Abstract
Objective:
This paper aimed to investigate the PI3K/Akt/mTOR signal-pathway regulator factor-related lncRNA signatures (PAM-SRFLncSigs), associated with regulators of the indicated signaling pathway in patients with lung adenocarcinoma (LUAD) undergoing immunotherapy.
Materials and Methods:
In this retrospective study, we employed univariate Cox, multivariate Cox, and least absolute shrinkage and selection operator (LASSO) regression analyses to identify prognostically relevant long non-coding RNAs (lncRNAs), construct prognostic models, and perform Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses. Subsequently, immunoassay and chemotherapy drug screening were conducted. Finally, the prognostic model was validated using the Imvigor210 cohort, and tumor stem cells were analyzed.
Results:
We identified seven prognosis-related lncRNAs (AC084757.3, AC010999.2, LINC02802, AC026979.2, AC024896.1, LINC00941 and LINC01312). We also developed prognostic models to predict survival in patients with LUAD. KEGG enrichment analysis confirmed association of LUAD with the PI3K/Akt/mTOR signaling pathway. In the analysis of immune function pathways, we discovered three good prognostic pathways (Cytolytic_activity, Inflammation-promoting, T_cell_co-inhibition) in LUAD. Additionally, we screened 73 oncology chemotherapy drugs using the "pRRophetic" algorithm.
Conclusion:
Identification of seven lncRNAs linked to regulators of the PI3K/Akt/mTOR signaling pathway provided valuable insights into predicting the prognosis of LUAD, understanding the immune microenvironment and optimizing immunotherapy strategies.
Keywords: Immunotherapy, LncRNAs, Lung Adenocarcinoma, Prognosis, Tumor Microenvironment
Introduction
Lung cancer is the malignancy with the highest morbidity and mortality worldwide. The main type of non-small cell lung cancer (NSCLC) is LUAD. PI3K/ Akt/mTOR signaling pathway, as a crucial regulator of various cellular processes including cell proliferation, growth, survival, migration, apoptosis, translation, glucose metabolism and DNA repair, plays a pivotal role in essential cellular activities (1). The primary objective of regulating PI3K-AKT pathway is to stimulate cellular growth and proliferation. However, excessive activation of this signaling pathway leads to an overstimulation of cells, resulting in abnormal cell proliferation, such as tumorigenesis, as well as involvement in tumor erosion and metastasis. Dysregulation of this pathway is commonly observed in cancer, exerting control over multiple cancer-related features. It has been shown that aberrant activation or inhibition of the intracellular PI3K/Akt/mTOR pathway has been identified as a major contributor to cancer cell resistance against antitumor therapies (2). Notably, patients with EGFR mutations accompanied with PI3K pathway activation exhibited shorter progression-free survival (PFS) and overall survival (OS) (3). Activation of the PI3K/Akt/mTOR signaling pathway has been demonstrated to be prevalent in diffuse gliomas and escalated with tumor grade (4).
Excessive activation of the PI3K/Akt/mTOR pathway can detrimentally impact the patient’s prognosis. In the selected preclinical and initial clinical trials, targeted therapeutic intervention through inhibition of PI3K/ Akt/mTOR signaling has been contemplated as a viable strategy due to its intimate correlation with tumorigenesis and disease progression (5).
Many individuals diagnosed with lung cancer are unfortunately diagnosed in the advanced stages of the disease, either with locally progressed tumors or metastatic spread. This delayed diagnosis, coupled with the absence of suitable therapeutic targets, contributed to the unfavorable prognosis experienced by patients (6, 7). Consequently, it becomes imperative to investigate potential biomarkers that can aid in the early detection and treatment of LUAD, enabling intervention at an earlier stage. Through our research, we identified some lncRNAs as diagnostic biomarkers for LUAD patients (7-9). Despite their inability to encode proteins, lncRNAs play a pivotal role in gene transcription and expression. Numerous studies have demonstrated the involvement of lncRNAs in the various diseases, influencing critical cellular functions, such as chromatin modification, gene expression regulation, cell differentiation, and cell cycle progression. Additionally, lncRNAs can exert their effects by acting as competing RNAs, thereby impacting expression of the target genes (10, 11). In general, lncRNAs carry out their functions by interacting with various biomolecules, including DNA, RNA and proteins. They play a crucial role in regulation of gene expression, which significantly impacts tumorigenesis and disease progression. Nevertheless, specific roles of these lncRNAs in lung adenocarcinoma (LUAD) and their precise association with the PI3K/AKT/mTOR signaling pathway remained largely unknown.
Overall, the PI3K/AKT/mTOR pathway serves as a vital coordinator of cellular responses to internal and external stimuli, thereby influencing key processes associated with carcinogenesis (12). Hence, investigating the expression patterns of lncRNAs associated with the PI3K/AKT/mTOR signaling pathway in LUAD, in addition to comprehending their diverse expressions and functions in LUAD can furnish us with a more profound comprehension of LUAD. Furthermore, this exploration can unveil pivotal regulatory factors in the mechanism of LUAD development, as well as the biological mechanisms of drug resistance, thereby offering novel prospects for prevention, diagnosis and treatment of LUAD. In our investigation, we meticulously scrutinized and sieved through PAM-SRFLncSig to construct a prognostic risk model tailored specifically for LUAD patients. Through this approach, we delved into the intricate interplay between PAM-SRFLncSig and LUAD prognosis, as well as its impact on the immune microenvironment and potential implications for immunotherapy. Additionally, we identified promising molecular markers and potential drug targets, thus presenting a fresh research strategy for LUAD immunotherapy.
Materials and Methods
Databases
The flow chart of this study is shown in Figure S1A (See Supplementary Online Information at www.celljournal. org). The Molecular Signature Database (MSigDB) is a comprehensive resource housing numerous annotated gene sets, meticulously categorized into human and mouse collections. In this investigation, we initially procured four gene sets associated with the PI3K/Akt/mTOR signaling pathway from MSigDB. The database can be accessed at https://www.gsea-msigdb.org/gsea/msigdb/index.jsp website. Specifically, we focused on the WikiPathways gene set labeled as CP (canonical pathways) within the human gene C2 (curated gene sets) in MSigDB. The downloaded gene sets were as follows: "WP_FOCAL_ ADHESIONPI3KAKTMTORSIGNALING_PATHWAY (n=309)","WP_PI3KAKTMTOR_SIGNALING_ PATHWAY_AND_THERAPEUTIC_OPPORTU NITIES (n=30)" and "WP_PI3KAKTMTOR_VITD3_ SIGNALLING (n=22)". The "HALLMARK_PI3K_ AKT_MTOR_SIGNALING" gene set (n=105) was subsequently acquired from the H (hallmark gene sets) category, focusing on the genes up-regulated upon activation of the PI3K/AKT/mTOR pathway. After eliminating duplicate genes from the four gene sets, a final set of 399 genes associated with the PI3K/Akt/ mTOR signaling pathway was obtained and included in the subsequent analysis (Fig .S1B, Table S1, See Supplementary Online Information at www.celljournal. org). The data for this study encompassed mRNA transcription data (n=19508), lncRNA transcription data (n=13481), corresponding clinical information files (Table S2, See Supplementary Online Information at www.celljournal.org), and prognostic data from 494 patients with LUAD, which were obtained from The Cancer Genome Atlas (TCGA; https://portal.gdc. cancer.gov/), as a collaborative resource established by the National Cancer Institute and the National Human Genome Research Institute, which provides comprehensive cancer genomic data, including mutation, mRNA expression and methylation data. Patient data from the IMVigor210 clinical trial were also downloaded from the "IMvigor210CoreBiologies" R package.
Extracting expression matrix of lncRNAs co-expressed with the gene sets
For data analysis, we utilized the “R software (version 4.1.3)”. Initially, we extracted expression matrix of the lncRNAs co-expressed with the PI3K/AKT/mTOR gene set using the "limma" package. The data were processed and subjected to a Pearson correlation test, and a total of 3610 lncRNA associated with LUAD were identified (|cor| > 0.4, P<0.0001). The resulting genome-lncRNA associations were visualized using a Sankey diagram. Then, we constructed a prognostic model for genomeassociated lncRNAs using the “limma” package.
Construction of the correlated lncRNAs prognostic model with Cox regression analysis
We conducted univariate Cox regression analysis to identify significant lncRNAs (P<0.05) and established correlation of lncRNA expression with patient survival. To streamline our model, we employed LASSO regression, which incorporated a penalty function into the commonly used multiple linear regression, continuously compressing the coefficients to prevent covariance and overfitting (13). Risk factor for each gene was calculated using the following formula (7-9):
PAM-SRFLncSig=ΣI=1nCoef (i)×Expr (i)
Subsequently, the data was divided, and a total of 494 LUAD patients were randomly assigned to the training cohort (n=330) and testing cohort (n=164). If all p-values between the two groups were non-significant, it indicated that there was no statistical difference in the various clinical indicators of the randomized groups, ensuring well-grouped data without any statistical bias.
Analysis and validation of a clinically independent prognostic model for patients with LUAD
The LUAD patients in the training cohort were divided into high-risk and low-risk subgroups, based on the median risk score. OS in the both subgroups was analyzed using Kaplan-Meier survival curves (P<0.001). Furthermore, a Cox regression (P<0.001) risk model was established to determine prognosis of clinicopathological factors, including age, gender, race, American Joint Committee on Cancer (AJCC) stage, primary tumor (T), distant metastasis (M), regional lymph nodes (N), and risk score. We developed a mathematical model using the Cox regression risk model. Clinical receiver operating characteristic (ROC) curves and survival time ROC curves were used for this assessment. In addition, we constructed column line plots to predict one, three and five year(s) OS in LUAD patients. These column line plots were created using the “rms” package in the “R software”. Nomograms were employed to visualize parameters related to patient survival. Consistency index (C-index) and calibration curves were used to explore performance of the column line plots and the predicted versus actual survival probabilities.
GO and KEGG enrichment analysis
To unravel the profound biological implications of PAMSRFLncSig in LUAD, we embarked on an enrichment analysis employing the GO database and the KEGG pathway to delve into the genes encoding co-expressed proteins of prognostic lncRNAs. By comparing expression of lncRNAs in LUAD patients, we meticulously screened and identified 418 differentially expressed lncRNAs (logFC filter=1, FDR <0.05), employing the false discovery rate (FDR), as a stringent filter. Subsequently, we subjected these lncRNAs to Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses, unveiling the enriched p-values for specific GO annotations and KEGG pathways, thereby illuminating the synergistic correlation of the functions of all lncRNAs. We deemed a corrected P value FDR <0.1, as the significance index for GO enrichment, while P<0.5 was considered as the significance index for KEGG.
Immunoassay of PAM-SRFLncSig high- and low-risk subgroups
To assess relative differences in immune profiles between high- and low-risk subgroups, we obtained immunocompetent gene set files and analyzed differential expression of immune checkpoints in these populations. In addition, we performed a single-sample gene set enrichment analysis (ssGSEA) using the "limma", "GSVA" and "GSEABase" packages in the "R software" to assess the infiltrating immune cells and immune-related functions in LUAD patients. Role of PAM-SRFLncSig in 13 immune functions was assessed and presented in a heat map (14).
Analysis of TMB and TIDE scores
Tumor mutation burden (TMB), defined as the number of cellular mutations per one million bases, serves as a biomarker for predicting the efficacy of immunotherapy (15). We analyzed differences in TMB and survival between high- and low-risk groups using the “limma” and “ggpubr” packages, and performed a KM survival analysis to explore associations between TMB, risk subgroups, and survival outcomes (P<0.001). To predict patients response to immunotherapy and identify factors related to tumor immune escape mechanisms, we utilized the tumor immune dysfunction and exclusion (TIDE) algorithm. TIDE scoring file was obtained from http://tide.dfci.harvard. edu/ (16). Furthermore, we analyzed myeloid-derived suppressor cells (MDSC), cancer-associated fibroblasts (CAF), tumor-associated macrophages M2 (TAMM2), microsatellite instability (MSI), Merck18, IFNG, CD274 and cluster of differentiation (CD) (17-20).
Screening of potential chemotherapeutic agents for oncology
Drug resistance poses a significant obstacle in the field of oncology treatment. We used the “pRRophetic “software package in the “R software” to predict the maximum half inhibitory concentration (IC50) of the applicable drugs to high- and low-risk subgroups, in order to evaluate drug sensitivity of the prediction model. This algorithm, which has been extensively employed in the various studies, has been previously published (20).
IMvigor210 model validated PAM-SRFLncSig model for immunotherapy
We validated prognostic value of the PAM-SRFLncSig model, using an immunotherapy dataset (Imvigor210). This dataset encompassed expression data from human metastatic uroepithelial carcinoma (mUC) samples, specifically associated with patient response to anti- PD-L1 immunotherapy. Relevant data was obtained from the "IMvigor210CoreBiologies" package (21). While IMvigor210 focused primarily on mUC, it encompassed a broad cohort of patients, many of whom have concurrently suffered from different types of malignancy, including lung cancer, providing valuable insights into the feasibility of immunotherapy in LUAD (22). By examining efficacy of immunotherapy and influence of specific variables on treatment response, this study extends its implications beyond mUC. LUAD patients may have similar expression patterns of immune checkpoint-related genes and TMB as mUC. Consequently, discoveries of IMvigor210 may serve as pivotal cues, inspiring further investigation into the realm of immunotherapeutic approaches for LUAD. Initially, we matched the genes of LUAD patients, obtained through LASSO regression analysis, with the immunotherapy data in the Imvigor210 model. Subsequently, risk scores were assigned for the patients in Imvigor210, using the same formula employed for risk scoring. Finally, based on these risk scores, the patients were categorized into high- and low-risk subgroups, and the differences in survival and immunotherapy response between high- and low-risk patients in IMvigor210 were analyzed using the "survival", "survminer", "caret", "timeROC", "limma" and "ggpubr" packages (14).
Differential analysis of tumor stem cells
RNA expression-based stemness index (mRNAsi), is a metric that captures the likeness of tumor cells to cancer stem cells (CSCs). To investigate association between genetic characteristics and tumor stemness, we employed the one-class logistic regression (OCLR) algorithm, in order to calculate mRNAsi to measure the degree of similarity between tumor cells and stem cells based on gene expression levels (23). Subsequently, we calculated the mRNAsi of each LUAD patient and categorized them into high- and low-mRNAsi subgroups and performed survival analyses on these subgroups (P<0.001). Lastly, we examined correlation between mRNAsi and clinicopathological factors in the LUAD patients (P<0.001).
Ethical approval
The work was approved by the Guangdong Medical University committee (Zhanjiang, China, YS2021159).
Statistical analysis
Statistical analysis of this study was utilized in the "R software (version 4.1.3)". The "limma" package was employed to screen relevant lncRNAs, followed by regression analysis utilizing one-way COX, multi-way COX, and LASSO. Kaplan-Meier (KM) curves were used to compare survival times between high- and lowrisk subgroups, as determined by the median risk score, while ROC curves were used to assess quality of clinically independent prognostic models for the LUAD patients. Subsequently, a nomogram was constructed using the "rms" package of the TCGA dataset, incorporating significant variables identified through multivariate Cox regression. Prognostic accuracy of the nomogram was assessed using the c-index. Prognostic lncRNA coexpression protein coding genes were enriched using the GO database and KEGG pathway. Infiltrating immune cells and immune-related functions in the LUAD patients were evaluated using the "limma", "GSVA", and "GSEABase" packages in the “R software”. PAMSRFLncSig role in immune function was assessed through ssGSEA. Impact of immunotherapy was then evaluated using TMB and TIDE scores. Potential drugs for the LUAD patients were identified using the pRRophetic package in the “R software”. Finally, the OCLR algorithm was employed to analyze the correlation of mRNAsi with clinicopathological factors in the LUAD patients.
Results
Co-expression relationship between the gene sets and lncRNAs
The Sankey diagram outcome is visually depicted in the Figure S2 (See Supplementary Online Information at www.celljournal.org). Through univariate Cox regression analysis, a total of 339 lncRNAs exhibited significance (P<0.05), with 31 lncRNAs displayed an even more remarkable P<0.0001 (Table S3, See Supplementary Online Information at www.celljournal.org). The latter lncRNAs included AL606489.1, DEPDC1-AS1, LINC02323, AC084757.3, LINC01116, LINC01537, BZW1-AS1, AC021066.1, AC034223.1, TMPO-AS1, LINC02178, AP000695.2 LINC00519, LINC02320, LINC02802, SMILR, AP005137.2, MIR193BHG, NKILA, AC025419.1, LINC00941, AC009743.1, AC108136.1, LINC01117, ABCA9-AS1, LINC00707, AC090023.2, FLG-AS1, AC079949.2, AL049836.1, LINC01312 and OGFRP1.
Construction of a clinical prognostic model for gene set-associated lncRNAs
A total of 494 patients with LUAD were randomly divided into a training cohort (n=330) and a testing cohort (n=164). Following validation, comparison of the clinical indicators between the two groups of patients, the difference was not statistically significant (P>0.05), indicating a good grouping. Subsequently, we analyzed the independent clinical prognosis of the training group. We employed one-way Cox analysis and LASSO regression to screen lncRNAs (P<0.05, Table S4, See Supplementary Online Information at www.celljournal.org, Fig.1A, B). Seven differentially expressed PAM-SRFLncSigs were then identified using multivariate Cox regression analysis (significance filtering criteria P<0.05), and the results are presented in Table S5 (See Supplementary Online Information at www.celljournal.org). In addition, we calculated risk scores for the training cohort and divided them into high-risk (n=165) and low-risk (n=165) subgroups, based on median risk scores. We also plotted KM survival curves for patients in both subgroups (Fig .1C). The low-risk subgroup exhibited significantly longer survival time compared to the high-risk subgroup (P<0.001). Heat map of the training cohort demonstrated differential expression of the seven risk-related lncRNAs in the high- and low-risk subgroups. Blue color indicated low expression and red color indicated high expression. AC010999.2 [hazard ratios (HR)<1] and AC026979.2 (HR<1) appeared as red in the low-risk region, indicating high gene expression and a lower risk of tumor death (Fig .1D). Risk score distribution of the LUAD patients revealed significantly lower survival rate in the high-risk subgroup, compared to the low-risk subgroup (Fig.1E, F). Results of the testing cohort and the entire cohort are depicted in the Fig.S3A-F (See Supplementary Online Information at www.celljournal.org). KM survival curve plot for the testing cohort patients demonstrated a significantly lower probability of survival in the high-risk subgroup compared to the low-risk subgroup (P<0.05). Consistency between results of the training and testing cohorts indicated accurate prediction of the patient survival by the clinical prognostic model for LUAD.
Fig.1.
Target genes were identified using lasso Cox regression analysis for the construction of seven lncRNA signatures in the training cohort. A. Log(λ) versus change in regression coefficient. B. Partial-likelihood deviance curve with Log(λ) in Lasso regression. C. The probability of survival was significantly lower in the high-risk subgroup than the low-risk subgroup by the Kaplan-Meier test. D. AC010999.2 and AC026979.2 were highly expressed in the lowrisk subgroup, and AC084757.3, LINC02802, AC024896.1, LINC00941 and LINC01312 were highly expressed in the high-risk subgroup. E, F. Training cohort survival status and risk score for each case.
Association of PAM-SRFLncSig with clinical profiles of the LUAD patients
To comprehend relationship of PAM-SRFLncSig and the clinical profile of the LUAD patients, we visually represented the patient clinical profile and risk scores. The outcomes revealed that M (P=0.77), age (P=0.59), gender (P=0.83), race, stage, T and N (P>0.05) exhibited no association with risk scores (Fig .S4A-G, See Supplementary Online Information at www.celljournal. org). However, the survival status was correlated with the risk score (Fig .S4H, See Supplementary Online Information at www.celljournal.org). Univariate Cox regression analysis demonstrated that AJCC stage (HR=1.552, 95% confidence interval (CI): 1.303-1.849, P<0.001), T (HR=1.481, 95% CI: 1.162 ~ 1.888, P=0.001), N (HR=1.827, 95% CI: 1.454 ~ 2.296, P<0.001), and risk score (HR=1.269, 95% CI: 1.185 ~ 1.358, P<0.001) significantly influenced the survival time of the LUAD patients (Fig .S4I, See Supplementary Online Information at www.celljournal.org). Furthermore, multifactorial Cox regression analysis revealed that only the risk score (HR=1.210, 95% CI: 1.124 ~ 1.303, P<0.001) exhibited a significant association with the survival time of the LUAD patients (Fig .S4J, See Supplementary Online Information at www.celljournal.org).
Evaluation of the clinically independent prognostic model for patients with LUAD
We assessed the efficacy of patient clinical prognostic models, through the utilization of clinical ROC curves and survival time ROC curves. Prognostic capacity of these predictive models was evaluated by calculating the area under the curve (AUC) of time-dependent ROC curves. ROC curves were plotted for 1-, 3- and 5-year(s) intervals, with AUC values of 0.717, 0.670 and 0.686, respectively (Fig .S5A, See Supplementary Online Information at www.celljournal.org). All AUC values exceeded 0.5, indicating that the model played a critical role in predicting outcomes in the patients with LUAD (24). Figure S5B (See Supplementary Online Information at www.celljournal.org) displays the clinicopathological indicators for 1-year survival probability, including risk score (AUC=0.717), age (AUC=0.470), gender (AUC=0.554), race (AUC=0.497), AJCC stage (AUC=0.698), T stage (AUC=0.633), M stage (AUC=0.497) and N stage (AUC=0.652). These clinical indicators, except for age, AJCC stage and M stage, were found to be potential predictors of 1-year survival probability for the LUAD patients (AUC>0.5). Clinicopathological indicators for 3- and 5-years survival probabilities are depicted in the Figure S5C and D (See Supplementary Online Information at www.celljournal. org). Overall, risk scores exhibited a relatively high AUC value, making them superior predictors of prognosis for the LUAD patients.
Clinical C-index curves can be used to evaluate predictive efficacy and differentiation of clinically independent prognostic models for patients (25). A C-index curve closer to one indicates greater accuracy in the predicted values of the Cox regression model (Fig. S5E, See Supplementary Online Information at www. celljournal.org). We employed a nomogram based on univariate and multifactorial Cox regression analyses to visually predict patients OS at 1-, 3- and 5-year intervals. The predictive performance of the nomogram was further assessed using the calibration curve method, which demonstrated favorable prediction outcomes (Fig. S5F, G, See Supplementary Online Information at www. celljournal.org).
Model validation of clinical grouping data for the LUAD patients
We categorized patients with LUAD into high-risk subgroups and low-risk subgroups, based on their clinical characteristics (age, gender, race, etc.). Our findings revealed that the OS rate of deceased patients was not associated with the risk score (P=0.311, Fig .S6A, See Supplementary Online Information at www.celljournal. org). Survival probability of the patients with LUAD over time was 100% (Fig .S6B, See Supplementary Online Information at www.celljournal.org). Notably, age ≤65 years (P=0.004) and age >65 (P=0.005), as well as male patients (P=0.022) and female patients (P=0.001), black or African American patients (P=0.041) and white patients (P<0.001), stage I patients (P=0.011) and stage II patients (P=0.014), stage T2 patients (P=0.005) and stage T3 patients (P=0.003), stage M0 patients (P<0.001), stage N0 patients (P=0.003), and stage N1 patients (P=0.044) exhibited significant associations with survival (Fig.S6C-J, N, O, Q, S, T, See Supplementary Online Information at www.celljournal.org). The high-risk subgroup of patients experienced a more rapid decline in survival probability compared to the low-risk subgroup. However, there was no significant correlation between risk scores and OS rates in the LUAD patients with stage III (P=0.053), stage IV (P=0.743), T1 (P=0.379), T4 (P=0.131), M1 (P=0.743), and N2 (P=0.160, Fig.S6K-M, P, R, U, See Supplementary Online Information at www. celljournal.org). Furthermore, we employed principal component analysis (PCA) to evaluate distinguishability of the all genes, mRNA, lncRNA, and risk lncRNA between the high and low-risk groups (Fig .S7A-D, See Supplementary Online Information at www.celljournal. org). The results suggested that samples from the two risk subgroups typically had different statuses that can be characterized by lncRNA signatures.
Functional enrichment analysis of lncRNA-related pathways in the LUAD patients with a prognosis
We conducted GO enrichment analysis (n=158), based on lncRNAs expression in the LUAD patients and identified lncRNAs (P<0.1) that were closely associated with specific pathways. Among these, a majority of the differentially expressed lncRNAs were found to be particularly relevant to pathways such as collagencontaining extracellular matrix (GO: 0062023), epidermis development (GO: 0008544), cell-cell junction (GO: 0005911) and skin development (GO: 0043588; Fig.2A). Notable examples include LAMA1, LAMA3, COL7A1 for collagen-containing extracellular matrix, GJB5, LAMA3, COL7A1 for epidermal development, LAMA1, COL7A1, GJB3 for cell-cell junction, and TP63, SPRRA1, GJB3 for skin development. GO analysis of the differentially expressed genes revealed that 20 genes exhibited significant enrichment in the biological process category at a P value of 0.1 (Fig .2B). Furthermore, at P<0.05, KEGG pathway maps also demonstrated significant enrichment of the majority of differentially expressed lncRNAs in pathways such as (hsa04060 cytokinecytokine receptor interactions) including IL22RA1, MSTN, IL1R2, (hsa04510 foci adhesion) and (hsa05206 microRNAs in cancer; Fig.2C). Functional enrichment chord plot of KEGG highlighted a notable differential expression pattern in the LAMA1 gene (Fig .2D).
Fig.2.
GO/KEGG pathway was used for the all differential lncRNAs between the high-risk and low-risk subgroups (n=158) and for 13 immune function pathways in the high- and low-risk subgroups. A, B. Results of GO pathway analysis. C, D. Results of KEGG pathway analysis. E. Thirteen immune function pathways in the training cohort, F. Thirteen immune function pathways in the testing cohort and G. Thirteen immune function pathways in the entire cohort. GO; Gene ontology and KEGG; Kyoto Encyclopedia of Genes and Genomes.
Immune function study of PAM-SRFLncSig high-risk and low-risk subgroups
The high-risk and low-risk subgroups were visually represented through a heat map, showcasing the expression levels of 13 immune functional pathways. In the training cohort, the low-risk subgroup exhibited high expression levels of Cytolytic_activity, Inflammation-promoting, and T_cell_co-inhibition pathways, while the highrisk subgroup displayed low expression levels (P<0.05, Fig .2E). This suggests that these pathways are indicators of a low-risk state for LUAD with a better prognosis and lower risk of cancer recurrence. Consistency in immune function pathways was observed in both the testing cohort and the entire cohort, mirroring the findings of the training cohort (Fig .2F, G).
Analysis of tumor mutation burden of PAMSRFLncSig
Somatic mutation data from the TCGA database were downloaded to analyze disparities in values and subgroups between the high-risk and low-risk prognostic models. It should be noted that there was no significant difference (P>0.05) in TMB between both of the high-risk and low-risk subgroups (Fig .3A-C). Subsequently, survival analysis was conducted using the “survminer” package, and survival curves were plotted (Fig .3D-F). The combined survival curves of TMB and high/low risk revealed that the H-TMB+ lowrisk subgroup exhibited the longest survival in both of the training cohort and the entire cohort (P<0.001). In contrast, the L-TMB+ high-risk subgroup had the shortest survival (Fig .3G-I).
Fig.3.
Analysis of TMB of PAM-SRFLncSig. Box plots show no significant difference in TMB between the high-risk and low-risk subgroups of the A. Training cohort, B. Testing cohort and C. Entire cohort. KM survival curves show differences in TMB between the high-risk and low-risk subgroups of the D. Training cohort, E. Testing cohort, F. Entirecohort. Survival curve results for TMB combined with high-risk and low-risk of the G. Training cohort, H. Testing cohort, and I. Entire cohort. TMB; Tumor mutation burden.
Analysis of tumor immune escape and immunotherapy in the high-risk and low-risk subgroups of PAMSRFLncSig
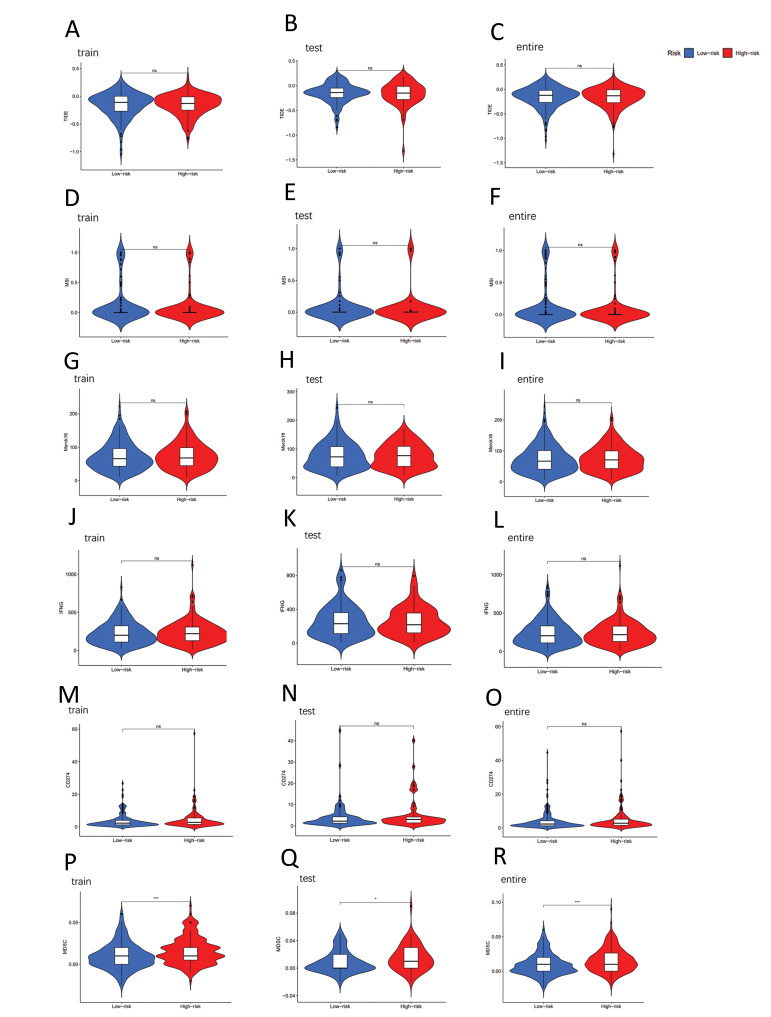
In the high-risk and low-risk subgroups, analyses of tumor immune escape and immunotherapy were performed using PAM-SRFLncSig, with TIDE scoring showing no significant difference in the training cohort, testing cohort and entire cohort (P>0.05, Fig .4A-C). However, statistically significant differences were observed in MDSC (Fig .4P-R), Exclusion (Fig .5A-C), CAF (Fig .5J-L) and TAMM2 (Fig .5M-O) between the high-risk and low-risk subgroups (P<0.001). On the other hand, other immunomarkers, such as MSI, Merck8, IFNG, CD274 (Fig .4D-O), Dysfunction and CD8 (Fig .5D-I), did not exhibit statistically significant difference between the high-risk and low-risk subgroups (P>0.05).
Fig.4.
Tumor microenvironmental results of PAM-SRFLncSig. Differences in A-C. TIDE, D-F. MSI, G-I. Merck18, J-L. IFNG, M-O. CD274 and P-R. MDSI were analyzed in high- and low-risk subgroups. *; P<0.05, **; P<0.01, ***; P<0.001, and ns; No significance.
Fig.5.
Tumor microenvironmental results of PAM-SRFLncSig. Differences in A-C. Exclusion, D-F. Dysfunction, G-I. CD8, J-L. CAF and M-O. TAMM2 were analyzed in high-risk and low-risk subgroups. *; P<0.05, **; P<0.01, ***; P<0.001, and ns; No significance.
Screening of tumor chemotherapy drugs
The pRRophetic algorithm was employed to screen cancer chemotherapy agents within the highrisk and low-risk subgroups of PAM-SRFLncSig. A total of 73 drugs were identified through this algorithm. Upon comparing drug sensitivity, notable variations in IC50 values were observed between the low-risk and high-risk subgroups for multiple drugs (P<0.05). Notably, the LUAD patients in the highrisk subgroup had increased sensitivity to the various drugs (Bicalutamide, Bleomycin, BMS.509744, BMS.754807, Bortezomib, Bryostatin.1, CGP.082996, CGP.60474, CI.1040, CMK, Dasatinib, Docetaxel and Embelin). Conversely, the low-risk subgroup demonstrated lower IC50 values for drugs, such as BIRB.0796, CCT007093, and EHT.1864, indicating that the low-risk subgroup was better treated. In this regard, Figure S8A-P (See Supplementary Online Information at www.celljournal.org) illustrates the top 16 drugs that show potential for dosing in LUAD patients.
IMvigor210 model validated PAM-SRFLncSig model for immunotherapy
By conducting LASSO regression analysis and aligning the genes obtained from the LUAD patients with immunotherapy data isolated from the Imvigor210 model, we identified the LINC00941 and LINC01312 genes. Utilizing a risk scoring formula, patients in the Imvigor210 dataset were assigned scores and subsequently classified into the high-risk and low-risk subgroups (P<0.001). Unfortunately, there was no statistically significant survival between the two subgroups of IMvigor210 bladder cancer target genes (P=0.440, Fig. S9A, See Supplementary Online Information at www. celljournal.org), indicating poor predictive capability. Subsequently, the PAM-SRFLncSig model was validated using ROC curves, demonstrating improved prediction of 5-years survival in bladder cancer patients (AUC=0.514, Fig .S9B, See Supplementary Online Information at www. celljournal.org). Furthermore, no significant difference in target gene risk scores, derived from LASSO regression, was observed among different drug responses to IMvigor210 bladder cancer immunotherapy in the LUAD patients (P=0.092, Fig .S9C, See Supplementary Online Information at www.celljournal.org).
Differential analysis of tumor stem cells
Utilizing the OCLR algorithm, we computed the mRNAsi for each LUAD patient based on their gene expression profiles, yielding indices ranging from 0 to 1. A higher index value signified less differentiated cells, heightened stem cell characteristics and significantly increased aggressiveness. Regrettably, our Kaplan-Meier analysis failed to reveal a significant correlation in OS between the LUAD patients with low mRNAsi and those with high mRNAsi (P=0.182, Fig .6A). However, we observed significant differences in mRNAsi levels between the lung cancer tissue group and the normal control group (P<0.05, Fig .6B). Furthermore, our correlation analysis between mRNAsi and clinicopathological factors in the LUAD patients unveiled a noteworthy association (P<0.05, Fig .6C-F). Notably, male patients exhibited a higher mRNAsi value compared to female patients, suggesting that males with stronger stem cell characteristics and heightened tumor malignancy tend to possess elevated mRNAsi levels during the later stages of LUAD development.
Fig.6.
Differential analysis of tumor stem cells. A. The KM curve showed no significant correlation between OS in the LUAD patients with low mRNAsi and high mRNAsi. B. There is a significant difference in mRNAsi between normal and tumor tissues. C-F. Significant differences in mRNAsi between clinicopathological factors. KM; Kaplan-Meier, OS; Overall survival, LUAD; Lung adenocarcinoma, M; Metastasis, and T; Primary tumor.
Discussion
Lung cancer, as the most prevalent and deadliest form of cancer worldwide, claims over one million lives annually (26). Despite the advances in early detection and treatment options for LUAD, the prognosis remains grim, with less than 15% of patients surviving beyond five years (27). Therefore, it is imperative to identify biomarkers and develop targeted therapies specific to LUAD patients to enhance their prognosis.
Numerous studies have demonstrated potential prognostic significance of immune-related lncRNAs in lung adenocarcinoma, highlighting the growing interest in utilizing lncRNAs as prognostic indicators for malignancies (28). lncRNAs may affect the onset and progression of LUAD by regulating the PI3K/Akt/mTOR signaling pathway. The results suggested that lncRNAs could regulate cancer development at many levels, such as TME, tumor growth, invasion, metastasis and recurrence (29). Through a series of bioinformatics analyses, we developed a gene set-associated PAM-SRFLncSig model and identified seven lncRNAs, namely AC084757.3, AC010999.2, LINC02802, AC026979.2, AC024896.1, LINC00941 and LINC01312, using univariate Cox, LASSO and multivariate Cox regression analyses. Among these lncRNAs, previous studies demonstrated upregulation of LINC00941 in tumor tissues and plasma of NSCLC patients (30). LINC00941 is associated with infiltration and lymphatic metastasis of gastric cancer. It also promotes metastasis of papillary thyroid and colorectal cancers (31, 32). Combined with our study, we believe that LINC00941 plays a crucial role in LUAD and it is highly correlated with tumor development and metastasis. Therefore, LINC00941 may serve as an important biological marker targeted for immunotherapy and chemotherapy. Survival analysis of the training cohort using these lncRNAs revealed a significant difference in OS between the high-risk and low-risk subgroups, while LUAD patients in the high-risk subgroup exhibiting lower OS rates. Additionally, we observed higher expression levels of AC010999.2 and AC026979.2 in the low-risk subgroup, suggesting their potential as protective genes. C-index value, calibration curve verification and ROC curve analysis showed that the model had good predictive ability. Notably, KEGG pathway analysis revealed significant enrichment of the cytokine-cytokine receptor interaction (hsa04060), focal adhesion (hsa04510) and microRNAs in cancer (hsa05206) pathways. Focal adhesion, known to play a crucial role in tumor metastasis and aggressiveness, is primarily influenced by focal adhesion kinase (FAK) and its phosphorylation. FAK serves as a mediator of multiple signaling pathways in cellular signaling and it can activate intracellular PI3K/ Akt signaling pathways to regulate cell growth. The PI3K-Akt signaling pathway has emerged as a pivotal pathway for cancer cell survival, orchestrating crucial processes, such as cell proliferation, apoptosis, invasion, metastasis and angiogenesis by modulating activation status of downstream signaling molecules (33). KEGG enrichment analysis further corroborated association between LUAD and PI3K/Akt/mTOR signaling pathway. Therefore, PI3K/Akt/mTOR signaling pathway is closely related to the pathogenesis and prognosis of LUAD.
In recent years, immunotherapy has exhibited promising initial results in treatment of the various malignancies, including melanoma, lung cancer and others (34). Exploring specific biomarkers for LUAD can facilitate diagnosis and rational treatment choices. In our immune function pathway analysis, we identified Cytolytic_ activity, Inflammation-promoting and T_cell_coinhibition, as good prognostic pathways in LUAD. Notably, our analysis of TMB revealed no statistically significant difference between the high- and low-risk subgroups. Furthermore, when assessing tumor immune escape and immunotherapy using the PAM-SRFLncSig approach, we found no significant difference in TIDE scoring between the high- and low-risk subgroups. However, we did observe statistically significant distinctions in MDSC, Exclusion, CAF and TAMM2 between these subgroups. It is worthy to note that CAF-specific therapy has emerged as a valuable adjunct to immunotherapy, offering substantial clinical benefits for cancer patients (35). Some studies revealed the important regulatory role of the PI3K/Akt/ mTOR pathway in transformation of lung cancer growth patterns and chemotherapy resistance (36). In our study, we employed the pRRophetic algorithm to screen 16 potential oncologic chemotherapeutic drugs for LUAD patients. Potential therapeutic agents in the low-risk subgroup are BIRB.0796, CCT007093 and EHT.1864, while potential therapeutic agents in the high-risk subgroup are Bicalutamide, Bleomycin, BMS.509744, BMS.754807, Bortezomib, Bryostatin.1, CGP.082996, CGP.60474, CI.1040, CMK, Dasatinib, Docetaxel and Embelin.
Nevertheless, it is important to acknowledge the limitations of our study. Primarily, utilization of data solely from the TCGA database to construct the PAMSRFLncSig model introduced the possibility of sample selection bias, potentially stemming from a specific population or region. Consequently, the reliability of our results may be compromised, and the applicability may be limited. Furthermore, absence of alternative databases for additional validation further exacerbated this issue. Additionally, inconsistencies and missing data pertaining to the various molecular features, such as gene expression, mutations or methylation, may also be present. Lastly, exploration of therapeutic efficacy in LUAD patients necessitated implementation of animal and clinical trials. By scrutinizing expression levels of these lncRNAs in patient tissue samples, we can evaluate their impact on survival and disease progression, thereby enabling clinicians to devise more tailored treatment strategies. Further studies are still needed to verify the therapeutic reliability and clinical applicability of PAM-SRFLncSig in LUAD treatment, and the therapeutic modality of combining PI3K/Akt/mTOR signaling pathway inhibitors with other therapeutic agents or targeted drugs is a future direction of research.
Overall, multivariate COX regression analysis identified seven PAM-SRFLncSigs for the construction of a model for LUAD patients and confirmed the importance of PAM-SRFLncSig-associated lncRNAs in assessment of immunotherapy and prognosis of LUAD patients. The model can also be used to screen chemotherapeutic drugs for lung cancer, which has important clinical applications.
Conclusion
Based on the samples obtained from 494 LUAD patients, seven differentially expressed PAM-SRFLncSigs with prognostic value were screened, and the PAM-SRFLncSig model was constructed and validated. Validation of LUAD association with the PI3K/Akt/mTOR signaling pathway related lncRNAs through KEGG enrichment analysis. We found that these lncRNAs were associated with tumor immune function, and performed TMB, tumor immune escape and immunotherapy analyses on the high- and lowrisk subgroups. Finally, we screened 16 relevant drugs to guide clinical application.
Supplementary PDF
Acknowledgments
There is no financial support and conflict of interest in this study.
Author’s Contributions
L.C., X.Z.; Conceptualization, Methodology, Software, and Supervision. J.Z.; Data curation and Writing- Original draft preparation. J.Z., M.F., L.L.; Visualization and Investigation. Y.K., R.L.; Software and Validation. J.Z., Y.K., X.Z., L.C.; Writing-reviewing and editing. All authors read and approved the final version of the manuscript.
References
- 1.Zou Z, Tao T, Li H, Zhu X. mTOR signaling pathway and mTOR inhibitors in cancer: progress and challenges. Cell Biosci. 2020;10:31–31. doi: 10.1186/s13578-020-00396-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ren B, Liu H, Yang Y, Lian Y. Effect of BRAF-mediated PI3K/Akt/ mTOR pathway on biological characteristics and chemosensitivity of NSCLC A549/DDP cells. Oncol Lett. 2021;22(2):584–584. doi: 10.3892/ol.2021.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blakely CM, Watkins TBK, Wu W, Gini B, Chabon JJ, McCoach CE, et al. Evolution and clinical impact of co-occurring genetic alterations in advanced-stage EGFR-mutant lung cancers. Nat Genet. 2017;49(12):1693–1704. doi: 10.1038/ng.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mohamed E, Kumar A, Zhang Y, Wang AS, Chen K, Lim Y, et al. PI3K/AKT/mTOR signaling pathway activity in IDH-mutant diffuse glioma and clinical implications. Neuro Oncol. 2022;24(9):1471–1481. doi: 10.1093/neuonc/noac064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duan Y, Haybaeck J, Yang Z. Therapeutic potential of PI3K/AKT/ mTOR pathway in gastrointestinal stromal tumors: rationale and progress. Cancers (Basel) 2020;12(10):2972–2972. doi: 10.3390/cancers12102972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang W, Wei C, Huang F, Huang W, Xu X, Zhu X. A tumor mutational burden-derived immune computational framework selects sensitive immunotherapy/chemotherapy for lung adenocarcinoma populations with different prognoses. Front Oncol. 2023;13:1104137–1104137. doi: 10.3389/fonc.2023.1104137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiong Z, Han Z, Pan W, Zhu X, Liu C. Correlation between chromatin epigenetic-related lncRNA signature (CELncSig) and prognosis, immune microenvironment, and immunotherapy in non-small cell lung cancer. PLoS One. 2023;18(5):e0286122–e0286122. doi: 10.1371/journal.pone.0286122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin Q, Zhang M, Kong Y, Huang Z, Zou Z, Xiong Z, et al. Risk score = LncRNAs associated with doxorubicin metabolism can be used as molecular markers for immune microenvironment and immunotherapy in non-small cell lung cancer. Heliyon. 2023;9(3):e13811–e13811. doi: 10.1016/j.heliyon.2023.e13811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu Y, Tao T, Li S, Tan S, Liu H, Zhu X. Prognostic model and immunotherapy prediction based on molecular chaperone-related lncRNAs in lung adenocarcinoma. Front Genet. 2022;13:975905–975905. doi: 10.3389/fgene.2022.975905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu T, Wang Y, Chen D, Liu J, Jiao W. Potential clinical application of lncRNAs in non-small cell lung cancer. Onco Targets Ther. 2018;11:8045–8052. doi: 10.2147/OTT.S178431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu S, Liu D, Chang T, Wen X, Ma S, Sun G, et al. Cuproptosis- associated lncRNA establishes new prognostic profile and predicts immunotherapy response in clear cell renal cell carcinoma. Front Genet. 2022;13:938259–938259. doi: 10.3389/fgene.2022.938259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bossler F, Hoppe-Seyler K, Hoppe-Seyler F. PI3K/AKT/mTOR signaling regulates the virus/host cell crosstalk in HPV-positive cervical cancer cells. Int J Mol Sci. 2019;20(9):2188–2188. doi: 10.3390/ijms20092188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou D, Liu X, Wang X, Yan F, Wang P, Yan H, et al. A prognostic nomogram based on LASSO Cox regression in patients with alpha-fetoprotein-negative hepatocellular carcinoma following non-surgical therapy. BMC Cancer. 2021;21(1):246–246. doi: 10.1186/s12885-021-07916-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ye W, Wu Z, Gao P, Kang J, Xu Y, Wei C, et al. Identified gefitinib metabolism-related lncRNAs can be applied to predict prognosis, tumor microenvironment, and drug sensitivity in nonsmall cell lung cancer. Front Oncol. 2022;12:939021–939021. doi: 10.3389/fonc.2022.939021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai D, Tan L, Zuo X, Liu D, Jiao D, Wan G, et al. Prognostic ferroptosis-related lncRNA signatures associated with immunotherapy and chemotherapy responses in patients with stomach cancer. Front Genet. 2021;12:798612–798612. doi: 10.3389/fgene.2021.798612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu J, Li K, Zhang W, Wan C, Zhang J, Jiang P, et al. Large-scale public data reuse to model immunotherapy response and resistance. Genome Med. 2020;12(1):21–21. doi: 10.1186/s13073-020-0721-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu Z, Li S, Zhu X. The mechanism of stimulating and mobilizing the immune system enhancing the anti-tumor immunity. Front Immunol. 2021;12:682435–682435. doi: 10.3389/fimmu.2021.682435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu P, Luo H, Kong Y, Lai WF, Cui L, Zhu X. Cancer neoantigen: Boosting immunotherapy. Biomed Pharmacother. 2020;131:110640–110640. doi: 10.1016/j.biopha.2020.110640. [DOI] [PubMed] [Google Scholar]
- 19.Tan S, Li D, Zhu X. Cancer immunotherapy: Pros, cons and beyond. Biomed Pharmacother. 2020;124:109821–109821. doi: 10.1016/j.biopha.2020.109821. [DOI] [PubMed] [Google Scholar]
- 20.Yu J, Lan L, Liu C, Zhu X. Improved prediction of prognosis and therapy response for lung adenocarcinoma after identification of DNA-directed RNA polymerase-associated lncRNAs. J Cancer Res Clin Oncol. 2023;149(14):12737–12754. doi: 10.1007/s00432-023-05118-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Powles T, Eder JP, Fine GD, Braiteh FS, Loriot Y, Cruz C, et al. MPDL3280A (anti-PD-L1) treatment leads to clinical activity in metastatic bladder cancer. Nature. 2014;515(7528):558–562. doi: 10.1038/nature13904. [DOI] [PubMed] [Google Scholar]
- 22.Zhang W, Yao S, Huang H, Zhou H, Zhou H, Wei Q, et al. Molecular subtypes based on ferroptosis-related genes and tumor microenvironment infiltration characterization in lung adenocarcinoma. Oncoimmunology. 2021;10(1):1959977–1959977. doi: 10.1080/2162402X.2021.1959977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sokolov A, Paull EO, Stuart JM. One-class detection of cell states in tumor subtypes. Pac Symp Biocomput. 2016;21:405–416. [PMC free article] [PubMed] [Google Scholar]
- 24.Gu X, Li H, Sha L, Zhao W. A prognostic model composed of four long noncoding RNAs predicts the overall survival of Asian patients with hepatocellular carcinoma. Cancer Med. 2020;9(16):5719–5730. doi: 10.1002/cam4.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou ZR, Wang WW, Li Y, Jin KR, Wang XY, Wang ZW, et al. In-depth mining of clinical data: αthe construction of clinical prediction model with R. Ann Transl Med. 2019;7(23):796–796. doi: 10.21037/atm.2019.08.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen X, Zhi Q, Wang Y, Li Z, Zhou J, Huang J. Hypoxia induces multidrug resistance via enhancement of epidermal growth factor-like domain 7 expression in non-small lung cancer cells. Chemotherapy. 2017;62(3):172–180. doi: 10.1159/000456066. [DOI] [PubMed] [Google Scholar]
- 27.Gong T, Cui L, Wang H, Wang H, Han N. Knockdown of KLF5 suppresses hypoxia-induced resistance to cisplatin in NSCLC cells by regulating HIF-1α-dependent glycolysis through inactivation of the PI3K/Akt/mTOR pathway. J Transl Med. 2018;16(1):164–164. doi: 10.1186/s12967-018-1543-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qi X, Chen G, Chen Z, Li J, Chen W, Lin J, et al. Construction of a novel lung adenocarcinoma immune-related LncRNA pair signature. Int J Gen Med. 2021;14:4279–4289. doi: 10.2147/IJGM.S325240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021;22(2):96–118. doi: 10.1038/s41580-020-00315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ren MH, Chen S, Wang LG, Rui WX, Li P. LINC00941 promotes progression of non-small cell lung cancer by sponging miR- 877-3p to regulate VEGFA expression. Front Oncol. 2021;11:650037–650037. doi: 10.3389/fonc.2021.650037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo C, Tao Y, Zhang Y, Zhu Y, Minyao DN, Haleem M, et al. Regulatory network analysis of high expressed long non-coding RNA LINC00941 in gastric cancer. Gene. 2018;662:103–109. doi: 10.1016/j.gene.2018.04.023. [DOI] [PubMed] [Google Scholar]
- 32.Wu N, Jiang M, Liu H, Chu Y, Wang D, Cao J, et al. LINC00941 promotes CRC metastasis through preventing SMAD4 protein degradation and activating the TGF-beta/SMAD2/3 signaling pathway. Cell Death Differ. 2021;28(1):219–232. doi: 10.1038/s41418-020-0596-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen J, Cao B, Wang Y, Ma C, Zeng Z, Liu L, et al. Hippo component YAP promotes focal adhesion and tumour aggressiveness via transcriptionally activating THBS1/FAK signalling in breast cancer. J Exp Clin Cancer Res. 2018;37(1):175–175. doi: 10.1186/s13046-018-0850-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohaegbulam KC, Assal A, Lazar-Molnar E, Yao Y, Zang X. Human cancer immunotherapy with antibodies to the PD-1 and PD-L1 pathway. Trends Mol Med. 2015;21(1):24–33. doi: 10.1016/j.molmed.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saw PE, Chen J, Song E. Targeting CAFs to overcome anticancer therapeutic resistance. Trends Cancer. 2022;8(7):527–555. doi: 10.1016/j.trecan.2022.03.001. [DOI] [PubMed] [Google Scholar]
- 36.Li X, Li C, Guo C, Zhao Q, Cao J, Huang HY, et al. PI3K/Akt/ mTOR signaling orchestrates the phenotypic transition and chemo-resistance of small cell lung cancer. J Genet Genomics. 2021;48(7):640–651. doi: 10.1016/j.jgg.2021.04.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.