Pan et al. describe that the stability of cyclin D1 mRNA fluctuates throughout the cell cycle, controlled by PC4, a small RNA binding protein modified by cell cycle–dependent phosphorylation and ubiquitination. This regulation impacts cell cycle progression and tumorigenesis, suggesting PC4 as a promising therapeutic target for hepatocellular carcinoma treatment.
Abstract
The cell cycle is a highly regulated process in which proteins involved in cell cycle progression exhibit periodic expression patterns, controlled by specific mechanisms such as transcription, translation, and degradation. However, the precise mechanisms underlying the oscillations of mRNA levels in cell cycle regulators are not fully understood. In this study, we observed that the stability of cyclin D1 (CCND1) mRNA fluctuates during the cell cycle, with increased stability during interphase and decreased stability during the M phase. Additionally, we identified a key RNA binding protein, positive coactivator 4 (PC4), which plays a crucial role in stabilizing CCND1 mRNA and regulating its periodic expression. Moreover, the binding affinity of PC4 to CCND1 mRNA is modulated by two cell cycle–specific posttranslational modifications: ubiquitination of K68 enhances binding and stabilizes the CCND1 transcript during interphase, while phosphorylation of S17 inhibits binding during the M phase, leading to degradation of CCND1 mRNA. Remarkably, PC4 promotes the transition from G1 to S phase in the cell cycle, and depletion of PC4 enhances the efficacy of CDK4/6 inhibitors in hepatocellular carcinoma, suggesting that PC4 could serve as a potential therapeutic target. These findings provide valuable insights into the intricate regulation of cell cycle dynamics.
Introduction
The cell cycle, which is crucial for cell growth, is tightly controlled by a complex and multilayered system (Cho et al., 2001; Stumpf et al., 2013; Liu et al., 2017; Fischer et al., 2022). Any disruption in this finely tuned system can lead to uncontrolled cell proliferation and contribute to the development of cancer (Otto and Sicinski, 2017). A fundamental aspect of this control system is the regulation of periodic changes in the abundance of proteins involved in the cell cycle. This regulation ensures a precise and irreversible phase transition necessary for cell cycle progression. While the degradation of certain cell cycle proteins through the ubiquitin system provides one level of control (Dang et al., 2021), the majority of proteins are regulated by the periodic expression of their mRNA (Fischer and Müller, 2017; Emanuele et al., 2020). However, the precise molecular mechanisms underlying the fluctuations in mRNA expression of cell cycle regulators are still not fully understood.
One important signaling pathway involved in cell cycle progression is the cyclin D-cyclin-dependent kinase (CDK)–retinoblastoma protein (RB) pathway. While CDKs remain constant throughout the cell cycle, the expression of cyclin D is dynamically regulated at multiple levels. Numerous studies have shown that both the ubiquitin-proteasome-mediated pathway (Lin et al., 2006; Fasanaro et al., 2010) and transcriptional program (Liu et al., 2017) can drive the periodic expression of cyclin D and influence cell cycle progression. However, the contribution of posttranscriptional modulation, particularly through RNA binding proteins (RBPs), in the fluctuating expression of cyclin D and the plasticity of the cell cycle remains largely unknown.
Similar to the oscillation of cyclin D expression, the activation of a series of cell cycle–related proteins through posttranslational modifications (PTMs) also fluctuates throughout the cell cycle. The timely activation of cell cycle executors by PTMs allows for cell cycle entry and ensures precisely coordinated cell cycle progression. For instance, phosphorylation of PLK1 by Aurora A during the G2 phase is necessary for PLK1 activation and entry into mitosis (Seki et al., 2008a, 2008b). Therefore, it would be interesting to investigate whether PTMs periodically direct RBPs to regulate cyclin D oscillation and reshape cell cycle progression during cell proliferation.
In this study, we conducted a comprehensive and unbiased screening and identified the human positive cofactor 4 (PC4) as a novel RBP that regulates the periodic change in cyclin D1 (CCND1) expression by mediating its mRNA stability, ultimately affecting the transition from G1 to S phase. Furthermore, we found that the mRNA binding affinity of PC4 is regulated by two cell cycle–dependent PTMs: K68 ubiquitination facilitates PC4 binding and stabilizes the CCND1 transcript during interphase, whereas S17 phosphorylation inhibits PC4–mRNA binding affinity in the M phase, leading to CCND1 degradation. Importantly, our study revealed that PC4 promotes the proliferation of hepatocellular carcinoma (HCC) cells, and depletion of PC4 increases sensitivity to CDK4/6 inhibitors, suggesting that PC4 may be a potential target for HCC treatment. Overall, our findings highlight the role of PC4 in regulating cyclin D1 mRNA stability and add another layer of complexity to cell cycle regulation.
Results
PC4 acts as an RBP that stabilizes CCND1 mRNA
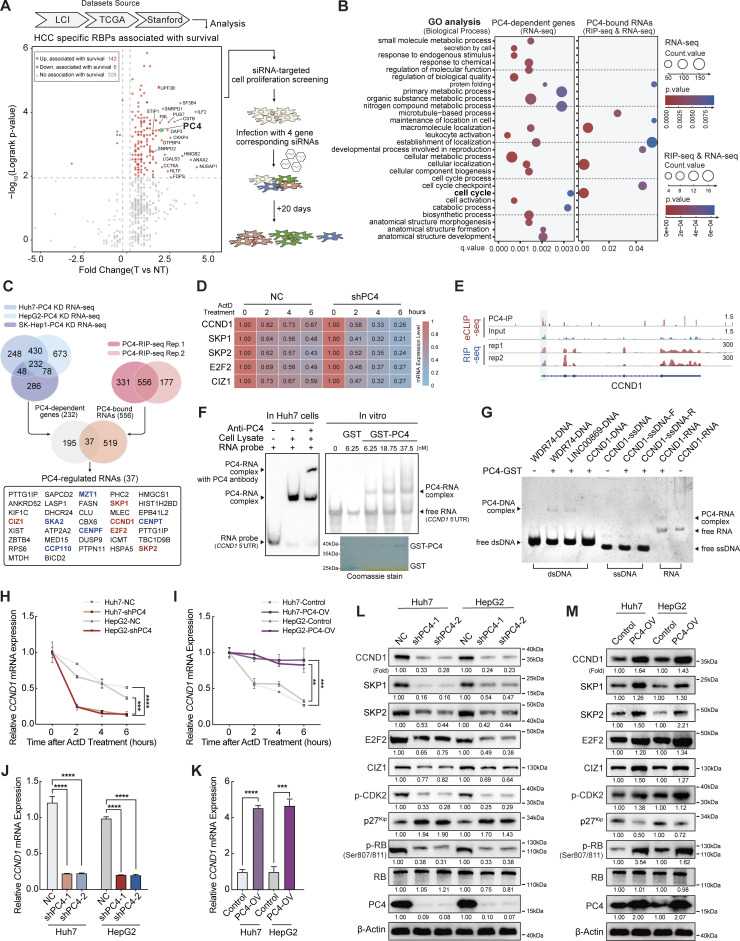
To identify RBPs that promote and maintain HCC and may serve as potential therapeutic targets, we developed a two-step screening system. In the first step, we utilized the dataset from Dang’s research (Dang et al., 2017), which consisted of 672 RBP expressions and associated patient outcomes in 1,225 clinical samples of HCC, to investigate whether the expression of any RBPs correlated with HCC prognosis. Through this analysis, we validated that the differential expression of 148 RBPs was significantly associated with overall patient survival. Among the top 20 RBPs that were highly expressed in tumors with poor prognosis, four candidates had undefined roles in HCC and were selected based on their essentiality in liver cancer cell proliferation experiments. After the second cell proliferation screening, PC4 knockdown had the most significant inhibitory effect on HCC cell growth (Fig. 1 A). Thus, in the present study, we focused on this molecule, which may be the most promising candidate target for HCC development.
Figure 1.
PC4 acts as an RBP that stabilizes CCND1 mRNA. (A) Schematic representation showing the screening process used to identify PC4 as an essential RBP that promotes HCC. In the initial step, a comprehensive analysis of differential expression and survival data was conducted using the Liver Cancer Institute (LCI), TCGA-Liver Hepatocellular Carcinoma (LIHC), and Stanford datasets, as described by Dang et al. (2017). This analysis led to the identification of 148 RBPs that were clinically relevant in HCC. Among these RBPs, the top 20 candidates with the highest clinical relevance were selected. Notably, four of these candidates had previously unknown roles in HCC, making them particularly interesting for further investigation. To evaluate the potential impact of these four RBP candidates on cell proliferation, an siRNA screening was performed. Huh7 cells were treated with siRNAs targeting each candidate, and their effects on cell growth were assessed at day 1 and day 20. (B) Dot plot of gene ontology (GO) enrichment showing significant GO terms for differentially expressed genes after PC4 knockdown (left) and RNAs that are bound with and regulated by PC4 (right) in Huh7 cells. Color indicates P value and dot size denotes the number of genes enriched. Dots are not shown for terms with no statistically significant (P < 0.05) enrichment. (C) Venn diagrams showing the overlapping sets of data from three different categories related to PC4-bound RNA transcripts with a significant alternation upon PC4 knockdown. The first category represents differential expression transcripts identified by RNA-seq upon PC4 knockdown in the three cell lines, with the number of genes indicated. The second category represents PC4-bound RNA transcripts identified by RIP-seq in two independent experiments. The third category represents the intersection of both datasets. In the Venn diagrams, five PC4-upregulated RNA targets that are related to the cell cycle are shown in red, while five PC4-downregulated RNAs involved in the cell cycle are represented in blue. Furthermore, among the five RNAs shown in red, they belong to G1/S genes, whereas the other five RNAs in blue are categorized as G2/M genes. (D) Heatmap representing the mRNA half-life of five indicated genes in Huh7 cells with knockdown of NC or PC4, following treatment with actinomycin D (ActD) at different timepoints (h). The color bar represents the level of mRNA expression. Data are generated from n = 3 biological replicates. (E) Distribution of PC4-binding peaks across CCND1 mRNA from RIP-seq and eCLIP-seq, which were performed by anti-PC4 antibody in Huh7 cells. Data are generated from n = 2 biological replicates. (F) EMSA analysis of the association of PC4-CCND1 5′UTR in Huh7 cells and in vitro. In the in vitro assay, the transcribed 5′UTR fragment of CCND1 mRNA was incubated with different concentrations of recombinant GST-tagged PC4 protein and separated on a non-denaturing PAGE. The recombinant GST-PC4 proteins were examined by Coomassie brilliant blue staining. (G) EMSA analysis showing the association of various DNA and RNA molecules with recombinant GST-tagged PC4 protein including (1) the association of WDR74, LINC00869, and CCND1 dsDNA with PC4 protein. Lane 1–4 corresponds to synthesized DNA sequences of WDR74, LINC00869, and CCND1, which were derived from PC4 ChIP-seq peaks for WDR74 and LINC00869, and H3K27ac ChIP-seq peak for CCND1. (2) The association of CCND1-5′UTR ssDNA with PC4 protein. Lane 5–7 represents the interaction between PC4 protein and CCND1-5′UTR-forward sequence (CCND1-ssDNA-F) and CCND1-5′UTR-reverse sequence (CCND1-ssDNA-R). (3) The association of CCND1-5′UTR RNA with PC4 protein. Lane 8–9 indicates the binding of PC4 protein with CCND1-5′UTR RNA. (H) qPCR showing the mRNA stability of CCND1 in Huh7 and HepG2 cells with stable knockdown of PC4 upon treatment with ActD. Data are generated from n = 3 biological replicates. (I) qPCR showing the mRNA stability of CCND1 in Huh7 and HepG2 cells with stable overexpression of PC4 upon treatment with ActD. Data are generated from n = 3 biological replicates. (J) qPCR showing CCND1 mRNA expression in Huh7 and HepG2 cells with stable knockdown of PC4. Data in each group were normalized to that in NC. Data were generated from n = 3 biological replicates. (K) qPCR showing CCND1 mRNA expression in Huh7 and HepG2 cells with stable overexpression of PC4. Data in each group were normalized to that in NC. Data are generated from n = 3 biological replicates. (L) Western blot showing the indicated protein expressions in Huh7 and HepG2 cells with stable knockdown of PC4. The protein expressions are quantified and normalized, and the values are listed below each band. The PC4 and β-actin blots are duplicated in Fig. S1 L. (M) Western blot showing the indicated protein expressions in Huh7 and HepG2 cells with stable overexpression of PC4. The protein expressions are quantified and normalized, and the values are listed below each band. All the data were shown as means ± SD (one-way ANOVA test); error bars represent SD. **P < 0.01, ***P < 0.001, ****P < 0.0001. F, G, L, and M are representative of three independent experiments. Source data are available for this figure: SourceData F1.
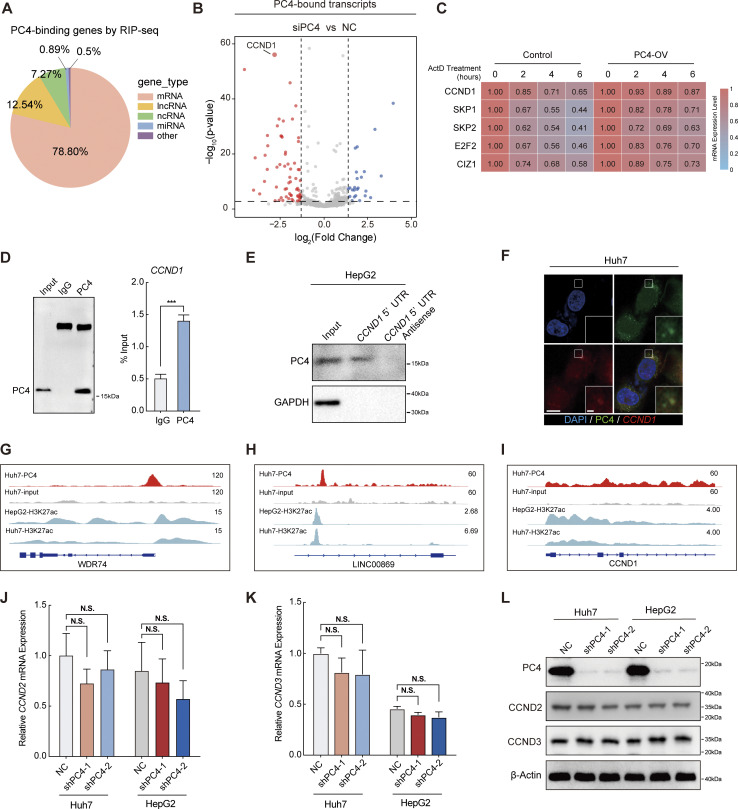
Since PC4 has been recognized as a potential RBP, we next determined the RNA transcripts that were directly bound by PC4 in Huh7 cells using PC4 RNA immunoprecipitation sequencing (RIP-seq) analysis. Notably, PC4 predominantly bound to protein-coding transcripts (Fig. S1 A). To precisely examine the effect of PC4 on its bound transcripts, we performed RNA sequencing (RNA-seq) analysis of PC4-knockdown Huh7 cells. Interestingly, the majority of genes bound to and regulated by PC4 exhibited a significant decrease in mRNA expression in PC4-knockdown cells (Fig. S1 B). More importantly, these genes were enriched in the cell cycle pathway (Fig. 1 B), suggesting that PC4 functions as an RBP to regulate cell cycle–related mRNA targets, control cell cycle progression, and promote cell proliferation. To further understand whether PC4 is involved in cell cycle machinery control, we performed RNA-seq analysis in three liver cancer cell lines (Huh7, HepG2, and SK-Hep-1) with or without PC4 knockdown and two biological replicates of PC4 RIP-seq in Huh7 cells. Among the 37 transcripts that were posttranscriptionally regulated by PC4, 10 were involved in the cell cycle pathway (Fig. 1 C and Data S1), confirming that PC4 may exert its effects on the cell cycle by directly binding to RNA. We carefully considered five transcripts showing downregulation upon PC4 knockdown that participated in RB:E2F activation and the cell cycle G1/S phase transition. Using an mRNA half-life assay, we found that the mRNA stabilities of these five genes were much lower after PC4 knockdown (Fig. 1 D). In contrast, the reverse was true in PC4-overexpressing (OV) cells (Fig. S1 C). Among these five genes, CCND1 stood out because of its important role in cell cycle progression and it showed the highest differential expression upon PC4 depletion (Fig. S1 B); therefore, we selected CCND1 as the downstream target of PC4 for further study.
Figure S1.
PC4 acts as an RBP that stabilizes CCND1 mRNA. Related to Fig. 1. (A) Pie chart showing the distribution of the PC4 RIP-seq reads in RNA classes. Data are generated from n = 2 biological replicates. (B) Volcano plot of the average difference of PC4-bound transcripts in NC and siPC4 groups determined by two-tailed Student’s t test from a linear model fit (x-axis). The y-axis indicates the P values. The genes of significantly downregulated (FC < −1, P < 0.05) are shown in red and upregulated (FC > 1, P < 0.05) genes are shown in blue. Vertical dashed lines indicate a cut-off of FC (1 or −1); horizontal dashed lines indicate a cut-off of P value (0.05). Data are generated from n = 2 biological replicates. (C) Heatmap representing the mRNA half-life of five indicated genes in Huh7 cells with control or overexpression of PC4, following treatment with ActD at different timepoints (hours). The color bar represents the level of mRNA expression. Data are generated from n = 3 biological replicates. (D) Western blot (left) and RIP-qPCR (right) showing the interaction between PC4 protein and CCND1 mRNA. Data were generated from n = 3 biological replicates. (E) RNA pull-down analysis showing the complex of PC4-CCND1 5′UTR in HepG2 cells. (F) Selected images of RNA-FISH. Huh7 cells expressing PC4-GFP were incubated with Cy5-labeled CCND1 5′UTR probe and stained with DAPI. PC4 protein is stained in green, CCND1 mRNA probe is stained in red, and DNA is stained with DAPI in blue. Scale bar = 40 μm; insert scale bar = 3.5 μm. (G) IGV browser tracks showing PC4 and H3K27ac ChIP-seq reads in WDR74 DNA in Huh7 and HepG2 cells. PC4 ChIP-seq data are generated from n = 2 biological replicates. H3K27ac ChIP-seq data are generated from the dataset of GSM2360941 and GSM646355. (H) IGV browser tracks showing PC4 and H3K27ac ChIP-seq reads in LINC00869 DNA in Huh7 and HepG2 cells. PC4 ChIP-seq data are generated from n = 2 biological replicates. H3K27ac ChIP-seq data are generated from the dataset of GSM2360941 and GSM646355. (I) IGV browser tracks showing PC4 and H3K27ac ChIP-seq reads in CCND1 DNA in Huh7 and HepG2 cells. PC4 ChIP-seq data are generated from n = 2 biological replicates. H3K27ac ChIP-seq data are generated from the dataset of GSM2360941 and GSM646355. (J) qPCR showing CCND2 mRNA expression in Huh7 and HepG2 cells with PC4 knockdown. Data in each group was normalized to that in NC. Data are generated from n = 3 biological replicates. (K) qPCR showing CCND3 mRNA expression in Huh7 and HepG2 cells with PC4 knockdown. Data in each group was normalized to that in NC. Data are generated from n = 3 biological replicates. (L) Western blot showing CCND2 and CCND3 protein expression in Huh7 and HepG2 cells with PC4 knockdown. The PC4 and β-actin blots are duplicated in Fig. 1 L. All graphed data are shown as means ± SD (one-way ANOVA test); error bars represent SD. ***P < 0.001. D–F and L are representative of three independent experiments. Source data are available for this figure: SourceData FS1.
CCND1 is a core component of the cyclin–CDK complex that phosphorylates the tumor suppressor RB and releases the transcription factors (TFs) E2F1-3 to initiate the G1/S transition (Sherr, 1995; Morgan, 1997). To determine how PC4 regulates CCND1 mRNA metabolism, we first validated the specific binding between the PC4 protein and CCND1 mRNA using enhanced crosslinking and immunoprecipitation followed by next-generation sequencing (eCLIP-seq). PC4 preferred to bind to the 5′UTR of CCND1 mRNA (Fig. 1 E). Furthermore, both the results from Huh7 cells and in vitro RNA-electrophoretic mobility shift assay (EMSA) confirmed this binding (Fig. 1 F). Consistent with this observation, RIP-quantitative polymerase chain reaction (qPCR), RNA pull-down, and RNA-fluorescence in situ hybridization (FISH) analyses revealed that this interaction occurred in distinct liver cancer cells (Fig. S1, D–F). Additionally, we found that the PC4 protein bound only to CCND1 mRNA with high affinity, but not to double-strand DNA (dsDNA) or single-strand DNA (ssDNA) (Fig. 1 G and Fig. S1, G–I). Finally, we verified that the decay rate of CCND1 was much higher following PC4 knockdown, whereas PC4 overexpression significantly extended the half-life of CCND1 mRNA in both Huh7 and HepG2 cells (Fig. 1, H and I). Moreover, PC4 depletion considerably reduced the CCND1 mRNA (Fig. 1 J) and protein levels (Fig. 1 L). However, the opposite effects were observed in PC4-OV cells (Fig. 1, K and M), indicating that PC4 interacts with and stabilizes CCND1 mRNA, leading to enhanced CCND1 protein expression. Notably, PC4 depletion did not alter the mRNA expression of cyclin D2 (CCND2) or cyclin D3 (CCND3) (Fig. S1, J–L), suggesting that PC4 specifically regulates CCND1 expression. Taken together, these results indicate that PC4 posttranscriptionally regulates CCND1 gene expression, independent of its classical function as a co-TF (Ge and Roeder, 1994; Kretzschmar et al., 1994).
Ubiquitination and phosphorylation of PC4 show periodic fluctuations and are associated with the stability of CCND1 mRNA
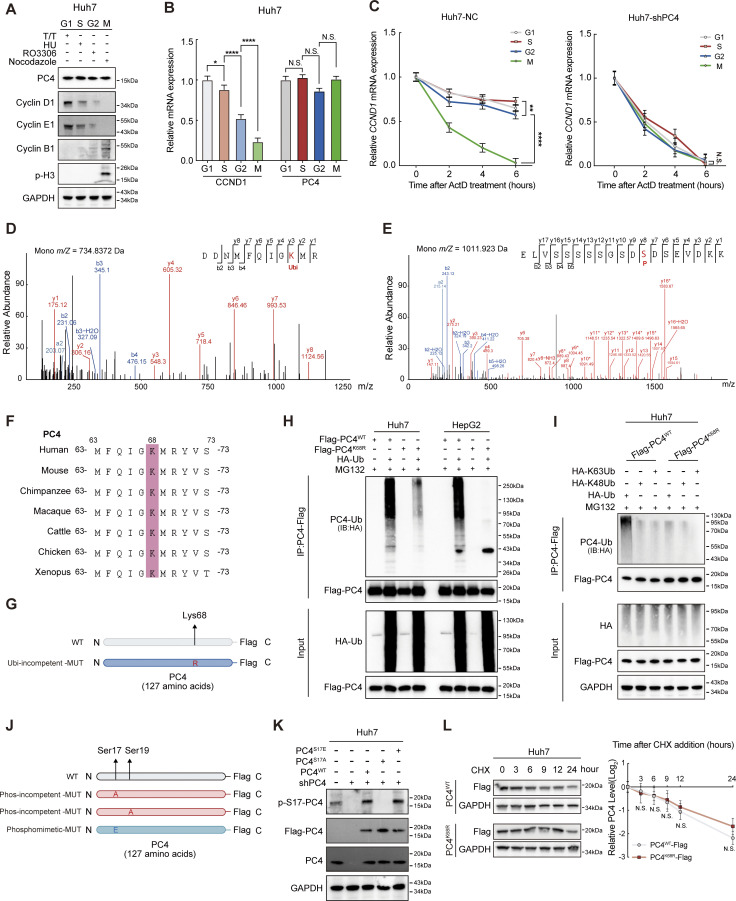
CCND1 is expressed periodically throughout the cell cycle (Baldin et al., 1993). Both its mRNA and protein levels peak in the G1 phase and gradually decline from the S phase to the M phase (Fig. S2, A and B), as shown previously. This cell cycle–dependent oscillation is, in part, driven by the transcription network (Liu et al., 2017) and the ubiquitin–proteasome system (Lin et al., 2006; Fasanaro et al., 2010). However, it is unclear whether CCND1 expression is periodically regulated at the posttranscriptional level. Interestingly, the stability of CCND1 mRNA also fluctuated over the cell cycle, increasing in the G1 and S phases, gradually decreasing at the end of the G2 phase, and decreasing to a sufficiently low level in the M phase (Fig. S2 C). Notably, in cells where PC4 was depleted, the fluctuations in CCND1 mRNA stability during the cell cycle were completely abolished (Fig. S2 C), indicating that PC4 may posttranscriptionally control the stability of CCND1 mRNA during different cell cycle phases.
Figure S2.
Ubiquitination and phosphorylation of PC4 show periodic fluctuations and are associated with the stability of CCND1 mRNA. Related to Fig. 2. (A) Western blot showing CCND1 and PC4 proteins expression at different cell cycle phases in Huh7 cells. (B) qPCR showing CCND1 and PC4 mRNA expression at different cell cycle phases in Huh7 cells. Data are generated from n = 3 biological replicates. (C) CCND1 mRNA stability upon ActD treatment in Huh7 cells with or without PC4 knockdown at different cell cycle phases. Data are generated from n = 3 biological replicates. (D) Liquid chromatography (LC)-MS/MS of PC4 ubiquitinated conjugation site. m/z, mass/charge ratio. (E) LC-MS/MS of PC4 phosphorylated conjugation site. m/z, mass/charge ratio. (F) Sequence alignment of PC4 residues aa 63–73 between different species using the Uniport alignment tool. (G) Schematic amino acid sequence of WT or Lys/Arg-mutant (K68R) PC4 protein. (H) Western blot showing different ubiquitination levels of PC4-Flag in Huh7 and HepG2 cells transfected with HA-tagged ubiquitin and indicated Flag-tagged PC4 variants. IB, immunoblot. (I) Western blot showing different ubiquitination levels of PC4-Flag in Huh7 cells transfected with indicated HA-tagged ubiquitin mutants and Flag-tagged PC4 variants. (J) Schematic amino acid sequence of WT or mutant (S17A/S19A/S17E) PC4 protein. (K) Western blot showing the phosphorylation level of PC4 in Huh7 cells expressing exogenous Flag-tagged PC4-WT, S17A, or S17E with endogenous PC4 knockdown. The p-Ser signal and Flag-tagged PC4 were examined with p-S17-PC4 and other indicated antibodies. (L) Western blot and statistical analysis showing PC4 protein stability upon cycloheximide (CHX) chase treatment at different timepoints in Huh7 cells expressing Flag-tagged PC4 variants. Data are generated from n = 3 biological replicates. All quantifications are shown as mean ± SD (one-way ANOVA test); error bars represent SD. *P < 0.05, **P < 0.01, ****P < 0.0001. A, H, I, and K are representative of three independent experiments. Source data are available for this figure: SourceData FS2.
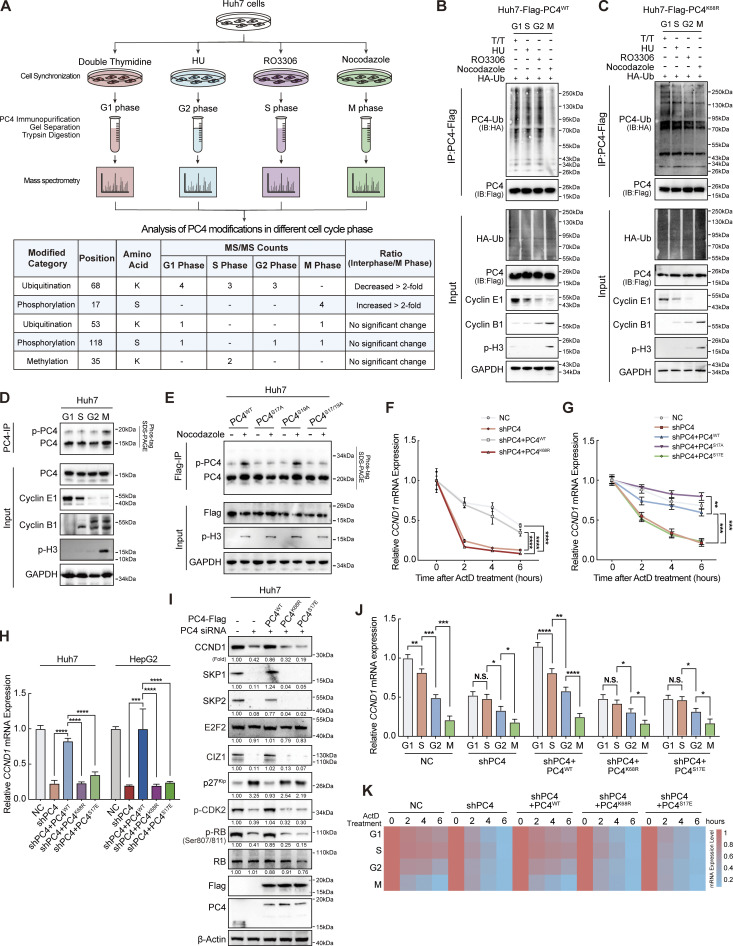
To uncover the mechanism by which PC4 manipulates the periodic stability of CCND1 mRNA, we monitored the PC4 levels as the cells progressed through the cell cycle. Because the mRNA and protein levels of PC4 remained stable in different cycle phases (Fig. S2, A and B), we wondered whether the PTMs of PC4 exhibited a fluctuating pattern across the cell cycle. We immunoprecipitated PC4 from synchronized Huh7 cells and identified PC4 modifications using mass spectrometry (MS) (Fig. 2 A). Five modifications of PC4 were detected in different cell cycle phases. Among them, two periodically changing modifications grabbed our attention: (1) ubiquitination of lysine at position 68 (K68) that appears in interphase and disappears in the M phase, and (2) phosphorylation of serine at position 17 (S17) that only emerges in the M phase (Fig. 2 A and Fig. S2, D and E).
Figure 2.
Ubiquitination and phosphorylation of PC4 show periodic fluctuations and are associated with the stability of CCND1 mRNA. (A) Workflow for the identification of PC4 modifications during different cell cycle phases in Huh7 cells. Briefly, cells were treated with specific reagents to synchronize them in different cell cycle phases. After that, media was removed and cells were lysed. Then, the cell lysates were subjected to IP using the PC4 antibody. Subsequently, PC4 protein and its modifications were separated by SDS-PAGE before MS analysis. (B) Western blot showing the alternations of PC4 ubiquitination level at different cell cycle phases in PC4WT Huh7 cells. T/T, double-thymidine. HU, hydroxyurea. (C) Western blot showing the alternations of PC4 ubiquitination level at different cell cycle phases in PC4K68R Huh7 cells. (D) Western blot showing the changes of endogenous PC4 phosphorylation level at different cell cycle phases in Huh7 cells. (E) Western blot showing the presence of PC4 phosphorylation at asynchronous or synchronized M phase in Huh7 cells expressing Flag-tagged PC4 variant. (F) qPCR showing CCND1 mRNA stability upon ActD treatment in PC4-knockdown Huh7 cells stably expressing indicated Flag-tagged PC4 variants. Data are generated from n = 3 biological replicates. (G) qPCR showing CCND1 mRNA stability upon ActD treatment in PC4-knockdown Huh7 cells stably expressing indicated Flag-tagged PC4 variants. Data are generated from n = 3 biological replicates. (H) qPCR showing CCND1 mRNA expressions in PC4-knockdown Huh7 cells stably expressing indicated Flag-tagged PC4 variants. Data are generated from n = 3 biological replicates. (I) Western blot showing the indicated protein expressions in PC4-knockdown Huh7 cells stably expressing indicated Flag-tagged PC4 variants. The protein expressions are quantified and normalized, and the values are listed below each band. (J) qPCR showing CCND1 mRNA expressions in PC4-knockdown Huh7 cells stably expressing indicated Flag-tagged PC4 variants at different cell cycle phases. Data are generated from n = 3 biological replicates. (K) Heatmap showing CCND1 mRNA stability upon ActD treatment in PC4-knockdown Huh7 cells stably expressing indicated Flag-tagged PC4 variants at different cell cycle phases. The color bar represents the level of mRNA expression. Data are generated from n = 3 biological replicates. All quantifications are shown as mean ± SD (one-way ANOVA test); error bars represent SD. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. B–E and I are representative of three independent experiments. Source data are available for this figure: SourceData F2.
The K68 site of PC4 was highly conserved among vertebrates (Fig. S2 F). To confirm K68 ubiquitination, we created a K68R mutant of PC4 (PC4K68R) by replacing the Lys residue with Arg (Fig. S2 G). We found that in both Huh7 and HepG2 cells, PC4K68R led to a prominent reduction in ubiquitination (Fig. S2 H), confirming K68 ubiquitination of PC4. In addition, PC4 was mainly modified with K63-, but not with K48-linked chains at the K68 residue (Fig. S2 I). We then investigated the fluctuations in K68 ubiquitination throughout the cell cycle. PC4 ubiquitination levels increased markedly in the G1, S, and G2 phases, but were nearly undetectable in the M phase (Fig. 2 B). In contrast, the PC4K68R ubiquitination level remained the same throughout the cell cycle (Fig. 2 C), suggesting that PC4 undergoes interphase K68 ubiquitination.
Next, we focused on another modification: S17 phosphorylation. To verify the existence of S17 phosphorylation, we established a series of PC4 mutants, including phosphorylation-incompetent mutants (PC4S17A and PC4S19A, which were constructed as negative control, and PC4S17/19A) and the phosphomimetic mutant PC4S17E (Fig. S2 J), and generated a specific antibody against PC4-S17 phosphorylation (p-S17-PC4). We detected this phosphorylation signal in PC4-depleted cells with re-expression of PC4WT or PC4S17E, but no phosphorylation signal was observed in cells with re-expression of PC4S17A (Fig. S2 K). We also monitored changes in S17 phosphorylation during the cell cycle. Consistent with the MS observations, PC4 phosphorylation peaked in the M phase and gradually decreased in the other phases (Fig. 2 D). Conversely, this oscillation was abolished in both PC4S17A and PC4S17/19A, but not in PC4S19A (Fig. 2 E), demonstrating S17 phosphorylation of PC4 during mitosis.
We then determined whether PC4 uses these two dynamic modifications to manipulate CCND1 mRNA stability. The stability of CCND1 mRNA was rescued by re-expression of PC4WT in PC4-knockdown cells. In comparison, PC4K68R lost this ability (Fig. 2 F), suggesting that K68 ubiquitination mediates the effects of PC4 in stabilizing CCND1 mRNA. In contrast, while overexpression of PC4S17A reversed the decrease in CCND1 transcript stability upon PC4 knockdown, PC4S17E failed to exert this effect (Fig. 2 G), indicating that S17 phosphorylation blocks PC4 from stabilizing CCND1 mRNA. Likewise, further experiments suggested that neither PC4K68R nor PC4S17E was capable of inducing CCND1 mRNA or protein expression (Fig. 2, H and I). Notably, the fluctuations in CCND1 mRNA expression and stability across the cell cycle were reversed by overexpressing PC4WT, but not PC4K68R or PC4S17E, in PC4-knockdown cells (Fig. 2, J and K). These data suggest that PC4 is a cell cycle–regulated protein that is ubiquitinated at K68 during interphase and phosphorylated at S17 in the M phase. These two modifications have opposite effects on PC4-mediated modulation of CCND1 mRNA stability, which may have contributed to the fluctuation of CCND1 expression in the cell cycle.
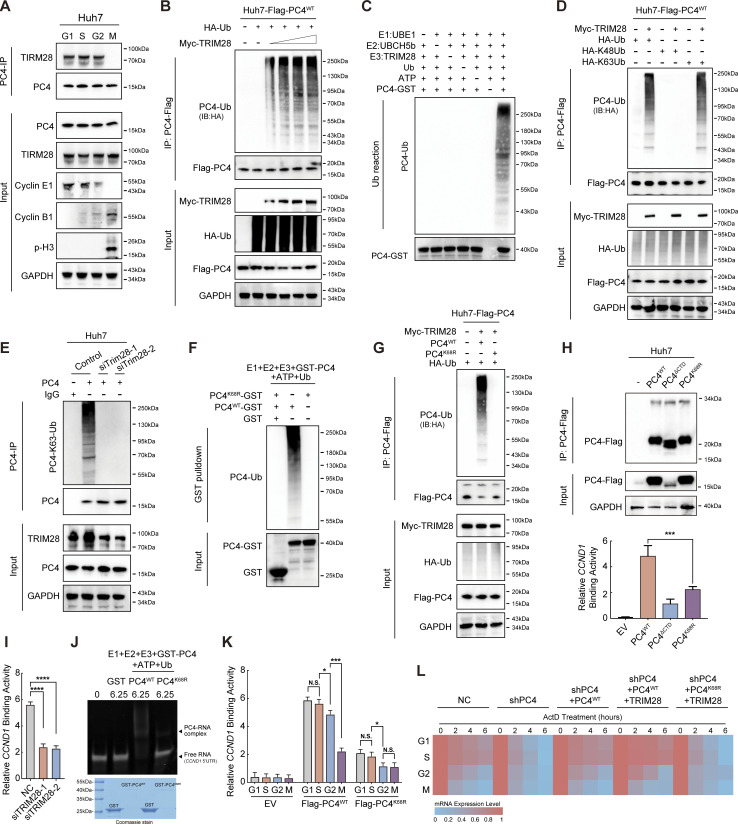
During interphase, ubiquitination of PC4 at K68 by TRIM28 enhances the interaction with and stabilization of CCND1 mRNA
To further explore the mechanism by which PC4 ubiquitination at K68 increased CCND1 mRNA stability, we aimed to identify the ubiquitin ligase responsible for this modification. We examined the PC4 interactome in synchronized Huh7 cells and found that TRIM28, a RING domain–containing E3 ubiquitin ligase, was specifically associated with PC4 during interphase (Data S2). This cell cycle–dependent interaction was validated using an immunoprecipitation (IP) assay (Fig. 3 A). Furthermore, Myc-tagged TRIM28 strongly promoted PC4 ubiquitination in a dose-dependent manner (Fig. 3 B). The in vitro ubiquitylation assay using recombinant proteins provided additional evidence by demonstrating that the polyubiquitylation of PC4 occurred exclusively when TRIM28 was present (Fig. 3 C). Additionally, TRIM28-mediated K63-linked polyubiquitination of PC4 (Fig. 3 D) was significantly reduced by TRIM28 knockdown (Fig. 3 E). TRIM28 was unable to ubiquitinate K68R-mutated PC4 both in vitro and in Huh7 cells (Fig. 3, F and G), indicating that TRIM28 is critical for PC4 ubiquitination at K68.
Figure 3.
During interphase, ubiquitination of PC4 at K68 by TRIM28 enhances the interaction with and stabilization of CCND1 mRNA. (A) Western blot showing the interaction between PC4 and TRIM28 at different cell cycle phases in Huh7 cells. (B) Western blot showing the ubiquitination level of PC4-Flag in Huh7 cells transfected with Flag-tagged PC4, HA-tagged Ub, and increasing amounts of Myc-TRIM28 as indicated. (C) Western blot showing the ubiquitination level of GST-PC4 in vitro ubiquitylation assay. The recombinant proteins GST-tagged PC4, TRIM28 along with their required cofactors were incubated at 30°C for 1 h. Blots were probed with the indicated antibodies. (D) Western blot showing the ubiquitination level of PC4-Flag in Huh7 cells transfected with Flag-tagged PC4 and Myc-tagged TRIM28 together with HA-tagged wild-type ubiquitin or K48-linked ubiquitin or K63-linked ubiquitin. IB, immunoblot. (E) Western blot showing the effect of TRIM28 on PC4 ubiquitination in Huh7 cells transduced with control or TRIM28 siRNA. (F) Western blot showing the ubiquitination levels of GST-PC4 and GST-PC4K68R in vitro ubiquitylation assay. Recombinant proteins (GST-PC4 and GST-PC4K68) were incubated with the E1, E2, and E3 for 1 h. Blots were probed with the indicated antibodies. (G) Western blot showing the ubiquitination levels of Flag-PC4 and Flag-PC4K68R in Huh7 cells transfected with Myc-tagged TRIM28, HA-tagged Ub. Ub, ubiquitin. (H) RIP-qPCR and Western blot showing the association of indicated Flag-tagged PC4 variants and CCND1 mRNA in Huh7 cells. Data are generated from n = 3 biological replicates. (I) RIP-qPCR showing the effect of TRIM28 on the association of PC4 and CCND1 mRNA. Data were generated from n = 3 biological replicates. (J) In vitro EMSA analysis showing the interaction between indicated GST-tagged recombinant PC4 variants and CCND1 5′UTR. The Ub-reaction products were incubated with CCND1 5′UTR mRNA, followed by EMSA. Data are generated from n = 3 biological replicates. (K) RIP-qPCR showing the association of indicated Flag-tagged PC4 variants and CCND1 mRNA at different cell cycle phases. Data are generated from n = 3 biological replicates. (L) Heatmap showing CCND1 mRNA stability upon ActD treatment in PC4-depleted Huh7 cells stably expressing indicated Flag-tagged PC4 variants and Myc-tagged TRIM28 at different cell cycle phases. The color bar represents the level of mRNA expression. Data are generated from n = 3 biological replicates. All quantifications are shown as mean ± SD (one-way ANOVA test); error bars represent SD. *P < 0.05, ***P < 0.001, ****P < 0.0001. A–G are representative of three independent experiments. Source data are available for this figure: SourceData F3.
Since the C-terminal domain (aa 63–106) of PC4 contributes to nucleotide binding (Wang et al., 2004; Mortusewicz et al., 2016), we reckoned that TRIM28-mediated K68 ubiquitination could affect PC4–CCND1 mRNA association. The first hint came from the observation that when K68 ubiquitination was disrupted in Huh7 cells, PC4K68R exhibited a substantially weaker binding capacity to the CCND1 transcript, resembling the depletion of the C-terminal domain (PC4ΔCTD) to some extent (Fig. 3 H), but did not affect PC4 proteasomal degradation (Fig. S2 L). Consistently, the PC4–CCND1 interaction was significantly suppressed when TRIM28 was silenced in Huh7 cells (Fig. 3 I). Moreover, recombinant PC4K68R had a much lower binding affinity for mRNA in vitro (Fig. 3 J), highlighting the importance of K68 ubiquitination in supporting the interaction between PC4 with CCND1 mRNA. Notably, K68 ubiquitination could serve as an important factor that periodically controls the PC4–CCND1 mRNA interaction. During interphase, the K68 ubiquitination level was relatively high and the interaction was relatively stable. In the M phase, the interaction weakened as K68 ubiquitination sufficiently decreased (Fig. 3 K). Therefore, TRIM28 overexpression further increased the K68 ubiquitination level, PC4–CCND1 mRNA binding, and CCND1 mRNA stability in PC4WT cells with PC4 knockdown during interphase; however, this effect was compromised in PC4K68R cells (Fig. 3 L).
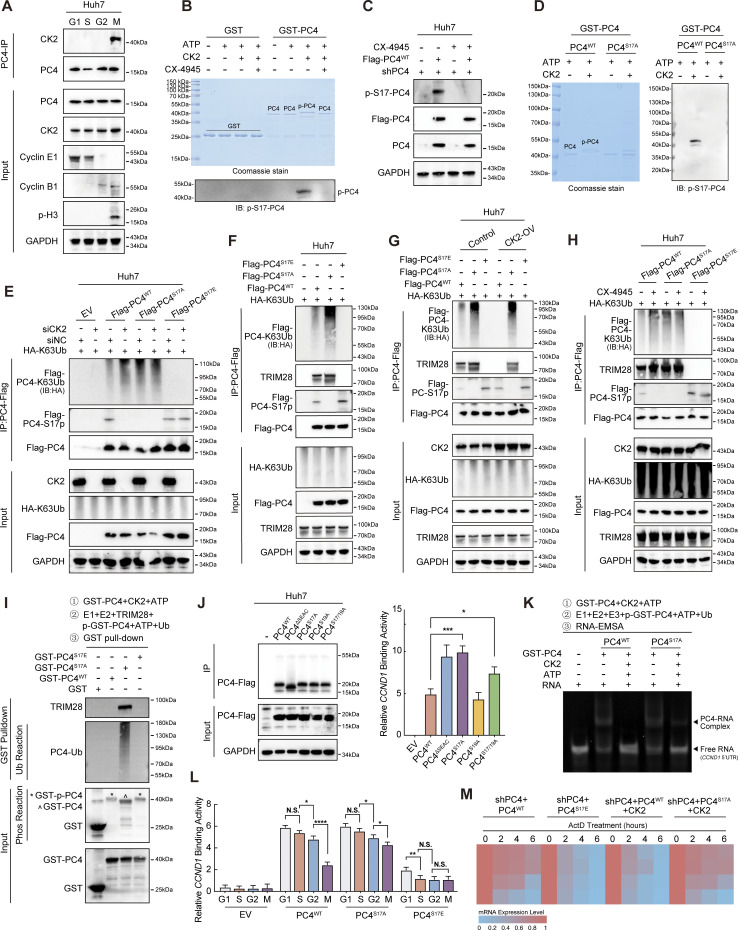
In the M phase, phosphorylation of PC4 at S17 by casein kinase 2 (CK2) hampers its interaction with TRIM28 and subsequent ubiquitination, leading to the degradation of CCND1 mRNA
We investigated the biological role of PC4 S17 phosphorylation. Since CK2 mediates PC4 phosphorylation within the N-terminal serine-rich acidic stretch (SEAC) domain (Ge et al., 1994), we investigated whether S17 could be a target of CK2 phosphorylation. As expected, CK2 specifically interacted with PC4 in the M phase (Fig. 4 A and Data S2) and efficiently phosphorylated PC4 at S17 residue (Fig. 4, B–D). This observation was verified by the addition of a CK2 inhibitor (CX-4945), which significantly reduced the S17 phospho-signal of PC4 in vitro and in Huh7 cells (Fig. 4, B and C).
Figure 4.
In the M phase, phosphorylation of PC4 at S17 by CK2 hampers its interaction with TRIM28 and subsequent ubiquitination, leading to the degradation of CCND1 mRNA. (A) Western blot showing the interaction between PC4 and CK2 at different cell cycle phases in Huh7 cells. (B) Coomassie staining and western blot showing the phosphorylation of PC4 in vitro phosphorylation assay with the recombinant proteins GST-PC4, CK2, and CK2 inhibitor CX-4945. (C) Western blot showing the S17 phosphorylation of PC4 in Huh7 cells with stable PC4 knockdown and re-expression of Flag-tagged PC4WT upon CK2 inhibitor CX-4945 treatment. (D) Coomassie staining and western blot showing S17 phosphorylation of PC4 in vitro phosphorylation assay with the recombinant CK2 and GST-PC4 variants proteins. (E) Western blot showing the ubiquitination of PC4 in Huh7 cells transfected with K63-linked HA-tagged ubiquitin and indicated Flag-tagged PC4 variants with or without CK2 knockdown. EV, empty vector; IB, immunoblot. (F) Western blot showing the ubiquitination of PC4 and the interaction of PC4 and TRIM28 in Huh7 transfected with K63-linked HA-tagged ubiquitin and indicated Flag-tagged PC4 variants. (G) Western blot showing the ubiquitination of PC4 and the interaction of PC4 and TRIM28 in Huh7 transfected with K63-linked HA-tagged ubiquitin and indicated Flag-tagged PC4 variants with or without CK2 overexpression. (H) Western blot showing the ubiquitination of PC4 and the interaction of PC4 and TRIM28 in Huh7 transfected with K63-linked HA-tagged ubiquitin and indicated Flag-tagged PC4 variants with or without CK2 inhibitor CX-4945. (I) Western blot showing the ubiquitination of PC4 and the interaction of PC4 and TRIM28 in phosphorylation assay and ubiquitination assay with recombinant proteins GST-PC4, GST-PC4S17A and GST-PC4S17E, followed by GST pull-down. * represents phosphorylated GST-tagged recombinant PC4. ^ represents non-phosphorylated GST-tagged recombinant PC4. (J) RIP-qPCR and western blot showing the association of indicated Flag-tagged PC4 variants and CCND1 mRNA in Huh7 cells. Data were generated from n = 3 biological replicates. (K) In vitro EMSA analysis showing the interaction between GST-tagged recombinant PC4 variant proteins and CCND1 5′UTR after phosphorylation assay and ubiquitination assay. (L) RIP-qPCR showing the association of indicated Flag-tagged PC4 variants and CCND1 mRNA in Huh7 cells at different cell cycle phases. Data were generated from n = 3 biological replicates. (M) Heatmap showing CCND1 mRNA stability upon ActD treatment in PC4-depleted Huh7 cells stably expressing indicated Flag-tagged PC4 variants and CK2 at different cell cycle phases. The color bar represents the level of mRNA expression. Data are generated from n = 3 biological replicates. All quantifications are shown as mean ± SD (one-way ANOVA test); error bars represent SD. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. A–I are representative of three independent experiments. Source data are available for this figure: SourceData F4.
Considering the possibility of crosstalk between phosphorylation and ubiquitination pathways and the potential of these processes to influence each other functionally (Hunter, 2007; Beltrao et al., 2013; Swaney et al., 2013), we aimed to determine the interplay between S17 phosphorylation and K68 ubiquitination. Strikingly, in comparison with PC4WT, PC4S17A moderately increased the K63-linked ubiquitination level of PC4, whereas PC4S17E significantly decreased this level. Notably, silencing CK2 enhanced K63-linked ubiquitination in PC4WT cells but did not affect the abundance of ubiquitination in PC4S17A cells. In addition, silencing CK2 failed to reverse the reduction of PC4 ubiquitination induced by the S17E mutation (Fig. 4 E). These results suggested that PC4 ubiquitination occurs after S17 dephosphorylation. Next, we determined whether S17 phosphorylation inhibited the association between TRIM28 and PC4. Co-IP analyses revealed that S17 phosphorylation (PC4S17E) precluded the TRIM28–PC4 interaction and suppressed downstream ubiquitination (Fig. 4 F). In PC4WT cells, inhibition of CK2 enhanced this interaction and TRIM28-mediated PC4–K68-linked polyubiquitination, while CK2 overexpression blocked these events. However, in PC4S17A cells, CK2 had no influence on the interaction between TRIM28 and PC4S17A nor on the K63-linked polyubiquitination of PC4S17A (Fig. 4, G and H). Consistent with these findings, in vitro experiments showed that CK2-directed PC4 S17 phosphorylation disrupted the binding between TRIM28 and PC4, thereby inhibiting subsequent K68 ubiquitination (Fig. 4 I).
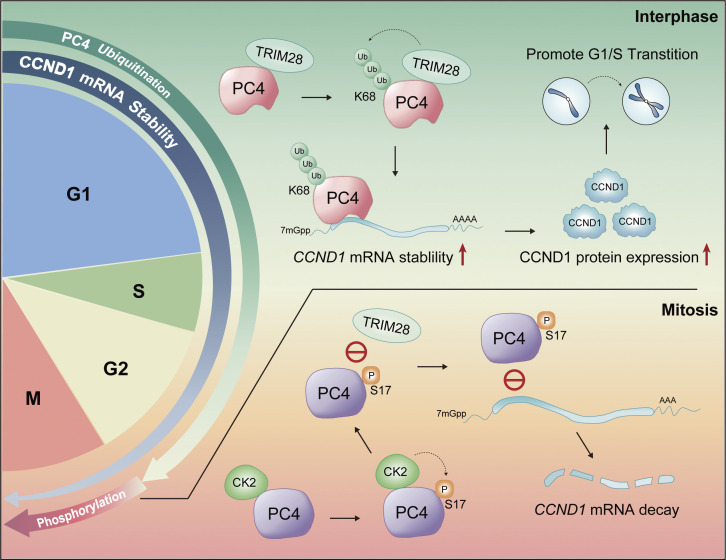
On the basis of the aforementioned results, we hypothesized that CK2-mediated S17 phosphorylation interferes with the binding between PC4 and CCND1 mRNA. Indeed, PC4S17A markedly strengthened CCND1 mRNA binding affinity in comparison with PC4WT or the PC4S19A mutant (Fig. 4 J). Furthermore, CK2 diminished the mRNA binding capacity of PC4WT, but not PC4S17A (Fig. 4 K). More importantly, PC4S17A maintained the cell cycle–related fluctuation of the PC4–CCND1 mRNA association, whereas PC4S17E failed to exert this effect (Fig. 4 L). In addition, PC4S17E antagonized the fluctuations of CCND1 mRNA stability during the cell cycle. Also, CK2 interrupted the periodically changing CCND1 transcript stability in PC4WT cells, but not in PC4S17A cells (Fig. 4 M). Thus, PC4 is a cell cycle–dependent RBP whose RNA binding function is determined by a phosphorylation–ubiquitination switch across the cell cycle phases. During interphase, TRIM28-promoted K68 ubiquitination switches PC4 function “on” to enhance the interaction with and stabilization of CCND1 mRNA. However, after mitotic entry, CK2-catalyzed S17 phosphorylation turns PC4 function “off” by completely dissociating PC4 from TRIM28 and CCND1 mRNA, resulting in CCND1 mRNA degradation. These two dynamically controlled modifications further highlight PC4 as an RBP involved in cell cycle progression and are likely critical factors that allow PC4 to control CCND1 mRNA stability in the cell cycle.
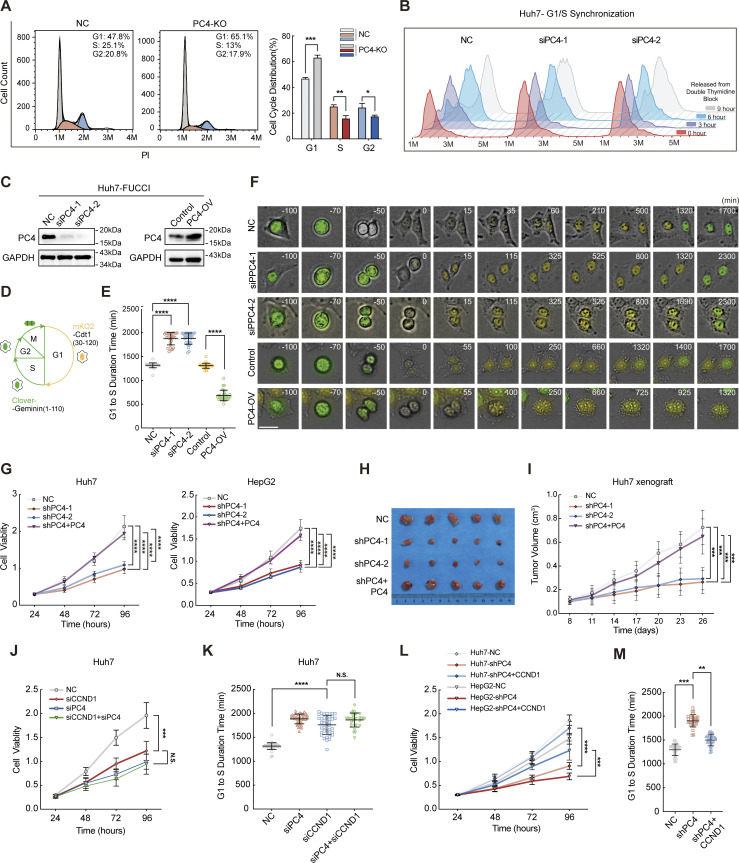
PC4 promotes G1–S transition and cell proliferation partially through a CCND1-dependent manner
Inspired by emerging evidence that PC4 regulates CCND1 mRNA stability, we wondered whether PC4 could promote the G1–S phase transition and cell proliferation by stabilizing the levels of the CCND1 transcript. To study the overall effect of PC4 on cell cycle dynamics, we first performed flow cytometry on non-synchronized cells. As expected, PC4 deficiency induced an elevation of cells in the G1 phase, accompanied by a reduction in the non-G1 phases (G2/M/S), linking PC4 to the cell cycle transition (Fig. 5 A). We further synchronized the cell cycle at the G1/S phase boundary and released the cells to assess the number of cycling cells passing through the G1 phase. PC4 depletion markedly impaired the G1/S transition since a lower rate of cells entered the S phase in comparison with the control cells (Fig. 5 B and Fig. S3 G). We also constructed a model using Huh7-FUCCI (a fluorescent ubiquitination-based cell cycle indicator) and performed time-lapse microscopy to evaluate the G1/S transition (Fig. 5 D). PC4 knockdown resulted in a substantial increase in the duration of the G1 phase after a prolonged 10-h period. In contrast, PC4 overexpression resulted in a shorter G1 phase by reducing the duration time by half (Fig. 5, C, E, and F; and Videos 1, 2, and 3).
Figure 5.
PC4 promotes G1–S transition and cell proliferation partially through a CCND1-dependent manner. (A) Asynchronized cell cycle analysis of Huh7 cells stably expressing control or PC4-KO. The number on the upper right corner represents the percentage of cells in different cell cycle phases. Data are generated from n = 3 biological replicates. (B) Synchronized cell cycle analysis of Huh7 cells with control or PC4 knockdown. Cells were treated with double-thymidine block, followed by release for indicated period of time. Data were generated from n = 3 biological replicates. (C) Western blot showing PC4 expression level in Huh7-FUCCI cells transfected with indicated siRNAs or plasmids. (D) Illustration of FUCCI-expressing Huh7 cells at each cell cycle phase. Cells at G1 display yellow fluorescence from mKO2, while cells at non-G1 phases (S, G2, and M) display green fluorescence from Clover. (E) The time length of the G1 phase in F. n = 30 cells in each group. Open circle represents an individual cell. (F) Representative frames of cell cycle tracking in FUCCI-expressing Huh7 cells transfected with indicated siRNAs or plasmids via time-lapse imaging. Scale bar = 50 μm. (G) Cell viability analysis by CCK8 assay in Huh7 and HepG2 cells with PC4 stable knockdown. Data are generated from n = 5 biological replicates. (H) Photographs of dissected tumors in the nude mice subcutaneously injected with Huh7 cell stably expressing NC, shPC4-1, shPC4-2, or shPC4+PC4 (the sequence for PC4 induction is optimized and resistant to shRNA). Data are generated from n = 5 biological replicates. (I) Tumor volume in the nude mice subcutaneously injected with Huh7 cell stably expressing NC, shPC4-1, shPC4-2, or shPC4+PC4. Data are generated from n = 5. (J) Cell viability analysis by CCK8 assay in Huh7 cells with NC, siPC4, siCCND1, or siPC4+siCCND1. Data are generated from n = 5 biological replicates. (K) The lengths of time from G1 to S phase in FUCCI-expressing Huh7 cells treated with NC, siPC4, siCCND1, or siPC4+siCCND1. Data are generated from n = 30 for each group. (L) Cell viability analysis by CCK8 assay in Huh7 and HepG2 cells with empty vector (NC), PC4 stable knockdown (shPC4), or cyclin D1 complemented PC4 knockdown (shPC4+CCND1). Data are generated from n = 5 biological replicates. (M) The lengths of time from G1 to S phase in FUCCI-expressing Huh7 cells treated with NC, shPC4, or shPC4+CCND1. Data are generated from n = 30 for each group. All quantifications were shown as means ± SD (one-way ANOVA test); error bars represent SD. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. C is representative of three independent experiments. Source data are available for this figure: SourceData F5.
Figure S3.
PC4 knockdown inhibits liver cancer cell growth in vitro and in vivo. Relative to Fig. 5. (A) qPCR and western blot showing PC4 protein and mRNA level of Huh7 and HepG2 cells stably expressing control shRNA (NC), human PC4-targeting shRNA (shPC4-1 and shPC4-2), or shRNA with PC4 expression (shPC4+PC4; the sequence for PC4 induction is optimized and resistant to shRNA). β-actin, loading control. Data were generated from n = 3 biological replicates. (B) Colony assay performed in Huh7 and HepG2 cells after PC4 stable knockdown. Data were generated from n = 3 biological replicates. (C) Soft agar assay performed in Huh7 and HepG2 cells with PC4 stable knockdown. Scale bar = 300 μm. Data are generated from n = 4 biological replicates. (D) EdU immunofluorescent staining performed in Huh7 and HepG2 cells with PC4 stable knockdown. Scale bar = 75 μm. Data were generated from n = 3 biological replicates. (E) Tumor weight in the nude mice subcutaneously injected with Huh7 cell stably expressing NC, shPC4-1, shPC4-2, or shPC4+PC4. Data were generated from n = 5 biological replicates. (F) Representative images of PC4, Ki67, and CCND1 immunostaining in tumor tissue of indicated xenograft mice. Scale bar = 75 μm. (G) Histograms show the cell cycle profiles in NC or PC4-knockdown Huh7 cells treated with double-thymidine block, followed by releasing for an indicated period of time. Data are generated from n = 3 biological replicates. All the data were shown as means ± SD (one-way ANOVA test); error bars represent SD. **P < 0.01, ***P < 0.001, ****P < 0.0001. Source data are available for this figure: SourceData FS3.
Video 1.
The real-time cell-cycle transition of Huh7 cells under different conditions: control, PC4 knockdown, and PC4-OV. Frame rate, 29.97 fps.
Video 2.
The real-time cell-cycle transition of Huh7 cells under different conditions: control, PC4 knockdown, and PC4-OV. Frame rate, 29.97 fps.
Video 3.
The real-time cell-cycle transition of Huh7 cells under different conditions: control, PC4 knockdown, and PC4-OV. Frame rate, 29.97 fps.
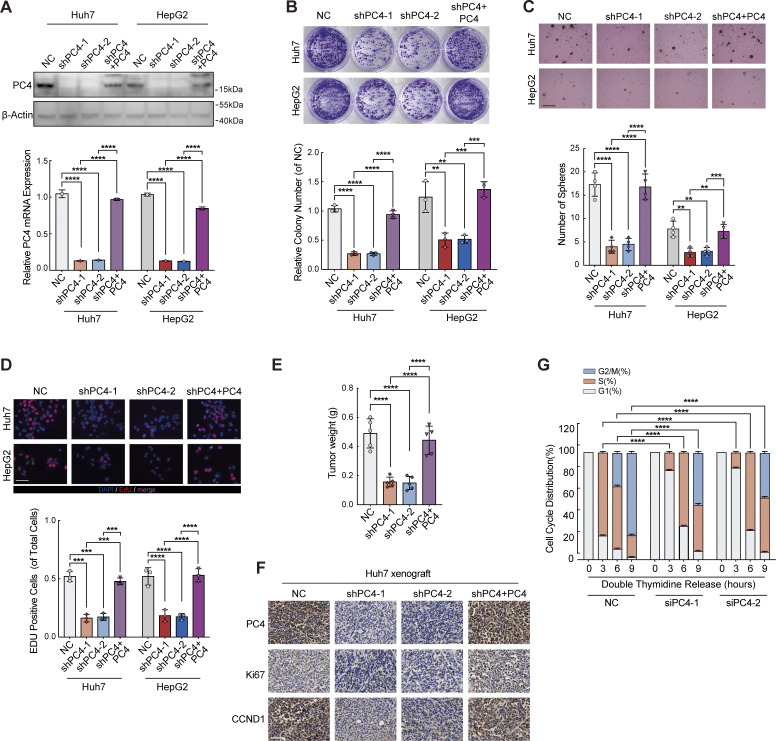
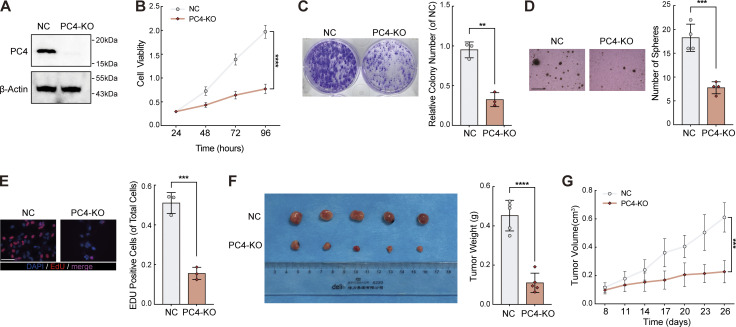
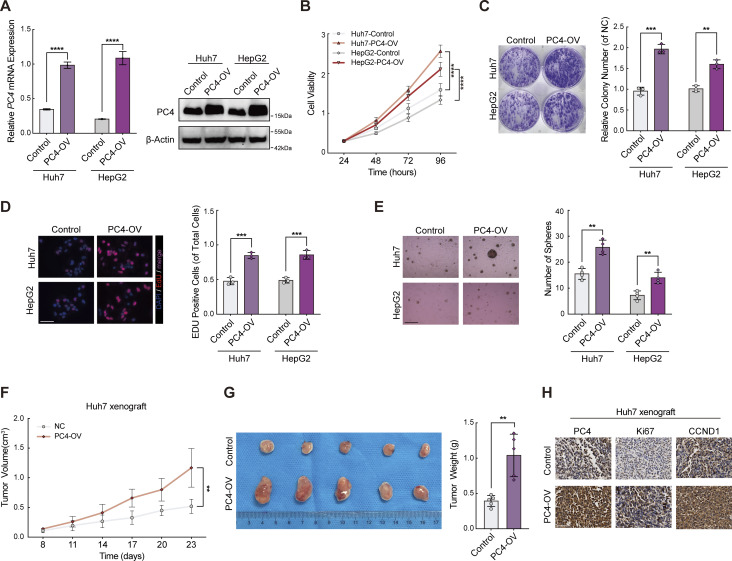
Next, we determined the effect of PC4 on the development of liver cancer. In agreement with the aforementioned results, PC4 knockdown or knockout (KO) significantly impaired the proliferation of Huh7 and HepG2 cells both in vitro and in vivo (Fig. 5, G–I; Fig. S3, A–F; and Fig. S4, A–G). Specificity was further substantiated by the re-expression of PC4 in PC4-silenced HCC cells, which reversed this phenotype. In contrast, PC4 overexpression substantially enhanced HCC cell growth (Fig. S5, A–H). These results highlight that PC4 is a cell cycle regulator that drives the G1–S transition and promotes liver cancer cell proliferation.
Figure S4.
PC4-KO inhibits liver cancer cell growth in vitro and in vivo. Relative to Fig. 5. (A) Western blot showing PC4 protein expression levels in Huh7 cells with control or PC4-KO. Data are generated from n = 3 biological replicates. (B) Cell viability analysis by CCK8 assay in Huh7 cells with control or PC4-KO. Data were generated from n = 5 biological replicates. (C) Colony assay in Huh7 cells with control or PC4-KO. Data are generated from n = 3 biological replicates. (D) Soft agar assay in Huh7 cells with control or PC4-KO. Scale bar = 300 μm. Data are generated from n = 4 biological replicates. (E) EdU assay in Huh7 cells with control or PC4 knockout. scale bar = 75 μm. Data are generated from n = 3 biological replicates. (F) Photographs of dissected tumors and tumor weight in the nude mice subcutaneously injected with Huh7 cell with control or PC4-KO. Data are generated from n = 5 biological replicates. (G) Tumor volume in the nude mice subcutaneously injected with Huh7 cell with control or PC4-KO. Data were generated from n = 5 biological replicates. All quantifications were shown as means ± SD (unpaired Student’s t test); error bars represent SD. **P < 0.01, ***P < 0.001, ****P < 0.0001. A is representative of three independent experiments. Source data are available for this figure: SourceData FS4.
Figure S5.
PC4-OV promotes liver cancer cell growth in vitro and in vivo. Relative to Fig. 5. (A) Western blot and qPCR showing PC4 expression in Huh7 and HepG2 cells stably overexpressing control or PC4. Data are generated from n = 3 biological replicates. (B) Cell viability analysis by CCK8 assay in Huh7 cells and HepG2 cells with control or PC4 stable overexpression. Data are generated from n = 3 biological replicates. (C) Colony assay in Huh7 and HepG2 cells with control or PC4 stable overexpression. Data are generated from n = 3 biological replicates. (D) EdU immunofluorescent staining performed in cells stably expressing control or PC4-OV. Scale bar = 75 μm. Data are generated from n = 3 biological replicates. (E) Soft agar assay in cells with control or PC4 stable overexpression. Scale bar = 300 μm. Data are generated from n = 4 biological replicates. (F) Tumor volume in the nude mice subcutaneously injected with Huh7 cell stably expressing control or PC4. Data are generated from n = 5 biological replicates. (G) Photographs of dissected tumors and tumor weight in the nude mice subcutaneously injected with Huh7 cell stably expressing control or PC4. (H) Representative images of Ki67 (top) and PC4 (bottom) immunostaining in the nude mice subcutaneously injected with Huh7 cell stably expressing control or PC4. Scale bar = 75 μm. Data are generated from n = 5 biological replicates. All quantifications were shown as means ± SD (one-way ANOVA test); error bars represent SD. **P < 0.01, ***P < 0.001, ****P < 0.0001. Source data are available for this figure: SourceData FS5.
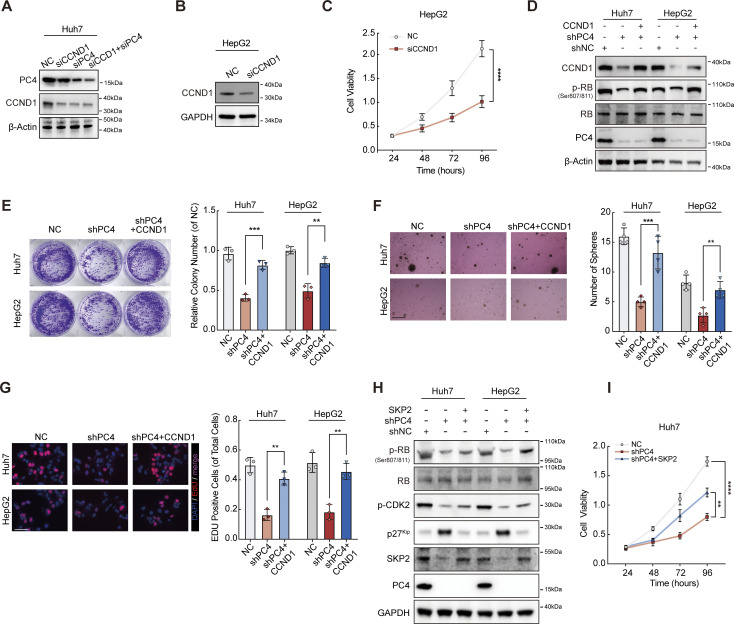
If this hypothesis is true, we expect that CCND1 depletion may phenocopy PC4 depletion. Indeed, CCND1 knockdown in liver cancer cells led to extended duration of the G1 phase and impaired cell growth, though the phenotypic effects were not as pronounced as the significant impact observed upon PC4 knockdown (Fig. 5, J and K; and Fig. S6, A–C). Notably, loss of CCND1 alone or in combination with PC4 suppressed cell proliferation to a similar extent, suggesting that CCND1 and PC4 are involved in the same pathway (Fig. 5, J and K). We also found that complementation of CCND1 partially alleviated the extension of the G1 phase (Fig. 5 M) and the cell growth defects observed in PC4-knockdown cells (Fig. 5 L and Fig. S6, D–G). Thus, PC4 accelerated the G1/S phase transition, which was partially dependent on CCND1. Given the potential role of PC4 in promoting cell cycle progression through the regulation of other critical G1/S regulators like SKP2 (Fig. 1, L and M), we conducted rescue experiments by reintroducing SKP2 into cells that were depleted of PC4. Similar to CCND1, complementing SKP2 partially elevated the expression of p-CDK2 and p-RB (Fig. S6 H) and restored the impaired cell growth observed in PC4-knockdown cells (Fig. S6 I). These results further support our hypothesis that PC4 plays a vital role in cell cycle progression by influencing the expression of factors critical for the G1/S phase transition.
Figure S6.
PC4 promotes G1–S transition and cell proliferation partially through a CCND1-dependent manner. Relative to Fig. 5. (A) Western blot showing PC4 and CCND1 protein expressions in Huh7 cells with control (NC), PC4 knockdown (siPC4), CCND1 knockdown (siCCND1), or combination knockdown of PC4 and CCND1 (siPC4+siCCND1). (B) Western blot showing CCND1 protein expression in HepG2 cells with control (NC) or CCND1 knockdown (siCCND1). (C) Cell viability analysis by CCK8 assay in HepG2 cells with NC or siCCND1. Data are generated from n = 5 biological replicates. (D) Western blot showing indicated protein expressions in Huh7 and HepG2 cells with NC, shPC4, or shPC4+CCND1. (E) Representative data and quantification of colony formation in colony assay in Huh7 and HepG2 cells transfected with NC, shPC4, or shPC4+CCND1. Data are generated from n = 3 biological replicates. (F) Representative data and quantification of soft agar colonies formation in soft agar assay in Huh7 and HepG2 cells transfected with NC, shPC4, or shPC4+CCND1. Scale bar = 300 μm. Data are generated from n = 4 biological replicates. (G) Representative data and quantification of EdU-positive cells in EdU assay in Huh7 and HepG2 cells transfected with NC, shPC4, or shPC4+CCND1. Scale bar = 75 μm. Data are generated from n = 3 biological replicates. (H) Western blot showing indicated protein expressions in Huh7 and HepG2 cells with NC, shPC4, or shPC4+SKP2. (I) Cell viability analysis by CCK8 assay in Huh7 cells with NC, shPC4, and shPC4+SKP2. Data were generated from n = 5 biological replicates. All quantifications were shown as means ± SD (one-way ANOVA test); error bars represent SD. **P < 0.01, ***P < 0.001, ****P < 0.0001. A, B, D, and H are representative of three independent experiments. Source data are available for this figure: SourceData FS6.
Collectively, our results reveal a previously unrecognized role of PC4 in stabilizing the mRNA of cell cycle key regulators, which in turn promotes G1/S transition and accelerates liver cancer cell proliferation. Here, our focus has been on understanding how PC4 specifically regulates the stability of CCND1 mRNA, shedding light on the mechanisms behind this critical step in cell cycle control. Briefly, PC4 physically interacted with the CCND1 transcript, posttranscriptionally enhancing its mRNA stability and eventually increasing its protein abundance (Fig. S3 F and Fig. S5 H). As a result, high expression of CCND1 facilitates cyclin D-Cdk4/6 complex–mediated effects on RB phosphorylation and E2F dissociation, which further promotes E2F-driven G1/S gene expression, leading to a shorter G1 phase duration and faster S phase entry.
Phosphorylation of PC4 at S17 or deubiquitylation at K68 prolongs G1–S transition and suppresses cell proliferation
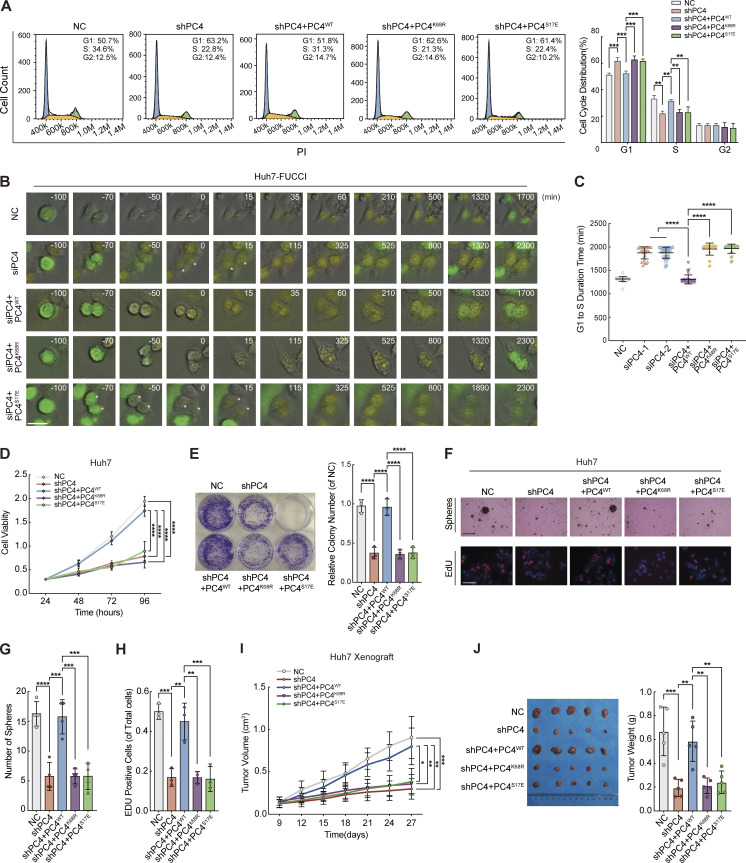
To further verify whether PC4 uses these two modifications to manipulate the cell division cycle, we performed several rescue experiments by knocking down endogenous PC4 and replacing it with PC4WT or one of the PC4–RNA binding-deficient mutants, PC4K68R or PC4S17E. PC4 depletion–mediated G1 phase lengthening was significantly overcome by PC4WT but not by the two mutants (Fig. 6, A–C). Similarly, the two mutants did not induce cell proliferation (Fig. 6, D–H), indicating that these two modifications orchestrate S phase entry and cell growth. Further investigation using a xenograft mouse model showed that cell growth was significantly reduced by S17 phosphorylation of PC4 or deubiquitination of K68 (Fig. 6, I and J). These data indicate that S17 dephosphorylation and K68 ubiquitination are required for PC4–RNA binding, which allows PC4 to modulate the stability and expression of CCND1 mRNA, thereby maintaining cell cycle progression and proliferation.
Figure 6.
Phosphorylation of PC4 at S17 or deubiquitylation at K68 prolongs the G1–S transition and suppresses cell proliferation. (A) Asynchronized cell cycle analysis in PC4-knockdown Huh7 cells stably expressing indicated Flag-tagged PC4 variants. The number in the upper right corner represents the percentage of cells in different cell cycle phases. Data are generated from n = 3 biological replicates. (B) Representative frames of FUCCI-expressing Huh7 transfected with siRNA and PC4-mutant plasmid via time-lapse imaging. Scale bar = 50 μm. (C) The time length of the G1 phase in FUCCI-expressing Huh7 transfected with siPC4 and PC4-mutant plasmid via time-lapse imaging. Data are generated from n = 30 cells in each group. (D) Cell viability analysis by CCK8 assay in PC4-knockdown Huh7 cells transfected with indicated Flag-tagged PC4 variants. Data are generated from n = 5 biological replicates. (E) Representative images and quantification of colony formation in colony assays in PC4-knockdown Huh7 cells transfected with indicated Flag-tagged PC4 variants. Data are generated from n = 3 biological replicates. (F) Representative images of soft agar colony formation in soft agar assays and EdU-positive cells in EdU assays in PC4-knockdown Huh7 cells transfected with indicated Flag-tagged PC4 variants. Soft agar scale bar = 300 μm, EdU scale bar = 75 μm. (G) Quantification of soft agar colony formation in soft agar assays in PC4-knockdown Huh7 cells transfected with indicated Flag-tagged PC4 variants. Data are generated from n = 4 biological replicates. (H) Quantification of EdU-positive cells in EdU assays in PC4-knockdown Huh7 cells transfected with indicated Flag-tagged PC4 variants. Data are generated from n = 3 biological replicates. (I) Tumor volume in the nude mice subcutaneously injected with PC4-knockdown Huh7 cells stably expressing indicated Flag-tagged PC4 mutants. Data are generated from n = 3 biological replicates. (J) Representative photographs of dissected tumors and tumor weight in the nude mice subcutaneously injected with PC4-knockdown Huh7 cell stably expressing indicated Flag-tagged PC4 mutants. Data are generated from n = 5 biological replicates. All quantifications are shown as means ± SD (one-way ANOVA test); error bars represent SD. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Expression of phosphorylated PC4 S17 and ubiquitinated K68 in HCC
Considering the important role of PC4 in the cell cycle, we propose that PC4 has a potential clinical significance in HCC. Our preliminary data showed that PC4 was overexpressed in HCC tissues and was an unfavorable factor for patient survival (Pan et al., 2023). Next, we evaluated the levels of PC4 S17 phosphorylation and K68 ubiquitination in HCC cell lines and cancer tissues. Interestingly, S17 phosphorylation was much lower in cancer cells and tissues than in normal controls. In contrast, K68 ubiquitination was higher in HCC samples (Fig. 7 A and Fig. S7, A and B). These results demonstrated that the switch is “on,” so PC4 acts as an RBP to posttranscriptionally regulate cell cycle gene expression in HCC. We found that the interaction between CK2 and PC4 was reduced, whereas the interaction between TRIM28 and PC4 was enhanced in HCC tissues (Fig. 7 A). These observations suggest that S17 phosphorylation mediated by CK2 and K68 ubiquitination induced by TRIM28 are clinically relevant processes in patients with HCC and indicate that modified PC4 could be an effective biomarker and therapeutic target for liver cancer.
Figure 7.
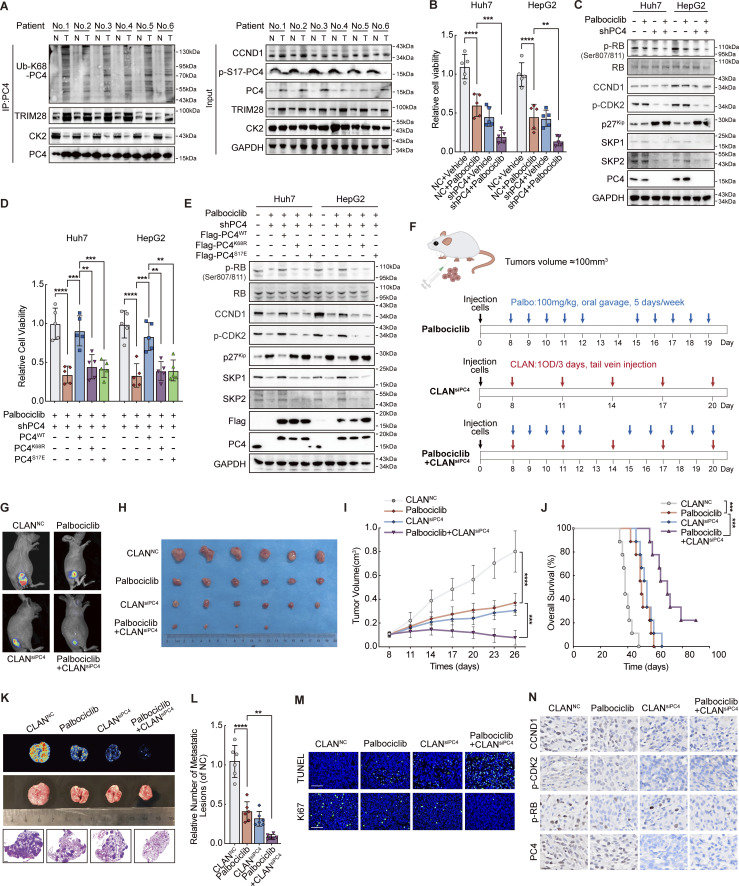
PC4 exhibits distinct modifications in HCC, and PC4 loss sensitizes HCC cells to CDK4/6 inhibitors. (A) Western blot showing the indicated protein expressions between six paired human HCC and adjacent tissue samples. N, normal. T, tumor. (B) Cell viability analysis by CCK8 assay in Huh7 and HepG2 cells treated with Palbociclib, siPC4, or a combination of Palbociclib and siPC4. Data are generated from n = 5 biological replicates. (C) Western blot shows the indicated protein expressions in Huh7 and HepG2 cells with Palbociclib, siPC4, or a combination of Palbociclib and siPC4. (D) Cell viability analysis by CCK8 assay in PC4-knockdown Huh7 and HepG2 cells treated with a combination of Palbociclib and PC4WT, PC4K68R, or PC4S17E. Data are generated from n = 5 biological replicates. (E) Western blot showing the indicated protein expressions in PC4-knockdown Huh7 and HepG2 cells treated with a combination of Palbociclib and PC4WT, PC4K68R, or PC4S17E. (F) Schema for Palbociclib, CLANsiPC4, or combination of Palbociclib and CLANsiPC4-treated Huh7-Luc xenografts model. (G) Bioluminescence from Huh7-Luc xenografts mice treated in F. (H) Photograph of tumors from Huh7-Luc xenografts mice treated in F. (I) Quantification of the tumor volume in Huh7-Luc xenografts mice treated in F. Data were generated from n = 6 in each group. (J) Overall survival from Huh7-Luc xenografts mice treated in F. Data are generated from n = 9 in each group. (K) Bioluminescence (top), photographs (middle), and H&E staining (bottom) of lungs from CLANNC, Palbociclib, CLANsiPC4, or Palbociclib+CLANsiPC4-treated Huh7-Luc xenografts mice. Scale bar = 1 mm. (L) Quantification of metastatic lesions to lung in K. Data are generated from n = 6 in each group. (M) Tunnel and Ki67 staining from Huh7-Luc xenografts mice treated in F. Data are generated from n = 6 in each group. Scale bar = 75 μm. (N) Representative images of CCND1, p-CDK2, p-RB, and CCND1 immunostaining from Huh7-Luc xenografts mice treated in F. Scale bar = 20 μm. Data are generated from n = 6 in each group. All quantifications are shown as means ± SD (one-way ANOVA test); error bars represent SD. **P < 0.01, ***P < 0.001, ****P < 0.0001. A, C, and E are representative of three independent experiments. Source data are available for this figure: SourceData F7.
Figure S7.
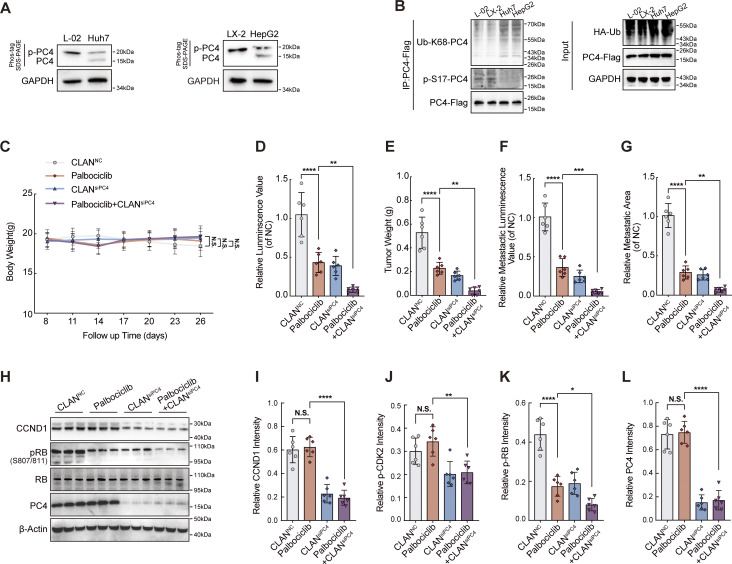
PC4 exhibits distinct modifications in HCC, and PC4 loss sensitizes HCC cells to CDK4/6 inhibitors. Related to Fig. 7. (A) Western blot showing the phosphorylated and unphosphorylated PC4 expressions in four liver cells (Huh7, HepG2, L-02, and LX-2). (B) Western blot showing ubiquitination and phosphorylation levels of endogenous PC4 in four liver cells (Huh7, HepG2, L-02, and LX-2). (C) Body weight from CLANNC(n = 6), Palbociclib (n = 6), CLANsiPC4(n = 6), or Palbociclib+CLANsiPC4 (n = 6) -treated Huh7-Luc xenografts mice at each time point in Fig. 7 F. (D) Luminescence value from Huh7-Luc xenografts mice treated in Fig. 7 G. Data were generated from n = 6 in each group. (E) Quantification of tumor weight from Huh7-Luc xenografts mice treated in Fig. 7 F. Data were generated from n = 6 in each group. (F) Quantification of metastatic luminescence value in Fig. 7 L. Data are generated from n = 6 in each group. (G) Quantification of metastatic area in Fig. 7 L. Data are generated from n = 6 in each group. (H) Western blot showing the indicated protein expressions in CLANNC (n = 3), Palbociclib (n = 3), CLANsiPC4 (n = 3), and Palbociclib+CLANsiPC4 (n = 3) -treated Huh7-Luc xenografts. (I) Quantifications of CCND1 intensity in immunostaining from Huh7-Luc xenografts mice treated in Fig. 7 F. (J) Quantifications of p-CDK2 intensity in immunostaining from Huh7-Luc xenografts mice treated in Fig. 7 F. (K) Quantifications of p-RB intensity in immunostaining from Huh7-Luc xenografts mice treated in Fig. 7 F. (L) Quantifications of PC4 intensity in immunostaining from Huh7-Luc xenografts mice treated in Fig. 7 F. All quantifications were shown as means ± SD (one-way ANOVA test); error bars represent SD. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. A, B, H, and J–L are independent experiments. Source data are available for this figure: SourceData FS7.
PC4 loss sensitizes HCC cells to CDK4/6 inhibitors
Recent clinical studies have revealed the success of CDK4/6 inhibitors in cancer therapy, and Palbociclib, a well-tolerated and selective CDK4/6 inhibitor, has been shown to have effective prolonged progression-free survival (Bollard et al., 2017). Since PC4 depletion causes cell cycle arrest, we sought to explore whether PC4 deficiency sensitizes liver cancer cells to CDK4/6 inhibitors. Consistent with our hypothesis, both Huh7 and HepG2 cells with PC4 knockdown were more sensitive to Palbociclib treatment (Fig. 7 B). Next, we assessed whether blocking the PC4–RNA binding capacity could facilitate HCC cell sensitivity to Palbociclib. As expected, while re-expressing PC4WT reduced the sensitivity of PC4-knockdown cells to Palbociclib, PC4K68R or PC4S17E restored this sensitivity (Fig. 7 D). In addition, depleting PC4 or inhibiting its RNA binding capacity resulted in a reduction in protein levels of CCND1 and SKP2 (Fig. 7, C and E). This prevented RB hyperphosphorylation-mediated E2F activation and subsequent gene expressions related to the S phase entry.
To further evaluate whether PC4 acts as a regulator of the response to CDK4/6 inhibitors in vivo, we created a cationic lipid-assisted PEG-b-PLA nanoparticle (CLAN) that encapsulated PC4 siRNAs inside the aqueous core (Xu et al., 2019). Nude mice harboring Huh7-Luc xenografts were treated with the CDK4/6 inhibitor Palbociclib, CLANsiPC4, or a combination of Palbociclib and CLANsiPC4 once the tumor reached a volume of 100 mm3 (Fig. 7 F). All treatments were well tolerated without obvious side effects or weight loss (Fig. S7 C). Treatment with Palbociclib or CLANsiPC4 showed a remarkable therapeutic effect (Fig. 7 G), which is consistent with previous observations. Surprisingly, in comparison with either treatment alone, the combination of Palbociclib with CLANsiPC4 further reduced tumor burden, improved overall survival, and suppressed lung metastasis (Fig. 7, H–L; and Fig. S7, D–G), suggesting increased sensitivity to Palbociclib upon PC4 knockdown. This enhanced therapeutic efficacy of the combination treatment was mainly due to the suppression of cell proliferation and promotion of cell apoptosis, as indicated by the results of Ki67 and TUNEL staining (Fig. 7 M). Moreover, PC4 knockdown combined with Palbociclib treatment markedly reduced the expressions of CCND1 and SKP2 and the phosphorylation of CDK2 and RB (Fig. 7 N and Fig. S7, H–L). These observations suggest that, in contrast to Palbociclib treatment, which blocked the activity of cyclin-dependent kinases, PC4 knockdown limits cell cycle progression by downregulating the protein expressions of G1/S key regulators, which further prevents excessive phosphorylation of RB. Thus, PC4 deficiency and CDK4/6 inhibition cooperatively induce cell cycle dysregulation, leading to a vulnerability that can potentially be exploited in liver cancer therapy.
Taken together, our results show that PC4 participates in cell cycle regulation, in part, by affecting the mRNA stability of several cell cycle regulators, particularly CCND1, which plays a critical role in cell cycle progression and cell proliferation. More importantly, TRIM28-directed K68 polyubiquitination during interphase and CK2-mediated S17 phosphorylation in the M phase were defined as critical PTMs that regulate the biological functions of PC4 and fluctuations in CCND1 mRNA stability in the cell cycle (Fig. 8). In addition, PC4 depletion exacerbated the inhibitory effects of CDK4/6 inhibitors on HCC growth.
Figure 8.
PC4 modifications regulate the dynamic of CCND1 mRNA stability across the cell cycle. Model summarizing: PC4 plays a posttranscriptional regulatory role in controlling the stability of CCND1 mRNA during the cell cycle. The modifications of PC4 undergo periodic changes and they regulate the fluctuations in CCND1 mRNA stability. In interphase, PC4 interacts with TRIM28 and is ubiquitinated (Ub) at residue K68 by TRIM28. This ubiquitination enhances PC4’s binding affinity with CCND1 mRNA, leading to increased stability and protein expression of CCND1. This promotes the transition from G1 to S phase. On the other hand, during mitosis, PC4 is phosphorylated at residue S17 by CK2. This phosphorylation inhibits PC4’s interaction with TRIM28, preventing K68 ubiquitination and impairing PC4’s binding to CCND1 mRNA. Consequently, the degradation of CCND1 mRNA increases.
Discussion
One of the molecular characteristics of HCC is the transcriptomic imbalance that drives rapid cell cycle progression and uncontrolled cell proliferation (Lee et al., 2004; Boyault et al., 2007; Calderaro et al., 2019; Rebouissou and Nault, 2020). This imbalanced transcriptional program generally requires (1) a DNA binding, TF-driven, cell cycle–related transcriptional profile to globally increase pre-RNA synthesis and (2) RBPs to modulate the posttranscriptional profile to regulate cell cycle–related RNA metabolism and protein production. Although recent studies have mainly focused on how TFs support high oncogenic growth rates (Park et al., 2022; Wang et al., 2022), the mechanism by which RBPs reset the posttranscriptional program that drives rapid mitotic division remains poorly understood. RBPs may induce a significant transcriptomic imbalance and accelerated cell cycle progression in HCC because they are aberrantly expressed and capable of quickly recognizing and determining the fates of a wide range of RNA transcripts (Gebauer et al., 2021). In this study, we analyzed the differentially expressed RBPs associated with a poor prognosis in HCC and identified PC4 as an oncogenic RBP in HCC. This is the first report showing that PC4, a previously identified DNA binding protein (Ge and Roeder, 1994; Kretzschmar et al., 1994; Das et al., 2006; Mortusewicz et al., 2016), is also a novel RBP that drives the G1–S transition by stabilizing cell cycle–related mRNAs, increasing their gene expressions, and subsequently inducing HCC cell division. Thus, we defined PC4 as a cell cycle–regulated gene.
CCND1 is one of the central regulators of the cell division cycle (Sherr, 1995; Morgan, 1997), and its expression level exhibits periodic changes during the cell cycle (Baldin et al., 1993), which is probably due to the ubiquitin proteasome–mediated pathway (Lin et al., 2006; Fasanaro et al., 2010; Chaikovsky et al., 2021) and transcriptional networks (Lim and Kaldis, 2013). Nevertheless, it is worth investigating whether CCND1 mRNA is regulated at the posttranscriptional level across the cell cycle. Here, we found that the stability of CCND1 mRNA fluctuated during the cell cycle and underwent a downshift from the G1 phase to the M phase. Interestingly, PC4 stabilizes CCND1 mRNA during interphase but destabilizes it in the M phase. The dynamic fluctuation in CCND1 mRNA stability during the cell cycle serves a crucial purpose in ensuring accurate cell cycle progression. One possible explanation is that, during transitions between cell cycle phases, it becomes imperative to eliminate components from the previous phase prior to entering the next phase. Failure to do so can disrupt the orderly progression of the cell cycle. Therefore, posttranscriptional regulation by PC4, coupled with proteasome-mediated degradation, finely tunes CCND1 expression and precisely drives cell cycle progression. In HCC, the expression of CCND1 was commonly increased, partially because its mRNA stability is increased by PC4 throughout the cell cycle; thus, PC4 inhibition could be a highly effective strategy for disrupting the stabilization of CCND1 mRNA, decreasing its expression during the cell cycle, and limiting HCC proliferation.
Although the mRNA and protein levels of PC4 did not show periodic changes, we surprisingly found two cell cycle–specific PTMs on PC4, namely, K68 ubiquitination and S17 phosphorylation, which affected the binding of PC4 to CCND1 mRNA and the cell cycle outcome. Furthermore, CK2 is the key kinase that catalyzes S17 phosphorylation, and TRIM28 appears to be the main ubiquitin E3 ligase for K68 ubiquitination. During interphase, TRIM28-mediated K68 ubiquitination of PC4 enhances the binding and stabilization of CCND1 mRNA. In the M phase, CK2-catalyzed S17 phosphorylation interferes with the interaction of PC4 with TRIM28, which blocks downstream K68 ubiquitination and disassociates PC4 from CCND1 mRNA, eventually leading to CCND1 degradation (Fig. 8). We speculated that S17 phosphorylation confers a negative charge on the SEAC region, which may lead to a conformational change in PC4 that inhibits its interaction with TRIM28. Further investigations are required to clarify how S17 phosphorylation interrupts the interaction between PC4 and TRIM28. Evidence from a series of studies has indicated that the two PTMs of PC4 are strictly regulated during the cell cycle, which in turn controls the biological functions of PC4 in posttranscriptionally regulating cell cycle–related gene expression and manipulating cell cycle progression. Another topic of interest is the identity of the specific protein phosphatase responsible for removing the PC4 phosphorylation mark at S17 at the entry into the G1 phase. More research is warranted to fully understand the regulatory networks that control cell cycle progression and posttranscriptional and posttranslational regulation of the cell cycle transcriptome.
CDK4/6 inhibitors are under investigation for the treatment of HCC (Bollard et al., 2017). Currently, studies are underway to identify genes whose loss may increase the efficacy of CDK4/6 inhibitors in HCC. In this study, we found that PC4 deficiency increased the sensitivity to the CDK4/6 inhibitor Palbociclib. This may be due to the fact that targeting PC4 can reduce the expressions of CCND1 and SKP2. Decreased protein level of CCND1 can impede the phosphorylation of RB by reducing the expression of the cyclin D-Cdk4/6 complex. Additionally, reduced SKP2 expression can inhibit the degradation of p27, resulting in suppressed CDK2 activation and preventing excessive phosphorylation of RB. Therefore, combining PC4 inhibition with CDK4/6 inactivation has the potential to induce synthetic lethality in HCC.
In summary, our results revealed that posttranslationally modified PC4, as a master regulator of RNA metabolism, controls the periodic changes in CCND1 mRNA stability and drives rapid cell cycle progression to promote HCC cell proliferation. These findings highlight the importance of RBP in driving cell cycle progression, and the selective targeting of functional RBP represents a promising strategy for liver cancer therapy.
Materials and methods
Mice
All animal experiments complied with ethical policies regarding animal research and were approved by the Animal Care and Use Committee Guidelines of the Third Military Medical University.
For the in vivo tumor growth model, 6–8-wk-old male BALB/c nude mice (Cavens Biogle) were fed in a specific pathogen–free room and inoculated subcutaneously with each group of Huh7 cells (5 × 106 in 200 μl of phosphate-buffered saline [PBS]) at a dorsal site. Tumor size was monitored using calipers every 3 days, and tumor volumes were calculated as length × (width)2/2. At the endpoint, the mice were sacrificed and xenografts from the animals were isolated and weighed. Subsequently, the tumors and organs were dissected, fixed with 4% paraformaldehyde for immunohistochemistry/immunofluorescence analysis, and frozen in liquid nitrogen for RNA and protein analyses.
To investigate drug efficacy, Huh7-Luc cells were used to establish in vivo models. CLANsiPC4 was generated by a CLAN that encapsulated PC4 siRNAs inside the aqueous core (Xu et al., 2019). After inoculation with Huh7-Luc cells, mice were weighed and tumor size was monitored by calipers every 3 days. Once the tumor reached a volume of 100 mm3, the mice were randomized and treated with vehicle (NC), Palbociclib (100 mg/kg, 5 days/wk), CLANsiPC4 (1 OD/3 d), or a combination of Palbociclib and CLANsiPC4. The mice were then injected with DTZ-Luc1 (Meisen CTCC) and evaluated weekly using bioluminescence imaging to quantify the tumor burden. The endpoint of the survival study was defined as mouse death or a tumor volume of 2,000 mm3. Once the mice were euthanized, the organ tissues were fixed in 4% paraformaldehyde for further analysis.
Plasmids
Vectors used in the study and their source are listed in Data S3. PC4-OV lentiviral plasmid was generated by cloning corresponding coding sequences of PC4 into a Ubi-MCS-3FLAG-SV40-EGFP-IRES-puromycin vector. For PC4 point mutation plasmids, the sequences of Flag-tagged PC4 point mutants (WT, S17A, S17E, S19A, S17/19A, and K68R) were generated and cloned into pLenti-Blast (CMV-EF-1a-MCS-3XFLAG-Blasticidin-SV40-BleoR) vectors. For PC4 domain deletion plasmids, the sequences of Flag-tagged PC4 domain deletions were generated and cloned into pLenti-Puro (CMV-EF-1a-MCS-3FLAG-Puromycin-SV40-BleoR). For GST-PC4 point mutation plasmids, the sequences of GST-tagged PC4 point mutants (WT, S17A, S17E, and K68R) were generated and cloned into pgex-6p-1 vectors.
The CCND1-expressing plasmid was constructed by cloning the CCND1 ORF into the CMV-MCS-EGFP-SV40-neomycin vector. The plasmid expressing CK2 was constructed by cloning the CK2 ORF into the pcDNA3.1 vector. The plasmid expressing MYC-tagged TRIM28 was generated by cloning the MYC-tagged TRIM28 cDNAs into the pcDNA3.1 vector. All vectors constructed in this study were verified using Sanger sequencing before use.
The following plasmids were purchased from Addgene: HA-Ubiquitin-WT (17608; gift from Ted Dawson), HA-Ubiquitin-K48 (17605), HA-Ubiquitin-K63 (17606; gifts from Ted Dawson), FU-H2B-GFP-IRES-Puro (69550; gift from Charles Gersbach), and FUCCI-reporter (pLL3.7m-Clover-Geminin-IRES-mKO2-Cdt; 83841; gift from Michael Lin). The pLV2-EF1a-SKP2(human)-3×Myc-Puro plasmid was purchased from MIAOLING BIOLOGY (P48609).
Cells
The sources of each cell line are listed in Data S3. All cells were cultured in Dulbecco’s modified Eagle medium (Gibco) supplemented with 10% fetal bovine serum (Biological Industries) and 1% penicillin/streptomycin (Beyotime) and maintained at 37°C in a 5% CO2 atmosphere and routinely checked for mycoplasma contamination.
Huh7-PC4-KO cells were generated using CRISPR-Cas9. Briefly, gene-specific sgRNAs against PC4 were cloned into the pLentiCRISPRv2 lentiviral vector (52961; Addgene, gift from Feng Zhang). The lentiviral vector was cotransfected with the lentiviral packaging plasmids psPAX2 (12260; Addgene) and pMD2.G (12259; Addgene; gifts from Didier Trono) at a ratio of 6:4:1.5. This DNA mixture was then diluted in 600 μl of OptiMEM (Sigma-Aldrich). In addition, 1 μg/μl poly-ethylenimine dissolved into 1.2 ml of OptiMEM was combined with the DNA mixture and transfected into HEK293T cells. After 8 h of incubation, fresh culture medium was replaced. The viral particles were collected 48 h after the medium was refreshed. Eventually, the collected viral liquid was centrifuged at 500 × g for 10 min followed by filtration with a 0.45-μm polyvinylidene difluoride (PVDF) syringe filter. For transduction, Huh7 cells were plated in 6-well plates at 360,000 cells per well and transfected with 500 μl of lentivirus supplemented with 10 μg/ml polybrene (Sigma-Aldrich). After 2 days, positive selection of transfected cells was performed with 1.4 μg/ml puromycin. The cells were then diluted and individually isolated in 96-well plates. Monocolonies were selected and grown for several weeks. PC4-KO efficiency was assessed by Sanger sequencing and protein disruption was evaluated by western blotting. The guide RNA sequences used are listed in Data S3.
Stable PC4-knockdown cells were generated using shPC4 virus. First, shRNA targeting PC4 was cloned into the hU6-MCS-CBh-gcGFP-IRES-puromycin vector. The lentiviral vector was packaged in the psPAX2 and pMD2.G plasmids and incubated with HEK293T cells for 48 h. Viral particles were collected, filtered, and transfected into the expected cell lines to knock down PC4. The cells were selected and maintained in the presence of puromycin. PC4-KD efficiency was evaluated by western blotting. The shRNAs used in this study are listed in Data S3.
Cells stably overexpressing PC4 were generated using the PC4-OV virus. The PC4-OV lentiviral vector was cotransduced into HEK293T cells using the lentiviral packaging plasmids psPAX2 and PMD2.G. The viral particles were produced, collected, and filtered. Cells were transduced and stable polyclonal populations of cells were selected using puromycin.
RNA interference
To transiently and specifically knock down a gene, cells were transfected with the indicated siRNA using Lipofectamine RNAiMAX or 3000 (Thermo Fisher Scientific) according to the manufacturer’s instructions. The knockdown efficiency of the target gene was determined by western blotting. The sequence of siRNAs used is listed in Data S3.
Colony formation
Approximately 1,000 Huh7 or HepG2 cells transfected with the indicated shRNAs or vectors were seeded in triplicate in 6-well plates. Colonies were formed after 3–4 wk of culture. The cells were fixed in 4% paraformaldehyde for 5 min at room temperature, stained with 0.5% crystal violet for 15 min, and washed with PBS. Images were automatically captured and colonies were counted.
Cell proliferation assay
A total of 2,000 Huh7 or HepG2 cells transfected with the indicated shRNAs or vectors were seeded into 96-well plates and incubated for 4 days. Subsequently, at every 24 h, cells were washed with PBS and incubated in the dark with CCK-8 (WST-8, [2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium, monosodium salt]) (10 μl per well; Dojindo) for 2 h. Subsequently, cell viability was evaluated by measuring the absorbance of the dye at 450 nm.
EdU assay
A total of 5,000 Huh7 or HepG2 cells transfected with the indicated shRNAs or vectors were seeded into 12-well plates and grown for a couple of days. A Click-iT EdU Imaging Kit (900584; Sigma-Aldrich) was used to label proliferating cells, in accordance with the manufacturer’s instructions. Images were acquired using the EVOS M5000 imaging system (Invitrogen). EdU- and 4′,6-diamidino-2-phenylindole (DAPI)–stained cells were counted manually. The growth rate was calculated as the percentage of EdU-positive cells among the total number of cells.
Cell synchronization
For G1 phase arrest, the cells were subjected to double-thymidine (2 mM) treatment. Briefly, cells were treated for 16–18 h (Huh7, 16 h; LX-2, 18 h) with thymidine (T3763; Sigma-Aldrich) arrest and released for 8 h, which was followed by 16–18 h (Huh7,16 h; LX-2,18 h) of thymidine arrest; subsequently, the cells were harvested. At this point, the cells were synchronized in the G1 phase.
Cell cycle analysis
For cell cycle analysis, the cells were trypsinized and washed twice with PBS before being fixed in 70% ethanol overnight. The next day, cells were washed twice with PBS and resuspended in a PBS solution containing propidium iodide/RNaseA (10 μg/ml and 0.1 mg/ml, respectively) and incubated for 15 min in the dark. Samples were tested using an Attune NxT flow cytometer (Life Technologies) and the data were analyzed using FlowJo software.
Live cell microscopy
For live imaging of the G1/S transition progression, fluorescent ubiquitination-based cell cycle indicator (FUCCI)–report fluorescent cells were seeded in a 96-well plate in an environmental control system at 27°C. The imaging was performed using an MD Image Xpress Micro Confocal microscope equipped with an sCMOS sensor, a 100× objective, and >3 log dynamic range intensity detection. The images were acquired every 20 min for a total duration of 48 h using a two-color epifluorescence imaging protocol. Images were acquired and analyzed with MetaXpress PowerCore software. The duration of the G1 phase was considered as the time in which the green fluorescence completely changed to yellow.
Immunohistochemistry
For tissue immunohistochemistry staining, tissue sections were dewaxed, rehydrated, antigen retrieved, and incubated with normal goat serum for 1 h at room temperature to block nonspecific binding, followed by incubation with human PC4 antibody (Rabbit Polyclonal to PC4, 1:200, HPA001311; Sigma-Aldrich) at 4°C overnight. Subsequently, the slides were incubated with a biotinylated secondary antibody (ab207995; Abcam) at 37°C for 30 min, and positive staining was visualized by using 3,3′-diaminobenzidine.
RNA-seq
Total RNA was extracted using TRIzol Reagent (15596018; Invitrogen) according to the manufacturer’s instructions. Next, 500 ng of total RNA was obtained to generate sequencing libraries using the TruSeq RNA Sample Prekit V2 (Illumina). Next, the libraries were loaded onto an Illumina HiSeq 2500 for 2 × 100 bp paired-end read sequencing. Fastq files were produced and the quality of the reads was assessed using FastQC software. After alignment of the reads to the human genome (ver. hg19) using the Bowite-Tophat2 algorithm, BAM files were generated and quantified for differential transcript analysis using Cyfflinks. The differentially expressed transcripts were defined as a statistical cut-off of P < 0.001 and fold change (FC) (log2) > 1 and < −1. The complete differentially expressed transcripts are listed in Data S1.
qPCR
Total RNA was isolated using the QIAGEN RNAeasy kit. Subsequently, RNA reverse transcription and cDNA synthesis were performed using a PrimeScript RT reagent kit (Takara). qPCR was performed using TB Green Fast qPCR Mix (Takara) and the primers listed in Data S3 on a StepOnePlus thermocycler (Applied Biosystem). The FC in gene expression was calculated by the 2−ΔΔCt method and normalized to levels of the housekeeping gene glyceraldehyde 3-phosphate dehydrogenase.
Chromatin immunoprecipitation (ChIP)
Cells were crosslinked at room temperature with formaldehyde (1% final concentration) for 15 min and quenched with 0.125 M glycine for 10 min. Then, cells were washed with PBS twice, lysed with 200 μl of cell lysis buffer (50 mM Tris-Cl, pH 8.0, 10 mM EDTA, 1% sodium dodecyl sulfate [SDS]) with protease inhibitor, and incubated for 10 min at 4°C. The lysed chromatin was sonicated for 30 s with 30-s intervals of rest for 30 cycles, yielding DNA fragments of 150–250 bp. After centrifugation at 14,000 rpm for 10 min, the supernatants were precipitated using 5 μg of anti-PC4 antibody overnight at 4°C with rotation. The next day, 20 μl of protein A/G magnetic beads (88802; Thermo Fisher Scientific) were incubated with the complex for 1 h and washed once with IP Wash I buffer, twice with high salt buffer, once with IP Wash II buffer, and finally washed twice with Tris-EDTA buffer. The washed immunocomplexes were eluted with 200 μl of elution buffer (1% SDS, 0.1 M NaHCO3) at 45°C for 30 min and reverse-crosslinked overnight in a 65°C water bath. The next day, the eluted DNA was incubated with RNase A (1 mg/ml) for 2 h at 37°C and treated with proteinase K (20 mg/ml; 25530049; Invitrogen) for 2 h at 55°C. The eluted DNA was purified using a QIAquick gel extraction kit in 55 ml of EB elution buffer.
ChIP-seq libraries were generated using the TruSeq ChIP Sample Prep Kit (Illumina) according to the manufacturer’s instructions and sequenced using an Illumina Novaseq platform with paired-end reads of 150 bases.
RIP
The cells were crosslinked with 0.3% formaldehyde and incubated with 0.125 M glycine. The cells were collected and incubated in 200 μl of cell lysis buffer for 30 min on ice. Then, the RNA–protein complexes were enriched using 5 μg of anti-PC4 or anti-Flag antibodies conjugated with protein A/G beads at 4°C rotation overnight. The RNA immunocomplex was washed and incubated with proteinase K to release RNAs from PC4–RNA immunocomplexes. Subsequently, the RNA was isolated and extracted using the phenol-chloroform method.
For RIP-seq, RIP-seq libraries were generated using TruSeq Stranded mRNA (Illumina) according to the manufacturer’s instructions and sequenced using an Illumina HiSeq 2500 platform with paired-end reads of 75 bases. For RIP-qPCR, the primers used to validate target gene expression are listed in Data S3.
eCLIP
The eCLIP experiments were performed as described previously (Van Nostrand et al., 2016). Briefly, cells were UV-crosslinked at a constant energy of 400 mJ/cm2, followed by lysis in eCLIP lysis buffer on ice and sonication by BioRuptor. To fragment RNA, cell lysates were treated with RNase I (AM2295; Thermo Fisher Scientific). Next, 2% of the lysate was allocated as input, whereas the rest was used as the IP sample. IP samples were immunoprecipitated with anti-PC4 antibody conjugated with Dynabeads protein A/G (10004D; Thermo Fisher Scientific) at 4°C rotation overnight and stringently washed. Subsequently, the IP samples were dephosphorylated with FastAP (EF0652; Thermo Fisher Scientific) and T4 PNK (M021L; NEB), followed by on-bead ligation of barcoded RNA adapters to the 30 ends (M0437M; T4 RNA Ligase High Conc; NEB). Both IP and input samples were run on NuPAGE Bis-Tris protein gels (NP0321BOX; Invitrogen) and transferred to nitrocellulose membranes, where the RNA in the 15–90-kD region was cut off on the membrane. To digest the proteins, proteinase K (P1807S; NEB) was added to the membrane until the RNA was released. RNA was then reverse-transcribed using Superscript III (600107; Agilent), followed by treatment with ExoSAP-IT (78201.1.ML; Thermo Fisher Scientific) to remove excess oligonucleotides. The samples were cleaned with Dynabeads MyOne Silane (37002D; Thermo Fisher Scientific) and subjected to qPCR to determine the appropriate number of PCR cycles. Libraries were generated using the Q5 PCR Master Mix (M0492L; NEB) and sequenced using the Illumina NextSeq 550 platform with paired-end reads of 100 bases. FASTQ files were processed using the eCLIP pipeline version 0.2.1a (https://github.com/YeoLab/eclip/releases/tag/0.2.1a). Briefly, the adapter and adapter dimers were trimmed. PCR duplicates were removed and reads were mapped to the GRCh37 human reference genome. Peaks were called, annotated by gene, and visualized using the IGV software.
RNA pull-down assay
The RNA pull-down assay was performed using an RNA pull-down kit (Bersin Bio) according to the manufacturer’s instructions. The biotin-labeled probe sequences are listed in Data S3. In brief, template PCR fragments containing the T7 RNA polymerase promoter and the region of interest were purified for in vitro transcription. Subsequently, biotinylated RNA transcripts were generated using T7 transcription buffer, rNTPs, RNasin, Biotin-14-CTP, a T7 DNA template, and T7 RNA polymerase (BersinBio RNA pulldown kit). The reaction mixture was incubated at 37°C for 2 h, and RNA was purified using RNA easy spin columns (74104; Qiagen). Next, DNAse (79254; Qiagen) treatment was performed to remove the template DNA. To form RNA secondary structures, biotinylated RNA was denatured at 90°C for 2 min and incubated with precooled RNA structure buffer. Next, streptavidin magnetic beads were incubated with the RNAs at 25°C for 30 min.
The cells were lysed on ice and centrifuged at 15,000 × g for 15 min. The supernatant fractions were collected and mixed with a biotinylated RNA-bead mixture at 25°C for 2 h. Subsequently, the beads were washed and the bound protein (PC4) was eluted at 37°C for western blot analysis.
RNA-EMSA
RNA-EMSA was performed using an RNA EMSA Kit (BersinBio) according to the manufacturer’s instructions. The biotin-labeled probe sequences are listed in Data S3. Briefly, biotin-labeled RNA probes were incubated with PBS, protein lysates, or protein lysates with an anti-PC4 antibody for 30 min to allow the binding of PC4 to the biotin-labeled RNA probes. PAGE was performed, followed by electrophoretic transfer of the RNA probes to nylon membranes. The membranes were washed and blocked. Images of RNA probes were captured in the dark.
RNA-FISH
RNA-FISH was performed using an RNA FISH Kit (Bersinbio) in accordance with the manufacturer’s instructions. The specific probe sequences are listed in Data S3. Briefly, Huh7-PC4-GFP cells were seeded in 20-mm glass-bottom dishes and fixed with 4% paraformaldehyde for 30 min at room temperature. After two PBS washes, cells were digested with proteinase K for 15 min at 37°C, washed twice with PBS for 5 min, and mixed with 1% fixation solution for 10 min at room temperature. Fixed samples were dehydrated in 70% ethanol for 5 min, 85% ethanol for 5 min, and 100% ethanol for 5 min at room temperature. Next, the samples were incubated in the prehybridization solution for 30 min at 37°C, followed by incubation in the hybridization buffer with the indicated probes at 37°C overnight. The following day, the samples were washed with SSC buffer and stained with DAPI to mark the nucleus. Fluorescent images were acquired using a Leica SP-8 STED 3X microscope.
Protein expression and purification
The His-GST-tagged-PC4 or His-GST-tagged-PC4 mutants expressed in the BL21(DE3) strain of E. coli were induced by treatment with 0.2 mM isopropyl-β-d-thiogalactoside at 18°C for 16 h. Bacteria were collected by centrifugation, resuspended in a buffer containing 20 mM Tris-HCl (pH 8.0), 500 mM NaCl, and 20 mM imidazole (pH 8.0), and lysed under high pressure. Cell debris was removed by centrifugation at 18,000 rpm for 1 h at 4°C. The supernatant of the His-tagged protein was loaded onto a Ni-NTA column (GE) and washed with lysis buffer. The fusion protein was eluted with a buffer containing 20 mM Tris-HCl (pH 8.0), 500 mM NaCl, and 500 mM imidazole. Ulp1 protease was added to remove the His-tag from the recombinant protein, which was followed by dialysis with the lysis buffer for 3 h at 4°C. The mixture was applied to another Ni-NTA resin to remove the proteases and uncleaved proteins. Next, the eluted proteins were concentrated and loaded onto a pre-equilibrated HiLoad 16/60 Superdex 200-pg column and eluted at a flow rate of 1 ml/min with a buffer containing 10 mM Tris-HCl (pH 8.0) and 100 mM NaCl or 10 mM Tris-HCl (pH 8.0) and 100 mM KCl. Peak fractions were analyzed using SDS-PAGE and stained with Coomassie Brilliant Blue R-250. High-purity fractions were pooled, concentrated, and stored in PBS.
IP assay
Cells were collected and lysed using IP Lysis Buffer (50 mM Tris-HCl [pH 7.5], 1 mM EDTA, 150 mM NaCl, 1 mM EGTA, 5 mM MgCl2, 10% glycerol, and 0.2% NP-40) supplemented with protease inhibitor (Thermo Fisher Scientific), Phosphatase Inhibitors Cocktail 2 (P5726; Sigma-Aldrich), and 1 mM 1,4-dithiothreitol (DTT) (Sigma-Aldrich). The cell lysate was then centrifuged at 20,000 × g for 15 min at 4°C, and the supernatant was incubated with the primary antibody overnight at 4°C. The next day, the antibody-bound protein was incubated with Protein A/G Beads (88802; Thermo Fisher Scientific) for 2 h at 4°C. After six washes with the Wash Buffer (0.5 M Tris-HCl pH 7.4, 1.5 M NaCl), immunoprecipitated complexes were denatured with the loading buffer (Fdbio science) for 10 min at 95°C. The samples were then stored at −20°C or readied for SDS-PAGE.
Cycloheximide chase assay
Cells were treated with 100 μg/ml cycloheximide and lysed at the indicated time points, which was followed by Western blot analysis with the indicated antibodies.
In vitro kinase assay
For this assay, 2 mg of recombinant GST-tagged-PC4 proteins (PC4-WT or PC4-S17A) was incubated with recombinant CK2 holoenzyme (New England Biolabs) in ATP-containing NEB PK buffer (New England Biolabs) at 37°C for 45 min. To test whether the PC4 protein was phosphorylated by CK2, the final product was denatured and further analyzed by SDS-PAGE Coomassie Brilliant Blue staining.
In vitro ubiquitylation assay
Substrates were incubated in a reaction buffer containing 25 mM Tris-HCl (pH 7.4), 2 mM ATP, 5 mM MgCl2, and 0.1 mM DTT for 1 h for ubiquitylation at 30°C. The ubiquitylated products were purified using beads. The final product was denatured and analyzed by Western blotting.
Western blot
The antibodies used for Western blots are listed in Data S3. Briefly, protein extracts were separated by SDS-PAGE or Phospho-tag SDS-PAGE and electrophoretically transferred to a 0.45-μm PVDF membrane (EMD Millipore). The membrane was incubated in 5% milk/phosphate-buffered saline Tween-20 (PBST) for 1 h at room temperature and then cut into strips corresponding to the molecular mass of the target. The strips were then incubated with the indicated primary antibodies at 4°C overnight. After three washes with PBST, the membrane strips were incubated with the appropriate secondary antibodies. Membranes were visualized using the Immobilon Western Chemiluminescent HRP Substrate (EMD Millipore) on an ImageQuant LAS 4000 platform or Bio-Rad ChemiDoc. The densitometric ratio of protein bands was calculated by ImageJ.
MS
Cells or tissues were lysed in lysis buffer (8 M urea, 1% protease inhibitor, and 1% phosphorylase inhibitor) on ice for 30 min. After sonication and centrifugation, the supernatant was collected and the protein concentration was determined using the bicinchoninic acid assay (Beyotime). Extracted protein (200 μg) of each sample was mixed with 20% TCA (Sigma-Aldrich) at 4°C for 2 h and centrifuged at 4,500 × g for 5 min. The supernatant was discarded and the precipitate was washed with chilled acetone two to three times. After drying, 200 mM TEAB (Sigma-Aldrich) was added to the precipitate, followed by sonication. Next, the peptides were reduced with 50 mM DTT (Sigma-Aldrich) at 56°C for 30 min. The proteins were alkylated with 11 mM iodoacetamide (Acros) at room temperature in the dark for 15 min. Subsequently, the precipitate was digested by incubation with 0.01 μg/μl trypsin (Promega) at 37°C overnight. Then, the peptides were separated with a gradient of 2–60% acetonitrile in 10 mM ammonium bicarbonate (pH 10) for 80 min into 80 fractions by high-pH reverse-phase high-performance liquid chromatography using the Agilent 300 Extend C18 column (particle size, 5 μm; internal diameter, 4.6 mm; length, 250 mm). The peptides were eventually combined into 12 fractions.
Statistical analysis
All data are presented as means ± SD; error bars in figures represent SD; for at least triplicate experiments. Statistical analysis was performed using SPSS (version 22.0) and GraphPad Prism (version 9.5.0). No statistical method was used to predetermine sample size and no data were excluded from the analyses. The data distribution was tested for normality. Comparisons between groups were performed using two-side unpaired Student’s t test. Comparisons among three or more groups were performed using one-way ANOVA followed by Tukey’s post hoc test. In the in vivo experiments, mice were age- and weight-randomized appropriately. Survival data were analyzed using the Kaplan–Meier method. Significance is indicated by asterisks: n.s., not significant, P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001. P values <0.05 are considered as statistically significant. The immunoblots shown are representative of three independent experiments with similar results.
Reagents
Information on all reagents used in this study is provided in Data S3.
Online supplemental material
Fig. S1 shows that PC4 acts as an RBP that stabilizes CCND1 mRNA. Fig. S2 shows that ubiquitination and phosphorylation of PC4 show periodic fluctuations and are associated with the stability of CCND1 mRNA. Fig. S3 shows that PC4 knockdown inhibits liver cancer cell growth in vitro and in vivo. Fig. S4 shows that PC4 KO inhibits liver cancer cell growth in vitro and in vivo. Fig. S5 shows that PC4 overexpression promotes liver cancer cell growth in vitro and in vivo. Fig. S6 shows that PC4 promotes G1–S transition and cell proliferation partially through a CCND1-dependent manner. Fig. S7 shows that PC4 exhibits distinct modifications in HCC, and PC4 loss sensitizes HCC cells to CDK4/6 inhibitors. Videos 1, 2, and 3 depict the real-time cell cycle transition of Huh7 cells under different conditions: control, PC4 knockdown, and PC4 overexpression. Data S1 shows the gene datasets obtained from RNA-seq, RIP-seq, and ChIP-seq. Data S2 displays the modified peptides of PC4 observed in different cell cycle phases within Huh7 cells. Data S3 presents the oligos, reagents, and resources utilized in this study.
Supplementary Material
shows the gene datasets obtained from RNA-seq, RIP-seq, and ChIP-seq.
displays the modified peptides of PC4 observed in different cell cycle phases within Huh7 cells.
presents the oligos, reagents, and resources utilized in this study.
is the source file for Fig. 1.
is the source file for Fig. 2.
is the source file for Fig. 3.
is the source file for Fig. 4.
is the source file for Fig. 5.
is the source file for Fig. 7.
is the source file for Fig. S1.
is the source file for Fig. S2.
is the source file for Fig. S3.
is the source file for Fig. S4.
is the source file for Fig. S5.
is the source file for Fig. S6.
is the source file for Fig. S7.
Acknowledgments
We thank Professor H. Lu for reviewing the manuscript and Y.Q. Wang and L.M. Xie for assistance with live cell imaging.
This work was supported by grants from the National Natural Science Foundation of China (82192884 and 82073067) and Guangdong Science and Technology Department (2023B1212060013).
Author contributions: Q. Pan and P. Luo conceived, designed, and performed most of the in vitro and in vivo experiments, analyzed the data, and wrote and revised the manuscript. K. Hu and Y. Qiu provided the FUCCI system and performed eCLIP-seq and microscopy experiments. G. Liu carried out cellular and biochemical experiments. S. Dai performed animal experiments and tissue immunohistochemistry. B. Cui collected the clinical data. C. Shi and D. Yin conceived, supervised, and supported the project and reviewed and revised the manuscript. All authors have read and approved the final manuscript.
Footnotes
C. Shi is the lead contact.
Data availability
All unique/stable reagents generated in this study are available from the corresponding author, Dr. Chunmeng Shi, upon reasonable request. Source data for Figs. 1, 2, 3, 4, 5, 7, S1, S2, S3, S4, S5, S6, and S7 are provided online. The sequencing data (RNA-seq, eCLIP-seq, RIP-seq) underlying Fig. 1, C and E, are openly available on the GEO database with the accession number GSE202401. The MS data underlying Fig. 2 A is available on the PRIDE database with the accession number PXD033637.
Data availability
All unique/stable reagents generated in this study are available from the corresponding author, Dr. Chunmeng Shi, upon reasonable request. Source data for Figs. 1, 2, 3, 4, 5, 7, S1, S2, S3, S4, S5, S6, and S7 are provided online. The sequencing data (RNA-seq, eCLIP-seq, RIP-seq) underlying Fig. 1, C and E, are openly available on the GEO database with the accession number GSE202401. The MS data underlying Fig. 2 A is available on the PRIDE database with the accession number PXD033637.
References
- Baldin, V., Lukas J., Marcote M.J., Pagano M., and Draetta G.. 1993. Cyclin D1 is a nuclear protein required for cell cycle progression in G1. Genes Dev. 7:812–821. 10.1101/gad.7.5.812 [DOI] [PubMed] [Google Scholar]
- Beltrao, P., Bork P., Krogan N.J., and van Noort V.. 2013. Evolution and functional cross-talk of protein post-translational modifications. Mol. Syst. Biol. 9:714. 10.1002/msb.201304521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollard, J., Miguela V., Ruiz de Galarreta M., Venkatesh A., Bian C.B., Roberto M.P., Tovar V., Sia D., Molina-Sánchez P., Nguyen C.B., et al. 2017. Palbociclib (PD-0332991), a selective CDK4/6 inhibitor, restricts tumour growth in preclinical models of hepatocellular carcinoma. Gut. 66:1286–1296. 10.1136/gutjnl-2016-312268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyault, S., Rickman D.S., de Reyniès A., Balabaud C., Rebouissou S., Jeannot E., Hérault A., Saric J., Belghiti J., Franco D., et al. 2007. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology. 45:42–52. 10.1002/hep.21467 [DOI] [PubMed] [Google Scholar]
- Calderaro, J., Ziol M., Paradis V., and Zucman-Rossi J.. 2019. Molecular and histological correlations in liver cancer. J. Hepatol. 71:616–630. 10.1016/j.jhep.2019.06.001 [DOI] [PubMed] [Google Scholar]
- Chaikovsky, A.C., Li C., Jeng E.E., Loebell S., Lee M.C., Murray C.W., Cheng R., Demeter J., Swaney D.L., Chen S.H., et al. 2021. The AMBRA1 E3 ligase adaptor regulates the stability of cyclin D. Nature. 592:794–798. 10.1038/s41586-021-03474-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, R.J., Huang M., Campbell M.J., Dong H., Steinmetz L., Sapinoso L., Hampton G., Elledge S.J., Davis R.W., and Lockhart D.J.. 2001. Transcriptional regulation and function during the human cell cycle. Nat. Genet. 27:48–54. 10.1038/83751 [DOI] [PubMed] [Google Scholar]
- Dang, F., Nie L., and Wei W.. 2021. Ubiquitin signaling in cell cycle control and tumorigenesis. Cell Death Differ. 28:427–438. 10.1038/s41418-020-00648-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang, H., Takai A., Forgues M., Pomyen Y., Mou H., Xue W., Ray D., Ha K.C.H., Morris Q.D., Hughes T.R., and Wang X.W.. 2017. Oncogenic activation of the RNA binding protein NELFE and MYC signaling in hepatocellular carcinoma. Cancer Cell. 32:101–114.e8. 10.1016/j.ccell.2017.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, C., Hizume K., Batta K., Kumar B.R., Gadad S.S., Ganguly S., Lorain S., Verreault A., Sadhale P.P., Takeyasu K., and Kundu T.K.. 2006. Transcriptional coactivator PC4, a chromatin-associated protein, induces chromatin condensation. Mol. Cell. Biol. 26:8303–8315. 10.1128/MCB.00887-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuele, M.J., Enrico T.P., Mouery R.D., Wasserman D., Nachum S., and Tzur A.. 2020. Complex cartography: Regulation of E2F transcription factors by cyclin F and ubiquitin. Trends Cell Biol. 30:640–652. 10.1016/j.tcb.2020.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasanaro, P., Capogrossi M.C., and Martelli F.. 2010. Regulation of the endothelial cell cycle by the ubiquitin-proteasome system. Cardiovasc. Res. 85:272–280. 10.1093/cvr/cvp244 [DOI] [PubMed] [Google Scholar]
- Fischer, M., and Müller G.A.. 2017. Cell cycle transcription control: DREAM/MuvB and RB-E2F complexes. Crit. Rev. Biochem. Mol. Biol. 52:638–662. 10.1080/10409238.2017.1360836 [DOI] [PubMed] [Google Scholar]
- Fischer, M., Schade A.E., Branigan T.B., Müller G.A., and DeCaprio J.A.. 2022. Coordinating gene expression during the cell cycle. Trends Biochem. Sci. 47:1009–1022. 10.1016/j.tibs.2022.06.007 [DOI] [PubMed] [Google Scholar]
- Ge, H., and Roeder R.G.. 1994. Purification, cloning, and characterization of a human coactivator, PC4, that mediates transcriptional activation of class II genes. Cell. 78:513–523. 10.1016/0092-8674(94)90428-6 [DOI] [PubMed] [Google Scholar]
- Ge, H., Zhao Y., Chait B.T., and Roeder R.G.. 1994. Phosphorylation negatively regulates the function of coactivator PC4. Proc. Natl. Acad. Sci. USA. 91:12691–12695. 10.1073/pnas.91.26.12691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer, F., Schwarzl T., Valcárcel J., and Hentze M.W.. 2021. RNA-binding proteins in human genetic disease. Nat. Rev. Genet. 22:185–198. 10.1038/s41576-020-00302-y [DOI] [PubMed] [Google Scholar]
- Hunter, T. 2007. The age of crosstalk: Phosphorylation, ubiquitination, and beyond. Mol. Cell. 28:730–738. 10.1016/j.molcel.2007.11.019 [DOI] [PubMed] [Google Scholar]
- Kretzschmar, M., Kaiser K., Lottspeich F., and Meisterernst M.. 1994. A novel mediator of class II gene transcription with homology to viral immediate-early transcriptional regulators. Cell. 78:525–534. 10.1016/0092-8674(94)90429-4 [DOI] [PubMed] [Google Scholar]
- Lee, J.-S., Chu I.S., Heo J., Calvisi D.F., Sun Z., Roskams T., Durnez A., Demetris A.J., and Thorgeirsson S.S.. 2004. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 40:667–676. 10.1002/hep.20375 [DOI] [PubMed] [Google Scholar]
- Lim, S., and Kaldis P.. 2013. Cdks, cyclins and CKIs: Roles beyond cell cycle regulation. Development. 140:3079–3093. 10.1242/dev.091744 [DOI] [PubMed] [Google Scholar]
- Lin, D.I., Barbash O., Kumar K.G., Weber J.D., Harper J.W., Klein-Szanto A.J., Rustgi A., Fuchs S.Y., and Diehl J.A.. 2006. Phosphorylation-dependent ubiquitination of cyclin D1 by the SCF(FBX4-alphaB crystallin) complex. Mol. Cell. 24:355–366. 10.1016/j.molcel.2006.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., Chen S., Wang S., Soares F., Fischer M., Meng F., Du Z., Lin C., Meyer C., DeCaprio J.A., et al. 2017. Transcriptional landscape of the human cell cycle. Proc. Natl. Acad. Sci. USA. 114:3473–3478. 10.1073/pnas.1617636114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan, D.O. 1997. Cyclin-dependent kinases: Engines, clocks, and microprocessors. Annu. Rev. Cell Dev. Biol. 13:261–291. 10.1146/annurev.cellbio.13.1.261 [DOI] [PubMed] [Google Scholar]
- Mortusewicz, O., Evers B., and Helleday T.. 2016. PC4 promotes genome stability and DNA repair through binding of ssDNA at DNA damage sites. Oncogene. 35:761–770. 10.1038/onc.2015.135 [DOI] [PubMed] [Google Scholar]
- Otto, T., and Sicinski P.. 2017. Cell cycle proteins as promising targets in cancer therapy. Nat. Rev. Cancer. 17:93–115. 10.1038/nrc.2016.138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, Q., Luo P., and Shi C.. 2023. PC4-mediated Ku complex PARylation facilitates NHEJ-dependent DNA damage repair. J. Biol. Chem. 299:105032. 10.1016/j.jbc.2023.105032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, S., Mossmann D., Chen Q., Wang X., Dazert E., Colombi M., Schmidt A., Ryback B., Ng C.K.Y., Terracciano L.M., et al. 2022. Transcription factors TEAD2 and E2A globally repress acetyl-CoA synthesis to promote tumorigenesis. Mol. Cell. 82:4246–4261.e11. 10.1016/j.molcel.2022.10.027 [DOI] [PubMed] [Google Scholar]
- Rebouissou, S., and Nault J.-C.. 2020. Advances in molecular classification and precision oncology in hepatocellular carcinoma. J. Hepatol. 72:215–229. 10.1016/j.jhep.2019.08.017 [DOI] [PubMed] [Google Scholar]
- Seki, A., Coppinger J.A., Du H., Jang C.-Y., Yates J.R. III, and Fang G.. 2008a. Plk1- and beta-TrCP-dependent degradation of Bora controls mitotic progression. J. Cell Biol. 181:65–78. 10.1083/jcb.200712027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki, A., Coppinger J.A., Jang C.-Y., Yates J.R., and Fang G.. 2008b. Bora and the kinase Aurora a cooperatively activate the kinase Plk1 and control mitotic entry. Science. 320:1655–1658. 10.1126/science.1157425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr, C.J. 1995. D-type cyclins. Trends Biochem. Sci. 20:187–190. 10.1016/s0968-0004(00)89005-2 [DOI] [PubMed] [Google Scholar]
- Stumpf, C.R., Moreno M.V., Olshen A.B., Taylor B.S., and Ruggero D.. 2013. The translational landscape of the mammalian cell cycle. Mol. Cell. 52:574–582. 10.1016/j.molcel.2013.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaney, D.L., Beltrao P., Starita L., Guo A., Rush J., Fields S., Krogan N.J., and Villén J.. 2013. Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat. Methods. 10:676–682. 10.1038/nmeth.2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nostrand, E.L., Pratt G.A., Shishkin A.A., Gelboin-Burkhart C., Fang M.Y., Sundararaman B., Blue S.M., Nguyen T.B., Surka C., Elkins K., et al. 2016. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat. Methods. 13:508–514. 10.1038/nmeth.3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Zhang S., Zhang Y., Jia J., Wang J., Liu X., Zhang J., Song X., Ribback S., Cigliano A., et al. 2022. TAZ is indispensable for c-MYC-induced hepatocarcinogenesis. J. Hepatol. 76:123–134. 10.1016/j.jhep.2021.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J.-Y., Sarker A.H., Cooper P.K., and Volkert M.R.. 2004. The single-strand DNA binding activity of human PC4 prevents mutagenesis and killing by oxidative DNA damage. Mol. Cell. Biol. 24:6084–6093. 10.1128/MCB.24.13.6084-6093.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, C.-F., Iqbal, S., Shen, S., Luo, Y.-L., Yang, X. and Wang, J.. 2019. Development of “CLAN” nanomedicine for nucleic acid therapeutics. Small. 15:e1900055. 10.1002/smll.201900055 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows the gene datasets obtained from RNA-seq, RIP-seq, and ChIP-seq.
displays the modified peptides of PC4 observed in different cell cycle phases within Huh7 cells.
presents the oligos, reagents, and resources utilized in this study.
is the source file for Fig. 1.
is the source file for Fig. 2.
is the source file for Fig. 3.
is the source file for Fig. 4.
is the source file for Fig. 5.
is the source file for Fig. 7.
is the source file for Fig. S1.
is the source file for Fig. S2.
is the source file for Fig. S3.
is the source file for Fig. S4.
is the source file for Fig. S5.
is the source file for Fig. S6.
is the source file for Fig. S7.
Data Availability Statement
All unique/stable reagents generated in this study are available from the corresponding author, Dr. Chunmeng Shi, upon reasonable request. Source data for Figs. 1, 2, 3, 4, 5, 7, S1, S2, S3, S4, S5, S6, and S7 are provided online. The sequencing data (RNA-seq, eCLIP-seq, RIP-seq) underlying Fig. 1, C and E, are openly available on the GEO database with the accession number GSE202401. The MS data underlying Fig. 2 A is available on the PRIDE database with the accession number PXD033637.
All unique/stable reagents generated in this study are available from the corresponding author, Dr. Chunmeng Shi, upon reasonable request. Source data for Figs. 1, 2, 3, 4, 5, 7, S1, S2, S3, S4, S5, S6, and S7 are provided online. The sequencing data (RNA-seq, eCLIP-seq, RIP-seq) underlying Fig. 1, C and E, are openly available on the GEO database with the accession number GSE202401. The MS data underlying Fig. 2 A is available on the PRIDE database with the accession number PXD033637.