INTRODUCTION
When choosing medications and calculating doses in infants, children, and adolescents, it is common to consider the patient’s diagnosis, age, weight, and sometimes body surface area. Despite careful therapeutic decisions, patients may experience inadequate therapeutic effect, a spectrum of adverse effects from mild to severe, or hypersensitivity reactions. Pharmacogenomics (PGx, also referred to as pharmacogenetics), where genomic information is used to tailor medication management, is a strategy to maximize drug efficacy and minimize toxicity. An increasing number of Food and Drug Administration (FDA) medication labels contain PGx information but do not necessarily recommend or require pre-prescription genetic testing. The Clinical Pharmacogenetics Implementation Consortium (CPIC) is an international group of experts in pharmacology and PGx dedicated to creating expert reviewed, evidence-based, and regularly updated gene-drug clinical practice guidelines.1 A serial, cross-sectional study that included prescribing information from nearly 2.9 million pediatric patients from 16 sites, reported a conservative annual prescribing prevalence of 8000 to 11,000 per 100,000 patients who received prescriptions for CPIC level A medications (those with prescribing guidelines).2 However, the systematic implementation of these guidelines to inform medication selection and dosing in pediatrics remains limited.3
In some pediatric clinics and hospitals, the implementation of PGx started as early as 20044 and has gradually increased.5 Models for implementation vary between sites.6 With rising numbers of clinics and hospitals implementing PGx, pediatric providers may encounter past PGx results with therapeutic recommendations for specific medications but are unaware of the applicability of those results to new or additional medications. Patients with complex needs may be exposed to several concurrent medications, and providers may lack knowledge and/or time to consider the evidence for individualized dosing or titration.3 As other articles in this Special Issue describe, pediatric providers should expect increased use of exome sequencing (ES) and genome sequencing (GS) for diagnostic testing, which will enable opportunistic screening of genes that can influence medication response (pharmacogenes). Only two pharmacogenes (both related to malignant hyperthermia and discussed in Case 3a later in discussion) are currently recommended for opportunistic analysis from ES/GS due to analytical validity concerns for other pharmacogenes.7
Pharmacogenes can encode drug-metabolizing enzymes, transporters, drug targets, or immune response proteins. Most well-established pharmacogenes encode drug metabolism enzymes, particularly cytochrome P450 enzymes.8 The impact of variant pharmacogenes may depend on whether a drug is a pro-drug that is activated by drug-metabolizing enzymes, or an active drug that is metabolized into inactive or active metabolites (Fig. 1).
Fig. 1.
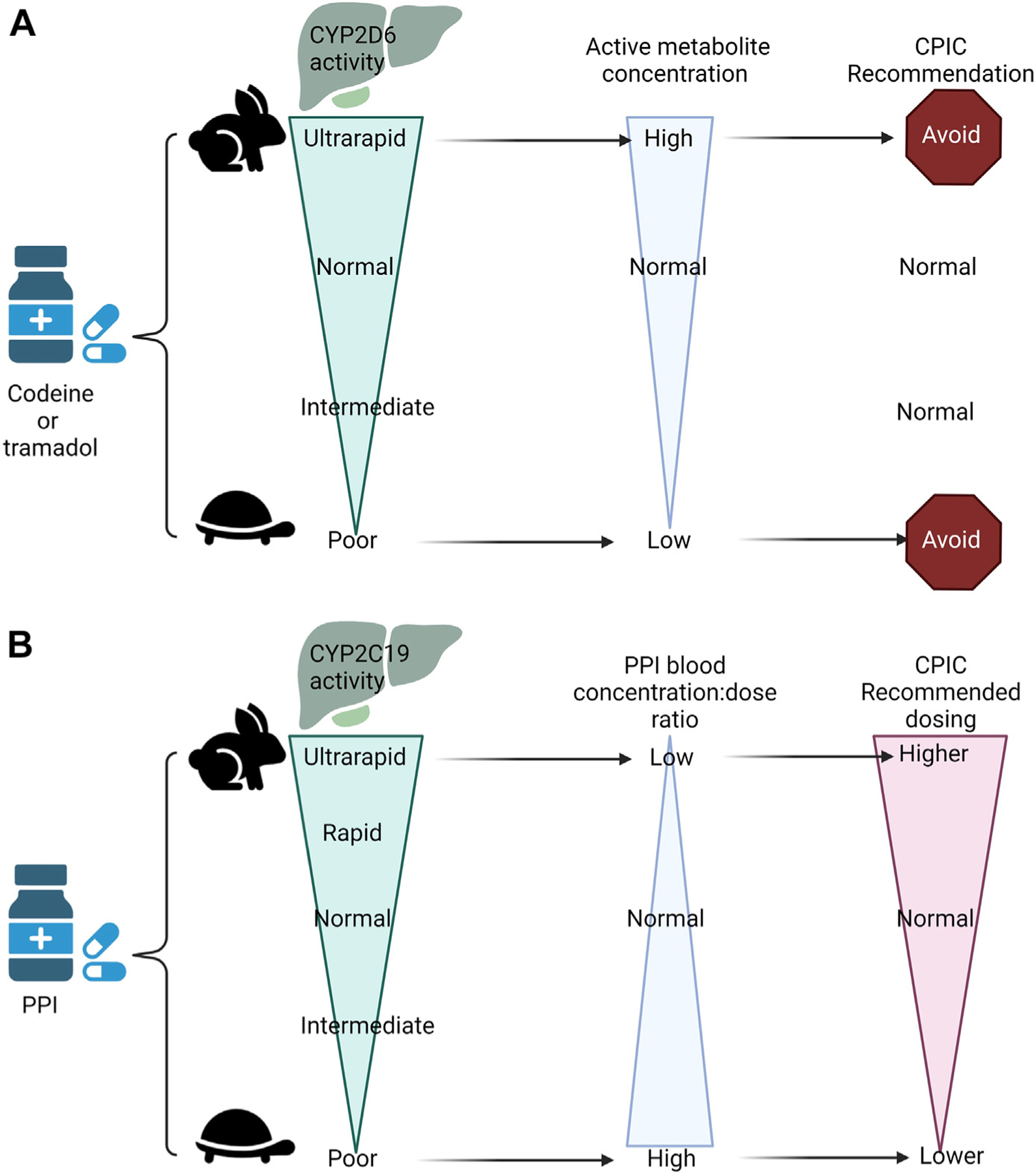
Examples of a pro-drug being metabolized to an active drug (codeine to morphine, A) and active drug being metabolized to an inactive drug (Proton pump inhibitors, (PPIs), B) with the blood concentrations and Clinical Pharmacogenetics Implementation Consortium (CPIC) recommendations.38,44 This figure was created using Biorender.com.
PGx uses a unique nomenclature system that catalogs alleles with star numbers (eg, *1, *2, *3). The *1 allele denotes a “normal” function allele. A star allele can be informed by one or more variants in a gene. The Pharmacogene Variation Consortium (PharmVar) catalogs the star allele nomenclature and functional characterization for many pharmacogenes.9 The functional consequence of a star allele that codes for a drug-metabolizing enzyme is categorized as: no function, decreased function, normal function, and increased function.10,11 The phenotype of an individual for each drug-metabolizing enzyme or transporter is determined by the combined functional activity of the maternal and paternal star alleles (called a diplotype). CPIC guidelines include an assessment of the activity of each allele, translation of diplotypes into phenotypes, and dosing recommendations.1 For genes associated with medication hypersensitivity (eg, HLA-A, HLA-B), a single risk allele is associated with hypersensitivity.10
To avoid repeating recent reviews in pediatric PGx,8,12 herein we use a case-based approach to illustrate the use of PGx data in pediatric clinical care. Pharmacogenes featured in cases and their potential phenotypes are in Table 1.
Table 1.
Genes tested in case examples, their potential phenotypes, and the availability of CPIC guidelines
| Genes Tested in Case Examples | Potential Reported Phenotypes | At Least 1 Gene-Drug CPIC Published Guideline (Yes/No) |
|---|---|---|
| Cytochrome P450 genes: | ||
| CYP2B6 | PM, IM, NM, RM, UM | Yes |
| CYP2C19 | PM, IM, NM, RM, UM | Yes |
| CYP2C9 | PM, IM, NM | Yes |
| CYP2D6 | PM, IM, NM, UM | Yes |
| CYP3A5 | PM, IM, NM | Yes |
| NUDT15 | PM, IM, NM | Yes |
| TPMT | PM, IM, NM | Yes |
| Transporter genes | ||
| SLC6A4 | Normal or low activity | No |
| Receptor genes | ||
| HTR2A | Normal, intermediate or low activity | No |
| Hypersensitivity genes | ||
| HLA-A*31:01 | Positive, negative | Yes |
| HLA-B*15:02 | Positive, negative | Yes |
| Other genes | ||
| CFTR | Cystic fibrosis | Yes |
| RYR1 | Malignant hyperthermia susceptibility | Yes |
| MT-RNR1 | MT-RNR1 aminoglycoside-induced hearing loss or nephrotoxicity susceptibility | Yes |
Abbreviations: IM, intermediate metabolizer; NM, normal metabolizer; PM, poor metabolizer; RM, rapid metabolizer; UM, ultrarapid metabolizer.
CASES AND DISCUSSION
Case 1: Medically Complex Patient
ST is a 9 year-old patient with medical complexity consequent to a non-fatal submersion event resulting in cardiac arrest, anoxic brain injury, severe neurologic impairment, and subsequent need for many medications. She has demonstrated persistent pain and anxiety behaviors attributed to visceral hyperalgesia and likely dysfunction of the neurologic-gut axis, despite formula and medication regimen changes. Parents ask about selective serotonin reuptake inhibitors (SSRIs) and share that they learned from other parents that genetic testing can be used to help manage these medications. What resources are available to determine if results of PGx testing are relevant for SSRIs?
Parents are increasingly finding information about PGx testing on the internet and through social networks. Some parents are initiating their child’s testing through direct-to-consumer laboratories. Pediatric providers should be prepared to answer parents’ questions about PGx testing and to use information from PGx reports.
ST’s mother specifically asks about genetic testing for SSRIs. The reader is referred to a pragmatic review of considerations whether or not to test13 and a short, useful guide for pharmacogenetic test selection.14 Briefly, the first step is to determine if a medication has PGx guidelines or drug label with PGx advice. The Pharmacogenomics Knowledge Database (PharmGKB) is a useful place to start (Table 2). Guidelines and recommendations as well as drug labels can be searched with a Pediatric filter (Fig. 2). If PGx information is in one or more drug labels, it is tagged as either PGx: testing required, testing recommended, actionable or informative. Specific SSRI guideline recommendations are addressed in Case 2.
Table 2.
Resources for common questions
| Clinical Question | Resources to Find Answers |
|---|---|
| How do I determine if my patient’s PGx test results are relevant for medications I want to prescribe? | Pharmacogenomics Knowledgebase (PharmGKB) Clinical Guideline Annotations15
|
| How do I find medications that can change a child’s genotype-predicted cytochrome P450 metabolizing phenotype? | FDA Drug Development and Drug Interactions Table of Substrates, Inhibitors, and Inducers17
|
| How do I find a laboratory that offers PGx testing? | First check with your clinic/hospital’s laboratory and use existing service if available.
|
| How do I find a genetic counselor near me? | Use the National Society of Genetic Counselors’ search tool: https://findageneticcounselor.nsgc.org/In-Person-FindaGC |
Fig. 2.
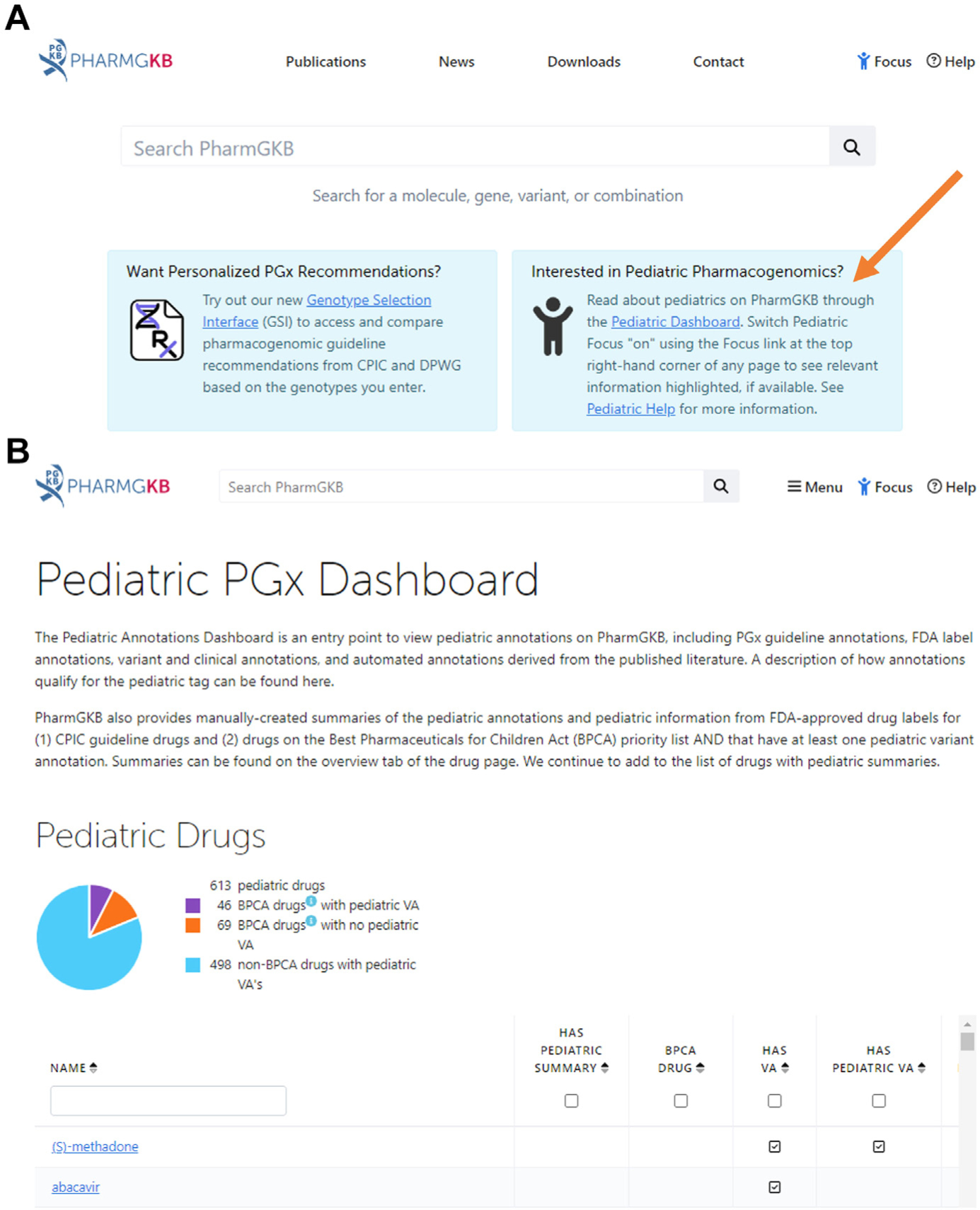
Find the Pharmacogenomics Knowledgebase (PharmGKB) pediatric website by going to pharmgkb.org and clicking the link for the pediatric website (indicated by the orange arrow in A) or go straight to what is shown in panel B at https://www.pharmgkb.org/pediatric/dashboard.
Drug-drug interactions must also be considered when a child is on many different medications.15 Phenoconversion is a type of drug-drug interaction that changes a child’s genotype-predicted cytochrome P450 metabolizing phenotype.15–17 Medications, supplements, or food that act as inducers can increase the activity of a particular drug-metabolizing enzyme, whereas inhibitors can reduce or eliminate enzyme activity (see Table 2 for resource).
Some pediatric hospitals reduce the burden on clinicians by providing PGx just-in-time information delivery models.4,6 Consulting with local pharmacists may also be helpful as pharmacy practices are increasingly incorporating PGx information into their medication therapy management frameworks.18 Newer PGx delivery models include genetic counselors, given their expertise in obtaining, evaluating, and communicating genetic test results.19
Case 2: Neurology Patient
JP is an 11-year-old nonverbal male with autism spectrum disorder, epilepsy, and worsening aggressive behavior. Current medications are clonidine (for sleep), olanza-pine (for agitation), and buspirone (for agitation). In the past, levetiracetam worsened behavior, risperidone caused fatigue and ataxia, and divalproex was ineffective. The patient is admitted to address behavior concerns, where oxcarbazepine and escitalopram are considered. Panel-based PGx testing is done on admission and results are reflected in Table 3. Which of these results are valuable, and how might they potentially explain prior medication responses and inform future drug choices?
Table 3.
Case 2 panel-based testing results
| Genotype Results | Phenotype | Relevant Medication(s) | Reference(s) |
|---|---|---|---|
| HLA-B*15:02 | Non-carrier | Oxcarbazepine | 24 |
| HLA-A*31:01 | Non-carrier | Oxcarbazepine | 24 |
| CYP2D6 *1/*4 | IM | Risperidone | 25 |
| CYP2C19 *1/*17 | RM | Escitalopram | 26 |
| CYP2C9 *1/*1 | NM | None | |
| CYP2B6 *1/*6 | IM | None | |
| SLC6A4 S/S | Low Activity | None | 27,28 |
| HTR2A–1438 G > A G/A | Intermediate Activity | None | 29 |
Relevant medication(s) refers to which medications are relevant to the case. Information about genes and results not featured in the case discussion can be found on PharmGKB.
Abbreviations: IM, intermediate metabolizer; NM, normal metabolizer; RM, rapid metabolizer.
Panel-based testing may or may not provide results relevant to a patient’s current treatment as illustrated in Table 3. This patient had panel-based testing for many genes that have no CPIC guidelines relevant to the patient at the time of evaluation, but may be useful in the future (eg, if this patient were prescribed NSAIDs the CYP2C9 result would be relevant).20 There are CPIC guidelines for oxcarbazepine21 and escitalopram,22 which are being considered during JP’s admission.
JP’s results for HLA-B*15:02 and HLA-A*31:01 inform his provider that oxcarbazepine is not contraindicated. Very serious cutaneous adverse reactions (eg, Stevens-Johnson Syndrome and toxic epidermal necrosis) can occur after treatment with carbamazepine or oxcarbazepine in patients carrying human leukocyte antigen (HLA) alleles HLA-B*15:02 and HLA-A*31:01, which have variable frequency based on ethnic and geographical ancestry. CPIC recommends avoiding carbamazepine and oxcarbazepine in patients carrying one or more copies of these alleles.21
The CPIC guideline for SSRIs currently includes recommendations for escitalopram, based on CYP2C19 metabolizer status (Fig. 3).22 Based on JP’s CYP2C19 rapid metabolizer phenotype, standard escitalopram dosing is recommended.
Fig. 3.
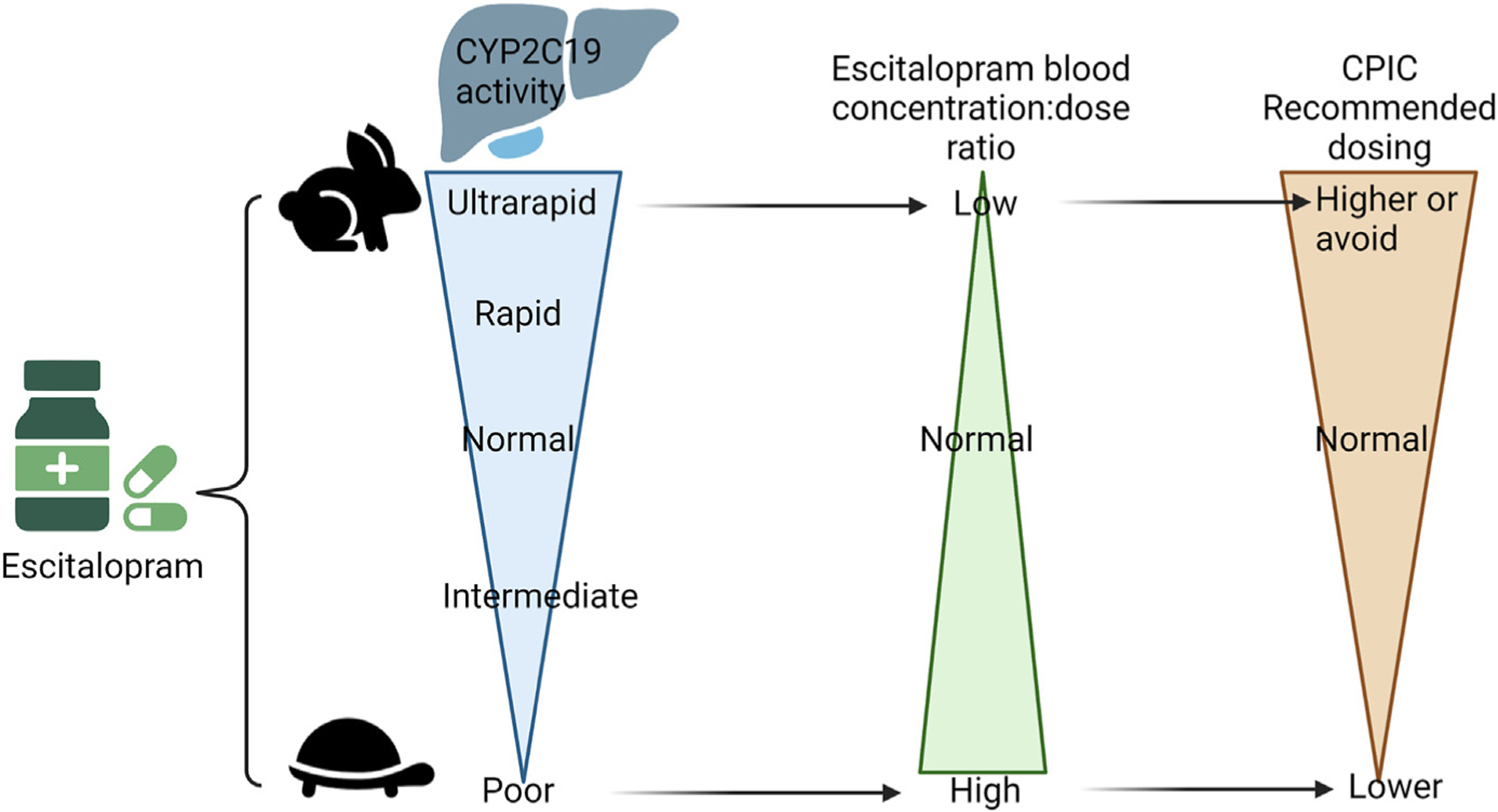
Escitalopram dosing recommendations are based on CYP2C19 phenotype.22 This figure was created using Biorender.com.
Risperidone is metabolized by CYP2D6 into an active metabolite (9-hydroxyrisper-idone or paliperidone). To determine if JP’s CYP2D6 intermediate metabolizer status explained his past adverse reactions, JP’s provider might search PharmGKB’s Clinical Guideline Annotations to discover that while there is no CPIC guideline for risperidone, a Dutch Pharmacogenetics Working Group guideline recommends decreased dosing for CYP2D6 poor metabolizers. For CYP2D6 ultrarapid metabolizers, an alternative drug or titrating risperidone to the maximum dose for the active metabolite (paliperidone) is recommended.23 Although dosing recommendations do not exist for CYP2D6 intermediate metabolizers, slower risperidone clearance may have contributed to JP’s previous side effects.
Case 3a: Surgical Patient
CW is a 14-year-old male scheduled for vertebral body tethering for progressive scoliosis. CW previously participated in a research study during which genomic screening by a Clinical Laboratory Improvement Amendment (CLIA) certified and College of American Pathologists (CAP) accredited laboratory found a pathogenic variant in RYR1. The finding was disclosed to the adolescent and his mother, and the PDF report was placed in CW’s electronic health record. During the preoperative clinic visit, CW’s mother shared the report with the anesthesiologist. There was no known family history of exposure to anesthesia. CW previously tolerated pressure equalizer tube placement at 18 months and tonsillectomy at 3 years of age. What precautions are indicated by the finding of a pathogenic variant in RYR1?
A pathogenic variant (previously referred to as mutation) in the RYR1 gene identifies CW as having malignant hyperthermia susceptibility (MHS), an autosomal dominant condition which may remain clinically silent until adequate exposure to a triggering drug agent. Pathogenic variants in the CACNA1S gene have been reported in ~ 1% of patients with malignant hyperthermia.24 Exposure to volatile anesthetics or depolarizing neuromuscular blockers, particularly succinylcholine, or the combination of succinylcholine and a volatile anesthetic, can trigger life-threatening malignant hyperthermia.25 Children can present differently than adults, delaying diagnosis.26–28 Clinical testing of RYR1 and CACNA1S for MHS is available but not routinely performed in clinical settings prior to surgeries.29
ES and GS are increasingly used for clinical diagnosis. The American College of Medical Genetics and Genomics (ACMG) recommends that regardless of indication for clinical ES/GS, analysis should routinely include specified genes known to inform genetic disorders with preventive or early treatment options, including RYR1 and CACNA1S (since 2013).30,31 Large scale ES/GS studies have analyzed the ACMG specified genes and provided results to study participants, including children.32–34 As the use of clinical and research ES/GS increases, pediatric clinicians are prudent to add questions about prior genetic testing for MHS to standard pre-anesthesia assessments.
In our case, CW previously tolerated anesthesia. Even in cases like this, the CPIC guideline that includes 48 RYR1 and 2 CACNA1S pathogenic variants states that succinylcholine and potent volatile anesthetics (sevoflurane, halothane, enflurane, isoflurane, methoxyflurane, and desflurane) are contraindicated in any patient with MHS.35 Studies have shown that RYR1 pathogenic variants are not 100% penetrant for malignant hyperthermia phenotypes, which may explain CW’s previous tolerance to anesthesia. A recent study estimated between 0.25 and 0.76 probability of developing malignant hyperthermia with one of the 9 known RYR1 pathogenic variants when exposed to a triggering agent.36 Importantly, trigger agents may be initially tolerated, with subsequent exposure resulting in malignant hyperthermia, indicating the need for continued vigilance.35
Case 3b: Surgical Patient, Continued
CW’s mother reported that after his prior tonsillectomy, oral codeine was ineffective. Of note, a prior hospitalization for major depression led to PGx panel testing, and he was found to be a CYP2D6 poor metabolizer (diplotype *3/*4) and CYP2C19 normal metabolizer (diplotype *1/*1). Current medications include sertraline daily for depression and cetirizine daily as needed for seasonal allergies. The orthopedic surgeon reviewed the psychiatry PGx report, noted recommendations for antidepressants and antipsychotics, and was uncertain about the implications for postoperative pain management. How do these PGx results inform postoperative management?
Opioids and nonsteroidal anti-inflammatory drugs (NSAIDs) are essential components of perioperative pain management.37 Ondansetron is commonly prescribed as opioids increase postoperative nausea and vomiting. Variable CYP2D6 function can impact effectiveness and risk for adverse reactions for some opioids and may impact ondansetron efficacy. In our case, CW has CYP2D6 *3/*4 genotype, with 2 no function alleles, consistent with a poor metabolizer phenotype. CW’s results for CYP2D6 and CYP2C19 do not inform the use of NSAIDs, which are metabolized by CYP2C9 and discussed further in Case 4.
Codeine and tramadol are prodrugs (inactive in their dosage forms) and are O-demethylated by CYP2D6 to their active (analgesic) metabolites, morphine, and O-desmethyltramadol, respectively. CYP2D6 poor metabolizers cannot convert codeine or tramadol to their active forms, and codeine and tramadol will not provide pain relief. Evidence-based guidelines recommend avoiding codeine and tramadol in any CYP2D6 poor metabolizer (see Fig. 1A). If an opioid is required for pain management, then a non-tramadol and a non-codeine containing analgesic should be considered.38
The FDA issued a contraindication for codeine products and tramadol in all children less than 12 years of age, children less than 18 years after tonsillectomy or who are obese, have obstructive sleep apnea or lung disease.39 Since then, the use of codeine has decreased and the use of oxycodone has increased in children.2 Oxycodone and hydrocodone are converted by CYP2D6 to their more active metabolites, oxymorphone and hydromorphone. Although lower active drug concentrations are observed in CYP2D6 poor metabolizers, a clear clinical difference in drug response has not been demonstrated. Because past studies had few participants identified as CYP2D6 ultrarapid metabolizers, there is insufficient evidence regarding the safety and efficacy of oxycodone and hydrocodone for these individuals.38 For CW, if opioids are required for post-operative pain management, standard dosing of oxycodone or hydrocodone would be appropriate.
Ondansetron, a 5-HT receptor antagonist, is used to prevent nausea and vomiting and is inactivated by CYP2D6. Evidence from clinical studies in adults indicates an alternative medication may be needed for CYP2D6 ultrarapid metabolizers due to inadequate effectiveness.40 Standard dosing was effective and well tolerated by adult patients who were CYP2D6 intermediate or poor metabolizers. Studies in children at risk for postoperative nausea and vomiting are needed.41,42 Further discussion of ondansetron is included in case 4 later in discussion.
Case 4: Oncology Patient
AR is a 16-year-old female diagnosed with acute lymphoblastic leukemia. Her tumor genetic testing indicated she had the BCR-ABL translocation, which means a tyrosine kinase inhibitor (eg, dasatinib or imatinib) is indicated. Her treatment regimen will also include mercaptopurine, ondansetron, opioids, and NSAIDs. She may receive voriconazole, tacrolimus, proton pump inhibitors (PPIs), and/or an SSRI. For these reasons, the oncology pharmacist orders a multi-gene PGx panel. A buccal swab was performed for her PGx panel, as a blood sample at the start of treatment would be largely leukemic cells that may have more mutations than her germline. The panel test results are shown in Table 4. The medications relevant to each gene on the medications she will receive are detailed later in discussion. How might these PGx results inform her chemotherapy and symptomatic management?
Table 4.
Case 4 Panel-based testing results
| Genotype Results | Phenotype | Relevant Medication(s) | Reference(s) |
|---|---|---|---|
| CYP2D6 *1/*1x2 | UM | Ondansetron, codeine, tramadol, paroxetine, fluvoxamine | 9,31,48 |
| CYP2C19 *1/*2 | IM | Voriconazole, omeprazole, lansoprazole, pantoprazole, dexlansoprazole, sertraline, escitalopram, citalopram | 10,31,51 |
| CYP2C9 *1/*1 | NM | Celecoxib, flurbiprofen, ibuprofen and lornoxicam | 30 |
| CYP3A5 *1/*3 | IM | Tacrolimus | 52 |
| TPMT *1/*1 | NM | Mercaptopurine | 53 |
| NUDT15 *1/*3 | IM | Mercaptopurine | 53 |
Relevant medication(s) refers to which medications are relevant to the case. Information about genes and results not featured in the case discussion can be found on PharmGKB.
Abbreviations: IM, intermediate metabolizer; NM, normal metabolizer; UM, ultrarapid metabolizer.
Thiopurines (eg, mercaptopurine) are inactive prodrugs that are metabolized into active immunosuppressants, which are then inactivated by the TPMT and NUDT15 enzymes. The CPIC guideline aligns with the FDA package insert for mercaptopurine, which indicates that TPMT or NUDT15 intermediate metabolizers often tolerate recommended standard mercaptopurine doses, but some require dose reduction based on toxicities.43 If AR had been found to be a TPMT or NUDT15 poor metabolizer, her dose would need to be reduced dramatically (~10-fold). As most patients with leukemia in the U.S. are enrolled in clinical trials, the recommended dose of mercaptopurine based on TPMT and NUDT15 will be dictated by the study protocol.
Ondansetron and opioids were addressed in the previous case. However, in this case, AR is a CYP2D6 ultrarapid metabolizer. Ondansetron is prescribed to prevent or reduce chemotherapy-induced nausea and vomiting. As a CYP2D6 ultrarapid metabolizer, she metabolizes ondansetron quickly, and CPIC recommends an alternative antiemetic not metabolized by CYP2D6 (eg, granisetron),40 though this is based on adult studies, and 2 recent studies in children could not replicate differences in response.41,42 If opioids are needed for pain control, codeine and tramadol should be avoided because CYP2D6 ultrarapid metabolizers produce more of the active metabolite than normal metabolizers (see Fig. 1A), which puts them at increased risk of respiratory depression and other adverse events.38 If pain can be controlled by NSAIDs, the patient’s CYP2C9 result is relevant for some options (see Table 4). The patient is a normal metabolizer for CYP2C9, so she should receive the typical doses of these medications (Fig. 4).
Fig. 4.
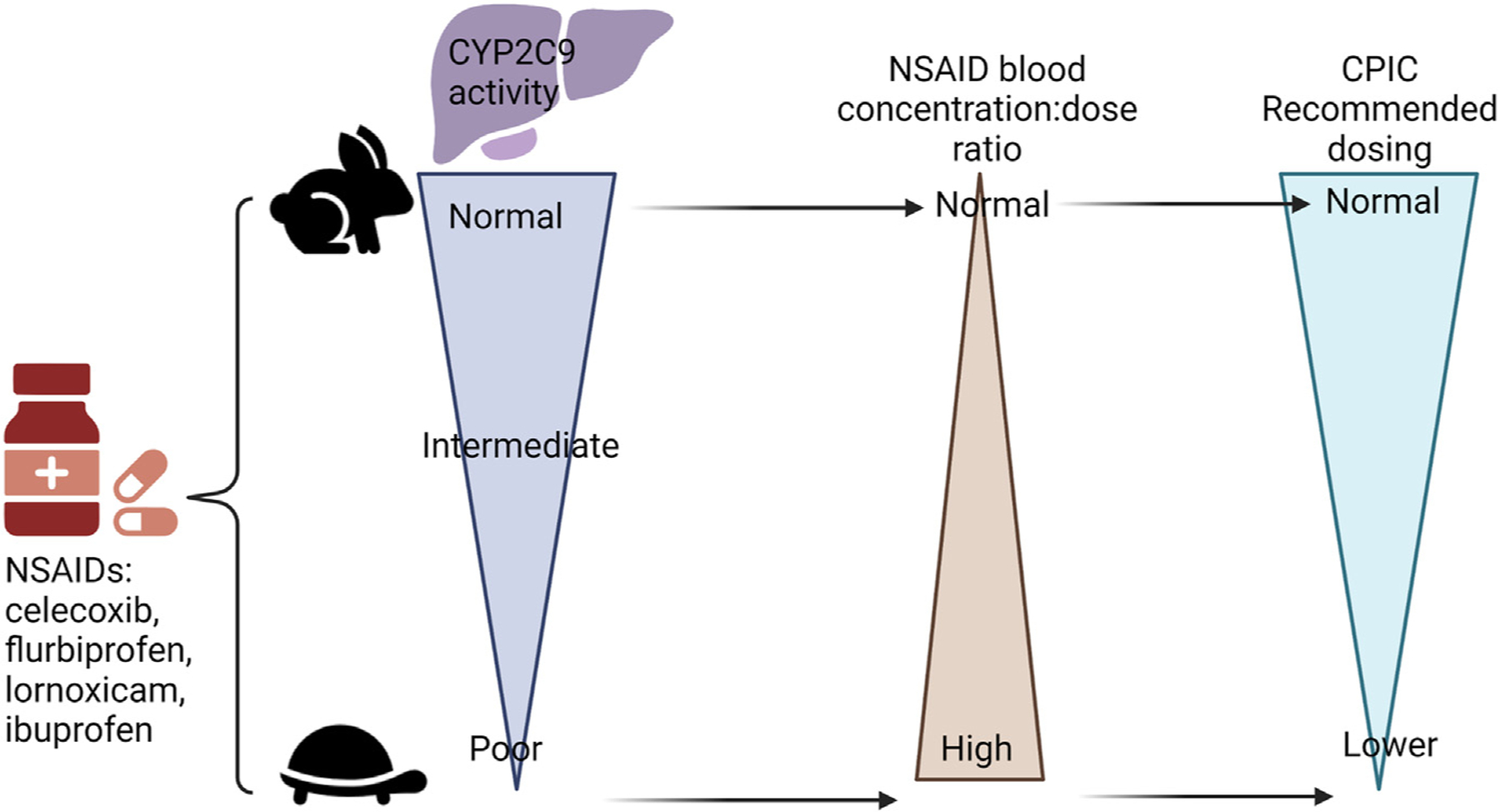
Nonsteroidal anti-inflammatory drugs (NSAIDs) dosing recommendations are based on CYP2C9 phenotype.20 Of note, CYP2C9 does not have any variants that would make a patient a rapid or ultrarapid metabolizer. This figure was created using Biorender.com.
The antifungal agent voriconazole may be used prophylactically or to treat a fungal infection. As this patient is a CYP2C19 intermediate metabolizer, CPIC recommends the usual starting dose with adjustments being made for clinical factors, such as drug interactions, hepatic function, renal function, fungal species, site of infection, therapeutic drug monitoring, and comorbidities (Fig. 5).20 The patient will likely have higher dose-adjusted concentrations than normal metabolizers so may need a decreased dose after therapeutic drug monitoring.
Fig. 5.
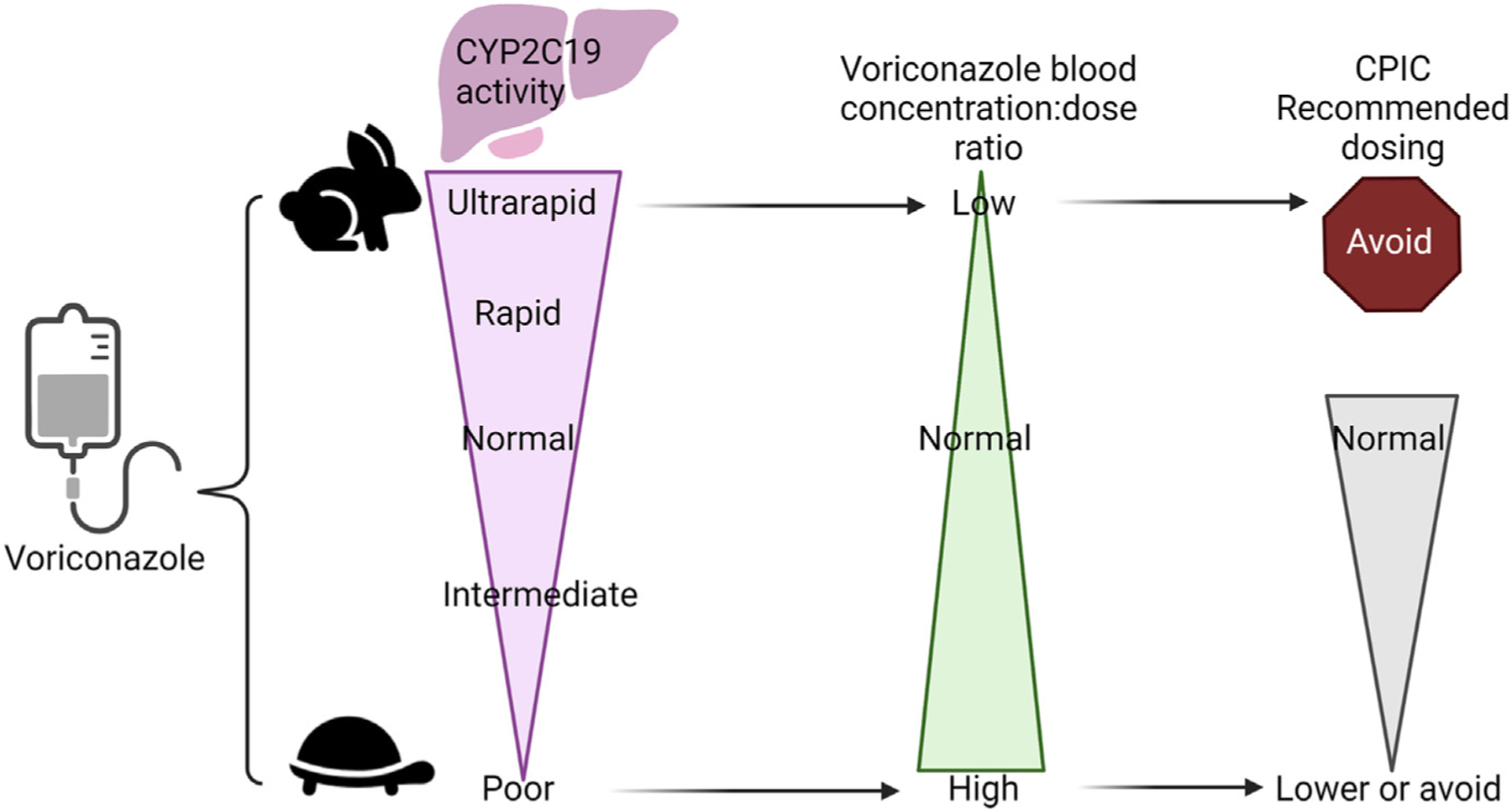
Voriconazole recommendations are based on CYP2C19 phenotype.59 This figure was created using Biorender.com.
PPIs (eg, omeprazole) may be prescribed to this patient prophylactically or for gastroesophageal reflux. As a CYP2C19 intermediate metabolizer, she should receive the usual dose at the start of therapy but consider a 50% reduced dose for chronic therapy more than 12 weeks due to increased risk for adverse events, including respiratory and gastrointestinal infections.44 CYP2C19 rapid and ultrarapid metabolizers are likely to have reduced blood concentrations and could need higher doses (see Fig. 1B).
Tacrolimus may be considered for AR if she requires a hematopoietic stem cell transplant. CYP3A5 variation explains ~ 50% of the variability in tacrolimus plasma concentrations.45 Individuals who carry at least one *1 normal function allele (intermediate and normal metabolizers) are called expressers and those without are called non-expressers or poor metabolizers. The most common phenotype among individuals of European ancestry is poor metabolizer, and this epidemiology is what the usual dosing regimens are based on in the U.S. For expressers, CPIC recommends starting doses 1.5–2x higher than normal (though not exceeding 0.3 mg/kg/d), which should be followed by routine therapeutic drug monitoring (Fig. 6).45 AR is an intermediate metabolizer so she would need a higher than normal starting dose of tacrolimus to prevent or treat graft-versus-host disease.
Fig. 6.
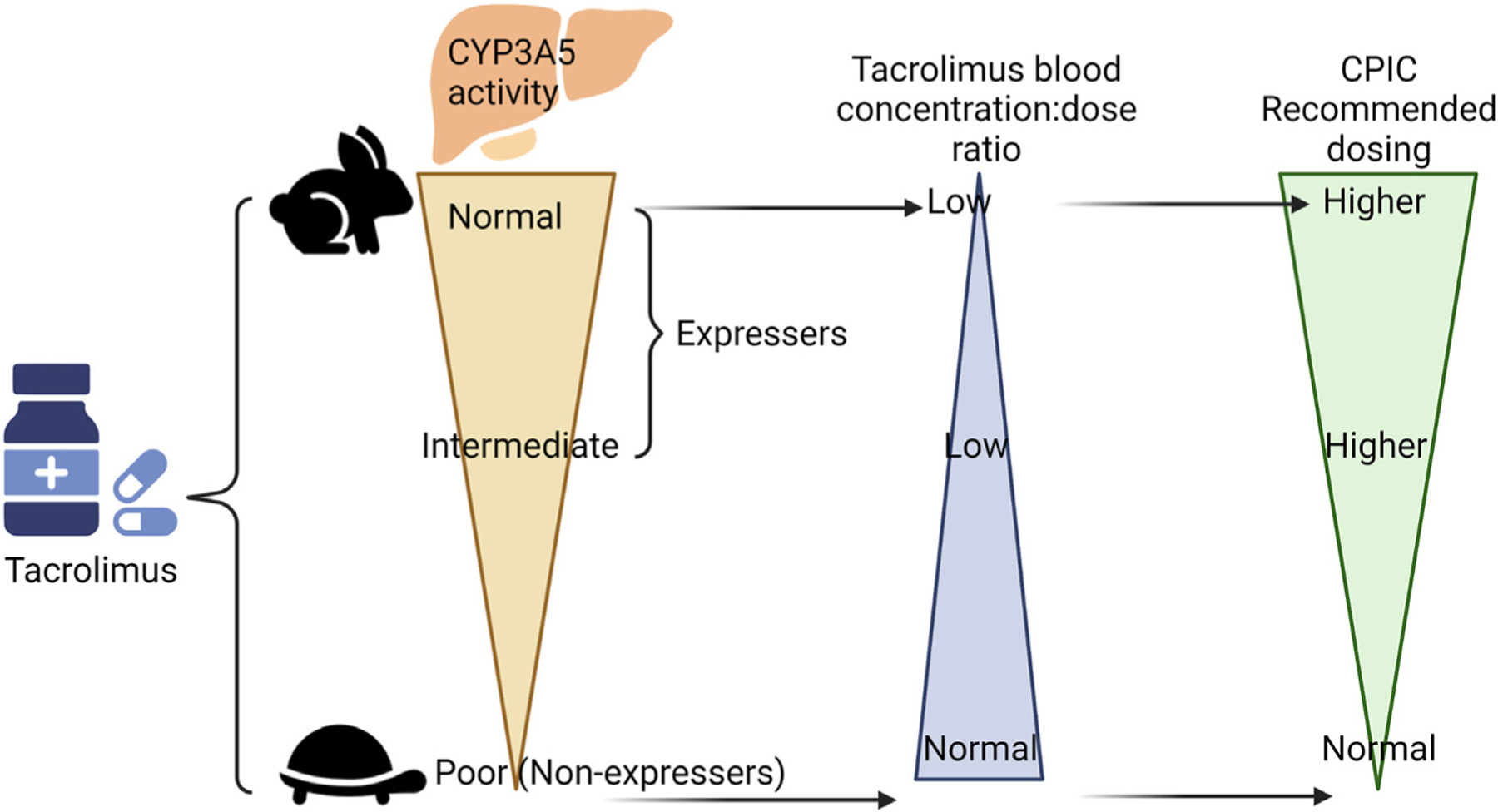
Tacrolimus recommendations are based on CYP3A5 phenotype.45 This figure was created using Biorender.com.
SSRIs are often prescribed to adolescent patients with cancer for depression and anxiety, and were previously discussed in Case 2.
Case 5: Patient with Rare Genetic Disease
ML is a 6-week-old infant who had a positive newborn screen for cystic fibrosis. After additional CFTR testing F508del/G542X and a confirmatory sweat test, ML is being evaluated by providers at an accredited CF center. ML’s parents feel overwhelmed by the lifelong care ML will require but feel hopeful when they learn that ML’s genetic results indicate ML will qualify for a new type of medication called CFTR modulators.
CFTR testing is recommended as part of the diagnostic process for cystic fibrosis (CF).46 Pathogenic variants in the CFTR gene result in dysfunction or absence of the chloride ion channel, cystic fibrosis transmembrane conductance regulator. People with CF have pathogenic variants in each of their CFTR genes resulting in multi-organ disease manifestations.47 Disease management had relied on symptom-based treatments with incremental improvement over the years in lifespan and quality of life.48 CFTR testing has become an essential first step before prescribing a new class of drugs called CFTR modulators, considered transformational therapies as they address the molecular cause of CF. CFTR modulators are small molecules designed to restore or improve the function of certain types of variant CFTR protein.47 There are over a 1000 pathogenic and likely pathogenic CFTR variants listed in ClinVar,49 and each codes for one of the 6 classes of variant CFTR proteins.50 Available modulators target Class II, protein processing, and Class III gating variant CFTR.47 The first FDA approved modulator, ivacaftor, is a potentiator. It helps the gating variant CFTR channel open at the cell membrane. Originally, it was approved for one specific pathogenic variant, G551D, found in less than 5% of patients with CF. Since then, its use has been expanded to include other gating variants in CFTR.51
ML has the most common pathogenic variant found in people with CF, F508del, and ML’s other variant results in absent CFTR protein. Three FDA-approved modulators contain at least one corrector small molecule essential for patients who have F508del. CFTR correctors augment the protein folding process and transit to the cell membrane.52 The most recently FDA-approved triple therapy modulator, elexacaftor/tezacaftor/ivacaftor, has 2 CFTR correctors to maximize stable folding and transport of the F508del CFTR protein to the cell membrane and it has the potentiator to improve the gating function of the F508del protein. It is approved in children 6 years and older,53 is considered lifelong, and costs over $300,000 per year in the U.S.54 Health Canada Guidelines indicate elexacaftor/tezacaftor/ivacaftor will be the primary modulator therapy for any CF patient over the age of 12 who has at least one F508del variant.55
Throughout ML’s life, multiple medications and therapies will be needed48 to manage symptoms that may include PPIs (discussed above) and aminoglycosides for which CPIC guidelines are available. Specific variants in the mitochondrial gene, MT-RNR1, predispose individuals to severe aminoglycoside-induced hearing loss. A point-of-care MT-RNR1 genotyping test has recently been developed for clinical use,56 but testing is not done routinely due to the complexity of genotyping mitochondrial DNA. It is recommended that aminoglycoside antibiotics be avoided in individuals with one of the high-risk MT-RNR1 variants unless the infection severity or lack of effective alternative therapies outweighs the risk of hearing loss.57 Mitochondrial genes are maternally inherited. Because each cell in the body has hundreds to thousands of mitochondria and each mitochondrion has many copies of mitochondrial DNA,58 genetic counseling should precede genetic testing.
CLINICS CARE POINTS.
Evidence-based gene-drug guidelines are freely available online through PharmGKB and CPIC; however, evidence specifically for children may be limited or absent.
- Potential for phenoconversion due to concomitant administration of inducers and inhibitors may need to be considered when using PGx test results to select or dose medications.
- FDA Drug Development and Drug Interactions Table of Substrates, Inhibitors, and Inducers is a helpful resource
Some pediatric health care facilities have or are building PGx clinical decision support into their EHRs, and these may or may not incorporate individualized phenoconversion adjustments to therapeutic recommendations.
- Some pediatric health care facilities have pharmacogenomic consultation services that may be managed by a pharmacy or a pharmacogenomic-specific multidisciplinary team.
- Pediatric prescribers should find out what is available at their facility or local children’s hospital.
- Pediatric providers should consider the following if they work at a facility that does not offer point-of-care clinical decision support or consultation services:
- Identify commonly prescribed medications and proactively identify which of those medications have evidence-based guidelines and have the potential to be impacted by phenoconversion.
- Identify a local or regional genetic counselor who might be able to help identify reputable laboratories offering PGx testing or who may be willing to schedule clinical encounters with patients interested in PGx testing. The National Society of Genetic Counselors is a good place to start (see Table 2).
KEY POINTS.
Pediatric patients are prescribed medications influenced by pharmacogenomics.
Guidelines are available for using pharmacogenomics to manage medications in pediatric patients.
Cases are presented to demonstrate the utility of pharmacogenomics in pediatric patients.
DISCLOSURE
L.B. Ramsey serves as a consultant and has received grant funding from BTG Specialty Pharmaceuticals. L.B. Ramsey receives funding from the National Institutes of Health, United States (R01HD099775 and R01HD089928). C.A. Prows receives funding from the National Institutes of Health (U01 HG011172, R01HG010166) and is an inventor on Patent US 10,662,476 B2 - Personalized pain management and anesthesia: preemptive risk identification and therapeutic decision support but has not received any associated royalties. SLV receives funding from the National Institutes of Health (R01GM132204, U01HG010232, P50 HD106446). S.T. Girdwood receives funding from the National Institute of General Medical Sciences, United States (1R35GM146701).
REFERENCES
- 1.Relling MV, Klein TE, Gammal RS, et al. The Clinical Pharmacogenetics Implementation Consortium: 10 Years Later. Clin Pharmacol Ther 2020;107(1):171–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ramsey LB, Ong HH, Schildcrout JS, et al. Prescribing Prevalence of Medications With Potential Genotype-Guided Dosing in Pediatric Patients. JAMA Netw Open 2020;3(12):e2029411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avello K, Bell M, Stein Q, et al. Perspectives of Pediatric Providers Regarding Clinical Use of Pharmacogenetics. S D Med 2021;74(7):294–301. Available at: https://pubmed.ncbi.nlm.nih.gov/34449988/. Accessed October 14, 2022. [PubMed] [Google Scholar]
- 4.Ramsey LB, Prows CA, Zhang K, et al. Implementation of Pharmacogenetics at Cincinnati Children’s Hospital Medical Center: Lessons Learned Over 14 Years of Personalizing Medicine. Clin Pharmacol Ther 2019;105(1):49–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown JT, Ramsey LB, Van Driest SL, et al. Characterizing Pharmacogenetic Testing Among Children’s Hospitals. Clin Transl Sci 2021;14(2):692–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gregornik D, Salyakina D, Brown M, et al. Pediatric pharmacogenomics: challenges and opportunities: on behalf of the Sanford Children’s Genomic Medicine Consortium. Pharmacogenomics J 2021;21(1):8–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller DT, Lee K, Gordon AS, et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2021 update: a policy statement of the American College of Medical Genetics and Genomics (ACMG). Genet Med 2021;23(8):1391–8. [DOI] [PubMed] [Google Scholar]
- 8.Ramsey LB, Brown JT, Vear SI, et al. Gene-based dose optimization in children. Annu Rev Pharmacol Toxicol 2020;60:311–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaedigk A, Ingelman-Sundberg M, Miller NA, et al. The Pharmacogene Variation (PharmVar) Consortium: Incorporation of the Human Cytochrome P450 (CYP) Allele Nomenclature Database. Clin Pharmacol Ther 2018;103(3):399–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caudle KE, Dunnenberger HM, Freimuth RR, et al. Standardizing terms for clinical pharmacogenetic test results: Consensus terms from the Clinical Pharmacogenetics Implementation Consortium (CPIC). Genet Med 2017;19(2):215–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caudle KE, Sangkuhl K, Whirl-Carrillo M, et al. Standardizing CYP2D6 Genotype to Phenotype Translation: Consensus Recommendations from the Clinical Pharmacogenetics Implementation Consortium and Dutch Pharmacogenetics Working Group. Clin Transl Sci 2020;13(1):116–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang Girdwood SC, Rossow KM, Van Driest SL, et al. Perspectives from the Society for Pediatric Research: pharmacogenetics for pediatricians. Pediatr Res 2022;91(3):529–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicholson WT, Formea CM, Matey ET, et al. Considerations When Applying Pharmacogenomics to Your Practice. Mayo Clin Proc 2021;96(1):218–30. [DOI] [PubMed] [Google Scholar]
- 14.Bousman CA, Zierhut H, Müller DJ. Navigating the Labyrinth of Pharmacogenetic Testing: A Guide to Test Selection. Clin Pharmacol Ther 2019;106(2):309–12. [DOI] [PubMed] [Google Scholar]
- 15.Cicali EJ, Elchynski AL, Cook KJ, et al. How to Integrate CYP2D6 Phenoconversion Into Clinical Pharmacogenetics: A Tutorial. Clin Pharmacol Ther 2021;110(3): 677–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malki MA, Pearson ER. Drug-drug-gene interactions and adverse drug reactions. Pharmacogenomics J 2020;20(3):355–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stout SM, Nemerovski CW, Streetman DS, et al. Interpretation of Cytochrome P-450 Inhibition and Induction Effects From Clinical Data: Current Standards and Recommendations for Implementation. Clin Pharmacol Ther 2021; 109(1):82–6. [DOI] [PubMed] [Google Scholar]
- 18.Hayashi M, Hamdy DA, Mahmoud SH. Applications for pharmacogenomics in pharmacy practice: A scoping review. Res Social Adm Pharm 2022;18(7): 3094–118. [DOI] [PubMed] [Google Scholar]
- 19.Gammal RS, Fieg E. Pharmacist and genetic counselor collaboration in pharmacogenomics. Am J Health Syst Pharm 2022;79(18). 10.1093/AJHP/ZXAC168. [DOI] [PubMed] [Google Scholar]
- 20.Theken KN, Lee CR, Gong L, et al. Clinical Pharmacogenetics Implementation Consortium Guideline (CPIC) for CYP2C9 and Nonsteroidal Anti-Inflammatory Drugs. Clin Pharmacol Ther 2020;108(2):191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Phillips EJ, Sukasem C, Whirl-Carrillo M, et al. Clinical Pharmacogenetics Implementation Consortium Guideline for HLA Genotype and Use of Carbamazepine and Oxcarbazepine: 2017 Update. Clin Pharmacol Ther 2018;103(4):574–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bousman CA, Stevenson JM, Ramsey LB, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6, CYP2C19, CYP2B6, SLC6A4, and HTR2A Genotypes and Serotonin Reuptake Inhibitor Antidepressants. Clin Pharmacol Ther 2023;114(1):51–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Risperidone. Available at: https://www.pharmgkb.org/chemical/PA451257/guidelineAnnotation/PA166104943. Accessed October 5, 2022.
- 24.Yang L, Dedkova EN, Allen PD, et al. T lymphocytes from malignant hyperthermia-susceptible mice display aberrations in intracellular calcium signaling and mitochondrial function. Cell Calcium 2021;93:102325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larach MG, Klumpner TT, Brandom BW, et al. Succinylcholine Use and Dantrolene Availability for Malignant Hyperthermia TreatmentDatabase Analyses and Systematic Review. Anesthesiology 2019;130(1):41–54. [DOI] [PubMed] [Google Scholar]
- 26.Kim KSM, Kriss RS, Tautz TJ. Malignant Hyperthermia: A Clinical Review. Adv Anesth 2019;37:35–51. [DOI] [PubMed] [Google Scholar]
- 27.Brandom BW, Bina S, Wong CA, et al. Ryanodine receptor type 1 gene variants in the malignant hyperthermia-susceptible population of the United States. Anesth Analg 2013;116(5):1078–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nelson P, Litman RS. Malignant hyperthermia in children: An analysis of the north american malignant hyperthermia registry. Anesth Analg 2014;118(2):369–74. [DOI] [PubMed] [Google Scholar]
- 29.Biesecker LG, Dirksen RT, Girard T, et al. Genomic Screening for Malignant Hyperthermia Susceptibility. Anesthesiology 2020;133(6):1277–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller DT, Lee K, Abul-Husn NS, et al. ACMG SF v3.1 list for reporting of secondary findings in clinical exome and genome sequencing: A policy statement of the American College of Medical Genetics and Genomics (ACMG). Genet Med 2022; 24(7):1407–14. [DOI] [PubMed] [Google Scholar]
- 31.Green RC, Berg JS, Grody WW, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med 2013;15(7):565–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ceyhan-Birsoy O, Ceyhan-Birsoy O, Murry JB, et al. Interpretation of Genomic Sequencing Results in Healthy and Ill Newborns: Results from the BabySeq Project. Am J Hum Genet 2019;104(1):76–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Myers MF, Martin LJ, Prows CA. Adolescents’ and Parents’ Genomic Testing Decisions: Associations With Age, Race, and Sex. J Adolesc Heal 2020;66(3): 288–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hart MR, Biesecker BB, Blout CL, et al. Secondary findings from clinical genomic sequencing: prevalence, patient perspectives, family history assessment, and health-care costs from a multisite study. Genet Med 2019;21(5):1100–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gonsalves SG, Dirksen RT, Sangkuhl K, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for the Use of Potent Volatile Anesthetic Agents and Succinylcholine in the Context of RYR1 or CACNA1S Genotypes. Clin Pharmacol Ther 2019;105(6):1338–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ibarra Moreno CA, Hu S, Kraeva N, et al. An Assessment of Penetrance and Clinical Expression of Malignant Hyperthermia in Individuals Carrying Diagnostic Ryanodine Receptor 1 Gene Mutations. Anesthesiology 2019;131(5):983–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cravero JP, Agarwal R, Berde C, et al. The Society for Pediatric Anesthesia recommendations for the use of opioids in children during the perioperative period. Paediatr Anaesth 2019;29(6):547–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crews KR, Monte AA, Huddart R, et al. Clinical Pharmacogenetics Implementation Consortium Guideline for CYP2D6, OPRM1, and COMT Genotypes and Select Opioid Therapy. Clin Pharmacol Ther 2021;110(4):888–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.FDA (Food and Drug Administration). Safety Review Update of Codeine Use in Children; New Boxed Warning and Contraindication on Use after Tonsillectomy and/or Adenoidectomy. Available at: https://www.fda.gov/media/85072/download. Accessed March 31, 2020.
- 40.Bell GC, Caudle KE, Whirl-Carrillo M, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 genotype and use of ondansetron and tropisetron. Clin Pharmacol Ther 2017;102(2). 10.1002/cpt.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edwards A, Teusink-Cross A, Martin LJ, et al. Influence of CYP2D6 metabolizer status on ondansetron efficacy in pediatric patients undergoing hematopoietic stem cell transplantation: A case series. Clin Transl Sci 2022;15(3):610–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Black K, Brenn B, Gaedigk A, et al. Pediatric CYP2D6 Metabolizer Status and Post-Tonsillectomy Nausea and Vomiting After Ondansetron. Clin Transl Sci 2023;16(2):269–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Relling MV, Schwab M, Whirl-Carrillo M, et al. Clinical Pharmacogenetics Implementation Consortium Guideline for Thiopurine Dosing Based on TPMT and NUDT15 Genotypes: 2018 Update. Clin Pharmacol Ther 2019;105(5):1095–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lima JJ, Thomas CD, Barbarino J, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2C19 and Proton Pump Inhibitor Dosing. Clin Pharmacol Ther 2021;109(6):1417–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Birdwell KA, Decker B, Barbarino JM, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for CYP3A5 genotype and tacrolimus dosing. Clin Pharmacol Ther 2015;98(1):19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Farrell PM, White TB, Ren CL, et al. Diagnosis of Cystic Fibrosis: Consensus Guidelines from the Cystic Fibrosis Foundation. J Pediatr 2017;181:S4–15.e1. [DOI] [PubMed] [Google Scholar]
- 47.Clancy JP, Cotton CU, Donaldson SH, et al. CFTR modulator theratyping: Current status, gaps and future directions. J Cyst Fibros 2019;18(1):22–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Borowitz D, Robinson KA, Rosenfeld M, et al. Cystic Fibrosis Foundation Evidence-Based Guidelines for Management of Infants with Cystic Fibrosis. J Pediatr 2009;155(6):S73–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.ClinVar. Available at: https://www.ncbi.nlm.nih.gov/clinvar/. Accessed October 5, 2022.
- 50.Bareil C, Bergougnoux A. CFTR gene variants, epidemiology and molecular pathology. Arch Pediatr 2020;27(Suppl 1):eS8–12. 10.1016/S0929-693X(20)30044-0. [DOI] [PubMed] [Google Scholar]
- 51.CPIC® Guideline for Ivacaftor and CFTR – CPIC. Available at: https://cpicpgx.org/guidelines/guideline-for-ivacaftor-and-cftr/. Accessed October 5, 2022.
- 52.Van Goor F, Hadida S, Grootenhuis PDJ, et al. Correction of the F508del-CFTR protein processing defect in vitro by the investigational drug VX-809. Proc Natl Acad Sci U S A 2011;108(46):18843–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.fda, cder. TRIKAFTA HIGHLIGHTS OF PRESCRIBING INFORMATION. Available at: www.fda.gov/medwatch. Accessed October 5, 2022.
- 54.Guo J, Garratt A, Hill A. Worldwide rates of diagnosis and effective treatment for cystic fibrosis. J Cyst Fibros 2022;21(3):456–62. [DOI] [PubMed] [Google Scholar]
- 55.Chilvers MA, Waters V, Anderson MR, et al. CANADIAN CLINICAL CONSENSUS GUIDELINE FOR INITIATION, MONITORING AND DISCONTINUATION OF CFTR MODULATOR THERAPIES FOR PATIENTS WITH CYSTIC FIBROSIS. Available at: https://hpr-rps.hres.ca/details.php?drugproductid54285&query5kalydeco. Accessed October 5, 2022.
- 56.McDermott JH, Mahaveer A, James RA, et al. Rapid Point-of-Care Genotyping to Avoid Aminoglycoside-Induced Ototoxicity in Neonatal Intensive Care. JAMA Pediatr 2022;176(5):486–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McDermott JH, Wolf J, Hoshitsuki K, et al. Clinical Pharmacogenetics Implementation Consortium Guideline for the Use of Aminoglycosides Based on MT-RNR1 Genotype. Clin Pharmacol Ther 2022;111(2):366–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Craven L, Alston CL, Taylor RW, et al. Recent Advances in Mitochondrial Disease. Annu Rev Genomics Hum Genet 2017;18:257–75. [DOI] [PubMed] [Google Scholar]
- 59.Moriyama B, Obeng AO, Barbarino J, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guidelines for CYP2C19 and Voriconazole Therapy. Clin Pharmacol Ther 2017;102(1):45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]


