Abstract
Accurately identifying neoantigens is crucial for developing effective cancer vaccines and improving tumor immunotherapy. Mass spectrometry-based immunopeptidomics has emerged as a promising approach to identifying human leukocyte antigen (HLA) peptides presented on the surface of cancer cells, but false-positive identifications remain a significant challenge. In this study, liquid chromatography-tandem mass spectrometry-based proteomics and next-generation sequencing were utilized to identify HLA-presenting neoantigenic peptides resulting from non-synonymous single nucleotide variations in tumor tissues from 18 patients with renal cell carcinoma or pancreatic cancer. Machine learning was utilized to evaluate Mascot identifications through the prediction of MS/MS spectral consistency, and four descriptors for each candidate sequence: the max Mascot ion score, predicted HLA binding affinity, aliphatic index and retention time deviation, were selected as important features in filtering out identifications with inadequate fragmentation consistency. This suggests that incorporating rescoring filters based on peptide physicochemical characteristics could enhance the identification rate of MS-based immunopeptidomics compared to the traditional Mascot approach predominantly used for proteomics, indicating the potential for optimizing neoantigen identification pipelines as well as clinical applications.
Keywords: Immunopeptidomics, Mass spectrometry, Machine learning, Aliphatic index
Graphical Abstract
1. Introduction
Correct identification of tumor specific antigens generated by genomic mutations in tumor cells, known as neoantigens, is essential for developing effective cancer vaccines and improving the efficacy of tumor immunotherapy [1], [2]. Mass spectrometry (MS)-based immunopeptidomics is a promising approach for analyzing peptides present on the surface of cancer cells and can provide direct and reliable evidence for neoantigen identification [3], [4], [5], [6]. However, conventional proteomics workflows face significant challenges, especially false-positive peptide identification, when applied to immunopeptidomics because of differences in analysis purposes and sample nature [7], [8]. First, the diversity of intracellular hydrolysis mechanisms leads to inherently diverse human leukocyte antigen (HLA) peptides, with a particular issue being the lack of a positive charge at the C-termini [9], [10], [11]. This leads to lower fragmentation and ionization efficiency of peptides than traditional tryptic peptides, increasing the spectral peaks of intermediate internal fragments in tandem MS spectra and causing a loss of paired y-ions [11], [12], [13]. This poses challenges for database searching based on spectral similarity, such as using Mascot [14], to identify correct peptide spectrum matches. Second, the property of specific binding to the major histocompatibility complex (MHC), also known as HLA in humans, makes HLA peptides more similar in amino acid length and conservation but lower in abundance compared to tryptic digested peptides in conventional shotgun proteomics. Therefore, false-positive peptide identification from a large number of similar peptides makes accurate peptide spectrum matching difficult [15]. Third, most contaminants carry a single charge, which makes it challenging to identify and distinguish signal peaks from noise [8]. In shotgun proteomics, achieving a reliable and identical result is typically associated with a Mascot ion score at 25–30 or False Discovery Rate (FDR) ≤ 1% in general [16]. Despite being mentioned above, the reduced ionization efficiency arising from the absence of inherent positive charges in immunopeptides frequently leads to lower Mascot ion scores in the obtained results, and employing high cutoffs with Mascot can result in a notable decline in co-determination efficiency. Therefore, some studies utilizing shotgun pipelines for immunopeptidomic research employed reduced Mascot cutoff values, such as Mascot at 22, and/or FDR of 5–9% [17], to attain the intended number of candidate results. Therefore, it remains necessary to evaluate the attainable accuracy when employing this method for analyzing real clinical tumor samples, while also crucially focusing on the development of new strategies to enhance precision [15].
In this study, Liquid chromatography-tandem mass spectrometry (LC-MS/MS)-based proteomics and next-generation sequencing (NGS) were used to identify HLA-presenting neoantigenic peptides resulting from non-synonymous single nucleotide variations (SNVs). Tumor tissues from 18 patients with renal cell carcinoma or pancreatic cancer were analyzed using a conventional proteomic workflow based on Mascot. To comprehensively assess the precision of this method in the examination of genuine clinical tumor samples, all candidate sequences with a maximum Mascot ion score exceeding 10 were synthesized as standard peptides. Their mass spectrometry spectra were then compared to those of the corresponding peptides identified in the tumor tissues. Additionally, a machine learning-based filter biased towards HLA peptides was developed by incorporating peptide physicochemical information and experimental information. The results showed that conventional proteomic methods have limited capabilities in identifying immunopeptides originating from SNVs in actual clinical tumor samples. Nonetheless, by applying post-processing filter based on machine learning, the identification performance can be significantly improved, and it can also help rescue reliable identifications with low Mascot ion scores.
2. Material and methods
2.1. Patient material and HLA-typing
Cancerous and normal tissues from patients with renal cell carcinoma or pancreatic cancer were used in this study (Table S1). Sequence data of the genomic DNA samples were analyzed for HLA haplotypes at the HLA Laboratory (Kyoto, Japan). This study was conducted in accordance with the provisions of the Declaration of Helsinki and was approved by the institutional review board of the Kanagawa Cancer Center, Kanagawa, Japan (approval number: 2017–11 and 2017–83). Informed consent was obtained from all patients involved in the study after explaining the nature of the study and its possible consequences.
2.2. Genomic analyses: Mutation discovery
Total RNA and genomic DNA were extracted from frozen pieces of tumor and normal tissues using the Prep DNA/RNA kit (Qiagen, Düsseldorf, Germany) according to the manufacturer’s instructions and subjected to RNA sequencing and whole-exome sequencing at BGI (Beijing, China). Exome sequencing was performed using the SureSelectXT Human All Exon Kit V6 (Agilent Technologies, Santa Clara, CA, USA). DNA/RNA sequence reads were mapped against the GRCh38 human genome reference using minimap2 [18] with a default paired-end setting. The splice option was enabled. The resulting BAM files were sorted and indexed using SAMtools, and potential somatic mutations were called from the normal and tumor BAM files using the Karkinos variant caller [19]. Candidate somatic mutation sites were filtered based on overlapping variants that were also observed in RNA sequence reads. Somatic variations in RNA sequences were extracted using samtools/bamtools with the pileup option. The gene code, GRCh38 (release 42), was used as the transcript database. We developed a script to modify transcript sequences to reflect somatic mutations in the SNVs detected above. BioPython API was used to translate the altered somatic RNA sequences into amino acid sequences. The somatic amino acid sequences were then compared against a reference amino acid sequence, and only non-synonymous mutations were used as the peptide database for peptide searching by MS. To minimize information loss, FASTA files focusing on 15 amino acids (15mer) centered around mutations were created and used for MS analysis.
2.3. Immunoprecipitation
Tumor tissues were sliced into pieces of approximately 100 mg in size in a frozen state, and then weighed. If the total amount of tumor tissues was significantly less than 100 mg, all available quantity was weighed and utilized (Table S1). The weighted tissues were homogenized in 1 mL of lysis buffer containing 20 mM Tris-HCl pH 8.0, 1 mM EDTA, 100 mM sodium chloride, 1% Triton X-100 (Roche, Basel, Switzerland), 60 mM n-octylglucoside (Dojin Chemicals Co., Kumamoto, Japan), 1 mM PMSF (Sigma-Aldrich, Tokyo, Japan), cOmplete Protease Inhibitor cocktail (Roche, Basel, Switzerland), PhosSTOP Phosphatase Inhibitor cocktail (Roche, Basel, Switzerland) and 10 U/mL Benzonase (Millipore, Burlington, MA) for 30 s using TissueRuptor homogenizer (Qiagen, Düsseldorf, Germany). After 30 min of incubation on ice, lysates were cleared by centrifugation at 16,000g for 10 min at 4 °C. The protein concentration in the supernatant was measured using a BCA protein kit (Nacalai, Kyoto, Japan), and the samples were diluted to 1 mg/mL with lysis buffer. 100 μg of anti-HLA-ABC mAb W6/32 (BioXCell, New Haven, CT, USA) was conjugated to 2 mg of FG NHS beads (Tamagawa Seiki, Nagano, Japan) according to the manufacturer’s instructions. 0.5 mg of conjugated beads were used for immunoprecipitation from 1 mL of lysate for 2 h at 4 °C on a rotator. Immunoprecipitates were washed with 1 mL of 50 mM Ammonium bicarbonate buffer three times, and peptides were eluted with 200 μL of 0.1 M Glycine-HCl pH 2.5 for 5 min at room temperature.
2.4. Sample preparation of LC-MS/MS analysis
Samples obtained by immunoprecipitation were desalted using a MonoSpin C18 column (GL Sciences, Tokyo, Japan). The resin was conditioned with 300 μL of buffer A (5% formic acid, 90% acetonitrile) (5,000g, 30 s, room temperature) and equilibrated with 300 μL of 5% formic acid (5,000g, 30 s, room temperature). An equal volume of 0.2 M NDSB was added to the sample, and the mixture was acidified with 2.5% trifluoroacetic acid (TFA). The sample was passed through a spin column (5,000g, 30 s, 10 °C) until all the samples had passed through the spin column. The spin column was rinsed three times with 300 μL of 5% formic acid (5,000g, 30 s, 10 °C). The peptides were eluted from the spin column twice with 50 μL of buffer B (5% formic acid, 30% acetonitrile) (5000g, 30 s, 10 °C).
2.5. LC-MS/MS analysis
The eluates were dried using a SpeedVac (Thermo Fisher Scientific, Waltham, MA, USA) and resuspended in 0.1% TFA in 2% acetonitrile. A capillary reverse-phase high-performance (HP) LC-MS/MS system (ZAPLOUS System; AMR, Tokyo, Japan), comprising of an advanced ultra-high-performance (UHP) LC instrument (Michrom Bioresources, Auburn, CA), an HTC PAL autosampler (CTC Analytics, Zwingen, Switzerland), an Orbitrap Fusion ETD, and a quadrupole linear ion trap Orbitrap mass spectrometer (Thermo Fisher Scientific) equipped with a Dream Spray ESI source (Dream Spray; AMR, Tokyo, Japan), was used for LC-MS/MS analysis. Aliquots of samples were automatically injected onto a C18 PepMap 100 Peptide Trap cartridge (5 × 0.3 mm I.D.; Thermo Fisher Scientific, Waltham, MA) attached to an injector valve for desalting and concentrating the peptides. After washing the trap with 0.1% TFA in 98% MilliQ water and 2% acetonitrile, the peptides were loaded into a separation capillary reverse phase column (L-column2 micro C18 column 3 µm, 200 Å, 150 × 0.2 mm I.D.; CERI, Tokyo, Japan) by switching the valve. The eluents used were 0.1% formic acid in water (A), and 100% acetonitrile (B). The column was developed at a flow rate of 1.0 μL/min, with a concentration gradient of acetonitrile: from 5% B to 30% B for 100 min, then from 30% B to 95% B for 2 min, maintained at 95% B for 8 min, from 95% B to 5% B for 2 min, and finally re-equilibrated with 5% B for 8 min. The effluents were introduced into the mass spectrometer via a nanoelectrospray ion interface that held the separation column outlet directly connected to a Dream Spray electrospray ion source. The electrospray ionization voltage was 1.8 kV, and the transfer capillary of the orbitrap inlet was heated to 280 °C. No sheath or auxiliary gases were used. The mass spectrometer was operated in a data-dependent acquisition mode, in which MS acquisition with a mass range of m/z 390–1590 was automatically switched to MS/MS acquisition under the automated control of the Xcalibur software. The top 20 precursor ions were selected by an MS scan with Orbitrap at a resolution of 240,000, and for the subsequent MS/MS scans through ion traps in the normal/centroid mode, using the automatic gain control (AGC) mode with AGC values of 2 × 105 and 1 × 104 for full MS and MS/MS, respectively. We also employed a dynamic exclusion capability that allowed the sequential MS/MS acquisition of abundant ions in the order of their intensities, with an exclusion duration of 5 s and exclusion mass widths of − 5 and + 5 ppm. The trapping time was 35 ms with an auto-gain control.
2.6. Data analysis of LC-MS/MS
Mass spectra were extracted using the Proteome Discoverer (version 2.5). All MS/MS samples were analyzed using the Mascot database search engine (version 2.6; Matrix Science, London, UK). Mascot was set up to search for an in-house database with the digestion enzyme none specific. Mascot was searched using a product ion mass tolerance of 0.60 Da and a precursor ion tolerance of 5.0 ppm by considerations of sensitivity, acquisition time, and the goal of obtaining more candidates through a more relaxed database search filter. Gln->pyro-Glu at the N-terminus, oxidation of methionine and acetyl at the N-terminus, and phospho-serine, threonine, and tyrosine were specified in Mascot as variable modifications. Scaffold (version 4, Proteome Software, https://www.proteomesoftware.com/) was used to validate the MS/MS-based peptide and protein identification. To include a broader range of peptide matches, a relatively low threshold of 10 was set for the Mascot ion scores of candidate peptides. To preserve the possibility of identifying additional false negatives, FDR-based cutoff was not employed to further adjust the Mascot search results [20]. Candidate sequences (CandiSeqs) were identified based on the following criteria: 1) maximum Mascot ion score (MaxMascotIonScore) higher than 10; 2) presence of mutation points; 3) length of 8–12mers.
2.7. Validation of identified candidate sequences against synthetic peptides
For validation the identifications by Mascot, synthetic peptides (SynPeps) were synthesized for all CandiSeqs, and their mass spectra were measured under the same conditions. The tandem MS spectra of each CandiSeq from the clinical samples were compared with those of the corresponding SynPeps, and the number of consistent peaks (MS2) was counted. MS2 revealed the consistency in MS/MS spectra between CandiSeqs and their corresponding SynPeps.
To assess and compare the identification capabilities of Mascot and the novo machine learning filter, a subset of reliable identifications with highly consistent MS/MS spectra (HCS) were further selected by two experienced MS spectral analysts by comprehensively evaluating MS2 information, peptide fragmentation patterns, and retention time.
3. Results
3.1. Experimental design
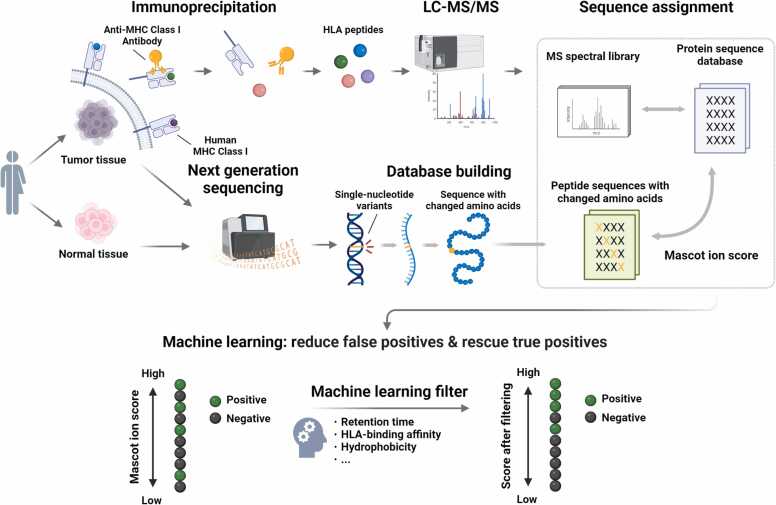
The workflow of this study is illustrated in Fig. 1. Tumor and normal tissues were collected from nine patients with renal cell carcinoma and nine patients with pancreatic cancer, and HLA-peptide complexes were extracted by immunoprecipitation. LC-MS/MS was used to analyze the peptides and obtain tandem MS fingerprints for each HLA peptide. NGS was performed on tumor and normal tissues from each patient to construct a database of peptide sequences containing SNV mutations by comparison with the reference gene sequence obtained from the corresponding normal tissue counterpart for each tumor tissue. Mascot-based tandem MS spectral searching was used to obtain sequence assignment results (the top-ranked sequence by Mascot) corresponding to each tandem MS spectrum. To validate the identification efficacy of Mascot, CandiSeqs with a length of 8–12 amino acids, Mascot score ≥ 10, and containing amino acid mutations were synthesized. The tandem MS spectra of each CandiSeq from the tumor samples were compared with those of the corresponding SynPep measured under the same conditions, and MS2 was counted. To evaluate the prediction performance, reliable identifications with highly consistent MS/MS spectra were further selected based on a combination of MS2, retention time, and MS/MS fragmentation pattern. To overcome the false-positive identification of HLA peptides by Mascot, a machine learning filter was developed, which includes physiochemical property-related factors such as retention time, HLA-binding affinity, and hydrophobicity. Our data showed that a substantial enhancement in immunopeptide identification performance can be achieved through the implementation of post-processing filters grounded in machine learning.
Fig. 1.
Workflow of the present study. Tumor and normal tissues were collected from nine patients with renal cell carcinoma and nine patients with pancreatic cancer; HLA-peptide complexes were extracted by immunoprecipitation; LC-MS/MS was used to analyze the peptides and obtain tandem MS fingerprints for each HLA peptide; NGS was performed on tumor and normal tissues from each patient to construct a database of peptide sequences containing SNV mutations by comparison with the reference gene sequence; Mascot-based tandem MS spectral searching was used to obtain sequence assignment results corresponding to each tandem MS spectrum; all candidate sequences were synthesized, MS analyzed and evaluated by MS/MS spectral consistency; a machine learning-based filter biased towards HLA peptides was developed by incorporating peptide physicochemical information and experimental information. HLA, human leukocyte antigen; LC-MS/MS, liquid chromatography-tandem mass spectrometry; NGS, next-generation sequencing; SNV, single nucleotide variations.
3.2. Descriptor collection for machine learning
1) Descriptors recorded during MS experiment and data analysis: MaxMascotIonScore, NL, ClinRT and TPM. The MaxMascotIonScore, actual retention time (ClinRT) and normalization level (NL) were recorded for each CandiSeq, where NL was defined as the actual intensity of the strongest peak in the mass spectrum, indicating the signal-to-noise ratio. The transcript per million (TPM) value determined by RNA-Seq was included as a descriptor regarding the expression level of the corresponding gene for machine learning.
2) Descriptors predicted by sequence information: PI, InstabilityIndex, IfStable, AliphaticIndex, Gravy, Hydrophobicity, MinRank and Entrp. Six physiochemical features of CandiSeqs were obtained by using their sequence information. Specifically, theoretical isoelectric point (PI), instability index (InstabilityIndex), stability (IfStable), aliphatic index (AliphaticIndex), and grand average hydropathicity (Gravy) were determined using the Expasy ProtParam tool [21] (https://web.expasy.org/protparam/). For all CandiSeqs, theoretical hydrophobicity (Hydrophobicity) was predicted using a web-based tool (Proteotypic Peptide Analyzing Tool from Thermo Fisher Scientific; https://www.thermofisher.com/jp/ja/home/life-science/protein-biology/peptides-proteins/custom-peptide-synthesis-services/peptide-analyzing-tool.html), analogous to the approach employed for HCS identifications (as summarized in Fig. S1). HLA-binding affinity was obtained using NetMHCpan-4.1[22] (https://services.healthtech.dtu.dk/services/NetMHCpan-4.1/) and the minimal percentile rank (MinRank) among the HLA class I types of the corresponding patients was selected. Shannon’s entropy value (Entrp) of the peptide sequences was calculated using the diversity R function in the vegan package.
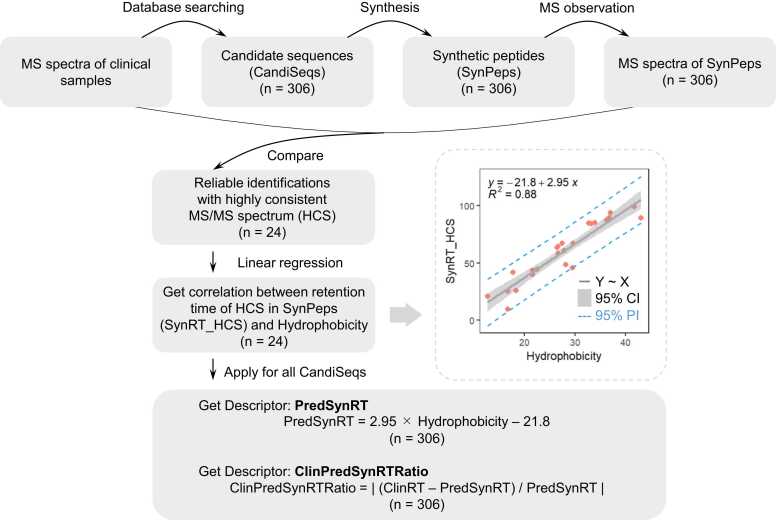
3) Descriptors derived from extended calculations: PredSynRT and ClinPredSynRTRatio. As shown in Fig. 2, descriptors for signifying retention time deviation were calculated based on the robust linear correlation existing between Hydrophobicity and standard retention time for reliable identifications of HCS. Each CandiSeq in the tissue samples had an actual retention time during MS observations. To enable a machine-learning filter that could directly filter the results of Mascot identification without synthesizing candidate peptides, the predicted retention time of each CandiSeq was calculated based on its sequence information, and the degree of deviation between the predicted and actual retention times was used as a descriptor for machine learning. The subset of HCS identifications was used to determine the correlation between the predicted Hydrophobicity and retention time under experimental conditions (The selection process for Hydrophobicity was summarized in Fig. S2). Simple linear regression was performed using the lm function in R (ver. 4.2.1; https://www.r-project.org/) to derive Eq. (1), which describes the relationship between the standard retention time of reliable identifications with highly consistent MS/MS spectra (SynRT_HCS) and Hydrophobicity under the experimental conditions:
| SynRT_HCS = 2·95 × Hydrophobicity – 21·8 | (1) |
Fig. 2.
Generation process of descriptors PredSynRT and ClinPredSynRTRatio for signifying retention time deviation. Standard retention time of CandiSeqs can be calculated using the robust linear correlation existing between hydrophobicity and retention time for reliable identifications of HCS. CI, confidence interval; ClinPredSynRTRatio, ratio between predicted standard and actual retention time; ClinRT, actual retention time; HCS, identifications with highly consistent MS/MS spectra; PI, prediction interval; PredSynRT, predicted standard retention time; R2, coefficient of determination.
We obtained the predicted standard retention times (PredSynRT) for all CandiSeqs using Eq. (2):
| PredSynRT = 2·95 × Hydrophobicity – 21·8 | (2) |
and Eq. (3) was used to calculate the degree of deviation between ClinRT and PredSynRT (ClinPredSynRTRatio).
| ClinPredSynRTRatio = | (ClinRT – PredSynRT) / PredSynRT | | (3) |
3.3. Calculations for machine learning
For CandiSeqs, 14 descriptors were collected and used as training features, whereas MS2 normalized by sequence length (MS2Norm) was used as the training target. MS2Norm was defined as MS2 divided by the length of the sequence (count of amino acids) Eq. (4):
| MS2Norm = MS2 / length of sequence (count of amino acids) | (4) |
All calculations were performed in R. CandiSeq data were randomly divided into a training set (80%, n = 245; Table S2) and a test set (20%, n = 61; Table S3). A random forest regression algorithm was used to build the model using the randomForest function in the R package of randomForest. Model performance was evaluated using k-fold (k = 5) cross-validation (CV) on the training set to average the biases introduced by sample split, and the predict function in the randomForest package was used to predict the completely independent test set. The model prediction results were obtained using the leave-one-out (LOO) CV method to provide each Candiseq with a distinct and consistent score within the current dataset used for comparing with Mascot. This approach maximizes the utilization of existing dataset resources with all CandiSeqs. DeLong’s test for two correlated receiver operating characteristic (ROC) curves was performed using roc.test function in pROC package with the method of “delong.” The two-sided Wilcoxon signed-rank test was performed using wilcox.exact R function, the Spearman’s rank correlation coefficient was calculated using cor R function, and the test for association between paired samples was calculated using cor.test function with the method of “spearman.”
3.4. Efficacy of Mascot-based immunopeptide identification
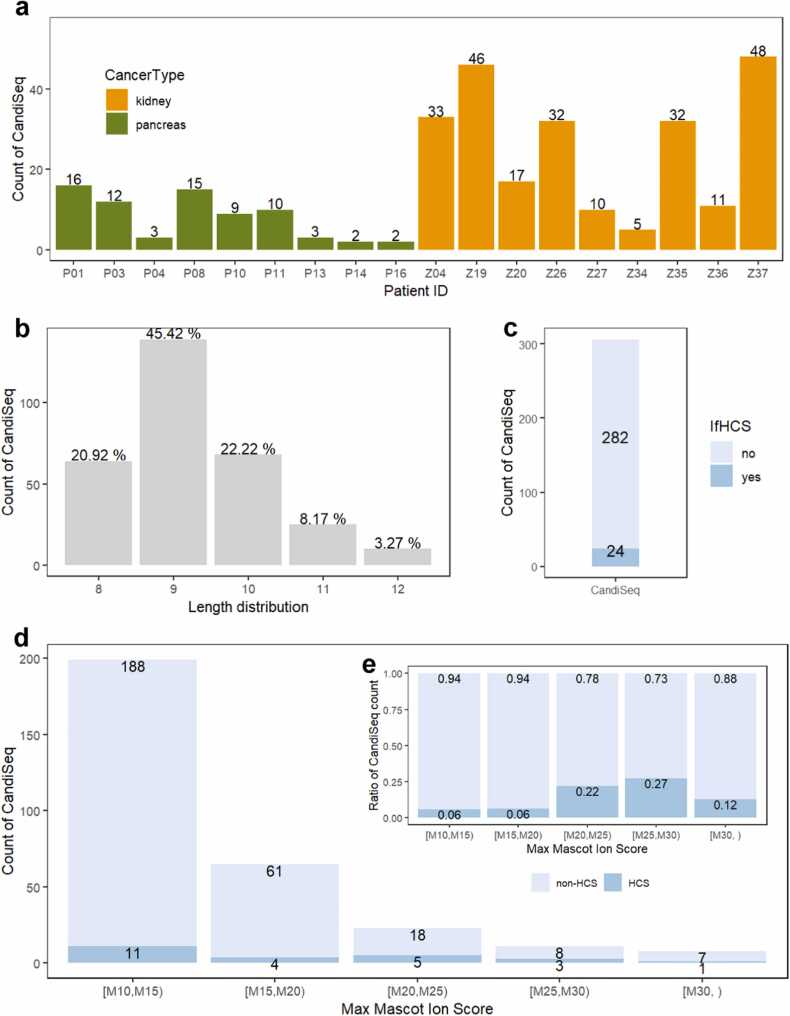
As shown in Fig. 3a, a total of 306 CandiSeqs containing SNV mutations were identified in the tumor tissues of 18 patients with cancer. The number of CandiSeqs per patient ranged from 2 to 16 for the nine patients with pancreatic cancer, and from 5 to 48 for the nine patients with renal cell carcinoma. According to the length distribution shown in Fig. 3b, 9mers constituted the majority, making up 45.42% of the total CandiSeq detections, whereas 12mers comprised the smallest proportion at 3.27%. To validate the identification efficacy of Mascot, 306 SynPeps were synthesized from all the CandiSeqs. Through a comparison of their tandem MS spectra, 24 reliable identifications with highly consistent MS/MS spectra, referred to as HCS, were selected among the total of 306 CandiSeqs (Fig. 3c). The amount and proportion of HCS at different Mascot levels are shown in Figs. 3d and 3e. In Fig. 3d, eight CandiSeqs were identified by Mascot at levels greater than 30. However, only one of these identifications was verified as an HCS, resulting in a recall of 0.12. In the Mascot range of 25 to 30, a set of 11 CandiSeqs was obtained, out of which three were validated as HCS, yielding the highest identification rate of 0.27. An increase in both the amount and proportion of false positives was observed within Mascot range below 25. However, it is crucial to highlight that even at lower Mascot range of 10 to 15, a noteworthy number of 11 HCS (accounting for 45.83% of all 24 HCS) persisted. This finding underscores the limited efficacy of Mascot-based immunopeptide identification for HLA peptides originating from SNVs. Information regarding the overall detected peptides was summarized in Fig. S3. As observed in Fig. S3, the CandiSeq detection count exhibited a significant correlation with both overall and 8–12mer detections, while showing no significant correlation with tumor tissue input, tumor purity or mutation number, which indicated that the variation in CandiSeq amount can be considered as a result of a comprehensive interplay with different factors, including the type and quantity of MHC-I, the number of mutations, as well as the amount of tumor tissue input.
Fig. 3.
Results of Mascot-based immunopeptide identification from 18 patients with cancer. The (a) individual count of detected CandiSeqs in different cancer types; (b) the length distribution of CandiSeqs; (c) count of HCS in CandiSeqs; (d) count and (e) ratio of HCS in CandiSeq at different cutoff values of Mascot ion score.
3.5. Machine learning-based filter of immunopeptide identification
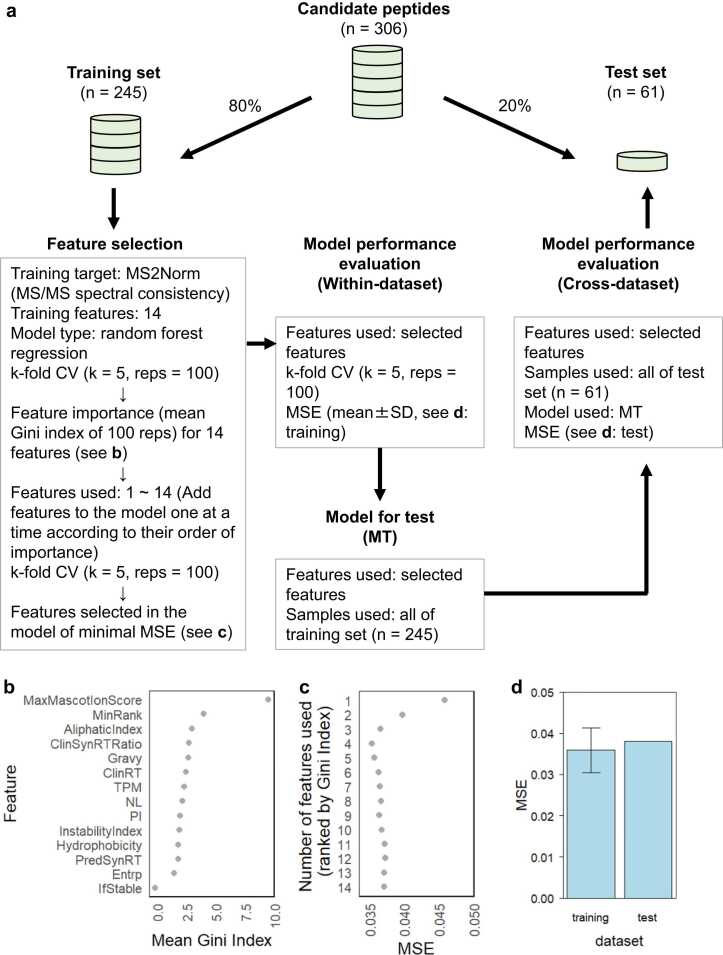
A machine learning-based filter was developed to address the limitation of Mascot in calculating the theoretical spectra for HLA peptides resulting from nonspecific protease cleavage. With details, 14 descriptors related to accurate identification and HLA peptide properties were included as training features and the normalized MS2 values, referred to as MS2Norm, as training targets to construct random forest regression models. To account for the effect of peptide sequence length on the number of peaks in tandem MS measurements, MS2Norm represents the relative consistency between the clinical sample spectra and the assigned sequence spectra, considering the proportion of peaks that may match. Peptides with higher MS2Norm values were more likely to be assigned correctly. Fig. 4a illustrates the process of constructing the machine learning filter. The entire dataset of 306 CandiSeqs was randomly divided into an 80% training set (n = 245) and a 20% test set (n = 61). The training set was used for feature selection, model performance evaluation, and building a model for test. A completely independent test set is used to validate the model obtained from the training set. Regarding feature selection, a two-step approach was developed: firstly, all features were ranked according to importance; secondly, the features were sequentially incorporated into the model based on their important ranking, and the impact on the model’s mean squared error (MSE) was monitored.
Fig. 4.
(a) Development of machine learning models. Important features were selected by considering (b) the Gini indices for 14 descriptors, and (c) the MSE of models with a sequentially increasing number of features used: the model achieved its minimum MSE when using the top four features (MaxMascotIonScore, MinRank, AliphaticIndex, and ClinSynRTRatio) based on importance; (d) the MSE of training (height, mean MSE values; error bars, standard deviation) and test (height, the MSE value) sets. CV, cross-validation; MSE, mean squared error; MT, model for test.
As illustrated in Fig. 4a, a training set with 14 features was used, and a 5-fold CV was repeated 100 times, recording the Gini indices for each feature during this process. We scored the importance of each feature in the model based on the average of the Gini indices and ranked the 14 features in descending order according to their importance (Fig. 4b). Starting with one feature and increasing one feature at a time in the order of Gini indices, we performed loop modeling and calculated the MSE for each model. If the addition of a feature results in an increase rather than a decrease in MSE, it is deemed unhelpful for prediction. Therefore, the combination of features yielding the minimum MSE will be selected as the ultimate set of the important features. As shown in Fig. 4c, four features (MaxMascotIonScore, MinRank, AliphaticIndex, and ClinPredSynRTRatio) used in the model with the minimal MSE were selected as critical features for building the prediction model and evaluating its prediction performance. A 5-fold CV was performed for all training set samples using the four critical features to evaluate the performance of the within-training set prediction. As shown in Fig. 4d, for training set, the mean MSE was 0.0363 ± 0.0057 (mean ± standard deviation: SD), demonstrating effective regression performance. Next, we used all the samples in the training set to establish a model (model for test: MT) for predicting the MS2Norm values of the other independent dataset (test set). As shown in Fig. 4d, the predicted MSE for the test set was 0.0381, suggesting high performance as well as reproducibility in both the training and the test sets. To assess the influence of sequence overlap within the datasets on the evaluation of model performance, all peptide sequences that exhibit an overlap exceeding 8 amino acid residues in the complete CandiSeq dataset (both training and test datasets) were summarized in Table S4. As shown in Table S4, a collective occurrence of 16 subsequences (with amino acid residues ≥ 8) repeated two or more times throughout the entire dataset, which involved a total of 30 peptides, constituting 9.80% of the overall peptide count of 306. In cases where the sequence is entirely replicated (Pair No. 3, 4, 6 and 7), they share identical AliphaticIndex and MinRank values, but exhibit notable differences in MaxMascotIonScore and/or ClinSynRTRatio values, indicating that while certain features are identical for the same sequence, they were considered as distinct samples in the models; in cases where the sequences are not identical but have more than 8 common subsequences, variations are observed not only in MaxMascotIonScore and ClinSynRTRatio but also in AliphaticIndex and MinRank predicted using the sequence information, which indicated that even with an overlap of more than 8 amino acid residues, specific segments continue to have a significant impact on the overall peptide’s aliphatic index and its affinity with HLA. In summary, our model incorporated both the sequence information of the CandiSeqs and the recorded parameters from the MS experiments and database searches, ensuring the absence of duplicate samples in either internal CV on the training set or when predicting the test set using the MT model built on the training set. While some sequence overlap may result in similar feature values, it can be considered that the performance evaluation of the current models was not significantly affected by repeated sequences.
In order to conduct a comparative analysis with Mascot, the training and test sets were consolidated, and the outcomes were obtained through LOO CV methodology for all CandiSeqs.
3.6. Comparison of prediction performance before and after machine learning-based filtering
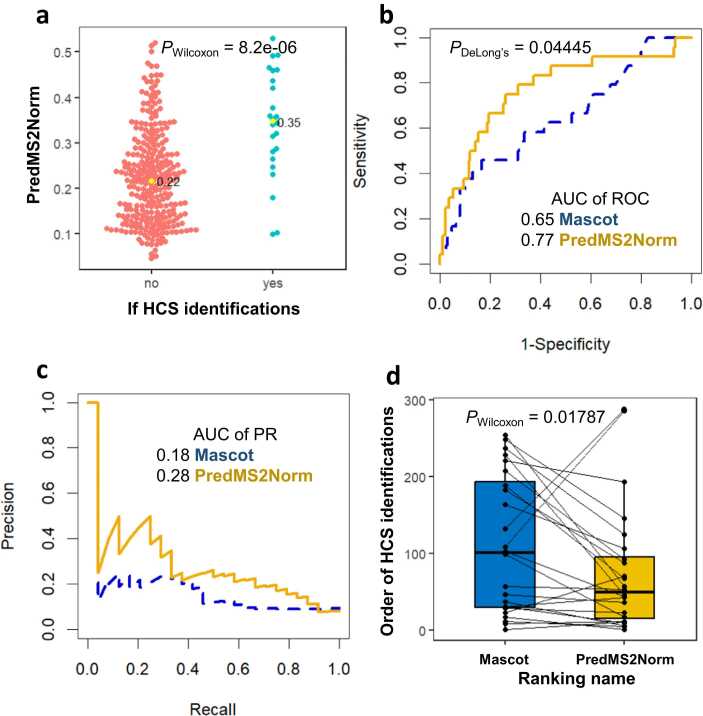
To further confirm the efficacy of machine learning-based filtering compared to Mascot in terms of filtering false positives and rescuing HCS, the distributions of predicted MS2Norm score in HCS and not-HCS identifications were illustrated in Fig. 5a. In Fig. 5a, 24 HCS identifications exhibited significantly higher predicted MS2Norm scores (0.35 vs. 0.22 at median; P value < 0.001). As shown in Figs. 5b and 5c, the area under the curve (AUC) values for ROC and precision-recall (PR) after machine learning-based filtering were 0.77 and 0.28, whereas those for identifications based on Mascot were 0.65 and 0.18. We also compared the rankings of the 24 HCS before and after machine learning-based filtering. As shown in Fig. 5d, the ranking of HCS identifications after filtering exhibited a notably higher position compared to the ranking before filtering. We further validated the filtering effect by varying the cutoff threshold. As shown in Table S5, to detect 20% of the HCS peptides (n = 5), Mascot would require 23 screenings (identification rate 21.7%), while ranking after machine learning-based filtering would require 10 screenings (identification rate 45.5%). To detect 50% of the HCS peptides (n = 12), Mascot would require 100 screenings (identification rate 12.0%), while ranking after filtering would require 46 screenings (identification rate 26.1%); 80% of the HCS peptides (n = 19) would have been identified by screening 208 Mascot candidate sequences (identification rate 10.9%), while 125 rescored candidate sequences (identification rate 15.2%). Therefore, under various cutoff thresholds, the positive rate after machine learning-based filtering exhibited an approximate range of 1.5 to twice the corresponding rate observed for Mascot, indicating that machine learning can effectively filter false positives and rescue positives such as HCS peptides in the identification of HLA peptides.
Fig. 5.
Comparison of prediction performance before and after machine learning-based filtering. (a) The distributions of predicted MS2Norm score in HCS and not-HCS identifications: P value was calculated with a Wilcoxon signed-rank test. The (b) ROC and (c) PR curves regarding HCS or not-HCS before and after the machine learning-based filtering: P value was calculated with DeLong’s test. (d) The order of HCS identifications in all CandiSeqs before and after the machine learning-based filtering: P value was calculated with a two-sample paired Wilcoxon signed-rank test. AUC, area under the curve; HCS, identifications with highly consistent MS/MS spectra; PR, precision-recall; PredMS2Norm score, predicted MS2Norm score; ROC, receiver operating characteristic.
3.7. Critical features and the direction of their bias correction effects
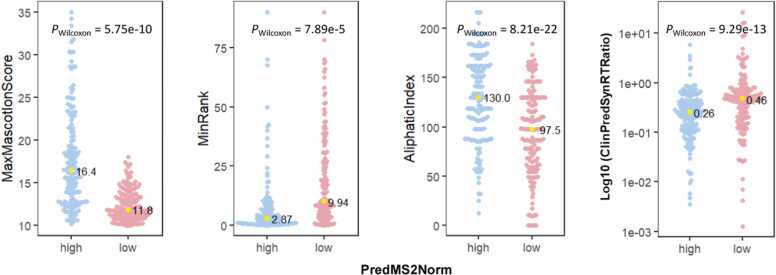
Finally, the relationship between critical features and the prediction scores after filtering was represented in Fig. 6. The median MaxMascotIonScore for the high prediction score group was 16.4, while the median value for the low prediction score group was 11.8 indicating a significant positive impact of Mascot on the prediction results; the median value of MinRank in the high score group was 2.87 indicating a relatively high ranking and robust affinity to HLA, whereas it was 9.94 in the low score group, which suggested a reverse effect on the prediction score; in the high score group, the aliphatic index was 130.0 at median, while it was 97.5 in the low score group, indicating a significant positive effect; the logarithm of ClinSynRTRatio in the high score group was 0.26, where a value closer to zero suggests a smaller deviation between the actual and predicted standard retention time, and it was 0.46 in the low score group, indicating a greater degree of deviation in retention time.
Fig. 6.
Level distribution of important features between groups with high or low predicted MS2Norm scores. The groups were divided based on the median value of predicted MS2Norm. Yellow dots indicate the median value of each group; P value was calculated with a Wilcoxon signed-rank test.
4. Discussion
MS-based immunopeptidomics play a central role in neoantigen identification. However, the conventional proteomics workflows face significant challenges in identifying HLA peptides from real clinical tumor samples.
As an after-processing method, the machine learning-based filter was developed by introducing predictive descriptors, including MaxMascotIonScore, MinRank, AliphaticIndex, and ClinPredSynRTRatio, to filter out false positives without complex modifications to the conventional MS-based peptidomics workflow. In the four critical features, the Mascot score, calculated based on the similarity between the experimental and theoretical MS spectra, remains an essential factor for positive identification, with a higher ion score indicating greater confidence in peptide identification [14]. Our results indicate that Mascot has reliable peptide identification ability for immunopeptides, where paired ion fragments are relatively fully detected.
However, for immunopeptides that cannot be well identified using the Mascot scoring algorithm, the combination of MinRank, AliphaticIndex, and ClinPredSynRTRatio provides a valuable addition to improve identification efficacy. MinRank was obtained from the predicted peptide-MHC binding affinity from NetMHCPan4.1 model, which was independent of the MS measurement and spectral searching process [22]. Previous reports have shown that evaluating the binding affinity of a candidate sequence to the corresponding HLA type of a patient may effectively filter out false positives [17], [23]. Our study showed that a combination of MinRank and other features enabled the filtering of false positives. Aliphatic index, which measures the relative volume occupied by aliphatic side chains in a peptide or protein sequence [24], is crucial for correctly identifying HLA peptides. Sequences that were correctly identified often had a relatively higher aliphatic index than negative sequences, indicating that sequences that were not sufficiently stable and could not resist hydrophobic environmental stress were more prone to false positives. Hydrophobicity and stability in hydrophobic environments have consistently been significant characteristics of HLA peptides, as the binding energy of protein-peptide interactions required to generate an immune response is directly related to the buried hydrophobic area [25], [26]. The significance of hydrophobic interactions is emphasized by a different investigation on T cell antigen receptor (TCR) sequence features, which found that the destiny of T cells during thymic selection is determined by the percentage of hydrophobic amino acids present in the third complementarity-determining region (CDR3β) [27]. Our data revealed that the strength of hydrophobic interactions and the stability of peptides in hydrophobic environments are crucial for correctly identifying HLA peptides in MS-based immunopeptidomics. ClinPredSynRTRatio represents the degree of deviation between the actual retention time of the peptides in the clinical samples and the predicted standard retention time of the assigned sequences. Retention time is an essential reference in MS identification but is not fully utilized in existing workflows. When a peptide is incorrectly assigned, there may be a significant difference between predicted and actual retention times. In our study, ClinPredSynRTRatio showed a significant negative correlation with the score after filtering, confirming and quantifying this trend.
Our results have some limitations, including a small sample size of participants and candidate peptides, a limited number of cancer types, and a single variation database. Therefore, further validation is required to identify HLA peptides with more human tumor samples. Moreover, considering the significance of HLA affinity in our prediction model, it is conceivable that degraded accuracy in NetMHCpan4.1 predictions for rare HLA-I alleles will impact the overall performance of our model. Additionally, because it is a filter based on Mascot identification results, it can only evaluate and correct the sequences proposed by Mascot, which means that the screening ability cannot be validated for non-tryptic peptides not included in the present MS spectral library or sequence database, or in cases where Mascot itself cannot provide optimal prediction. Several recent studies have reported improvements in workflows to enhance immunopeptide identification, e.g., approaches to create new specialized databases for immunopeptides, including non-tryptic peptides, and to develop more powerful MS spectral search engines to handle much larger search spaces [28], [29], [30], [31]. Nevertheless, there is still controversy about the enhancement of the new tools in identifying immunopeptides derived from actual physiological samples [32]. Additionally, optimization at the pipeline level can improve overall performance [33]. We anticipate that our machine learning-based approach, which integrated physicochemical factors and experimental parameters linked to HLA peptide properties, can enhance the precision of emerging immunopeptidomics identification databases and workflows, and ultimately, can provide advantages for the analysis of real clinical tumor samples.
5. Conclusions
Our study comprehensively assessed the precision of conventional proteomics method in the examination of genuine clinical tumor samples, and developed a machine learning-based and after-processing filter biased towards HLA peptides by incorporating peptide physicochemical information and experimental information. The machine learning-based filter with four critical features: MaxMascotIonScore, MinRank, AliphaticIndex, and ClinPredSynRTRatio, can effectively filter out false positive as well as rescue HCS peptides from Mascot identification results, resulting in a two-fold increase in the identification rate of HLA peptides compared to Mascot.
Funding
This work was supported by the Japan Society for the Promotion of Science (JSPS) Grant: Grant-in-Aid for Scientific Research (B):17H04278 and 21H02995 (T.S.) and Grant-in-Aid for Scientific Research (C):22K08724 (F.W.); AMED Grant Number 19ck0106360h0003 (T.S.); The Cell Science Research Foundation Fellowship (F.W.); and Brightpath Biotherapeutics Co., Ltd. S.I. and K.H were full time employees of Brightpath Biotherapeutics Co., Ltd. N.N. is the CSO of Brightpath Biotherapeutics Co., Ltd.
CRediT authorship contribution statement
Conception and design: F.W., N.N., T.K., and T.S. Provision of study materials or patients: T.K., S.M., and T.S. Collection and assembly of data: F.W., T.K., Y.N., H.U., S.I., K.H., Y.A, T.K., and T.S. Data analysis and interpretation: F.W., T.K, Y.N., H.U., S.I., K.H., Y.A, H.H., S.H., K.T., Y.M, N.N., T.K., and T.S. Manuscript preparation: All authors. Final approval of manuscript: All authors. Accountable for all aspects of the work: All authors. Declaration of Generative AI and AI-assisted technologies in the writing process. We did not use generative AI and AI-assisted technologies to analyse or draw insights in both the research and writing process.
Declaration of Competing Interest
Tetsuro Sasada received honoraria from Chugai and Bristol Myers Squibb, and research funds from Taiho.
Acknowledgment
We acknowledge the patients who participated in this study.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2024.01.023.
Contributor Information
Takeshi Kawamura, Email: kawamura@lsbm.org.
Tetsuro Sasada, Email: tsasada@kcch.jp.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
.
References
- 1.Xie N., Shen G., Gao W., Huang Z., Huang C., Fu L. Neoantigens: promising targets for cancer therapy. Signal Transduct Target Ther. 2023;8:9. doi: 10.1038/s41392-022-01270-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin M.J., Svensson-Arvelund J., Lubitz G.S., Marabelle A., Melero I., Brown B.D., Brody J.D. Cancer vaccines: the next immunotherapy frontier. Nat Cancer. 2022;3:911–926. doi: 10.1038/s43018-022-00418-6. [DOI] [PubMed] [Google Scholar]
- 3.Yadav M., Jhunjhunwala S., Phung Q.T., Lupardus P., Tanguay J., Bumbaca S., Franci C., Cheung T.K., Fritsche J., Weinschenk T., Modrusan Z., Mellman I., Lill J.R., Delamarre L. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature. 2014;515:572–576. doi: 10.1038/nature14001. [DOI] [PubMed] [Google Scholar]
- 4.Bassani-Sternberg M., Pletscher-Frankild S., Jensen L.J., Mann M. Mass spectrometry of human leukocyte antigen class I peptidomes reveals strong effects of protein abundance and turnover on antigen presentation. Mol Cell Proteom. 2015;14:658–673. doi: 10.1074/mcp.M114.042812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bassani-Sternberg M., Braunlein E., Klar R., Engleitner T., Sinitcyn P., Audehm S., Straub M., Weber J., Slotta-Huspenina J., Specht K., Martignoni M.E., Werner A., Hein R., D H.B., Peschel C., Rad R., Cox J., Mann M., Krackhardt A.M. Direct identification of clinically relevant neoepitopes presented on native human melanoma tissue by mass spectrometry. Nat Commun. 2016;7 doi: 10.1038/ncomms13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalaora S., Barnea E., Merhavi-Shoham E., Qutob N., Teer J.K., Shimony N., Schachter J., Rosenberg S.A., Besser M.J., Admon A., Samuels Y. Use of HLA peptidomics and whole exome sequencing to identify human immunogenic neo-antigens. Oncotarget. 2016;7:5110–5117. doi: 10.18632/oncotarget.6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aebersold R., Mann M. Mass-spectrometric exploration of proteome structure and function. Nature. 2016;537:347–355. doi: 10.1038/nature19949. [DOI] [PubMed] [Google Scholar]
- 8.Purcell A.W., Ramarathinam S.H., Ternette N. Mass spectrometry-based identification of MHC-bound peptides for immunopeptidomics. Nat Protoc. 2019;14:1687–1707. doi: 10.1038/s41596-019-0133-y. [DOI] [PubMed] [Google Scholar]
- 9.Olsen J.V., Ong S.E., Mann M. Trypsin cleaves exclusively C-terminal to arginine and lysine residues. Mol Cell Proteom. 2004;3:608–614. doi: 10.1074/mcp.T400003-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Biniossek M.L., Schilling O. Enhanced identification of peptides lacking basic residues by LC-ESI-MS/MS analysis of singly charged peptides. Proteomics. 2012;12:1303–1309. doi: 10.1002/pmic.201100569. [DOI] [PubMed] [Google Scholar]
- 11.Chen R., Li J. Enhanced mass spectrometry detection of MHC peptides. Methods Mol Biol. 2019:245–257. doi: 10.1007/978-1-4939-9597-4_15. [DOI] [PubMed] [Google Scholar]
- 12.Chen R., Fauteux F., Foote S., Stupak J., Tremblay T.L., Gurnani K., Fulton K.M., Weeratna R.D., Twine S.M., Li J. Chemical derivatization strategy for extending the identification of MHC class I immunopeptides. Anal Chem. 2018;90:11409–11416. doi: 10.1021/acs.analchem.8b02420. [DOI] [PubMed] [Google Scholar]
- 13.Pfammatter S., Bonneil E., Lanoix J., Vincent K., Hardy M.P., Courcelles M., Perreault C., Thibault P. Extending the comprehensiveness of immunopeptidome analyses using isobaric peptide labeling. Anal Chem. 2020;92:9194–9204. doi: 10.1021/acs.analchem.0c01545. [DOI] [PubMed] [Google Scholar]
- 14.Perkins D.N., Pappin D.J., Creasy D.M., Cottrell J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 15.Faridi P., Purcell A.W., Croft N.P. In immunopeptidomics we need a sniper instead of a shotgun. Proteomics. 2018;18 doi: 10.1002/pmic.201700464. [DOI] [PubMed] [Google Scholar]
- 16.Widbiller M., Schweikl H., Bruckmann A., Rosendahl A., Hochmuth E., Lindner S.R., Buchalla W., Galler K.M. Shotgun proteomics of human dentin with different prefractionation methods. Sci Rep. 2019;9:4457. doi: 10.1038/s41598-019-41144-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laumont C.M., Daouda T., Laverdure J.P., Bonneil E., Caron-Lizotte O., Hardy M.P., Granados D.P., Durette C., Lemieux S., Thibault P., Perreault C. Global proteogenomic analysis of human MHC class I-associated peptides derived from non-canonical reading frames. Nat Commun. 2016;7 doi: 10.1038/ncomms10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kakiuchi M., Nishizawa T., Ueda H., Gotoh K., Tanaka A., Hayashi A., Yamamoto S., Tatsuno K., Katoh H., Watanabe Y., Ichimura T., Ushiku T., Funahashi S., Tateishi K., Wada I., Shimizu N., Nomura S., Koike K., Seto Y., Fukayama M., Aburatani H., Ishikawa S. Recurrent gain-of-function mutations of RHOA in diffuse-type gastric carcinoma. Nat Genet. 2014;46:583–587. doi: 10.1038/ng.2984. [DOI] [PubMed] [Google Scholar]
- 20.Aggarwal S., Raj A., Kumar D., Dash D., Yadav A.K. False discovery rate: the Achilles' heel of proteogenomics. Brief Bioinform. 2022;23 doi: 10.1093/bib/bbac163. [DOI] [PubMed] [Google Scholar]
- 21.Wilkins M.R., Gasteiger E., Bairoch A., Sanchez J.C., Williams K.L., Appel R.D., Hochstrasser D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol Biol. 1999;112:531–552. doi: 10.1385/1-59259-584-7:531. [DOI] [PubMed] [Google Scholar]
- 22.Reynisson B., Alvarez B., Paul S., Peters B., Nielsen M. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 2020;48:W449–W454. doi: 10.1093/nar/gkaa379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith C.C., Selitsky S.R., Chai S., Armistead P.M., Vincent B.G., Serody J.S. Alternative tumour-specific antigens. Nat Rev Cancer. 2019;19:465–478. doi: 10.1038/s41568-019-0162-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ikai A. Thermostability and aliphatic index of globular proteins. J Biochem. 1980;88:1895–1898. [PubMed] [Google Scholar]
- 25.Chothia C., Janin J. Principles of protein-protein recognition. Nature. 1975;256:705–708. doi: 10.1038/256705a0. [DOI] [PubMed] [Google Scholar]
- 26.Kosmoliaptsis V., Chaudhry A.N., Sharples L.D., Halsall D.J., Dafforn T.R., Bradley J.A., Taylor C.J. Predicting HLA class I alloantigen immunogenicity from the number and physiochemical properties of amino acid polymorphisms. Transplantation. 2009;88:791–798. doi: 10.1097/TP.0b013e3181b4a9ff. [DOI] [PubMed] [Google Scholar]
- 27.Lagattuta K.A., Kang J.B., Nathan A., Pauken K.E., Jonsson A.H., Rao D.A., Sharpe A.H., Ishigaki K., Raychaudhuri S. Repertoire analyses reveal T cell antigen receptor sequence features that influence T cell fate. Nat Immunol. 2022;23:446–457. doi: 10.1038/s41590-022-01129-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Declercq A., Bouwmeester R., Hirschler A., Carapito C., Degroeve S., Martens L., Gabriels R. MS(2)Rescore: data-driven rescoring dramatically boosts immunopeptide identification rates. Mol Cell Proteom. 2022;21 doi: 10.1016/j.mcpro.2022.100266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilhelm M., Zolg D.P., Graber M., Gessulat S., Schmidt T., Schnatbaum K., Schwencke-Westphal C., Seifert P., de Andrade Kratzig N., Zerweck J., Knaute T., Braunlein E., Samaras P., Lautenbacher L., Klaeger S., Wenschuh H., Rad R., Delanghe B., Huhmer A., Carr S.A., Clauser K.R., Krackhardt A.M., Reimer U., Kuster B. Deep learning boosts sensitivity of mass spectrometry-based immunopeptidomics. Nat Commun. 2021;12:3346. doi: 10.1038/s41467-021-23713-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tran N.H., Zhang X., Xin L., Shan B., Li M. De novo peptide sequencing by deep learning. Proc Natl Acad Sci USA. 2017;114:8247–8252. doi: 10.1073/pnas.1705691114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tran N.H., Qiao R., Xin L., Chen X., Liu C., Zhang X., Shan B., Ghodsi A., Li M. Deep learning enables de novo peptide sequencing from data-independent-acquisition mass spectrometry. Nat Methods. 2019;16:63–66. doi: 10.1038/s41592-018-0260-3. [DOI] [PubMed] [Google Scholar]
- 32.Zhang L., Liu G., Hou G., Xiang H., Zhang X., Huang Y., Zhang X., Li B., Lee L.J. IntroSpect: motif-guided immunopeptidome database building tool to improve the sensitivity of HLA I binding peptide identification by mass spectrometry. Biomolecules. 2022;12 doi: 10.3390/biom12040579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shahbazy M., Ramarathinam S.H., Illing P.T., Jappe E.C., Faridi P., Croft N.P., Purcell A.W. Benchmarking bioinformatics pipelines in data-independent acquisition mass spectrometry for immunopeptidomics. Mol Cell Proteom. 2023;22 doi: 10.1016/j.mcpro.2023.100515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material