Abstract
Objective:
Motoric cognitive risk (MCR) syndrome, a predementia syndrome characterized by slow gait and subjective cognitive concerns, is associated with multiple age-related risk factors. We hypothesized that MCR is associated with biological age acceleration. We examined the associations of biological age acceleration with MCR, and mortality risk in MCR cases.
Methods:
Biological age was determined using proteomic and epigenetic clocks in participants aged 65 years and older in the LonGenity study (N = 700, females = 57.9%) and Health and Retirement Study (HRS; N = 1,043, females = 57.1%) cohorts. Age acceleration (AgeAccel) was operationally defined as the residual from regressing predicted biological age (from both clocks separately) on chronological age. Association of AgeAccel with incident MCR in the overall sample as well as with mortality risk in MCR cases was examined using Cox models and reported as hazard ratios (HRs).
Results:
AgeAccel scores derived from a proteomic clock were associated with prevalent MCR (odds ratio adjusted for age, gender, education years, and chronic illnesses [aOR] = 1.36, 95% confidence interval [CI] = 1.09–1.71) as well as predicted incident MCR (HR = 1.19, 95% CI = 1.00–1.41) in the LonGenity cohort. In HRS, the association of AgeAccel using an epigenetic clock with prevalent MCR was confirmed (aOR = 1.47, 95% CI = 1.16–1.85). Participants with MCR and accelerated aging (positive AgeAccel score) were at the highest risk for mortality in both LonGenity (HR = 3.38, 95% CI = 2.01–5.69) and HRS (HR = 2.47, 95% CI = 1.20–5.10).
Interpretation:
Accelerated aging predicts risk for MCR, and is associated with higher mortality in MCR patients.
Introduction
Motoric cognitive risk (MCR) syndrome is a predementia syndrome characterized by the presence of slow gait and subjective cognitive complaints.1,2 A multicountry study, which included ~27,000 participants, reported the prevalence of MCR to be 9.7%. Individuals with MCR are at high risk to transition to both Alzheimer disease (AD) and vascular dementia.1,2 MCR has better predictive validity for dementia compared to its individual components of slow gait or cognitive complaints.1,2 The MCR concept is based on extensive research that demonstrates that gait is impaired early in dementias.3 Although described only a decade ago, the high clinical utility has led to MCR garnering great clinical and research interest, especially in resource poor and low middle income country settings.4 MCR epidemiology has been independently described in more than a dozen countries.5 The predictive validity of MCR for both AD and vascular dementia was demonstrated in multiple cohorts.4,5 MCR is strongly associated with established dementia risk factors such as age, low education, low physical activity, depressive symptoms, and cardiovascular disease in multiple cohorts.4,5 MCR was associated with polygenic risk scores for obesity as well as AD in the Health and Retirement Study (HRS).6 MCR has been linked to multiple dementia-related pathologies. Presence of antemortem MCR was highly correlated with postmortem Alzheimer and Lewy body pathology.7 Other studies have shown that MCR was correlated with small vessel disease on imaging studies in different populations.4 Although much has been learned about the epidemiology of MCR, the underlying biology is still being elucidated.6,8
Aging is the biggest risk factor for dementia, and is described in either chronological or biological dimensions.9 Chronological aging (number of years a person has been alive) increases at the same rate for everyone, whereas biological aging (decline in function due to physiological and disease-related changes) does not.10 But chronological age by itself fails to explain the heterogeneity in health outcomes observed in individuals. For instance, individuals within the same chronological age range can be either functionally independent or frail with multiple chronic diseases. Biological age reflects an individual’s accumulated physiological condition.10 Changes at the epigenetic (DNA methylation [DNAm])11 as well as proteomic levels (protein expression)12 have been used to quantify biological aging. Individuals who were biologically older on epigenetic and proteomic biological aging biomarkers (accelerated aging) were reported to perform worse on cognitive and physical tests.12,13 Age acceleration (AgeAccel) is operationally defined as the residual that results from regressing predicted biological age on chronological age. A positive value indicates an individual’s biological age is greater than their chronological age (accelerated aging), whereas a negative value indicates that the individual is biologically younger than their chronological age (slower aging).14
The incidence of MCR increases with chronological age.13,15 Moreover, there is a higher prevalence of age-associated diseases like diabetes, hypertension, stroke, and heart disease in MCR cases.2,6,15 But the role of biological aging in the pathogenesis of MCR is not yet established. Hence, we studied the contribution of biological aging to MCR using 2 biological aging biomarkers (a.k.a. clocks) in 2 independent cohorts. We used a proteomic clock in the community-based LonGenity study, and then replicated our findings using a second-generation epigenetic clock in the nationally representative HRS cohort.6,12,16,17 We hypothesized that age acceleration (faster aging phenotype) would be over-represented in individuals with MCR. To establish the clinical relevance of this concept, we also examined whether age acceleration would increase mortality risk in MCR.18,19 Establishing the validity of biological age acceleration in MCR patients might open up opportunities to target biological aging pathways to slow down the incidence of dementia.
Subjects and Methods
Participants
LonGenity Cohort.
The LonGenity study is a prospective study that aims to discover genotypes that confer longevity and successful aging in a relatively homogenous cohort. The study, which was established in 2007 at Albert Einstein College of Medicine, New York, recruits Ashkenazi Jewish participants, aged 65 years and older, who were either offspring of parents with exceptional longevity (OPEL; defined by having at least one parent who lived to age 95 years or older) or offspring of parents with usual survival (OPUS; defined by having neither parent survive to age 95 years). Participants were recruited using public records such as voter registration lists or through contacts at community organizations, synagogues, and advertisements in Jewish newspapers in New York City and surrounding counties. Potential participants were first contacted by telephone to assess interest and eligibility. Exclusion criteria include the following: major cognitive impairment (determined using established dementia cutoff scores of >2 on the AD8 or >8 on the Blessed Information Memory Concentration test) at the initial screening interview, having a sibling in the study, or severe visual impairment.20,21 Participants who were potentially eligible on the telephone interview were invited to our research center for further evaluation. Participants received detailed medical history evaluation, cognitive testing, and functional evaluation at baseline as well as at annual follow-up visits. All participants signed written informed consent for study assessment and genetic testing prior to enrollment. The Einstein institutional review board (IRB) approved the parent study protocol as well as this investigation.
LonGenity participants were provided telephone reminders prior to their annual follow-up visits. We also did follow-up telephone calls every 3 months. If participants did not respond for an interview, we contacted a previously identified significant other. Also, a reminder letter was sent via post. Several outreach strategies approved by the local IRB are in place to optimize retention. Each subject is assigned a primary staff contact. At each visit, study aims and the beneficial nature of participation were reviewed. If participants could not attend visits at our center, we sent a packet with survey questions to fill out to their homes. We engaged participants and families in several other ways in between visits, including quarterly newsletters, sharing articles about new study findings, and holding periodic appreciation symposiums. Study personnel were available to answer any queries or concerns from participants or their families. These activities have resulted in high retention; 86% and 93% of participants returned for follow-up visits at Waves 2 and 3, respectively.
Health and Retirement Study.
Our findings were replicated in the HRS, a nationally representative USA-based cohort study of sociodemographic, economic, and psychosocial measures in individuals aged 50 years and older since 1992. For the present analysis, participants aged 65 years and older were included. The HRS is supported by the National Institute on Aging (NIA U01AG009740) and Social Security Administration, and is conducted at the University of Michigan. In 2016, as part of the HRS venous blood study,22 DNA methylation assays were done on a nonrandom sample of 4,018 individuals. We have previously reported the epidemiology of MCR in HRS.6,18,23 Racial distribution of HRS includes 82.0% Caucasian, 12.6% African American, and 5.4% others.
Ethical approval for the HRS was obtained from the University of Michigan IRB. The Einstein IRB approved this analysis.
MCR Syndrome
MCR syndrome was diagnosed based on criteria proposed by Verghese et al1,2,15 as the presence of subjective cognitive complaints and slow gait in older individuals without dementia or mobility disability. There is no requirement to do cognitive tests to define MCR.1,2,15 MCR diagnosis has been applied to both LonGenity and HRS cohorts at each study wave in our previous studies.2,6,8,15,18
In LonGenity, subjective cognitive complaints were based on responses by participants to standardized questions about memory as a part of the Health Self-Assessment Questionnaire and from the Geriatric Depression Scale.24 Subjective cognitive complaints were classified based on a positive response by the participant to any of the following 5 questions: (1) In the past year, how often did you have trouble remembering things: frequently, sometimes, rarely, or never? “Frequently” and “sometimes” responses were coded as positive. (2) Compared with 1 year ago, do you have trouble remembering things more often, less often, or about the same?” “More often” was coded as positive. (3) Compared with 10 years ago, do you have trouble remembering things more often, less often, or about the same? “More often” was coded as positive. (4) Has your memory change caused you any serious health problems (eg, forgetting to turn off the stove, getting lost, misplacing valuables)? “Yes” was coded as positive. (5) Do you feel that you have more problems with memory than most? “Yes” was coded as positive. Gait speed at normal pace was measured using an 8.5m-long computerized walkway with embedded pressure sensors (GAITRite; CIR Systems, Franklin, NJ). The GAITRite system is widely used in clinical and research settings, and excellent reliability has been reported in our and other centers.25,26 Slow gait was defined as walking speed 1 standard deviation (SD) or more below age and sex-specific means as described in previous MCR studies in the LonGenity cohort. Age- and sex-adjusted slow gait cut scores (m/s) were as follows: men <75 years old = 1.02, men ≥75 years old = 0.85, women <75 years old = 0.97, and women ≥75 years old = 0.77. Dementia was diagnosed at consensus case conferences using established Diagnostic and Statistical Manual of Mental Disorders, 4th edition criteria after reviewing all available clinical, neuropsychological, and medical information by study clinicians and a licensed neuropsychologist.27 Only 728 of 1,098 participants with baseline gait and subjective cognitive data were included in this study.
In HRS, subjective cognitive complaints were classified based on a positive response by the participant to either of the following 2 questions on the core HRS interview: (1) “How would you rate your memory at the present time? Would you say it is excellent, very good, good, fair, or poor?” “Fair” and “poor” responses were coded as positive. (2) “Compared with the previous interview, would you say your memory is better now, about the same, or worse now than it was then?” A response of “worse” was coded as positive.6 Participants aged 65 years and older were timed while walking at normal pace over a 98.5-inch (8 feet) course with start and stop points marked on the floor. The mean of 2 trials was calculated. Walking times were converted to gait speed (m/s). Slow gait was defined as gait speed 1 SD or more below age- and sex-specific means.28 Age- and sex-adjusted slow gait cut scores (m/s) were as follows: men <75 years old, 0.61; men ≥ 75 years old, 0.48; women <75 years old, 0.54; and women ≥ 75 years old, 0.42. In HRS, self-reported or proxy-reported physician diagnosis of AD or dementia was used.6 Only 1,074 of 4,018 participants with baseline gait and subjective cognitive data were included in this study.
Biological Clocks
Chronological age was calculated as the difference between date of birth and assessment date, up to 3 decimal places. We used a proteomic clock in LonGenity and DNAm GrimAge clock in HRS to quantify biological aging. Proteomic clocks are based on the expression level of a selected subset of proteins, facilitated by the advent of highly multiplexed platforms like Somascan assay that can measure thousands of proteins simultaneously.12,17 Epigenetic clocks, on other hand, are more established, and have successfully captured chronological age and age-associated phenotypes.11,29-32 AgeAccel is defined as the residual resulting from regressing predicted age (proteomic or DNAm GrimAge) on chronological age. A positive AgeAccel value indicates faster aging, and a negative value suggests slower aging.
Proteomic Prediction of Chronological Age in LonGenity.
LonGenity used the 5 k SomaScan Assay V4, which includes 5,209 SOMAmer reagents targeting human proteins. We have reported proteomic profiles of aging and frailty using the same data in LonGenity.16,17 SomaScan data standardization done by SomaLogic has been previously described.33 After implementing quality checks, 960 sequences that failed checks were removed. After exclusion of non-human proteins, deprecated markers, and noncleavable, non-biotin, as well as spuriomers, 4,265 SOMAmer reagents were available for proteomic analysis in 1025 participants (age = 65–95 years).17 Relative fluorescence unit values observed after data normalization procedure for each SOMAmer reagent were natural log transformed.
A proteomic chronological age predictor was constructed with elastic net regression using the glmnet R package.34 We selected 325 participants in the training set. The participants in the training set were individuals who were not part of MCR characterization in the cohort. Using the cv-glmnet function, the optimal lambda value to minimize cross-validation prediction error rate was selected based on 10-fold cross-validation using the training set. The program set an alpha value of 0.5 for elastic net regression. The age was predicted in the validation set consisting of 700 participants.
Epigenetic Prediction of Chronological Age in HRS.
DNAm GrimAge is a second-generation epigenetic clock developed by regressing time to all-cause mortality using DNAm surrogated for 7 biomarkers, methylation surrogate for pack years of smoking, gender, and age at the time of blood draw.29 The linear combination of the covariate values was linearly transformed into an age estimate (DNAm GrimAge).29 GrimAge has been shown to outperform other epigenetic clocks for predicting many health and cognitive outcomes as well as mortality.29,35
Epigenetic clocks were defined in 4,018 participants in the 2016 HRS venous blood substudy.31 GrimAge was developed based on the 7 DNAm surrogates of plasma proteins (adrenomedullin levels, beta-2 micro-globulin, cystatin C, growth differentiation factor 15, leptin, plasminogen activation inhibitor 1, tissue inhibitor metalloproteinase 1) and smoking pack-years in a 2-stage procedure.29 DNAm GrimAge is based on methylation level at 1,030 CpG sites. Further details on epigenetic clock construction in HRS can be accessed from the HRS website (https://hrsdata.isr.umich.edu/data-products/epigenetic-clocks).
Frequent interactions with participants and family made it possible to track life events well in the LonGenity cohort. In LonGenity, death of participants was ascertained from designated contacts, and supplemented by obituary and National Death Index (NDI) searches by our staff in participants who were lost to follow-up.36 In HRS, death date was obtained from the decedent’s spouse, family member, or friend during an exit interview. Reported death dates were then confirmed through the Social Security Death Index and Insight databases, and linked to the NDI. The HRS Tracker File (trk2018v2a) was used to obtain mortality data for this analysis.
Covariates
Age, sex, and education (years) data were based on self-report on standardized questionnaires administered by research assistants in both cohorts. Presence or absence of self-report of physician diagnoses of depression, diabetes, heart failure, hypertension, myocardial infarction, strokes, Parkinson disease (PD), chronic obstructive lung disease, and arthritis were summed to calculate a global health score (GHS; range = 0–9) as previously described.24,37 In HRS, PD diagnosis was not present, and the GHS range was 0 to 8.
Statistical Analysis
Proteomic Clock.
In LonGenity, we created a proteomic clock using elastic net regression. We first did a sensitivity analysis to check the association of proteomic age and AgeAccel measures with incident mortality in the overall sample in the validation set (regardless of MCR status). Cox proportional hazard models were used to compute hazard ratios (HRs) with 95% confidence intervals (CIs) to predict incident all-cause mortality based on proteomic age (predicted) and AgeAccel. All models were adjusted for age, gender, cohort status (OPUS or OPEL), education (years), and GHS. Time scale was follow-up time in years from baseline to date of death or final contact, whichever came first. Proportional hazard assumptions of all models were tested graphically and analytically and were adequately met.
Association Analysis with MCR.
Logistic regression analysis was used to test the association between the AgeAccel measure and prevalent MCR in LonGenity. Cox proportional hazard models were used to compute HRs with 95% CIs to predict incident MCR based on AgeAccel. Models were adjusted for age, gender, and education (years) in the LonGenity cohort. Models were further adjusted for GHS. Eleven participants developed dementia during follow-up, 4 of whom had MCR at baseline or at a study wave prior to incident dementia diagnosis. Participants who met criteria for incident dementia (n = 7) without an intermediary MCR stage were not counted as incident MCR. To graphically depict results using Kaplan–Meier survival analysis, we characterized individuals with positive AgeAccel values as fast agers and those with negative AgeAccel values as slow agers.
In HRS, we checked the association of the epigenetic age acceleration measure only with prevalent MCR, as there were no subsequent waves with gait data (to define incident MCR). All analysis were adjusted for age, gender, education years, and HRS sampling weights.31
Age Acceleration and Mortality in MCR.
In both cohorts, participants were divided into 4 groups based on MCR and AgeAccel status: (1) slow ager/no MCR, (2) slow ager/MCR, (3) fast ager/no MCR, and (4) fast ager/MCR. Cox proportional hazard models were used to compute HRs with 95% CIs to predict death based on the AgeAccel/MCR status at baseline in LonGenity and 2016 (baseline year of epigenetic/MCR assessment) in HRS. In HRS, median follow-up was 2 years, as we used death dates based on 2018 core data.
All statistical analyses were conducted using SPSS software (version 24; IBM, Armonk, NY) and R.
Sensitivity Analysis
We assessed the potential effect of unmeasured confounding by E-value methodology.38 E-Value summarizes the minimum strength of association that the unmeasured confounder would need to have with both the exposure and outcome to negate the observational result. We calculated E-value for estimates of MCR as well as mortality as outcome, and the corresponding lower limit of the 95% CI was also reported. The calculation of E-value was implemented using the E-value calculator.39
To address the association between frailty and biological aging, we repeated the MCR incidence analysis in the LonGenity cohort excluding individuals who met physical frailty40 criteria at baseline.
Results
Study Population
LonGenity Cohort.
Of the 728 participants (of a total sample of 1,098) without dementia at baseline who completed gait assessments and subjective cognitive complaint questionnaires, 700 participants had proteomic data available. There were 84 prevalent cases of MCR (12%), 135 cases of incident MCR, and 481 participants who remained free from MCR over follow-up (median = 4.03 years). In the overall sample, mean age was 74.88 years (SD = 6.39), 57.90% of participants were female, and mean years of education was 17.66 (SD = 2.82). Demographic and clinical characteristics by MCR status are summarized in Table 1. Among the chronic illnesses examined, hypertension, diabetes, and stroke were more prevalent in participants who had MCR than those who remained MCR free (see Table 1). There were no significant differences in age, sex, slow gait, and MCR prevalence between the 28 cases without proteomic data and the 700 with proteomic data.
TABLE 1.
Demographic Characteristics of LonGenity Cohort at Baseline by MCR Status
| Characteristic | No MCR, n = 481a | Prevalent MCR, n = 84 | Incident MCR, n = 135 |
|---|---|---|---|
| Age, mean yr ± SD | 74.7 ± 6.4 | 75.7 ± 6.9 | 75.3 ± 6.0 |
| Women, n (%) | 286 (59.5) | 46 (54.8) | 73 (54.1) |
| Education, mean yr ± SD | 17.68 ± 2.73 | 17.50 ± 3.26 | 17.69 ± 2.83 |
| Frailty index, mean ± SD | 0.14 ± 0.07 | 0.22 ± 0.09 | 0.17 ± 0.07 |
| Medical illnesses | |||
| Global health score, mean ± SDb | 1.08 ± 0.97 | 1.77 ± 1.18 | 1.45 ± 1.07 |
| Hypertension, % | 37.7 | 62 | 58.1 |
| Myocardial infarction, % | 4.4 | 3.6 | 9.7 |
| Congestive heart failure, % | 0.4 | 1.2 | 1.5 |
| Stroke, % | 1.7 | 6 | 5.2 |
| Cancer, % | 34.9 | 31 | 33.3 |
| Diabetes, % | 6 | 15.5 | 11.1 |
| Chronic lung disease, % | 2.5 | 3.6 | 2.2 |
| Arthritis, % | 38.6 | 56.6 | 40.6 |
| Parkinson disease, % | 1.3 | 2.4 | 0 |
| Depression, % | 18.5 | 32.1 | 24.1 |
| Cognitive tests, mean ± SD | |||
| FCSRT free recall | 33.70 ± 5.34 | 34.00 ± 4.86 | 33.76 ± 4.91 |
| DSST | 61.38 ± 14.34 | 58.29 ± 12.57 | 60.48 ± 13.73 |
No MCR: participants who were free from MCR at baseline as well as throughout the study follow-up.
Presence or absence of physician-diagnosed depression, diabetes, heart failure, hypertension, myocardial infarction, strokes, Parkinson disease, ohronic obstructive lung disease, and arthritis was used to calculate a global health score (range = 0–9) as previously described.37
Abbreviation: DSST = Digit Symbol Substitution Test; FCSRT = Free and Cued Selective Reminding Test; MCR = motoric cognitive risk synrome; SD = standard deviation.
Proteomic Clock in LonGenity Cohort
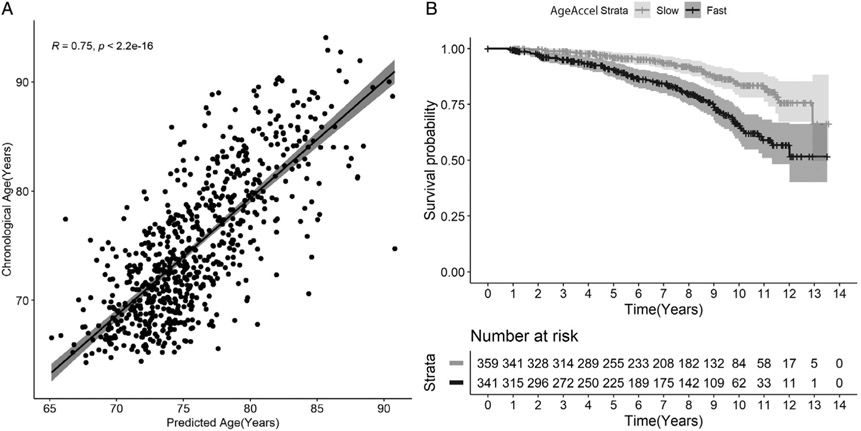
Elastic net regression selected 139 from 4,265 proteins to model the proteomic clock (Table S1). The correlation between proteomic age predicted by our model and chronological age was r = 0.75 (p < 2.2E−16; Fig 1A). We checked power of the clock by predicting mortality using the AgeAccel measure in the overall sample. The predicted proteomic age was associated with mortality even after adjustment for chronological age, gender, cohort, education years, and GHS (HR = 1.16, 95% CI = 1.10–1.24, p = 6.72E−07). Higher AgeAccel measure was also associated with incident mortality (HR = 1.56, 95% CI = 1.31–1.87, p = 6.72E−07). The E-value for this analysis was 2.49 for the point estimate and 1.95 for the lower 95% CIs. The observed HR of 1.56 may be explained by an unmeasured confounder that was associated with both the AgeAccel measure and MCR by a HR of 2.49 each, above and beyond the measured confounders, but a weaker confounding effect could not do so.
FIGURE 1: :
(A) Correlation of chronological age and predicted age using proteomic data. Age prediction was carried out using an elastic net regression method in 700 participants in the validation set. Correlation of predicted age using proteomic markers and chronological age was ~0.75. (B) Kaplan–Meier survival curves for time to death for participants who are slow (grey) and fast (black) agers based on age acceleration (AgeAccel).
Figure 1B shows that the 341 fast agers with positive AgeAccel measures at baseline had significantly reduced survival compared to the 359 slow agers with negative AgeAccel measure at baseline (log-rank test chi-squared = 21.927, p < 0.0001). There were 76 deaths in the 341 fast ager group (22.3%), whereas only 38 deaths were reported in the 359 slow agers (10.6%). Positive AgeAccel scores predicted mortality even after adjusting for age, gender, education status, cohort (OPEL or OPUS), and GHS (HR = 2.51, 95% CI = 1.68–3.76, p = 8.00E−06, E-value [CI] = 4.46 [2.75]).
Prevalent and Incident MCR: LonCenity
The AgeAccel measure was associated with prevalent MCR, with higher rate of age acceleration in MCR compared to non-MCR LonCenity participants at baseline (odds ratio [OR] = 1.36, 95% CI = 1.09–1.71, E-value [CI] = 2.06 [1.40]).
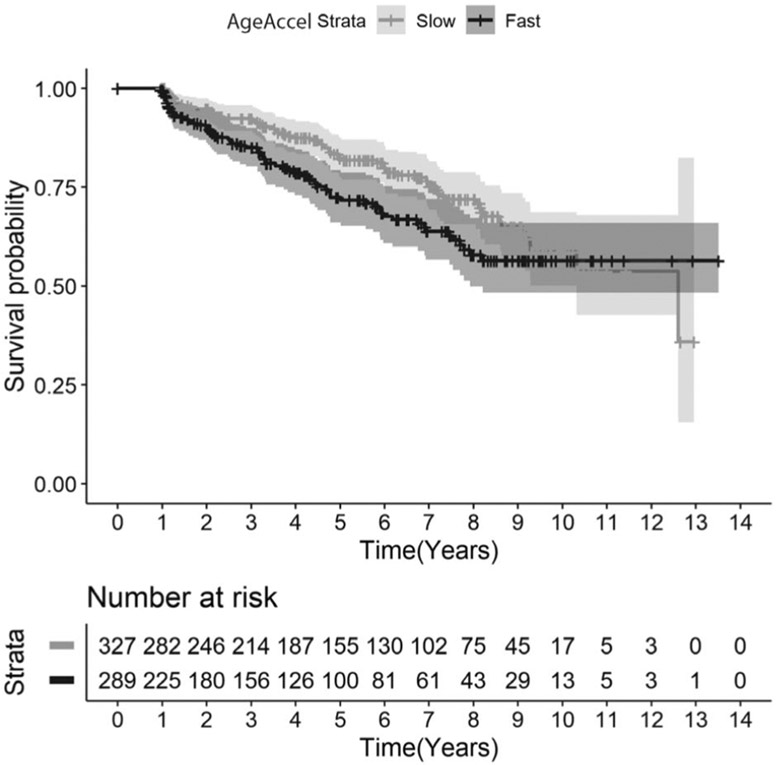
Of the 616 MCR-free LonGenity participants at baseline, 135 (21.9%) developed incident MCR. The median follow-up time was similar in non-MCR (4.10 years, SD = 3.56) and MCR cases (3.73 years, SD = 2.58). The baseline AgeAccel was 0.027 (SD = 0.988) in incident MCR cases and −0.07 (SD = 0.949) in non-MCR cases. Baseline AgeAccel was associated with increased risk of developing MCR (HR = 1.22, 95% CI = 1.03–1.46, p = 0.024, E-value [CI] = 1.74 [1.21]) adjusted for chronological age, gender, and education years. Further adjustment for GHS attenuated the association (HR = 1.19, 95% CI = 1.00–1.41, p = 0.049, E-value [CI] = 1.67 [1.00]). Participants with positive AgeAccel (fast agers) at baseline had higher risk of incident MCR (HR = 1.55, 95% CI = 1.10–2.19, p = 0.012, E-value [CI] = 2.47 [1.43]) compared to participants with negative AgeAccel measure (slow agers). Even after further adjustment for GHS, the association of fast agers with incident MCR remained (HR = 1.51, 95% CI = 1.07–2.13, p = 0.019, E-value [CI] = 2.39 [1.34]).
Figure 2 shows that the 289 fast agers with positive AgeAccel at baseline had higher risk of developing incident MCR compared to the 327 slow agers with negative AgeAccel measure at baseline (log-rank test chi-squared = 4.3944, p = 0.036). Of the 289 fast agers, 67 developed incident MCR (23.2%). Of the 327 slow agers (20.7%), 68 developed incident MCR.
FIGURE 2: :
Kaplan–Meier survival curves for time to motoric cognitive risk syndrome for participants who are slow (grey) and fast (black) agers based on age acceleration (AgeAccel).
Of the 341 fast agers, 78 (22.9%) met criteria for physical frailty40 as well as 50 (13.9%) of 359 slow agers (p = 0.002). After excluding 128 participants with physical frailty at baseline, fast ager (AgeAccel+) was still associated with incident MCR (HR adjusted for age, gender, education years, and CHS = 1.57, 95% CI = 1.07–2.29, p = 0.019, E-value [CI] = 2.52 [1.34]).
MCR: Health and Retirement Study
Demographic and clinical characteristics by MCR status in the HRS sample are summarized in Table 2. There were 83 prevalent MCR cases (7.73%) in the 1,074 HRS participants. Thirty-one participants had dementia at baseline and were excluded from the analysis. Hypertension, diabetes, and stroke were more common in MCR participants compared to participants without MCR (see Table 2).
TABLE 2.
Demographic Characteristics of Health and Retirement Study at Baseline by MCR Status
| Baseline Characteristic | No MCR, n = 960a |
Prevalent MCR, n = 83 |
|---|---|---|
| Age, mean yr ± SD | 75.5 ± 7.0 | 76.4 ± 8.3 |
| Women, n (%) | 564 (57.5) | 50 (53.8) |
| Education, mean yr ± SD | 12.99 ± 2.86 | 11.3 ± 4.19 |
| Medical illnesses, % | ||
| Hypertension | 67.9 | 78.3 |
| Heart conditionb | 31.9 | 41.5 |
| Myocardial infarction | 2.4 | 3.8 |
| Congestive heart failure | 5.0 | 12.7 |
| Stroke | 7.7 | 19.3 |
| Cancer | 19.5 | 24.1 |
| Diabetes | 28 | 47.6 |
| Lung disease | 10.7 | 13.3 |
| Arthritis | 69.3 | 81.9 |
| Depression | 19.4 | 39.8 |
| Angina | 18.9 | 41.9 |
No MCR: participants who were free from MCR at baseline as well as throughout the study follow-up.
Any one of myocardial infarction, coronary heart disease, angina, congestive heart failure, or other heart problems.
Abbreviation: MCR = motoric cognitive risk syndrome; SD = standard deviation.
The epigenetic biological aging biomarker (DNAm GrimAge AgeAccel) was associated with prevalent MCR (OR = 1.47, 95% CI = 1.16–1.85, p = 0.001, E-value [CI] = 2.30 [1.59]) when adjusted for chronological age, gender, and education years. The association remained after further adjustment for GHS (OR = 1.30, 95% CI = 1.02–1.66, p = 0.031, E-value [CI] = 1.92 [1.16]). There were 460 participants who were fast agers and 583 slow agers. Of the 83 MCR cases, 49 were fast agers (OR = 1.76, 95% CI = 1.08–2.87, p = 0.023, E-value [CI] = 1.98 [1.24]).
Mortality: LonGenity
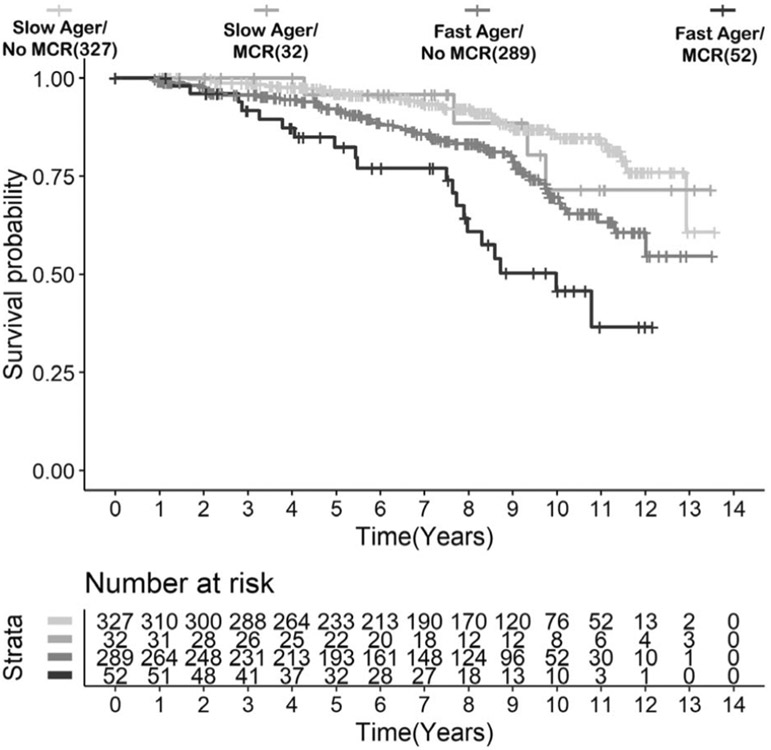
Prevalent MCR predicted mortality (HR = 2.00, 95% CI = 1.26–3.18, p = 3.47E−03) in LonGenity. Of the 84 LonGenity participants with prevalent MCR, 52 (61.9%) were fast agers (AgeAccel+/MCR+) and 20 (38.4%) in this subgroup died during follow-up. Mortality rates in the other groups were as follows: 56 deaths in 289 (19.4%) participants who were fast agers without MCR (AgeAccel+/MCR−), 4 deaths in 32 participants (12.5%) in the MCR slow ager subgroup (AgeAccel−/MCR+), and 34 deaths in 327(10.4%) non-MCR slow ager subgroup (AgeAccel−/MCR−). Figure 3 shows that the 52 participants who had MCR and were fast agers at baseline had shorter survival compared to the other groups (HR = 3.38, 95% CI = 2.01–5.69, p = 4.00E−06, E-value [CI] = 6.22 [3.43]).
FIGURE 3: :
Kaplan–Meier survival curves for time to death for LonGenity participants based on motoric cognitive risk syndrome (MCR) and age acceleration status.
Mortality: Health and Retirement Study
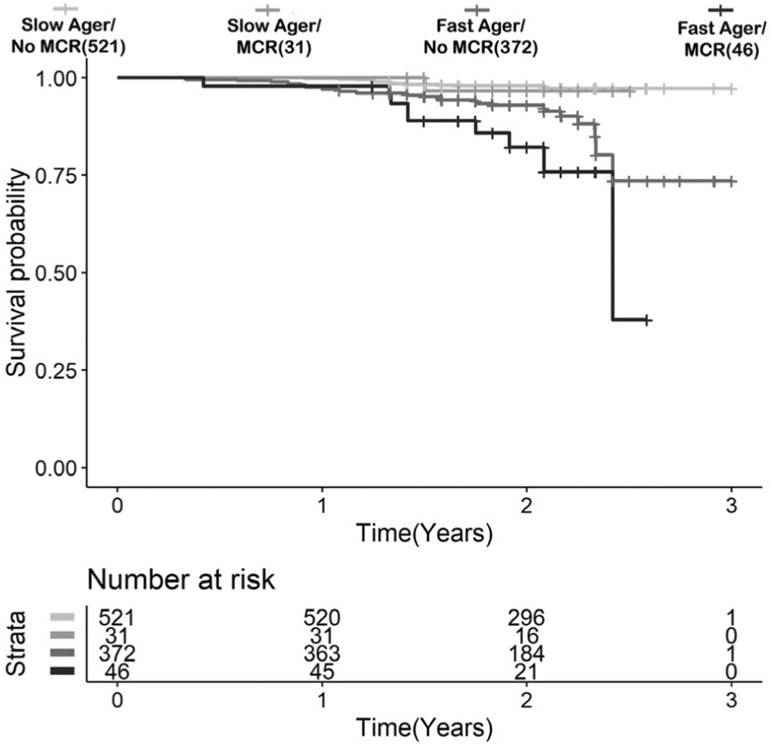
A subset of 970 participants who participated in the 2018 interview of HRS had mortality data: 418 fast agers and 552 slow agers. There were 54 deaths reported over a median follow-up of 2.00 years: 41 in fast agers (GrimAge AgeAccel > 0; 76%) and 13 (24%) in slow agers. Both DNAm GrimAge AgeAccel (HR = 2.08, 95% CI = 1.56–2.78, p = 5.53E−07, E-value [CI] = 3.58 [2.49]) and MCR (HR = 2.07, 95% CI = 1.01–4.26, p = 0.047, E-value [CI] = 3.56 [1.11]) were associated with mortality adjusted for age, gender, education years, and HRS sampling weight. In the 46 MCR participants who were fast agers (MCR+/AgeAccel+), 9 deaths were reported (19.6%). Only 1 death occurred among the 31 participants (3.2%) in the MCR and slow ager subgroup (MCR+/AgeAccel−). Thirty-two deaths were reported in the 372 participants (8.6%) who were faster agers with no MCR (MCR−/AgeAccel+), whereas only 12 deaths were reported in 521 participants who were MCR−/AgeAccel− (2.3%). Figure 4 shows that the 55 MCR and fast agers at baseline had increased risk of death compared to the other groups (HR = 2.47, 95% CI = 1.20–5.10, p = 0.014, E-value [CI] = 4.38 [1.69]).
FIGURE 4: :
Kaplan–Meier survival curves for time to death for Health and Retirement Study participants based on motoric cognitive risk syndrome (MCR) and age acceleration status.
Discussion
This study showed that accelerated biological aging, characterized using both proteomic and epigenetic clocks, was associated with prevalence and incidence of MCR in community-residing older adults from 2 independent cohorts. We identified a subgroup of MCR participants with faster aging phenotype who had a >2-fold increased risk for mortality in both cohorts compared to older adults without this phenotype. This study, to our knowledge, is the first to address the relationship of biological age acceleration to MCR. Our observations point toward an effect of age acceleration on influencing the initiation as well as progression of dementia in individuals.
The relationship between biological aging and MCR may result from shared biological mechanisms.41 Aging and dementia phenotypes, for example, have common biological derangements such as abnormal neuronal network activity, inflammation, oxidative damage, decreased proteasome activity, cellular senescence, reduced DNA repair, impaired molecular waste disposal, stem cell exhaustion, neuronal calcium dysregulation, mitochondrial dysfunction, gut microbiota alterations, and impaired adaptive stress response.42 Aging and MCR phenotypes also overlap with clinical disorders such as frailty, which in turn is linked to risk of dementia.24 In the LonGenity cohort, frailty status at baseline predicted incident MCR.24 Individuals without frailty or with low frailty were reported to better tolerate Alzheimer pathology compared to those with higher frailty.43 Although there was minor clinical overlap between frailty and faster biological aging phenotypes in the LonGenity cohort, our findings suggest that each condition makes independent contributions to risk of MCR. Furthermore, the majority of individuals with the fast aging phenotype did not meet frailty criteria. The biological derangements mapping to faster aging may, hence, precede the clinical onset of physical frailty, and in turn, MCR. In both cohorts, some chronic illnesses were over-represented in individuals with prevalent MCR as well as at baseline in those who went on to develop MCR. The pathways leading to MCR may be differentially influenced by chronic illness profiles in high-risk individuals. Epidemiological studies have shown MCR to be associated with higher mortality.18,19 Many of the abnormal biological and clinical pathways cited above have also been implicated in mortality, and may explain the lower survival in individuals who met criteria for MCR as well as accelerated aging in both our cohorts.44
Biological clocks are associated with complex traits such as angina, diabetes, frailty, and depression that were also associated with MCR in our epidemiological studies.15,45,46 DNAm age acceleration has been linked to mortality,14 frailty,46 physical limitations,30 self-rated health,30 and complex diseases in older adults.13 In 5,100 participants in the Generation Scotland study, an association was reported between an epigenetic age acceleration measure and body mass index, total cholesterol/high-density lipoprotein ratio, socioeconomic status, high blood pressure, and smoking behavior.47 Age acceleration was not associated with genetic risk (AD polygenic score and apolipoprotein E status) and AD family history in the same study.47 There is a paucity of studies that have directly examined the association of DNAm clocks with dementia.48,49 One study reported an association between methylation age and dementia risk in 52 participants.50 In a prospective study with up to 15 years of follow-up, accelerated GrimAge at baseline was associated with greater declines on multiple cognitive domains.51 But other investigators have found no association of DNAm age acceleration with either incident dementia48,49 or mild cognitive impairment.52 These mixed results may be due to smaller sample sizes in prior studies as well as the variation in age range and gender representation across cohorts. Another factor influencing results might be the varied and evolving forms of methylation clocks. Most studies to date have focussed on epigenetic clocks, but the present study is the first one to explore association of a proteomic age acceleration measure with a dementia-associated trait. Our findings point toward a role for age acceleration as a catalyst in dementia pathogenesis via the MCR pathway.
The current study has many strengths, including using established operational definitions of MCR in both LonGenity and HRS cohorts.6,18,24 Despite slight differences in the individual operational criterion for MCR, the converging findings regarding the role of accelerated aging with MCR in 2 independent cohorts using both proteomic and epigenetic clocks is another strength. Our proteomic clock derived from >4,000 proteins was developed in the well-characterized racially homogenous LonGenity cohort. There is considerable variability in how cognitive and motor function decline with aging, with individuals appearing younger or older than their chronological age. Biological clocks offer a more objective method of molecularly quantifying how well an individual is aging beyond their chronological age. DNAm GrimAge is one the most successful biological clocks. It outperforms chronological age as well as other epigenetic clocks in the prediction of age-related clinical phenotypes such as coronary heart disease, cancer, diabetes, comorbidity count, and physical functioning as well as all-cause mortality.29 Higher DNAm GrimAge was also associated with brain vascular lesions and lower cognitive ability in older age, independent of early life cognitive ability.53 LonGenity was among the first cohorts to establish a proteomic aging clock.12,17 These studies using the proteomic clock to define biological aging showed that individuals who were biologically younger than their chronological age performed better on cognitive and physical tests, whereas age acceleration predicted mortality.12,17 In the present study, we used the same biological age prediction strategy to understand the relationship of a proteomic clock with MCR.
Limitations
Limitations include the absence of longitudinal analysis of MCR in HRS. Expanding the number of proteins in proteomic assays might further improve the prediction model for biological age. The limited follow-up period in HRS may have underestimated mortality effects. The lower bound of the chronological age limit in both cohorts was set by the parent studies (no upper bound in both cohorts). The effect of biological age acceleration on MCR needs to be studied in younger chronological ages to better understand the early pathological stages of MCR. Discrepancies in self-report of age, especially reporting younger age than actual, may lead to underestimating the association of biological age with MCR. Loss of participants due to death is inevitable. The relatively high retention in the LonGenity cohort minimizes but does not exclude the effect of attrition on the observed findings. Frequent interactions with participants and family made it possible to track life events well in this cohort. Residual or unmeasured confounding is a possibility with any observational study. E-values are a relatively new statistical method to address the issue of unmeasured confounding.38,39 The high E-values for the different analyses indicates that our observations are robust and less likely to be effected by unmeasured confounding. The association of biological aging with incident MCR needs to be replicated in other independent samples using the same as well as new biological clocks. In the meantime, replicating the association of biological aging with prevalent MCR in an independent cohort after accounting for major dementia risk factors and with a completely different biological aging clock is a reassuring first step.
In conclusion, accelerated aging is a hallmark of MCR. Older individuals with MCR and accelerated aging are at highest risk of mortality. Biological age acceleration may be an important modifiable dementia risk factor. Our analyses show that accelerated aging has incremental value over several established dementia risk factors such as chronological age, gender, education, and chronic medical illnesses as well as frailty in predicting MCR. This novel finding opens the possibility of discovering specific accelerated biological aging pathways (independent of established dementia risk factors) that may lead to dementia via MCR. Accelerated biological aging might also be investigated as a target of intervention, although more research is needed in this area. Lifestyle changes such as physical exercise and healthy dietary intervention including caloric restriction show promise in preliminary studies in staving off the effects of accelerated aging on health outcomes.
Supplementary Material
Acknowledgments
This work was supported by grants from the NIH, National Institute of Aging (R01AG057548, J.V.; R56AG044829, S.M. and J.V.; P01AG021654, N.B.; R01AG046949, N.B.; R01AG057909, N.B.; R01AG061155, S.M.), Nathan Shock Center of Excellence for the Biology of Aging (P30AG038072, N.B.), American Federation for Aging Research (S.M.), and Glenn Center for the Biology of Human Aging Paul Glenn Foundation Grant (N.B.). The HRS was supported by an NIH National Institute on Aging grant (1R56AG057548-01). The sponsors had no role in the design or conduct of the study; collection, management, analysis, or interpretation of the data; or preparation, review, or approval of the manuscript.
Footnotes
Potential Conflicts of Interest
Nothing to report.
Data Availability Statement
HRS data used in the current study were obtained from the HRS database (https://hrs.isr.umich.edu/). Derived data (MCR definition) are available from the corresponding author on request by any qualified investigator subject to a data use agreement. LonGenity data used in this study are available upon request. Please contact the corresponding author for further information.
References
- 1.Verghese J, Wang C, Lipton RB, Holtzer R. Motoric cognitive risk syndrome and the risk of dementia. J Gerontol, Ser A 2013;68:412–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Verghese J, Annweiler C, Ayers E, et al. Motoric cognitive risk syndrome: multicountry prevalence and dementia risk. Neurology 2014;83:718–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Montero-Odasso M, Verghese J, Beauchet O, Hausdorff JM. Gait and cognition: a complementary approach to understanding brain function and the risk of falling. J Am Geriatr Soc 2012;60:2127–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mullin DS, Cockburn A, Welstead M, et al. Mechanisms of motoric cognitive risk—hypotheses based on a systematic review and meta-analysis of longitudinal cohort studies of older adults. Alzheimers Dement 2022;18:2413–2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meiner Z, Ayers E, Verghese J. Motoric cognitive risk syndrome: a risk factor for cognitive impairment and dementia in different populations. Ann Geriatr Med Res 2020;24:3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sathyan S, Wang T, Ayers E, Verghese J. Genetic basis of motoric cognitive risk syndrome in the health and retirement study. Neurology 2019;92:e1427–e1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kravatz NL, Ayers E, Bennett DA, Verghese J. Olfactory dysfunction and incidence of motoric cognitive risk syndrome: a prospective clinical-pathologic study. Neurology 2022;99:e1886–e1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sathyan S, Barzilai N, Atzmon G, et al. Association of anti-inflammatory cytokine IL10 polymorphisms with motoric cognitive risk syndrome in an Ashkenazi Jewish population. Neurobiol Aging 2017;58:e231–e238. e238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.López-Otín C, Blasco MA, Partridge L, et al. The hallmarks of aging. Cell 2013;153:1194–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamczyk MR, Nevado RM, Barettino A, et al. Biological versus chronological aging: JACC focus seminar. J Am Coll Cardiol 2020;75:919–930. [DOI] [PubMed] [Google Scholar]
- 11.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol 2013;14:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lehallier B, Gate D, Schaum N, et al. Undulating changes in human plasma proteome profiles across the lifespan. Nat Med 2019;25:1843–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jylhävä J, Pedersen NL, Hägg S. Biological age predictors. EBioMedicine 2017;21:29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen BH, Marioni RE, Colicino E, et al. DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging 2016;8:1844–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verghese J, Ayers E, Barzilai N, et al. Motoric cognitive risk syndrome: multicenter incidence study. Neurology 2014;83:2278–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sathyan S, Ayers E, Gao T, et al. Plasma proteomic profile of frailty. Aging Cell 2020;19:e13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sathyan S, Ayers E, Gao T, et al. Plasma proteomic profile of age, health span, and all-cause mortality in older adults. Aging Cell 2020;19:e13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ayers E, Verghese J. Motoric cognitive risk syndrome and risk of mortality in older adults. Alzheimers Dement 2016;12:556–564. [DOI] [PubMed] [Google Scholar]
- 19.Beauchet O, Sekhon H, Launay C, et al. Motoric cognitive risk syndrome and mortality: results from the EPIDOS cohort. Eur J Neurol 2019;26:794–e756. [DOI] [PubMed] [Google Scholar]
- 20.Galvin J, Roe C, Powlishta K, et al. The AD8: a brief informant interview to detect dementia. Neurology 2005;65:559–564. [DOI] [PubMed] [Google Scholar]
- 21.Blessed G, Tomlinson BE, Roth M. The association between quantitative measures of dementia and of senile change in the cerebral grey matter of elderly subjects. Br J Psychiatry 1968;114:797–811. [DOI] [PubMed] [Google Scholar]
- 22.Crimmins E, Faul J, Thyagarajan B, Weir D. Venous blood collection and assay protocol in the 2016 Health and Retirement Study. Ann Arbor, MI: Survey Research Center, Institute for Social Research, University of Michigan, 2017. [Google Scholar]
- 23.Callisaya ML, Ayers E, Barzilai N, et al. Motoric cognitive risk syndrome and falls risk: a multi-center study. J Alzheimer’s Dis 2016;53:1043–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sathyan S, Ayers E, Gao T, et al. Frailty and risk of incident motoric cognitive risk syndrome. J Alzheimer’s Dis 2019;71:S85–S93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holtzer R, Wang C, Verghese J. The relationship between attention and gait in aging: facts and fallacies. Motor Control 2012;16:64–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brach JS, Perera S, Studenski S, Newman AB. The reliability and validity of measures of gait variability in community-dwelling older adults. Arch Phys Med Rehabil 2008;89:2293–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lipton RB, Katz MJ, Kuslansky G, et al. Screening for dementia by telephone using the memory impairment screen. J Am Geriatr Soc 2003;51:1382–1390. [DOI] [PubMed] [Google Scholar]
- 28.Verghese J, Wang C, Allali G, et al. Modifiable risk factors for new-onset slow gait in older adults. J Am Med Dir Assoc 2016;17:421–425. [DOI] [PubMed] [Google Scholar]
- 29.Lu AT, Quach A, Wilson JG, et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging 2019;11:303–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Belsky DW, Moffitt TE, Cohen AA, et al. Eleven telomere, epigenetic clock, and biomarker-composite quantifications of biological aging: do they measure the same thing? Am J Epidemiol 2018;187:1220–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crimmins EM, Thyagarajan B, Levine ME, et al. Associations of age, sex, race/ethnicity, and education with 13 epigenetic clocks in a nationally representative US sample: the health and retirement study. J Gerontol, Ser A 2021;76:1117–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gale CR, Marioni RE, Harris SE, et al. DNA methylation and the epigenetic clock in relation to physical frailty in older people: the Lothian birth cohort 1936. Clin Epigenetics 2018;10:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Candia J, Cheung F, Kotliarov Y, et al. Assessment of variability in the SOMAscan assay. Sci Rep 2017;7:14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Friedman J, Hastie T, Tibshirani R. glmnet: Lasso and elastic-net regularized generalized linear models. R package version 2009;1. [Google Scholar]
- 35.McCrory C, Fiorito G, Hernandez B, et al. GrimAge outperforms other epigenetic clocks in the prediction of age-related clinical phenotypes and all-cause mortality. J Gerontol, Ser A 2021;76:741–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Milman S, Atzmon G, Huffman DM, et al. Low insulin-like growth factor-1 level predicts survival in humans with exceptional longevity. Aging Cell 2014;13:769–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Verghese J, Wang C, Lipton RB, et al. Quantitative gait dysfunction and risk of cognitive decline and dementia. J Neurol Neurosurg Psychiatry 2007;78:929–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.VanderWeele TJ, Ding P. Sensitivity analysis in observational research: introducing the E-value. Ann Intern Med 2017;167:268–274. [DOI] [PubMed] [Google Scholar]
- 39.Mathur MB, Ding P, Riddell CA, VanderWeele TJ. Website and R package for computing E-values. Epidemiology 2018;29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fried LP, Tangen CM, Walston J, et al. Frailty in older adults: evidence for a phenotype. J Gerontol, Ser A 2001;56:M146–M157. [DOI] [PubMed] [Google Scholar]
- 41.Franceschi C, Garagnani P, Morsiani C, et al. The continuum of aging and age-related diseases: common mechanisms but different rates. Front Med 2018;5:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mattson MP, Arumugam TV. Hallmarks of brain aging: adaptive and pathological modification by metabolic states. Cell Metab 2018;27:1176–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wallace LM, Theou O, Godin J, et al. Investigation of frailty as a moderator of the relationship between neuropathology and dementia in Alzheimer’s disease: a cross-sectional analysis of data from the rush memory and aging project. Lancet Neurol 2019;18:177–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wahl D, Solon-Biet SM, Cogger VC, et al. Aging, lifestyle and dementia. Neurobiol Dis 2019;130:104481. [DOI] [PubMed] [Google Scholar]
- 45.Hillary RF, Stevenson AJ, McCartney DL, et al. Epigenetic measures of ageing predict the prevalence and incidence of leading causes of death and disease burden. Clin Epigenetics 2020;12:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Breitling LP, Saum K-U, Perna L, et al. Frailty is associated with the epigenetic clock but not with telomere length in a German cohort. Clin Epigenetics 2016;8:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCartney DL, Stevenson AJ, Walker RM, et al. Investigating the relationship between DNA methylation age acceleration and risk factors for Alzheimer’s disease. Alzheimer’s Dement 2018;10:429–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fransquet P, Lacaze P, Saffery R, et al. Accelerated epigenetic aging in peripheral blood does not predict dementia risk. Curr Alzheimer Res 2021;18:443–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sibbett RA, Altschul DM, Marioni RE, et al. DNA methylation-based measures of accelerated biological ageing and the risk of dementia in the oldest-old: a study of the Lothian birth cohort 1921. BMC Psychiatry 2020;20:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Degerman S, Josefsson M, Adolfsson AN, et al. Maintained memory in aging is associated with young epigenetic age. Neurobiol Aging 2017;55:167–171. [DOI] [PubMed] [Google Scholar]
- 51.Zheng Y, Habes M, Gonzales M, et al. Mid-life epigenetic age, neuroimaging brain age, and cognitive function: coronary artery risk development in young adults (CARDIA) study. Aging 2022;14:1691–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shadyab AH, McEvoy LK, Horvath S, et al. Association of Epigenetic age acceleration with incident mild cognitive impairment and dementia among older women. J Gerontol, Ser A 2022;77:1239–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hillary RF, Stevenson AJ, Cox SR, et al. An epigenetic predictor of death captures multi-modal measures of brain health. Mol Psychiatry 2021;26:3806–3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
HRS data used in the current study were obtained from the HRS database (https://hrs.isr.umich.edu/). Derived data (MCR definition) are available from the corresponding author on request by any qualified investigator subject to a data use agreement. LonGenity data used in this study are available upon request. Please contact the corresponding author for further information.