Abstract
Vibrio parahaemolyticus is a significant cause of foodborne illness, and its incidence worldwide is on the rise. It is thus imperative to develop a straightforward and efficient method for typing strains of this pathogen. In this study, we conducted a pangenome analysis of 75 complete genomes of V. parahaemolyticus and identified the core gene mtlA with the highest degree of variation, which distinguished 44 strains and outperformed traditional seven-gene-based MLST when combined with aer, another core gene with high degree of variation. The mtlA gene had higher resolution to type strains with a close relationship compared to the traditional MLST genes in the phylogenetic tree built by core genomes. Strong positive selection was also detected in the gene mtlA (ω > 1), representing adaptive and evolution in response to the environment. Therefore, the panel of gene mtlA and aer may serve as a tool for the typing of V. parahaemolyticus, potentially contributing to the prevention and control of this foodborne disease.
Keywords: Vibrio parahaemolyticus, Pan-genome, Core genome, Typing, mtlA, aer, Positive selection
1. Introduction
Vibrio parahaemolyticus, first identified by Tsunesaburo Fujino during a foodborne disease outbreak in Japan in 1950 [1], is predominantly found in temperate and tropical sea areas worldwide [2]. These pathogenic bacteria can be easily identified in estuarine, and coastal waters and even seafood [3]. Due to the increasing consumption of seafood and cross-border travel, the incidence of foodborne disease caused by V. parahaemolyticus has garnered significant global attention. Therefore, developing a rapid typing system capable of typing isolates and preventing outbreaks is highly necessary.
Approximately 50 years ago, serovar typing (based on somatic O antigen and capsular K antigen) successfully differentiated V. parahaemolyticus isolates with diverse antigenic backgrounds [4]. This method was later replaced by pulse field gel electrophoresis (PFGE), which became the standard tool for typing the origin of isolates [5]. Subsequently, with the development of molecular biotechnological techniques, restriction fragment length polymorphism (RFLP), random amplified polymorphic DNA (RAPD), enterobacterial repetitive intergenic-consensus sequence PCR (ERIC-PCR) and ribotyping based on polymerase chain reaction (PCR) were employed as complementary measures to estimate the relevance among isolates [6]. The previously mentioned methods were found to be time-consuming. As a result, multilocus sequence typing (MLST) has emerged as a frequently used method for molecular typing of bacteria, as it targets seven housekeeping genes of each species [7]. González-Escalona et al. constructed a modified MLST scheme for V. parahaemolyticus, which analyzed 100 isolates by including the general seven housekeeping genes located on two chromosomes [8]. The MLST analysis consisted of three genes located on chromosome Ⅰ: dnaE (DNA polymerase III, alpha subunit), gyrB (DNA gyrase subunit B), and recA (RecA protein), and four housekeeping genes on chromosome II: dtdS (threonine 3-dehydrogenase), pntA (transhydrogenase, alpha subunit), pyrC (dihydro-oratase) and tnaA (tryophanase). The conventional MLST scheme has been used to study the evolutionary distance among V. parahaemolyticus strains. Theethakaew et al. applied this method to type isolates recovered from clinical, human carrier and environmental sources in Thailand [9]. Additionally, Muangnapoh et al. employed MLST to identify novel V. parahaemolyticus strains derived from aquatic bird feces [10]. However, interspecies recombination been reported in the recA gene which results in an undefined MLST type [11]. Moreover, recombination in recA affected the topology of the MLST phylogenetic tree, which might bury true evolution [11]. Whole genome sequencing (WGS) analysis has the potential to identify subtle nucleotide changes among isolates [12]. Based on WGS, both core-genome MLST (cgMLST) and whole-genome MLST (wgMLST) were developed for precise typing [13]. However, the high cost and requirement for bioinformatics analysis limit their application in pathogen typing and traceability. Therefore, utilizing fewer genes can be more cost-effective, making large-scale studies or routine surveillance more feasible within budget constraints. Moreover, cost-effective options often streamline the experimental and analytical processes, leading to quicker results. This efficiency was particularly beneficial in scenarios requiring timely and high-throughput analyses. In this study, we identified the mtlA and aer genes with high variation in V. parahaemolyticus genomes, and validated their distinguishing effect using all complete genomes downloaded from National Center for Biotechnology Information (NCBI), which were isolated from different regions of the world at various times. This set of genomes with complete background information allowed for more thorough analysis of pathogen typing and tracing the origin of pandemic disease.
2. Materials and methods
2.1. Data source
The complete genomes of Vibrio parahaemolyticus were downloaded from NCBI before September 2022. The background information of the strains, such as isolation time and location, was recorded simultaneously.
2.2. ANI calculation and genomic conservation analysis
To identify the conservation of V. parahaemolyticus complete genomes, we used the Fast Average Nucleotide Identity (FastANI) tool to calculate the whole-genome average nucleotide identity (ANI) value through alignment-free approximate sequence mapping (fragLen 1000) [14]. The alignment fraction (the ratio of the count of bidirectional fragment mappings and total query fragments) was also calculated. Mauve2.3.1 [15] running under Java 1.8.0_361-b09 was simultaneously used to identify changes from the perspective of genomic structure, such as translocations, deletions, insertions and others.
2.3. High variable gene screening
To determine a set of variable core genes, the prokaryotic Pan-Genome Analysis Pipeline (PGAP) [16] was implemented to perform a pangenome analysis of V. parahaemolyticus genomes. The complete genomes downloaded from NCBI were first converted into three input files, including protein sequences, nucleotide sequences and annotation files, using the CONVERTER command in the PGAP package. Then, the prepared files of genomes were clustered by running PGAP commands with 95% identity and coverage cutoff. Gene clusters present in all the genomes were defined as core genes. The core genes with the highest variation were searched by BLASTN to determine orthologous genes with the lowest identity. The screening core genes were abstracted from complete genomes by ABSTRACT and then the number of typing clusters was calculated by BLASTCLUST. In addition, the traditional MLST genes were also abstracted and analyzed using the same command. The specific fragments of these seven housekeeping genes were obtained by cutting with primers from the pubMLST website [17]. The primers of genes were designed by Primer3 [18]. Eventually, a Venn diagram of Jvenn (http://bioinfo.genotoul.fr/jvenn) [19] was used to display the differences between the traditional MLST typing method and ours.
2.4. Selective pressure analysis
To identify the positive selection sites of gene mtlA and aer, we calculated the nonsynonymous (dN) and synonymous (dS) substitution rates using the preset running mode of EasyCodeML v1.4 [20]. The analysis was calculated based on the site model including four null models: M0, M1a, M7 and M8a and three alternative models: M3, M2a and M8. Four nest models (M0 vs. M3, M1a vs. M2a, M7 vs. M8 and M8a vs. M8) were formed by comparing the null model with the alternative model. If the alternative model was significantly superior to the null model (likelihood ratio test (LRT) threshold of p<0.05), the gene is considered to be under positive selection [21].
2.5. Construction of phylogenetic tree
To ascertain the phylogenetic position of strains typed by traditional MLST genes, mtlA and mtlA combined with aer (mtlA/aer), a phylogenetic tree of 75 V. parahaemolyticus strains was constructed using core genome. The core genes were first concatenated and aligned by MAFFT v6.864b with normal model [22]. Then, the neighbor-joining algorithm was chosen to build a phylogenetic tree via MEGA11 [23]. Finally, the phylogenetic tree was visualized and decorated using Interactive Tree of Life [24].
3. Results
3.1. Genome background information
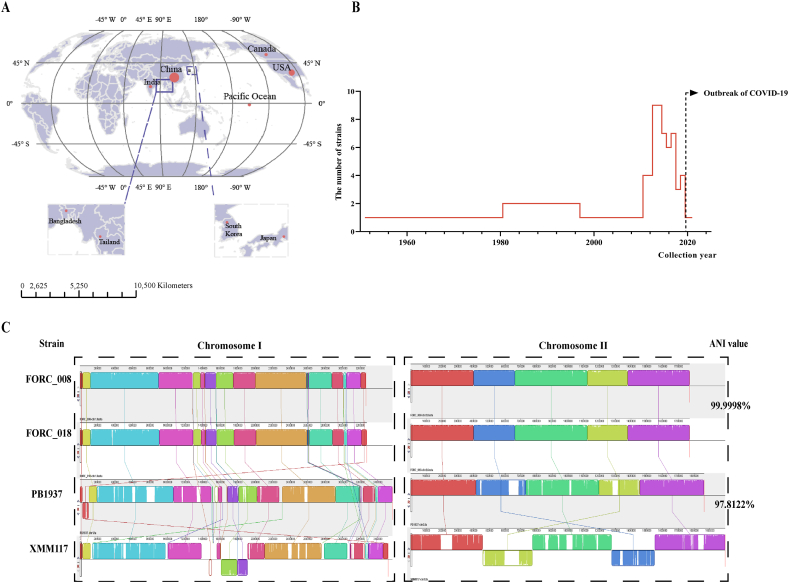
A total of 76 complete genomes of V. parahaemolyticus were downloaded from NCBI, before September 2022. The BTXS2 strain (GCA_015172915.1) was excluded from the study after quality checks because its sole PacBio sequencing might contain insufficient coverage and random errors. Upon reviewing the details of the 75 V. parahaemolyticus strains, we observed that they were predominantly isolated from China, South Korea and the United States (Fig. 1A). Additionally, the 75 V. parahaemolyticus strains were collected over a considerable period of time, spanning from 1951 to 2021, providing a representative range of both temporal and regional factors (Fig. 1B). As a result, these strains were well-suited for typing analysis (Supplementary Table S1).
Fig. 1.
The spatiotemporal distribution of V. parahaemolyticus and genome conservation analysis. (A) The main collection regions of V. parahaemolyticus strains. The red circles represent the separation regions of V. parahaemolyticus, and their size represents the number of strains collected in that region. (B) The year range of V. parahaemolyticus isolation. (C) Genetic collinearity analysis of maximum and minimum ANI value genomes. Left: The chromosome Ⅰ structure of the four strains. Right: The chromosome Ⅱ structure of the four strains. Strains FORC_018 and FORC_008 had the highest ANI values, and exhibited the highest similarity in genetic structure (the above two chromosome structure diagrams). The genome structure of the other two strains showed slight changes with XMM117 mainly having two inversions in two chromosomes while PB1937 had translocations in chromosome Ⅰ (the two below ones). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.2. Genome conservation analysis
The genome of V. parahaemolyticus consists of two chromosomes, with chromosome Ⅰ spanning 3.39 (3.49–3.29) Mb and chromosome Ⅱ spanning 1.83 (1.89–1.78) Mb. ANI values were calculated for all the genomes, and it was discovered that strains PB1937 (GCF_003351885.1) and XMM117 (GCF_023205935.1) had the lowest ANI value (97.8122%). These two strains were detected in Fujian Province of China and submitted in 2018 and 2022, respectively. Strains FORC_018 (GCF_001887055.1, Nov 28, 2016) and FORC_008 (GCF_0012244315.1, Aug 5, 2015) detected in South Korea, had the highest ANI value of 99.9998%, and exhibited the highest similarity in genetic structure. The genome structure of the other two strains showed slight changes with strain XMM117 mainly having two inversions in two chromosomes while PB1937 had translocations in chromosome Ⅰ (Fig. 1C). Therefore, ANI prediction and genomic structure comparison revealed that the genome of V. parahaemolyticus was highly conserved.
3.3. Highly variable gene screening
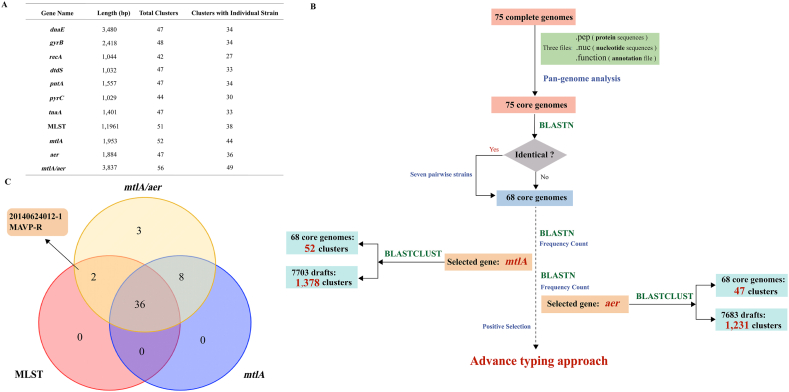
The pangenome analysis revealed the presence of 2532 core genes in 75 genomes with seven pairwise strains showing completely identical core genomes (Table 1). This reflected a close relationship between the pairs and was validated by the fact that they were isolated from the same location at a close time. Therefore, only one strain was selected from each pair and 68 genomes were chosen for further study.
Table 1.
Seven pairwise strains with identical core genome.
| Strain | Alignment fraction | ANI value | |
|---|---|---|---|
| 20-082A3 | 20-082E4 | 98.77 | 99.9999 |
| FDAARGOS_51 | 10329 | 99.19 | 99.9999 |
| FORC_008 | FORC_018 | 99.19 | 99.9999 |
| MAVP-Q PDT000221686.2 | MAVP-Q PDT000138751.1 | 99.35 | 99.9999 |
| R14 | R13 | 98.85 | 99.9999 |
| TJ-20 | VP120 | 99.16 | 99.9999 |
| TJ-187 | LH24 | 98.50 | 99.9999 |
The MLST analysis of the 68 genomes revealed that the seven housekeeping genes combined together could differentiate them into 51 clusters, with 38 clusters harboring only one strain (Fig. 2A). A single strain formed its own cluster based on the analysis of its nucleotide sequence, with at least one nucleotide differing from other strains. The accumulation of these nucleotide differences results in the presence of specific SNP (single nucleotide polymorphism) sites, enabling complete differentiation from other strains and the formation of an independent cluster. Hence, further analysis was performed to identify new genes with higher resolution (Fig. 2B).
Fig. 2.
The process of highly variable core gene screening and the comparison of typing resolution between traditional MLST and the set of selected core genes. (A) The resolution of the mtlA, aer and seven traditional housekeeping genes. (B) The process of the highly variable gene screening. (C) Venn diagram depicting strains that can be separated by traditional MLST, the sole mtlA gene and mtlA/aer. The numbers in each shape represent the number of strains that are unique to this typing marker and the numbers in the overlapping region are the number of strains common to the corresponding typing markers.
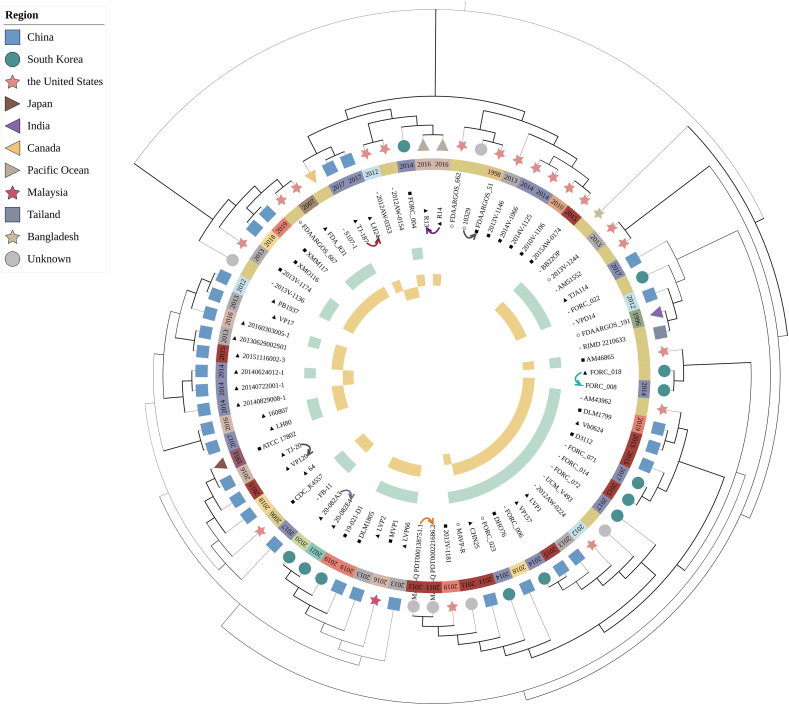
The gene mtlA (1953 bp), which encodes PTS (phosphotransferase system) mannitol transporter subunit IICBA, showed the lowest identity among the 68 genomes, and could differentiate the 68 strains into 52 clusters, with 44 clusters harboring only one strain (Fig. 2A and B). However, the mtlA gene failed to distinguish strains 20140624012-1 and MAVP-R (Fig. 2C), which could be separated into two clusters by MLST. So, using the same screening method, we discovered another core gene, aer, which could segregate these two strains. The typing result obtained by combining mtlA and aer exceeds the resolution of traditional MLST (Fig. 3). From the tree, compared to MLST typing, we found that the gene mtlA can completely differentiate strains from the same source that were temporally adjacent and had close genetic relationship, thus achieving traceability. For example, the mtlA gene can distinguish between strains XMM117 and XMO116, which were separately isolated in China in 2018 and 2019 and could not be distinguished using traditional MLST. Furthermore, the combined use of mtlA and aer genes can completely differentiate these strains (strains 20140829008-1, 20140722001-1 and 20140624012-1) isolated in China in the same year (in 2014) that could not be distinguished using traditional MLST. These results demonstrated that the combination of mtlA and aer completely exceeded the segregation effect of MLST.
Fig. 3.
Phylogenetic tree built with concatenated core genes of 75 strains. The pairs of strains connected by arrows indicate identical core genomes. The strip between the strain name and isolated region indicates the isolation time with the exact year in the text. The green outer strip represents the traditional MLST resolution, while the two yellow strips denoted the segregation of the mtlA and aer genes. These strips made of rectangles representing typing clusters, each cluster containing only one strain. ○ represents an isolate from a clinal source, ▲represents a food source, ■ is an environment and - means an unclear isolate source. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
To further validate the differentiation effectiveness of the mtlA gene and mtlA/aer, we downloaded the 9700 draft genomes of V. parahaemolyticus from NCBI and extracted the relevant genes. Among them 7703 draft genomes contained the mtlA gene, 7613 draft genomes contained both mtlA and aer genes, while only 2504 draft genomes contained all seven traditional MLST genes. Our analysis discovered that the mtlA gene could divide the 7703 draft genomes into 1378 clusters with 965 of which containing only one strain, mtlA/aer could divide the 7613 draft genomes into 2384 clusters with 2084 of which containing only one strain, while the seven MLST genes could only divide the 2504 draft genomes into 125 clusters (Table 2). Since the genes used for traditional MLST typing were usually 700 bp, we designed two pairs of primers targeted to the highly variable region of the mtlA and aer genes to obtain equivalent lengths (Table 3). We then revealed that the 721 bp fragment of the mtlA gene could distinguish 7725 drafts into 689 clusters, and 329 clusters contained one strain (Table 2). Combined with the 885 bp fragment of the aer gene, these two genes produced 1593 clusters from 7657 drafts and were able to identify some genomes that could not be separated individually by traditional MLST typing. In particular, there were 5854 drafts that included the variable regions of the mtlA, aer and seven housekeeping genes simultaneously, and the resolution of the mtlA gene was significantly superior to that of each of the seven MLST type genes (Table 4). Because the mtlA gene had comparable discriminatory power to traditional MLST and the cost-effective advantage of being a single gene, it has the potential to serve as a promising marker for future typing of V. parahaemolyticus, especially when combining with aer.
Table 2.
The resolution of gene mtlA, aer and traditional MLST in drafts genomes.
| Gene name | Full gene |
Specific fragment |
||||||
|---|---|---|---|---|---|---|---|---|
| Length (bp) | Drafts | Totala | Individualb | Length (bp) | Drafts | Totala | Individualb | |
| mtlA | 1953 | 7703 | 1378 | 965 | 721 | 7725 | 689 | 329 |
| aer | 1884 | 7683 | 1231 | 842 | 885 | 7715 | 551 | 224 |
| dnaE | 3480 | 7723 | 1897 | 1604 | 596 | 7756 | 419 | 154 |
| dtdS | 1032 | 3051 | 116 | 53 | 497 | 7693 | 533 | 208 |
| gyrB | 2418 | 4503 | 390 | 254 | 629 | 7619 | 390 | 254 |
| pntA | 1557 | 7742 | 1062 | 661 | 470 | 7747 | 304 | 114 |
| pyrC | 1029 | 6497 | 737 | 350 | 533 | 6596 | 515 | 248 |
| recA | 1044 | 7260 | 571 | 251 | 773 | 7307 | 460 | 185 |
| tnaA | 1401 | 7715 | 1089 | 646 | 463 | 7747 | 305 | 115 |
| MLST | 11960 | 2504 | 125 | 114 | 3961 | 5977 | 1841 | 1594 |
| mtlA/aer | 3837 | 7613 | 2384 | 2084 | 1606 | 7657 | 1593 | 1108 |
Means total clusters were divided by relevant gene, individual.
Means individual clusters contained one strained were divided by relevant gene.
Table 3.
The primers of mtlA and aer gene.
| Primer | Length (bp) | |
|---|---|---|
| mtlA-Forward | 721 | ATTCAGCAAGCGTCTGAG |
| mtlA-Reverse | TACGTGCACGACCCGTCA | |
| aer-Forward | 885 | CTGGCTGAGAACTTCAAC |
| aer-Reverse | TTACAGTTTAAAACGTTTGATCTC |
Table 4.
The resolution of gene mtlA, aer and traditional MLST in 5854 drafts genomes contained all these genes.
| Gene name | Total cluster | Individual cluster |
|---|---|---|
| mtlA | 495 | 206 |
| aer | 442 | 182 |
| dnaE | 362 | 139 |
| dtdS | 446 | 183 |
| gyrB | 468 | 176 |
| pntA | 249 | 98 |
| pyrC | 393 | 51 |
| recA | 381 | 149 |
| tnaA | 247 | 94 |
| MLST | 1752 | 1512 |
| mtlA/aer | 1061 | 672 |
3.4. Positive selection of mtlA
We ran the preset mode of EasyCodeML on the mtlA and aer genes and found that ω values of M2a and M8 alternative models, which support positive selection, were 1.21486 and 1.23796 of analysis in gene mtlA (ω > 1), respectively. However, the ω value of M8 in gene aer was 1.00000 (ω = 1) which meant the gene aer was under the neutral selection. The LRT test of M7 vs. M8, a powerful test for positive selection, also indicated a strong evidence of codon sites of gene mtlA involved in positive Darwinian selection (p<0.05). Based on these parameters, we concluded that the mtlA gene might be more adaptive to adverse environmental condition. Detailed parameter estimates and LRTs can be found in Supplementary Table S2 and Table S3.
4. Discussion
V. parahaemolyticus is a major food-borne pathogen that can cause severe gastroenteritis and septicemia [25,26], it can survive in marine ecosystems, including seawater, creatures [27] and even sediments [28], which might serve as vehicles for V. parahaemolyticus transmission [29]. In recent years, the detection rate of V. parahaemolyticus has increased in China, possibly due to the rise in international trade and travel. Therefore, effective surveillance and control measures are necessary to prevent the spread of this pathogen and safeguard public health.
We constructed a phylogenetic tree of 75 V. parahaemolyticus using concatenated core genes (Fig. 3). To some extent, the close relationship among isolates from China, South Korea and the United States (USA) may be due to the movement of people and trade. For instance, FORC_22 detected in South Korea in 2016, had a relatively close relationship with TJA114, which was isolated from Tianjin Province in China in 2020. Through analyzing the variation in core genes, we revealed that the mtlA gene can segregate 68 V. parahaemolyticus into 52 clusters, showing a higher resolution than seven housekeeping gene-based MLST typing.
MtlA is a component of the phosphoenolpyruvate (PEP): carbohydrate phosphotransferase system (PTS) and functions as a mannitol transporter subunit, IICBA [30]. In addition to sugar transfer, the PTS also plays a role in carbon storage, carbon metabolism, nitrogen embolism, gene expression, biofilm formation and even bacterial virulence [31]. For instance, Klebsiella pneumoniae, a gram-negative bacterium that causes pneumonia, also harbored the PTS enzyme II subunit-C gene celB, similar to the gene mtlA. The deletion of the celB gene significantly influenced the biofilm formation and increased the survival rate in intragastric infection [32]. This system also plays an important role in the surface attachment and virulence of many strains of V. cholerae [33]. Aer is one of the methyl-accepting chemotaxis proteins (MCPs), which played a crucial role in cell survival, pathogenesis, and biodegradation. Bacterial adaptation to a variety of environmental conditions contributes to the diversity observed among MCPs [34]. Moreover, Escherichia coli exhibited chemotaxis towards PTS sugars through the capability of sensing the redox state by Aer [35]. So, the association with pathogenesis and host adaption might explain the high variation of mtlA and aer genes in V. parahaemolyticus genomes.
With the development of high-throughput sequencing technologies, more and more draft genomes of V. parahaemolyticus were obtained and used for MLST analysis directly. The combination of the seven gene fragments of traditional MLST indeed has a higher resolution than mtlA/aer in the 5854 drafts that contained all these gene fragments (Table 4). But this application is limited by the absence of some MLST genes in the draft genomes (Table 2). As we can see, only 2504 out of 9700 draft genomes (25.81%) contained all the complete CDS of the seven genes, and 5977 out of 9700 draft genomes (61.62%) contained all the seven selected fragments. But if we use the panel of mtlA and aer as typing markers, the ration increased to 78.48% (7613/9700) for complete CDS and 78.94% (7657/9700) for selected fragments, respectively. So in both the 75 complete genomes and the 9700 drafts, the resolution of mtlA/aer can significantly cover the discriminatory power of traditional MLST genes. In conclusion, our findings suggested that the mtlA and aer genes may serve as powerful typing markers for V. parahaemolyticus strains in future.
There are several limitations in our study. First, only 75 complete genomes were available in the NCBI database and were mainly distributed in China, the USA and Korea, which limited origin-tracing function. Information on draft genomes might play important roles in future applications. Second, the length of the mtlA and aer genes is longer than 1000 bp, which might limit its application using Sanger sequencing. However, with the development of third-generation sequencing, this might be resolved.
5. Conclusion
Using core genes as typing markers, V. parahaemolyticus strains isolated from different regions can be effectively distinguished, making them a powerful tool for origin-typing. The mtlA gene distinguished the majority of the 75 genomes and completely exceed the segregation effect of MLST when combined with aer. Collinearity analysis indicated that the genome of V. parahaemolyticus is relatively stable, emphasizing the significance of highly variable typing genes. Furthermore, positive selection acting on the mtlA gene suggested its crucial role in the growth of V. parahaemolyticus.
Data availability statement
The data underlying this article are available at National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/assembly/?term=Vibrio%20parahaemolyticus).
Funding
This research was funded by National Key R&D Program of China (2022YFC2302800) and Shanghai Science and Technology Innovation Action Plan (21142202100).
CRediT authorship contribution statement
Lei Zhou: Writing – original draft, Methodology, Investigation, Formal analysis, Data curation. Danlei Liu: Visualization, Validation, Methodology. Yongqiang Zhu: Visualization, Software. Zilong Zhang: Writing – review & editing, Visualization, Validation. Shiwen Chen: Visualization, Validation. Guoping Zhao: Writing – review & editing, Project administration, Conceptualization. Huajun Zheng: Writing – review & editing, Supervision, Project administration, Methodology, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e25642.
Contributor Information
Guoping Zhao, Email: gpzhao@sibs.ac.cn.
Huajun Zheng, Email: zhenghj@chgc.sh.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Joseph S.W., Colwell R.R., Kaper J.B. Vibrio parahaemolyticus and related halophilic Vibrios. Crit. Rev. Microbiol. 1982;10:77–124. doi: 10.3109/10408418209113506. [DOI] [PubMed] [Google Scholar]
- 2.Martinez-Urtaza J., Baker-Austin C. Vibrio parahaemolyticus. Trends Microbiol. 2020;28:867–868. doi: 10.1016/j.tim.2020.02.008. [DOI] [PubMed] [Google Scholar]
- 3.Alipour M., Issazadeh K., Soleimani J. Isolation and identification of Vibrio parahaemolyticus from seawater and sediment samples in the southern coast of the Caspian Sea. Comp. Clin. Pathol. 2014;23:129–133. doi: 10.1007/s00580-012-1583-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Montague T.S., Le Clair R.A., Zen-Yoji H. Typing of O antigens of Vibrio parahaemolyticus by a slide agglutination test. Appl. Microbiol. 1971;21:949–950. doi: 10.1128/am.21.5.949-950.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wong H.C., Lu K.T., Pan T.M., Lee C.L., Shih D.Y. Subspecies typing of Vibrio parahaemolyticus by pulsed-field gel electrophoresis. J. Clin. Microbiol. 1996;34:1535–1539. doi: 10.1128/jcm.34.6.1535-1539.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong H.C., Lin C.H. Evaluation of typing of Vibrio parahaemolyticus by three PCR methods using specific primers. J. Clin. Microbiol. 2001;39:4233–4240. doi: 10.1128/JCM.39.12.4233-4240.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maiden M.C.J. Multilocus sequence typing of bacteria. Annu. Rev. Microbiol. 2006;60:561–588. doi: 10.1146/annurev.micro.59.030804.121325. [DOI] [PubMed] [Google Scholar]
- 8.González-Escalona N., Martinez-Urtaza J., Romero J., Espejo R.T., Jaykus L.-A., DePaola A. Determination of molecular phylogenetics of Vibrio parahaemolyticus strains by multilocus sequence typing. J. Bacteriol. 2008;190:2831–2840. doi: 10.1128/JB.01808-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Theethakaew C., Feil E.J., Castillo-Ramírez S., Aanensen D.M., Suthienkul O., Neil D.M., Davies R.L. Genetic relationships of Vibrio parahaemolyticus isolates from clinical, human carrier, and environmental sources in Thailand, determined by multilocus sequence analysis. Appl. Environ. Microbiol. 2013;79:2358–2370. doi: 10.1128/AEM.03067-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muangnapoh C., Tamboon E., Supha N., Toyting J., Chitrak A., Kitkumthorn N., Ekchariyawat P., Iida T., Suthienkul O. Multilocus sequence typing and virulence potential of Vibrio parahaemolyticus strains isolated from aquatic bird feces. Microbiol. Spectr. 2022;10 doi: 10.1128/spectrum.00886-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jesser K.J., Valdivia-Granda W., Jones J.L., Noble R.T. Clustering of Vibrio parahaemolyticus isolates using MLST and whole-genome phylogenetics and protein Motif Fingerprinting. Front. Public Health. 2019;7:66. doi: 10.3389/fpubh.2019.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petronella N., Kundra P., Auclair O., Hébert K., Rao M., Kingsley K., De Bruyne K., Banerjee S., Gill A., Pagotto F., Tamber S., Ronholm J. Changes detected in the genome sequences of Escherichia coli, Listeria monocytogenes, Vibrio parahaemolyticus, and Salmonella enterica after serial subculturing. Can. J. Microbiol. 2019;65:842–850. doi: 10.1139/cjm-2019-0235. [DOI] [PubMed] [Google Scholar]
- 13.Stevens E.L., Carleton H.A., Beal J., Tillman G.E., Lindsey R.L., Lauer A.C., Pightling A., Jarvis K.G., Ottesen A., Ramachandran P., Hintz L., Katz L.S., Folster J.P., Whichard J.M., Trees E., Timme R.E., McDERMOTT P., Wolpert B., Bazaco M., Zhao S., Lindley S., Bruce B.B., Griffin P.M., Brown E., Allard M., Tallent S., Irvin K., Hoffmann M., Wise M., Tauxe R., Gerner-Smidt P., Simmons M., Kissler B., Defibaugh-Chavez S., Klimke W., Agarwala R., Lindsay J., Cook K., Austerman S.R., Goldman D., McGARRY S., Hale K.R., Dessai U., Musser S.M., Braden C. Use of whole genome sequencing by the Federal Interagency Collaboration for genomics for food and feed safety in the United States. J. Food Protect. 2022;85:755–772. doi: 10.4315/JFP-21-437. [DOI] [PubMed] [Google Scholar]
- 14.Jain C., Rodriguez-R L.M., Phillippy A.M., Konstantinidis K.T., Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018;9:5114. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Darling A.C.E., Mau B., Blattner F.R., Perna N.T. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao Y., Wu J., Yang J., Sun S., Xiao J., Yu J. PGAP: pan-genomes analysis pipeline. Bioinforma. Oxf. Engl. 2012;28:416–418. doi: 10.1093/bioinformatics/btr655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jolley K.A., Bray J.E., Maiden M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018;3:124. doi: 10.12688/wellcomeopenres.14826.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Untergasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B.C., Remm M., Rozen S.G. Primer3--new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bardou P., Mariette J., Escudié F., Djemiel C., Klopp C. jvenn: an interactive Venn diagram viewer. BMC Bioinf. 2014;15:293. doi: 10.1186/1471-2105-15-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao F., Chen C., Arab D.A., Du Z., He Y., Ho S.Y.W. EasyCodeML: a visual tool for analysis of selection using CodeML. Ecol. Evol. 2019;9:3891–3898. doi: 10.1002/ece3.5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Z., Nielsen R., Goldman N., Pedersen A.M. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics. 2000;155:431–449. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katoh K., Asimenos G., Toh H. Multiple alignment of DNA sequences with MAFFT. Methods Mol. Biol. Clifton NJ. 2009;537:39–64. doi: 10.1007/978-1-59745-251-9_3. [DOI] [PubMed] [Google Scholar]
- 23.Tamura K., Stecher G., Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021;38:3022–3027. doi: 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Letunic I., Bork P. Interactive Tree of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nair G.B., Ramamurthy T., Bhattacharya S.K., Dutta B., Takeda Y., Sack D.A. Global dissemination of Vibrio parahaemolyticus serotype O3:K6 and its serovariants. Clin. Microbiol. Rev. 2007;20:39–48. doi: 10.1128/CMR.00025-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoon J.-H., Lee S.-Y. Characteristics of viable-but-nonculturable Vibrio parahaemolyticus induced by nutrient-deficiency at cold temperature. Crit. Rev. Food Sci. Nutr. 2020;60:1302–1320. doi: 10.1080/10408398.2019.1570076. [DOI] [PubMed] [Google Scholar]
- 27.Powell A., Baker-Austin C., Wagley S., Bayley A., Hartnell R. Isolation of pandemic Vibrio parahaemolyticus from UK water and shellfish produce, Microb. Ecol. 2013;65:924–927. doi: 10.1007/s00248-013-0201-8. [DOI] [PubMed] [Google Scholar]
- 28.de Jesús Hernández-Díaz L., Leon-Sicairos N., Velazquez-Roman J., Flores-Villaseñor H., Guadron-Llanos A.M., Martinez-Garcia J.J., Vidal J.E., Canizalez-Roman A. A pandemic Vibrio parahaemolyticus O3:K6 clone causing most associated diarrhea cases in the Pacific Northwest coast of Mexico. Front. Microbiol. 2015;6:221. doi: 10.3389/fmicb.2015.00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cabrera-García M.E., Vázquez-Salinas C., Quiñones-Ramírez E.I. Serologic and molecular characterization of Vibrio parahaemolyticus strains isolated from seawater and fish products of the Gulf of Mexico. Appl. Environ. Microbiol. 2004;70:6401–6406. doi: 10.1128/AEM.70.11.6401-6406.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deutscher J., Aké F.M.D., Derkaoui M., Zébré A.C., Cao T.N., Bouraoui H., Kentache T., Mokhtari A., Milohanic E., Joyet P. The bacterial phosphoenolpyruvate:carbohydrate phosphotransferase system: regulation by protein phosphorylation and phosphorylation-dependent protein-protein interactions. Microbiol. Mol. Biol. Rev. MMBR. 2014;78:231–256. doi: 10.1128/MMBR.00001-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barabote R.D., Saier M.H. Comparative genomic analyses of the bacterial phosphotransferase system. Microbiol. Mol. Biol. Rev. MMBR. 2005;69:608–634. doi: 10.1128/MMBR.69.4.608-634.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu M.-C., Chen Y.-C., Lin T.-L., Hsieh P.-F., Wang J.-T. Cellobiose-specific phosphotransferase system of Klebsiella pneumoniae and its importance in biofilm formation and virulence. Infect. Immun. 2012;80:2464–2472. doi: 10.1128/IAI.06247-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Houot L., Chang S., Absalon C., Watnick P.I. Vibrio cholerae phosphoenolpyruvate phosphotransferase system control of carbohydrate transport, biofilm formation, and colonization of the germfree mouse intestine. Infect. Immun. 2010;78:1482–1494. doi: 10.1128/IAI.01356-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salah Ud-Din A.I.M., Roujeinikova A. Methyl-accepting chemotaxis proteins: a core sensing element in prokaryotes and archaea. Cell. Mol. Life Sci.: CM. 2017;74:3293–3303. doi: 10.1007/s00018-017-2514-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greer-Phillips S.E., Alexandre G., Taylor B.L., Zhulin I.B. Aer and Tsr guide Escherichia coli in spatial gradients of oxidizable substrates. Microbiology (Reading, England) 2003;149:2661–2667. doi: 10.1099/mic.0.26304-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available at National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/assembly/?term=Vibrio%20parahaemolyticus).