Abstract
Mammographic density (MD), after accounting for age and body mass index (BMI), is a strong heritable risk factor for breast cancer. Genome-wide association studies (GWAS) have identified 64 SNPs in 55 independent loci associated with MD in women of European ancestry. Their associations with MD in Asian women, however, are largely unknown.
Using linear regression adjusting for age, BMI, and ancestry-informative principal components, we evaluated the associations of previously reported MD-associated SNPs with MD in a multi-ethnic cohort of Asian ancestry. Area and volumetric mammographic densities were determined using STRATUS (N=2,450) and Volpara™ (N=2,257). We also assessed the associations of these SNPs with breast cancer risk in an Asian population of 14,570 cases and 80,870 controls.
Of the 61 SNPs available in our data, 21 were associated with MD at a nominal threshold of P value < 0.05, all in consistent directions with those reported in European ancestry populations. Of the remaining 40 variants with a P-value of association > 0.05, 29 variants showed consistent directions of association as those previously reported. We found that nine of the 21 MD-associated SNPs in this study were also associated with breast cancer risk in Asian women (P < 0.05), seven of which showed a direction of associations that was consistent with that reported for MD.
Our study confirms the associations of 21 SNPs (19/55 or 34.5% out of all known MD loci identified in women of European ancestry) with area and/or volumetric densities in Asian women, and further supports the evidence of a shared genetic basis through common genetic variants for MD and breast cancer risk.
Keywords: Mammographic density, Asian, Common genetic variation, SNPs
INTRODUCTION
Mammographic Density (MD) refers to the radiologically dense or “white” parts of a mammogram and consists of stroma and glandular tissue. There are population-specific differences in MD. Pre-menopausal Asian women are reported to have higher MD compared to pre-menopausal European women, whereas among post-menopausal women, the converse has been observed (1). MD phenotypes have high heritability, and twin studies have suggested that 60-70% of the variability is genetic, with age, body mass index (BMI), hormone replacement therapy use, menopausal status, parity, and other lifestyle and reproductive factors accounting for the remaining variation (2-4). Recently, two large-scale MD genome-wide association studies (GWAS) identified more than 40 SNPs associated with MD phenotypes, including the absolute dense, percentage dense and non-dense regions on a mammogram, increasing the number of known SNPs by more than four-fold, from 15 to 64 (5-10). Together, the known common genetic variants associated with MD currently account for at least 12% of SNP-based heritability for MD (5).
Mammographic density is a strong risk factor for breast cancer. After accounting for age and BMI, women in the highest quintile of MD are four to five times more likely to develop breast cancer compared to women in the lowest quintile (11-13). Notably, there is shared heritability between MD and breast cancer risk - in women of European descent, at least 40% of the reported MD-associated loci are also known to be associated with breast cancer risk (5), and conversely, 18% of the variants associated with breast cancer risk are also associated with MD (14).
To date, the majority of MD GWAS have involved only women of European ancestry (5-10). Differences in the genetic architecture of MD might contribute the differences in MD distributions among populations. In this study, using a cross-sectional study of healthy women of Asian ancestry, we evaluated the association of MD-associated genetic variants previously identified in women of European descent for their association with MD in Asian women.
METHODS
Study Participants and Data Collection
This study included women recruited into the Malaysian Mammography Study (MyMammo) from Subang Jaya Medical Centre (SJMC) and University Malaya Medical Centre (UMMC) between October 2011 and December 2016. MyMammo, described previously in (15), is a subsidized opportunistic screening mammography programme for Malaysian women aged between 40 and 74 years old without prior history of breast cancer. Women enrolled in the programme donated blood and completed a questionnaire on anthropometric, lifestyle, and reproductive factors, and family history of cancers. MyMammo was approved by the Independent Ethics Committee Ramsay Sime Darby Health Care (reference number: 21109.4) and the Medical Ethics Committee University Malaya Medical Centre (reference number: 1030.8) and all participants provided written informed consent.
Mammographic Density (MD) Assessments
All mammographic images were full-field digital mammograms (FFDM) performed on Hologic (87.5%), GE (7.6%), or Siemens (4.9%) systems. MD measurements were derived using two algorithms: STRATUS (16) derives area-based estimates of total dense area (DA), non-dense area (NDA) and percent density (PDA: dense area/total breast area) from processed images; and Volpara™ (version 1.5.4) (17) derives volumetric measures of dense volume (DV), non-dense volume (NDV) and percent density (PDV: dense volume/total breast volume) from raw images. There were no significant differences between left and right breast side measurements (data not shown) and for this study, we used measurements from the left Cranio-caudal (CC) view mammograms.
Genotyping and Imputation
Genotype calling and quality control procedure and imputation have been described previously by (18). In brief, genotyping was done using one of the two arrays: OncoArray (19) and Illumina HumanOmniExpress v1.1 (20). All data were imputed using the 1000 Genomes Project (Phase 3) data as the reference panel (21), except BioBank Japan, for which the HapMap Phase II (release 22) (22) was used. SNPs with an overall minor allele frequency > 1% and imputation r2 > 0.3 for the studies genotyped on the OncoArray and imputation r2 > 0.7 for the Biobank Japan study were included in this analysis.
Association of SNPs with Mammographic Density (MD) Phenotypes
MD measurements were transformed using Box-Cox transformations, deriving the transformations that gave close approximation to a normal distribution. Cube-root, square-root, and 6th root transformations were used on STRATUS DA, PDA, and NDA, respectively. Natural log transformations were used for Volpara™ DV and PDV, and 4th root transformation for NDV measurements.
A literature search was conducted in April 2022 to identify genetic loci associated at genome-wide significance level (P < 5 x 10−8) with absolute density, absolute non-density, and percent density in previous studies. Linear regression, adjusting for age, BMI, and the first 10 ancestry-informative principal components, was used to determine the associations of the 61 SNPs with the transformed MD phenotypes. The principal components analysis for this study was performed by the OncoArray Consortium and the methods used to compute the ancestry-informative principal components have been described (23). Briefly, ancestry analysis was performed using ancestry informative markers and samples from 1000 Genomes Project, and the contribution of European, Asian, and African ancestry to each individual was derived. In the second stage, a separate principal components analysis was conducted on dataset used in this study to define the Chinese, Malay and Indian subpopulations.
The MyMammo study was genotyped in two batches, hence analyses were conducted by batch and the results were meta-analyzed using the fixed effect inverse variance weighted method with the METAL software (24). For comparability across different MD phenotypes, effect estimates were standardised by dividing the estimated betas and standard errors by the standard deviation of age and BMI adjusted transformed MD phenotypes. SNPs were considered significant if at least one of the following, the corresponding reported MD phenotype, or the equivalent area or volumetric MD phenotype i.e., absolute density (DA/DV), percent density (PDA/PDV) or non-density (NDA/NDV), was significant at P < 0.05 and consistent in direction with those previously reported.
To assess if the lead variant in our study is different from that previously reported in women of European descent, regional association plots were generated using LocusZoom (25) and the LD pattern, as measured by correlation coefficient r2, was estimated using the 1000 Genomes Project ASN dataset (Nov2014, hg19). When no usable LD information was available from LocusZoom, patterns of LD were investigated using LDlink (26), where LD was calculated using the 1000 Genomes Project Asian datasets.
To estimate the proportion of the variance in MD phenotypes explained by significant MD-associated SNPs, we computed the adjusted R-squared for a linear regression model with covariates including age, BMI, and ancestry informative principal components, and subtracted this value from the adjusted R-squared for a model which included all significant MD SNPs and the above co-variates.
Association with Breast Cancer Risk
For association analyses of SNPs and breast cancer risk, the data included (a) 8,245 invasive cases and 7,645 controls of Asian ancestry participating in 2 multi-ethnic case-control studies: the Malaysian Breast Cancer Genetics study (MyBrCa) (15) and the Singapore Breast Cancer Cohort study (SGBCC) (27); and (b) 6,325 invasive cases and 73,225 controls of Asian ancestry from the BioBank Japan (28). The MyBrCa study is a study of sequentially recruited incident and prevalent breast cancer cases recruited from University Malaya Medical Centre and Sime Darby Medical Centre starting from October 2002. MyBrCa was approved by the Medical Ethics Committee of University Malaya Medical Centre (application number: 842.9) and the Independent Ethics Committee of Sime Darby Medical Centre (application numbers: 201109.4 and 201208.1). The SGBCC is a breast cancer cohort with both retrospective and prospective components, recruited from six tertiary hospitals, starting from April 2010. SGBCC was approved by the National Healthcare Group Domain Specific Review Board (reference number: 2009/00501) and the SingHealth Centralised Institutional Review Board (reference number: 2019/2246 [2010/632/B]). The BioBank Japan (BBJ) is a multi-institutional hospital-based registry of incident and prevalent patients identified with any of 47 target diseases. Participants were recruited between June 2003 and March 2008 from 66 hospitals located throughout Japan. Study participants from all the above studies provided written informed consent. We meta-analyzed the GWAS summary statistics using fixed effects inverse variance meta-analysis (METAL). SNPs with minor allelic frequencies (MAFs) < 1% were excluded.
Sensitivity Analyses
Since the Indian population is genetically distinct from the East Asian population, we repeated the association analyses stratified by these two ancestry groups and combined the results using meta-analysis. Indian population was defined as those with ≥ 80% Indian ancestry (N=511 and 505 for the STRATUS and Volpara datasets, respectively), and East Asian population was defined as those with ≤ 20% Indian ancestry (N=1,908 and 1,707 for STRATUS and Volpara datasets, respectively). Heterogeneity between populations was assessed using Cochran’s Q test computed by METAL.
We also performed the regression analyses in the subset of women that were included in both the area-based (STRATUS) and volumetric density (Volpara) studies (N=1,757) and compared with results from the STRATUS (N=2,450) and Volpara (N=2,257) analyses.
RESULTS
Validation of Mammographic Density (MD) Associated Common Genetic Variants
After excluding women who were diagnosed with breast cancer following mammography, had breast implants, had missing raw or processed CC views mammograms, had missing data for age at mammography, BMI, ethnicity, or genotype data, 2,951 women were included in subsequent analyses [Table 1]. The majority of participants were Chinese (57%), postmenopausal (59%), and parous (83%) [Table 1]. We found high correlations between measurements of dense area and dense volume, percent dense area and percent dense volume, and non-dense area and non-dense volume, with correlation coefficients, r of 0.73, 0.70, and 0.91, respectively. Figure S1 shows the distribution of untransformed and transformed area and volumetric mammographic densities.
Table 1.
Characteristics of Study Participants
| Variable | All (N = 2,951) | STRATUS (N = 2,450) | Volpara (N = 2,257) | STRATUS vs. Volpara | |||
|---|---|---|---|---|---|---|---|
| Mean (SD) or N (%) | Mean (SD) or N (%) | Mean (SD) or N (%) | P value | ||||
| Age at mammography (years) | 53.2 | (8.2) | 52.7 | (8.1) | 54.0 | (8.4) | <0.001 |
| BMI (kg/m2) | 25.2 | (4.6) | 25.1 | (4.5) | 25.5 | (4.7) | 0.005 |
| Menopausal status | <0.001 | ||||||
| Premenopausal | 1,203 | (40.8) | 1,047 | (42.7) | 851 | (37.7) | |
| Postmenopausal | 1,748 | (59.2) | 1,403 | (57.3) | 1,406 | (62.3) | |
| Parity status | 0.176 | ||||||
| Nulliparous | 500 | (16.9) | 400 | (16.3) | 402 | (17.8) | |
| Parous | 2,451 | (83.1) | 2050 | (83.7) | 1,855 | (82.2) | |
| Number of live birthsa | 2.8 | (1.2) | 2.3 | (1.5) | 2.3 | (1.5) | 0.949 |
| Age at 1st child birth (years)a | 27.3 | (4.7) | 27.4 | 4(.7) | 27.2 | (4.7) | 0.118 |
| Ethnicityb | <0.001 | ||||||
| Chinese | 1,682 | (57.0) | 1,476 | (60.2) | 1,214 | (53.8) | |
| Indian | 648 | (22.0) | 453 | (18.5) | 529 | (23.4) | |
| Malay | 621 | (21.0) | 521 | (21.3) | 514 | (22.8) | |
| Recruitment centre | <0.001 | ||||||
| Subang Jaya Medical Centre (SJMC) | 1,668 | (56.5) | 1,668 | (68.1) | 988 | (43.8) | |
| University Malaya Medical Centre (UMMC) | 1,283 | (43.5) | 782 | (31.9) | 1,269 | (56.2) | |
| Mammogram system | <0.001 | ||||||
| Hologic | 2,582 | (87.5) | 2,338 | (95.4) | 1,892 | (83.8) | |
| GE | 224 | (7.6) | 112 | (4.6) | 220 | (9.7) | |
| Siemens | 145 | (4.9) | 0 | (0) | 145 | (6.4) | |
Among parous women only.
Self-reported ethnicity.
Sixty-four SNPs from 55 independent MD-associated loci were selected for assessment (5-10). Of the 64 SNPs, 24, 2, 17, 1 and 14 SNPs were previously reported to be associated with DA, DV, PDA, PDV and NDA, respectively, whereas 5 were previously reported to be associated with both DA and PDA and one was reported to be associated with both NDA and PDA. Of these, one SNP (rs150249911) was excluded as the MAF was < 1%, and two SNPs (rs492602 and rs1704773) were excluded as the MAFs were < 1% from the first genotyping batch, and the genotype frequencies deviated from Hardy-Weinberg equilibrium (HWE) (P < 5x10−7) in the second genotyping batch, with departures from HWE observed in Indian women but not East Asian women when tested separately, leaving 61 SNPs from 53 independent loci available for association analyses. The list of SNPs and the corresponding reported MD-phenotype are shown in Table S1(a).
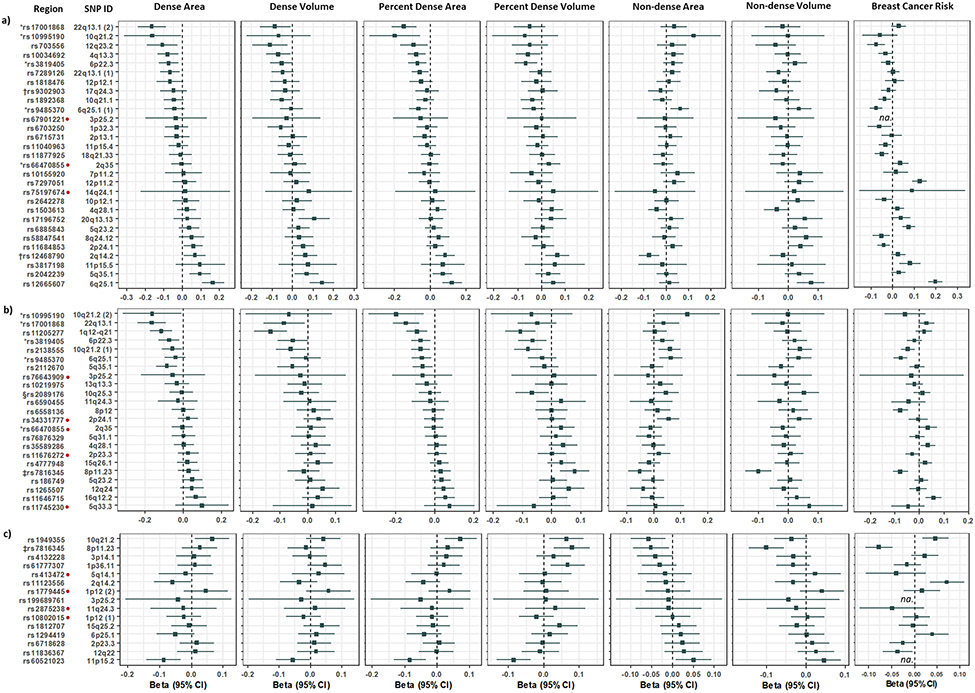
The associations of these SNPs with area (N=2,450) and volumetric densities (N=2,257), adjusting for age, BMI, and ancestry-informative principal components are reported in Figure 1, Table S1(a) and S1(b). Of the 24 testable SNPs that were reported to be associated with absolute density (DA/DV), eight SNPs [33%] were significantly associated with DA/DV in this study [Figure 1(a)]. Of the 17 and 14 testable SNPs that were reported to be associated with percent density (PDA/PDV) or non-density (NDA), respectively, six [35%] and two SNPs [14%], respectively, were significantly associated with the corresponding MD phenotypes [Figures 1(b) and (c)]. Out of the five testable SNPs reported to be associated with both DA and PDA, three were significantly associated with both DA and PDA [60%] while one SNP was significantly associated with PDA but not DA in this study. One testable SNP that was reported to be associated with NDA and PDA, was associated with NDA, NDV and PDV, but not PDA in our study. Of the 61 SNPs evaluated, the strongest associations with MD were observed for variants at CCDC170/ESR1 with DA/PDA/DV, ZNF365 with PDV, SV2A/SF384 with DA/PDA/DV/PDV, KCNU1 with NDV, ARNTL with PDV, and SGSM3 with DA/PDA, and the associations were consistent in direction with those previously reported [Table S1(a), Figure S3]. All of the 21 validated SNPs and 29 [72.5%] of the remaining 40 SNPs with P value > 0.05 showed associations in the same direction as those reported in European women. Together, significant MD SNPs explained 3.5% of the variance in DA, 2.3% of DV, 2.1% of PDA, 1.4% of PDV, 0.3% of NDA, and 0.5% of NDV for women in this study.
Figure 1. Association of SNPs associated with mammographic density in women of European ancestry with MD and breast cancer risk in Asian women.
Associations for SNPs reported to be associated with a) Dense area or volume, b) Percent dense area or volume, c) Non-dense area, in women of European ancestry with area and volumetric densities, and breast cancer risk in Asian women. Associations for SNPs excluded from the breast cancer risk analysis are not available and marked ‘na.’ in the forest plots.
Note: SNPs in each section a), b) and c), are ordered by Beta values of the respective reported association phenotype. For example, forest plots in a) are ordered according to Beta values for dense area in this study. SNPs associated with volumetric densities are annotated for reference. Area measurements were performed using STRATUS and volumetric density measurements using Volpara™.
* SNP reported to be associated with both dense area or percent dense area; † SNP is associated with dense volume; ‡ SNP is associated with non-dense area and percent dense area; § SNP is associated with percent dense volume; Direction of association with MD in this study is not consistent with reported association in women of European ancestry.
We inspected the regional associations for all 21 validated MD-associated SNPs and observed that for 20 of these, the lead variant that was significantly associated with MD in our study is different from that previously reported in women of European descent (Table S4(c)). For associations at seven loci (8 SNPs) , the lead SNP for at least one validated MD phenotype in our study is in LD with the published SNP [4q13.3 (AREG), 5q35.1 (DOCK2/FOXI1), 6q25.1 (CCDC/ESR1), 8p11.23 (KCNU1), 2 SNPs at 10q21.2 (ZNF365), 20q13.13 (SMIM25) and 22q13.1 (SGSM3) , r2 > 0.2 at < 200kb] [Figure S2]. In regions where 11 of the remaining SNPs are located, the lead SNP was not in LD or weakly correlated with the reported SNP in European women (r2 < 0.2), and where 1 of the remaining SNPs is located, the LD information of the lead SNP in Asians was not available.
Association with Breast Cancer Risk
We assessed the associations of 20 out of the 21 validated MD SNPs with breast cancer risk in Asian women, adjusting for age and ancestry-informative principal components [Figure 2 and Table S2]. Nine (45%) out of the 20 SNPs evaluable MD SNPs were also associated with breast cancer risk in Asians (rs11684853 at 2p24.1, rs10034692 at 4q13.3, rs1949355 at 10q21.2 and rs2138555 at 10q21.2 at P < 0.05; rs12665607 at 6q25.1 at P = 2 x 10−31; rs9485370 at 6q25.1 at P = 2 x 10−7; rs7816345 at 8p11.23 at P = 5 x 10−7; rs11646715 at 16q12.2 at P = 4 x 10−4; rs703556 at 12q23.2 at P = 5 x 10−4), seven of which were in directions that were consistent with those for MD [Table S2]. Of these seven, six are within known breast cancer susceptibility loci (29, 30).
Sensitivity Analyses
As our analyses included women of Indian and East Asian ancestry, we conducted a separate analysis by ancestry followed by a meta-analysis, and found that all of the 21 SNPs validated in our original analysis were in consistent directions of association in the meta-analysis. Twenty out of 21 SNPs were significant at P < 0.05 and one had a P value of 0.056 [Table S3(b)]. We also found that in addition to the 21 SNPs, two SNPs, rs1892368 (PRKG1) and rs3817198 (LSP1), were associated with MD in both Indian and East Asian women in the meta-analysis but not in the stratified analyses. Both SNPs did not show significant heterogeneity across the two ancestry groups. In the Indian only subgroup, three additional SNPs were significant at P < 0.05, rs10155920 (ELDR), rs11040963 (MRPL17/OR2AG2), rs4132228 (ADAMTS9), and in the East Asian subgroup one additional SNP was significant at P < 0.05, rs6715731 (TET3/BOLA3). All four SNPs showed considerable heterogeneity across the two ancestry groups (P < 0.10).
Finally, as STRATUS and Volpara measures were available for an overlapping cohort of women, we repeated the meta-analyses in 1,757 women where both the area and volumetric density measures were available. Twenty of the 21 validated SNPs were associated with MD in the same direction as in the all women analysis, 17 of which were at P < 0.05 [Table S4(a), Table S4(b)].
DISCUSSION
MD is a strong and highly heritable risk factor for breast cancer (2-4). In this study, we evaluated the association of 61 common genetic variants previously reported to be associated with MD in women of European ancestry in Asian women. We found that 34.4% (21/61) of the variants tested were associated with MD in consistent directions with the reported associations (P < 0.05). Furthermore, we found that 9 variants associated with MD in this study were also associated with breast cancer risk in Asian women.
The strongest associations with MD for women in this study were observed for variants at CCDC170/ESR1, ZNF365, SV2A/SF384, KCNU1, ARNTL, and SGSM3, and the associations were consistent in direction with those previously reported [Figure S3]. We have previously showed the association of the 6q25.1 (CCDC170/ESR1) locus with area-based density measured using ImageJ in 865 women of Chinese ancestry and our current results show that, in addition to DA and PDA, variants in the 6q25.1 (ESR1/CCDC170) region are also associated with DV. We also validated the association of one of the strongest signals for DA/PDA MD reported in European women, rs10995190 at ZNF365 with DA/PDA/NDA in our study. Notably, the MAFs for this SNP in Chinese and Malay women were lower than that observed for Indian women in our cohort (1.8% and 1.7% compared to 8.6%). The evaluation of this variant in Asian populations has not previously been performed: the minor allele was absent in the smaller all-Chinese study (31). The variant at the SV2A/SF384 locus has been reported to be associated with breast cancer risk, adult height, birth length and PDA in previous studies (32-34), and in this study, we showed that it is associated with four MD phenotypes, namely DA, DV, PDA and PDV. The variant at 8q11.23 (KNCU1) was previously reported to be associated with breast size, breast cancer risk, PDA and NDA in women of European ancestry (35), and in this study, we showed that it is associated with PDV, NDA and NDV. The variant at SGSM3 was previously showed to be associated with breast cancer risk in European and Chinese women, and with DA in European women (36, 37), and in this study, we showed that it is associated with DA and PDA.
The findings from our study support previous evidence of a shared genetic basis for MD and breast cancer risk (5, 7, 8, 10, 38). We found nearly half (9/20) of the SNPs associated with MD in our study were also associated, with breast cancer risk in Asian women (P < 0.05). For 7/9 variants, the association is in the direction predicted from the overall association of MD phenotypes and breast cancer risk. In particular, the directions of association for the 6q25.1 and 10q21.2 variants were consistent for MD and breast cancer risk, supporting the findings of previous studies that the associations of these two loci with breast cancer risk could be mediated through MD (14, 39). Notably, fine mapping of the ESR1 locus shows that there are multiple independent signals for breast cancer risk, only some of which are associated with MD (40).
Surprisingly, the association for the variant at 8q11.23 is in the opposite direction as might be expected for MD and breast cancer risk, with the effect allele of the 8q11.23 variant being associated with lower non-dense volume but reduced breast cancer risk. The same pattern of association has been observed in women of European descent. Further investigations exploring the functional impact of SNPs at this locus may help us understand the conflicting associations observed.
The findings from this study demonstrate the relevance of conducting studies in diverse populations. At seven loci, namely AREG, DOCK2/FOXI1, ESR1/CCDC170, KCNU1, ZNF365, SMIM25 and SGSM3, we found that the lead SNPs in our study were different, albeit correlated to that of the previously reported lead SNPs in Europeans, while in 11 regions, the lead SNPs are weakly or not correlated with the reported SNPs. As the LD pattern may be different between Asians and European populations, fine-mapping and functional studies, utilising data from populations of different ancestries, help to determine the causal variants at these loci.
We could not evaluate three MD-associated SNPs reported at 5q11.2 (ITGA1) and 19q13.33 (FUT2/MAMSTR). The variant at 5q11.2 had very low MAF in our population (0.2%), similar to MAFs reported in East Asians (0.1%) and South Asians (0%) in the 1000 Genomes Project, and variants at 19q13.33 showed deviations from HWE in one genotyping batch which had a multi-ethnic set of samples, and low MAFs in another genotyping batch which comprised of only samples from Chinese women. However, variants correlated with the reported SNPs in these regions did not show significant associations with MD in this study and none of these three SNPs are known to be associated with breast cancer risk (data not shown). These observations suggest that the two abovementioned loci may not play an important role in MD and its associated disease risk in the Asian population.
A potential limitation of this study is that our analyses were based on two fully automated methods for measuring density, STRATUS and Volpara™, whereas the previous analyses in European ancestry women were largely based the semi-automated Cumulus method. However, Cumulus, STRATUS and Volpara™ scores are strongly correlated, and both STRATUS and Volpara™ have been shown to be associated with breast cancer risk with effect sizes that are comparable to those from Cumulus (16, 39, 41-43). Another limitation of this study is the considerably smaller sample size in comparison to the GWASs conducted in women of European ancestry. This may have resulted in insufficient power to validate a larger number of SNPs (44). Notably, there was a small number of women of Indian ancestry in our study (N=505 and N=511 in the STRATUS and Volpara studies, respectively) to enable us to reliably validate the MD SNPs in this population exclusively. The results from our sensitivity analysis suggest that there may be ethnic differences in at least a proportion of the MD-associated SNPs.
Although previous studies in both Western and Asian populations have confirmed a strong inherited component for MD (2, 4, 45, 46), the proportion of variation explained by reported MD-associated SNPs in this study was low, ranging from 0.3-3.5%, depending on the MD phenotype. Breast cancer risk GWAS have benefited from collaborative efforts resulting in very large sample sizes. Further research involving larger studies with good representation of diverse populations is warranted to uncover additional inherited factors influencing MD.
CONCLUSIONS
This Asian study confirmed the associations of 34.5% (19/55) of known MD loci at consistent directions of associations with those reported in European women and furthermore found that nearly half of the evaluable variants were also associated with breast cancer risk in Asian women. However, collectively, these variants account for a small proportion of the heritable component of MD in Asian women and future studies could focus on genome-wide approaches in large studies.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank all study participants and for their participation, and the radiologists, radiographers, support staff and research assistants of Subang Jaya Medical Centre, University Malaya Medical Centre and Cancer Research Malaysia for their contribution and support throughout the duration of this study. The Malaysian Mammographic Density Study and the Malaysian Breast Cancer Genetic Study was supported by grants from Newton-Ungku Omar Fund (grant no: MR/P012930/1), Wellcome Trust (grant no: v203477/Z/16/Z), Malaysian Ministry of Higher Education High Impact Research Grant (grant number: UM.C/HIR/ MOHE/06), and donations from the Sime Darby LPGA Tournament, Estee Lauder Group of Companies, Yayasan Sime Darby, Yayasan PETRONAS, and other donors of Cancer Research Malaysia. SGBCC is funded by NUS Start Up Grant, National University Cancer Institute Singapore (NCIS) Centre Grant [grant no: MRC/CG/NCIS/2010, NMRC/CG/012/2013, CGAug16M005, CG21APR1005], NMRC Clinical Scientist Award [grant no: NMRC/CSA/0048/2013], NMRC Clinician Scientist Award-Senior Investigator [grant no: NMRC/CSA-SI/0015/2017], Asian Breast Cancer Research Fund, Breast Cancer Prevention Programme under Saw Swee Hock School of Public Health, and Breast Cancer Screening and Prevention Programme under Yong Loo Lin School of Medicine. WKH and SM are recipients of the L’Oreal-UNESCO For Women in Science National Fellowship, SM is a recipient of the Ong Hin Tiang and Ong Sek Pek Foundation Postgraduate Scholarship, and JL is the recipient of a National Research Foundation Singapore Fellowship (NRF-NRFF2017-02) and supported by the Agency for Science, Technology and Research (A*STAR). SL was supported by CA244670 from the National Institute of Health.
Footnotes
COMPETING INTERESTS
The Authors declare no Competing Non-Financial Interests but the following Competing Financial Interests: Mikael Eriksson and Per Hall report research grants and a patent on system and method for assessing breast cancer risk using imagery with a license to iCAD, Inc.
DATA AVAILABILITY
The BioBank Japan dataset used in this manuscript is publicly available. All other genotype and phenotype datasets used to support the findings in this manuscript is available from the corresponding author on reasonable request.
REFERENCES
- 1.Rajaram N, Mariapun S, Eriksson M, Tapia J, Kwan PY, Ho WK, et al. Differences in mammographic density between Asian and Caucasian populations: a comparative analysis. Breast Cancer Res Treat. 2017;161(2):353–62. [DOI] [PubMed] [Google Scholar]
- 2.Boyd NF, Dite GS, Stone J, Gunasekara A, English DR, McCredie MR, et al. Heritability of mammographic density, a risk factor for breast cancer. N Engl J Med. 2002;347(12):886–94. [DOI] [PubMed] [Google Scholar]
- 3.Holowko N, Eriksson M, Kuja-Halkola R, Azam S, He W, Hall P, et al. Heritability of Mammographic Breast Density, Density Change, Microcalcifications, and Masses. Cancer Res. 2020;80(7):1590–600. [DOI] [PubMed] [Google Scholar]
- 4.Evans DG, van Veen EM, Howell A, Astley S. Heritability of mammographic breast density. Quant Imaging Med Surg. 2020;10(12):2387–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sieh W, Rothstein JH, Klein RJ, Alexeeff SE, Sakoda LC, Jorgenson E, et al. Identification of 31 loci for mammographic density phenotypes and their associations with breast cancer risk. Nat Commun. 2020;11(1):5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stevens KN, Lindstrom S, Scott CG, Thompson D, Sellers TA, Wang X, et al. Identification of a novel percent mammographic density locus at 12q24. Hum Mol Genet. 2012;21(14):3299–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindström S, Thompson DJ, Paterson AD, Li J, Gierach GL, Scott C, et al. Genome-wide association study identifies multiple loci associated with both mammographic density and breast cancer risk. Nat Commun. 2014;5:5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen H, Fan S, Stone J, Thompson DJ, Douglas J, Li S, et al. Genome-wide and transcriptome-wide association studies of mammographic density phenotypes reveal novel loci. Breast Cancer Res. 2022;24(1):27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernandez-Navarro P, González-Neira A, Pita G, Díaz-Uriarte R, Tais Moreno L, Ederra M, et al. Genome wide association study identifies a novel putative mammographic density locus at 1q12-q21. Int J Cancer. 2015;136(10):2427–36. [DOI] [PubMed] [Google Scholar]
- 10.Brand JS, Li J, Humphreys K, Karlsson R, Eriksson M, Ivansson E, et al. Identification of two novel mammographic density loci at 6Q25.1. Breast Cancer Res. 2015;17:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Habel LA, Lipson JA, Achacoso N, Rothstein JH, Yaffe MJ, Liang RY, et al. Case-control study of mammographic density and breast cancer risk using processed digital mammograms. Breast Cancer Res. 2016;18(1):53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCormack VA, dos Santos Silva I. Breast density and parenchymal patterns as markers of breast cancer risk: a meta-analysis. Cancer Epidemiol Biomarkers Prev. 2006;15(6):1159–69. [DOI] [PubMed] [Google Scholar]
- 13.Boyd NF, Guo H, Martin LJ, Sun L, Stone J, Fishell E, et al. Mammographic density and the risk and detection of breast cancer. N Engl J Med. 2007;356(3):227–36. [DOI] [PubMed] [Google Scholar]
- 14.Stone J, Thompson DJ, Dos Santos Silva I, Scott C, Tamimi RM, Lindstrom S, et al. Novel Associations between Common Breast Cancer Susceptibility Variants and Risk-Predicting Mammographic Density Measures. Cancer Res. 2015;75(12):2457–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan MM, Ho WK, Yoon SY, Mariapun S, Hasan SN, Lee DS, et al. A case-control study of breast cancer risk factors in 7,663 women in Malaysia. PLoS One. 2018;13(9):e0203469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eriksson M, Li J, Leifland K, Czene K, Hall P. A comprehensive tool for measuring mammographic density changes over time. Breast Cancer Res Treat. 2018;169(2):371–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.H R, B M, Y MJ, K N, H J, editors. Robust Breast Composition Measurement - Volpara IWDM’10: Proceedings of the 10th international workshop on Digital Mammography; 2010; Berlin, Heidelberg: Springer-Verlag [Google Scholar]
- 18.Ho WK, Tai MC, Dennis J, Shu X, Li J, Ho PJ, et al. Polygenic risk scores for prediction of breast cancer risk in Asian populations. Genet Med. 2022;24(3):586–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michailidou K, Lindström S, Dennis J, Beesley J, Hui S, Kar S, et al. Association analysis identifies 65 new breast cancer risk loci. Nature. 2017;551(7678):92–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Low SK, Chin YM, Ito H, Matsuo K, Tanikawa C, Matsuda K, et al. Identification of two novel breast cancer loci through large-scale genome-wide association study in the Japanese population. Sci Rep. 2019;9(1):17332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449(7164):851–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amos CI, Dennis J, Wang Z, Byun J, Schumacher FR, Gayther SA, et al. The OncoArray Consortium: A Network for Understanding the Genetic Architecture of Common Cancers. Cancer Epidemiol Biomarkers Prev. 2017;26(1):126–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015;31(21):3555–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ho PJ, Yeoh YS, Miao H, Lim SH, Tan EY, Tan BKT, et al. Cohort profile: The Singapore Breast Cancer Cohort (SGBCC), a multi-center breast cancer cohort for evaluation of phenotypic risk factors and genetic markers. PLoS One. 2021;16(4):e0250102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagai A, Hirata M, Kamatani Y, Muto K, Matsuda K, Kiyohara Y, et al. Overview of the BioBank Japan Project: Study design and profile. J Epidemiol. 2017;27(3S):S2–S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shu X, Long J, Cai Q, Kweon SS, Choi JY, Kubo M, et al. Identification of novel breast cancer susceptibility loci in meta-analyses conducted among Asian and European descendants. Nat Commun. 2020;11(1):1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Key TJ, Appleby PN, Reeves GK, Roddam AW, Group EHaBCC. Insulin-like growth factor 1 (IGF1), IGF binding protein 3 (IGFBP3), and breast cancer risk: pooled individual data analysis of 17 prospective studies. Lancet Oncol. 2010;11(6):530–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mariapun S, Ho WK, Kang PC, Li J, Lindström S, Yip CH, et al. Variants in 6q25.1 Are Associated with Mammographic Density in Malaysian Chinese Women. Cancer Epidemiol Biomarkers Prev. 2016;25(2):327–33. [DOI] [PubMed] [Google Scholar]
- 32.Gudbjartsson DF, Walters GB, Thorleifsson G, Stefansson H, Halldorsson BV, Zusmanovich P, et al. Many sequence variants affecting diversity of adult human height. Nat Genet. 2008;40(5):609–15. [DOI] [PubMed] [Google Scholar]
- 33.Paternoster L, Howe LD, Tilling K, Weedon MN, Freathy RM, Frayling TM, et al. Adult height variants affect birth length and growth rate in children. Hum Mol Genet. 2011;20(20):4069–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang B, Shu XO, Delahanty RJ, Zeng C, Michailidou K, Bolla MK, et al. Height and Breast Cancer Risk: Evidence From Prospective Studies and Mendelian Randomization. J Natl Cancer Inst. 2015;107(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eriksson N, Benton GM, Do CB, Kiefer AK, Mountain JL, Hinds DA, et al. Genetic variants associated with breast size also influence breast cancer risk. BMC Med Genet. 2012;13:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan T, Zhang K, Chen W. Genetic variants of ESR1 and SGSM3 are associated with the susceptibility of breast cancer in the Chinese population. Breast Cancer. 2017;24(3):369–74. [DOI] [PubMed] [Google Scholar]
- 37.Michailidou K, Hall P, Gonzalez-Neira A, Ghoussaini M, Dennis J, Milne RL, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet. 2013;45(4):353–61, 61e1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brand JS, Humphreys K, Li J, Karlsson R, Hall P, Czene K. Common genetic variation and novel loci associated with volumetric mammographic density. Breast Cancer Res. 2018;20(1):30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lindström S, Vachon CM, Li J, Varghese J, Thompson D, Warren R, et al. Common variants in ZNF365 are associated with both mammographic density and breast cancer risk. Nat Genet. 2011;43(3):185–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dunning AM, Michailidou K, Kuchenbaecker KB, Thompson D, French JD, Beesley J, et al. Breast cancer risk variants at 6q25 display different phenotype associations and regulate ESR1, RMND1 and CCDC170. Nat Genet. 2016;48(4):374–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Astley SM, Harkness EF, Sergeant JC, Warwick J, Stavrinos P, Warren R, et al. A comparison of five methods of measuring mammographic density: a case-control study. Breast Cancer Res. 2018;20(1):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eng A, Gallant Z, Shepherd J, McCormack V, Li J, Dowsett M, et al. Digital mammographic density and breast cancer risk: a case-control study of six alternative density assessment methods. Breast Cancer Res. 2014;16(5):439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jeffreys M, Harvey J, Highnam R, Martí J, Oliver A, Freixenet J, et al. Comparing a New Volumetric Breast Density Method (VolparaTM) to Cumulus. International Workshop on Digital Mammography: Springer Berlin Heidelberg; 2010. p. 408–13. [Google Scholar]
- 44.Brentnall AR, Warren R, Harkness EF, Astley SM, Wiseman J, Fox J, et al. Mammographic density change in a cohort of premenopausal women receiving tamoxifen for breast cancer prevention over 5 years. Breast Cancer Res. 2020;22(1):101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stone J, Dite GS, Gunasekara A, English DR, McCredie MR, Giles GG, et al. The heritability of mammographically dense and nondense breast tissue. Cancer Epidemiol Biomarkers Prev. 2006;15(4):612–7. [DOI] [PubMed] [Google Scholar]
- 46.Sung J, Song YM, Stone J, Lee K, Jeong JI, Kim SS. Genetic influences on mammographic density in Korean twin and family: the Healthy Twin study. Breast Cancer Res Treat. 2010;124(2):467–74. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The BioBank Japan dataset used in this manuscript is publicly available. All other genotype and phenotype datasets used to support the findings in this manuscript is available from the corresponding author on reasonable request.