Abstract
Apoptosis is a key phenomenon in the regulation of the life span of terminally differentiated leukocytes. Human gingiva represents an established model to study immune responses to bacterial infection. In this investigation, we used the TUNEL (terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling) technique to evaluate presence and topographic location of apoptosis-associated DNA damage in human gingival biopsies along with the expression of the p53 and Bcl-2 apoptosis-regulating proteins. Qualitative data analysis showed high densities of cells expressing DNA damage and p53 both within the epithelial attachment to the tooth and in the perivascular infiltrate (infiltrated connective tissue [ICT]) immediately underlying the site of chronic bacterial aggression. Topographic consistency between DNA damage- and p53-positive cells was consistently observed. Quantitative analysis of the ICT showed mean densities of DNA damage- and p53-positive cells of 345 ± 278 and 403 ± 182 cells/mm2, respectively. Numerical consistency was confirmed by multivariate regression analysis: densities of DNA damage-positive cells were significantly predicted by densities of p53-positive cells (P = 0.001, r2 = 0.84). In the ICT, cells displaying biotinylated DNA nicks were 3.8% ± 2.7% of total cellularity, while p53- and Bcl-2-positive cells represented 4.4% ± 1.7% and 15.4% ± 6.7% of total cells, respectively. It is suggested that p53 expression associated with DNA damage is a prevalent phenomenon in chronically inflamed human gingiva, and that apoptosis may be a relevant process for the maintenance of local immune homeostasis at sites of chronic bacterial challenge in vivo.
Superficial periodontal tissues are constantly exposed to a mixed anaerobic gram-negative flora which can induce inflammatory responses leading to destruction of the tooth-supporting apparatus, i.e., periodontal diseases. Preservation of periodontal health is thus dependent on the establishment and the maintenance over time of a local host-bacterium equilibrium (9). In this respect, the existence of a high rate of epithelial cell turnover and a highly regulated local immune response are thought to concur in limiting the penetration of pathogenic microorganisms into the gingival tissues (8, 9, 27). Recent investigations have indicated that the recruitment of inflammatory cells at this site of bacterial challenge is dependent on the selective activation of subepithelial capillary loops to express specific leukocyte adhesion molecules (26, 41). Following diapedesis, polymorphonuclear leukocytes migrate into the gingival junctional epithelium along gradients of chemotactic and haptotactic molecules to reach the front of bacterial challenge (43, 44). The majority of mononuclear cells, on the other hand, enter the perivascular connective tissue to form an inflammatory infiltrate composed mainly of a specific subset of T cells, B cells, and macrophages (37, 42, 45). Despite the chronic nature of the bacterial stimuli, in a healthy person, the size of the inflammatory infiltrate has been shown to remain fairly constant over the life of the individual (28, 35, 36). A specific mechanism(s) should therefore account for the observed stability in the size of the inflammatory infiltrate in spite of the continuous influx of leukocytes. One such mechanism may be programmed cell death of tissue-infiltrating leukocytes.
Apoptosis is a programmed form of cell death which results in the elimination of specific cells without disturbance of tissue structure or function (3, 17, 48). It is implicated in a wide variety of biological phenomena, including inflammatory responses (3, 12).
The apoptotic process can be modulated by various stimuli, including hormones, cytokines, growth factors, bacterial or viral infections, and immune responses. Among other factors, the products of two genes, those encoding the p53 and the Bcl-2 proteins, have been shown to play a fundamental regulatory role in this process (18, 49). The Bcl-2 protein can prevent or markedly reduce cell death induced by a wide variety of stimuli (31). Under physiologic conditions, Bcl-2 expression seems to be associated with a pool of less differentiated cells and with cells undergoing terminal differentiation. In these cells, Bcl-2 will prevent apoptotic cell death and thus play a pivotal role in tissue development, cell maturation, and terminal differentiation (13, 20). Conversely, the p53 tumor suppressor gene, whose expression can induce apoptosis, has been implicated in almost all forms of inhibition of cell replication (19). Its expression has been found to be essential for the apoptotic response to the accumulation of DNA damage. p53 expression is also implicated in the regulation of tissue dynamics via its induction of apoptosis in terminally differentiated cells, including inflammatory cells (30).
Emerging evidence indicates that bacterium-modulated apoptosis appears to be an important phenomenon in the pathogenesis of infectious diseases (2). Specific pathogens or their exocellular products may directly induce apoptosis of host cells (23, 51). Conversely, phagocytosis of bacteria or exposure to bacterial components such as lipopolysaccharide may delay programmed cell death of terminally differentiated polymorphonuclear leukocytes (PMN) (1, 4).
The aim of this investigation was to evaluate in situ the presence of cells with apoptosis-associated DNA breaks in the marginal portion of healthy human gingiva. Furthermore, we compared the distribution of cells with damaged DNA with the topographic location of cells expressing the Bcl-2 and p53 apoptosis-regulating proteins.
MATERIALS AND METHODS
Reagents.
Terminal deoxynucleotidyltransferase (TdT; EC 2.7.7.31), DNase I (EC 3.1.21.1), and biotinylated dUTP (biotin-16-dUTP, a dUTP analogue carrying a biotin molecule linked via a 16-C spacer arm to the 5 position of the pyrimidine ring) were obtained from Boehringer GmbH (Mannheim, Germany). The Bcl-2/124 monoclonal antibody (mouse immunoglobulin G1 [IgG1]) was kindly provided by David Mason (Radcliffe Hospital, Oxford, England) (29). The PAb240 and PAb248 anti-wild-type p53 protein monoclonal antibodies (mouse IgG) were the generous gift of David Lane (University of Dundee, Dundee, Scotland) (30). Monoclonal antibodies reacting with the Ki67 proliferating cell nuclear antigen and with bromodeoxyuridine (clone BMC 9318) were from Boehringer. The T3-4B5 anti-CD3 antigen, MT310 anti-CD4, DK25 anti-CD8, 4KB128 anti-CD22, UCHL1 anti-CD45RO, My31 anti-CD56, EBM11 anti-CD68, and NP57 anti-PMN elastase monoclonal antibodies were obtained from Dakopatts AB (Roskilde, Denmark). The L48 anti-CD45RA monoclonal antibody was obtained from Becton Dickinson (Mountain View, Calif.). Horse anti-mouse IgG biotinylated secondary antibody, horse preimmune serum, and avidin-biotin-horseradish peroxidase macromolecular complexes (Vectastain ABC Elite kit) were from Vector Laboratories (Burlingame, Calif.). Rabbit anti-mouse Ig antibodies, alkaline phosphatase–anti-alkaline phosphatase (APAAP) complexes, and the new fuchsin substrate kit were obtained from Dakopatts.
Clinical specimens.
After informed consent was obtained, soft tissue gingival biopsy samples were taken from systemically healthy volunteers who were taking no medications. Subjects were free from periodontal disease as determined by the absence of clinical attachment loss or increased probing pocket depths. Specimens were harvested from sites without clinically detectable inflammation and without visible bacterial plaque. Soft tissue biopsy specimens of the junctional epithelium, the sulcular epithelium, part of the orogingival epithelium, and a portion of the supracrestal connective tissues were harvested as previously described (46). Specimens were immediately embedded (tissue freezing medium; Jung, Nussloch, Germany) and snap-frozen in liquid nitrogen slurry. Cryostat sections (6 μm thick) were obtained, briefly prefixed in acetone, and stored desiccated at −70°C until use.
DNA nick end labeling of tissue sections.
The presence of cell death-associated DNA fragmentation was assessed in situ by terminal TdT-mediated dUTP-biotin nick end labeling (TUNEL) essentially as described previously (7). After fixation in acetone for 15 min at 4°C, nonspecific tissue binding of biotin was blocked by sequential incubation with avidin and biotin blocking solutions (blocking kit; Vector Laboratories). All rinsing steps were performed with 15 mM phosphate-buffered saline (PBS), pH 7.2. Following preincubation for 10 min in TdT buffer (30 mM Tris-HCl [pH 7.2], 140 mM sodium cacodylate, 1 mM cobalt chloride) at room temperature, tissue sections were incubated for 60 min at 37°C in a humidified chamber with 30 μl of TdT buffer containing 0.4 U of TdT per μl and 4 μM biotinylated-dUTP (10). Optimal TdT and biotin-16-dUTP concentrations had been previously determined on human tonsil sections. Incorporation of biotin-16-dUTP at the 3′ end of DNA breaks was stopped by incubation for 15 min in 2× SSC buffer (300 mM sodium chloride, 30 mM sodium citrate) at room temperature. Sections were then covered with 2% bovine serum albumin in PBS for 10 min at room temperature to decrease nonspecific binding. After exposure to 3% H2O2 in methanol for 30 min to block endogenous peroxidase activity, specimens were incubated for 60 min with preformed biotin-avidin-horseradish peroxidase macromolecular complexes to detect the biotinylated-dUTP incorporated at the 3′ end of the DNA breaks. A black color was developed by exposure for 6 to 8 min to 0.5 mg of the chromogen 3′,3′-diaminobenzidine tetrahydrochloride per ml in the presence of H2O2 and nickel ions (15). Sections were lightly counterstained with methyl green, dehydrated, and permanently mounted.
For each biopsy, one positive control, three experimental slides, and one negative control were run in the same experiment. Positive controls with DNA breaks were obtained by incubating the specimens with DNase I (1 μg/ml) for 10 min at room temperature before incubation with TdT (7). For negative controls, TdT was omitted during incubation.
Immunocytochemistry.
Immunohistochemical staining was performed essentially as previously described (43). Sections were fixed for 15 min in acetone at 4°C. All rinsing steps were performed with 15 mM PBS (pH 7.2) for the detection of Bcl-2 and with 0.9% NaCl for the detection of p53 and Ki67. A standard three-stage immunoperoxidase ABC technique was used to detect Bcl-2 antigens (14) CD22, CD3, CD4, CD8, CD45RA, CD45RO, CD56, and PMN elastase, while a five-stage technique was used for visualization of the p53 protein. In brief, nonspecific tissue binding was blocked by incubation with 1.5% horse serum for 30 min. Optimal primary antibody titers had been previously determined on human tonsil sections. Specimens were incubated for 60 min with 40 μl of diluted primary antibody. Biotinylated horse anti-mouse IgG secondary antibodies were used at a 1:200 dilution for 30 min. Endogenous peroxidase activity was quenched by exposure for 30 min to 0.3% H2O2 in methanol. Sections were then incubated for 45 min with preformed avidin biotin-horseradish peroxidase macromolecular complexes. A black color was developed by exposure for 6 to 8 min to 0.5 mg of the chromogen 3′,3′-diaminobenzidine tetrahydrochloride per ml and 0.01% H2O2 in the presence of nickel ions (15). Sections where lightly counterstained with methyl green, dehydrated, and permanently mounted. Primary antibodies against Ki67 and CD68 antigens were detected by an APAAP technique (5). Sections were incubated for 30 min with a 1:25 dilution of rabbit anti-mouse Ig antibodies (Dakopatts), washed, and exposed for 30 min to a 1:50 dilution of APAAP complexes (Dakopatts). Color was developed with the new fuchsin substrate kit (Dakopatts) according to the recommended standard procedure in the presence of 4 mM levamisole to inhibit endogenous alkaline phosphatase. Slides were counterstained with hematoxylin and permanently mounted in Aquatex (Merck).
Controls.
Positive and negative controls were processed with each series. Standard sections of human tonsils were used as positive controls. The irrelevant antibromodeoxyuridine monoclonal antibody was used as a negative control. Specificity and sensitivity of each immunohistochemical staining were determined by comparison to the appropriate positive and negative controls included in each series. Sections with unsatisfactory signal-to-noise ratio were excluded from subsequent analysis, and the staining was repeated.
Data analysis.
The topographic distribution of cells positive for the presence of DNA breaks and of Bcl-2, p53, and Ki67 proteins was comparatively assessed in the junctional epithelium, sulcular epithelium, oral epithelium, and infiltrated connective tissue using consecutive section series. The degree of histologic inflammation in the connective tissue under the junctional epithelium was evaluated on hematoxylin-and-eosin-stained sections and assigned a Tagge inflammation score (38). Quantitative data analysis was performed with a computerized image analysis system. Images were acquired with a cooled CF8 RCC camera (KAPPA, Gleichen, Germany) and grabbed with a modular frame grabber (Imaging Technology Inc., Bedford, Mass.) installed on a personal computer. The digitized image was analyzed with customized BioScan OPTIMAS software (BioScan Inc., Edmonds, Wash.). After calibration of the system, the surface densities of inflammatory cells and marker-positive cells were evaluated within the defined infiltrated connective tissue (ICT) area at an original magnification of ×250. In this area, a standard lattice was overlaid on the computer screen, and marker-positive cells were identified by comparison to both positive and negative controls and then counted. Data were expressed as numbers of positive cells per millimeter squared and plotted as box plots (47). The association between the number of TUNEL-, p53-, and Bcl-2-positive cells in the perivascular inflammatory infiltrate was evaluated by regression analysis. The tested model predicted the dependent variable (the number of TUNEL-positive cells per area unit) as a function of the number of the cells expressing the apoptosis-regulating proteins p53 and Bcl-2 in the same surface area. Statistical analysis was performed with the SAS GLM procedure (Statistical Application Software version 6.09; SAS Institute, Cary, N.C.).
RESULTS
Degree of histologic inflammation and characterization of the infiltrate.
The presence of an inflammatory infiltrate was consistently detected in all specimens. The infiltrate was located in a perivascular position subjacent to the junctional epithelium, near the expected site of bacterial plaque aggression. All specimens demonstrated a Tagge inflammation score of 1, indicating the presence of a well-defined inflammatory lesion that did not extend deeply into the gingival connective tissue. The cell density within the inflammatory lesion was 8,764 ± 2,934 cells/mm2. CD3-, CD4-, CD8-, CD22-, CD45RA-, CD45RO-, CD56-, CD68-, and PMN elastase-positive cells were all observed in the ICT. Table 1 presents the observed densities of cells positive for the different leukocyte phenotypes. T cells predominated over B cells (CD3/CD22 ratio = 11 ± 2.3), and cells displaying the T-helper phenotype predominated over cells with the suppressor/cytotoxic phenotype (CD4/CD8 ratio = 2.9 ± 0.9). No significant difference was observed between the densities of mononuclear cells displaying the memory/activated phenotype and the naive/quiscent phenotype (CD45RO/CD45RA ratio = 1.1 ± 0.4).
TABLE 1.
Phenotypic characterization of inflammatory infiltrate
| Marker | Cells/mm2 (mean ± SD) | Ratio |
|---|---|---|
| CD22 (B cells) | 475 ± 403 | CD3/CD22, 11 ± 2.3 |
| CD3 (T cells) | 4,434 ± 2,173 | |
| CD4 (T helper cells) | 3,359 ± 2,585 | CD4/CD8, 2.9 ± 0.9 |
| CD8 (T suppressor cells) | 1,281 ± 1,131 | |
| CD45RA (naive cells) | 2,999 ± 1,771 | CD45RO/CD45RA, 1.1 ± 0.4 |
| CD45RO (memory cells) | 3,153 ± 1,567 | |
| CD56 (NK cells) | 152 ± 79 | |
| CD68 (macrophages) | 1,556 ± 973 | |
| PMN elastase (neutrophils) | 1,013 ± 754 |
Topographic distribution of cells with detectable DNA damage.
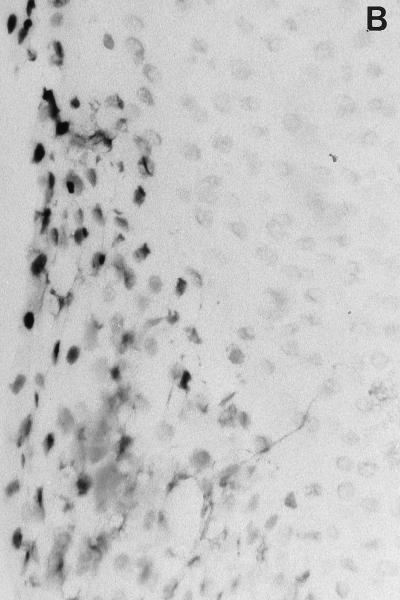
Incorporation of biotinylated-dUTP at the 3′ end of DNA breaks was detectable both in the epithelia and in the ICT (Table 2). Particularly strong signals were present (i) in the suprabasal cells of the junctional epithelium, in close proximity with bacterial plaque aggression (Fig. 1B), and (ii) in the perivascular inflammatory infiltrate. Sporadic positive cells were present in the upper layer cells of the sulcular and orogingival epithelia.
TABLE 2.
Tissue distribution of the markers studied
| Marker | Detection in:
|
|||
|---|---|---|---|---|
| Junctional epithelium | ICT | Sulcular epithelium | Orogingival epithelium | |
| DNA breaks | +++ | ++ | + | + |
| Bcl-2 expression | − | ++ | +/− (basal cells) | +/− (basal cells) |
| p53 expression | +++ | + | +/− (suprabasal) | +/− (suprabasal) |
| Ki67 expression | ++ (basal cells) | − | ++ (basal cells) | ++ (basal cells) |
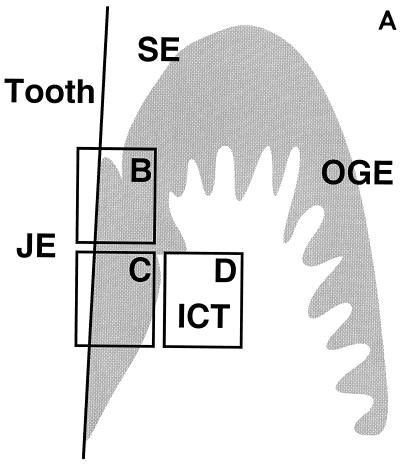
FIG. 1.
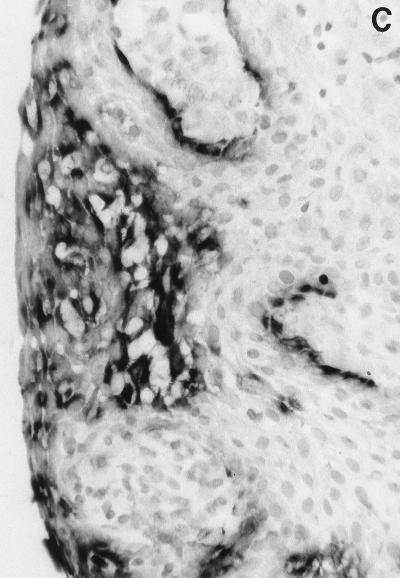
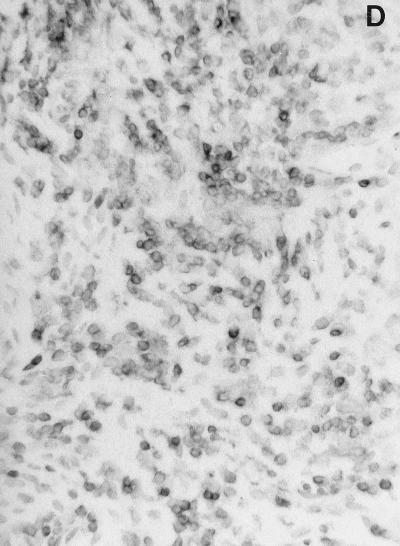
In situ detection of DNA damage, wild-type p53, and Bcl-2 in human gingiva. (A) Topographical orientation of the sections displayed in panels B to D. In all sections the tooth surface is on the left. Abbreviations: JE, junctional epithelium; SE, sulcular epithelium; OGE, orogingival epithelium; ICT, infiltrated connective tissue. (B) TUNEL-positive cells located in the most superficial portion of the JE (area B in panel A). The staining pattern that represents detection of DNA damage is located in the nucleus. Note the absence of positive cells in the SE. (C) Presence of p53-positive cells, preferentially located in the deeper layers of the JE (area C in panel A). (D) Bcl-2 staining pattern of the mononuclear cells present in the ICT (area D in panel A). Magnification for panels B to D, ×250.
Topographic distribution of p53-, Bcl-2-, and Ki67-positive cells.
Immunoreactive p53 was detectable in all areas where DNA damage could be demonstrated. Strong p53 reactivity was shown within the junctional epithelium, and, in particular, in its more basal layers (Fig. 1C). High numbers of positive cells were also detectable within the ICT, while only rare positive cells could be found in the suprabasal layers of the sulcular and orogingival epithelia. Similar patterns of reactivity were observed with both PAb240 and PAb248 monoclonal antibodies; the signal appeared to be located mainly in a perinuclear (PAb240) or cytoplasmic (PAb248) position.
Bcl-2-positive cells were located mainly within the perivascular inflammatory infiltrate (Fig. 1D); rare cells were observed in the basal layer of the gingival epithelia. No Bcl-2-positive cells could be detected within the junctional epithelium or the suprabasal layers of the other gingival epithelia.
Detection of the proliferating cell-associated Ki67 nuclear antigen was confined to the epithelia. Strong reactivity was found in association with the epithelial basal layers.
Quantitative evaluation.
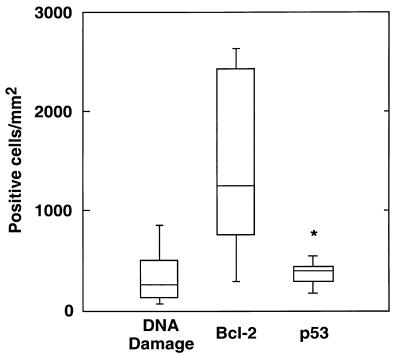
Box plots of the densities of ICT cells displaying biotinylated DNA nicks and detectable p53 and Bcl-2 expression are shown in Fig. 2. Scoring of adjacent sections revealed similar densities of DNA damage- and p53-positive cells (345 ± 278 and 403 ± 182 cells/mm2, respectively). Significantly more Bcl-2-positive cells, however, were observed (1,457 ± 898 cells/mm2). The association between DNA damage-positive cells and the expression of the Bcl-2 and p53 apoptosis-regulating proteins was further evaluated by least-squares regression analysis. The association was highly significant and explained 84% of the observed variability in the number of DNA damage-positive cells per millimeter squared (F = 21.142; P = 0.001 [Table 3]). Results indicated that the density of DNA damage-positive cells in the inflammatory infiltrate could be significantly predicted by a model including the density of cells expressing the p53 apoptosis-inducing protein (P = 0.017 [Table 3]). Conversely, the density of Bcl-2-positive cells did not significantly contribute to the regression equation (P = 0.781). Furthermore, the calculated regression coefficient was close to 1 (βi = 0.92), indicating a strong numerical consistency between DNA damage-positive cells and cells expressing the p53 protein.
FIG. 2.
Box plots summarizing the densities of DNA damage-, p53-, and Bcl-2-positive cells in the ICT of clinically normal human gingiva. As a comparison, total cellularity for that area was 8,764 ± 2,934 cells/mm2. Note the numerical consistency of DNA damage and p53-positive cells.
TABLE 3.
Regression model showing that density of cells with DNA breaks = β1 density of Bcl-2-positive cells + β2 density of p53-positive cellsa
| Parameter (βi) | Estimate | t for H0b | P |
|---|---|---|---|
| Bcl-2-positive cells | 0.021 | 0.288 | 0.781 |
| p53-positive cells | 0.919 | 3.008 | 0.017 |
Model (error): df, 2 (8); sum of squares, 1,903,299.294 (360,090.706); mean square, 951,649.647 (45,011.338); F value, 21.142; P = 0.001; r2 = 0.841.
Parameter estimate = 0.
Comparison of labeled DNA damage-, p53-, and Bcl-2-positive cells with the inflammatory cell density in the ICT indicated that positive cells represented a small yet significant fraction of the infiltrate. Cells displaying biotinylated DNA nicks were 3.8% ± 2.7% of total cells; similarly, p53 and Bcl-2 positive cells represented 4.4% ± 1.7% and 15.4% ± 6.7%, respectively.
DISCUSSION
The results of this investigation indicated that apoptosis-associated DNA damage and expression of the p53 and Bcl-2 apoptosis-regulating genes were prevalent phenomena in human clinically healthy gingival tissues exposed to chronic, low-grade bacterial challenge and inflammation. This represents, to our knowledge, the first in situ study indicating the relevance of the apoptotic process in chronic, low-grade, bacterially induced inflammation. Cells positive for DNA damage, p53, or Bcl-2 were selectively found in precise topographical locations: much of the expression was observed in the subepithelial inflammatory infiltrate and within the junctional epithelium and thus close to the area exposed to the oral microflora.
In situ detection of DNA damage at these sites of inflammation is an important observation since it may relate to a variety of biological phenomena, including programmed cell death. Use of the TUNEL technique allows the in situ detection of cells with DNA damage in a variety of tissues (7). Some investigations, however, have suggested that DNA damage evidenced with the TUNEL technique is not specific for the detection of apoptotic cell death but may also give positive results in areas of tissue necrosis (11). In this respect, it is important to underline that (i) in our material no section showed the characteristic histopathological signs of necrosis; (ii) the selective and consistent tissue distribution of DNA damage-positive cells, as well as the appearance of positive and negative controls, strongly indicated the nonartifactual nature of the signal; and (iii) the topographic consistency of p53 expression with the areas displaying DNA damage, as well as the strong statistical association between the density of p53-positive cells and the density of TUNEL-positive cells, supports the conclusion that at least some of the cells with detectable DNA damage may be apoptotic.
The presence of DNA damage-positive cells associated with the expression of the wild-type p53 apoptosis-inducing protein in the subepithelial inflammatory infiltrate suggests that apoptotic cell death may be an important phenomenon in the regulation of the inflammatory response to a chronic bacterial challenge. About 4% of the cells present in the subepithelial mononuclear inflammatory infiltrate displayed apoptosis-associated changes. Such a high prevalence is striking since in vitro the apoptotic process has been shown to be quite rapid and leading to cell fragmentation in a few hours (16). The high percentages of apoptotic cells in the inflammatory infiltrate detected in this study may speak for a significant role of apoptosis in preventing increases in cellularity and topographic extension of the infiltrate. In this respect, it should be observed that in this investigation the infiltrate consisted of macrophages as well as B and T cells and some PMN (Table 1). The lesion was a T-cell-dominated one with T-helper cell-to-suppressor ratios of 3 and substantially equal densities of activated/memory and naive/quiescent T cells. These observations are consistent with the results of previous studies and suggest that the analyzed material could be considered representative of the chronic inflammatory reaction consistently found in clinically healthy gingiva.
The current understanding of the limited permeability of junctional epithelium to bacterial plaque and its products under the clinically normal conditions evaluated in this study would speak against a direct bacterial effect leading to apoptosis within the inflammatory infiltrate. Possible hypotheses on the molecular regulation of this phenomenon come from in vitro experiments indicating that when deprived of certain cytokines or bacterial challenges, different leukocytes undergo apoptosis. In general, exposure to proinflammatory cytokines, such as interleukin-1β and tumor necrosis factor alpha, or bacterial lipopolysaccharide seems to prevent apoptosis (24) and to be associated with Bcl-2 expression (4). Conversely, other cytokines such as interleukin-4 and transforming growth factor β have been associated with an increase in p53 expression and apoptotic changes (22). A variety of ex vivo investigations have determined that the gingival mononuclear cell infiltrate expresses both apoptosis-preventing (proinflammatory) and apoptosis-inducing (anti-inflammatory) cytokines (25, 50, 6). The detection of both Bcl-2- and p53-positive cells within the inflammatory infiltrate is therefore not unexpected and suggests the presence of a finely regulated cytokine network, the balance of which may determine onset or inhibition of the apoptotic process and thus both the fate of individual cells and eventual variations in the size and cellularity of the infiltrate. Further investigations are needed in this area.
High numbers of DNA damage-positive cells were also found in the superficial layers of the junctional epithelium. This stratified epithelium is particular since it provides both a seal to restore mucosal continuity around erupted teeth and a compartment for peripheral defense (34). It is constantly exposed to a mixed bacterial flora which includes gram-negative anaerobes, and it has an exceptionally high rate of turnover. According to one estimation, epithelial cell desquamation in this tissue is 50 to 100 times faster than in the adjacent oral mucosa (21). High levels of DNA damage are presently considered to be a characteristic of the uppermost layers of rapidly renewing epithelia such as the intestinal mucosa or epidermal epithelium (7, 39), while wild-type p53 expression has been detected in the parabasal cells of these epithelia (30). In the present study, cells showing DNA damage and cells expressing the p53 apoptosis-inducing protein were found in topographically distinct regions of the junctional epithelium. These localizations are consistent with the sequential development of the apoptotic process, i.e., the initial expression of the p53 apoptosis-inducing protein in the parabasal layers, and consequent detection of DNA damage in the more superficial layers (Fig. 1B and C). These observations may be interpreted as the result of several, possibly correlated phenomena: (i) the interaction between specific bacteria present in dental plaque and junctional epithelium keratinocytes; (ii) the production of specific intraepithelial autocrine and paracrine stimuli; and/or (iii) the effect of paracrine stimuli generated by the subepithelial connective tissue infiltrate. It has been shown that a variety of bacterial pathogens are able to induce apoptosis in the infected cells: the leukotoxin of a periodontal pathogen, Actinobacillus actinomycetemcomitans, has been shown to induce apoptosis on human T cells (23); similarly, the intestinal pathogen Shigella flexneri induces apoptosis in infected macrophages (51). In this respect, it is generally agreed that some dental plaque bacteria are able to penetrate within the junctional epithelium, and they have been detected in an intracellular location (32, 33). Further investigations are in progress in this area. Also of interest is the recent recognition that junctional epithelium, like most body epithelia, plays an active role in the maintenance of surface integrity. Gingival keratinocytes, in fact, have been shown to produce, and respond to, a variety of cytokines and other inflammatory molecules (40) that may play a pivotal role in the homeostasis of this epithelium, possibly through induction and prevention of programmed cell death.
In summary, the results of this study indicate that apoptosis-associated cell damage is a prevalent phenomenon at sites of chronic bacterially induced inflammation in human gingiva and may play a role in the regulation of mucosal inflammation.
ACKNOWLEDGMENTS
This work was supported by Swiss National Science Foundation grant 32-37763.93, by the Clinical Research Foundation for the Promotion of Oral Health, and by a fellowship to D.C. from the Italian Society of Periodontology.
REFERENCES
- 1.Baran J, Guzik K, Hryniewicz W, Ernst M, Flad H, Pryjma J. Apoptosis of monocytes and prolonged survival of granulocytes as a result of phagocytosis of bacteria. Infect Immun. 1996;64:4242–4248. doi: 10.1128/iai.64.10.4242-4248.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen Y, Zychlinsky A. Apoptosis induced by bacterial pathogens. Microb Pathog. 1994;17:203–212. doi: 10.1006/mpat.1994.1066. [DOI] [PubMed] [Google Scholar]
- 3.Cohen J, Duke R. Apoptosis and programmed cell death in immunity. Annu Rev Immunol. 1992;10:267–293. doi: 10.1146/annurev.iy.10.040192.001411. [DOI] [PubMed] [Google Scholar]
- 4.Colotta F, Re F, Polentarutti N, Sozzani S, Mantovani A. Modulation of granulocyte survival and programmed cell death by cytokines and bacterial products. Blood. 1992;80:2012–2020. [PubMed] [Google Scholar]
- 5.Cordell J, Falini B, Erber W, et al. Immunoenzymatic labelling of monoclonal antibodies using immune complexes of alkaline phosphatase and monoclonal anti-alkaline phosphatase (APAAP) complexes. J Histochem Cytochem. 1984;32:219–229. doi: 10.1177/32.2.6198355. [DOI] [PubMed] [Google Scholar]
- 6.Fujihashi K, Beagley K, Kono K, Aicher W, Yamamoto M, DiFabio S, Xu-Amano J, McGhee J, Kiyono H. Gingival mononuclear cells from chronic inflammatory periodontal tissues produce interleukin (IL)-5 and Il-6 but not IL-2 and IL-4. Am J Pathol. 1993;142:1239–1250. [PMC free article] [PubMed] [Google Scholar]
- 7.Gavrieli Y, Sherman Y, Ben-Sasson S. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J Cell Biol. 1992;119:493–501. doi: 10.1083/jcb.119.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Genco R J. Host responses in periodontal diseases: current concepts. J Periodontol. 1992;63:338–355. doi: 10.1902/jop.1992.63.4s.338. [DOI] [PubMed] [Google Scholar]
- 9.Genco R J, Mergenhagen S E, editors. Host-parasite interactions in periodontal diseases. Washington, D.C: American Society for Microbiology; 1982. [Google Scholar]
- 10.Gorczyca W, Gong J, Darzynkiewicz Z. Detection of DNA strand breaks in individual apoptotic cells by the in situ terminal deoxynucleotidyl-transferase and nick translation assays. Cancer Res. 1993;53:1945–1951. [PubMed] [Google Scholar]
- 11.Grasl-Kraupp B, Ruttkay-Nedecky B, Koudelka H, Bukowska K, Bursch W, Schulte-Herman R. In situ detection of fragmented DNA (TUNEL assay) fails to discriminate among apoptosis, necrosis and autolytic cell death. A cautionary note. Hepatology. 1995;21:1465–1468. doi: 10.1002/hep.1840210534. [DOI] [PubMed] [Google Scholar]
- 12.Haslett C, Savill J, Whyte M, Stern M, Dransfield I, Meagher L. Granulocyte apoptosis in the control of inflammation. Philos Trans R Soc Lond Ser B. 1994;345:327–333. doi: 10.1098/rstb.1994.0113. [DOI] [PubMed] [Google Scholar]
- 13.Hockenbery D, Zutter M, Hickey W, Nahm M, Korsmeyer S. Bcl-2 protein is topographically restricted in tissues characterized by apoptotic cell death. Proc Natl Acad Sci USA. 1991;88:6961–6965. doi: 10.1073/pnas.88.16.6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu S M, Raine L, Fanger H. The use of antiavidin antibody and avidin biotin peroxidase complex in immunoperoxidase technics. Am J Clin Pathol. 1981;75:734–738. doi: 10.1093/ajcp/75.6.816. [DOI] [PubMed] [Google Scholar]
- 15.Hsu S M, Soban E. Color modification of diaminobenzidine precipitation by metallic ions and its application for double immunohistochemistry. J Histochem Cytochem. 1982;30:1079–1082. doi: 10.1177/30.10.6182185. [DOI] [PubMed] [Google Scholar]
- 16.Kerr J, Searle J, Harmon B, Bishop C. Apoptosis. In: Potten C, editor. Perspectives on mammalian cell death. Oxford, England: Oxford University Press; 1987. pp. 93–128. [Google Scholar]
- 17.Kerr J, Wyllie A, Currie A. Apoptosis: basic biologic phenomenon with wide ranging implications in tissue kinetics. Br J Cancer. 1972;26:239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lane D, Crawford L. T-antigen is bound to host protein in SV40-transformed cells. Nature. 1979;278:261–263. doi: 10.1038/278261a0. [DOI] [PubMed] [Google Scholar]
- 19.Lane D, Lu X, Hupp T, Hall P. The role of p53 protein in the apoptotic response. Philos Trans R Soc Lond Ser B. 1994;345:277–280. doi: 10.1098/rstb.1994.0106. [DOI] [PubMed] [Google Scholar]
- 20.LeBrun D, Warnke R, Cleary M. Expression of Bcl-2 in fetal tissues suggests a role in morphogenesis. Am J Pathol. 1993;142:743–753. [PMC free article] [PubMed] [Google Scholar]
- 21.Listgarten M. Normal development, structure, physiology and repair of gingival epithelium. Oral Sci Rev. 1972;1:3–67. [PubMed] [Google Scholar]
- 22.Mangan D, Robertson B, Wahl S. IL-4 enhances programmed cell death (apoptosis) in stimulated human monocytes. J Immunol. 1992;148:1812–1816. [PubMed] [Google Scholar]
- 23.Mangan D, Taichman N, Lally E, Wahl S. Lethal effects of Actinobacillus actinomycetemcomitans leukotoxin on human T lymphocytes. Infect Immun. 1991;59:3267–3272. doi: 10.1128/iai.59.9.3267-3272.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mangan D, Welch G, Wahl S. Lipopolysaccharide, tumor necrosis factor alpha and IL-1 beta prevent programmed cell death (apoptosis) in human peripheral blood monocytes. J Immunol. 1991;146:1541–1546. [PubMed] [Google Scholar]
- 25.Matsuki Y, Yamamoto T, Hara K. Localization of interleukin-1 (IL-1) mRNA expressing macrophages in human inflamed gingiva and IL-1 activity in gingival crevicular fluid. J Periodontal Res. 1993;28:35. doi: 10.1111/j.1600-0765.1993.tb01048.x. [DOI] [PubMed] [Google Scholar]
- 26.Moughal N A, Adonogianaki E, Thornhill M H, Kinane D F. Endothelial leukocyte adhesion molecule-1 (ELAM-1) and intercellular adhesion molecule-1 (ICAM-1) expression in gingival tissue during health and experimentally induced gingivitis. J Periodontal Res. 1992;27:623–630. doi: 10.1111/j.1600-0765.1992.tb01746.x. [DOI] [PubMed] [Google Scholar]
- 27.Page R C. The role of inflammatory mediators in the pathogenesis of periodontal disease. J Periodontal Res. 1991;26:230–242. doi: 10.1111/j.1600-0765.1991.tb01649.x. [DOI] [PubMed] [Google Scholar]
- 28.Page R C, Schroeder H E. Periodontitis in man and other animals. A comparative review. S. Basel, Switzerland: Karger; 1982. [Google Scholar]
- 29.Pezzella F, Cordell J, Pulford K, Gatter K, Mason D. Expression of the Bcl-2 oncogene protein is not specific for the 14;18 chromosomal translocation. Am J Pathol. 1990;137:225–232. [PMC free article] [PubMed] [Google Scholar]
- 30.Pezzella F, Micklem K, Turley H, Pulford K, Jones M, Kocialkowski S, Delia D, Aiello A, Bicknell R, Smith K, Harris A, Gatter K, Mason D. Antibody for detecting p53 protein by immunohistochemistry in normal tissues. J Clin Pathol. 1994;47:592–596. doi: 10.1136/jcp.47.7.592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reed J. Bcl-2 and the regulation of programmed cell death. J Cell Biol. 1994;124:1–6. doi: 10.1083/jcb.124.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saglie F R. Bacterial invasion and its role in the pathogenesis of periodontal disease. In: Hamada S, Holt S C, McGhee J R, editors. Periodontal disease: pathogens and host immune responses. Tokyo, Japan: Quintessence Publishing Co.; 1991. pp. 27–40. [Google Scholar]
- 33.Saglie F R, Carranza F A, Jr, Newman M G. The presence of bacteria within the oral epithelium in periodontal disease. I. A scanning and transmission electron microscopic study. J Periodontol. 1985;56:618–624. doi: 10.1902/jop.1985.56.10.618. [DOI] [PubMed] [Google Scholar]
- 34.Schroeder H. The junctional epithelium: origin, structure and significance. Acta Med Dent Helv. 1996;1:155–167. [Google Scholar]
- 35.Schroeder H, Graf-de-Beer M. Stereological analysis of chronic lymphoid cell infiltrates in human gingiva. Arch Oral Biol. 1976;21:527–537. doi: 10.1016/0003-9969(76)90018-2. [DOI] [PubMed] [Google Scholar]
- 36.Schroeder H E. Transmigration and infiltration of leukocytes in human junctional epithelium. Helv Odontol Acta. 1973;17:10–18. [PubMed] [Google Scholar]
- 37.Seymour G J, Gemmell E, Reinhardt R A, Eastcott J, Taubman M A. Immunopathogenesis of chronic inflammatory periodontal disease: cellular and molecular mechanisms. J Periodontal Res. 1993;28:478–86. doi: 10.1111/j.1600-0765.1993.tb02108.x. [DOI] [PubMed] [Google Scholar]
- 38.Tagge D, O’Leary T, El-Kafrawy A. The clinical and histological response of periodontal pockets to root planing and oral hygiene. J Periodontol. 1975;46:527–533. doi: 10.1902/jop.1975.46.9.527. [DOI] [PubMed] [Google Scholar]
- 39.Tamada Y, Takama H, Kitamura T, Yokochi K, Nitta Y, Ikeya T, Matsumoto Y. Identification of programmed cell death in normal human skin tissue by using specific labelling of fragmented DNA. Br J Dermatol. 1994;131:521–524. doi: 10.1111/j.1365-2133.1994.tb08553.x. [DOI] [PubMed] [Google Scholar]
- 40.Tonetti M. Molecular factors associated with compartmentalization of gingival immune responses and transepithelial neutrophil migration. J Periodontal Res. 1997;32:104–109. doi: 10.1111/j.1600-0765.1997.tb01389.x. [DOI] [PubMed] [Google Scholar]
- 41.Tonetti M, Gerber L, Lang N. Vascular adhesion molecules and initial development of inflammation in clinically healthy human keratinized mucosa around teeth and osseointegrated implants. J Periodontal Res. 1994;29:386–392. doi: 10.1111/j.1600-0765.1994.tb01239.x. [DOI] [PubMed] [Google Scholar]
- 42.Tonetti M, Imboden M, Gerber L, Lang N. Compartmentalization of inflammatory cell phenotypes in normal gingiva and peri-implant keratinized mucosa. J Clin Periodontol. 1995;22:735–742. doi: 10.1111/j.1600-051x.1995.tb00255.x. [DOI] [PubMed] [Google Scholar]
- 43.Tonetti M, Imboden M, Gerber L, Lang N, Laissue J, Mueller C. Localized expression of mRNA for phagocyte-specific chemotactic cytokines in human periodontal infections. Infect Immun. 1994;62:4005–4014. doi: 10.1128/iai.62.9.4005-4014.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tonetti M, Imboden M, Lang N. Neutrophil migration into the gingival sulcus is associated with transepithelial gradients of interleukin-8 and ICAM-1. J Periodontol. 1998;69:1140–1148. doi: 10.1902/jop.1998.69.10.1139. [DOI] [PubMed] [Google Scholar]
- 45.Tonetti M, Straub A, Lang N. Expression of the cutaneous lymphocyte antigen and the alphaIELbeta7 integrin by intraepithelial lymphocytes in healthy and diseased human gingiva. Arch Oral Biol. 1995;40:1125–1132. doi: 10.1016/0003-9969(95)00084-4. [DOI] [PubMed] [Google Scholar]
- 46.Tonetti M S, Schmid J, Haemmerle C H, Lang N P. Intraepithelial antigen presenting cells in the keratinized mucosa around teeth and osseointegrated implants. Clin Oral Implant Res. 1993;4:177–186. [Google Scholar]
- 47.Tukey J. Exploratory data analysis. Reading, Mass: Addison-Wesley; 1977. [Google Scholar]
- 48.Vaux D. Toward an understanding of the molecular mechanisms of physiological cell death. Proc Natl Acad Sci USA. 1993;90:786–789. doi: 10.1073/pnas.90.3.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vaux D, Cory S, Adams J. Bcl-2 gene promotes hematopoietic cell survival and co-operates with c-myc to immortalise pre-B cells. Nature. 1988;335:440–442. doi: 10.1038/335440a0. [DOI] [PubMed] [Google Scholar]
- 50.Wahl S M, Costa G L, Mizel D E, Allen J B, Skaleric U, Mangan D F. Role of transforming growth factor beta in the pathophysiology of chronic inflammation. J Periodontol. 1993;64:450–455. [PubMed] [Google Scholar]
- 51.Zychlinsky A, Prevost M, Sansonetti P. Shigella flexneri induces apoptosis in infected macrophages. Nature. 1992;358:167–169. doi: 10.1038/358167a0. [DOI] [PubMed] [Google Scholar]