Yano Y, Terrell M, Liang Y, Barr DB, Miller GW, Jones DP, Marcus M, Pennell KD, 2019. Multigenerational metabolic profiling in the Michigan PBB registry. Environ. Res 172, 182–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild CP, 2005. Complementing the genome with an “exposome”: the outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol. Biomark. Prev 14 (8), 1847–1850. [DOI] [PubMed] [Google Scholar]
- Wishart DS, 2015. Identifying putative drug targets and potential drug leads: starting points for virtual screening and docking. Methods Mol. Biol 1215, 425–444. [DOI] [PubMed] [Google Scholar]
- Wishart D, Arndt D, Pon A, Sajed T, Guo AC, Djoumbou Y, Knox C, Wilson M, Liang Y, Grant J, Liu Y, Goldansaz SA, Rappaport SM, 2015. T3DB: the toxic exposome database. Nucleic Acids Res. 43, D928–D934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, Cheng D, Jewell K, Arndt D, Sawhney S, Fung C, Nikolai L, Lewis M, Coutouly M-A, Forsythe I, Tang P, Shrivastava S, Jeroncic K, Stothard P, Amegbey G, Block D, Hau DD, Wagner J, Miniaci J, Clements M, Gebremedhin M, Guo N, Zhang Y, Duggan GE, MacInnis GD, Weljie AM, Dowlatabadi R, Bamforth F, Clive D, Greiner R, Li L, Marrie T, Sykes BD, Vogel HJ, Querengesser L, 2007. HMDB: the Human Metabolome Database. Nucleic Acids Res. 35, D521–D526. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.