ABSTRACT
The COVID-19 pandemic spurred the development of innovative solutions for specimen collection and molecular detection for large-scale community testing. Among these developments is the RHINOstic nasal swab, a plastic anterior nares swab built into the cap of a standard matrix tube that facilitates automated processing of up to 96 specimens at a time. In a study of unsupervised self-collection utilizing these swabs, we demonstrate comparable analytic performance and shipping stability compared to traditional anterior nares swabs, as well as significant improvements in laboratory processing efficiency. The use of these swabs may allow laboratories to accommodate large numbers of sample collections during periods of high testing demand. Automation-friendly nasal swabs are an important tool for high-throughput processing of samples that may be adopted in response to future respiratory viral pandemics.
KEYWORDS: scaled testing, specimen collection, respiratory pathogens
INTRODUCTION
The COVID-19 pandemic underscored the critical need for widespread, accessible, and affordable testing to combat an ongoing pandemic and monitor the community to prepare for a future epidemic or pandemic. The gold standard for diagnostic testing for COVID-19 and other respiratory pathogens is RT-qPCR, a molecular test that detects viral genetic material. As such, the pandemic also made clear that all the required supplies—from the swabs to the primers—can be severely impacted by supply chain disruptions. In addition, labor shortages exacerbated by pandemic conditions limited the number of trained personnel.
The nasal swab is a key component of specimen collection for RT-qPCR testing. While nasopharyngeal (NP) or oropharyngeal (OP) swabs were initially the standard for sample collection, several laboratories subsequently validated mid-turbinate (MT) and anterior nares (ANS) swabs as alternatives (1, 2). This allowed for faster and less invasive testing, circumvented supply chain bottlenecks, and, of particular importance, allowed participants to collect their own swabs in an unsupervised manner and send them to a laboratory without an in-person visit. These studies, along with the need for mass testing, spurred alternative swab designs that were mass-producible, easy-to-use, and laboratory-efficient. Among these was the RHINOstic automation-ready nasal swab, which became available in the summer of 2022 (3). The swab is made with an injection-molded polypropylene, as opposed to polyester or foam; the cap-integrated anterior nares swab and the collection tube are also designed to be automation-friendly, fitting into standard laboratory 96-well format. As such, the RHINOstic swabs may serve as an important alternative to standard swab collection methods.
Here, we describe the use of RHINOstic swabs to facilitate large-scale SARS-CoV-2 testing at a public university and in a community study in Seattle, Washington, USA, during the COVID-19 pandemic. We assessed these swabs on campus and within the greater Seattle community in terms of their ease of use and ability to collect robust specimens in unsupervised swabbing. We compared the analytical performance of RHINOstic swabs to US Cotton #3 ANS, a spun polyester swab, in an extraction-free RT-qPCR workflow, assess winter and summer shipping stability, assess their real-world performance by addressing their failure rates for RT-qPCR and sequencing pipelines, and evaluate their time-savings in a high-throughput SARS-CoV-2 testing facility. Lastly, we show how we integrated RHINOstic swabs into our workflow, including modifications of laboratory equipment and our accessioning process to allow for increased flexibility to adapt to massive swings in demand.
MATERIALS AND METHODS
Usability study: recruitment and sample collection
We recruited study participants to test the usability of RHINOstic swabs and provide user-experience feedback. We first recruited participants via the University of Washington-sponsored SARS-CoV-2 testing platform, the Husky Coronavirus Testing (HCT) research program, at on-campus collection locations on the Seattle, WA campus (protocol STUDY00011148). The second recruitment method occurred within the greater Seattle-area community via email, Facebook, and Twitter, as part of the ongoing Seattle Coronavirus Assessment Network (SCAN) study (protocol STUDY00010432). Eligibility criteria included participants with priority codes for enrollment or who had been previously identified as eligible to participate in the study through word-of-mouth, and the ability to understand and comply with study procedures, including a video call, in English. Exclusion criteria included participants (or both parents of a participating child) who had prior medical or laboratory training, and COVID-like symptoms (fever, cough, or shortness of breath) that could not be explained by another condition. Once recruited, study staff provided informed consent, and participants (or their guardians) completed an enrollment and illness questionnaire (Fig. S1) to ensure they met eligibility criteria. All eligible participants received two swab kits consisting of the necessary items needed to collect a specimen with a US Cotton #3 ANS swab and a RHINOstic ANS swab. US Cotton #3 swabs were used as the baseline due to their use in our laboratory’s clinical COVID-19 testing protocol. The US Cotton swabbing supplies included an individually packaged US Cotton #3 polyester swab and an empty, prelabeled 10 mL sample tube for transport; the RHINOstic swabbing supplies included an individually packaged RHINOstic polypropylene swab and an empty, prelabeled 1 mL sample tube (Thermo Fisher, #3740) for transport. Participants were provided with printed instructions on how to correctly collect both swab specimens (Fig. S2 and S3) themselves or from their child. Participants were additionally instructed to collect the US Cotton #3 swab first, as it was the clinically validated test at the time. All collections were observed by study staff either in person (HCT) or by teleconference (SCAN) but participants were not given further instructions or corrections during the collection process. Mistakes in specimen collection or problems encountered by a participant during the collection were recorded by observers.
After finishing the swab collection, all participants completed a Swab Collection Survey in REDCap (4) in which they were asked about their swabbing experience and to report any adverse events. In addition, participants were required to register their specimens in REDCap by entering the barcodes on the sides of each tube. Kits were returned to the laboratory by study staff (for HCT participants) or by courier (for SCAN), unpackaged, accessioned into our Laboratory Inventory Management System (LIMS), and processed. Specimens were transported at ambient temperatures, typically returned the same day or the following day, and processed within 2 days from the time of collection. Any irregularities in packaging or specimens (e.g., blood or bent swabs) were recorded by laboratory staff.
Usability study: specimen processing
All specimen processing occurred in a Class II biosafety cabinet. Specimens were accessioned by a technician in a class II biosafety cabinet using our custom Laboratory Information Management System (LIMS); for identification, the US Cotton swabs utilized two barcode stickers, while the RHINOstic swab utilized one barcode sticker and the integrated 2D barcode on the bottom of the 1 mL sample tube.
Each sample was suspended in TRIS EDTA [(low-TE; 10 mM Tris-HCl pH 7.5 (T2319-1L, Sigma), 0.1 mM EDTA (15575020, Invitrogen)] within its transport tube; the US Cotton swabs received 1 mL of solution due to the absorbency of the swab and to allow for two aliquots, while the RHINOstic swab received 300 uL. Samples were then agitated at room temperature on a plate shaker (Bio-Rad, Benchtop Shaking Incubator) at top speed for 1 minute, then allowed to sit for 10 minutes. The US Cotton swab eluate was aliquoted into two 1 mL sample tubes—one for testing, one kept as a backup for repeat testing or research—to work with our automated processes; each aliquot tube was also entered into the LIMS to link to the original sample (Fig. S4). Both the aliquoted US Cotton samples and RHINOstic samples were kept in a latch rack (ThermoFisher, catalog #4898-BR) at 4°C until they were prepared for proteinase digestion and heat treatment (5), followed by RT-qPCR.
General laboratory workflow: RHINOstic swab accessioning and processing
Specimens were delivered to the laboratory in latch racks from on-campus, observed testing locations or in clear biohazard bags (Associated Bag, 14–93) collected in drop boxes located around campus for participants electing to swab unobserved. Specimens were accessioned by a technician in a class II biosafety cabinet using our custom Laboratory Information Management System (LIMS) and were examined for viability. Discard criteria for specimens include broken or missing swabs, timed-out/expired specimens (any specimen that was collected more than 48 hours before accessioning), foreign material in the tubes, excessive blood, or failure of participants to link their university ID to the collection ID barcode (Fig. S4). Once cleared for processing, the specimen was then placed into a labeled latch rack. The latch rack was stored at 4°C until filled.
A full latch rack consisting of 96 RHINOstic swabs (94 clinical samples and two low TE controls) was decapped using a Capit-All Screw Cap Tube Capper/Decapper (Thermo Scientific, 4111MAT), fitted with a custom 3D-printed adapter for handling the RHINOstic swabs. Using a 96-well pipettor (Liquidator 96, Mettler Toledo), each specimen in the rack was eluted with 300 µL of low-TE, and then recapped. Each latch rack was agitated on a vortex for 1 minute and incubated for 10 minutes at room temperature before going into proteinase digestion and heat inactivation.
Proteinase K digestion and heat inactivation
After elution, specimens were again decapped with the Capit-All. Using the Liquidator, 50 uL of the specimens was added to a premade 96-well plate (Bio-Rad, HSP9601) containing 5 uL of proteinase K (Thermo Fisher, A42363) per well. The plate was sealed with foil (Eppendorf 0030127854 and 5392000013), incubated at 37°C for 15 minutes in a convection oven (Across International 0853924003042), and then transferred to a second oven for heat inactivation at 95°C for 15 minutes.
RT-qPCR
Each RT-qPCR reaction was performed at a final volume of 10 uL, and each assay was performed in technical duplicate on a 384-well plate (Thermo Fisher, 4309849) for a total of 4 RT-qPCR wells per sample. The reaction mix contained 1 uL TaqPath RT-qPCR MasterMix (Life Technologies, PN A15300), 0.125 uL RNase P TaqMan VIC assay (Life Technologies, A30064), 1 uL SARS-CoV-2 ORF1b FAM assay (PN 4332079, Life Technologies assay no. APGZJKF) or 1 uL spike (S) gene (PN 4332079, Life Technologies assay no. APXGVC4), and nuclease-free water (Thermo Fisher, 1907076). Primer sequences were designed against the Wuhan-Hu-1 sequence (MN908947.3) and are proprietary to Thermo Fisher. Using a Liquidator, 5 uL of specimen was added to each well. Plates were sealed using optically clear microseal B (Biorad). RT-qPCR was then performed on the Applied Biosystems QuantStudio 6 Pro (25°C for 2 minutes, 50°C for 15 minutes, 98°C for 3 minutes, followed by 40 cycles of 98°C for 3 seconds and 60°C for 30 seconds). Reported cycle threshold (Ct) values were obtained from the onboard analysis using predetermined thresholds. Positive controls contained purified nucleic acid with a sequence that was amplified by the ORF1b and spike gene assays.
Capit-all modifications for RHINOstic swabs
To immediately accommodate the RHINOstic swabs in our testing pipeline, we required that the swabs be compatible with the Thermo Scientific Capit-All Screw Cap Tube Capper/Decapper. Decapping a rack of RHINOstic swabs using the original decapper platform resulted in the tips of the swabs remaining in the tubes (Fig. S5A). This prevented the rack of tubes from being removed from the Capit-All. To solve this problem, a modified decapper platform was designed to work with a jack to lower the tubes for removal from the instrument and to raise it again for decapping (Fig. S5B). The final modified decapper platform and support were designed in SolidWorks and 3D printed using white PLA filament (HATCHBOX) (Fig. S5C). The modified decapper platform was then combined with the removable base and an adjustable metal scissor jack for toy cars (Mekek, B08Y1MPW6Y). A support was added to be used during decapping and recapping to prevent damaging the jack. The design and material of the RHINOstic swabs ensure that drippage from the swabs is extremely unlikely; however, to clinically validate the modified decapper platform and ensure no cross-contamination took place, a plate with known negatives and positives in a checkerboard pattern was prepared for RT-qPCR using the modified decapper platform. The results showed no cross-contamination between wells (Fig. S6).
RHINOstic limit of detection
We tested the analytical sensitivity of our laboratory-developed test by spiking the UV-inactivated SARS-CoV-2 virus (Zeptometrix, 0810587UV) onto RHINOstic swabs. Six healthy volunteers collected clinical matrix swabs by self-swabbing. One swab from each volunteer was tested to confirm SARS-CoV-2 negativity. Using a micropipette, 12 swabs (two swabs from each volunteer) were spiked at six different viral concentrations with the UV-inactivated SARS-CoV-2 virus. Spiked swabs were allowed to dry for approximately 30 minutes, and then were processed as described above.
Shipping stability study
We validated the use of RHINOstic swabs for home-based specimen collection across a range of temperatures and days, mimicking a winter and summer season shipping profile according to the International Air Transport Association (IATA) guidelines. For each season, 120 RHINOstic swabs were removed from their packaging and screwed into a matrix tube. A synthetic clinical matrix was prepared by mixing total nucleic acids extracted from a human cell line with artificial nasal mucus (Biochemazone, BZ253) and held on ice. Each RHINOstic swab was then dipped and swirled into a microfuge tube containing the synthetic clinical matrix. Swabs were allowed to dry prior to reinserting them into the matrix tube. Dipped swabs were stored at 4°C until spiked with the virus.
Next, two previously identified SARS-CoV-2-positive clinical specimens were removed from 4°C storage. Specimens were chosen based on their respective Ct values to create “high positive” swabs, targeting final Ct values > 30, and “low positive” swabs, targeting final Ct values < 27. A composite SARS-CoV-2-negative specimen was made by combining 25 µL each from previously identified negative SARS-CoV-2 clinical specimens. RHINOstic™ swabs were spiked with 10 µL of either “high positive,” “low positive,” or negative specimen in a class II biosafety cabinet with a micropipette and allowed to dry for approximately 30 minutes. 20 high-positive swabs, 10 low-positive swabs, and 10 negative swabs were treated for five and seven heat or cold cycles or immediately eluted to generate baseline Ct values by RT-qPCR for the stability experiment (Fig. S7 ; Tables S1 to S6). Ct values were analyzed for each set of specimens assayed throughout the experiment.
Comparing laboratory processing efficiency of US Cotton #3 vs RHINOstic swabs
Start and end times for specific steps to accession and process swabs were compared between US Cotton and RHINOstic. For the US Cotton swabs, steps included hydration in low TE, incubation time of 10 minutes, accessioning of samples into the LIMS and immediately aliquoting into matrix tubes, and loading of samples onto a 96-well plate; for the RHINOstic swabs, steps including accessioning into the LIMS, hydration in low TE, incubation time of 10 minutes, and loading of samples onto a 96-well plate. The number of specimens was recorded and used to calculate the total handling time. Three separate technicians were timed for each step in both workflows and the average times of each step were compared to evaluate the difference in handling time.
Extraction of RNA and genomic sequencing
Viral genome sequencing was attempted on SARS-CoV-2-positive specimens with a high quantity of SARS-CoV-2 RNA, Orf1b Ct ≤30. Total nucleic acids were extracted (Magna Pure 96, Roche) from 200 uL of the eluted specimen, and sequencing libraries were prepared (Illumina COVIDSeq kit) and sequenced (Illumina NextSeq2000 P200 kit). Consensus genomes were assembled against the SARS-CoV-2 reference genome Wuhan/Hu-1/2019 (Genbank accession MN908947, https://www.ncbi.nlm.nih.gov/nuccore/mn908947) using a modified iVar pipeline42 and deposited to GenBank and GISAID.
RESULTS
RHINOstic swabs are compatible with unsupervised anterior nares specimen collection
We chose to adopt the RHINOstic swabs because they are automation-friendly and cost-effective (3). However, a key concern in unsupervised testing is the ability of participants to collect their own specimens comfortably and robustly. The self-collection of ANS swabs has been demonstrated to be effective compared to swabs collected by a healthcare professional (6); however, while the RHINOstic swabs are clinically viable (3), their use in a large-scale unsupervised testing platform and real-world performance had not been demonstrated. Therefore, we assessed the RHINOstic swabs across two COVID-19 research studies, employing either supervised self-collection or unsupervised, home-based self-collection, to evaluate ease of use and specimen quality. We assessed usability metrics and specimen quality from 69 participants between 6 months and 67 years of age, and across sex, household income, and race (Table S7 to S10). Participants swabbed themselves or their child (if under the age of 13 years) while being observed either in-person or virtually by clinical study coordinators. After collection, all study participants or their guardians completed a survey reporting their level of confidence, kit ease of use, and level of discomfort experienced while swabbing.
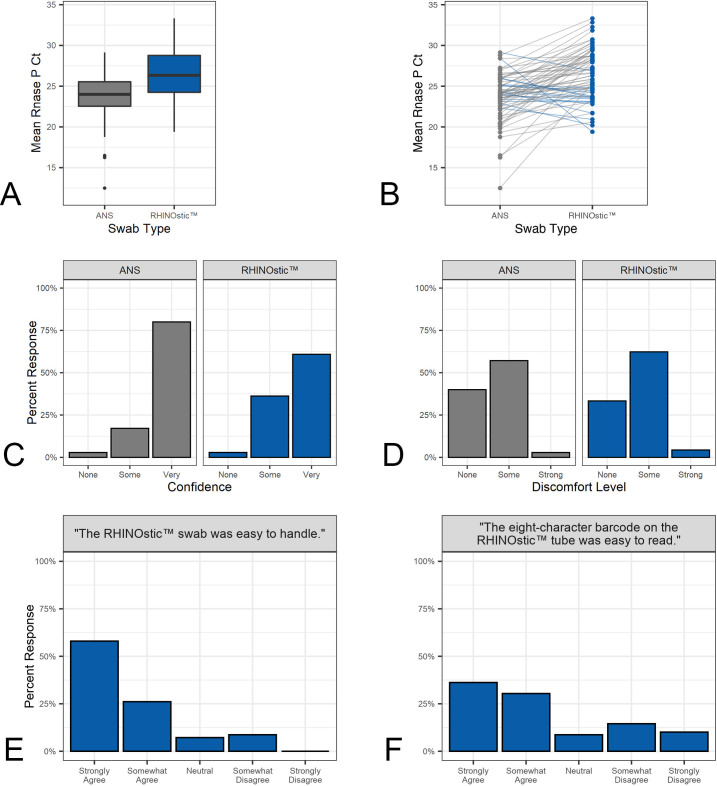
We found that at-home participants were able to collect high-quality specimens with RHINOstic swabs. Directions for specimen collection for the RHINOstic swab consisted of three steps: (1) insert the swab into the nostril and rotate it three times, (2) move the swab up and down once, and (3) hold it against the side of the nostril for 10 seconds. This procedure was completed for both nostrils. Most participants followed the instructions correctly (65/69); three participants failed to move the swab up and down and only one participant failed to swab both nostrils. Participants were most likely to miss or shorten the “hold for 10 seconds” step, with 4/69 participants failing to do this properly (Table S11). Despite these variations, no negative effect on specimen collection was demonstrated. Every specimen contained sufficient human material for SARS-CoV-2 testing based on measurements by RNase P RT-qPCR assay (Fig. 1A and B), in which a specimen was deemed acceptable based on a RNase P Ct value under 36. Likewise, there was no correlation between the self-reported demographics of age, sex, household income, or race and the ability of participants to collect a sufficient swab.
Fig 1.
Comparison of usability and analytical performance metrics between US Cotton #3 anterior nares swabs and RHINOstic swabs. (A) Boxplot of Rnase P Ct values from the 69 US Cotton and RHINOstic swabs. (B) Dotplot comparison of RNase P Ct values. (C) Barplot comparison of participants’ self-reported confidence that they had collected the swab correctly. (D) Barplot comparison of participants’ self-reported discomfort levels using the swab for collection. (E) Barplot distribution of participants’ self-reported overall ease handling the RHINOstic swab. (F) Barplot distribution of participants self-reported ease reading the eight-character barcode on the RHINOstic tube.
In addition to the sample quality, participants found the swabs relatively comfortable and easy to use (Fig. 1C through 1E). Only 2/69 participants reported “no confidence” that they collected the specimen correctly. A chi-squared contingency test revealed no significant difference in discomfort levels in RHINOstic swab users from those reported by US Cotton swab users (χ2 = 0.527, df = 2, P = 0.77). 62% and 4% of participants found the RHINOstic swabs somewhat or very uncomfortable, respectively, compared to 57% and 3% for US Cotton swab users. The most significant problem was that nearly 25% of participants (17/69) found the eight-character barcode on the tube difficult to read due to a small type size (Fig. 1F); the small size of the tube limits the size of the print for barcodes and other identifiers. To address the barcode size problem for a household-based study that our laboratory supports, we provided an inexpensive magnifying glass with their “welcome kit” (7).
Analytical performance of RHINOstic swabs
To test the performance of the RHINOstic swabs with our clinical assay, we performed a limit of detection (LoD) analysis. The LoD is the minimum number of SARS-CoV-2 RNA molecules that could be detected in greater than 95% of RT-qPCR reactions, wherein each reaction was performed 24 times in replicate at varying concentrations. To generate contrived specimens to test the LoD, we inoculated RHINOstic swabs with clinical matrix collected from a healthy volunteer using dilutions of UV-inactivated SARS-CoV-2. These experiments determined the RT-qPCR analytical sensitivity to be 15.63 molecules/reaction for the Orf1b assay and 31.25 molecules/reaction for the S-gene (spike gene) assay (Tables S1 and S2). This LoD is comparable to the LoD of many other RT-qPCR-based tests that have been issued Emergency Use Authorization from the FDA (8), as well as that of RHINOstic’s own validation study (3).
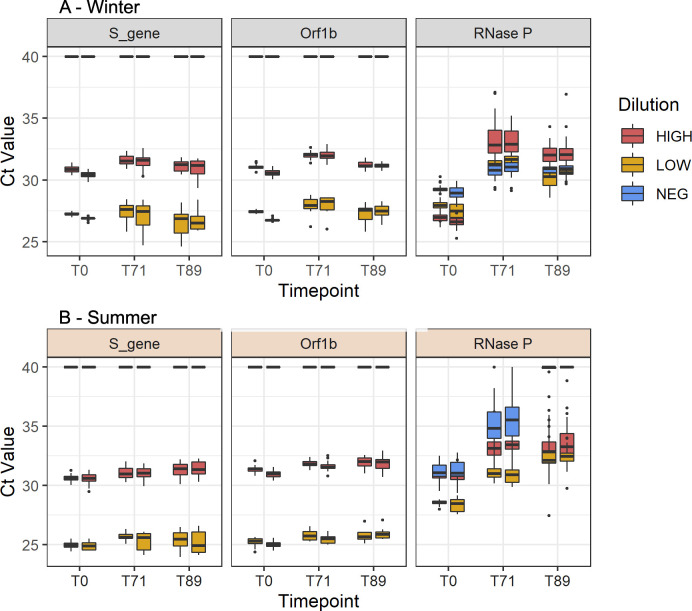
Stability of specimens on RHINOstic swabs is acceptable through mock winter and summer conditions
Our testing laboratory supports studies that employ a swab-and-send mechanism in which specimens are collected at home and then shipped to the laboratory (7, 9, 10). Thus, we tested the stability of SARS-CoV-2 and human RNase P on RHINOstic swabs through the winter and summer temperature profiles outlined by IATA. The temperature profiles mimicked the more extreme temperatures that a specimen may experience in the Pacific Northwest when being shipped from a home collection site to the testing laboratory (see Materials and methods). Overall, SARS-CoV-2 targets remained relatively stable when cycled through winter and summer shipping conditions for up to 89 hours with a maximum ΔCt of 1.17 (Fig. 2; Tables S3to S6). The detection of the human RNase P gene was variable, with increased Ct values after cycling through various temperature regimes. However, we were still able to reliably detect RNase P within standard ranges (Ct <36) for winter experimental samples at the 71 and 89 hours post-collection timepoint. During the summer stability experiment, RNase P detection at the 71-hour timepoint was within standard ranges and acceptable. However, at the 89-hour timepoint, RNase P amplification was poor, with many samples failing RNase P detection. Thus, with the summer stability timepoint as a limiting factor, the stability of the swabs was determined to be 71 hours post-collection.
Fig 2.
Performance of SARS-CoV-2 and RNase P assays during the winter (A) and summer (B) stability experiments.
Real-world performance of RHINOstic swabs for RT-qPCR and viral genome sequencing in comparison to US Cotton #3
We have used RHINOstic swabs in the University of Washington’s Husky Testing Research Program (9) and CASCADIA (7), a large household-based vaccine efficacy study, collecting and testing 184,368 specimens between 22 August 2021 and 31 March 2023. These specimens have demonstrated a low failure rate for both RT-qPCR testing and sequencing. Specimens were considered to have failed for insufficient specimen collection on the RT-qPCR assay when the RNase P control was above a detection threshold of Ct >36. Specimens that failed were automatically rerouted for repeat testing; if PCR failed again, the samples were considered failed overall as we could not discriminate between inhibition and insufficient collection. Failure rates on the RT-qPCR assay were compared retrospectively for over 200,000 US Cotton or RHINOstic specimens tested in our laboratory from 2020-11-20 to 2022-05-18. We saw that the failure remained low for RHINOstic swabs, 20 out of 73281 (0.03%) US Cotton swabs failed the assay compared to 227 out of 107131 (0.21%) RHINOstic swabs (Table 1). Across studies, we attempt to sequence SARS-CoV-2 genomes from positive specimens. Among positives, the number of specimens that met sequencing criteria (mean SARS-CoV-2 Ct <30) was comparable between those collected with US Cotton (60%) vs RHINOstic (57%) swabs as was the quality of genomes. We consider genome assemblies with >10% Ns or less than 20× coverage to be low quality. Of 3,224 specimens for which sequencing was attempted, 53 out of 421 (12.6%) of US Cotton swabs failed to generate high-quality genomes compared to 210 out of 2,803 (7.4%) RHINOstic swabs (Table 1).
TABLE 1.
Failure rate for specimens collected with each swab type from November 2020 to June 2022
| US cotton | RHINOstic | US cotton | RHINOstic | |
|---|---|---|---|---|
| RT-qPCR | Viral genome Sequencing | |||
| # Failed | 20 | 227 | 53 | 210 |
| # Passed | 73261 | 106904 | 368 | 2593 |
| % Failure | 0.03% | 0.21% | 12.59% | 7.49% |
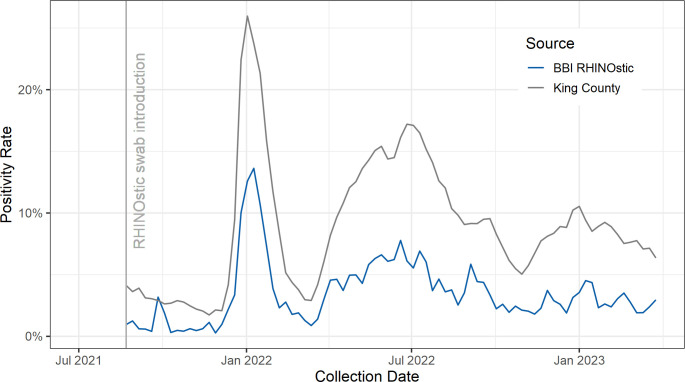
RHINOstic swabs have performed as expected for collecting specimens with detectable SARS-CoV-2. The SARS-CoV-2 positivity rate from RHINOstic swabs has closely followed the trend in our region of King County, Washington (Fig. 3) (11). As our laboratory supports community-based research testing programs rather than a clinical setting, we have historically reported a lower positivity rate compared to the broader population (9).
Fig 3.
Comparison of the weekly positivity rate for SARS-CoV-2 between our laboratory’s RHINOstic specimens and King County, Washington, beginning from deployment of RHINOstic swabs the week of 2021-08-22.
RHINOstic swabs allow a 3.5X gain in laboratory processing efficiency per specimen
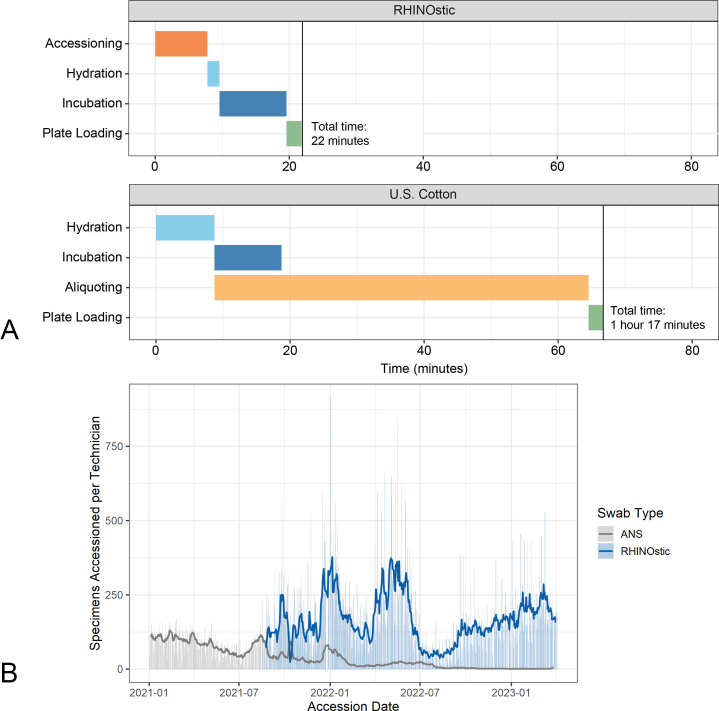
Demand for testing swung wildly during the COVID-19 pandemic. Our laboratory’s adoption of automation-friendly RHINOstic swabs allowed us to gain the efficiency required to be able to flex our capacity to serve our community during periods of high demand. RHINOstic swabs fit the 96-well format that is standard in high-throughput molecular biology laboratories. However, the integrated cap-swab combination required some innovation to use our automated decapping instrument. We devised a workaround using a 3D-printed block and a jack for miniature model cars to modify a standard Thermo Scientific Capit-All Decapper to function with RHINOstic swabs (Fig. S5). Most other equipment only required minor tweaks to incorporate these RHINOstic swabs.
Once we adapted our instruments to RHINOstic swabs, we directly compared hands-on accessioning and processing times for US Cotton swabs vs RHINOstic to illustrate the time-saving benefits (Fig. 4A). Using our established workflow, our technicians were able to process 3.5 racks or 329 RHINOstic swabs, in the time that it takes to process a single rack of 94 US Cotton #3 swabs. The efficiency gains from this workflow resulted in a dramatic increase in specimens processed per technician per day and allowed our laboratory the flexibility to intake thousands of specimens per day at the height of the Omicron waves in the winter of 2021–22 and spring 2022 (Fig. 4B).
Fig 4.
(A) Gantt chart demonstrating the average time to complete technician steps prior to heat inactivation of 96 samples, measured in minutes. Where the US Cotton swabs require an aliquoting step, which is done concurrently with the accessioning step (tubes are scanned and then aliquoted one at a time), the RHINOstic tubes do not require aliquoting and accessioning is done as a single process. (B) Comparison of specimens accessoned per technician per day between swab types as a measure of processing efficiency.
RHINOstic swabs result in overall labor and cost savings
In addition to the high demand for and limited supply of materials and professional staff, cost has been a large hurdle to overcome during the pandemic. Dry collection protocols have helped reduce both material and labor costs, but the US Cotton swabs, with their slower rehydration step and the need for aliquoting, still represent a significant amount of labor.
While the RHINOstic swab itself costs more than the US Cotton #3, we found it provides an overall cost reduction when factoring in both the materials and the labor in our laboratory (Table 2). The US Cotton #3 swabs are procured at a cost of $0.29 per swab from Steripack, while 10 mL tubes for the swabs are procured independently from VWR at a cost of $0.22 per tube, and the 1 mL sample tubes for storage are procured from Thermo Fisher for $0.54 each. Our accessioning protocol prescribes two storage aliquots for each specimen, totalling $1.59 per sample; a protocol with only one aliquot would cost $1.05 per sample. Meanwhile, the RHINOstic swab RH-S000001 unit, which includes a cap-integrated swab, 1 mL sample tube, and 1 mL sample tube cap, costs $1.24 per unit. Based on our laboratory’s hourly wage of $28.40 for a technician, the labor costs equals approximately $0.11 per RHINOstic swab and $0.38 per US Cotton swab. Therefore, when materials and labor are considered together, the RHINOstic swab workflow is less costly.
TABLE 2.
A comparison of the material, labor, and total costs of the US Cotton #3 swab protocol and the RHINOstic swab protocol. a
| Protocol | Materials | Labor | Total per swab |
|---|---|---|---|
| US Cotton (two aliquots) | $1.59 | $0.38 | $1.97 |
| US Cotton (single aliquot) | $1.05 | $0.38 | $1.43 |
| RHINOstic | $1.24 | $0.11 | $1.35 |
A “single aliquot” version of the US Cotton swab protocol has been calculated to more directly compare to the RHINOstic protocol in terms of materials (labor costs for a second aliquot are miniscule and have been excluded).
DISCUSSION
Laboratory methods for diagnostic testing during current and future pandemics require flexibility and the ability to rapidly validate new methods to meet fluctuating demands in testing. Here we describe our process to scale up our testing programs, increase efficiency, and maintain our program in times of supply shortages while keeping costs low. The ability to increase sample processing without increasing the cost of testing or further burdening the supply chain is vital for both the ongoing pandemic response and considering future pandemic events.
Our experience demonstrated that incorporating RHINOstic swabs greatly improved the efficiency of specimen accessioning. Although each swab is more expensive than the US Cotton #3, each technician can process more specimens per hour, allowing us to more than recoup the cost difference. The use of RHINOstic swabs in our clinical laboratory reduced the amount of technician processing time by a factor of 3.5, which also reduced the overhead cost of testing. The dry shipping and improved temperature stability to up to 71 hours have made shipping to a wider area feasible and allowed us to consider less expensive shipping modalities (US Postal Service vs private courier). The time and cost savings are critical for implementing widespread testing at large events, schools, and workplaces as we respond to the testing surges that COVID-19 or any future pandemic presents.
Our usability studies targeted the specific age groups that are most likely to participate in our studies, for example, a university population and families with young children. Given the small size of the RHINOstic swabs, we would recommend that others expand the usability studies to participants over the age of 65 and those who may have vision or dexterity challenges before adoption. Several of our study participants noted that the small type size on the barcode used to register their swabs was difficult to read. While all the kit boxes do include a larger barcode on them, CLIA regulations require the clinical identifier on the specimen tube itself. This is particularly necessary for unobserved collection; it is not uncommon for participants to swap tubes with other household members or send them back in the incorrect boxes, which can cause discrepancies when determining exactly who collected the swab. Because our laboratory workflow requires the smaller 1 mL sample tube, with a printed 2D barcode on each for accessioning, we elected to mitigate this issue in a later study by providing participants with a small, inexpensive magnifying glass. Reading barcodes electronically with a smartphone camera could be an alternative solution for at-home participants.
The analytical sensitivity of RHINOstic swabs is comparable to the US Cotton #3 anterior nares swabs our laboratory employed in the past. A direct comparison between US Cotton and RHINOstic swabs in our extraction-free workflow indicates that RNase P Cts are slightly higher for RHINOstic swabs, though the slight increase in Cts is still well within the comfortable bounds of our assay’s control metrics (Ct <36 for RNase P). In addition, stability studies of RHINOstic swabs spiked with SARS-CoV-2 indicated that the swabs have robust shipping characteristics. SARS-CoV-2 Cts remained stable for at least 71 hours after a simulated nasal swab-to-test scenario in both extreme winter and summer conditions.
Alongside widespread testing, genomic sequencing for SARS-CoV-2 is vital to epidemic public health response to understand what variants are currently circulating in a population. Not only were RHINOstic swabs comparable in sensitivity for RT-qPCR testing but they also showed excellent viability for genomic sequencing compared to US Cotton #3. RHINOstic swabs had comparable rates of positives with a sufficient viral load to be eligible for sequencing while also having a higher rate of sequencing success than specimens collected by US Cotton #3 in the set analyzed.
Lastly, it may be appropriate for researchers and clinical laboratories to consider the use of RHINOstic swabs or other automation-friendly swab designs for use in non-pandemic scenarios, such as seasonal flu and respiratory syncytial virus testing. Typically, nasopharyngeal swabs are used for testing for the presence of other respiratory pathogens; a self-collected, automation-friendly, extraction-free-compatible swab could expand availability and potential for respiratory pathogen surveillance at a larger scale.
Contributor Information
Lea M. Starita, Email: lstarita@uw.edu.
Elitza S. Theel, Mayo Clinic Minnesota, Rochester, Minnesota, USA
The Seattle Flu Alliance Investigators:
Trevor Bedford, Michael Boeckh, Helen Y. Chu, Janet A. Englund, Christina M. Lockwood, Barry R. Lutz, Robin Prentice, Jay Shendure, Lea M. Starita, Alpana Waghmere, and Ana A. Weil
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/jcm.01285-23.
Supplementary Tables S1-S11 and Supplementary Figures S1-S7.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. McCulloch DJ, Kim AE, Wilcox NC, Logue JK, Greninger AL, Englund JA, Chu HY. 2020. Comparison of unsupervised home self-collected midnasal swabs with clinician-collected nasopharyngeal swabs for detection of SARS-CoV-2 infection. JAMA Netw Open 3:e2016382. doi: 10.1001/jamanetworkopen.2020.16382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tu YP, Jennings R, Hart B, Cangelosi GA, Wood RC, Wehber K, Verma P, Vojta D, Berke EM. 2020. Patient-collected tongue, nasal, and mid-Turbinate Swabs for SARS-Cov-2 yield equivalent sensitivity to health care worker collected nasopharyngeal Swabs. Infectious diseases (except HIV/AIDS). doi: 10.1101/2020.04.01.20050005 [DOI]
- 3. Pettit ME, Boswell SA, Qian J, Novak R, Springer M. 2021. Accessioning and automation compatible anterior nares swab design. J Virol Methods 294:114153. doi: 10.1016/j.jviromet.2021.114153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Harris PA, Taylor R, Minor BL, Elliott V, Fernandez M, O’Neal L, McLeod L, Delacqua G, Delacqua F, Kirby J, Duda SN, REDCap Consortium . 2019. The REDcap consortium: building an international community of software platform partners. J Biomed Inform 95:103208. doi: 10.1016/j.jbi.2019.103208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Srivatsan S, Heidl S, Pfau B, Martin BK, Han PD, Zhong W, van Raay K, McDermot E, Opsahl J, Gamboa L, Smith N, Truong M, Cho S, Barrow KA, Rich LM, Stone J, Wolf CR, McCulloch DJ, Kim AE, et al. , Seattle Flu Study Investigators . 2021. Swabexpress: an end-to-end protocol for extraction-free COVID-19 testing. Clin Chem 68:143–152. doi: 10.1093/clinchem/hvab132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kim AE, Brandstetter E, Wilcox N, Heimonen J, Graham C, Han PD, Starita LM, McCulloch DJ, Casto AM, Nickerson DA, Van de Loo MM, Mooney J, Ilcisin M, Fay KA, Lee J, Sibley TR, Lyon V, Geyer RE, Thompson M, Lutz BR, Rieder MJ, Bedford T, Boeckh M, Englund JA, Chu HY. 2021. Evaluating specimen quality and results from a community-wide, home-based respiratory surveillance study. J Clin Microbiol 59:e02934-20. doi: 10.1128/JCM.02934-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Babu TM, Feldstein LR, Saydah S, Acker Z, Boisvert CL, Briggs-Hagen M, Carone M, Casto A, Cox SN, Ehmen B, Englund JA, Fortmann SP, Frivold CJ, Groom H, et al. 2023. CASCADIA: a prospective community-based study protocol for assessing SARS-Cov-2 vaccine effectiveness in children and adults using a remote nasal swab collection and web-based survey design. BMJ open 13:e071446. doi: 10.1136/bmjopen-2022-071446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. U.S. Department of health and human services food and drug administration. policy for coronavirus disease-2019 tests during the public health emergency. 2020. US food and drug administration. Available from: https://www.fda.gov/media/135659/download
- 9. Weil AA, Sohlberg SL, O’Hanlon JA, Casto AM, Emanuels AW, Lo NK, Greismer EP, Magedson AM, Wilcox NC, Kim AE, Back L, Frazar CD, Pelle B, Sibley TR, Ilcisin M, et al. 2021. SARS-Cov-2 epidemiology on a public University campus in Washington state. doi: 10.1101/2021.03.15.21253227 [DOI] [PMC free article] [PubMed]
- 10. Chu HY, Englund JA, Starita LM, Famulare M, Brandstetter E, Nickerson DA, Rieder MJ, Adler A, Lacombe K, Kim AE, Graham C, Logue J, Wolf CR, et al. , Seattle Flu Study Investigators . 2020. Early detection of COVID-19 through a citywide pandemic surveillance platform. N Engl J Med 383:185–187. doi: 10.1056/NEJMc2008646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Public health — Seattle & king County. 2023. COVID-19 Data. Available from: https://kingcounty.gov/depts/health/covid-19/data.aspx
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables S1-S11 and Supplementary Figures S1-S7.