Abstract
The Tpr protease of Porphyromonas gingivalis W83 is a membrane-associated enzyme capable of hydrolyzing chromogenic substrates for trypsin and bacterial collagenases. A previous study by us indicated that Tpr expression was increased under conditions of nutrient limitation. In the present study, we further characterized expression of the tpr gene using a tpr::lacZ reporter gene construct under a range of nutrient conditions. In P. gingivalis, transcription of tpr was initiated 215 bp upstream of the coding region and regulation of tpr expression was at the level of transcription. Deletion mutations in the tpr upstream region identified the promoter region immediately upstream of the transcription start site, determined by primer extension analysis. Three identical 17-bp direct repeats identified within the 5′ end of tpr mRNA were involved in tpr regulation. In an Escherichia coli background, tpr transcription was initiated after an AT-rich region upstream of tpr but not at the P. gingivalis start site. Tpr expression in P. gingivalis was suppressed by the addition of peptide and protein nutrients to a peptide-limited growth medium but was only slightly affected by addition of free amino acids. Low-molecular-weight fractions of brain heart infusion rich in phenylalanine, proline, and alanine had the greatest inhibitory effects on expression of the tpr::lacZ construct. Addition of the dipeptide phenylalanyl-phenylalanine to the growth medium resulted in a 10-fold decrease in tpr expression. This suggests that specific phenylalanine-containing peptides are a major factor controlling Tpr expression. Neither hemin starvation, heat shock, nor pH change had significant effects on Tpr expression.
Bacterial growth in nature is affected by environmental conditions such as the availability of essential nutrients and cofactors, the accumulation of toxic metabolites, and changes in pH, Eh, or temperature. Porphyromonas gingivalis responds to environmental changes by modifying its physiology and expression of potential virulence factors. These responses include induction of expression of DnaK and GroEL homologues by heat stress (21), as well as changes in growth rate and specific enzymatic activities in response to changes in pH (28) and the availability of hemin (25, 42) and collagen (30, 38).
P. gingivalis possesses numerous distinct proteases and peptidases, many of which are membrane associated or secreted (2, 9, 12, 22, 32). This finding is consistent with its peptide- and amino-acid-dependent metabolism (39). This proteolytic activity is recognized as an important virulence factor in periodontal diseases (15). Characterization of one of these proteases, the membrane-associated Tpr protease, indicated that it was a thiol-dependent protease whose proteolytic activity is activated by reducing conditions (5, 33). Tpr activity was significantly increased in cells cultured under nutrient-limited conditions, suggesting that expression of Tpr was regulated (34). In an analysis of the collagenase-like Pz-peptidase activity of Tpr, the membrane fraction of P. gingivalis W83 cells grown in Trypticase-yeast extract (TYE) medium in which the Tryticase Peptone content was reduced from 17 to 5 mg/ml (34) (0.5TYE) had twice the Pz-peptidase activity of cells grown in TYE and five times the activity of cells grown in brain heart infusion (BHI). Northern blot analysis suggested that the regulation of tpr expression occurred primarily at the transcriptional level (34).
The mechanisms of gene regulation and expression in this organism are not well understood, and little is known about the promoter structures of P. gingivalis genes. One study suggested that the RNA polymerase of P. gingivalis was structurally different from that of Escherichia coli (19). Putative promoter sequences of a number of cloned P. gingivalis genes have been identified, based only on their limited homology with the consensus sequences of E. coli promoters and without evidence of their promoter activity in P. gingivalis. To better understand the structures of P. gingivalis genes and their regulation, it is necessary to analyze native regulatory sequences.
Our previous study showing that Tpr peptidolytic activity (Pz-peptidase) and tpr mRNA expression were influenced by nutrient conditions (34) prompted us to analyze the noncoding region directly upstream of the tpr locus. The gene encoding the Tpr protease has been cloned and sequenced and consists of an open reading frame of 1,446 nucleotides (5, 33). The finding that the tpr protease gene was regulated by growth conditions provided a model for studying gene regulation in this important periodontopathogen. This model is especially relevant to the study of P. gingivalis virulence genes, since membrane-associated and extracellular proteases of this organism are recognized as key to its role in periodontitis (15). In the present study, we determine the transcriptional start site of the tpr gene, analyze the tpr promoter, and using a tpr::lacZ reporter gene, characterize tpr regulation by quantitating β-galactosidase activity in P. gingivalis transconjugants.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
P. gingivalis W83 was grown anaerobically in BHI broth (33) or TYE broth (11). Nutrient-limited medium was 0.5TYE (34). Cultures were incubated in an anaerobic chamber (Coy Manufacturing, Ann Arbor, Mich.) in a 5% CO2–10% H2–85% N2 atmosphere at 37°C. P. gingivalis transconjugants were selected on BHI blood agar plates containing gentamicin (200 μg/ml) and erythromycin (10 μg/ml) or tetracycline (10 μg/ml) as described previously (34). E. coli was grown in Luria-Bertani (LB) broth (3) or minimal A medium (3). E. coli XL-1 Blue (Stratagene, La Jolla, Calif.) was used as the host for plasmid construction and for some expression studies. For selection purposes, ampicillin (50 μg/ml), kanamycin (50 μg/ml), trimethoprim (200 μg/ml), and tetracycline (10 μg/ml), as appropriate, were used unless stated otherwise. Stocks of bacteria were stored at −70°C in 15% glycerol. Bacterial strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| P. gingivalis strains | ||
| W83 | Wild type, Gmr | Lab collection |
| W83/PM | P. gingivalis W83 tpr mutant, Emr | 32 |
| W83/lacZ | P. gingivalis W83 with tpr::lacZ fusion, Emr | This study |
| Plasmids | ||
| pTZ19R | Apr | 43 |
| pBluescript II SK(−) | Apr | Stratagene |
| pYS307 | Apr, tpr cloned in pTZ18R | 33 |
| pXCA601 | Promoterless lacZ gene, Tcr | 1 |
| pBLU-1 | BamHI-PstI tpr fragment of pBY307 cloned into pBluescript II SK(−) | This study |
| pBX-1 | BamHI-HindIII 3.5-kb lacZ fragment of pXCA601 ligated to EcoRI- and HindIII-digested pBLU-1 | This study |
| R751 | Tpr Tra+ IncP plasmid mobilizes vectors from E. coli to Bacteroides recipients | 40 |
| pBY2-IN | Kmr Emr, tpr insertional mutation | 32 |
| pBYZ | BamHI-HindIII 4.3-kb tpr::lacZ fragment of pBX-1 ligated to pBY2-IN | This study |
| pNJR12 | Kmr Smr Mob+ Tcr, replicable in P. gingivalis | 23 |
| pNTX | PstI-HindIII 3.5-kb lacZ fragment of pTXZ19R cloned in pNJR12 | This study |
| pNTX-400 | BamHI-HindIII 4.3-kb tpr::lacZ fragment of pBX-1 cloned in pNJR12 | This study |
| pNTX-100 | BamHI-HindIII 4.0-kb tpr::lacZ fragment of pTXZ19R-100 cloned in pNJR12 | This study |
| pNTX-30 | KpnI-HindIII 3.93-kb tpr::lacZ fragment of pTXZ19R-30 cloned in pNJR12 | This study |
| pNTX20 | BamHI-HindIII 3.88-kb tpr::lacZ fragment of pTXZ19R20 cloned in pNJR12 | This study |
| pNTX52 | KpnI-HindIII 3.85-kb tpr::lacZ fragment of pTXZ19R52 cloned in pNJR12 | This study |
| pNTX137 | BamHI-HindIII 3.76-kb tpr::lacZ fragment of pTXZ19R137 cloned in pNJR12 | This study |
| pNTX-100Δ | EcoRI-HindIII 3.9-kb tpr::lacZ fragment of pTXZ19R-100Δ cloned in pNJR12 | This study |
| pNTX-30Δ | KpnI-HindIII 3.81-kb tpr::lacZ fragment of pTXZ19R-30Δ cloned in pNJR12 | This study |
| pTXZ19R | PstI-HindIII 3.5-kb lacZ of pBX-1 cloned in pTZ19R | This study |
| pTXZ19R-400 | BamHI-HindIII 4.3-kb tpr::lacZ fragment of pBX-1 cloned in pTZ19R | This study |
| pTXZ19R-100 | BamHI-PstI 500-bp PCR product XZ-100 cloned in pTXZ19R | This study |
| pTXZ19R-30 | KpnI-PstI 430-bp PCR product XZ-30 cloned in pTXZ19R | This study |
| pTXZ19R20 | BamHI-PstI 380-bp PCR product XZ20 cloned in pTXZ19R | This study |
| pTXZ19R52 | KpnI-PstI 348-bp PCR product XZ52 cloned in pTXZ19R | This study |
| pTXZ19R137 | BamHI-PstI 263-bp PCR product XZ137 cloned in pTXZ19R | This study |
| pTXZ19R-100Δ | EcoRI-BamHI 120-bp PCR product XZ-100Δ cloned in pTXZ19R137 | This study |
| pTXZ19R-30Δ | KpnI-BamHI 50-bp PCR product XZ-30Δ cloned in pTXZ19R137 | This study |
Smr, Tpr, and Apr, resistance to streptomycin, trimethoprim, and ampicillin, respectively. Mob+, capable of being mobilized; Tra+, capable of self-transfer.
Primer extension analysis and DNA sequencing.
Total RNA was isolated from P. gingivalis and E. coli strains with TRIzol Reagent (Gibco BRL, Gaithersburg, Md.) by the manufacturer’s protocol. To map the 5′ terminus of tpr mRNA in P. gingivalis and E. coli, primer extension analysis was conducted as described previously (3). Three primers, tpr293, tpr170, and tpr64 (Table 2), were used for primer extension analysis. The primers were labeled with [γ-32P]ATP as described previously (3). Hybridization and primer extension were carried out as described previously (3) with Avian myoblastosis virus reverse transcriptase (Gibco BRL). Primer extension products were heated for 2 min at 95°C before being loaded on a sequencing gel. Dideoxy sequencing reaction mixtures with the same primer were run alongside the primer extension products. DNA sequencing reaction experiments were conducted with Sequenase version 2.0 DNA polymerase by following the protocols provided by the manufacturer (United States Biochemicals, Cleveland, Ohio).
TABLE 2.
Oligonucleotide primers used to amplify tpr upstream sequences
| Primer | Nucleotide sequencea | Location (nt)b |
|---|---|---|
| Bm300 | 5′-ATTCGGATCCTCGGGTCTCGTCTG-3′ | 291–314 |
| BamHI | ||
| Kpn365 | 5′-TTCAGGTACCAATTGTCAATT-3′ | 360–380 |
| KpnI | ||
| Bm200 | 5′-TAATGGATCCTAACGGTTTTTCATGC-3′ | 410–435 |
| BamHI | ||
| Kpn452 | 5′-AAATGGTACCTTAATTCG-3′ | 447–464 |
| KpnI | ||
| Bm100 | 5′-CCTTAATGGATCCCTTCATTTGTG-3′ | 529–552 |
| BamHI | ||
| Eco300 | 5′-CACGAATTCGGCTGTTCG-3′ | 286–303 |
| EcoRI | ||
| Bm424 | 5′-CGTTAGGATCCATTATTTCAA-3′ | 424–404 |
| BamHI | ||
| tprPst | 5′-CATCCCTGCAGGGCTGC-3′ | 801–785 |
| PstI | ||
| tpr293 | 5′-GCTTTCGCTTCCTCTTGTTGAGGA-3′ | 710–687 |
| tpr170 | 5′-AACTGTGACTTTAGGCTCTTAC-3′ | 587–566 |
| tpr64 | 5′-TGTGTACAAAAAAACTAACGAATTA-3′ | 482–458 |
New restriction sites engineered into the sequence are indicated.
Numbers correspond to the DNA sequence shown in Fig. 2B. nt, nucleotides.
Recombinant DNA methods.
Plasmid DNA was isolated by the alkaline lysis method (3). Chromosomal DNA was extracted from bacterial cells grown to early stationary phase by a miniprep method (3). Restriction enzyme digestion of DNA samples was carried out according to the manufacturer’s recommendations. Subcloning of DNA fragments and PCR products was done by restriction digestion and electrophoresis of agarose gels which were prepared with and run in a Tris-acetate-EDTA buffer (40 mM Tris · acetate, 2 mM Na2 · EDTA · 2H2O [pH 8.5]). The desired DNA fragments were excised from the gel and recovered by Glass milk purification as described by the manufacturer (GeneClean kit; Bio 101, Inc., La Jolla, Calif.). Transformation of E. coli was done by electroporation (3), except that mobilization plasmid R751 was introduced into E. coli strains by conjugation, as described previously (32). Both replicating and nonreplicating plasmids were introduced into P. gingivalis from E. coli by conjugation (32). Transconjugants were grown on BHI-blood agar containing antibiotics. Gentamicin was used to inhibit growth of E. coli donor cells, and erythromycin or tetracycline was used to select for P. gingivalis transconjugants containing a chromosomally integrated erythromycin resistance (Emr) gene or a plasmid-borne tetracycline resistance (Tcr) gene. Transconjugants were passaged twice on BHI-blood agar containing antibiotics before analysis.
PCR and primers.
To analyze the effects of deletion mutations on the expression of tpr, fragments containing the tpr 5′ end and upstream regions were generated by PCR with pYS307 DNA as a template (33). Primers used for PCR amplification are shown in Table 2. DNA fragment XZ-100 was generated with primers Bm300 and tprPst, fragment XZ-30 was generated with primers Kpn365 and tprPst, fragment XZ20 was generated with primers Bm200 and tprPst, fragment XZ52 was generated with primers Kpn452 and tprPst, and fragment XZ137 was generated with primers Bm100 and tprPst. Fragment XZ-100Δ was generated with primers Eco300 and Bm424, and fragment XZ-30Δ was generated with primers Kpn452 and Bm424. PCR amplification was carried out with TaqI polymerase (Gibco BRL). Twenty-five cycles were carried out at a denaturing temperature of 95°C for 1 min, an annealing temperature of 55°C for 1 min, and an extension temperature of 72°C for 1.5 min in a model 480 thermal cycler (Perkin-Elmer Cetus, Norwalk, Conn.).
Construction of tpr::lacZ fusion plasmids.
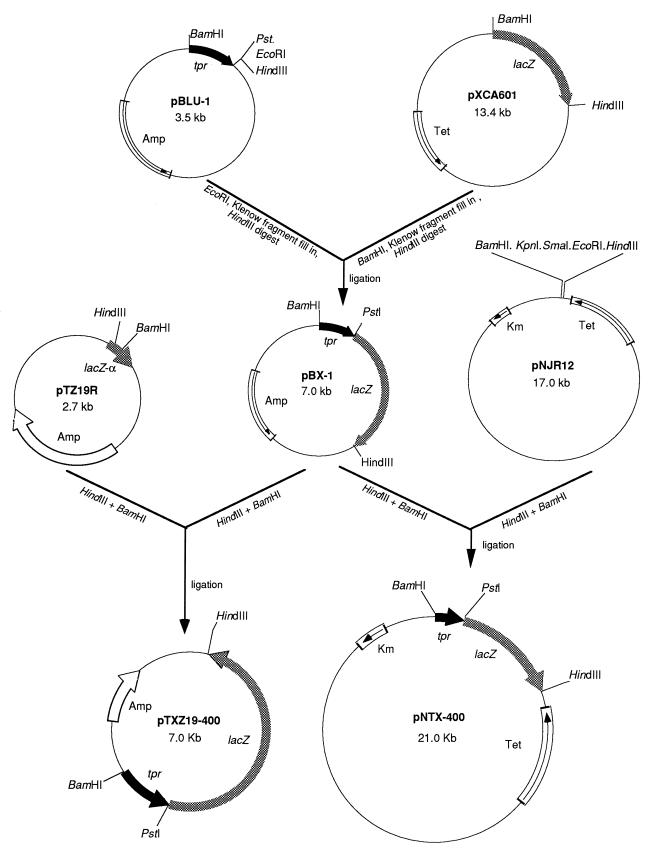
Plasmids pNJR12 (23) and pTZ19R (43) were used as vectors for construction of tpr::lacZ reporter plasmids for analysis in P. gingivalis and E. coli, respectively. To construct a tpr::lacZ reporter cassette, the 0.8-kb BamHI-PstI fragment of pYS307 (33) containing DNA upstream of tpr and the first 183 bp of the tpr coding region was first ligated to pBluescript II SK(−) (Stratagene) which had been digested with the same enzymes, yielding pBLU-1. Then, as shown in Fig. 1, the 3.5-kb BamHI-HindIII lacZ fragment of pXCA601 (1) was ligated to pBLU-1, yielding pBX-1, in which the promoterless lacZ gene was immediately preceded by the 5′ end of tpr and its upstream region. The 4.3-kb BamHI-HindIII fragment of pBX-1 was cloned into the corresponding sites of both pTX19R and pNJR12, yielding pTXZ19R-400 and pNTX-400, respectively.
FIG. 1.
Construction of tpr::lacZ fusion plasmids pTXZ19-400 and pNTX-400 for assay of β-galactosidase activity under control of the tpr promoter in E. coli and P. gingivalis, respectively. Relevant restriction enzyme sites are shown. The 0.8-kb fragment containing DNA upstream of tpr and the first 183 bp of the tpr coding region are shown as a black arrow. The promoterless lacZ gene is shown as a gray arrow. Plasmid vector DNAs that encode the alpha fragment of β-galactosidase (lacZ-α), ampicillin resistance (Amp), Tcr (Tet), and kanamicin resistance (Km) are shown by arrows.
To construct a series of deletions in the region upstream of tpr for analysis in the lacZ reporter system, a PstI-HindIII fragment of pBX-1 containing the promoterless lacZ gene was first ligated to pTZ19R (43), yielding pTXZ19R. Subsequently, PCRs were carried out with the primers shown in Table 2, yielding products containing several different tpr upstream regions. The PCR products were digested with restriction enzymes whose sites were engineered into their termini, and they were cloned into the corresponding sites on pTXZ19R upstream of the lacZ gene (Table 1). For analysis in P. gingivalis, the resulting tpr::lacZ fusions were subsequently cloned in pNJR12 (Table 1).
To construct a suicide plasmid carrying the TX-400 tpr::lacZ fusion construct, the 4.3-kb BamHI-HindIII fragment of pBX-1 was ligated to pBY2-IN (32) directly upstream of the ermF gene, in the same orientation as and replacing the 5′ end of tpr carried on pBY2-IN. The resulting plasmid, pBYZ, carries a promoterless lacZ gene and ermF in place of a 0.6-kb internal fragment of tpr.
Southern blotting.
Southern hybridizations were carried out as described previously (32). Hybridization bands were detected by the BluGENE Nonradioactive Nucleic Acid Detection System (Gibco BRL).
Characterization of nutrient factors.
Powdered BHI (Difco Laboratories, Detroit, Mich.) was made up as a 20% solution in H2O, autoclaved, and passaged sequentially through membranes (Amicon Inc., Beverly, Mass.) having molecular weight cutoffs of 50, 10, 5, and 3 kDa. The flowthrough was then fractionated over a Sephadex G-10 column. The amino acid compositions of low-molecular-weight fractions of interest were determined with a model 420 amino acid hydrolyzer and derivatizer (Applied Biosystems, Inc., Foster City, Calif.) at the Nucleic Acids and Protein Sequencing Unit, Biotechnology Laboratory, University of British Columbia. For assays, P. gingivalis was grown in 0.5TYE supplemented with individual BHI fractions at a final concentration of 20 mg/ml. The effects of l amino acids (Sigma Chemical Co., St. Louis, Mo.) and di- and tripeptides (BACHEM California, Torrance, Calif.) were determined similarly at concentrations of 1 to 5 mM.
Stress conditions and hemin limitation.
For assays of responses to heat shock and pH change, P. gingivalis cells were first grown in 0.5TYE to mid-log phase and the cells were transferred to 42°C or were incubated in 0.5TYE at pH 5.5 or 8.5 for 4 h before being assayed. For the hemin starvation response assay, P. gingivalis cells at a 1:10 inoculum were grown in 0.5TYE without the supplementation of hemin for two passages.
β-Galactosidase assays.
To analyze P. gingivalis β-galactosidase activity, P. gingivalis strains were grown in various nutritional media to logarithmic-growth phase (optical density at 660 nm = 0.5), unless stated otherwise. Cells were harvested, washed twice with phosphate-buffered saline, and incubated on ice for 10 min in 20 mM Na-p-tosyl-l-lysine chloromethyl ketone. Cells were then resuspended in the reporter lysis buffer and analyzed for β-galactosidase activity by a β-galactosidase enzyme assay system (Promega Co., Madison, Wis.) as described by the manufacturer. Assays were done in 96-well microplates, and β-galactosidase activity was measured at 405 nm in a microplate reader (model 3550; Bio-Rad Laboratories, Richmond, Calif.). A standard curve for purified β-galactosidase was prepared for each assay. Protein concentration was measured by using the Bradford reagent supplied by Bio-Rad, with bovine serum albumin (BSA) as the standard. For P. gingivalis, 1 U of β-galactosidase activity was equivalent to hydrolysis of 1 nmol of o-nitrophenyl-β-d-galactopyranoside (ONPG) min−1 mg of total cellular protein−1. Assay of β-galactosidase activity in E. coli was done as described previously, and β-galactosidase activity was expressed in Miller units (29). At least four independent experiments using triplicate samples were performed for each β-galactosidase assay, and the results were averaged for display as bar graphs.
Nucleotide sequence accession number.
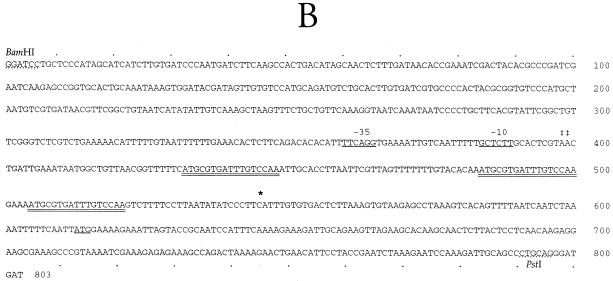
The GenBank accession number for the DNA sequence shown in Fig. 2 is AF022499.
FIG. 2.
Identification of the tpr promoter and transcription initiation site. (A) Primer extension analysis of tpr transcription in P. gingivalis W83. A DNA sequence ladder of the promoter region is shown. PE, primer extension reaction product. Oligonucleotide tpr64 was used as the primer for both the DNA sequence and primer extension reactions. Transcription initiation sites (‡) at two A residues are indicated. (B) Nucleotide sequence of the BamHI-PstI fragment containing DNA upstream of the tpr gene and the 5′ end of tpr. The transcription start site determined by primer extension (‡‡) is shown at nucleotides 398 to 399. Putative −35 and −10 promoter regions are underlined. Three 17-bp direct repeats in the 5′ region of the transcript are indicated by double underlining. An asterisk at nucleotide 545 indicates the tpr transcription start site when it is expressed in E. coli. The ATG start codon of Tpr at nucleotide 613 is underlined.
RESULTS
Primer extension and sequence analysis of the tpr promoter region.
The 5′ end of tpr mRNA was mapped by primer extension analysis of total RNA isolated from P. gingivalis W83. The primer extension product obtained with tpr293 was much larger than expected, suggesting that the transcription initiation site was more than 200 bp upstream of the tpr coding region (data not shown). Subsequently, using primer tpr64 (Table 2), we identified the transcription initiation site at two A residues 215 bp upstream of the coding region of tpr (Fig. 2). The primer extension product could be detected in P. gingivalis cells grown in 0.5TYE but not in cells grown in BHI. This result was consistent with the results of our previous study showing that P. gingivalis grown in BHI had no detectable tpr mRNA (34).
The nucleotide sequence of the region upstream of tpr was determined and is shown in Fig. 2B. The transcription start site determined by primer extension is labeled. Analysis of the region upstream of the transcription start site found no sequences closely resembling the −35 and −10 regions of E. coli consensus promoters. Interestingly, the region between the transcription initiation site and the tpr coding region contained three identical direct repeats of 17 bp (Fig. 2B).
Analysis of Tpr expression with a tpr::lacZ reporter gene.
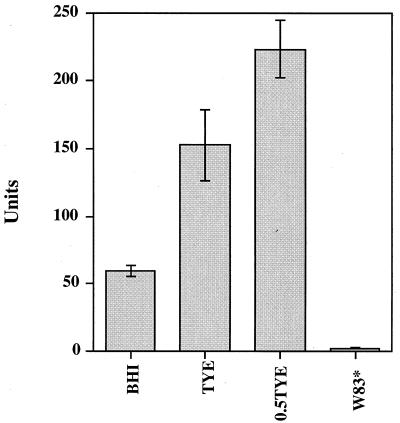
To facilitate analysis of tpr expression in P. gingivalis, we constructed the tpr::lacZ reporter plasmid pNTX-400, a pNJR12 derivative that replicates in P. gingivalis. This plasmid carried the 612-bp upstream region and the first 183 bp of the tpr coding sequence fused to a promoterless lacZ gene (Fig. 1). The lacZ gene lacked the first eight codons of the complete gene and could not be expressed by itself. The lacZ gene was fused in frame to the 5′ end of tpr so that expression of β-galactosidase was controlled by the tpr promoter. Plasmid pNTX-400 was introduced into P. gingivalis W83 by conjugation from an E. coli donor, and β-galactosidase activity was expressed in W83/pNTX-400 (Fig. 3). β-Galactosidase activity could also be detected in W83/pNTX-400 grown on agar plates containing the substrate 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (data not shown). These results indicated that the 612-bp upstream fragment carried sufficient information for initiation of tpr expression.
FIG. 3.
Effects of growth media on β-galactosidase expression in P. gingivalis W83/pNTX-400. Cells were grown in BHI, TYE, or 0.5TYE. W83* is P. gingivalis W83 grown in TYE, which served as the negative control. One unit of β-galactosidase activity is equivalent to 1 nmol of ONPG hydrolyzed min−1 mg of total cellular protein−1.
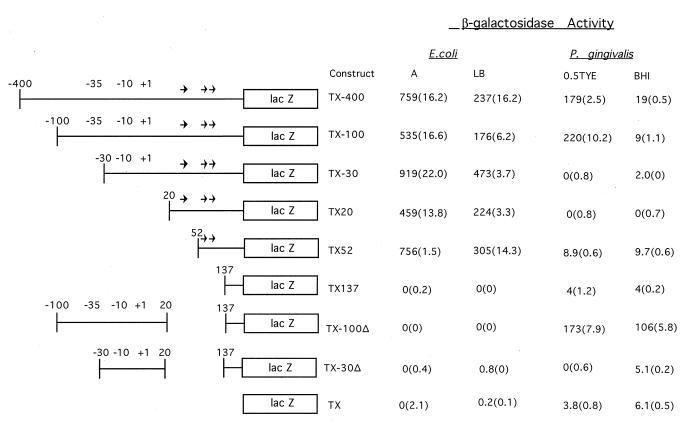
To analyze potential tpr regulatory sequences, we constructed plasmids pNTX-100, pNTX-30, pNTX20, pNTX52, pNTX137, pNTX-100Δ, and pNTX-30Δ (Table 1), all of which could replicate in P. gingivalis. Each plasmid contained a portion of the sequence upstream of tpr and the 5′ end of tpr fused in frame to the promoterless lacZ gene. We then measured β-galactosidase activity in P. gingivalis transconjugants carrying these plasmids (Fig. 4). Further deletions downstream, including deletions of the TX20, TX152, and TX137 constructs carried on pNTX20, pNTX52, and pNTX137, respectively, also resulted in loss of β-galactosidase activity. The promoterless lacZ gene in pNTX, which carried no tpr DNA, expressed minimal levels of β-galactosidase activity that were unaffected by the nutritional conditions tested (Fig. 4). On the other hand, P. gingivalis carrying pNTX-400 or pNTX-100, both of which retained the entire tpr promoter region, expressed much higher levels of β-galactosidase activity in 0.5TYE medium than in BHI (Fig. 4). These results were in agreement both with the results of primer extension analysis showing that the tpr transcript began 215 bp upstream of the coding region and with the regulated expression of tpr in the wild-type W83 (34). Deletion of all or part of the putative P. gingivalis promoter region (pNTX-30 and pNTX20) abolished β-galactosidase activity in P. gingivalis transconjugants. The important role of the three direct repeats in the 5′ region of tpr was suggested by pNTX-100Δ, which contained the putative promoter region but lacked the repeat region. In this construct, high levels of β-galactosidase activity were constitutively expressed, with scant evidence of regulation by growth conditions. This result strongly implicated the three direct repeats in control of tpr expression.
FIG. 4.
β-Galactosidase activities in P. gingivalis and E. coli carrying plasmids with the TX-400 tpr::lacZ construct and various deletion derivatives. Schematic maps of the tpr::lacZ constructs show the tpr locus DNA present in each construct (single line) and lacZ (open box). The locations of the 17-bp direct repeats are indicated by arrows, and putative −35 and −10 regions are shown. β-Galactosidase activities of E. coli grown in minimal A medium (A) or LB medium and of P. gingivalis grown in 0.5TYE or BHI medium are shown as means (standard deviations). β-Galactosidase activity in E. coli is expressed in Miller units. For P. gingivalis, 1 U of β-galactosidase activity is equivalent to hydrolysis of 1 nmol of ONPG min−1 mg of total cellular protein−1.
The effects of the same tpr::lacZ reporter gene constructs were also analyzed in E. coli, with a high-copy-number plasmid vector (Fig. 1). As shown in Fig. 4, β-galactosidase was expressed in E. coli cells carrying the TX-400, TX-100, TX-30, TX20, and TX52 constructs on a multicopy plasmid (Table 1). Expression was not enhanced by addition of isopropyl-1-thio-β-d-galactopyranoside, and it was not affected by the orientation of the insert in the vector (data not shown). Generally, cells grown in minimal A medium had two to three times more β-galactosidase activity than cells grown in LB medium. Control of tpr expression appeared to be quite different in P. gingivalis than in E. coli. The putative tpr promoter regions had no apparent effect on expression in an E. coli background, but deletion of all three 17-bp repeats abolished β-galactosidase activity in E. coli. Primer extension analysis of total RNA from E. coli/pTX-400 total RNA with primer tpr170 (data not shown) indicated that transcription started at a C nucleotide 68 bp upstream of the tpr translation start codon (Fig. 2B). This site, 147 bp downstream of the tpr transcription start site in P. gingivalis, is immediately upstream of the −35 promoter region suggested by Bourgeau et al. (5) and is 24 bp downstream of the last direct repeat sequence preceding tpr. These data show that E. coli RNA polymerase recognized as a transcription promoter a region upstream of tpr that was different from that recognized in P. gingivalis. This finding accounted for the differences observed between β-galactosidase expression in P. gingivalis and that in E. coli strains carrying the same tpr::lacZ fusion construct.
Analysis of a chromosomal tpr::lacZ fusion.
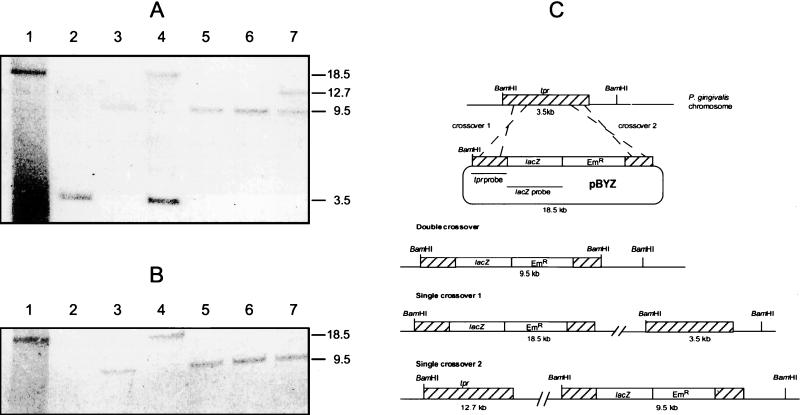
Unlike the multicopy pNTX-400 construct, the tpr gene exists in a single copy in P. gingivalis W83. To more accurately model native expression of the tpr gene, we constructed the suicide vector pBYZ (Table 1) and introduced it into P. gingivalis W83 by conjugation. Since this plasmid cannot replicate in P. gingivalis, the tpr::lacZ construct could be maintained only by integrating it into the P. gingivalis chromosome. Plasmid pBYZ carried the TX-400 tpr::lacZ construct described in the previous section inserted in place of the 5′ end of tpr carried on pBY2-IN (32). As shown in Fig. 5C, homologous recombination between pBYZ and chromosomal DNA resulted in either duplication of tpr (single-crossover event) or allelic exchange of the wild-type tpr gene with the tpr::lacZ reporter gene (double-crossover event). Analysis of a total of 75 Emr and gentamicin-resistant (Gmr) transconjugants indicated that all had lacZ integrated into their chromosomes (data not shown) and that all but three were the result of single-crossover events. Southern blot analysis of both single- and double-crossover constructs is shown in Fig. 5A and B. When P. gingivalis chromosomal DNA was digested with BamHI and probed with a 0.8-kb BamHI-PstI fragment of tpr, a single 9.5-kb hybridizing band was detected in clones 29, 55, and 64, indicating an allelic exchange event resulting in the replacement of the tpr gene with the tpr::lacZ reporter gene construct. In clones 17 and 65, the tpr probe hybridized with 18.5- and 3.5- or 12.7- and 9.5-kb bands, respectively, indicating single-crossover homologous recombination and duplication of tpr.
FIG. 5.
Southern blot analysis of BamHI-digested chromosomal DNA from P. gingivalis W83 strains showing integration of the tpr::lacZ construct at the tpr locus by single- or double-crossover homologous recombination. (A) Southern blot probed with a 0.8-kb BamHI-PstI 5′ fragment of tpr. (B) Southern blot probed with a 3.5-kb lacZ gene. Lanes: 1, plasmid pBYZ; 2, chromosomal DNA from P. gingivalis W83; 3 to 7, chromosomal DNA from P. gingivalis transconjugants 29, 17, 55, 64, and 65, respectively. The positions and sizes in kilobases of hybridizing bands are indicated. (C) Possible single- and double-crossover homologous recombination events in the tpr locus between pBYZ and the P. gingivalis chromosome. The locations of tpr DNA (hatched boxes), the promoterless lacZ reporter gene (open boxes), and the Emr determinant (open boxes) are shown. The locations of the BamHI fragments hybridizing with the tpr and lacZ probes are shown. The numbers below each construct indicate the sizes of the fragments generated by BamHI digestion.
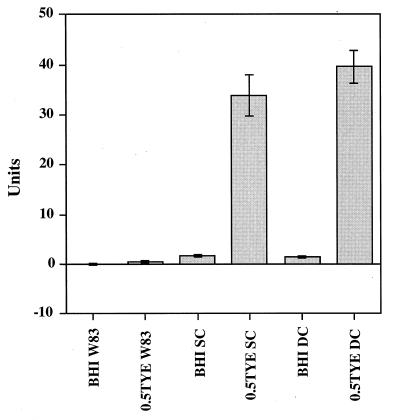
β-Galactosidase activity assays performed on these mutants showed that both single- and double-crossover mutants carrying tpr::lacZ had the same pattern of β-galactosidase activity when they were grown in 0.5TYE as when they were grown in BHI (Fig. 6), and this pattern was similar to that of W83/pNTX-400 (Fig. 3). These data suggest that tpr regulation in these mutants was the same as in the wild-type strain and that expression of tpr does not require the intact tpr gene product.
FIG. 6.
β-Galactosidase activities in P. gingivalis tpr::lacZ chromosomal mutants. P. gingivalis W83, a single-crossover (SC) transconjugant, and a double-crossover (DC) transconjugant (P. gingivalis W83/lacZ) were grown in BHI or 0.5TYE. One unit of β-galactosidase activity is equivalent to hydrolysis of 1 nmol of ONPG min−1 mg of total cellular protein−1.
Nutrient conditions regulate tpr expression.
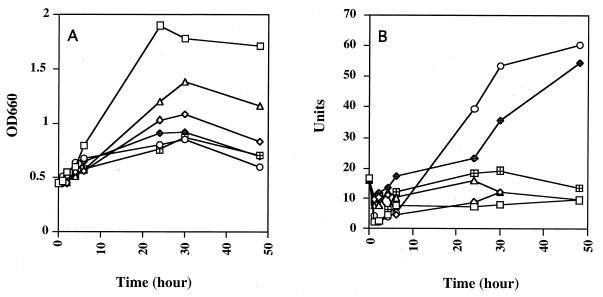
By using the tpr::lacZ reporter gene, we analyzed tpr expression under various nutrient and growth conditions. Clone 55, a tpr::lacZ double-crossover allelic exchange mutant, was designated P. gingivalis W83/lacZ. Results of growth studies and β-galactosidase activity assays with W83/lacZ are shown in Fig. 7. Growth rate and final optical density were highest when W83/lacZ was grown in BHI. The highest β-galactosidase activity in W83/lacZ was found in 0.5TYE at stationary-growth phase. Growth in BHI, TYE, or 0.5TYE supplemented with 1% BSA or 1% gelatin suppressed lacZ expression throughout growth. The pattern of β-galactosidase activity in 0.5TYE supplemented with 1% Casamino Acids was of particular interest. Under these conditions, β-galactosidase activity was low for the first 24 h of incubation and then increased rapidly over the next 24 h until it was at nearly the same level as that of cells grown in 0.5TYE.
FIG. 7.
Growth (A) and β-galactosidase activity (B) of P. gingivalis W83/lacZ. Cells were first grown in BHI to mid-log phase and resuspended in BHI (□); TYE (◊); 0.5TYE (○); or 0.5TYE supplemented with 1% BSA (▵), 1% gelatin (⊞), or 1% Casamino Acids (◊+) and incubated for the time intervals indicated. One unit of β-galactosidase activity is equivalent to hydrolysis of 1 nmol of ONPG min−1 mg of total cellular protein−1. OD600, optical density at 600 nm.
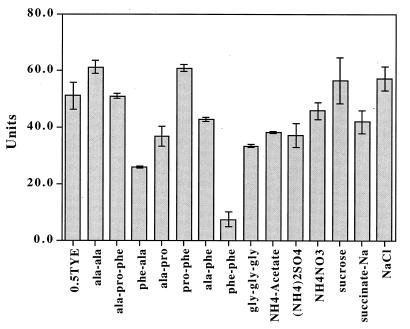
Since β-galactosidase activity in W83/lacZ was greatly suppressed in BHI compared with that in 0.5TYE, we size fractionated BHI and analyzed individual fractions for their effects on β-galactosidase activity. Low-molecular-mass (<700 Da) fractions rich in phenylalanine, proline, and alanine had the most suppressive effect on β-galactosidase activity in W83/lacZ grown in 0.5TYE supplemented with the individual BHI fractions (data not shown). To determine whether tpr expression was related to the presence of free amino acids in the medium, the β-galactosidase activity of P. gingivalis W83/lacZ was assayed in 0.5TYE supplemented with individual l amino acids. The supplementation of 0.5TYE with concentrations of free amino acids up to 5 mM did not significantly reduce β-galactosidase activity, suggesting that tpr expression was not induced by depletion of a single free amino acid (data not shown). The effects of various peptides and chemicals on β-galactosidase activity in W83/lacZ were then tested, and the results are shown in Fig. 8. The dipeptide phenylalanyl-phenylalanine suppressed β-galactosidase activity by approximately 10-fold compared to the activity of cells grown in 0.5TYE without supplementation. The dipeptides phenylalanyl-alanine and alanyl-proline also suppressed β-galactosidase activity to a lesser degree. The potential nitrogen sources ammonium acetate, ammonium nitrate, and ammonium sulfate had no significant effect on tpr expression.
FIG. 8.
Effects of peptides and several chemicals on β-galactosidase expression in P. gingivalis W83/lacZ. Cells were grown in BHI broth to mid-log phase, resuspended in 0.5TYE or 0.5TYE supplemented with 5 mM concentrations of peptides or 40 mM concentrations of other chemicals, and incubated overnight, after which β-galactosidase activity was analyzed. One unit of β-galactosidase activity is equivalent to hydrolysis of 1 nmol of ONPG min−1 mg of total cellular protein−1. ala-ala, alanine-alanine; ala-pro-phe, alanine-proline-phenylalanine; phe-ala, phenylalanine-alanine; ala-pro, alanine-proline; pro-phe, proline-phenylalanine; ala-phe, alanine-phenylalanine; phe-phe, phenylalanyl-phenylalanine; gly-gly-gly, glycine-glycine-glycine.
Effects of other environmental conditions on tpr expression.
To analyze whether tpr expression was influenced by heat shock or other stress conditions, β-galactosidase activity was measured in W83/lacZ cells which had been heat shocked, incubated at pH 5.5, incubated at pH 8.5, or hemin starved. The results suggested that tpr expression was not affected by pH changes or hemin starvation and was suppressed by approximately 25% by heat shock at 42°C (data not shown). Succinate, which can replace hemin as a required growth factor (27), had no significant effect on tpr expression.
DISCUSSION
The present study represents an initial attempt to characterize regulation of a potential virulence factor of P. gingivalis. There have been several reports stating that P. gingivalis proteases are regulated by the growth environment (6, 25, 28, 38), but little is known about how this regulation is achieved at the genetic level. Our previous report that tpr proteolytic activity (Pz-peptidase) was influenced by growth conditions made this an inviting model for investigating gene regulation in this organism (34). In that study, Northern blot analysis suggested that tpr expression was regulated by nutritional conditions and that this regulation occurred at the transcriptional level.
Primer extension analysis identified the tpr transcription initiation site, and DNA sequencing revealed three direct repeats in the 5′ noncoding region of the transcript that appear to act as regulatory elements. Initial primer extension reactions were carried out with primer tpr293 (Table 2). This primer would have been appropriate if the promoter sequence proposed by Bourgeau et al. (5) were correct. The location of the transcription start site 215 bp upstream of the tpr coding region may not be unusual for this organism. In the few studies of transcription in P. gingivalis, the transcription start sites were located similar distances upstream of the coding regions (18, 22). Further studies are needed for a better understanding of promoter structure in P. gingivalis and to determine whether promoters similar to that of tpr are a common feature in this organism. There have been a number of reports of expression of cloned P. gingivalis genes in E. coli. Some of these cloned genes were proposed to be transcribed from their own promoters, and regions homologous to E. coli promoters were identified (5, 7, 8, 16, 17, 31, 36). On the other hand, Klimpel and Clark, using antisera to the E. coli RNA polymerase core enzyme and ς70, found no proteins in P. gingivalis extracts that cross-reacted with either antiserum (19). This result suggested that E. coli and P. gingivalis RNA polymerases may be significantly different, which is supported by our data showing that E. coli and P. gingivalis RNA polymerases initiate tpr transcription at different sites. Thus, these data underscore the importance of analyzing gene expression in native systems and the limitations of predicting gene structure based on canonical E. coli studies.
P. gingivalis, which produces numerous membrane-associated and secreted proteases, has the ability to degrade proteins to short peptides and transport them into the cell for metabolism (10). Our results suggest that the availability of these peptides regulates expression of at least one of these proteases. A variety of nutrients, including those found in complex growth media as well as BSA, gelatin, and Casamino Acids, could suppress tpr expression. Of particular interest were results of a time course growth and expression study of the effects of supplementation with Casamino Acids (Fig. 7). Under these conditions, transcription of tpr, represented by β-galactosidase activity, was initially suppressed. However, after 48 h of incubation, tpr transcription increased to the level seen in cells grown in 0.5TYE without supplementation. Casamino Acids is an acid hydrolysate of casein in which, according to the manufacturer, free amino acids and small peptides are present in a ratio of 82 to 18%, respectively. This ratio suggests that the initial suppressive effect on tpr expression was due to the peptide component of the hydrolysate. Once the peptides were exhausted, transcription of tpr (and β-galactosidase activity) increased. While none of the 20 essential amino acids had a significant effect, this fact does not rule out the possibility that tpr expression might be influenced by combinations of free amino acids. Considered together, these results suggested that peptides, rather than free amino acids, regulate tpr expression.
The effects of allelic exchange and tpr gene duplication on β-galactosidase expression indicated that the Tpr protein is not involved in its own regulation. Our results also showed that a plasmid-borne tpr::lacZ fusion and a chromosomally integrated tpr::lacZ fusion exhibited the same regulation pattern, suggesting that shuttle vectors can be used to analyze the effects of 5′ deletion mutations on tpr expression. We observed higher levels of tpr expression in P. gingivalis cells in stationary-growth phase than in cells in logarithmic-growth phase. This is similar to the rpoS regulon in which 30 or more genes are expressed in response to starvation and during the transition to stationary phase (14). Proteins in this regulon can enhance long-term survival in nutrient-deficient medium and have diverse functions, including protection against DNA damage, determination of morphological changes, mediation of virulence, osmoprotection, and thermotolerance. Differential levels of expression of families of genes within this regulon are affected by supplementary regulatory factors, working individually and in combination to modulate activities of different promoters. At present, it is not known whether a similar regulon exists in P. gingivalis or whether expression of genes other than tpr is also regulated by the availability of peptides.
The identity of the factor(s) that controls tpr expression has not yet been determined. However, our results suggest that a short peptide or peptides containing phenylalanine are responsible. Low-molecular-weight fractions of BHI suppressed tpr expression to various extents. Analysis of BHI showed that a low-molecular-weight fraction rich in phenylalanine, alanine, and proline had the most suppressive effect on tpr expression as indicated by β-galactosidase activity. Attempts to further characterize this fraction in order to identify the specific factor that suppressed tpr expression were not successful, probably due to the inability to obtain sufficient quantities of individual peptides. When it was used to supplement 0.5TYE, the dipeptide phenylalanyl-phenylalanine had the most effect on tpr expression, as measured by β-galactosidase activity. Supplementation with other peptides and some chemicals and challenge by heat shock, pH change, or hemin limitation had little or no effect on tpr expression.
In an analogous system, Marugg et al. showed that the PrtP and PrtM proteases of Lactococcus lactis were regulated at the transcriptional level by leucine-containing peptides but not by free amino acids (26). Peptide content of the growth media had no effect on transcription of prtP and prtM in an Opp− strain of L. lactis, indicating that peptide uptake was required for this regulation to take place. In P. gingivalis, acquisition of peptide nutrients is certain to be an extremely important process. At present, the molecular mechanisms of peptide uptake in P. gingivalis are not known. Further studies are required to address this issue as well as to determine the specific molecular factors that control tpr transcription.
The actual mechanism of tpr regulation remains unclear, but our results are suggestive of the involvement of specific DNA binding factors. We identified three identical direct repeats between the transcription start site and the coding region of tpr. Deletion of these repeats abolished nutrient-dependent tpr regulation in P. gingivalis, even though the promoter region remained intact. The potential involvement of direct repeats in gene regulation has been reported in other studies. The regulatory region of the torCAD operon of E. coli contains four decameric direct repeats. These repeats, designated tor boxes, were found to be the targets of TorR, which regulates torC expression (41). In P. gingivalis, four direct repeats of 41 bp were identified upstream of the hagB gene (37). Transcription of hagB was greatly reduced when cells were grown in the absence of hemin, suggesting a possible regulatory role for these repeats (20). Three 12-bp direct repeats, tentatively proposed as transcription termination attenuators, were found within the putative transcription termination region of the prtT protease gene of P. gingivalis (22). The ospD genes of various Lyme disease-associated Borrelia spp. are preceded by between 1 and 12 copies of a 17-bp direct repeat that contains a potential −35 promoter sequence (24). While the functions of the direct repeats listed above are not known at this time, they may represent binding sites for specific regulatory proteins.
Most DNA binding proteins that act as transcription factors bind to the 5′ region of the promoter to exert effects on RNA transcription initiation. The locations of the three direct repeats within the untranslated region of the tpr transcript indicated that they are not involved in the initiation of transcription. However, if the three direct repeats are a regulatory protein binding site, they may influence tpr mRNA synthesis at the transcript elongation and termination stages. The genes in the TyrR regulon of E. coli are regulated by the TyrR protein, whose binding site is located downstream of the putative RNA polymerase binding site (35). He and Zalkin (13) found that the operator (PurR binding site) of the purB gene of E. coli was 242 bp downstream of the transcriptional start site and overlapped codons 62 to 67 in the structural gene. PurR-mediated repression of purB occurred by a transcriptional “roadblock” mechanism, and they identified a truncated purB mRNA species in a Northern blot (13). In our system, such a truncated mRNA would be extremely difficult to detect due to its small size. Furthermore, our Northern blot analysis of W83/PM, a tpr-deficient mutant, suggested that truncated tpr mRNA was unstable (34).
Another possible role of the 17-bp direct repeat in the tpr locus may be in site-specific genetic recombination, which can contribute to both genomic plasticity and antigenic variability. The genes encoding the major cysteine proteases and hemagglutinins of P. gingivalis contain large direct repeat regions that appear to contribute to such recombination-based heterogeneity in this gene family (4). The Tpr protease is distinct from this group of enzymes, and there have as yet been no studies of its conservation in different strains. There are 17-bp direct repeats associated with the gene encoding the highly variable surface-expressed VlsE protein of Borellia burgdorferi. The vlsE gene contains a cassette region flanked by 17-bp direct repeats. Recombination between the cassette and up to 15 silent cassette sequences resulted in antigenic variation of the VlsE protein (44).
The mechanism(s) by which nutrients regulate tpr expression in P. gingivalis W83 remains to be determined. Our current understanding of tpr expression is as follows. Transcription of the tpr gene begins 215 bp upstream of the coding region. Regulation appears to be at the transcript elongation and termination stages. A regulatory trans-acting factor may directly or indirectly sense the presence of certain short, phenylalanine-containing peptides and act on the three direct repeats to modulate tpr expression. Future studies will focus on the overall significance of Tpr among the array of proteolytic enzymes of this organism and on the specific role of Tpr in processing and acquisition of essential peptide nutrients.
ACKNOWLEDGMENTS
We gratefully acknowledge the assistance of Wen Luo. We also thank Christopher Fenno for assistance in preparation of the manuscript and Pauline Hannam for comments and suggestions throughout this study.
This study was supported by the Medical Research Council of Canada.
REFERENCES
- 1.Adams C W, Forrest M E, Cohen S N, Beatty J T. Structural and functional analysis of transcriptional control of the Rhodobacter capsulatus puf operon. J Bacteriol. 1989;171:473–482. doi: 10.1128/jb.171.1.473-482.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aduse-Opoku J, Muir J, Slaney J M, Rangarajan M, Curtis M A. Characterization, genetic analysis, and expression of a protease antigen (PrpRI) of Porphyromonas gingivalis W50. Infect Immun. 1995;63:4744–4754. doi: 10.1128/iai.63.12.4744-4754.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F A, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene Publishing and Wiley Interscience; 1993. [Google Scholar]
- 4.Barkocy-Gallagher G A, Han N, Patti J M, Whitlock J, Progulske-Fox A, Lantz M S. Analysis of the prtP gene encoding porphypain, a cysteine proteinase of Porphyromonas gingivalis. J Bacteriol. 1996;178:2734–2741. doi: 10.1128/jb.178.10.2734-2741.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourgeau G, Lapointe H, Péloquin P, Mayrand D. Cloning, expression, and sequencing of a protease gene (tpr) from Porphyromonas gingivalis W83 in Escherichia coli. Infect Immun. 1992;60:3186–3192. doi: 10.1128/iai.60.8.3186-3192.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carman R J, Ramakrishnan M D, Harper R H. Hemin levels in culture medium of Porphyromonas (Bacteroides) gingivalis regulate both hemin binding and trypsinlike protease production. Infect Immun. 1990;58:4016–4019. doi: 10.1128/iai.58.12.4016-4019.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi J-I, Takahashi N, Kato T, Kuramitsu H K. Isolation, expression, and nucleotide sequence of the sod gene from Porphyromonas gingivalis. Infect Immun. 1991;59:1564–1566. doi: 10.1128/iai.59.4.1564-1566.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dickinson D P, Kubiniec M A, Yoshimura F, Genco R J. Molecular cloning and sequencing of the gene encoding the fimbrial subunit protein of Bacteroides gingivalis. J Bacteriol. 1988;170:1658–1665. doi: 10.1128/jb.170.4.1658-1665.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fletcher H M, Schenkein H A, Macrina F L. Cloning and characterization of a new protease gene (prtH) from Porphyromonas gingivalis. Infect Immun. 1994;62:4279–4286. doi: 10.1128/iai.62.10.4279-4286.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grenier D, Mayrand D. Proteinases. In: Shah H N, Mayrand D, Genco R J, editors. Biology of the species Porphyromonas gingivalis. Boca Raton, Fla: CRC Press, Inc.; 1993. pp. 227–243. [Google Scholar]
- 11.Grenier D, Mayrand D, McBride B C. Further studies on the degradation of immunoglobulins by black-pigmented Bacteroides. Oral Microbiol Immunol. 1989;4:12–18. doi: 10.1111/j.1399-302x.1989.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 12.Grenier D, McBride B C. Isolation of a membrane-associated Bacteroides gingivalis glycylprolyl protease. Infect Immun. 1987;55:3131–3136. doi: 10.1128/iai.55.12.3131-3136.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He B, Zalkin H. Repression of Escherichia coli purB is by a transcriptional roadblock mechanism. J Bacteriol. 1992;174:7121–7127. doi: 10.1128/jb.174.22.7121-7127.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hengge-Aronis R. Survival of hunger and stress: the role of rpoS in early stationary phase gene regulation in E. coli. Cell. 1993;72:165–168. doi: 10.1016/0092-8674(93)90655-a. [DOI] [PubMed] [Google Scholar]
- 15.Holt S C, Bramanti T E. Factors in virulence expression and their role in periodontal disease pathogenesis. Crit Rev Oral Biol Med. 1991;2:177–281. doi: 10.1177/10454411910020020301. [DOI] [PubMed] [Google Scholar]
- 16.Jackson C A, Kirszbaum L, Dashper S, Reynolds E C. Cloning, expression and sequence analysis of the genes encoding the heterodimeric methylmalonyl-CoA mutase of Porphyromonas gingivalis W50. Gene. 1995;167:127–132. doi: 10.1016/0378-1119(95)00682-6. [DOI] [PubMed] [Google Scholar]
- 17.Joe A, Murray C S, McBride B C. Nucleotide sequence of a Porphyromonas gingivalis gene encoding a surface-associated glutamate dehydrogenase and construction of a glutamate dehydrogenase-deficient isogenic mutant. Infect Immun. 1994;62:1358–1368. doi: 10.1128/iai.62.4.1358-1368.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karunakaran T, Madden T, Kuramitsu H. Isolation and characterization of a hemin-regulated gene, hemR, from Porphyromonas gingivalis. J Bacteriol. 1997;179:1898–1908. doi: 10.1128/jb.179.6.1898-1908.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klimpel K W, Clark V L. The RNA polymerase of Porphyromonas gingivalis and Fusobacterium nucleatum are unrelated to the RNA polymerase of Escherichia coli. J Dent Res. 1990;69:1567–1572. doi: 10.1177/00220345900690090601. [DOI] [PubMed] [Google Scholar]
- 20.Lepine G, Progulske-Fox A. Duplication and differential expression of hemagglutinin genes in Porphyromonas gingivalis. Oral Microbiol Immunol. 1996;11:65–78. doi: 10.1111/j.1399-302x.1996.tb00339.x. [DOI] [PubMed] [Google Scholar]
- 21.Lu B, McBride B C. Stress response of Porphyromonas gingivalis. Oral Microbiol Immunol. 1994;9:166–173. doi: 10.1111/j.1399-302x.1994.tb00054.x. [DOI] [PubMed] [Google Scholar]
- 22.Madden T E, Clark V L, Kuramitsu H K. Revised sequence of the Porphyromonas gingivalis PrtT cysteine protease/hemagglutinin gene: homology with streptococcal pyrogenic exotoxin B/streptococcal proteinase. Infect Immun. 1995;63:238–247. doi: 10.1128/iai.63.1.238-247.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maley J, Shoemaker N B, Roberts I S. The introduction of colonic-Bacteroides shuttle plasmids into Porphyromonas gingivalis: identification of a putative P. gingivalis insertion-sequence element. FEMS Microbiol Lett. 1992;93:75–82. doi: 10.1016/0378-1097(92)90492-7. [DOI] [PubMed] [Google Scholar]
- 24.Marconi R T, Samuels D S, Landry R K, Garon C F. Analysis of the distribution and molecular heterogeneity of the ospD gene among the Lyme disease spirochetes: evidence for lateral gene exchange. J Bacteriol. 1994;176:4572–4582. doi: 10.1128/jb.176.15.4572-4582.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marsh P D, McKee A S, McDermid A S. Effect of haemin on enzyme activity and cytotoxin production by Bacteroides gingivalis W50. FEMS Microbiol Lett. 1988;55:87–92. [Google Scholar]
- 26.Marugg J D, Meijer W, van Kranenburg R, Laverman P, Bruinenberg P G, de Vos W M. Medium-dependent regulation of proteinase gene expression in Lactococcus lactis: control of transcription initiation by specific dipeptides. J Bacteriol. 1995;177:2982–2989. doi: 10.1128/jb.177.11.2982-2989.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mayrand D, McBride B C. Ecological relationships of bacteria involved in a simple, mixed anaerobic infection. Infect Immun. 1980;27:44–50. doi: 10.1128/iai.27.1.44-50.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McDermid A S, McKee A S, Marsh P D. Effect of environmental pH on enzyme activity and growth of Bacteroides gingivalis W50. Infect Immun. 1988;56:1096–1100. doi: 10.1128/iai.56.5.1096-1100.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 30.Minhas T, Greenman J, Schaffer A G. Effect of mucin, haemoglobin and collagen on the maximum specific growth rate, biomass and hydrolytic enzyme production of Porphyromonas gingivalis in continuous culture. Microb Ecol Health Dis. 1991;4:311. [Google Scholar]
- 31.Nakayama K. The superoxide dismutase-encoding gene of the obligately anaerobic bacterium Bacteroides gingivalis. Gene. 1990;96:149–150. doi: 10.1016/0378-1119(90)90357-w. [DOI] [PubMed] [Google Scholar]
- 32.Park Y, McBride B C. Characterization of the tpr gene product and isolation of a specific protease-deficient mutant of Porphyromonas gingivalis W83. Infect Immun. 1993;61:4139–4146. doi: 10.1128/iai.61.10.4139-4146.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park Y, McBride B C. Cloning of a Porphyromonas (Bacteroides) gingivalis protease and characterization of its product. FEMS Microbiol Lett. 1992;92:273–278. doi: 10.1016/0378-1097(92)90721-y. [DOI] [PubMed] [Google Scholar]
- 34.Park Y S, Lu B, Mazur C, McBride B C. Inducible expression of a Porphyromonas gingivalis W83 membrane-associated protease. Infect Immun. 1997;65:1101–1104. doi: 10.1128/iai.65.3.1101-1104.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pittard A J, Davidson B E. TyrR protein of Escherichia coli and its role as repressor and activator. Mol Microbiol. 1991;5:1585–1592. doi: 10.1111/j.1365-2958.1991.tb01904.x. [DOI] [PubMed] [Google Scholar]
- 36.Progulske-Fox A, Tumwasorn S, Holt S C. The expression and function of a Bacteroides gingivalis hemagglutinin gene in Escherichia coli. Oral Microbiol Immunol. 1989;4:121–131. doi: 10.1111/j.1399-302x.1989.tb00238.x. [DOI] [PubMed] [Google Scholar]
- 37.Progulske-Fox A, Tumwasorn S, Lepine G, Whitlock J, Savett D, Ferretti J J, Banas J A. The cloning, expression and sequence analysis of a second Porphyromonas gingivalis gene that codes for a protein involved in hemagglutination. Oral Microbiol Immunol. 1995;10:311–318. doi: 10.1111/j.1399-302x.1995.tb00160.x. [DOI] [PubMed] [Google Scholar]
- 38.Robertson P B, Lantz M, Marucha P T, Kornman K S, Trummel C L, Holt S C. Collagenolytic activity associated with Bacteroides species and Actinobacillus actinomycetemcomitans. J Periodontal Res. 1982;17:275–283. doi: 10.1111/j.1600-0765.1982.tb01154.x. [DOI] [PubMed] [Google Scholar]
- 39.Shah H N, Gharbia S E. Batch culture and physiological properties. In: Shah H N, Mayrand D, Genco R J, editors. Biology of the species Porphyromonas gingivalis. Boca Raton, Fla: CRC Press, Inc.; 1993. pp. 85–103. [Google Scholar]
- 40.Shoemaker N B, Guthrie E P, Salyers A A, Gardner J F. Evidence that the clindamycin-erythromycin resistance gene of Bacteroides plasmid pBF4 is on a transposable element. J Bacteriol. 1985;162:626–632. doi: 10.1128/jb.162.2.626-632.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simon G, Jourlin C, Ansaldi M, Pascal M C, Chippaux M, Mejean V. Binding of the TorR regulator to cis-acting direct repeats activates tor operon expression. Mol Microbiol. 1995;17:971–980. doi: 10.1111/j.1365-2958.1995.mmi_17050971.x. [DOI] [PubMed] [Google Scholar]
- 42.Smalley J W, Birss A J, McKee A S, Marsh P D. Haemin-restriction influences haemin-binding, haemagglutination and protease activity of cells and extracellular membrane vesicles of Porphyromonas gingivalis W50. FEMS Microbiol Lett. 1991;90:63–68. doi: 10.1016/0378-1097(91)90647-s. [DOI] [PubMed] [Google Scholar]
- 43.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 44.Zhang J R, Hardham J M, Barbour A G, Norris S J. Antigenic variation in Lyme disease borreliae by promiscuous recombination of VMP-like sequence cassettes. Cell. 1997;89:275–285. doi: 10.1016/s0092-8674(00)80206-8. [DOI] [PubMed] [Google Scholar]