ABSTRACT
Candida glabrata is one of the most common causes of systemic candidiasis, often resistant to antifungal medications. To describe the genomic context of emerging resistance, we conducted a retrospective analysis of 82 serially collected isolates from 33 patients from population-based candidemia surveillance in the United States. We used whole-genome sequencing to determine the genetic relationships between isolates obtained from the same patient. Phylogenetic analysis demonstrated that isolates from 29 patients were clustered by patient. The median SNPs between isolates from the same patient was 30 (range: 7–96 SNPs), while unrelated strains infected four patients. Twenty-one isolates were resistant to echinocandins, and 24 were resistant to fluconazole. All echinocandin-resistant isolates carried a mutation either in the FKS1 or FKS2 HS1 region. Of the 24 fluconazole-resistant isolates, 17 (71%) had non-synonymous polymorphisms in the PDR1 gene, which were absent in susceptible isolates. In 11 patients, a genetically related resistant isolate was collected after recovering susceptible isolates, indicating in vivo acquisition of resistance. These findings allowed us to estimate the intra-host diversity of C. glabrata and propose an upper boundary of 96 SNPs for defining genetically related isolates, which can be used to assess donor-to-host transmission, nosocomial transmission, or acquired resistance.
IMPORTANCE
In our study, mutations associated to azole resistance and echinocandin resistance were detected in Candida glabrata isolates using a whole-genome sequence. C. glabrata is the second most common cause of candidemia in the United States, which rapidly acquires resistance to antifungals, in vitro and in vivo.
KEYWORDS: Candida glabrata, antifungal, resistance, genomic, WGS, epidemiology
INTRODUCTION
Candida glabrata is a yeast normally found in commensal microbial communities in the human gastrointestinal tract (1, 2), and it is also an opportunistic pathogen (3) that has become the second most common cause of invasive candidiasis in the United States, followed by C. albicans (4). The rise of C. glabrata and other non-Candida albicans Candida (NCAC) species is concerning due to their increased resistance to antifungal medications (5 – 7).
Currently, only three major classes of medications (polyenes, azoles, and echinocandins) are approved for treating invasive fungal infections. Amphotericin B is a polyene antifungal agent with a broad range of activity against yeasts and mold. Toxicity remains a drawback, despite the development of novel, less-toxic, lipid-based polyene formulations. Azoles are active against Candida spp., with fluconazole being the most widely prescribed antifungal in the class, but fluconazole often has reduced activity against C. glabrata (8, 9). Treatments with echinocandins have shown improved survival and greater clinical success (10), and their fungicidal activity makes them the first choice against Candida species (11).
Approximately 10% of C. glabrata isolates are resistant to azoles (6). The predominant mechanism of azole resistance in this pathogen is overexpression of genes such as CgCDR1, CgCDR2, and CgSNQ2 that encode ATP-binding cassette (ABC) efflux pumps (7, 12). In addition, azole resistance is associated with mutations in the PDR1 gene, which is a transcription factor for ABC-transporter expression (13, 14).
Resistance to echinocandins is due to substitutions in the FKS1 and FKS2 genes that encode subunits of the β-(1,3)-D-glucan synthase enzyme complex. This enzyme catalyzes the production of glucan, the major component in Candida cell walls (15). In these proteins, two hotspots for mutation, HS1 and HS2, located in the regions of the enzymes that form a binding pocket for echinocandins, are most frequently associated with resistance in Candida species. In C. glabrata, mutations in the HS1 hotspot of FKS2 are the most common mutations (16) and occur at nearly three times the rate of FKS1 mutations; S663 (FKS2) and equivalent residue S629 (FKS1) are the most predominant HS1 hotspot mutations (17).
The emergence of echinocandin resistance in C. glabrata in response to treatment has been well-documented (18 – 20). Numerous in vitro (21) and in vivo (2, 18, 19, 22) studies have confirmed the ability of C .glabrata to develop resistance in the presence of echinocandins. Acquired resistance to fluconazole is well-documented (23), and multidrug resistance (MDR), understood here as resistance to at least two classes of antifungal drugs, has been recorded in numerous clinical cases (22, 24, 25). In addition, acquired MDR in isolates of C. glabrata has been reported in studies in vitro (26).
Most previous studies showing the acquisition of resistance in C. glabrata have relied on multilocus sequence typing (MLST), which incorporates only a small fraction of the genome in the analysis to identify isolates with the same genotype (27). A few whole-genome sequencing (WGS) studies have been performed on serial isolates of C. glabrata (28) to investigate the acquisition of echinocandin (29) and azole (30) resistance during therapy. However, these previous studies included few patients (28 – 30) and did not evaluate the genetic relationships among isolates in a systematic way by using a larger collection of serially collected isolates with different genetic backgrounds. Here, we conducted a retrospective analysis of serially collected clinical isolates of C. glabrata among patients with >1 case of candidemia as part of the Emerging Infections Program (EIP) population-based candidemia surveillance by the Centers for Disease Control and Prevention (CDC) in the United States. We used WGS to describe the genetic relationships between resistant and susceptible isolates from the same patient, proposed an SNP threshold for defining the clonal relationship between isolates, and demonstrated the acquisition of resistance in response to treatment. These results provide a genomic framework for future outbreak investigations and for studies investigating the nosocomial transmission of C. glabrata.
MATERIALS AND METHODS
Isolate collection
Since 2008, the CDC has conducted active, population-based surveillance for candidemia through the Emerging Infections Program in select counties within the United States (https://www.cdc.gov/ncezid/dpei/eip/index.html). For surveillance purposes, a case was defined as a positive blood culture for any Candida species collected from a patient who resided within the surveillance areas. If collected at autopsy, cultures must have been collected ≤12 hours from death. A patient with a positive Candida blood culture collected >30 days after the initial positive culture was considered to have a new case. Cases were identified through the inpatient and outpatient laboratories serving the surveillance population. For each case, a trained surveillance officer reviewed medical records to gather demographic information and clinical characteristic data. Candida isolates collected from the incident culture for the cases were sent to the CDC for species confirmation and antifungal drug susceptibility testing. For C. glabrata, subsequent isolates within the 30-day case period were also collected.
Human subjects
This activity was reviewed by the CDC and was conducted consistent with the applicable federal law and CDC policy (see, e.g., 45 CFR part 46, 21 CFR part 56; 42 USC §241(d); 5 USC §552 a; 44 USC §3501 et seq).
Antifungal susceptibility testing
Antifungal susceptibility testing (AFST) was performed by broth microdilution as described in Clinical and Laboratory Standards (CLSI) M27 ed3 (31). Microdilution plates were custom-made by ThermoFisher and were stored frozen until use. Breakpoints for resistance for Candida glabrata were defined according to CLSI as follows: the breakpoint for anidulafungin was ≥0.5 µg/mL, that for caspofungin was ≥0.5 µg/mL, and that for micafungin was ≥0.25 µg/mL. For fluconazole, the breakpoint was ≥64 µg/mL.
WGS prioritization
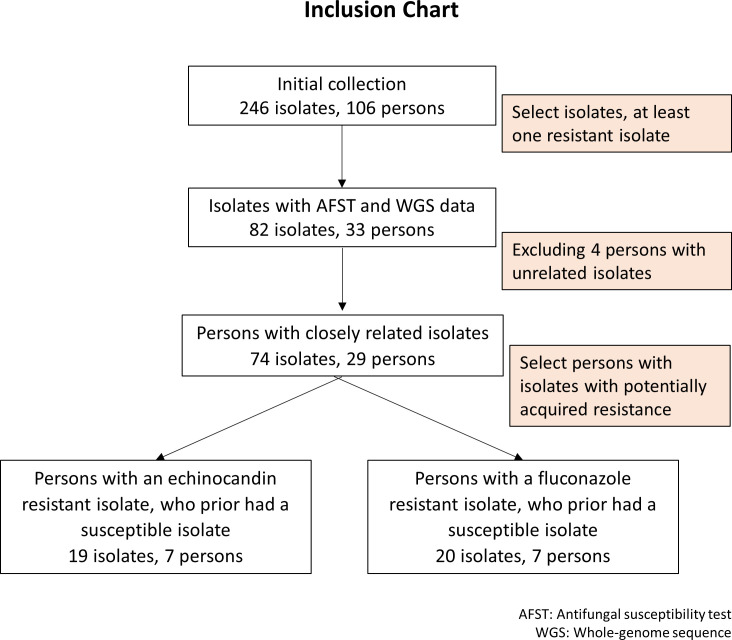
Patients with the >1 C. glabrata case (as defined above) between 2008 and 2015 were identified through a CDC candidemia surveillance database query. Using incident isolates only (i.e., only the first isolate for each case), isolates for patients of which at least one was found to be resistant to fluconazole or an echinocandin with MIC values above the defined breakpoints were submitted for whole-genome sequencing to determine the relatedness of strains across cases within each patient. Fourteen patients (42%) with all incident-susceptible isolates were submitted for WGS. Following WGS, patients with unrelated C. glabrata strains were excluded (criteria for relatedness among isolates are defined in the results). For patients with >2 cases, if at least two isolates were related, the patient was included based on only the related isolates (Fig. 1).
Fig 1.
Inclusion chart
We also defined a subgroup of patients with susceptible isolates followed by at least one resistant isolate, referred to as “patient with susceptible-to-resistant pattern.” This sub-dataset of 11 patients with related isolates serves to assess if resistance was acquired after treatment with antifungal drugs.
WGS library preparation
DNA was extracted using the DNeasy Blood and Tissue kit (Qiagen, Gaithersburg, MD, USA) according to the manufacturer’s recommendations. Genomic libraries were constructed using the NEBNext Ultra DNA Library Prep kit (New England Biolabs, Ipswich, MA, USA) for Illumina sequencing. They were sequenced on either the Illumina HiSeq 2500 platform using the HiSeq Rapid SBS kit v2 (500 cycles) or the NovaSeq platform using Illumina Nova-Seq using the 6000 Sp reagent kit (500 cycles). Read data were deposited into the Sequence Read Archive (SRA) database from the National Center for Biotechnology Information (NCBI), BioProject PRJNA1009065.
SNP calling and phylogenetic reconstruction
An assembly of C. glabrata CBS138 comprising 14 chromosomes, including the mitochondrial chromosome, was used as the reference for read mapping and SNP calling. This reference sequence had a length of 12.3 Mb, GC content of 38.6%, and N50 of 1.1 Mb. For the whole-genome SNP analysis, the MycoSNP workflow (v 0.21) was used (32).
A masked reference was used for phylogenetic reconstruction, and an unmasked reference was used for SNP annotations. The reference genome was masked for repeats using the nucmer command from MUMmer (v 4.0) (33) and Bedtools (v 2.29.2) (34). The reference genome was indexed for read alignment using the BWA index command and for variant calling using Samtools (v 1.10) (35) faidx and Picards GATK (v 2.22.9) [http://broadinstitute.github.io/picard/]. Trimming and filtering of low-quality data were performed using FaQCs (v 2.10) (36). The trimmed reads were used for alignment using the BWA (0.7.17) MEM command (37). Furthermore, the aligned BAM files from each sample were pre-processed using Samtools and Picard commands and made ready for variant calling. The Genome Analysis Toolkit (GATK) (v 4.1.4.1) (38) was used for variant calling using the haploid mode. The GATK VariantFiltration tool was used to filter sites based on the filtering expressions “QD <2.0,” “FS >60.0,” and “MQ <40.0.” Using a customized script, genotypes were filtered if the minimum genotype quality was <50, the percentage alternate allele was <0.80, or the depth was <10.
Variable positions in FASTA format were recovered from the VCF files using a Python script “vcfSnpsToFasta.py” (https://github.com/broadinstitute/broad-fungalgroup/tree/master/scripts/SNPs). For the phylogenetic analysis, 243,415 sites were concatenated. All positions containing gaps and missing data were eliminated. For the resulting sequences, the pairwise distance, neighbor-joining tree (NJ), and an additional phylogenetic tree with bootstrapping were calculated with the MEGA-X program (39). Parameters for the NJ tree were 1) 1,000 bootstraps, 2) nucleotide substitution transversion model, and 3) unequal base frequency (TVM). For tree visualization, we used the web-based JavaScript application, Microreact (40).
The median SNP differences were calculated for all pairwise comparisons between isolates from the same patient and among different patients using the MEGA-X pairwise distance matrix (39) and in-house R scripts (v 4.0.3) (41).
SNP annotations
The regular VCF output from MycoSNP did not allow us to identify all the mutations in the FKS2 gene since the gene sequence was masked. To address this issue, the MycoSNP code was modified not to execute the masking step on the reference genome. The VCF file obtained with the modified MycoSNP workflow was used by SnpEff (v 5.0) to predict the effects of associated mutations within genes (42) (Fig. S4). The SNPs were annotated using the database available in SnpEff for C. glabrata GCA 000002545. To identify mutations only in the coding regions, we used the parameters “-no-downstream,” “-no-upstream,” and “-no-intergenic.” Several filters were applied to the annotated VCF to identify the mutations present in some protein-encoded genes associated with resistance to echinocandins (FKS1 and FKS2) and a gene associated with resistance to fluconazole (PDR1) at specific positions previously reported in the literature. R in-house scripts were used to compare groups of samples, perform a Wilcoxon statistic test, and construct all R plots.
For the sub-dataset of patients with the susceptible-to-resistant pattern, we used R data frame filters to combine information on susceptibility profile, presence of mutations, collection dates, and clinical data on the treatment with antifungals to identify potential patients with acquired resistance.
RESULTS
Specimen collection and antifungal susceptibility testing
Initially, 246 C. glabrata isolates were available from 106 patients. Of those, 82 isolates from 33 patients were prioritized for WGS using the prioritization methods previously described (Fig. 1). The included isolates were obtained from surveillance sites across four states, mainly from Georgia (n = 33 isolates) and Maryland (n = 45 isolates), plus one more patient from Oregon (n = 2 isolates) and another from Tennessee (n = 2 isolates). Most (78%) of these 33 patients had two isolates, but two patients had a maximum of five isolates (Table S1).
SNP calling and phylogenetic reconstruction
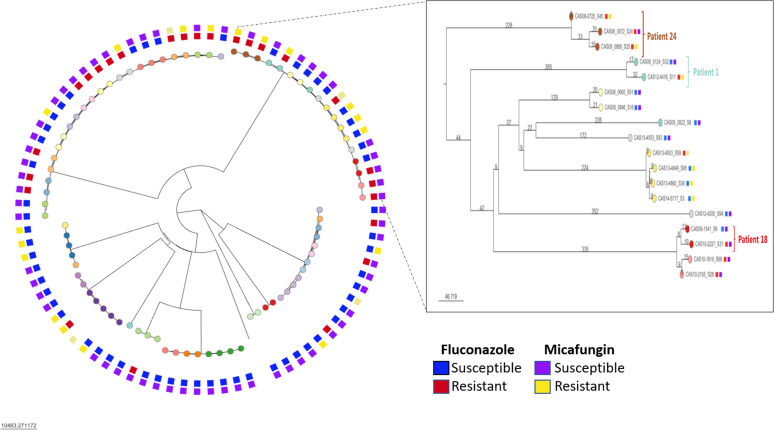
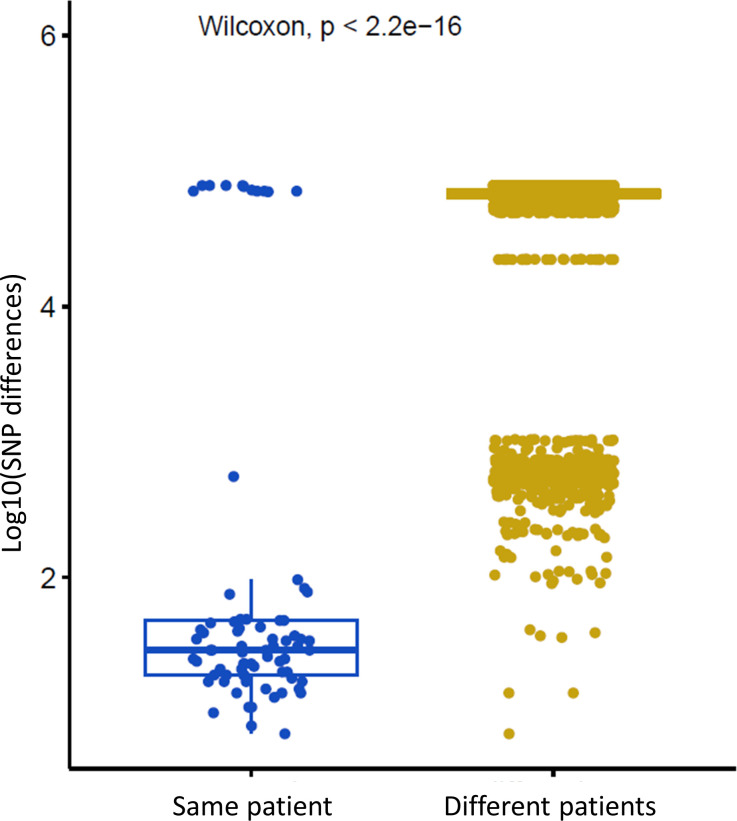
Phylogenetic analysis revealed that most isolates were clustered by patient (Fig. 2). A total of 243,415 nucleotide positions were used for the neighbor-joining tree method, and the median SNP difference observed between isolates from the same patient was 30 SNPs (range 7–96 SNPs), which was significantly lower than the median SNP difference observed between isolates from different patients (70,448 SNPs; P-value < 0.001; Fig. 3).
Fig 2.
Neighbor-joining phylogeny of C. glabrata. The final data set included a total of 243,415 nucleotides for 82 isolates. Each leaf node color represents a patient, and the external squares indicate susceptible/resistant phenotype. Box: zoom in shows examples of patients with isolates that acquired resistance to echinocandins (Patient 24), azoles (Patient 18), or both (Patient 1). Branch length indicates the number of SNPs.
Fig 3.
Genetic diversity of C. glabrata isolates. Left: SNP differences among several isolates collected from the same patient in 33 patients. Right: SNP differences for each pairwise comparison among all isolates from different patients in 106 patients. Y-axis shows the log 10 of SNP distances. Boxplot R version 4.0.3
Genetic clustering by patient was confirmed using a maximum likelihood tree with 1,000 bootstraps (Fig. S1). Isolates from Patient 12 and Patient 18 were grouped in two different clusters (Fig. S2). Only the related isolates were included in the median SNP difference calculation.
Four patients had large SNP differences between isolates (70,294–76,707 SNPs), and one patient (Patient 33) had 555 SNPs (Fig. S2). They were deemed to have multiple genotypes and were excluded from SNP annotation analysis. The remaining 29 patients had smaller SNP differences between isolates (7–96 SNPs) and were considered to have a single genotype (Fig. 2). Of the 29 patients with a single genotype, 11 patients exhibited the susceptible-to-resistant (SR) pattern, including four with a fluconazole-susceptible isolate followed by a fluconazole-resistant isolate (Table 2), four with an echinocandin-susceptible isolate followed by an echinocandin-resistant isolate (Table 1), and three with a susceptible isolate to both followed by an isolate resistant to echinocandins and fluconazole (Fig. 1).
TABLE 1.
SR pattern of echinocandin c
| ID | Patient ID | Date collected | Anid MIC value | Cas MIC value | Mutations FKS1 | Mutations FKS2 | Treatment b |
|---|---|---|---|---|---|---|---|
| CAS10-2614_S20 | 22 | 27-Oct-10 | 0.03 | 0.03 | Mica a | ||
| CAS12-3684_S68 | 22 | 21-Nov-11 | 1 | 2 | Ser663Pro | Mica | |
| CAS13-4530_S71 | 22 | 1-Nov-12 | 4 | >16 | Ser663Pro | Mica; Flu; Ampho | |
| CAS13-4778_S42 | 22 | 13-Mar-13 | 0.5 | 0.5 | Ser663Pro | Mica; Flu; Ampho | |
| CAS13-5011_S55 | 22 | 16-Jun-13 | 2 | 8 | Ser663Pro | Mica; Flu | |
| CAS08_0572_S24 | 24 | 13-Oct-08 | 0.125 | 0.125 | Unknown | ||
| CAS08-0725_S45 | 24 | 3-Dec-08 | 1 | 0.5 | Phe659Try | Unknown | |
| CAS09_0869_S25 | 24 | 5-Jan-09 | 1 | 2 | Phe659Try | Unknown | |
| CAS11-2902_S56 | 29 | 31-Jan-11 | 0.03 | 0.06 | Mica | ||
| CAS11-2996_S80 | 29 | 4-Mar-11 | 4 | >16 | Ser663Pro | Flu; Mica | |
| CAS156404_S48 | 31 | 18-Mar-15 | 0.03 | 0.03 | Mica | ||
| CAS156677_S67 | 31 | 6-Aug-15 | 2 | 8 | Ser629Pro | None | |
| CAS08_0124_S32 | 1 | 28-May-08 | 0.03 | 0.03 | |||
| CAS12-4416_S11 | 1 | 3-Nov-12 | 2 | 2 | Ser629Pro | Flu; Mica | |
| CAS13-5111_S2 | 14 | 9-Oct-13 | 0.03 | 0.03 | Mica | ||
| CAS13-5257_S26 | 14 | 3-Dec-13 | 2 | 8 | Ser663Pro | Mica | |
| CAS14-5368_S62 | 14 | 8-Jan-14 | 2 | 8 | Ser663Pro | Ampho; Mica | |
| CAS14-5293_S38 | 21 | 24-Nov-13 | 0.06 | 0.06 | Mica; Flu | ||
| CAS14-5468_S77 | 21 | 10-Feb-14 | 2 | 4 | Phe625Ser | Mica; Flu |
Mica = micafungin, Flu = fluconazole, Anid= anidulafungin, Ampho = amphotericin B.
Patient received systemic antifungal medication to treat candidemia on or after positive culture date.
Presence of the main reported mutations in HS1 FKS1 and FKS2. Shaded MIC values represent resistant isolates. Patient IDs in bold font represent patients in common with SR pattern for fluconazole.
Phylogenetic reconstruction showed no evidence of clustering by geographic site. In addition, time between isolation dates and the genetic distance between serially collected isolates were not correlated (Fig. S3).
SNP annotation related to echinocandin resistance
We analyzed sequences of the FKS1 and FKS2 genes extracted from the WGS data in 74 isolates representing 29 patients with a single genotype among all isolates. Twenty-one isolates were resistant to an echinocandin. Twenty of the echinocandin-resistant isolates carried mutations in the hotspot regions of FKS1 or FKS2, while none of the 53 susceptible isolates had mutations in the hotspot regions of these genes. (Table S2).
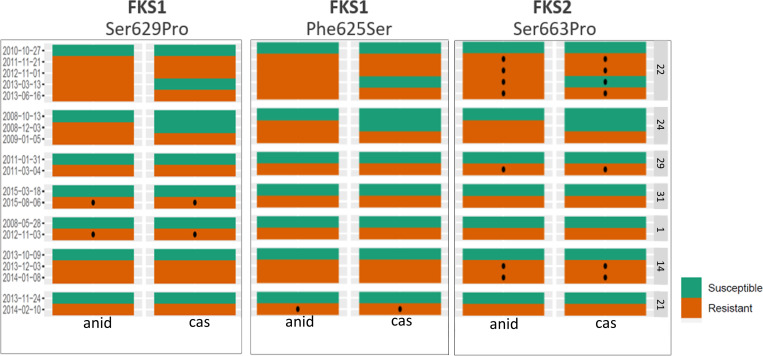
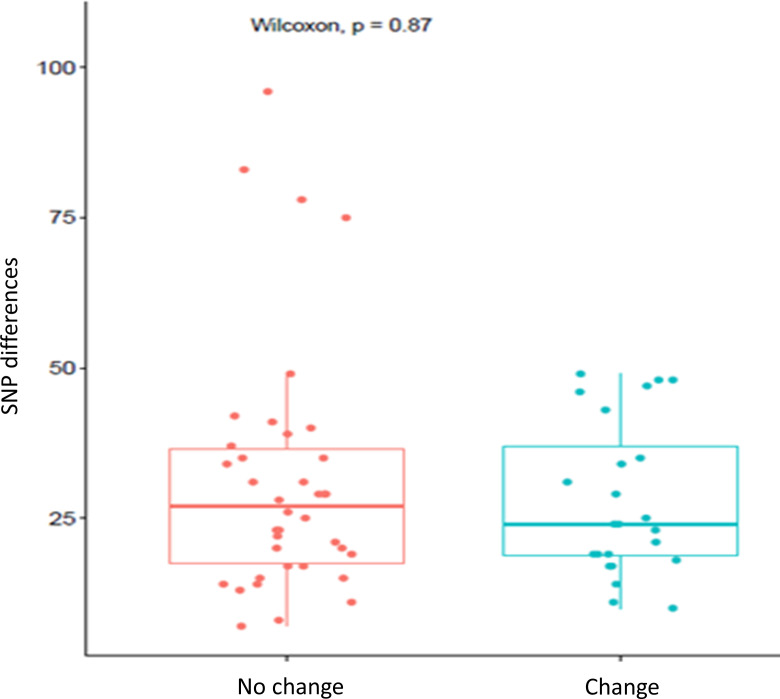
Of the seven patients in which echinocandin-resistant isolates with the same genotype were isolated after first recovering echinocandin-susceptible isolates (susceptible-to-resistant pattern), all had echinocandin-resistant isolates with mutations either in the FKS1 or FKS2 HS1 region. Seven echinocandin-resistant isolates from three cases had an S663P substitution in FKS2. Two echinocandin-resistant isolates from one case had an F659Y substitution in FKS2. Two echinocandin-resistant isolates from two different cases had an S629P mutation in FKS1, and one echinocandin-resistant isolate had an F625S mutation in FKS1 (Table 1; Fig. 4). The median number of SNPs between susceptible and echinocandin-resistant isolates from the same patient was not significantly different from the median number of SNPs between isolates when the resistance pattern did not change (24 SNPs vs 27 SNPs, P-value = 0.87; Fig. 5).
Fig 4.
FSK2 and FKS1 common mutations and susceptible/resistant phenotype (graphic representation of Table 1). Red and green boxes represent resistant and susceptible isolates, respectively. Black dots represent the presence of the mutation indicated on the top of each panel. Collection dates are displayed on the right side of the panels and Patient ID on the left. cas: caspofungin, anid: anidulafungin.*echinocandin-resistant isolate from Patient 24 carried the mutation Phe659Try in the FKS2 gene, not shown in the figure
Fig 5.
Left: SNP differences for each pairwise comparison among isolates from the same patient, where the resistant pattern did not change (S->S and R->R). Right: SNP differences for each pairwise comparison among isolates from the same patient where the resistant pattern changed (S->R). Boxplot R version 4.0.3
SNP annotation related to fluconazole resistance
Twenty-five isolates were resistant to fluconazole (MIC ≥64 µg/mL). We identified nine different non-synonymous polymorphisms in the PDR1 gene; three of them have been previously reported in the literature to be related to azole resistance (E714D, N109D, and R376Q). These changes were found in 16 (Table S2) of the 25 resistant isolates and were absent in all susceptible isolates.
In seven patients, fluconazole-resistant isolates were isolated after first recovering fluconazole-susceptible isolates (susceptible-resistant pattern). Five of those patients had isolates with different non-synonymous substitutions in the PDR1 gene. The amino acids differed between the susceptible and resistant paired isolates (Table 2).
TABLE 2.
SR pattern azoles a
| ID | Patient ID | Date collected | Flu MIC value | Mutations PDR1 | Treatment c |
|---|---|---|---|---|---|
| CAS10-2614_S20 | 22 | 27-Oct-10 | 8 | Mica b | |
| CAS12-3684_S68 | 22 | 21-Nov-11 | 4 | Mica | |
| CAS13-4530_S71 | 22 | 1-Nov-12 | 16 | Mica; Flu; Ampho | |
| CAS13-4778_S42 g | 22 | 13-Mar-13 | Mica; Flu; Ampho | ||
| CAS13-5011_S55 | 22 | 16-Jun-13 | 64 | p.Glu714Asp d | Mica; Flu |
| CAS08_0124_S32 | 1 | 28-May-08 | 32 | ||
| CAS12-4416_S11 | 1 | 3-Nov-12 | 64 | Flu; Mica | |
| CAS12-4158_S70 | 21 | 7-Jul-12 | 4 | None | |
| CAS13-4494_S47 | 21 | 19-Nov-12 | 8 | Flu | |
| CAS14-5293_S38 | 21 | 24-Nov-13 | 64 | Mica; Flu | |
| CAS14-5468_S77 | 21 | 10-Feb-14 | 32 | Mica; Flu | |
| CAS11-3025_S92 | 8 | 17-Mar-11 | 8 | Mica | |
| CAS12-3743_S58 | 8 | 3-Jan-12 | 128 | p.Gly1079Glu f | Flu; Mica |
| CAS10-2486_S79 | 15 | 28-Aug-10 | 4 | Flu | |
| CAS10-2596_S8 | 15 | 17-Oct-10 | 64 | p.Gly1099Ser | Flu; Mica |
| CAS11-3232_S57 | 15 | 16-May-11 | 8 | Mica | |
| CAS09-1541_S6 | 18 | 16-Sep-09 | 8 | Unknown | |
| CAS10-2227_S31 | 18 | 26-May-10 | 64 | p.Asn1091Asp d | Unknown |
| CAS09-1361_S41 | 19 | 19-Jul-09 | 8 | Unknown | |
| CAS09-1513_S89 | 19 | 27-Aug-09 | 256 | p.Arg376Gln e | Unknown |
Shaded MIC values represent resistant isolates. Patient IDs in bold font represent patients in common with SR pattern for echinocandins.
Mica = micafungin, Flu = fluconazole, Ampho = amphotericin B.
Patient received systemic antifungal medication to treat candidemia on or after positive culture date.
Previously reported by Won E. et al 2021.
Change in the same position R376W previously reported by Ferrari et al 2009 (43).
Change in the same position G1079R previously reported by Won E. et al 2021.
Fluconazole MIC value missing for this isolate.
Overall, serial isolates from 11 patients showed a susceptible-to-resistant pattern, four for echinocandins, four for fluconazole, and three for both. All 11 patients were treated with antifungals before or after recovery of the first susceptible isolate. Of the four patients whose isolates demonstrated acquired resistance to fluconazole, two received treatment with fluconazole and two were treated with antifungals of an unknown drug class. Of the four patients whose isolates demonstrated acquired resistance to echinocandins, three received treatment with micafungin and one was treated with an antifungal of an unknown drug class. All three cases with acquired MDR were treated with both fluconazole and micafungin (Tables 1 and 2).
DISCUSSION
We analyzed serial isolates of C. glabrata from patients with multiple episodes of candidemia and used genomic and epidemiological data to assess the genetic diversity among these isolates. We showed that in most cases, multiple isolates from the same patient were genetically related and could be clearly distinguished from isolates obtained from other patients. We also identified known mutations in FKS and PDR1 genes associated with acquired resistance to echinocandins and fluconazole, respectively. We found that isolates with different antifungal susceptibility profiles may be part of the same clonal population of C. glabrata.
The median SNP difference between isolates from the same patient was 30 SNPs, ranging from 7–96 SNPs. These SNP differences are similar to those reported in other studies that quantified the intra-host diversity of C. glabrata isolates using WGS. For example, Helmstetter et al. (28) detected 64–140 SNPs and insertions or deletions (Indels) between pairs of related strains (28). In addition, several studies compared the relatedness among C. glabrata isolates from the same patient using other molecular techniques (29, 30, 44, 45), and in general, they found similarity among isolates. Our findings are consistent with those reported by others and suggest that most patients are colonized by clonal populations of C. glabrata that contribute endogenous isolates to infection (45). However, several patients with heterogenous populations of unrelated isolates were identified.
Understanding how isolates from the same patient are related is essential for understanding nosocomial transmission and investigating outbreaks. Previous studies on C. auris showed that the SNP difference observed between isolates collected from the same patient can be used as a reference to assess transmission (46). However, these distances are likely to differ for different species and depend on the genome’s mutation rate and stability. The hypervariability of C. glabrata genomes has been shown previously (47). Here, we propose to use an upper boundary of 96 SNPs for defining genetically related isolates of C. glabrata. This SNP number is an approximation and may vary if new isolates are added. Although C. glabrata is not known to cause widespread nosocomial outbreaks, several cases of nosocomial acquisition (48, 49) and donor-to-host transmission of this pathogen have been reported (50, 51). We propose that the observed SNP difference within a patient could be used as a point of reference to assess donor-to-host transmission or nosocomial acquisition of C. glabrata.
In our study, 82% of patients were infected with isolates that were more closely related to each other than to isolates from other patients (7–96 SNPs), which suggests that these infections most likely originated from the endogenous populations of C. glabrata that are associated with each patient. However, six (18%) patients had infections with two or more genetically distinct strains (70,294–76,707 SNPs; Fig. S2). Previous studies based on MLST reported unrelated isolates from the same patient (2, 52). The most likely explanation for detecting isolates with different genotypes is that these patients were colonized by two or more genetically distinct populations of isolates. However, a secondary nosocomial infection with an exogenous strain cannot be ruled out (45). Detailed epidemiological data that were unavailable in our study are needed to distinguish between these two hypotheses.
To better understand the emergence of resistance and the relationships between resistant and susceptible isolates within the same patient, we investigated genetic substitutions in genes linked to resistance. Twenty echinocandin-resistant isolates had non-synonymous mutations in FKS genes (25% in FKS1; 75% in FKS2). The most common mutation observed in this study was the FKS2 gene in position Ser-663 (53). We did not find mutations in HS2 of either FKS1 or FKS2 (8, 16, 24). The main fluconazole resistance mechanism in C. glabrata is upregulation of the expression of the ABC-transporter genes. Although assessing gene expression levels was outside the scope of our study, we screened the fluconazole-resistant isolates for non-synonymous changes in the sequence of the gene PDR1, which serves as the main transcription factor that modulates the gene expression of the ABC transporter (13), and we found nine different polymorphisms. Three of the amino acid changes that we found in PDR1 (E714D, N109D, and R376Q) had been previously reported (12). Three patients had isolates with the corresponding mutations that were resistant to both echinocandins and fluconazole, suggesting in vivo acquisition of resistance to both fluconazole and echinocandins in response to treatment.
Our study demonstrated that isolates from the same patient were genetically related, independent of changes in susceptibility to antifungal drugs. Our data show that the acquisition of resistance to an antifungal drug did not have a significant impact on the SNP distances between serial isolates that remained susceptible or resistant over time compared with SNP distances between serial isolates that acquired resistance, confirming that the resistant phenotype could be achieved with very few genetic changes (26). A further implication of this finding is that the proposed boundary for defining genetically related isolates of C. glabrata is not affected by the susceptibility profile. Additional analysis of the number of SNP differences versus the time between isolate collection (Fig. S3) confirmed that the isolates were randomly pulled from the C. glabrata population within the host. Therefore, the observed genetic diversity reflects the inherent diversity in the population rather than accumulation of changes over time.
Our study has several limitations. First, SNP cutoffs are difficult to compare with those found in other studies since differences in bioinformatic methods and the selected reference genome can impact the SNP count. Second, our study included candidemia isolates only. To better define the diversity of isolates within patients, commensal isolates from patients’ natural microbiomes should also be included. Third, since only a single isolate was recovered at any given timepoint, the diversity defined here likely does not represent the entire pooled C. glabrata population within the patient. Future analyses using colonization isolates from the same patient may help validate these results further. Additionally, the SNP variation described may be influenced by several factors. Avenues for future research include the effect of the MLST sequence type (STs) of the C. glabrata isolates in the observed SNP diversity (27) and the presence of mutation in MSH2 genes that lead to a hyper-mutable phenotype (54).
In conclusion, the comparison of serial isolates of C. glabrata from the same patient at different times during the course of an infection allowed estimating the expected intra-host diversity. It can be instrumental for investigating 1) cases of donor-to-host transmission, 2) nosocomial acquisition, and 3) microevolutionary processes leading to the emergence of drug resistance in C. glabrata. One of the main contributions of this work was the sequencing of 82 genomes of clinical isolates of C. glabrata. The WGS data were submitted to the NCBI (BioProject accession PRJNA1009065) and together with MIC value susceptibility data (Table S2) published here could be included in future studies aimed at identifying new mechanisms of resistance in C. glabrata and new pathways of antifungal resistance evolution. In addition, we present here the use of an existing bioinformatics tool, SnpEff, for identifying mutations in human pathogenic fungi to facilitate further studies of genomic epidemiology of emerging resistance to antifungal drugs. WGS has become an essential part of epidemiological research to differentiate outbreak-associated illnesses from unrelated illnesses occurring at the same time. Our study provides a genomic framework for these future investigations.
ACKNOWLEDGMENTS
We thank the MDB laboratory team for continuing in providing support to perform all the fungal susceptibility testing procedures as part of our routine Emerging Infections Program (EIP) candidemia surveillance. We give special thanks to CDC’s Office of Advanced Molecular Detection (OAMD). The authors acknowledge the support of Lindsay Bonner of the Maryland Emerging Infections Program; Alexia Zhang and Abiele Ariane Ebuen of the Oregon Emerging Infections Program; Caroline Graber, Sherry Hillis and Sandra Hardin of the Tennessee Emerging Infections Program; and the participating Emerging Infections Program Laboratories.
We acknowledge Oak Ridge Institute for Science and Education (ORISE) for financial support for EM.
The findings and the conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention. The use of trade names is for identification only and does not imply endorsement.
Contributor Information
A. P. Litvintseva, Email: frq8@cdc.gov.
Kimberly E. Hanson, University of Utah, Salt Lake City, Utah, USA
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/jcm.01140-23.
Neighbor-joining tree with 1000 bootstrap replicates.
Neighbor-joining phylogeny of C. glabrata, patient with isolates that exhibited at least two unrelated isolates.
Number of SNPs between pairs of isolates from the same patient vs. number of days between collection samples.
Number of variant effects of each by functional class and by type and region from SnpEff report.
Supplementary tables and supplementary figure legends.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Barantsevich N, Barantsevich E. 2022. Diagnosis and treatment of invasive candidiasis. Antibiotics (Basel) 11:718. doi: 10.3390/antibiotics11060718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jensen RH, Johansen HK, Søes LM, Lemming LE, Rosenvinge FS, Nielsen L, Olesen B, Kristensen L, Dzajic E, Astvad KMT, Arendrup MC. 2015. Posttreatment antifungal resistance among colonizing candida isolates in candidemia patients: results from a systematic multicenter study. Antimicrob Agents Chemother 60:1500–1508. doi: 10.1128/AAC.01763-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arastehfar A, Daneshnia F, Salehi M, Yaşar M, Hoşbul T, Ilkit M, Pan W, Hagen F, Arslan N, Türk-Dağı H, Hilmioğlu-Polat S, Perlin DS, Lass-Flörl C. 2020. Low level of antifungal resistance of Candida glabrata blood isolates in Turkey: fluconazole minimum inhibitory concentration and FKS mutations can predict therapeutic failure. Mycoses 63:911–920. doi: 10.1111/myc.13104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pfaller MA, Andes DR, Diekema DJ, Horn DL, Reboli AC, Rotstein C, Franks B, Azie NE. 2014. Epidemiology and outcomes of invasive candidiasis due to non-albicans species of candida in 2,496 patients: data from the prospective antifungal therapy (PATH) registry 2004-2008. PLoS One 9:e101510. doi: 10.1371/journal.pone.0101510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Berkow EL, Lockhart SR. 2017. Fluconazole resistance in Candida species: a current perspective. Infect Drug Resist 10:237–245. doi: 10.2147/IDR.S118892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pfaller MA, Diekema DJ, Turnidge JD, Castanheira M, Jones RN. 2019. Twenty years of the SENTRY antifungal surveillance program: results for Candida species from 1997-2016. Open Forum Infect Dis 6:S79–S94. doi: 10.1093/ofid/ofy358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rodrigues CF, Silva S, Henriques M. 2014. Candida glabrata: a review of its features and resistance. Eur J Clin Microbiol Infect Dis 33:673–688. doi: 10.1007/s10096-013-2009-3 [DOI] [PubMed] [Google Scholar]
- 8. Zimbeck AJ, Iqbal N, Ahlquist AM, Farley MM, Harrison LH, Chiller T, Lockhart SR. 2010. FKS mutations and elevated echinocandin MIC values among Candida glabrata isolates from U.S. population-based surveillance. Antimicrob Agents Chemother 54:5042–5047. doi: 10.1128/AAC.00836-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pappas PG, Kauffman CA, Andes D, Benjamin DK, Calandra TF, Edwards JE, Filler SG, Fisher JF, Kullberg B-J, Ostrosky-Zeichner L, Reboli AC, Rex JH, Walsh TJ, Sobel JD. 2009. Clinical practice guidelines for the management of candidiasis: 2009 update by the infectious diseases society of America. Clin Infect Dis 48:503–535. doi: 10.1086/596757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Andes DR, Safdar N, Baddley JW, Playford G, Reboli AC, Rex JH, Sobel JD, Pappas PG, Kullberg BJ. 2012. Impact of treatment strategy on outcomes in patients with acndidemia and other forms of invasive candidiasis: a patient-level quantitative review of randomized trials. Clin Infect Dis 54:1110–1122. doi: 10.1093/cid/cis021 [DOI] [PubMed] [Google Scholar]
- 11. Nivoix Y, Ledoux MP, Herbrecht R. 2020. Antifungal therapy: new and evolving therapies. Semin Respir Crit Care Med 41:158–174. doi: 10.1055/s-0039-3400291 [DOI] [PubMed] [Google Scholar]
- 12. Won EJ, Choi MJ, Kim M-N, Yong D, Lee WG, Uh Y, Kim TS, Byeon SA, Lee SY, Kim SH, Shin JH. 2021. Fluconazole-resistant Candida glabrata bloodstream isolates, South Korea, 2008-2018. Emerg Infect Dis 27:779–788. doi: 10.3201/eid2703.203482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vermitsky J-P, Edlind TD. 2004. Azole resistance in Candida glabrata: coordinate upregulation of multidrug transporters and evidence for a PDR1-like transcription factor. Antimicrob Agents Chemother 48:3773–3781. doi: 10.1128/AAC.48.10.3773-3781.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Paul S, McDonald WH, Moye-Rowley WS. 2018. Negative regulation of Candida glabrata PDR1 by the deubiquitinase subunit Bre5 occurs in a ubiquitin independent manner. Mol Microbiol 110:309–323. doi: 10.1111/mmi.14109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pais P, Galocha M, Viana R, Cavalheiro M, Pereira D, Teixeira MC. 2019. Microevolution of the pathogenic yeasts Candida albicans and Candida glabrata during antifungal therapy and host infection. Microb Cell 6:142–159. doi: 10.15698/mic2019.03.670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Katiyar SK, Alastruey-Izquierdo A, Healey KR, Johnson ME, Perlin DS, Edlind TD. 2012. FKS1 and FKS2 are functionally redundant but differentially regulated in Candida glabrata: implications for echinocandin resistance. Antimicrob Agents Chemother 56:6304–6309. doi: 10.1128/AAC.00813-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Perlin DS. 2018. MSGERC Biennial Meeting 2018: antifungal resistance among Candida Spp. Available from: https://msgerc.org/resources/Documents/2018%20Speaker%20Handouts/Candida_2_Perlin.pdf
- 18. Cleary JD, Garcia-Effron G, Chapman SW, Perlin DS. 2008. Reduced Candida glabrata susceptibility secondary to an FKS1 mutation developed during candidemia treatment. Antimicrob Agents Chemother 52:2263–2265. doi: 10.1128/AAC.01568-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Thompson GR, Wiederhold NP, Vallor AC, Villareal NC, Lewis JS, Patterson TF. 2008. Development of caspofungin resistance following prolonged therapy for invasive candidiasis secondary to Candida glabrata infection. Antimicrob Agents Chemother 52:3783–3785. doi: 10.1128/AAC.00473-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Costa-de-Oliveira S, Marcos Miranda I, Silva RM, Pinto E Silva A, Rocha R, Amorim A, Gonçalves Rodrigues A, Pina-Vaz C. 2011. FKS2 mutations associated with decreased echinocandin susceptibility of Candida glabrata following anidulafungin therapy. Antimicrob Agents Chemother 55:1312–1314. doi: 10.1128/AAC.00589-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bordallo-Cardona MÁ, Sánchez-Carrillo C, Bouza E, Muñoz P, Escribano P, Guinea J. 2019. Detection of echinocandin-resistant Candida glabrata in blood cultures spiked with different percentages of FKS2 mutants. Antimicrob Agents Chemother 63:e02004-18. doi: 10.1128/AAC.02004-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chapeland-Leclerc F, Hennequin C, Papon N, Noël T, Girard A, Socié G, Ribaud P, Lacroix C. 2010. Acquisition of flucytosine, azole, and caspofungin resistance in Candida glabrata bloodstream isolates serially obtained from a hematopoietic stem cell transplant recipient. Antimicrob Agents Chemother 54:1360–1362. doi: 10.1128/AAC.01138-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Borst A, Raimer MT, Warnock DW, Morrison CJ, Arthington-Skaggs BA. 2005. Rapid acquisition of stable azole resistance by Candida glabrata isolates obtained before the clinical introduction of fluconazole. Antimicrob Agents Chemother 49:783–787. doi: 10.1128/AAC.49.2.783-787.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Garcia-Effron G, Lee S, Park S, Cleary JD, Perlin DS. 2009. Effect of Candida glabrata FKS1 and FKS2 mutations on echinocandin sensitivity and kinetics of 1,3-beta-D-glucan synthase: implication for the existing susceptibility breakpoint. Antimicrob Agents Chemother 53:3690–3699. doi: 10.1128/AAC.00443-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Corcione S, D’Avolio A, Pasero D, Trentalange A, Pagani N, Sanguinetti M, De Rosa FG. 2019. Acquisition of FKS2 mutation after echinocandin treatment of infective endocarditis by Candida glabrata. Infez Med 27:328–331. [PubMed] [Google Scholar]
- 26. Ksiezopolska E, Schikora-Tamarit MÀ, Beyer R, Nunez-Rodriguez JC, Schüller C, Gabaldón T. 2021. Narrow mutational signatures drive acquisition of multidrug resistance in the fungal pathogen Candida glabrata. Curr Biol 31:5314–5326. doi: 10.1016/j.cub.2021.09.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dodgson AR, Pujol C, Denning DW, Soll DR, Fox AJ. 2003. Multilocus sequence typing of Candida glabrata reveals geographically enriched clades. J Clin Microbiol 41:5709–5717. doi: 10.1128/JCM.41.12.5709-5717.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Helmstetter N, Chybowska AD, Delaney C, Da Silva Dantas A, Gifford H, Wacker T, Munro C, Warris A, Jones B, Cuomo CA, Wilson D, Ramage G, Farrer RA. 2022. Population genetics and microevolution of clinical Candida glabrata reveals recombinant sequence types and hyper-variation within mitochondrial genomes, virulence genes, and drug targets. Genetics 221:iyac031. doi: 10.1093/genetics/iyac031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Barber AE, Weber M, Kaerger K, Linde J, Gölz H, Duerschmied D, Markert A, Guthke R, Walther G, Kurzai O. 2019. Comparative genomics of serial Candida glabrata isolates and the rapid acquisition of echinocandin resistance during therapy. Antimicrob Agents Chemother 63:e01628-18. doi: 10.1128/AAC.01628-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vale-Silva L, Beaudoing E, Tran VDT, Sanglard D. 2017. Comparative genomics of two sequential Candida glabrata clinical isolates. G3 (Bethesda) 7:2413–2426. doi: 10.1534/g3.117.042887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Clinical and Laboratory Standard Institute, W., PA . 2017. Reference method for broth dilution antifungal susceptibility testing of Yeast- fourth edition, CLSI document M27-A4 [Google Scholar]
- 32. Bagal UR, Phan J, Welsh RM, Misas E, Wagner D, Gade L, Litvintseva AP, Cuomo CA, Chow NA. 2022. MycoSNP: a portable workflow for performing whole-genome sequencing analysis of Candida auris. Methods Mol Biol 2517:215–228. doi: 10.1007/978-1-0716-2417-3_17 [DOI] [PubMed] [Google Scholar]
- 33. Delcher AL, Salzberg SL, Phillippy AM. 2003. Using mummer to identify similar regions in large sequence SETS. Curr Protoc Bioinformatics Chapter 10:Unit doi: 10.1002/0471250953.bi1003s00 [DOI] [PubMed] [Google Scholar]
- 34. Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. doi: 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lo C-C, Chain PSG. 2014. Rapid evaluation and quality control of next generation sequencing data with FaQCs. BMC Bioinformatics 15:366. doi: 10.1186/s12859-014-0366-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM
- 38. McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. 2010. The genome analysis toolkit: a mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. doi: 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kumar S, Stecher G, Li M, Knyaz C, Tamura K, Battistuzzi FU. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. doi: 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Argimón S, Abudahab K, Goater RJE, Fedosejev A, Bhai J, Glasner C, Feil EJ, Holden MTG, Yeats CA, Grundmann H, Spratt BG, Aanensen DM. 2016. Microreact: visualizing and sharing data for genomic epidemiology and phylogeography. Microb Genom 2:e000093. doi: 10.1099/mgen.0.000093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. R_Core_Team . 2021. R: a language and environment for statistical computing
- 42. Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6:80–92. doi: 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ferrari S, Ischer F, Calabrese D, Posteraro B, Sanguinetti M, Fadda G, Rohde B, Bauser C, Bader O, Sanglard D. 2009. Gain of function mutations in CgPDR1 of Candida glabrata not only mediate antifungal resistance but also enhance virulence. PLoS Pathog 5:e1000268. doi: 10.1371/journal.ppat.1000268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Singh-Babak SD, Babak T, Diezmann S, Hill JA, Xie JL, Chen Y-L, Poutanen SM, Rennie RP, Heitman J, Cowen LE. 2012. Global analysis of the evolution and mechanism of echinocandin resistance in Candida glabrata. PLoS Pathog 8:e1002718. doi: 10.1371/journal.ppat.1002718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Carreté L, Ksiezopolska E, Gómez-Molero E, Angoulvant A, Bader O, Fairhead C, Gabaldón T. 2019. Genome comparisons of Candida glabrata serial clinical isolates reveal patterns of genetic variation in infecting clonal populations. Front Microbiol 10:112. doi: 10.3389/fmicb.2019.00112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chow NA, Gade L, Tsay SV, Forsberg K, Greenko JA, Southwick KL, Barrett PM, Kerins JL, Lockhart SR, Chiller TM, Litvintseva AP. 2018. Multiple introductions and subsequent transmission of multidrug-resistant Candida auris in the USA: a molecular epidemiological survey. Lancet Infect Dis 18:1377–1384. doi: 10.1016/S1473-3099(18)30597-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ahmad KM, Kokošar J, Guo X, Gu Z, Ishchuk OP, Piškur J. 2014. Genome structure and dynamics of the yeast pathogen Candida glabrata. FEMS Yeast Res 14:529–535. doi: 10.1111/1567-1364.12145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vazquez JA, Dembry LM, Sanchez V, Vazquez MA, Sobel JD, Dmuchowski C, Zervos MJ. 1998. Nosocomial Candida glabrata colonization: an epidemiologic study. J Clin Microbiol 36:421–426. doi: 10.1128/JCM.36.2.421-426.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Paluchowska P, Tokarczyk M, Bogusz B, Skiba I, Budak A. 2014. Molecular epidemiology of Candida albicans and Candida glabrata strains isolated from intensive care unit patients in Poland. Mem Inst Oswaldo Cruz 109:436–441. doi: 10.1590/0074-0276140099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tappeiner C, Goldblum D, Zimmerli S, Fux C, Frueh BE. 2009. Donor-to-host transmission of Candida glabrata to both recipients of corneal transplants from the same donor. Cornea 28:228–230. doi: 10.1097/ICO.0b013e318183a3e3 [DOI] [PubMed] [Google Scholar]
- 51. Al-Assiri A, Al-Jastaneiah S, Al-Khalaf A, Al-Fraikh H, Wagoner MD. 2006. Late-onset donor-to-host transmission of Candida glabrata following corneal transplantation. Cornea 25:123–125. doi: 10.1097/01.ico.0000164777.80879.07 [DOI] [PubMed] [Google Scholar]
- 52. Odds FC, Hanson MF, Davidson AD, Jacobsen MD, Wright P, Whyte JA, Gow NAR, Jones BL. 2007. One year prospective survey of candida bloodstream infections in Scotland. J Med Microbiol 56:1066–1075. doi: 10.1099/jmm.0.47239-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Aldejohann AM, et al. 2021. Emergence of resistant Candida glabrata in Germany. JAC Antimicrob Resist 3. doi: 10.1093/jacamr/dlab122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Healey KR, Zhao Y, Perez WB, Lockhart SR, Sobel JD, Farmakiotis D, Kontoyiannis DP, Sanglard D, Taj-Aldeen SJ, Alexander BD, Jimenez-Ortigosa C, Shor E, Perlin DS. 2016. Prevalent mutator genotype identified in fungal pathogen Candida glabrata promotes multi-drug resistance. Nat Commun 7:11128. doi: 10.1038/ncomms11128 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Neighbor-joining tree with 1000 bootstrap replicates.
Neighbor-joining phylogeny of C. glabrata, patient with isolates that exhibited at least two unrelated isolates.
Number of SNPs between pairs of isolates from the same patient vs. number of days between collection samples.
Number of variant effects of each by functional class and by type and region from SnpEff report.
Supplementary tables and supplementary figure legends.