Abstract
Shiga toxin type 1 (Stx1) belongs to the Shiga family of bipartite AB toxins that inactivate eukaryotic 60S ribosomes. The A subunit of Stxs are N-glycosidases that share structural and functional features in their catalytic center and in an internal hydrophobic region that shows strong transmembrane propensity. Both features are conserved in ricin and other ribosomal inactivating proteins. During eukaryotic cell intoxication, holotoxin likely moves retrograde from the Golgi apparatus to the endoplasmic reticulum. The hydrophobic region, spanning residues I224 through N241 in the Stx1 A subunit (Stx1A), was hypothesized to participate in toxin translocation across internal target cell membranes. The TMpred computer program was used to design a series of site-specific mutations in this hydrophobic region that disrupt transmembrane propensity to various degrees. Mutations were synthesized by PCR overlap extension and confirmed by DNA sequencing. Mutants StxAF226Y, A231D, G234E, and A231D-G234E and wild-type Stx1A were expressed in Escherichia coli SY327 and purified by dye-ligand affinity chromatography. All of the mutant toxins were similar to wild-type Stx1A in enzymatic activity, as determined by inhibition of cell-free protein synthesis, and in susceptibility to trypsin digestion. Purified mutant or wild-type Stx1A combined with Stx1B subunits in vitro to form a holotoxin, as determined by native polyacrylamide gel electrophoresis immunoblotting. StxA mutant A231D-G234E, predicted to abolish transmembrane propensity, was 225-fold less cytotoxic to cultured Vero cells than were the wild-type toxin and the other mutant toxins which retained some transmembrane potential. Furthermore, compared to wild-type Stx1A, A231D-G234E Stx1A was less able to interact with synthetic lipid vesicles, as determined by analysis of tryptophan fluorescence for each toxin in the presence of increasing concentrations of lipid membrane vesicles. These results provide evidence that this conserved internal hydrophobic motif contributes to Stx1 translocation in eukaryotic cells.
Enterohemorrhagic Escherichia coli (EHEC) consists of multiple serotypes, among which O157:H7 is the most commonly linked to epidemic and sporadic disease in humans in North America and parts of Europe (25). E. coli O157:H7 infections are a primary cause of hemorrhagic colitis and its extracolonic sequelae, the hemolytic-uremic syndrome and thrombotic thrombocytopenic purpura (25). The pathogenesis of EHEC infections is associated with the production of Shiga toxins (Stxs; formerly called Shiga-like toxins) which are similar to the type 1 Stx produced by Shigella dysenteriae (for reviews, see refereces 1 and 44). Stxs produced by EHEC include Stx type 1 (Stx1), Stx2, and Stx2 variants designated Stx2c (from human isolates) and Stx2e (from porcine isolates) (1).
The E. coli and S. dysenteriae Stxs make up the Shiga toxin family (1). These holotoxins are bipartite molecules composed of a single enzymatically active 32-kDa A subunit noncovalently associated with a pentamer of 7.5-kDa B subunits. The A subunit is an N-glycosidase that cleaves a specific adenine residue on 28S rRNA in 60S ribosomal subunits (1, 17). Stx and Stx1 are virtually identical molecules differing in only one amino acid in the A chain and, not surprisingly, are immunologically cross-reactive (20–22). The A chains of Stx2 and its variants Stx2c and Stx2e share approximately 60% nucleotide sequence homology and 56% amino acid sequence homology with Stx1 (1, 20, 21, 45).
The pentamer of B subunits mediates holotoxin binding to receptors on eukaryotic cells. The B subunits of Stx, Stx1, Stx2, and Stx2c bind globotriaosylceramide, while Stx2e binds globotetraosylceramide (26). Following receptor binding, Stx is internalized by clathrin-dependent endocytosis, delivered to an endosomal compartment, and transported to the trans-Golgi network (TGN) (for reviews, see references 34 and 38). It has been hypothesized that an active portion of Stx translocates from the TGN to the endoplasmic reticulum (ER) and to the nuclear envelope by retrograde transport (30, 34, 38). Evidence suggests that during intracellular routing, Stx is cleaved at a protease-sensitive loop (4, 13, 15), the disulfide bond located between Cys242 and Cys261 is reduced, and the A chain is separated into the enzymatically active 27.5-kDa A1 fragment and the 4-kDa A2 fragment (14, 15).
All A chains in the Stx family are functionally, mechanistically, and structurally similar to ricin and some other ribosomal inactivating proteins (RIPs) which share N-glycosidase activity (17). Site-directed mutagenesis of catalytic sites in Stx1 and ricin reveals that the amino acids required for enzymatic activity have been conserved (1, 20). Furthermore, the X-ray diffraction structure solutions for Stx and RIPs such as ricin reveal that these toxins contain conserved protein folding motifs that similarly orient the conserved amino acids in the active-site cleft (11). In addition to similar catalytic sites, Stxs, ricin, and several other RIPs all contain an internal hydrophobic region that shows strong transmembrane propensity. In ricin, mutations made in this hydrophobic region result in reduced cytotoxicity, suggesting a possible role for the region in toxin translocation across the ER membrane into the cytosol (6, 39). Ricin, like Stx, also undergoes toxin retrograde transport from the TGN to the ER (27, 38).
The work presented here was undertaken to determine if the internal hydrophobic sequences conserved among the bacterial Stxs (see Table 1) have a function similar to that of the internal hydrophobic region in ricin. To this end, we used Stx1A as a representative model of the bacterial Stxs. In Stx1A, the internal potential transmembrane-spanning segment is located at residues I224 through N241 and precedes a protease-sensitive loop located between C242 and C261. Proteolytic processing and disulfide bond reduction during intracellular transport may expose this Stx hydrophobic region to mediate Stx1A1 translocation across the ER membrane. We designed a series of site-specific mutations in the I224-through-N241 region of Stx1A that disrupted transmembrane propensity to various degrees. One mutation, A231D-G234E, that was predicted to abolish transmembrane propensity greatly reduced toxin cytotoxicity without affecting enzymatic activity. Additionally, this mutation reduced the ability of Stx1A to interact with lipid vesicles. These results suggest that this conserved internal hydrophobic region contributes to Stx translocation in eukaryotic cells.
TABLE 1.
Comparison of potential transmembrane regions of Shiga toxins and mutant Stx1As
| Toxin | Expressiona
|
Subset A hydrophobic motifb | Transmembrane propensityc
|
|||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WC | PPE | Out to in | In to out | |||||||||||||||||||||||
| Ricin | NA | NA | K | F | S | V | Y | D | V | S | I | L | I | P | I | I | A | L | M | V | Y | R | C | A | 1,467 | 1,251 |
| Stx2cd | NA | NA | R | I | S | F | N | S | L | S | A | I | L | G | S | V | A | V | I | L | N | C | H | S | 1,097 | 963 |
| Stx2 or Stx2e | NA | NA | R | I | S | F | N | N | I | S | A | I | L | G | T | V | A | V | I | L | N | C | H | H | 1,456 | 1,421 |
| Stx or Stx1 | ++ | ++ | R | I | S | F | G | S | I | N | A | I | L | G | S | V | A | L | I | L | N | C | H | H | 1,603 | 1,346 |
| F226Y | ++ | ++ | Y | 1,350 | 1,116 | |||||||||||||||||||||
| A231D | ++ | ++ | D | 777 | 697 | |||||||||||||||||||||
| I232E | + | − | E | 669 | 336 | |||||||||||||||||||||
| L233E | + | − | E | 724 | 332 | |||||||||||||||||||||
| G234E | ++ | ++ | E | 874 | 518 | |||||||||||||||||||||
| A231D-L233E | + | − | D | E | None | None | ||||||||||||||||||||
| A231D-G234E | ++ | ++ | D | E | 187 | None | ||||||||||||||||||||
| L240K | ++ | ++ | K | 1,407 | 902 | |||||||||||||||||||||
| N241D | ++ | ++ | D | 1,364 | 690 | |||||||||||||||||||||
| Deletion | + | − | R | —e | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | C | H | H | None | None |
Whole cell lysates and periplasmic extracts are designated WC and PPE, respectively. Approximate amounts of toxin in whole cell lysates and periplasmic extracts are as follows: ++, comparable to that of wild type; +, reduced amount; −, none. NA, not applicable.
The deduced amino acid sequences of the relevant hydrophobic regions of ricin (24), Stx2 (21, 22), Stx2c (12), Stx2e (45), Stx (41), and Stx1 (5) are from previously published data.
Transmembrane propensity was determined by the TMpred computer program, and a value above 500 is a significant indicator of a potential transmembrane-spanning domain (31). The orientation of the potential transmembrane-spanning region is designated as an outside-to-inside or an inside-to-outside helix.
The hydrophobic motif shown is from the toxin previously designated Stx2va.
Residue deleted.
MATERIALS AND METHODS
Strains and bacterial growth.
The E. coli strains used in this study were SY327 [F− araD Δ(lac-proAB) argE(Am) rif nalA recA56] (16), DH5αF′ [F′ endA1 hsdR17 (rk−mk+) supE44 thi-1 recA1 gyrA (Nalr) relA1 Δ(lacIZYA-argF)U169 deoR φ80dlacΔ(lacZ)M15], and JM105 [F′ traD36 lacIq Δ(lacZ)M15 proA+B+ thi apsL (Strr) endA sbcC hsdR4(rk− mk+) Δ(lac-proAB)] (33). Bacteria were grown in L broth (33), and E. coli cells containing plasmids encoding ampicillin resistance were selected with LB with ampicillin at 100 μg/ml.
Basic molecular biological techniques.
All restriction enzyme reactions were done in accordance with the manufacturer’s directions. Restriction enzymes were purchased from Gibco-BRL (Gaithersburg, Md.), except for SacI, which was purchased from Promega Corp. (Madison, Wis.). Standard procedures were used for DNA electrophoresis and plasmid construction (33). Plasmid DNA was extracted by an alkaline lysis procedure, and large-scale preparations were purified by using columns purchased from Qiagen (Santa Clarita, Calif.). DNA fragments were purified from electrophoretic agarose gel by electroelution (33) or by using DNA purification kits purchased from Qiagen.
Discontinuous sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) was done by standard procedures using 12.5% separating gels and 3.9% stacking gels, unless stated otherwise (33). Proteins were stained with silver as described previously (3). Immunoblotting was performed by transferring protein to Hybond ECL nitrocellulose (Amersham, Arlington Heights, Ill.) (3). The primary antibodies used throughout this study were either anti-Stx1A polyclonal serum used at a dilution of 1:20,000 or anti-Stx1B monoclonal antibody 13C4 (42) used undiluted from hybridoma (ATCC CRL1794) culture medium. The secondary antibody was anti-rabbit or anti-mouse immunoglobulin G linked to horseradish peroxidase and used at a dilution of 1:1,500. Proteins were detected by using ECL chemiluminescence procedures (Amersham).
Plasmid construction and site-directed mutagenesis.
Unique HindIII and NsiI sites in stx1A are located 239 bp apart and at opposite sides of the nucleotides encoding the hydrophobic region. To facilitate cloning of the HindIII/NsiI-digested mutant Stx1A DNA fragments, plasmid pUCΔH25 was constructed. The HindIII site located in the pUC19 polylinker was destroyed by cutting the plasmid with HindIII, filling in the 5′ overhangs with deoxynucleoside triphosphates by using Klenow enzyme (Gibco-BRL), and ligating the blunt ends by using T4 DNA ligase (Gibco-BRL). Successful removal of the HindIII site was confirmed by DNA sequencing (see below). The full-length Stx1A-encoding gene was removed from plasmid pSC25 (5) after digesting the plasmid DNA with SacI and PstI, and it was ligated into pUC19ΔH to make pUCΔH25.
The TMpred computer program was used to analyze and predict the transmembrane potential of various sequences (31). Point mutations were synthesized in the Stx1A-encoding gene by PCR overlap extension techniques (18). Two independent PCRs utilized overlapping mutagenic primers in conjunction with a primer outside of the mutation region. This initial PCR synthesized identical mutations in the overlapping region of the two PCR products. For use as template DNA in successive PCRs, products from the two independent reactions were combined and annealed, and the strands were extended with Pfu polymerase. The second PCR contained two primers located outside of and at opposite ends of the mutation region. Primers were purchased from Integrated DNA Technologies, Inc. (Coralville, Iowa), and nucleotide sequences were determined from a previously published sequence (5). Nucleotides that differ from the wild-type Stx1A sequence are italicized and underlined. Primers with a PR or PL designation are either sense or antisense strand sequences, respectively. Primers located outside of the mutation region are StxPR (5′-CTTTCCAGTTACACAATCAGG-3′; nucleotides [nt] 1372 to 1352), and StxPL (5′-CAGATAAATCGCCATTCGTTG-3′; nt 782 to 802). The overlapping mutagenic primers are StxF226Y-PR (5′-AATGCTTCCATAAGAAATTC-3′; nt 1084 to 1066), StxF226Y-PL (5′-GAATTTCTTATGGAAGCATT-3′; nt 1066 to 1084), StxA231D-PR (5′-GCTTCCCAGAATGTCATTAATGCTTC-3′; nt 1077 to 2002), StxA231D-PL (5′-GAAGCATTAATGACATTCTGGGAAGC-3′; nt 1077 to 2002), StxG234E-PR (5′-ATGCCACGCTCTCGAGAATTGCATT-3′; nt 1085 to 1109), StxG234E-PL (5′-AATGCAATTCTCGAGAGCGTGGCAT-3′; nt 1085 to 1109), StxA231D-G234E-PR (5′-TAATGCCACGCTCTCGAGGATATCATTAATGCTTC-3′; nt 1077 to 1111), and StxA231D-G234E-PL (5′-GAAGCATTAATGATATCCTCGAGAGCGTGGCATTA-3′; nt 1077 to 1111). Products from the first round of PCR were annealed, extended, and used as template DNAs for the second round of PCR that contained primers StxPL and StxPR.
Pfu polymerase (Stratagene, La Jolla, Calif.) was used in all PCRs as directed by the manufacturer. The cycling conditions for both rounds of PCR consisted of 94°C for 2 min followed by five cycles of 94°C for 1 min, 45°C for 1 min, and 72°C for 1 min and then 20 cycles of 94°C for 1 min, 52°C for 1 min, and 72°C for 1 min, with a final 2 min at 72°C. PCR products were purified from the reaction mixture by using Qiagen PCR purification techniques, digested with HindIII and NsiI, and then ligated into the HindIII-NsiI sites of the Stx1A-encoding gene in pUCΔH25.
Both DNA strands of entire HindIII-NsiI mutant fragments were sequenced by the dideoxy method with Sequenase version 2.0 (USB, Cleveland, Ohio) with [35S]dATP purchased from USB or from Amersham to confirm mutations and eliminate any possibility of fortuitous mutations.
Expression and purification of mutant and wild-type StxAs.
Each mutation designation has the wild-type amino acid, its position in Stx1A, and the amino acid substitution. Wild-type and mutant Stx1As, prepared by periplasmic extraction as previously described (9, 19), were used for both the trypsin sensitivity analyses and the protein synthesis inhibition assays (19). Purified toxin was used for holotoxin preparation, cytotoxicity assays, and lipid-toxin fluorescence studies. Mutant and wild-type Stx1As were purified by a previously described dye-ligand affinity chromatography procedure (2, 3), except that batch washes with buffers containing increasing molarities of NaCl, rather than an NaCl gradient, were used to elute proteins. Briefly, cells from 1-liter cultures with an optical density at 600 nm between 3 and 6 were harvested by centrifugation, and periplasmic extracts were prepared with polymyxin B sulfate (Sigma Chemical Co., St. Louis, Mo.) (19). Periplasmic proteins were concentrated by ammonium sulfate precipitation and dialyzed (3). Proteins were applied to a Matrex Gel Green A agarose column (1.6 by 43 cm; Amicon Corp., Lexington, Mass.) equilibrated with 10 mM phosphate buffer (pH 7.4), and the periplasmic proteins were allowed to bind to the Matrex column for 90 min before elution. Fractions were analyzed by SDS-PAGE, immunoblotting, and silver staining.
Expression and purification of Stx1B.
Stx1B was purified from E. coli JM105(pSCB32) periplasmic extracts by isoelectric focusing with a Rotofor preparative cell (Bio-Rad Laboratories, Hercules, Calif.) as described previously (2, 3). Fractions containing Stx1B were judged for purity by silver-stained SDS–18% PAGE.
Susceptibility of mutant Stx1A to trypsin digestion.
The trypsin sensitivities of mutant and wild-type Stx1As were compared by using a previously described technique (8, 19). Periplasmic extracts prepared from SY327 containing a mutant or wild-type plasmid were incubated for 15 min at 37°C with a final l-1-tosylamido-2-phenylethyl chloromethyl ketone-treated trypsin (Sigma) concentration of 5, 0.5, or 0.05 μg/ml. Digestions were terminated by the addition of phenylmethylsulfonyl fluoride to a final concentration of 1 mM. The extent of toxin digestion was analyzed by SDS-PAGE.
Protein synthesis inhibition assays.
Periplasmic extracts from E. coli SY327 producing wild-type or mutant toxin were analyzed for inhibition of protein synthesis in cell-free assays as previously described (19). Tenfold serial dilutions of periplasmic extracts were preincubated with reticulocyte lysate (Promega Corp.) in 93 mM potassium acetate (pH 7.4) at 30°C for 20 min to inactivate ribosomes. Also added, to give a final 25.5-μl protein synthesis reaction volume, were a 0.04 mM amino acid mixture (no methionine), 20 μCi of [35S]methionine (NEN, Boston, Mass.), and template mRNA consisting of either 1 μg of luciferase mRNA or 0.5 μg of brome mosaic virus mRNA (Promega Corp.). Mixtures were incubated at 30°C for 30 min, and incorporation of the radiolabel into protein was determined as described by the manufacturer (Promega Corp.). Activity was expressed as the percentage of [35S]methionine in trichloroacetic acid-precipitable material compared to that in the reticulocyte lysate positive control that was preincubated with buffer only.
In vitro holotoxin assembly.
Purified wild-type or mutant Stx1A was combined with purified Stx1B and dialyzed in 10 mM Tris-Cl (pH 7.0) by using a technique previously described to assemble holotoxin from recombinant Stx1A and Stx1B (2, 3). Holotoxin assembly was determined by electrophoresis in discontinuous nondenaturing polyacrylamide gels (native PAGE) using an 8% polyacrylamide resolving gel and a 5% polyacrylamide stacking gel and examined by silver staining or immunoblotting.
Cytotoxicity of mutant Stx1A for Vero cells.
In vitro-prepared wild-type and mutant holotoxins were examined for toxicity to African green monkey kidney (Vero) cells (CCL81; American Type Culture Collection, Rockville, Md.) by the method of Konowalchuk and colleagues (23). Vero cells were grown in 96-well microtiter plates (Falcon Labware, Lincoln Park, N.J.) for 24 h in medium 199 (Gibco-BRL), pH 7.4, supplemented with 5% heat-inactivated fetal bovine serum (HyClone Laboratories Inc., Logan, Utah), 1% (wt/vol) penicillin, and 1% (wt/vol) streptomycin (Gibco-BRL) at 37°C in a 6% CO2 atmosphere. Each toxin was prepared in Tris-Cl (pH 7.0) at approximately 0.25 μM. Serial 10-fold dilutions of toxin were added to Vero cells and observed microscopically for cytotoxicity at 1, 2, 3, and 4 days postaddition of toxin. Cell death was expressed as an average percentage (± the standard error [SE]) calculated from three separate experiments, each done in triplicate.
Synthetic lipid vesicle preparation and fluorescence spectroscopy.
Model membranes composed of synthetic lipid vesicles were prepared by sonication. Synthetic diolyl-phosphatidylcholine (DOPC) and diolyl-phosphatidylglycerol (DOPG) (Sigma) were mixed to give 70% DOPC–30% DOPG (mol/mol), the chloroform was removed under nitrogen, and the remaining lipid film was desiccated under a vacuum for 8 to 14 h. Lipids were hydrated in 10 mM HEPES containing 100 mM NaCl (pH 7.4), and the 20 mM lipid solution was sonicated in a bath sonicator for 1 to 1.5 h and used immediately. Purified toxin was mixed with various concentrations of vesicles to yield 18-μg/ml toxin in lipid concentrations of 0.125 to 3 mM. Samples were incubated at room temperature for 10 to 30 min prior to spectral measurements in a Photon Technology International Spectrofluorometer with the 710 Photomultiplier Detection System using a 2-mm path length quartz cuvette (Starna Cells, Inc.). Fluorescence intensities were measured at a 290-nm excitation wavelength and a 323-nm emission wavelength. The background fluorescence of vesicles without protein was subtracted. Fluorescence intensity was expressed as a percentage of that of toxin without lipid.
RESULTS
Site-directed mutagenesis of Stx1A.
A computer program designed to predict transmembrane propensity (31) was used to examine hydrophobic residues 224 through 241 of Stx1A and to design point mutations that disrupt transmembrane capacity to various degrees. The hydrophobic motifs and the predicted transmembrane potentials of the Stxs, ricin A, and the mutant Stx1As are shown in Table 1. The ricin A motif, although different in amino acid sequence, had a predicted transmembrane capability similar to those of the Stxs. PCR overlap extension techniques were used to synthesize point mutations (18). Chain termination DNA sequencing confirmed each mutation (results not shown). Ten mutant stx1As were cloned into pUC19-derived plasmids and expressed from a lacZ promoter in E. coli DH5αF′ and SY327 (Table 1). The I232E, L233E, A231D-L233E, and ΔI224-N241 mutant proteins were not stable in periplasmic extracts and could only be detected at low concentrations in whole cell lysates (Table 1). The F226Y, A231D, G234E, A231D-G234E, L240K, and N241D mutant Stx1As were expressed and stable in the periplasm. We selected the F226Y, A231D, G234E, and A231D-G234E mutant proteins, which gave three degrees of transmembrane disruption, for further characterization in this study. The F226Y mutation only slightly reduced transmembrane propensity, the A231D and G234E mutations reduced membrane-spanning capacity to nearly one-half of that of the wild type, and the A231D-G234E mutation eliminated transmembrane proclivity (Table 1).
Comparison of the enzymatic activities of mutant and wild-type Stx1As.
To investigate whether residue substitutions in the hydrophobic region of Stx1A altered toxin enzymatic activity, periplasmic extracts from E. coli SY327 expressing wild-type or mutant Stx1A were compared for the ability to inhibit protein synthesis in vitro. As expected, periplasmic extracts prepared from E. coli SY327(pUC19ΔH) (vector-only control), which did not encode Stx1A, did not inhibit protein synthesis (results not shown). Stx1A mutations F226Y, A231D, G234E, and A231D-G234E inhibited protein synthesis at levels similar to that of wild-type Stx1A. Figure 1 shows the results of a typical experiment: twofold less A231D or A231D-G234E and the same amount of G234E or F226Y, compared to wild-type Stx1A, were required to inhibit protein synthesis by 50%. These results provide evidence that mutations F226Y, A231D, G234E, and A231D-G234E in Stx1A did not affect the enzymatic activity of the toxin and suggest that the mutations did not alter subunit A structure at sites distant from the mutations.
FIG. 1.
Inhibition of protein synthesis by wild-type or mutant Stx1As. Rabbit reticulocyte lysates were preincubated with periplasmic extracts containing wild-type Stx1A (○) or F226Y (▵), A231D (▿), G234E (◊), or A231D-G234E (□) mutant Stx1A at the dilutions indicated. Background activity (omitted mRNA) was subtracted from each value, and percent protein synthesis was determined by comparison to the control reaction preincubated with buffer only. Datum points are the means in one representative experiment ± the SE.
Susceptibility of mutant Stx1A to trypsin digestion.
As a measure of protein folding disruption, mutant and wild-type Stx1As were exposed to 10-fold-increasing amounts of trypsin and analyzed by SDS-PAGE and immunoblotting. Periplasmic extracts containing F226Y, A231D, G234E, or A231D-G234E Stx1A digested with trypsin showed protein degradation patterns similar to that of trypsin-digested wild-type Stx1A (Fig. 2). In the absence of trypsin, two proteins, approximately 34 and 32 kDa, from mutant and wild-type Stx1A extracts reacted strongly with the anti-Stx1A polyclonal sera (Fig. 2, lanes 2, 6, 10, 14, and 18). The presence of the two Stx1A forms was confirmed in multiple immunoblots (results not shown). Presumably, these correspond to the precursor Stx1A (Mr, 34,804), which contains the intact signal sequence and the mature toxin (Mr, 32,217) which had the signal sequence cleaved during toxin transport into the bacterial cell periplasm. These results indicate that minor amounts of cytoplasmic proteins were present in the extracts, probably due to the activity of polymyxin B, which was used to extract the periplasmic proteins. All periplasmic extracts, including the vector-only control E. coli SY327(pUCΔH19), showed two proteins (approximately 36 and 20 kDa) that reacted weakly with anti-Stx1A serum (Fig. 2, lanes 1 to 21). Similar to previous reports of unrelated immunoreactive contaminants in recombinant Stx1A preparations, these proteins presumably also represent immunoreactive host proteins unrelated to toxin (8).
FIG. 2.
Immunoblot of wild-type (wt) or mutant Stx1A following trypsin digestion. Periplasmic extracts of mutant and wild-type Stx1As were incubated at 37°C for 15 min with no trypsin (lanes 2, 6, 10, 14, and 18) or with trypsin at 0.05 (lanes 3, 7, 11, 15, and 19), 0.5 (lanes 4, 8, 12, 16, and 20), or 5 ng/ml (lanes 5, 9, 13, 17, and 21) ng/ml. Lanes: 1, pUC19ΔH (vector-only control); 2 to 5, wild-type Stx1A; 6 to 9, F226Y Stx1A; 10 to 13, A231D Stx1A; 14 to 17, G234E Stx1A; 18 to 21, A231D-G234E Stx1A (AG). The locations of Stx1A and the two primary trypsin degradation products (★) are shown on the left. The positions of the following molecular size standards are indicated on the right (from top to bottom: 95.5, 55, 43, 29, 18.4, and 12.4 kDa).
Trypsin digestion of wild-type and mutant Stx1As resulted in the disappearance of the 34- and 32-kDa Stx1As and the appearance of smaller proteins of approximately 27 kDa and less, and these varied in amount depending upon the concentration of trypsin in the reaction mixture. Digestion with 0.05- and 0.5-ng/ml trypsin resulted in the appearance of the 27-kDa degradation protein, which corresponds to the 27.5-kDa Stx1A1 fragment (Fig. 2, lanes 3, 4, 7, 8, 11, 12, 15, 16, 19, and 20). At 5-ng/ml trypsin, the 27-kDa protein was detected for the wild-type and all mutant Stx1As (Fig. 2, lanes 5, 9, 13, 17, and 21). Although the wild-type, A231D, and G234E Stx1As showed somewhat greater resistance to trypsin digestion than did the F226Y and A231D-G234E Stx1As, the digested proteins generated for each were similar (Fig. 2, compare lanes 5, 13, and 17 with lanes 9 and 21). These results provide evidence that the tertiary structure of the toxin was not grossly altered by the amino acid substitutions.
Association of mutant Stx1A with Stx1B subunits.
Wild-type and mutant Stx1As were combined in vitro with purified Stx1B to form holotoxins. All of the Stx1As purified by Matrex Gel Green A chromatography eluted with 0.4 M NaCl, except for the A231D-G234E mutant toxin, which eluted with 0.15 M NaCl. Stx1A was purified to >95% homogeneity, and Stx1B was purified to >80% homogeneity, as estimated by silver-stained SDS-PAGE (results not shown). Each mutant Stx1A associated with Stx1B in a manner similar to that of wild-type Stx1A, as determined by native PAGE and immunoblotting (Fig. 3). Immunoblotting with anti-Stx1A polyclonal serum (Fig. 3A, lanes 3, 5, 7, 9, and 11) or anti-Stx1B monoclonal antibody (Fig. 3B, lanes 3, 5, 7, 9, and 11) confirmed that the species migrating like the holotoxin contained both the A and B subunits. Electrophoresis of Stx1B alone did not show any immunoreactivity with anti-Stx1A serum but was detected with anti-Stx1B antibody (Fig. 3A and B, lanes 1). Since Stx1B was added in excess to make holotoxin, monomers of Stx1B were also detected in wild-type and mutant holotoxin samples (Fig. 3B, lanes 3, 5, 7, 9, and 11). As expected, in the absence of Stx1B, the wild-type, F226Y, A231D, and G234E Stx1As were not detected by native PAGE (Fig. 3A, lanes 2, 4, 6, 8, and 10). The inability of Stx1A, with its strong positive charge (pI 9.0), to migrate into this native gel has been previously reported and is most likely the reason why the mutants did not migrate into this gel (3, 48). However, the A231D-G234E Stx1A entered the stacking gel and was detected at the interface of the stacking and resolving gels (Fig. 3A, lane 10). Furthermore, the A231D-G234E holotoxin migrated more rapidly toward the anode in the native PAGE than did any other holotoxin (Fig. 3A and B, lanes 11). These results suggest that the substitution with two acidic amino acids increased the mobility of the A231D-G234E holotoxin toward the anode.
FIG. 3.
Wild-type and mutant holotoxins in native-PAGE immunoblots with stacking and resolving gels (indicated at the left) developed with anti-Stx1A serum (A) or an anti-Stx1B monoclonal antibody (B). Lanes: 1, Stx1B; 2, wild-type Stx1A; 3, wild-type Stx1A associated with Stx1B; 4, G234E Stx1A; 5, G234E Stx1A plus Stx1B; 6, A231D Stx1A; 7, A231D Stx1A plus Stx1B; 8, F226Y Stx1A; 9, F226Y Stx1A plus Stx1B; 10, A231D-G234E Stx1A; 11, A231D-G234E Stx1A plus Stx1B. The positions of the holotoxin and Stx1B and Stx1A monomers are indicated at the right.
Cytotoxicity of mutant Stx1A for Vero cells.
To investigate the effects on cytotoxicity, mutant holotoxins prepared as described above were examined in a standard Vero cell assay (23). A representative experiment is shown in Fig. 4. The cytotoxicity of the A231D-G234E holotoxin was significantly reduced in comparison with that of the wild-type or the other mutant holotoxins. As expected, cells incubated with individual subunits of Stx1B or Stx1A (mutant or wild type) remained viable throughout the experiment. The A231D and G234E holotoxins were nearly identical to the wild-type holotoxin in cytotoxicity, resulting in 50% Vero cell death with approximately 10−6 μM holotoxin at 72 h postaddition of toxin. The F226Y holotoxin demonstrated no more than a 10-fold difference in cytotoxicity from the wild-type holotoxin. However, a 225-fold higher concentration of the A231D-G234E holotoxin was required to kill 50% of the Vero cells. Unlike that of the other holotoxins tested, the cytotoxicity produced by the A231D-G234E holotoxin usually reached maximal levels within 24 h. This was faster than the rate of cell death observed with the wild-type holotoxin, which increased over the 4-day period (results not shown).
FIG. 4.
Cytotoxicity of wild-type and mutant holotoxins. Holotoxins or Stx1As, at the concentrations indicated, were incubated with Vero cells for 72 h. Cell viability was determined by microscopic analysis and expressed as a percentage. The datum points are the means ± the SE of one representative experiment. Holotoxins are indicated by shaded symbols, and Stx1As are indicated by open symbols as follows: wild type, • and ○; F226Y, ▴ and ▵; A231D, ▾ and ▿; G234E, ⧫ and ◊; A231D-G234E alone, ■ and □.
Interaction of the wild-type and A231D-G234E Stx1As with synthetic lipid vesicles.
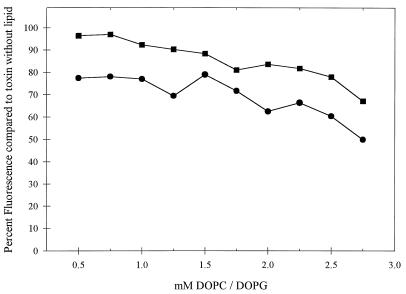
The interaction of the A231D-G234E Stx1A with model membranes was compared to that of wild-type Stx1A by examining the tryptophan fluorescence of toxins when incubated with increasing concentrations of lipid vesicles. Spectral emission scans of the wild-type and A231D-G234E Stx1As in aqueous solutions at pH 7.4 showed emission maximums at a wavelength of 323 nm with an excitation wavelength of 290 nm. These conditions were used in subsequent experiments. Reflecting the ability of Stx1A to interact with membranes, a concentration-dependent decrease in fluorescence intensity was observed in the presence of 70% DOPC–30% DOPG (mol/mol) synthetic lipid vesicles (Fig. 5). Similar findings were reported earlier for Stx and its interaction with synthetic lipid vesicles (43). In contrast to the results obtained with wild-type Stx1A, the fluorescence of A231D-G234E Stx1A did not decrease as rapidly. At all of the lipid concentrations tested, the mutant toxin was less able to interact with the synthetic vesicles than was the wild-type toxin. For example, the fluorescence of wild-type Stx1A was reduced by 20% in 0.5 mM lipid and by 51% in 2.75 mM lipid, while the fluorescence of A231D-G234E was reduced by 4 and 33% in those lipids, respectively (Fig. 5). At pH 5.2, both the mutant and wild-type Stx1As showed reduced association with lipid vesicles (data not shown). But even at pH 5.2, the mutant toxin associated with lipid vesicles less than did wild-type Stx1A.
FIG. 5.
Interaction of wild-type Stx1A and A231D-G234E Stx1A with synthetic lipid vesicles. Purified wild-type (•) or A231D-G234E (■) A subunits at 18 μg/ml were combined with increasing concentrations of 70% DOPC–30% DOPG (mol/mol) vesicles. Fluorescence intensities were obtained at an excitation wavelength of 290 nm and an emmission wavelength of 323 nm and expressed as a percentage of that of the toxin without lipid vesicles. The results of one representative experiment are shown.
DISCUSSION
The most significant finding of this study is that disruption of a conserved sequence of hydrophobic amino acids (I224 through N241) in Stx1 resulted in decreased cytotoxicity. We used site-specific mutation to systematically alter the membrane-spanning capability of the amino acids in this sequence and then compared mutant and wild-type toxins for protein structure stability, holotoxin formation, enzymatic activity, and cytotoxicity. In all of the analyses, each mutant toxin behaved similarly to the wild-type toxin, with one striking exception. The A231D-G234E mutation, which abolished the membrane-spanning capacity of the amino acid sequence, significantly reduced cytotoxicity toward Vero cells in culture. The reduced cytotoxicity could not be explained by changes in enzymatic activity, holotoxin formation, or in vitro toxin stability. As determined by fluorescence spectroscopy, A231D-G234E Stx1A was hampered in the ability to interact with synthetic lipid vesicles compared with wild-type Stx1A. This suggests that the mechanism of reduced cytotoxicity may be due to reduced or aberrant translocation of this mutant toxin. These results support a possible role for hydrophobic residues I224 through N241 in the translocation of Stx1A across the ER membrane. In addition, the time course of the reduced cell death due to the A231D-G234E mutant toxin was more rapid than wild-type toxin cytotoxicity rates. Interestingly, cytotoxicity was not altered among mutant toxins that reduced, but did not abolish, the membrane-spanning capacity of the region.
Hydrophobic sequences may be necessary for efficient toxin translocation in eukaryotic target cells (32). Following binding to eukaryotic cells, Stx1 is internalized into the endosome and undergoes retrograde translocation from the TGN to the ER (34, 36, 38). It is well known that brefeldin A, which blocks retrograde transport, protects susceptible cells from Stx intoxication (37). Toxin must traverse the ER membrane to reach the ribosomes on the cytosolic side of the ER (13, 15, 37). The conserved hydrophobic sequence may mediate this translocation.
Our first effort to determine the function of residues I224 through N241 was to create a deletion mutant construct. However, the toxin expressed from this construct was not stable in the periplasmic space of E. coli SY327 and was not characterized further. Computer predictions were used to design a series of site-specific mutations to systematically reduce the transmembrane propensity of the hydrophobic sequence. Replacing hydrophobic F226 with a polar Y residue to maintain the phenyl ring structure at that position was predicted to make only a slight change in the transmembrane-spanning capacity of the sequence. Single internal hydrophobic amino acids were replaced with charged acidic amino acids (A231D, I232E, L233E, and G234E) to reduce transmembrane propensity by about 50%. Replacing two hydrophobic amino acids with acidic side-chain residues, as in the A231D-L233E and A231D-G234E mutations, was predicted to abolish membrane-spanning ability. The I232E, L233E, and A231D-L233E mutant toxins were not stable when secreted into the periplasm of E. coli DH5α or SY327, suggesting that residues I232 and L233 are important for maintaining tertiary structure. Preliminary evidence suggested that I232E Stx1A was stable in E. coli strains inactivated for the periplasmic protease DegP (40). The stability of site-specific or deletion mutations must be determined empirically. Instability of mutant toxins is not uncommon and has been reported for ricin (39), heat-labile enterotoxin (46), and diphtheria toxin (47).
We characterized F226Y, A231D, G234E, and A231E-G234E, each representing one of three groups of mutations predicted to cause decreasing transmembrane-spanning capacity. We predicted that stepwise reductions in membrane-spanning capacity would result in stepwise decreases in cytotoxicity. However, this was not the case. Mutations that merely reduced the transmembrane-spanning ability of the I224-to-N241 region did not affect cytotoxicity. The mutation that abolished transmembrane potential was the only mutation that also affected cytotoxicity. It may be that the assay that measured cultured Vero cell death was not sensitive enough to detect subtle differences in cytotoxicity, or it may be that these mutations retained enough of the hydrophobic sequence to efficiently intoxicate cells.
Central to the interpretation of our results are several pieces of evidence showing that the mutations did not grossly alter the Stx1A tertiary structure. First, all of the mutant Stx1As were as enzymatically active as wild-type Stx1A. This enzymatic activity is dependent upon the integrity of the catalytic cleft, which has been highly conserved among all RIPs, and the results show that the mutations did not alter this architecture. Second, purified mutant Stx1As successfully associated with purified B subunits to form holotoxins in vitro. Since the mutations in the internal hydrophobic sequence are located adjacent to carboxy-terminal residues that are essential for Stx1A interaction with Stx1B (3), the successful holotoxin formation by these mutant toxins suggests that the folded structure of the C-terminal region was not significantly altered. Third, mutant Stx1As showed protease digestion patterns similar to that of wild-type Stx1A (Fig. 2) and were consistent with previously reported trypsin digestion patterns of Stx1A (8). Two mutant toxins, F226Y and A231D-G234E, appeared to be more sensitive to protease digestion at the highest concentration of trypsin used, suggesting that these mutant toxins were less stable than the wild-type toxin. However, F226Y was as cytotoxic as the wild-type holotoxin, even though it was more susceptible to trypsin digestion. Therefore, this subtle difference in trypsin sensitivity does not necessarily explain the more-than-200-fold decrease in cytotoxicity measured with the A231D-G234E mutant toxin.
Although this paper presents results consistent with the interpretation that the hydrophobic region in Stx1A plays a role in toxin translocation, several alternative explanations for the reduced cytotoxicity of A231D-G234E need to be addressed. First, if residues A231 and/or G234 interact with subunit B residues, the amino acid substitutions may have altered holotoxin formation or in vivo stability. Two pieces of evidence suggest that this is not the case: analysis of holotoxin formation and structural information from the highly similar Stx molecule. The native-PAGE immunoblot of the A231D-G234E holotoxin (Fig. 3) shows the formation of a complex that was immunoreactive with anti-A and anti-B immunoglobulins and migrated in a native-PAGE gel to a position similar to that of the holotoxin. Interestingly, the A231D-G234E holotoxin migrated slightly farther into the gel than the wild type or the other mutant holotoxins. Although unlikely, the association of A231D-G234E Stx1A with fewer than five Stx1B subunits could explain these results. However, even in the absence of Stx1B, A231D-G234E Stx1A also migrated farther into the gel than the other Stx1As, suggesting that the difference in mobility is intrinsic to the Stx1A molecule. Therefore, it is likely that the substitution with two acidic residues (D and E) in Stx1A resulted in the increased mobility toward the anode. In addition, X-ray diffraction analysis of Stx shows that residues A231 and G234 are positioned away from StxB and do not directly interact with the B pentamer (3, 10, 11). Residues A231 and G234 reside in an alpha helix (residues S228 through S235 in StxA) that extends toward the globular head structure of StxA and away from StxB. Interestingly, although the F226Y mutation did not affect holotoxin formation (Fig. 3) or cytotoxicity (Fig. 4), the F226 residue is located in a β-strand that may directly contact Stx1B (10, 11). The conservative Y substitution is likely to have retained the appropriate A-B interactions.
Second, excess B subunit in the holotoxin preparation may have inhibited toxin activity. However, native-PAGE analysis of each holotoxin preparation used in the cytotoxicity assays showed that similar amounts of excess B subunits were present in each. Furthermore, the cytotoxicities of all of the Stx1As examined were similar, except that of A231D-G234E. It is therefore unlikely that the excess B subunits interfered with the results of the cytotoxicity assay.
Third, since the Stx1A hydrophobic sequence is located adjacent to a protease labile loop and the disulfide bond (residues C242 and C261), mutations at A231 and G234 may have affected proteolytic processing and/or disulfide bond reduction of Stx1 during eukaryotic translocation. Investigations with both Stx and Stx1 holotoxins have shown that although these processes are not strictly required for cell intoxication, they are required for efficient translocation (14, 15). Although proteases other than trypsin likely function in the proteolysis of Stx1 in vivo, the A231D-G234E mutant toxin was degraded by trypsin, which suggests that the protease-sensitive region was accessible. Interestingly, deletions or point mutations disrupting the protease-sensitive region of Stx or Stx1 result in delayed, but not decreased, cytotoxicity (4, 13). This is in contrast to the pattern of Vero cell killing by the A231D-G234E holotoxin. Although reduced, the cytotoxicity induced by the mutant toxin occurred more rapidly than that induced by the wild-type toxin, reaching maximal levels about 24-h posttreatment with toxin. Incubations for 96 h did not result in increased cell death. Also, it has been shown that elimination of the disulfide bond in Stx results in increased cytotoxicity early during incubation with target cells and reduced cytotoxicity after longer incubations. Investigators have suggested that the disulfide bond stabilizes the A chain and prevents premature dissociation and degradation (14). Again, the pattern of Vero cell killing by the A231D-G234E holotoxin did not mimic this scenario, as cytotoxicity was reduced, compared to that of the wild-type toxin, for the duration of the experiment.
Although the A231D-G234E mutation is predicted to abolish transmembrane capacity in the hydrophobic region of Stx1A, cytotoxicity was not abolished. Supportive of these findings is the reduced but not abolished interaction of A231D-G234E Stx1A with synthetic lipid membrane vesicles compared to that of wild-type Stx1A. Strikingly similar results have been obtained by mutational analysis of the carboxy-terminal hydrophobic sequence of ricin A. Although the amino acid sequence in this region has not been conserved between bacterial and plant RIPs, the hydrophobic motif and its position within the A chains of these toxins have been conserved (Table 1). A P250A mutation in ricin A, which abolishes transmembrane potential, results in an enzymatically active holotoxin that has 170-fold-reduced cytotoxicity toward Vero cells (39). These results suggest that there are several routes by which a toxin can traffic though target cells to reach the ribosomes.
The complete mechanism by which Stx1A translocates through eukaryotic target cells remains to be defined. Unlike Pseudomonas exotoxin A, which has a KDEL-like carboxyl-terminal tetrapeptide signal that is essential for retrograde translocation and cytotoxicity (7), ricin and Stx1 do not have a KDEL-like sequence. Interestingly, Stx1B can translocate retrogradely through the Golgi to the ER in the absence of Stx1A, suggesting that Stx1B is involved in signaling translocation (26, 38). Evidence also suggests that during intracellular transport of Stx, the A subunit is proteolytically cleaved, producing the 27.5-kDa A1 and 4-kDa A2 fragments, and reduction of the disulfide bond connecting the two subunits would release StxA1 from StxA2 and the pentamer of StxB (14, 15). Therefore the A1 subunit may move across the membrane aided by StxA2 and Stx1B. It remains to be determined whether StxA1 interacts specifically with the ER machinery that could be involved in translocation. Resolution of the crystal structure of Stx1A1 may provide information on whether the hydrophobic region changes after Stx1A1 separates from Stx1A2. The internal hydrophobic sequence appears to be hidden or buried in full-length Stx1A (10, 11). The environmental change from an acidified endosome to the neutral pH of the ER, combined with the intracellular processing of Stx1A, could play a role in moving Stx1A across the ER membrane. Other toxins, such as diphtheria toxin, take advantage of the changing endosomal pH to expose a pH-sensitive hydrophobic domain that mediates translocation of the toxin out of the endosome (29, 35). However, lysosomotrophic agents such as NH4Cl do not inhibit the intoxication of cells by Stx, suggesting that Stx does not utilize pH changes in the same manner as other toxins (38). Synthetic peptides that include the conserved hydrophobic region of Stx (residues 220 through 246) readily associate with lipid membranes in vitro, and this association and the level of membrane insertion potential increase as the pH increases from 5 to 7 (28, 32). Our results suggested also that Stx1A has an increased ability to associate with lipid membranes under neutral pH conditions.
Ongoing investigations in our laboratory will further characterize intracellular trafficking of A231D-G234E and other mutations in an effort to determine the function of this conserved hydrophobic sequence.
ACKNOWLEDGMENTS
This work was supported, in part, by the Idaho Agriculture Experiment Station and Public Health Service grant AI33981 from the National Institutes of Health.
We acknowledge Paula Austin, Larry Evans, William Trumble, and Carla Wesson for technical support.
REFERENCES
- 1.Acheson D W K, Donohue-Rolfe A, Keusch G T. The family of Shiga and Shiga-like toxins. In: Alouf J E, Freer J H, editors. Sourcebook of bacterial toxins. 1st ed. Vol. 1. London, England: Academic Press, Inc.; 1991. pp. 415–433. [Google Scholar]
- 2.Austin P R, Hovde C J. Purification of recombinant Shiga-like toxin type I B subunit. Protein Expr Purif. 1995;6:771–779. doi: 10.1006/prep.1995.0008. [DOI] [PubMed] [Google Scholar]
- 3.Austin P R, Jablonski P E, Bohach G A, Dunker A K, Hovde C J. Evidence that the A2 fragment of Shiga-like toxin type I is required for holotoxin integrity. Infect Immun. 1994;62:1768–1775. doi: 10.1128/iai.62.5.1768-1775.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burgess B J, Roberts L M. Proteolytic cleavage at arginine residues within the hydrophilic disulphide loop of the Escherichia coli Shiga-like toxin I A subunit is not essential for cytotoxicity. Mol Microbiol. 1993;10:171–179. doi: 10.1111/j.1365-2958.1993.tb00913.x. [DOI] [PubMed] [Google Scholar]
- 5.Calderwood S B, Auclair F, Donohue-Rolfe A, Keusch G T, Mekalanos J J. Nucleotide sequence of the Shiga-like toxin genes of Escherichia coli. Proc Natl Acad Sci USA. 1987;84:4364–4368. doi: 10.1073/pnas.84.13.4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaddock J A, Roberts L M, Jungnickel B, Lord J M. A hydrophobic region of ricin A chain which may have a role in membrane translocation can function as an efficient noncleaved signal peptide. Biochem Biophys Res Commun. 1995;217:68–73. doi: 10.1006/bbrc.1995.2746. [DOI] [PubMed] [Google Scholar]
- 7.Chaudhary V K, Jinno Y, FitzGerald D, Pastan I. Pseudomonas exotoxin contains a specific sequence at the carboxyl terminus that is required for cytotoxicity. Proc Natl Acad Sci USA. 1990;87:308–312. doi: 10.1073/pnas.87.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deresiewicz R L, Austin P R, Hovde C J. The role of tyrosine-114 in the enzymatic activity of the Shiga-like toxin I A-chain. Mol Gen Genet. 1993;241:467–473. doi: 10.1007/BF00284701. [DOI] [PubMed] [Google Scholar]
- 9.Donohue-Rolfe A, Keusch G T. Shigella dysenteriae 1 cytotoxin: periplasmic protein release by polymyxin B and osmotic shock. Infect Immun. 1983;39:270–274. doi: 10.1128/iai.39.1.270-274.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fraser M E, Chernaia M M, Kozlov Y V, James M N G. Crystal structure of the holotoxin from Shigella dysenteriae at 2.5 angstrom resolution. Nat Struct Biol. 1994;1:59–64. doi: 10.1038/nsb0194-59. [DOI] [PubMed] [Google Scholar]
- 11.Fraser M E, Chernaia M M, Kozlov Y V, James M N G. Shiga toxin. In: Parker M W, editor. Protein toxin structure. R. G. Georgetown, Tex: Landes Co.; 1996. pp. 173–190. [Google Scholar]
- 12.Gannon V P J, Teerling C, Masri S A, Gyles C L. Molecular cloning and nucleotide sequence of another variant of the Escherichia coli Shiga-like toxin II family. J Gen Microbiol. 1990;136:1125–1135. doi: 10.1099/00221287-136-6-1125. [DOI] [PubMed] [Google Scholar]
- 13.Garred O, Dubinina E, Holm P K, Olsnes S, Vandeurs B, Kozlov J V, Sandvig K. Role of processing and intracellular transport for optimal toxicity of Shiga toxin and toxin mutants. Exp Cell Res. 1995;218(1):39–49. doi: 10.1006/excr.1995.1128. [DOI] [PubMed] [Google Scholar]
- 14.Garred O, Dubinina E, Polesskaya A, Olsnes S, Kozlov J, Sandvig K. Role of the disulfide bond in Shiga toxin A-chain for toxin entry into cells. J Biol Chem. 1997;272:11414–11419. doi: 10.1074/jbc.272.17.11414. [DOI] [PubMed] [Google Scholar]
- 15.Garred O, Van Deurs B, Sandvig K. Furin-induced cleavage and activation of Shiga toxin. J Biol Chem. 1995;270:10817–10821. doi: 10.1074/jbc.270.18.10817. [DOI] [PubMed] [Google Scholar]
- 16.Goldberg I, Mekalanos J J. Cloning of the Vibrio cholerae recA gene and construction of a Vibrio cholerae recA mutant. J Bacteriol. 1986;165:715–722. doi: 10.1128/jb.165.3.715-722.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartley M R, Chaddock J A, Bonness M S. The structure and function of ribosome-inactivating proteins. Trends Plant Sci. 1996;1:254–260. [Google Scholar]
- 18.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 19.Hovde C J, Calderwood S B, Mekalanos J J, Collier R J. Evidence that glutamic acid 167 is an active-site residue of Shiga-like toxin I. Proc Natl Acad Sci USA. 1988;85:2568–2572. doi: 10.1073/pnas.85.8.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson M. Structure-function analyses of Shiga toxin and the Shiga-like toxins. Microb Pathog. 1990;8:235–242. doi: 10.1016/0882-4010(90)90050-z. [DOI] [PubMed] [Google Scholar]
- 21.Jackson M P, Neill R J, O’Brien A D, Holmes R K, Newland J W. Nucleotide sequence analysis and comparison of the structural genes for Shiga-like toxin I and Shiga-like toxin II encoded by bacteriophages from Escherichia coli 933. FEMS Microbiol Lett. 1987;44:109–114. doi: 10.1016/0882-4010(87)90106-9. [DOI] [PubMed] [Google Scholar]
- 22.Jackson M P, Newland J W, Holmes R K, O’Brien A D. Nucleotide sequence analysis of the structural genes for Shiga-like toxin I encoded by bacteriophage 933J from Escherichia coli. Microb Pathog. 1987;2:147–153. doi: 10.1016/0882-4010(87)90106-9. [DOI] [PubMed] [Google Scholar]
- 23.Konowalchuk J, Speirs J I, Stavric S. Vero response to a cytotoxin of Escherichia coli. Infect Immun. 1977;18:775–779. doi: 10.1128/iai.18.3.775-779.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamb F I, Roberts L M, Lord J M. Nucleotide sequence of cloned cDNA coding for preproricin. FEBS Lett. 1985;148:265–270. doi: 10.1111/j.1432-1033.1985.tb08834.x. [DOI] [PubMed] [Google Scholar]
- 25.Lansbury L E, Ludlam H. Escherichia coli O157: lessons from the past 15 years. J Infect. 1997;34:189–193. doi: 10.1016/s0163-4453(97)94059-7. [DOI] [PubMed] [Google Scholar]
- 26.Lingwood C A. Role of verotoxin receptors in pathogenesis. Trends Microbiol. 1996;4:147–153. doi: 10.1016/0966-842x(96)10017-2. [DOI] [PubMed] [Google Scholar]
- 27.Lord J M, Roberts L M. The intracellular transport of ricin: why mammalian cells are killed and how Ricinus cells survive. Plant Physiol Biochem. 1996;34:253–261. [Google Scholar]
- 28.Menikh A, Saleh M T, Gariépy J, Boggs J M. Orientation in lipid bilayers of a synthetic peptide representing the c-terminus of the A1 domain of Shiga toxin. A polarized ATR-FTIR study. Biochemistry. 1997;36:15865–15872. doi: 10.1021/bi970944+. [DOI] [PubMed] [Google Scholar]
- 29.Papini E, Sandon D, Rappuoli R, Montecucco C. On the membrane translocation of diphtheria toxin: at low pH the toxin induces ion channels on cells. EMBO J. 1988;7:3353–3359. doi: 10.1002/j.1460-2075.1988.tb03207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pelham H R B, Roberts L M, Lord J M. Toxin entry: how reversible is the secretory pathway? Trends Cell Biol. 1992;2:183–185. doi: 10.1016/0962-8924(92)90230-k. [DOI] [PubMed] [Google Scholar]
- 31.Persson B, Argos P. Prediction of transmembrane segments in proteins utilising multiple sequence alignments. J Mol Biol. 1994;237:182–192. doi: 10.1006/jmbi.1994.1220. [DOI] [PubMed] [Google Scholar]
- 32.Saleh M T, Ferguson J, Boggs J M, Gariépy J. Insertion and orientation of a synthetic peptide representing the C-terminus of the A(1) domain of Shiga toxin into phospholipid membranes. Biochemistry. 1996;35:9325–9334. doi: 10.1021/bi960177z. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 34.Sandvig K, Garred O, Prydz K, Kozlov J V, Hansen S H, van Deurs B. Retrograde transport of endocytosed Shiga toxin to the endoplasmic reticulum. Nature (London) 1992;358:510–512. doi: 10.1038/358510a0. [DOI] [PubMed] [Google Scholar]
- 35.Sandvig K, Olsnes S. Rapid entry of nicked diphtheria toxin into cells at low pH. Characterization of the entry process and effects of low pH on the toxin molecule. J Biol Chem. 1981;256:9068–9076. [PubMed] [Google Scholar]
- 36.Sandvig K, Olsnes S, Brown J E, Petersen O W, van Deurs B. Endocytosis from coated pits of Shiga toxin: a glycolipid-binding protein from Shigella dysenteriae 1. J Cell Biol. 1989;108:1331–1343. doi: 10.1083/jcb.108.4.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sandvig K, Prydz K, van Deurs B. Endocytic uptake of ricin and Shiga toxin. In: Courtoy P J, editor. Endocytosis. From cell biology to health, disease and therapy. 1st ed. Vol. 1. New York, N.Y: Springer Verlag; 1992. pp. 405–412. [Google Scholar]
- 38.Sandvig K, van Deurs B. Endocytosis and intracellular sorting of ricin and Shiga toxin. FEBS Lett. 1994;346:99–102. doi: 10.1016/0014-5793(94)00281-9. [DOI] [PubMed] [Google Scholar]
- 39.Simpson J C, Lord J M, Roberts L M. Point mutations in the hydrophobic C-terminal region of ricin A chain indicate that Pro250 plays a key role in membrane translocation. Eur J Biochem. 1995;232(2):458–463. doi: 10.1111/j.1432-1033.1995.tb20831.x. [DOI] [PubMed] [Google Scholar]
- 40.Strauch K L, Johnson K, Beckwith J. Characterization of degP, a gene required for proteolysis in the cell envelope and essential for growth of Escherichia coli at high temperature. J Bacteriol. 1989;171:2689–2696. doi: 10.1128/jb.171.5.2689-2696.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strockbine N A, Jackson M P, Sung L M, Holmes R K, O’Brien A D. Cloning and sequencing of the genes for Shiga toxin from Shigella dysenteriae type 1. J Bacteriol. 1988;170:1116–1122. doi: 10.1128/jb.170.3.1116-1122.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strockbine N A, Marques L R, Holmes R K, O’Brien A D. Characterization of monoclonal antibodies against Shiga-like toxin from Escherichia coli. Infect Immun. 1985;50:695–700. doi: 10.1128/iai.50.3.695-700.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Surewicz W K, Surewicz K, Mantsch H H, Auclair F. Interaction of Shigella toxin with globotriaosyl ceramide receptor-containing membranes: a fluorescence study. Biochem Biophys Res Commun. 1989;160:126–132. doi: 10.1016/0006-291x(89)91630-6. [DOI] [PubMed] [Google Scholar]
- 44.Tesh V L, O’Brien A D. The pathogenic mechanisms of Shiga toxin and the Shiga-like toxins. Mol Microbiol. 1991;5:1817–1822. doi: 10.1111/j.1365-2958.1991.tb00805.x. [DOI] [PubMed] [Google Scholar]
- 45.Weinstein D L, Jackson M P, Samuel J E, Holmes R K, O’Brien A D. Cloning and sequencing of a Shiga-like toxin type II variant from an Escherichia coli strain responsible for edema disease of swine. J Bacteriol. 1988;170:4223–4230. doi: 10.1128/jb.170.9.4223-4230.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wülfing C, Rappuoli R. Efficient production of heat-labile enterotoxin mutant proteins by overexpression of dsbA in a degP-deficient Escherichia coli strain. Arch Microbiol. 1997;167:280–283. doi: 10.1007/s002030050444. [DOI] [PubMed] [Google Scholar]
- 47.Zdanovsky A G, Kulaeva O I, Yankovsky N K. Construction and expression of diphtheria toxin-encoding gene derivatives in Escherichia coli. Gene. 1992;116:81–86. doi: 10.1016/0378-1119(92)90632-y. [DOI] [PubMed] [Google Scholar]
- 48.Zollman T M, Austin P R, Jablonski P E, Hovde C J. Purification of recombinant Shiga-like toxin type I A1 fragment from Escherichia coli. Protein Expr Purif. 1994;5:291–295. doi: 10.1006/prep.1994.1044. [DOI] [PubMed] [Google Scholar]