Abstract
Scientific document classification is a critical task for a wide range of applications, but the cost of collecting human-labeled data can be prohibitive. We study scientific document classification using label names only. In scientific domains, label names often include domain-specific concepts that may not appear in the document corpus, making it difficult to match labels and documents precisely. To tackle this issue, we propose WanDeR, which leverages dense retrieval to perform matching in the embedding space to capture the semantics of label names. We further design the label name expansion module to enrich its representations. Lastly, a self-training step is used to refine the predictions. The experiments on three datasets show that WanDeR outperforms the best baseline by 11.9%. Our code will be published at https://github.com/ritaranx/wander.
Keywords: Scientific Document Classification, Weak Supervision, Retrieval
1. INTRODUCTION
Scientific document classification aims to assign scientific literature to pre-defined categories, supporting various applications [5, 27, 38]. Recently, pretrained language models (PTLMs) have demonstrated impressive performance in document classification [1, 7]. However, they often require a large number of annotations for fine-tuning, which restricts their deployment in real-world applications. While practitioners cannot afford to label many documents, it is often easier for them to provide category-descriptive label names as weak supervision for each class [17, 34]. Motivated by this, we focus on scientific document classification under the setting where only the label name for each class as well as the unlabeled corpus are available [18]. This task is challenging as the label names can be short and succinct, often containing a few words only. How to mine class-relevant knowledge with weak supervision is nontrivial.
There exist plenty of studies on automatic document categorization using class-relevant keywords [16–18, 25, 33]. These methods often leverage the keywords as input to extract relevant documents with hard matching for pseudo label generation. Although these methods achieve competitive performance, they mainly focus on tasks from general domains. For these tasks, the keywords can be commonly used words (e.g. ‘Good/Bad’ for reviews), and they can be matched with many examples. However, for scientific documents, the label names can either be too domain-specific, or contain multiple concepts [37]. As a result, they often have limited coverage over the corpus, which causes performance degradation when applying prior weakly-supervised techniques to the scientific domain.
In this work, we propose WanDeR (Weakly-supervised Scientific Text Classification using Dense Retrieval), a multi-stage training framework for weakly supervised text classification using dense retrieval (DR), as shown in Figure 1. In DR, both queries and documents are represented as dense vectors, and the relevance between them is calculated via similarity metrics (e.g. dot product) [14]. This makes DR an ideal choice to tackle the above challenges, as it captures the semantics for different classes and circumvents the mismatch issue. To incorporate DR into the framework, we regard label names as queries, and retrieve the most relevant documents from the unlabeled corpus for each class (Stage-I, Sec. 3.1) to create an initial set of pseudo-labeled documents, which can be used to fine-tune the PTLM for the target task.
Figure 1:
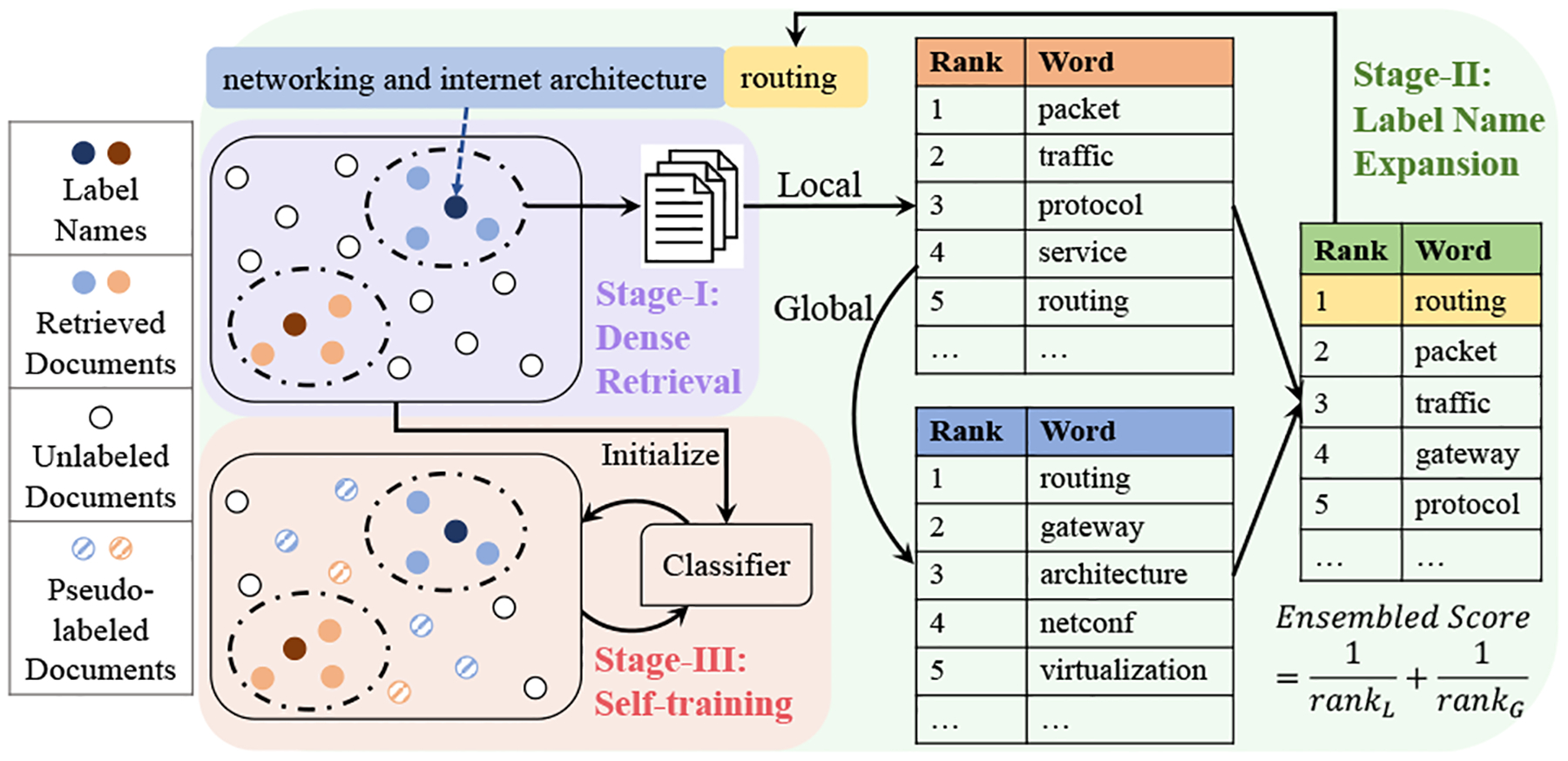
Framework of WanDeR.
Although Stage-I is able to extract relevant documents, their performance can be less satisfactory as label names are insufficient to capture all the class-specific information. To overcome this drawback, in Stage-II, we expand the label names with the extracted keywords using local and global information (Sec. 3.2). Specifically, we first adopt the TF-IDF algorithm [11] on the retrieved documents to select the top-ranked words. In addition, we use the PTLM to calculate the embedding similarity between the candidate words and the label names as the global score. The local and global information is connected via an ensemble ranking module, and we augment the label name for each class by selecting the word with the highest score. The above expansion step is repeated multiple times to enrich the query [8] and help the DR model retrieve more relevant documents from the corpus.
To leverage all unlabeled data to further improve the performance, an additional step is to harvest self-training [18, 28] (Stage-III, Sec. 3.3) to refine the PTLM classifier by bootstrapping over high-confident examples and improve its generalization ability.
We verify the effectiveness of WanDeR by conducting experiments on three datasets and show that our model outperforms the previous weakly-supervised approaches by a large margin. Our analysis further confirms the advantage of leveraging dense retrieval for tackling the limited coverage issue of label names as well as the efficacy of multi-stage training for improving the performance.
2. PRELIMINARIES
2.1. Problem Definition
Our weakly-supervised scientific document classification with C classes is defined as follows. The input is a training corpus 𝒳 = {d1,d2, …, d|𝒳|} of documents without any labels. In addition, for each class c (1 ≤ c ≤ C), a label-specific name wc is given, which consists of one or a few words. We aim to learn a classifier f(x;θ) : 𝒳 → 𝒴. Here 𝒳 denotes samples and 𝒴 = {1, 2, ⋯, C} is the label set. While there exist works on multi-label classification [22] or metadata-aware classification [9, 36], we focus on the basic setting by assuming (1) each document only belongs to one category and (2) no other metadata information are available.
2.2. Challenges for Scientific Text Classification
While existing weakly supervised methods [18, 25] achieve competitive performance on general-domain datasets, applying them directly to scientific datasets often causes performance degradation. To illustrate this, we use AGNews [35] as the general-domain dataset and MeSH [5] as the scientific dataset. The average precision (i.e. the portion of correctly matched examples) and coverage (i.e. the portion of examples that can be matched by label names) for label names are shown in Figure 2a. We observe that for the scientific domain, the precision and coverage decline by 6% and 37% respectively. Moreover, the results of per-class coverage (presented in Figure 2b) indicate that the label distribution is more imbalanced for scientific data. For MeSH, there are 4 out of 11 classes where the label name cannot match any examples from the unlabeled corpus.
Figure 2:

Pilot Studies. FS means fully-supervised model.
These two issues prevent the previous weakly-supervised models [18, 25] from performing well. As shown in Figure 2c, gaps to the performance of the fully-supervised model are much larger for scientific datasets (36%) than general-domain datasets (8%), which indicates that these advanced techniques cannot resolve the unique challenges that exist in the scientific domain.
3. METHOD
From the analysis in the above section, we conclude that it is necessary to propose techniques beyond hard matching to better harvest the semantic label name information. Towards this goal, we present our framework WanDeR in Figure 1, a multi-stage training scheme based on dense retrieval, to perform document classification using label names only. The three stages are detailed below.
3.1. Stage-I: Dense Retrieval with Label Names
Directly using the label-indicative keywords to extract documents is sub-optimal for scientific documents, due to their limited coverage and inferior ability to capture the class-related semantics. Motivated by this, we propose to leverage dense retrieval (DR) [14] to effectively retrieve the most relevant documents. Specifically, DR represents the input information (“query”) q and target corpus (“document”) d in the continuous embedding space as g(q;ϕ),g(d;ϕ) respectively, where g(·;ϕ) is the dense retrieval model with ϕ being the parameter of g. Then, DR matches queries and documents via approximate nearest neighbor (ANN) using the relevance score r(q,d;ϕ) = 〈g(q;ϕ),g(d;ϕ)〉, where 〈·,·〉is the cosine similarity. Next, we introduce the approach to train the DR model as well as leverage DR to extract documents from the corpus 𝒳.
Task-adaptive DR Model Pretraining.
To pretrain a DR model (·;ϕ) on the corpus 𝒳, we use the contrastive learning widely adopted in recent research [10, 12, 32]. Specifically, for each document di ∈ 𝒳, we sample two sentences di,1, di,2 as the positive pair. The training objective for di can be written as
| (1) |
where are the in-batch negatives, and τ = 0.01 is the parameter for temperature. Contrastive pretraining improves both the alignment and uniformity for embeddings [13, 24, 29, 30], which can better support the retrieval task in our framework.
Document Retrieval using Label Names.
With the DR model, we aim to extract an initial set of labeled data for each class by feeding the label names (as queries) to the DR model. The initial retrieved document set 𝒟i for the i-th class can be written as
| (2) |
where k is the number of retrieved examples, and the label of the retrieved document is determined by the category of the label name. In this way, we get rid of the challenge brought by those infrequent label names and provide a flexible way to encode the label-related semantics. All retrieved examples are then used for classification, which will be discussed in the following part.
Training Classifiers with Retrieved Text.
With the retrieved document set 𝒟, one can simply finetune a classifier f(·;θ) with the standard cross-entropy loss:
| (3) |
The fine-tuned model is used for target classification tasks.
3.2. Stage-II: Expand Label Names with Local and Global Information
One drawback of the above stage is that the label names are often too abstract to fully represent the semantics information for classes. As such, the retrieved documents still contain label noise, which hurts the downstream performance. To tackle this, we propose to automatically extract class-related keywords to expand the label name, by using both local information from the retrieved documents and global information from the general pretrained models.
Local Information for Keyword Extraction.
To identify the class-related keywords, we assume terms that appear frequently within documents from a specific class while infrequently for other classes are more likely to be class-indicative words for that class [16]. Inspired by TF-IDF [11], we measure the indicativeness of word w for class c from the retrieved document 𝒟 as
| (4) |
Here tfw,c, cntw,c stands for the frequency and occurrence time of word w within documents from class c and tfw, is the frequency of w in corpus, A is the average number of words per class. In this way, words appear commonly in the class-related documents while being less generic will receive higher score. For each class, we extract m words with the highest score as the candidate set C.1
Global Information for Keyword Semantics.
The above step only considers the word occurrence in the local corpus, without modeling the semantic information. An ideal keyword, however, should also have a closer meaning to the label name. Motivated by this, we leverage the PTLM to transfer the global knowledge from pretraining corpora and encode the contextual information for each word. We calculate the embeddings of both label names and candidate words by averaging the output of all tokens from the last layer of PTLM h(·;Ψ). For word w ∈ C from the candidate set of class c, the global score is calculated between w and the label name wc using the embedding similarity as
| (5) |
Ensemble Reranking.
To effectively combine the local and global information, we sort candidate words w ∈ Ci for i-th class using the score Lw,c,Gw,c, respectively. Then, each word w will have two ranks as rankL,c (w) and rankG,c (w). We rerank the words using the ensemble score based on Reciprocal Rank Fusion (RRF) [6]:
| (6) |
For each class, we add one word with the highest score to expand the label name. For expansion, we simply concatenate the previous label name and the newly identified word for enrichment [2, 19].
Iterative Label Name Expansion.
The above process can be conducted multiple times. In each iteration, we first use local and global scores to detect the expanded words using Eq. (4)–(6) and enrich the label names. Then, we use the expanded label names as queries to update the retrieved documents 𝒟 with Eq. (2) as we expect the quality of 𝒟 will improve by incorporating additional class-indicative words. With the updated 𝒟, more relevant words can be extracted to enrich the class information. The above iteration is repeated 5 times, and the retrieved documents after the final iteration can be used to train another classifier using Eq. (3).
3.3. Stage-III: Refine Classifier with Self-training
The pseudo-labeled samples in Stage-II are only from the top retrieved documents with the expanded label names. To generalize its current knowledge to the whole unlabeled corpus, self-training is adopted to bootstrap the model on the entire unlabeled corpus [15, 18, 31] as
| (7) |
where is the hard pseudo label, γ is the confidence threshold. With self-training, the model is refined by its high-confident predictions to improve generalization ability. Stage-III stops when less than 1% of samples change their labels.
4. EXPERIMENTS
4.1. Experiment Setups
Datasets.
We conduct experiments on three datasets from multiple domains including MeSH [5], arXiv-CS [4], arXiv-Math [4]. The statistics for each dataset are shown in Table 1. For arXiv-CS and arXiv-Math, we select papers from years 2017–2020 as the training set, 2021–2022 as the test set, and use the topic from the main category as the label.
Table 1:
Dataset statistics.
| Dataset | Domain | # Train | # Test | # Class | # OOV | Avg. Len. |
|---|---|---|---|---|---|---|
| MeSH | BioMedical | 16.3k | 3.5k | 11 | 4 (36%) | 254.3 |
| arXiv-Math | Mathematics | 62.5k | 6.3k | 16 | 3 (19%) | 214.4 |
| arXiv-CS | Computer Science | 75.7k | 5.1k | 20 | 5 (25%) | 188.2 |
Baselines.
We compare WanDeR with these baselines: (1) IR [23] leverages TF-IDF to assign labels for documents. (2) Dataless [3] uses Wikipedia to embed labels and documents. Each document is classified to the label with the highest similarity. (3) SentenceBERT [20] is trained on NLI data to embed labels and documents for classification. (4) LOTClass [18], (5) X-Class [25], and (6) FastClass [26] are three recent methods that use PTLMs for label-name-only text classification by using masked language modeling or pretrained representations.
Implementations.
We use the pre-trained SciBERT [1] as the backbone. The retrieval model g (Eq. (1)) and PTLM h (Eq. (5)) are initialized from SciBERT, and g is pretrained on the corpus 𝒳 for 5 epochs. The maximum length is set to 512. For Stage-I and II, we finetune f (·; θ) for 5 epochs with Adam as the optimizer and set the batch size and learning rate to 32 and 2e-5. Other hyperparameters include τ in Eq. (1), k for ANN in Eq. (2), γ in Eq. (7), m in Sec. 3.2. We set τ = 0.01, m = 100, k = 100, γ = 0.8, α = 0.5 without tuning. We study the effect of k,γ in Sec. 4.3.
4.2. Experiment Results
Main Experiments.
We report both Macro-F1 and Micro-F1 scores for WanDeR and baselines in Table 2. The mean and variance over 5 runs are calculated when fine-tuning is used. We observe that WanDeR consistently achieves the best performance on three datasets, with an average gain of 11.9%. In contrast, X-Class and LOTClass, which achieve strong results on general-domain tasks, fail to perform well on the scientific domain, as they cannot handle the challenges mentioned in Sec. 2.2. Moreover, traditional baselines, such as IR and Dataless, are inferior to other methods using PTLMs, indicating their limited ability for modeling scientific text. Although SentenceBERT and FastClass use extra labeled data for embedding learning, they fail to expand the label names for enriching representations, leading to sub-optimal performance.
Table 2:
Performance on three datasets. Bold and blue indicate the best and second-best results for each dataset. Macro-F1 is the main metric as the label distribution is imbalanced.
| Method | MeSH | arXiv-Math | arXiv-CS | |||
|---|---|---|---|---|---|---|
| Mi-F1 | Ma-F1 | Mi-F1 | Ma-F1 | Mi-F1 | Ma-F1 | |
| Fully Supervised | 90.5±0.3 | 90.3±0.2 | 80.6±0.4 | 79.1±0.3 | 83.0±0.2 | 78.2±0.4 |
| IR [23] | 40.6 | 37.6 | 27.8 | 22.9 | 24.5 | 22.8 |
| Dataless [3] | 36.1 | 26.8 | 18.9 | 13.4 | 20.5 | 18.2 |
| SentenceBERT [20] | 68.6 | 66.0 | 48.9 | 41.1 | 50.7 | 47.7 |
| LOTClass [18] | 57.9±1.7 | 44.9±1.6 | 43.8±2.0 | 35.2±1.5 | 51.5±1.4 | 47.1±1.8 |
| X-Class [25] | 55.2±1.4 | 54.4±1.8 | 46.5±1.4 | 39.1±1.4 | 60.6±1.2 | 51.6±1.3 |
| FastClass [26] | 78.5±1.3 | 78.1±1.1 | 53.5±1.3 | 44.5±1.2 | 59.8±0.8 | 50.5±0.9 |
| WanDeR | 82.0 ±0.4 | 81.9 ±0.4 | 58.0 ±0.8 | 51.9 ±0.7 | 65.6 ±0.8 | 58.9 ±0.6 |
| Gain Δ | 3.5 (4.4%) | 3.8 (4.9%) | 4.5 (8.4%) | 7.4 (16.6%) | 5.0 (8.2%) | 7.3 (14.1%) |
| WanDeR (Stage-I) | 76.6±1.0 | 75.6±0.8 | 56.4±1.4 | 49.8±0.9 | 61.8±1.1 | 54.7±1.2 |
| WanDeR (Stage-II) | 79.9±0.6 | 80.2±0.7 | 57.1±1.1 | 51.0±1.0 | 64.6±1.0 | 58.1±0.6 |
Effect of Multi-stage Training.
The bottom two rows in Table 2 show the performance of WanDeR after Stage-I and II, which justifies that all three stages contribute to the final performance. Moreover, WanDeR outperforms all baselines even without self-training (Stage-III), indicating that it can retrieve a small set of high-quality data to support downstream tasks sufficiently.
4.3. Ablation and Hyperparameter Studies
Study of DR Models.
To illustrate the effect of task-adaptive contrastive learning (TAPT) for DR model pretraining, we substitute (·) with other models including BM25 [21], SciBERT [1] without TAPT, the strong unsupervised DR model Contriever [12], and compare the performance in Figure 3a. Overall, our model achieves the best performance, which justifies the need for TAPT as it effectively reduces the distribution shifts and also produces better embeddings. Instead, using sparse retrieval model (BM25) yields undesirable performance as it cannot understand label names well.
Figure 3:

Studies of Different Retrieval Models and Hyperparameters (Best View in Colors).
Effect of Hyperparameters.
We study the effect of k and γ in WanDeR on MeSH and arXiv-CS, as shown in Figure 3b and 3c. We observe that the performance first increases with larger k as the model benefits from more retrieved examples. When k reaches 100, the performance remains stable, as too many retrieved examples introduce label noise and diminish the performance gain. We also run experiments with different thresholds γ. The result indicates that the model performance is insensitive to γ, and the self-training component leads to performance gain in most studied regions.
Effect of Local and Global Information.
Figure 4 illustrates the performance of WanDeR and its variants over 5 expansion iterations. Overall, we observe that removing local or global information hurts the performance, since these two modules provide complementary information. Combining these two terms together results in better pseudo labels and improves downstream performance.
Figure 4:
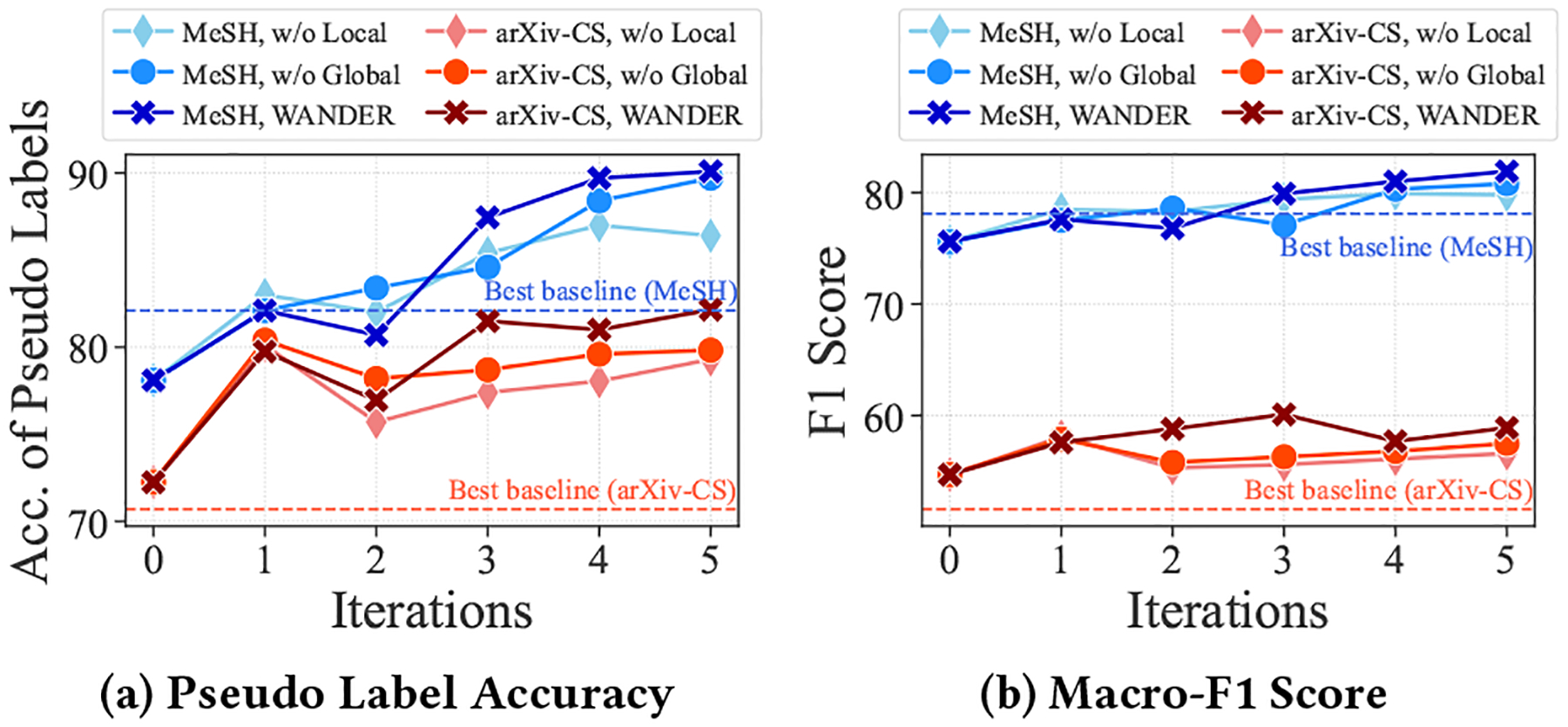
Study on Effects of Local and Global Information.
4.4. Case Studies
We present a case study in Table 3 to showcase that WanDeR is able to discover class-related keywords to expand label names. Take diabetes as an example, it is often related to high glucose level and glycemic index. Besides, insulin and metformin are used as treatments for diabetes. Moreover, take machine learning as another example, it is applied to classifiation tasks. Boosting, ensemble, tree are all techniques to tackle machine learning problems. These all indicate that WanDeR can enrich the semantics of label names.
Table 3:
Case Study on expanded keywords for three tasks.
| Dataset | Class | Expanded Keyword |
|---|---|---|
| MeSH | Diabetes | insulin, glucose, diabetic, metformin, glycemic |
| MeSH | Neoplasms | tumor, carcinoma, cell, tumour, chemotherapy |
| arXiv-Math | Combinatorics | graph, combinatorial, vertex, edge, bipartite |
| arXiv-Math | Statistics theory | estimation, sample, regression, treatment, inference |
| arXiv-CS | Information theory | entropy, channel, shannon, capacity, decoder |
| arXiv-CS | Game Theory | player, equilibrium, nash, payoff, strategy |
5. CONCLUSION
We propose WanDeR, a multi-stage training framework for weakly-supervised scientific document classification with label name only. We leverage dense retrieval to go beyond hard matching and harness the semantics of label names. In addition, we propose a label name expansion module to enrich its representations, and use self-training to improve the model’s generalization ability. Experiments on three datasets demonstrate that WanDeR outperforms the baselines by 11.9% on average. For future works, we plan to extend WanDeR to other scenarios such as multi-label classification.
CCS CONCEPTS.
• Computing methodologies → Natural language processing.
Footnotes
We omit words that already appeared in label names during the expansion (stage-II).
Contributor Information
Ran Xu, Emory University, Atlanta, GA, USA.
Yue Yu, Georgia Institute of Technology, Atlanta, GA, USA.
Joyce Ho, Emory University, Atlanta, GA, USA.
Carl Yang, Emory University, Atlanta, GA, USA.
REFERENCES
- [1].Beltagy Iz, Lo Kyle, and Cohan Arman. 2019. SciBERT: A Pretrained Language Model for Scientific Text. In EMNLP-IJCNLP. 3615–3620. [Google Scholar]
- [2].Cao Guihong, Nie Jian-Yun, Gao Jianfeng, and Robertson Stephen. 2008. Selecting good expansion terms for pseudo-relevance feedback. In SIGIR. 243–250. [Google Scholar]
- [3].Chang Ming-Wei, Ratinov Lev-Arie, Roth Dan, and Srikumar Vivek. 2008. Importance of Semantic Representation: Dataless Classification.. In AAAI. 830–835. [Google Scholar]
- [4].Clement Colin B, Bierbaum Matthew, O’Keeffe Kevin P, and Alemi Alexander A. 2019. On the Use of ArXiv as a Dataset. arXiv preprint arXiv:1905.00075 (2019). [Google Scholar]
- [5].Cohan Arman, Feldman Sergey, Beltagy Iz, Downey Doug, and Weld Daniel S. 2020. SPECTER: Document-level Representation Learning using Citation-informed Transformers. In ACL. 2270–2282. [Google Scholar]
- [6].Cormack Gordon V, Clarke Charles LA, and Buettcher Stefan. 2009. Reciprocal rank fusion outperforms condorcet and individual rank learning methods. In SIGIR. 758–759. [Google Scholar]
- [7].Devlin Jacob, Chang Ming-Wei, Lee Kenton, and Toutanova Kristina. 2019. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. In NAACL-HLT. [Google Scholar]
- [8].Diaz Fernando, Mitra Bhaskar, and Craswell Nick. 2016. Query Expansion with Locally-Trained Word Embeddings. In ACL. [Google Scholar]
- [9].Ganguly Soumyajit and Pudi Vikram. 2017. Paper2vec: Combining graph and text information for scientific paper representation. In ECIR. 383–395. [Google Scholar]
- [10].Gao Luyu and Callan Jamie. 2022. Unsupervised Corpus Aware Language Model Pre-training for Dense Passage Retrieval. In ACL. 2843–2853. [Google Scholar]
- [11].Grootendorst Maarten. 2022. BERTopic: Neural topic modeling with a class-based TF-IDF procedure. arXiv preprint arXiv:2203.05794 (2022). [Google Scholar]
- [12].Izacard Gautier, Caron Mathilde, Hosseini Lucas, Riedel Sebastian, Bojanowski Piotr, Joulin Armand, and Grave Edouard. 2022. Unsupervised Dense Information Retrieval with Contrastive Learning. TMLR (2022). [Google Scholar]
- [13].Kan Xuan, Cui Hejie, Lukemire Joshua, Guo Ying, and Yang Carl. 2022. Fbnetgen: Task-aware gnn-based fmri analysis via functional brain network generation. In MIDL. [PMC free article] [PubMed] [Google Scholar]
- [14].Karpukhin Vladimir, Oguz Barlas, Min Sewon, Lewis Patrick, Wu Ledell, Edunov Sergey, Chen Danqi, and Yih Wen-tau. 2020. Dense Passage Retrieval for Open-Domain Question Answering. In EMNLP. 6769–6781. [Google Scholar]
- [15].Liang Chen, Yu Yue, Jiang Haoming, Er Siawpeng, Wang Ruijia, Zhao Tuo, and Zhang Chao. 2020. Bond: Bert-assisted open-domain named entity recognition with distant supervision. In KDD. 1054–1064. [Google Scholar]
- [16].Mekala Dheeraj and Shang Jingbo. 2020. Contextualized weak supervision for text classification. In ACL. 323–333. [Google Scholar]
- [17].Meng Yu, Shen Jiaming, Zhang Chao, and Han Jiawei. 2018. Weakly-supervised neural text classification. In CIKM. 983–992. [Google Scholar]
- [18].Meng Yu, Zhang Yunyi, Huang Jiaxin, Xiong Chenyan, Ji Heng, Zhang Chao, and Han Jiawei. 2020. Text classification using label names only: A language model self-training approach. EMNLP (2020). [Google Scholar]
- [19].Naseri Shahrzad, Dalton Jeffrey, Yates Andrew, and Allan James. 2021. Ceqe: Contextualized embeddings for query expansion. In ECIR. 467–482. [Google Scholar]
- [20].Reimers Nils and Gurevych Iryna. 2019. Sentence-BERT: Sentence Embeddings using Siamese BERT-Networks. In EMNLP-IJCNLP. 3982–3992. [Google Scholar]
- [21].Robertson Stephen and Zaragoza Hugo. 2009. The probabilistic relevance framework: BM25 and beyond. Foundations and Trends in Information Retrieval 3, 4 (2009), 333–389. [Google Scholar]
- [22].Shen Jiaming, Qiu Wenda, Meng Yu, Shang Jingbo, Ren Xiang, and Han Jiawei. 2021. TaxoClass: Hierarchical multi-label text classification using only class names. In NAACL-HLT. 4239–4249. [Google Scholar]
- [23].Trstenjak Bruno, Mikac Sasa, and Donko Dzenana. 2014. KNN with TF-IDF based framework for text categorization. Procedia Engineering 69 (2014), 1356–1364. [Google Scholar]
- [24].Wang Tongzhou and Isola Phillip. 2020. Understanding contrastive representation learning through alignment and uniformity on the hypersphere. In ICML. 9929–9939. [Google Scholar]
- [25].Wang Zihan, Mekala Dheeraj, and Shang Jingbo. 2021. X-Class: Text Classification with Extremely Weak Supervision. In NAACL. 3043–3053. [Google Scholar]
- [26].Xia Tingyu, Wang Yue, Tian Yuan, and Chang Yi. 2022. FastClass: A Time-Efficient Approach to Weakly-Supervised Text Classification. EMNLP (2022). [Google Scholar]
- [27].Xie Yi, Sun Yuqing, and Bertino Elisa. 2021. Learning domain semantics and cross-domain correlations for paper recommendation. In SIGIR. 706–715. [Google Scholar]
- [28].Xu Ran, Yu Yue, Cui Hejie, Kan Xuan, Zhu Yanqiao, Ho Joyce, Zhang Chao, and Yang Carl. 2023. Neighborhood-Regularized Self-Training for Learning with Few Labels. In AAAI, Vol. 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Xu R, Yu Y, Zhang C, K Ali M, Ho JC, and Yang C. 2022. Counterfactual and factual reasoning over hypergraphs for interpretable clinical predictions on ehr. In Machine Learning for Health. PMLR, 259–278. [PMC free article] [PubMed] [Google Scholar]
- [30].Yang Yi, Cui Hejie, and Yang Carl. 2022. Pre-train Graph Neural Networks for Brain Network Analysis. In IEEE-Big Data. [Google Scholar]
- [31].Yu Yue, Kong Lingkai, Zhang Jieyu, Zhang Rongzhi, and Zhang Chao. 2022. AcTune: Uncertainty-Based Active Self-Training for Active Fine-Tuning of Pre-trained Language Models. In NAACL. 1422–1436. [Google Scholar]
- [32].Yu Yue, Xiong Chenyan, Sun Si, Zhang Chao, and Overwijk Arnold. 2022. COCO-DR: Combating the Distribution Shift in Zero-Shot Dense Retrieval with Contrastive and Distributionally Robust Learning. In EMNLP. 1462–1479. [Google Scholar]
- [33].Yu Yue, Zuo Simiao, Jiang Haoming, Ren Wendi, Zhao Tuo, and Zhang Chao. 2021. Fine-Tuning Pre-trained Language Model with Weak Supervision: A Contrastive-Regularized Self-Training Approach. In NAACL. 1063–1077. [Google Scholar]
- [34].Zhang Jieyu, Yu Yue, Li Yinghao, Wang Yujing, Yang Yaming, Yang Mao, and Ratner Alexander. 2021. WRENCH: A Comprehensive Benchmark for Weak Supervision. In NeurIPS. [Google Scholar]
- [35].Zhang Xiang, Zhao Junbo Jake, and LeCun Yann. 2015. Character-level Convolutional Networks for Text Classification. In NIPS. [Google Scholar]
- [36].Zhang Yu, Garg Shweta, Meng Yu, Chen Xiusi, and Han Jiawei. 2022. Motifclass: Weakly supervised text classification with higher-order metadata information. In WSDM. 1357–1367. [Google Scholar]
- [37].Zhang Yu, Meng Yu, Wang Xuan, Wang Sheng, and Han Jiawei. 2022. Seed-Guided Topic Discovery with Out-of-Vocabulary Seeds. In NAACL. 279–290. [Google Scholar]
- [38].Zhuang Yuchen, Li Yinghao, Zhang Junyang, Yu Yue, Mou Yingjun, Chen Xiang, Song Le, and Zhang Chao. 2022. ReSel: N-ary Relation Extraction from Scientific Text and Tables by Learning to Retrieve and Select. In EMNLP. 730–744. [Google Scholar]


