Abstract
Since their discovery 21 years ago, channelrhodopsins have come of age and have become indispensable tools for optogenetic control of excitable cells such as neurons and myocytes. Potential therapeutic utility of channelrhodopsins has been proven by partial vision restoration in a human patient. Previously known channelrhodopsins are either proton channels, non-selective cation channels almost equally permeable to Na+ and K+ besides protons, or anion channels. Two years ago, we discovered a group of channelrhodopsins that exhibit over an order of magnitude higher selectivity for K+ than for Na+. These proteins, known as “kalium channelrhodopsins” or KCRs, lack the canonical tetrameric selectivity filter found in voltage- and ligand-gated K+ channels, and use a unique selectivity mechanism intrinsic to their individual protomers. Mutant analysis has revealed that the key residues responsible for K+ selectivity in KCRs are located at both ends of the putative cation conduction pathway, and their role has been confirmed by high-resolution KCR structures. Expression of KCRs in mouse neurons and human cardiomyocytes enabled optical inhibition of these cells’ electrical activity. In this minireview we briefly discuss major results of KCR research obtained during the last two years and suggest some directions of future research.
Keywords: optogenetics, photocurrent, ion channels, ion selectivity
Significance
Potassium-selective channelrhodopsins known as “kalium channelrhodopsins” or KCRs are fundamentally important because they expand our understanding of how ion channels discriminate between different ionic species. These proteins are also important because they can be used as optogenetic silencing tools most closely matching endogenous repolarization processes.
Introduction
Electrogenic microbial rhodopsins (light-activated ion channels and pumps) have been recruited for optical control of excitable mammalian cells, such as neurons and myocytes [1]. This powerful approach, named optogenetics [2], is increasingly popular in neuroscience and cardiology labs, has already been used for partial vision restoration in a human patient [3] and is considered for gene therapy to treat other neurological, psychiatric, and cardiac disorders. Upon photostimulation, channelrhodopsins (ChRs) generate passive ion currents across the cell membrane and are therefore more efficient as optogenetic tools than ion-pumping rhodopsins that transport only one ion per absorbed photon [4,5]. The first ChRs were discovered in the chlorophyte alga Chlamydomonas reinhardtii [6] and shown to be primarily proton channels that also conduct Na+, K+ and, to a lesser extent, divalent metal cations [7,8]. ChRs with similar selectivity but different spectral sensitivity and photocurrent kinetics have been cloned from many other chlorophytes and streptophytes [9–11] and are known as cation channel rhodopsins or CCRs. Natural ChRs with strictly anion selectivity (anion channelrhodopsins or ACRs) were first identified in cryptophytes [4] and then in several other major eukaryotic lineages [12,13].
Cation channel function appears to have evolved independently in a different group of cryptophyte ChRs that show higher protein sequence homology with haloarchaeal proton-pumping rhodopsins than with other known ChRs [14]. In particular, the homologs of three residues (Asp85, Thr89 and Asp96) in the transmembrane helix 3 (TM3) required for H+ pumping in bacteriorhodopsin (BR) are conserved in these proteins, and yet they are capable of passive conductance of protons and metal cations, like chlorophyte CCRs [15–17]. A detailed patch clamp and flash photolysis study of a representative CCR from the cryptophyte Guillardia theta (GtCCR2) has revealed that it shares with haloarchaeal H+ pumps not only the sequence similarity, but also some functional characteristics. In GtCCR2, opening of the channel from the cytoplasmic side requires the homolog of BR’s Asp96 to be unprotonated [15], as has been proposed for BR itself [18]. To highlight the similarity of their sequences and function with haloarchaeal H+-pumping rhodopsins, cryptophyte CCRs have been named “BR-like cation channelrhodopsins” or BCCRs. This naming was further justified by cryo-EM studies on the BCCR known as ChRmine (derived from the cryptophyte Rhodomonas lens [17], although initially misattributed to the ciliate Tiarina fusus [19]). ChRmine structures show trimeric organization [20,21], typical of haloarchaeal rhodopsin pumps like BR [22,23].
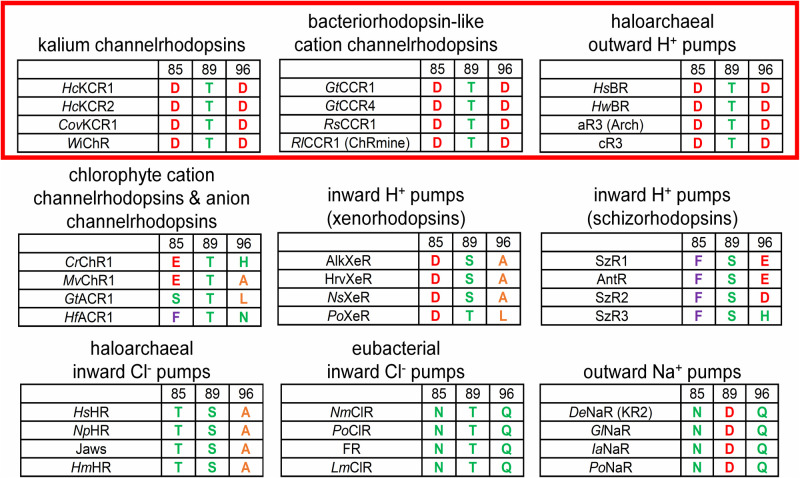
Recently, a new name, “pump-like channelrhodopsins” (PLCRs) has been proposed for BCCRs [20]. However, this name is highly confusing, because besides haloarchaeal H+ pumps, several other types of ion-pumping rhodopsins with different TM3 motifs are known, such as inward H+ pumps xenorhodopsins [24,25] and schizorhodopsins [26,27]; haloarchaeal [28] and eubacterial [29] inward Cl– pumps; and outward Na+ pumps [30–32]. (Also see recent reviews [33,34] for more examples and a more detailed discussion). However, only the DTD motif, typical of haloarchaeal outward H+ pumps first observed in BR but no other types of ion-pumping rhodopsins, is conserved in BCCRs (Figure 1), so the name BCCRs is more accurate than PLCRs and should be used in the literature.
Figure 1 .
TM3 residue motifs conserved in different families of microbial rhodopsins. Carboxylated residues are shown in red; polar residues in green; aromatic residues in violet; and small non-polar residues in orange. The red rectangle highlight that the DTD motif is only found in KCRs, BCCRs, and haloarchaeal H+ pumps. Abbreviations: HcKCR1 and HcKCR2, Hyphochytrium catenoides kalium channelrhodopsins 1 and 2, respectively; CovKCR1, Colponema vietmamica kalium channelrhodopsin 1; WiChR, Wobblia inhibitory channelrhodopsin; GtCCR1 and GtCCR4, Gulliardia theta cation channelrhodopsins 1 and 4, respectively; RsCCR1, Rhodomonas salina cation channelrhodopsin 1; RlCCR1, Rhodomonas lens cation channelrhodopsin 1; HsBR and HwBR, Halobacterium salinarum and Haloquadratum walsbyi bacteriorhodopsins, respectively; aR3, archaeorhodopsin 3; cR3, cruxrhodopsin 3; CrChR1, Chlamydomonas reinhardtii channelrhodopsin 1; MvChR1, Mesostigma viride channelrhodopsin 1; GtACR1, Gulliardia theta anion channelrhodopsin 1; HfACR1, Hondaea fermentalgiana anion channelrhodopsin 1; NsXeR, Nanosalina sp. xenorhodopsin; PoXeR, Parvularcula oceani xenorhodopsin; SzR1-3, schizorhodopsins 1-3, respectively; AntR, Antarctic rhodopsin; HsHR, Halobacterium salinarum halorhodopsin; NpHR, Natronomonas pharaonis halorhodopsin; HmHR, Haloarcula marismortui halorhodopsin; NmClR, Nonlabens marinus chloride-pumping rhodopsin; PoClR, Parvularcula oceani chloride-pumping rhodopsin; FR, Fulvimarina rhodopsin; LmClR, Lewinella maritima chloride-pumping rhodopsin; DeNaR, Dokdonia eikasta natrium rhodopsin; GlNaR, Gillisia limnaea natrium rhodopsin; IaNaR, Indibacter alkaliphilus natrium rhodopsin; PoNaR, Parvularcula oceani natrium rhodopsin.
All so far tested cryptophyte BCCRs and chlorophyte CCRs that conduct metal cations in addition to protons show a slightly lower permeability to K+ than to Na+ (PK/PNa) and are therefore used to stimulate neuronal activity by depolarizing the membrane [19,35]. Surprisingly, two ChRs from the fungus-like stramenopile Hyphochytrium catenoides showed much higher PK/PNa values (and lower PH/PNa values), hyperpolarized the membrane upon illumination and inhibited neuronal firing [36]. To reflect this unusual functionality, H. catenoides ChRs have been named “kalium channelrhodopsins” or KCRs (HcKCR1 and HcKCR2). The discovery of light-gated K+ channels in nature has attracted much attention from the research community [37] because of two reasons. First, KCRs lack the “K+ channel signature sequence” T(S)VGY(F)G that forms a tetrameric selectivity filter in previously known voltage- and ligand gated K+ channels [38] and therefore reveal an alternative mechanism for K+ selection. Second, the direction of KCR photocurrents is independent of the Cl– gradients that are reversed in the axonal terminals and immature neurons and therefore, unlike ACRs, KCRs are not compromised as optogenetic silencing tools in axons and embryonic neurons [39,40].
Out of all known ChRs, protein sequences of HcKCRs show the highest homology to those of BCCRs (including conservation of the DTD motif in TM3, Figure 1), although their source organism is phylogenetically very distant from cryptophytes. Close homologs of HcKCRs have been found in other stramenopile and colponemid protists, and in metagenomic samples [41,42], but not in cryptophytes. Close homologs of KCRs form a distinct branch of the phylogenetic tree together with KCRs, but most of these channels are not K+ selective [41,43], which provides a unique possibility to identify the residues involved in K+ selection by comparative analysis of their sequences. Only six K+ selective homologs are currently known: in addition to HcKCRs, these are WiChR from Wobblia lunata [42], B1ChR2 from Bilabrum sp. [42], and CovKCR1 and CovKCR2 from Colponema vietnamica [41]. As the first discovered, HcKCRs have been characterized in more detail than the other homologs. Recently obtained high-resolution cryo-EM structures of HcKCRs [44,45] help unravel their structure-function relationships. In this minireview we summarize and discuss so far published literature on KCRs, and outline future research directions.
Channel Gating and Intramolecular Proton Transfers
The kinetics of channel gating can only be estimated using single-turnover conditions, i.e., under ns laser flash excitation, when all ChR molecules are excited simultaneously. In HcKCR1, channel opening is biphasic with the time constants (τ) ~0.3 and 3 ms [36]. However, in contrast to other ChRs, in which channel closing was also biphasic, closing of the HcKCR1 channel could be fit with only one exponent (τ ~25–26 ms) [36,44].
As in earlier studied ChRs [46], passive channel current in HcKCR1 is preceded by active intramolecular charge displacement reflecting retinal isomerization [36]. The rise of the proton transfer current is biphasic, with τ values roughly corresponding to those of the Schiff base deprotonation monitored by flash photolysis in detergent-purified protein as the rise of the blue-shifted M intermediate. In contrast to chlorophyte CCRs [46], opening of the HcKCR1 channel is ~2-fold faster than active proton transfer. The voltage dependence of the proton transfer current is insensitive to the bath pH, indicating that the photoactive site is inaccessible to protons from outside [36]. Mutagenetic neutralization of the residue corresponding to BR’s Schiff base proton acceptor Asp85 (the D105N mutation) slowed the rise and decay of M relative to the wild type, suggesting that Asp105 is the proton acceptor [44], as is the corresponding residue (Asp87) in GtCCR2 [15]. A strong reduction of photocurrents in the GtCCR2_D87N [15] and HcKCR1_D105N [44] mutants suggests the importance of the proton transfers for channel opening in BCCRs and KCRs, in contrast to chlorophyte CCRs, in which replacement of the corresponding residue (Glu123 in CrChR2) with Thr caused acceleration of channel opening without reduction of the current amplitude [47]. In HcKCR1, mutagenetic neutralization of the 2nd carboxylate in the photoactive site (Asp229, corresponding to Asp212 in BR) also suppressed photocurrents and caused a large blue spectral shift [44] indicating deprotonation of the Schiff base at neutral pH.
Mutagenetic neutralization of Asp116 (the homolog of BRs’ Asp96) strongly decreased channel currents in HcKCR1 [41,42,44,45], but did not abolish them completely as the corresponding D98N mutation in GtCCR2 [15]. In the latter protein, deprotonation of Asp98 is required for channel opening and is >10-fold faster than reprotonation of the Schiff base, which occurs not from this residue but by proton return from earlier protonated Asp87. In HcKCRs, Asp116 controls the selectivity of the channel in addition to its gating (see the next section).
A characteristic feature of nearly all ChRs is the presence of a Cys in the position of BR’s Thr90 in the middle of TM3. In chlorophyte CCRs, this residue forms a hydrogen bond (H-bond) with a conserved Asp in TM4 (the homolog of BR’s Asp115), the so-called “DC gate” [48]. Disruption of this bond by mutation of either residue causes a dramatic slowing of photocurrent decay after the light is off [49,50]. Although Asp115 is replaced with non-polar Val (133) in HcKCRs, the C110T mutation slowed the decay >1500-fold in both channels [44]. The current view is that the homolog of Asp115 in CrChR2 (Asp156) serves as a proton donor to the Schiff base [51], and the effect of the DC gate mutations on photocurrent kinetics is attributed to retardation of the Schiff base reprotonation. However, in HcKCRs the effect of the Asp110 mutations is observed in the absence of the carboxylate H-bonding partner, which requires a different explanation. Slower decay kinetics was also found in the HcKCR1_T109V mutant [45], although the effect was less dramatic than in the C110T mutant.
In contrast to other KCRs, photocurrents generated by B1ChR2 exhibit strong inward rectification, i.e., outward currents are much smaller than inward currents recorded at the voltages equidistant from the reversal potential [42]. Most tested BCCRs show similar behavior [14,52], and most chlorophyte CCRs show weaker inward rectification [8,53,54]. In CrChR2 rectification was explained by a combination of a nonlinear transport function and asymmetric competition between several cation species [55]. In GtACR1 strong inward rectification was induced by Glu replacement of the residues near the extracellular entry into the channel pore [56]. Further research is needed to elucidate mechanisms of inward rectification in B1ChR2 and potentially to reduce it for optogenetic needs.
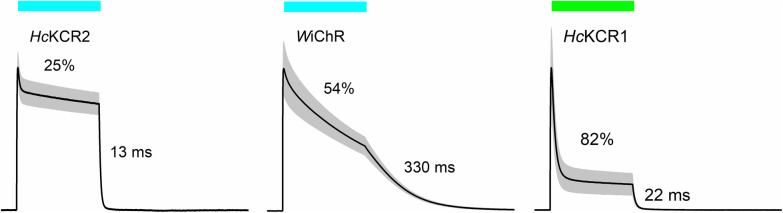
Photocurrents generated by all ChRs decrease under continuous illumination (a phenomenon known as desensitization). As other ChRs, KCR differ in the degree of desensitization (Figure 2). Mechanisms of desensitization are poorly understood and seem to involve different mechanisms in different ChRs. In Rhodomonas BCCRs desensitization is caused by accumulation of a long-lived UV-absorbing intermediate [17]. Under physiological (i.e., asymmetric) ionic conditions a decrease in positive KCR photocurrent during illumination is partially caused by a decrease in PK/PNa, as discussed in the next section.
Figure 2 .
Photocurrents generated by the indicated KCRs at –40 mV under physiological ionic conditions in response to a 1-s light pulse, the duration of which is shown by the colored bars. The current traces were normalized at the peak value. The numbers show desensitization (reduction of photocurrent at the end of the light pulse in % of the peak value) and the time of half-amplitude reduction of photocurrent after the light is turned off. The grey area shows the s.e.m. (n=5–7 cells).
Ion Selectivity
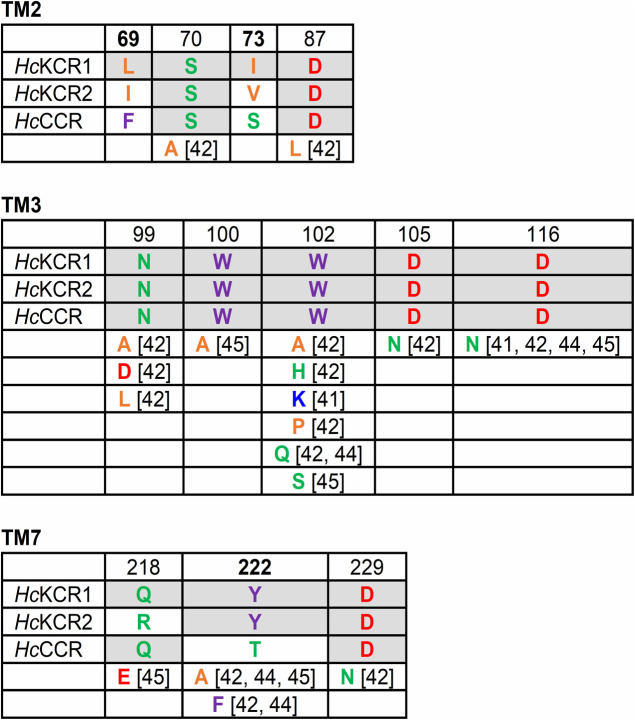
The third ChR encoded by the H. catenoides genome, designated HcCCR, is more selective for Na+ than K+ (PK/PNa 0.02) [41,43], although it shows 70–73% identity and 83–86% similarity with HcKCRs in the seven-transmembrane (7TM) domain at the primary structure level. Replacement of its individual helices with those of HcKCR1 revealed that only TM2 and TM7 confer K+ selectivity [41]. Replacement of individual residues showed that residues in the positions 69 and 73 (TM2), and 222 (TM7) are responsible for this effect, and a combination of the three mutations (F69L, S73I and T222Y) converted the Na+ channel into a K+ channel.
Individually, the S73I and T222Y mutations caused stronger effects than the F69L mutation. The latter mutation alone produced only a small increase in K+ selectivity, but its effect was increased in the S73I_T222Y background, indicating a synergistic action [41]. Ile73 corresponds to Glu90 of CrChR2, although there is little overall homology in TM2 between KCRs and chlorophyte CCRs. Replacement of Glu90 with Ala, Gln or His reduced permeability of CrChR2 to protons [57,58], and its replacement with Lys or Arg made CrChR2 permeable to anions [59]. Tyr222 is conserved in H. catenoides and C. vietnamica KCRs, but is replaced with Phe in WiChR and B1ChR2, which are more K+ selective. The WiChR_F240Y mutation decreased K+ selectivity, but so did the reverse mutation at the same position HcKCR1_Y222F [42,44], showing that the effect of the residue in this position is controlled by a wider protein matrix (i.e., interactions of the residue in this position with other residues).
The PK/PNa ratio of the triple HcCCR_F69L_S73I_T222Y mutant was only ~8 [41], indicating that there are other residues contributing to K+ selectivity in HcKCR1. Indeed, mutations of several other residues along the putative ion conduction pathway in HcKCR1 showed statistically significant shifts of the reversal potential revealing a decrease in PK/PNa, although these residues are conserved in HcKCR1 and HcCCR [36,42,44,45]. Such residues and their substitutions in TM2 (positions 70 and 87), TM3 (99, 100, 102, 105, and 116) and TM7 (218 and 229) that led to a decrease of K+ selectivity are shown in Figure 3. In addition, a small reduction of K+ selectivity was observed in the HcKCR1_N18D (the N terminus) and F144A (TM4) mutants [45].
Figure 3 .
The residues in TM2, TM3 and TM7 that determine K+ selectivity of KCRs, as demonstrated by mutation analysis. The lower rows show the substitutions of the residues conserved in HcKCR1 and HcCCR that nevertheless led to a decrease in K+ selectivity in the former. The color code is as in Fig. 1. For more detailed explanation see the text.
Out of the residues conserved in all three H. catenoides ChRs, mutations of Trp102 (the residue corresponding to Arg82 in BR) and Asp116 (Asp96 in BR) caused the greatest reduction of K+ selectivity in HcKCRs. Arg82 is conserved in most microbial rhodopsins, except BCCRs, which demonstrate high residue variability at this position (Lys in >50% of known sequences, Arg in ~35%, and Ala, Pro, Gln, Thr, Glu, Ile, or Tyr but not Trp in the remaining 15%). Metagenomic KCR homologs with Arg or Pro in this position are not K+ selective [41], which, together with the results of mutagenetic studies in HcKCRs suggests that this Trp is required for K+ selectivity. Among other microbial rhodopsins, Trp in BR’s Arg82 position is found in most xenorhodopsins that are inward H+ pumps, although Arg is conserved in the H+-pumping homolog from Parvularcula oceani (PoXeR) [24] suggesting that, in contrast to KCRs, in xenorhodopsins replacement of Arg with Trp does not change the nature of the transported ion. Asp116 is conserved not only in all known KCR homologs, but also in BCCRs, haloarchaeal H+-pumping rhodopsins, and non-electrogenic algal rhodopsins with unknown function. We conclude that its role in K+ selectivity of KCRs is determined by its interactions with other residues, some of which have been revealed by structural and computational analysis (see the next section).
Of the six currently known KCRs, WiChR shows the highest PK/PNa ratio. One molecular determinant responsible for it is Asp47 located in TM1 and corresponding to Cys29 of HcKCRs. The HcKCR1_C29D mutant showed an increased PK/PNa, and the reverse WiChR_D47C mutant, a decreased PK/PNa, compared to the respective wild types [42]. Several other HcKCR1 mutants exhibited slightly higher K+ selectivity than the wild type: Y81A, Y106A, F221A, and H225A/F/Y [44,45].
PK/PNa of HcKCR1 does not change during the single-turnover photocycle under physiological conditions [36]. However, all tested KCRs showed more depolarized reversal potentials after prolonged illumination than at the time of the peak current [36,41,42]. This was not observed under symmetrical K+ conditions or when Na+ in the bath was replaced with Ca2+ or Mg2+, which led to the conclusion that the reason for this behavior is accumulation of a late electrogenic photocycle intermediate with lower PK/PNa.
KCR Structures
Structures of both HcKCRs incorporated in nanodisks have been obtained by cryo-EM [44,45]. As those of the cryptophyte BCCR ChRmine [20,21], they form trimers characteristic of haloarchaeal ion-pumping rhodopsins with the space between the protomers filled with lipids. As in ChRmine, but not the pumps, TM3 is short (partially unwound at the extracellular side) creating a large extracellular vestibule, but in contrast to ChRmine, TM7 in HcKCRs protrudes longer into the cytoplasm, as in chlorophyte and cryptophyte ChRs.
Structural analysis has revealed that the photocurrent action spectra difference between HcKCR1 (λmax 540 nm) and HcKCR2 ((λmax 490 nm) arises from the difference in the chromophore ring/chain geometry [44]. The HcKCR2 electron density map could only be modeled using 6-s-cis retinal to avoid a steric clash of C17 of the chromophore with Ala140 (replaced with Gly in HcKCR1). In addition, Ala136 in HcKCR2 (replaced with Thr in HcKCR1) creates a cavity to accommodate C16. Rotation of the β-ionone ring shortens the π-conjugated system and induces a blue shift in HcKCR2 as compared to HcKCR1. This conclusion has been confirmed by HcKCR1_T136A_G140A and HcKCR2_A136T_A140G mutations.
The overall arrangement of the photoactive site (the Schiff base region) in HcKCRs is similar to that in ChRmine, although no water molecules have been resolved in the former. In both HcKCRs, the D229N mutation caused a larger blue shift at pH 7 than the D105N mutation, which suggests that Asp229 is unprotonated and acts as the primary counterion to the Schiff base [44]. In both HcKCRs, the side chain of Asp229 is stabilized by H-bonds with Tyr81 (TM2) and Tyr106 (TM3). These tyrosines are highly conserved in KCRs and BCCRs and form a similar H-bonding arrangement with the Asp229 homolog (Asp253) in ChRmine [20,21]. Intriguingly, Asp212 in BR and Asp234 in GtACR1, the homologs of Asp229 in HcKCRs, are also H-bonded to two Tyr residues [22,60], but the positions of the latter are different: Tyr57/72 (TM2, BR/GtACR1 numbering) corresponds to Cys77, and Tyr185/207 (TM6), to Phe202 in HcKCRs.
A fundamental difference between KCRs and canonical voltage- and ligand-gated K+ channels is that in the former the residues implicated in K+ selectivity are distributed along the putative conduction pathway between TM1, 2, 3 and 7 within each protomer [44,45] rather than form a single tetrameric selectivity filter contributed by two or four subunits, as in the latter [61]. In the cryo-EM HcKCR structures [44,45], Asp116 near the cytoplasmic entrance is H-bonded to Ser70 and Arg244, as predicted by homology models [41]. A loss in K+ selectivity in HcCCR by replacement of the nearby Ile73 with polar Ser, and of Leu69 with bulkier Phe is predicted to perturb this network, but this hypothesis needs to be verified by obtaining a high-resolution structure of HcCCR. Molecular dynamic (MD) simulations using the HcKCR1 structure show transient binding of partially dehydrated K+ to Asp116 and nearby Thr120 leading to breaking of the H-bond between Asp116 and Arg244, and reorientation of the latter towards the cytoplasm [44].
Trp102 and Tyr222 form a constriction near the extracellular entry and interact with two other aromatic residues, Phe/Tyr221 and His225 [44]. Disruption of this interaction by replacement of Tyr222 with Thr in HcCCR, and by mutations in HcKCRs decreases K+ selectivity. When measured under reversed physiological conditions (i.e., K+ outside and Na+ inside), the absolute magnitude of the reversal potentials in both HcKCRs was much smaller than under normal physiological conditions (Na+ outside and K+ inside) [44]. This observation is explained by the hydrophobic side chain cluster near the extracellular entry acting as a hydrophobic size exclusion filter that prevents flow of larger hydrated Na+ from outside but passes smaller partially dehydrated K+ and Na+ flowing from the cytoplasm.
Optogenetic Applications
Under physiological ionic conditions upon illumination, KCRs generate outward K+ fluxes and thus mimic endogenous repolarization processes in neurons mediated by voltage-gated K+ channels. Therefore, using KCRs as optogenetic inhibitors creates less undesired side effects, as compared to other classes of inhibitory tools, such as ACRs that may cause neurotransmitter release at axonal terminals and ion-pumping rhodopsins that may drive the membrane potential beyond physiological values. Illumination inhibited spiking in KCR-expressing mouse cortical [36] and hippocampal [42] neurons. Consistent with slow photocurrent decay in WiChR (Figure 2), a single 5-ms pulse results in complete cessation of firing for 500 ms in WiChR-expressing neurons upon holographic two-photon (2P) stimulation with infrared light [42]. Also because of its slower photocycle, the operational light sensitivity of WiChR was higher than that of HcKCR1 when probed with pulses of continuous light [42], as previously found in slow mutants of other ChRs [49,62]. Besides neurons, WiChR has been tested in human induced pluripotent stem cell-derived atrial cardiomyocytes (aCMs) [42]. Illumination fully suppressed spontaneous action potentials and contractions of aCM syncytia, which were restored after light was turned off. When high time-resolution is not required, the combination of a large photocurrent amplitude and the so-far largest PK/PNa value may make WiChR a preferred inhibitory tool. However, the faster HcKCRs are better suited for temporally precise silencing. Another advantage of HcKCR1 over WiChR is its more red-shifted absorption (540 nm vs. 490 nm).
Future Directions
First examples of their optogenetic applications described in the previous section have already shown that KCRs are promising neuronal silencing tools. Also, their unique selectivity mechanism, fundamentally different from that of canonical K+ channels, opens up a new avenue for molecular engineering. HcKCR dark (closed-state) structures discussed in this minireview have provided some clues to this mechanism, but better understanding requires more structure/function research. Time-resolved serial femtosecond X-ray crystallography has been instrumental in deciphering initial conformational changes following photoactivation of C1C2, a hybrid between CrChR1 and CrChR2 [63]. However, flash photolysis showed that a major conducting state (P520) was not formed in the crystals, which limits the utility of this method for analysis of channel gating. KCR mutants with long-lived open states (such as the HcKCR1_C110T mutant [44]) may provide an opportunity to freeze-capture the open state by cryo-EM.
Several strategies can be envisioned to optimize KCRs as optogenetic tools. First, net photocurrent amplitude tested by whole-cell patch clamp depends not only on the unitary conductance of a particular ChR, but also on the expression level of the transgene and targeting of the encoded protein to the cell membrane. HcKCRs and especially B1ChR2 can be made more efficient silencing tools by improving their suboptimal expression/targeting in neurons [36,42]. Second, the PK/PNa values of all known KCRs are lower than that of typical voltage-gated K+ channels. A systematic functional analysis of KCR homologs and mutants in combination with their structural analysis by cryo-EM and X-ray crystallography is needed to engineer better K+ selectivity. Alternatively, further screening of KCR homologs may identify natural variants with higher conductance and greater K+ selectivity. Finally, all known KCRs absorb blue or green light, which limits their utility in combinatorial applications such as all-optical electrophysiology – simultaneous optical perturbation and measurement of membrane voltage [64]. Also, red light penetrates more deeply through biological tissues, and therefore red-shifted optogenetic tools are desired. The spectral sensitivity of rhodopsins is mostly determined by the residues in the conserved retinal-binding pocket according to fairly well-understood quantum mechanical principles [65]. However, color-tuning mutations frequently lead to a loss of function, so a search for natural red-shifted KCR variants may be more promising than engineering the retinal pocket by mutagenesis, especially considering that the most red-shifted chlorophyte CCR [10] and cryptophyte ACR [12] have been found in this way.
The evolutionary relationship between KCRs and cryptophyte BCCRs is relatively close but remains unclear. BCCRs themselves are a very divergent family, only a few members of which have been functionally characterized [15,17,52], and the structure of only one member has been resolved [20,21]. Both functional and structural studies on other BCCRs are needed to clarify the differences between them and KCRs. It is also possible that future research will uncover some ChR variants intermediate between KCRs and BCCRs.
Chlorophyte CCRs are the only ChR family, the biological functions of which as photoreceptors in phototaxis in their source organisms have been proven by gene knockdown using RNA interference [6] and later, confirmed by RNA interference [66] and gene knockout [67,68]. Photoreceptor currents similar to those in C. reinhardtii have been recorded from all tested chlorophytes [69–72] and a freshwater cryptophyte [73]. The latter suggests that BCCRs (and possibly ACRs) also guide phototaxis. The biological functions of KCRs in their source organisms are currently unknown and need to be elucidated in the future.
To conclude, we would like to point out that KCRs are yet another example of nature being a better molecular engineer than us scientists, as we have already found earlier with ACRs, natural versions of which [4] are highly more conductive than engineered versions [74,75].
Conflict of Interest
The authors declare no conflict of interest.
Author Contributions
All authors contributed to writing of this review.
Data Availability
The evidence data generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
Note Added to Proof
While this Review was in press, a study by Fan et al. was published in Cell (https://doi.org/10.1016/j.cell.2022.12.035), in which the authors successfully used HcKCR1 to analyze hippocampal behavioral time scale plasticity in mice.
Acknowledgements
This work was supported by the National Institutes of Health grants R35GM140838 and U01NS118288, and Robert A. Welch Foundation Endowed Chair AU-0009 to JLS.
References
- [1].Emiliani, V., Entcheva, E., Hedrich, R., Hegemann, P., Konrad, K. R., Lüscher, C., et al. . Optogenetics for light control of biological systems. Nature Reviews Methods Primers 2, 55 (2022). https://doi.org/10.1038/s43586-022-00136-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Deisseroth, K., Feng, G., Majewska, A. K., Miesenböck, G., Ting, A., Schnitzer, M. J.. Next-generation optical technologies for illuminating genetically targeted brain circuits. J. Neurosci. 26, 10380–10386 (2006). https://doi.org/10.1523/JNEUROSCI.3863-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Sahel, J. A., Boulanger-Scemama, E., Pagot, C., Arleo, A., Galluppi, F., Martel, J. N., et al. . Partial recovery of visual function in a blind patient after optogenetic therapy. Nat. Med. 27, 1223–1229 (2021). https://doi.org/10.1038/s41591-021-01351-4 [DOI] [PubMed] [Google Scholar]
- [4].Govorunova, E. G., Sineshchekov, O. A., Liu, X., Janz, R., Spudich, J. L.. Natural light-gated anion channels: A family of microbial rhodopsins for advanced optogenetics. Science 349, 647–650 (2015). https://doi.org/10.1126/science.aaa7484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Mardinly, A. R., Oldenburg, I. A., Pegard, N. C., Sridharan, S., Lyall, E. H., Chesnov, K., et al. . Precise multimodal optical control of neural ensemble activity. Nat. Neurosci. 21, 881–893 (2018). https://doi.org/10.1038/s41593-018-0139-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sineshchekov, O. A., Jung, K.-H., Spudich, J. L.. Two rhodopsins mediate phototaxis to low- and high-intensity light in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U.S.A. 99, 8689–8694 (2002). https://doi.org/10.1073/pnas.122243399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Nagel, G., Ollig, D., Fuhrmann, M., Kateriya, S., Musti, A. M., Bamberg, E., et al. . Channelrhodopsin-1: A light-gated proton channel in green algae. Science 296, 2395–2398 (2002). https://doi.org/10.1126/science.1072068 [DOI] [PubMed] [Google Scholar]
- [8].Nagel, G., Szellas, T., Huhn, W., Kateriya, S., Adeishvili, N., Berthold, P., et al. . Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc. Natl. Acad. Sci. U.S.A. 100, 13940–13945 (2003). https://doi.org/10.1073/pnas.1936192100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zhang, F., Prigge, M., Beyriere, F., Tsunoda, S. P., Mattis, J., Yizhar, O., et al. . Red-shifted optogenetic excitation: A tool for fast neural control derived from Volvox carteri. Nat. Neurosci. 11, 631–633 (2008). https://doi.org/10.1038/nn.2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Klapoetke, N. C., Murata, Y., Kim, S. S., Pulver, S. R., Birdsey-Benson, A., Cho, Y. K., et al. . Independent optical excitation of distinct neural populations. Nat. Methods 11, 338–346 (2014). https://doi.org/10.1038/nmeth.2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Tashiro, R., Sushmita, K., Hososhima, S., Sharma, S., Kateriya, S., Kandori, H., et al. . Specific residues in the cytoplasmic domain modulate photocurrent kinetics of channelrhodopsin from the alga Klebsormidium nitens. Commun. Biol. 4, 235 (2021). https://doi.org/10.1038/s42003-021-01755-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Govorunova, E. G., Sineshchekov, O. A., Li, H., Wang, Y., Brown, L. S., Spudich, J. L.. RubyACRs, non-algal anion channelrhodopsins with highly red-shifted absorption. Proc. Natl. Acad. Sci. U.S.A. 117, 22833–22840 (2020). https://doi.org/10.1073/pnas.2005981117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Govorunova, E. G., Sineshchekov, O. A., Li, H., Wang, Y., Brown, L. S., Palmateer, A., et al. . Cation and anion channelrhodopsins: Sequence motifs and taxonomic distribution. MBio 12, e0165621 (2021). https://doi.org/10.1128/mBio.01656-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Govorunova, E. G., Sineshchekov, O. A., Spudich, J. L.. Structurally distinct cation channelrhodopsins from cryptophyte algae. Biophys. J. 110, 2302–2304 (2016). https://doi.org/10.1016/j.bpj.2016.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Sineshchekov, O. A., Govorunova, E. G., Li, H., Spudich, J. L.. Bacteriorhodopsin-like channelrhodopsins: Alternative mechanism for control of cation conductance. Proc. Natl. Acad. Sci. U.S.A. 114, E9512–E9519 (2017). https://doi.org/10.1073/pnas.1710702114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Shigemura, S., Hososhima, S., Kandori, H., Tsunoda, S. P.. Ion channel properties of a cation channelrhodopsin, Gt_CCR4. Appl. Sci. 9, 3440 (2019). https://doi.org/10.3390/app9173440 [Google Scholar]
- [17].Sineshchekov, O. A., Govorunova, E. G., Li, H., Wang, Y., Melkonian, M., Wong, G. K.-S., et al. . Conductance mechanisms of rapidly desensitizing cation channelrhodopsins from cryptophyte algae. mBio 11, e00657–00620 (2020). https://doi.org/10.1128/mBio.00657-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Wang, T., Sessions, A. O., Lunde, C. S., Rouhani, S., Glaeser, R. M., Duan, Y., et al. . Deprotonation of D96 in bacteriorhodopsin opens the proton uptake pathway. Structure 21, 290–297 (2013). https://doi.org/10.1016/j.str.2012.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Marshel, J. H., Kim, Y. S., Machado, T. A., Quirin, S., Benson, B., Kadmon, J., et al. . Cortical layer-specific critical dynamics triggering perception. Science 365, eaaw5202 (2019). https://doi.org/10.1126/science.aaw5202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kishi, K. E., Kim, Y. S., Fukuda, M., Inoue, M., Kusakizako, T., Wang, P. Y., et al. . Structural basis for channel conduction in the pump-like channelrhodopsin ChRmine. Cell 185, 672–689 (2022). https://doi.org/10.1016/j.cell.2022.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Tucker, K., Sridharan, S., Adesnik, H., Brohawn, S. G.. Cryo-EM structures of the channelrhodopsin ChRmine in lipid nanodiscs. Nat Commun 13, 4842 (2022). https://doi.org/10.1038/s41467-022-32441-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Luecke, H., Schobert, B., Richter, H. T., Cartailler, J. P., Lanyi, J. K.. Structure of bacteriorhodopsin at 1.55 A resolution. J. Mol. Biol. 291, 899–911 (1999). https://doi.org/10.1006/jmbi.1999.3027 [DOI] [PubMed] [Google Scholar]
- [23].Shibata, M., Inoue, K., Ikeda, K., Konno, M., Singh, M., Kataoka, C., et al. . Oligomeric states of microbial rhodopsins determined by high-speed atomic force microscopy and circular dichroic spectroscopy. Sci. Rep. 8, 8262 (2018). https://doi.org/10.1038/s41598-018-26606-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Inoue, K., Ito, S., Kato, Y., Nomura, Y., Shibata, M., Uchihashi, T., et al. . A natural light-driven inward proton pump. Nat. Commun. 7, 13415 (2016). https://doi.org/10.1038/ncomms13415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Shevchenko, V., Mager, T., Kovalev, K., Polovinkin, V., Alekseev, A., Juettner, J., et al. . Inward H+ pump xenorhodopsin: Mechanism and alternative optogenetic approach. Sci. Adv. 3, e1603187 (2017). https://doi.org/10.1126/sciadv.1603187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Inoue, K., Tsunoda, S. P., Singh, M., Tomida, S., Hososhima, S., Konno, M., et al. . Schizorhodopsins: A novel family of rhodopsins from Asgard archaea that function as light-driven inward H+ pumps. Sci. Adv. 6, eaaz2441 (2020). https://doi.org/10.1126/sciadv.aaz2441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Harris, A., Lazaratos, M., Siemers, M., Watt, E., Hoang, A., Tomida, S., et al. . Mechanism of inward proton transport in an antarctic microbial rhodopsin. J. Phys. Chem. B 124, 4851–4872 (2020). https://doi.org/10.1021/acs.jpcb.0c02767 [DOI] [PubMed] [Google Scholar]
- [28].Matsuno-Yagi, A., Mukohata, Y.. Two possible roles of bacteriorhodopsin; A comparative study of strains of Halobacterium halobium differing in pigmentation. Biochem. Biophys. Res. Commun. 78, 237–243 (1977). https://doi.org/10.1016/0006-291x(77)91245-1 [DOI] [PubMed] [Google Scholar]
- [29].Yoshizawa, S., Kumagai, Y., Kim, H., Ogura, Y., Hayashi, T., Iwasaki, W., et al. . Functional characterization of flavobacteria rhodopsins reveals a unique class of light-driven chloride pump in bacteria. Proc. Natl. Acad. Sci. U.S.A. 111, 6732–6737 (2014). https://doi.org/10.1073/pnas.1403051111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Inoue, K., Ono, H., Abe-Yoshizumi, R., Yoshizawa, S., Ito, H., Kogure, K., et al. . A light-driven sodium ion pump in marine bacteria. Nat. Commun. 4, 1678 (2013). https://doi.org/10.1038/ncomms2689 [DOI] [PubMed] [Google Scholar]
- [31].Balashov, S. P., Imasheva, E. S., Dioumaev, A. K., Wang, J. M., Jung, K. H., Lanyi, J. K.. Light-driven Na+ pump from Gillisia limnaea: A high-affinity Na+ binding site is formed transiently in the photocycle. Biochemistry 53, 7549–7561 (2014). https://doi.org/10.1021/bi501064n [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kajimoto, K., Kikukawa, T., Nakashima, H., Yamaryo, H., Saito, Y., Fujisawa, T., et al. . Transient resonance raman spectroscopy of a light-driven sodium-ion-pump rhodopsin from Indibacter alkaliphilus. J Phys Chem B 121, 4431–4437 (2017). https://doi.org/10.1021/acs.jpcb.7b02421 [DOI] [PubMed] [Google Scholar]
- [33].Inoue, K. Diversity, mechanism, and optogenetic application of light-driven ion pump rhodopsins. In Optogenetics: Light-Sensing Proteins and Their Applications in Neuroscience and Beyond. (Yawo, H., Kandori, H., Koizumi, A., Kageyama, R. eds.) vol. 1293, pp. 89–126 (Springer Singapore, 2021). [DOI] [PubMed] [Google Scholar]
- [34].Rozenberg, A., Inoue, K., Kandori, H., Béjà, O.. Microbial rhodopsins: The last two decades. Annu. Rev. Microbiol. 75, 427–447 (2021). https://doi.org/10.1146/annurev-micro-031721-020452 [DOI] [PubMed] [Google Scholar]
- [35].Boyden, E. S., Zhang, F., Bamberg, E., Nagel, G., Deisseroth, K.. Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 8, 1263–1268 (2005). https://doi.org/10.1038/nn1525 [DOI] [PubMed] [Google Scholar]
- [36].Govorunova, E. G., Gou, Y., Sineshchekov, O. A., Li, H., Lu, X., Wang, Y., et al. . Kalium channelrhodopsins are natural light-gated potassium channels that mediate optogenetic inhibition. Nat. Neurosci. 25, 967–974 (2022). https://doi.org/10.1038/s41593-022-01094-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Strack, R. Light-gated potassium channels from nature. Nat. Methods 19, 923 (2022). https://doi.org/10.1038/s41592-022-01582-4 [DOI] [PubMed] [Google Scholar]
- [38].Mironenko, A., Zachariae, U., de Groot, B. L., Kopec, W.. The persistent question of potassium channel permeation mechanisms. J. Mol. Biol. 433, 167002 (2021). https://doi.org/10.1016/j.jmb.2021.167002 [DOI] [PubMed] [Google Scholar]
- [39].Mahn, M., Prigge, M., Ron, S., Levy, R., Yizhar, O.. Biophysical constraints of optogenetic inhibition at presynaptic terminals. Nat. Neurosci. 19, 554–556 (2016). https://doi.org/10.1038/nn.4266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Messier, J. E., Chen, H., Cai, Z. L., Xue, M.. Targeting light-gated chloride channels to neuronal somatodendritic domain reduces their excitatory effect in the axon. eLife 7, e38506 (2018). https://doi.org/10.7554/eLife.38506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Govorunova, E. G., Sineshchekov, O. A., Brown, L. S., Bondar, A. N., Spudich, J. L. Structural foundations of potassium selectivity in channelrhodopsins. mBio, e0303922 (2022). https://doi.org/10.1128/mbio.03039-22 [DOI] [PMC free article] [PubMed]
- [42].Vierock, J., Peter, E., Grimm, C., Rozenberg, A., Chen, I. W., Tillert, L., et al. WiChR, a highly potassium selective channelrhodopsin for low-light one- and two-photon inhibition of excitable cells. Sci. Adv. eadd7729 (2022). https://doi.org/10.1126/sciadv.add7729 [DOI] [PMC free article] [PubMed]
- [43].Govorunova, E. G., Sineshchekov, O. A., Brown, L. S., Spudich, J. L.. Biophysical characterization of light-gated ion channels using planar automated patch clamp. Front. Mol. Neurosci. 15, 976910 (2022). https://doi.org/10.3389/fnmol.2022.976910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Tajima, S., Kim, Y. S., Fukuda, M., Byrne, E. F. X., Wang, P. Y., Paggi, J. M., et al. Structural basis for ion selectivity in potassium-selective channelrhodopsins. bioRxiv (2022). https://doi.org/10.1101/2022.10.30.514430 [DOI] [PMC free article] [PubMed]
- [45].Zhang, M., Shan, Y., Pei, D. Cryo-EM structures of kalium channelrhodopsins KCRs. bioRxiv (2022). https://doi.org/10.1101/2022.11.09.515798
- [46].Sineshchekov, O. A., Govorunova, E. G., Wang, J., Li, H., Spudich, J. L.. Intramolecular proton transfer in channelrhodopsins. Biophys. J. 104, 807–817 (2013). https://doi.org/10.1016/j.bpj.2013.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Gunaydin, L. A., Yizhar, O., Berndt, A., Sohal, V. S., Deisseroth, K., Hegemann, P.. Ultrafast optogenetic control. Nat. Neurosci. 13, 387–392 (2010). https://doi.org/10.1038/nn.2495 [DOI] [PubMed] [Google Scholar]
- [48].Radu, I., Bamann, C., Nack, M., Nagel, G., Bamberg, E., Heberle, J.. Conformational changes of channelrhodopsin-2. J. Am. Chem. Soc. 131, 7313–7319 (2009). https://doi.org/10.1021/ja8084274 [DOI] [PubMed] [Google Scholar]
- [49].Berndt, A., Yizhar, O., Gunaydin, L. A., Hegemann, P., Deisseroth, K.. Bi-stable neural state switches. Nat. Neurosci. 12, 229–234 (2009). https://doi.org/10.1038/nn.2247 [DOI] [PubMed] [Google Scholar]
- [50].Dawydow, A., Gueta, R., Ljaschenko, D., Ullrich, S., Hermann, M., Ehmann, N., et al. . Channelrhodopsin-2-XXL, a powerful optogenetic tool for low-light applications. Proc. Natl. Acad. Sci. U.S.A. 111, 13972–13977 (2014). https://doi.org/10.1073/pnas.1408269111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Lorenz-Fonfria, V. A., Resler, T., Krause, N., Nack, M., Gossing, M., Fischer von Mollard, G., et al. . Transient protonation changes in channelrhodopsin-2 and their relevance to channel gating. Proc. Natl. Acad. Sci. U.S.A. 110, E1273–1281 (2013). https://doi.org/10.1073/pnas.1219502110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Yamauchi, Y., Konno, M., Ito, S., Tsunoda, S. P., Inoue, K., Kandori, H.. Molecular properties of a DTD channelrhodopsin from Guillardia theta. Biophys. Physicobiol. 14, 57–66 (2017). https://doi.org/10.2142/biophysico.14.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Hou, S. Y., Govorunova, E. G., Ntefidou, M., Lane, C. E., Spudich, E. N., Sineshchekov, O. A., et al. . Diversity of Chlamydomonas channelrhodopsins. Photochem. Photobiol. 88, 119–128 (2012). https://doi.org/10.1111/j.1751-1097.2011.01027.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Watanabe, S., Ishizuka, T., Hososhima, S., Zamani, A., Hoque, M. R., Yawo, H.. The regulatory mechanism of ion permeation through a channelrhodopsin derived from Mesostigma viride (MvChR1). Photochem. Photobiol. Sci. 15, 365–374 (2016). https://doi.org/10.1039/c5pp00290g [DOI] [PubMed] [Google Scholar]
- [55].Gradmann, D., Berndt, A., Schneider, F., Hegemann, P.. Rectification of the channelrhodopsin early conductance. Biophys. J. 101, 1057–1068 (2011). https://doi.org/10.1016/j.bpj.2011.07.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Sineshchekov, O. A., Govorunova, E. G., Li, H., Wang, X., Spudich, J. L.. Opposite charge movements within the photoactive site modulate two-step channel closing in GtACR1. Biophys. J. 117, 2034–2040 (2019). https://doi.org/10.1016/j.bpj.2019.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Ruffert, K., Himmel, B., Lall, D., Bamann, C., Bamberg, E., Betz, H., et al. . Glutamate residue 90 in the predicted transmembrane domain 2 is crucial for cation flux through channelrhodopsin 2. Biochem. Biophys. Res. Commun. 410, 737–743 (2011). https://doi.org/10.1016/j.bbrc.2011.06.024 [DOI] [PubMed] [Google Scholar]
- [58].Eisenhauer, K., Kuhne, J., Ritter, E., Berndt, A. E., Wolf, S., Freier, E., et al. . In channelrhodopsin-2 E90 is crucial for ion selectivity and is deprotonated during the photocycle. J. Biol. Chem. 287, 6904–6911 (2012). https://doi.org/10.1074/jbc.M111.327700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Wietek, J., Wiegert, J. S., Adeishvili, N., Schneider, F., Watanabe, H., Tsunoda, S. P., et al. . Conversion of channelrhodopsin into a light-gated chloride channel. Science 344, 409–412 (2014). https://doi.org/10.1126/science.1249375 [DOI] [PubMed] [Google Scholar]
- [60].Li, H., Huang, C. Y., Govorunova, E. G., Schafer, C. T., Sineshchekov, O. A., Wang, M., et al. . Crystal structure of a natural light-gated anion channelrhodopsin. eLife 8, e41741 (2019). https://doi.org/10.7554/eLife.41741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].MacKinnon, R. Potassium channels. FEBS Lett. 555, 62–65 (2003). https://doi.org/10.1016/s0014-5793(03)01104-9 [DOI] [PubMed] [Google Scholar]
- [62].Govorunova, E. G., Sineshchekov, O. A., Hemmati, R., Janz, R., Morelle, O., Melkonian, M., et al. . Extending the time domain of neuronal silencing with cryptophyte anion channelrhodopsins. eNeuro 5, 0174–0118 (2018). https://doi.org/10.1523/ENEURO.0174-18.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Oda, K., Nomura, T., Nakane, T., Yamashita, K., Inoue, K., Ito, S., et al. . Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin. eLife 10, e62389 (2021). https://doi.org/10.7554/eLife.62389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Fan, L. Z., Kheifets, S., Bohm, U. L., Wu, H., Piatkevich, K. D., Xie, M. E., et al. . All-optical electrophysiology reveals the role of lateral inhibition in sensory processing in cortical layer 1. Cell 180, 521–535 (2020). https://doi.org/10.1016/j.cell.2020.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Katayama, K., Sekharan, S., Sudo, Y. Color tuning in retinylidene proteins. in Optogenetics: Light-Sensing Proteins and their Applications. (Yawo, H., Kandori, H., Koizumi, A. eds.) pp. 89–109 (Springer Japan, Tokyo 2015). [Google Scholar]
- [66].Berthold, P., Tsunoda, S. P., Ernst, O. P., Mages, W., Gradmann, D., Hegemann, P.. Channelrhodopsin-1 initiates phototaxis and photophobic responses in Chlamydomonas by immediate light-induced depolarization. Plant Cell 20, 1665–1677 (2008). https://doi.org/10.1105/tpc.108.057919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Greiner, A., Kelterborn, S., Evers, H., Kreimer, G., Sizova, I., Hegemann, P.. Targeting of photoreceptor genes in Chlamydomonas reinhardtii via zinc-finger nucleases and CRISPR/Cas9. Plant Cell 29, 2498–2518 (2017). https://doi.org/10.1105/tpc.17.00659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Baidukova, O., Oppermann, J., Kelterborn, S., Fernandez Lahore, R. G., Schumacher, D., Evers, H., et al. . Gating and ion selectivity of channelrhodopsins are critical for photo-activated orientation of Chlamydomonas as shown by in vivo point mutation. Nat. Commun. 13, 7253 (2022). https://doi.org/10.1038/s41467-022-35018-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Litvin, F. F., Sineshchekov, O. A., Sineshchekov, V. A.. Photoreceptor electric potential in the phototaxis of the alga Haematococcus pluvialis. Nature 271, 476–478 (1978). https://doi.org/10.1038/271476a0 [DOI] [PubMed] [Google Scholar]
- [70].Harz, H., Hegemann, P.. Rhodopsin-regulated calcium currents in Chlamydomonas. Nature 351, 489–491 (1991). https://doi.org/10.1038/351489a0 [Google Scholar]
- [71].Braun, F. J., Hegemann, P.. Two light-activated conductances in the eye of the green alga Volvox carteri. Biophys. J. 76, 1668–1678 (1999). https://doi.org/10.1016/S0006-3495(99)77326-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Govorunova, E. G., Sineshchekov, O. A., Li, H., Janz, R., Spudich, J. L.. Characterization of a highly efficient blue-shifted channelrhodopsin from the marine alga Platymonas subcordiformis. J. Biol. Chem. 288, 29911–29922 (2013). https://doi.org/10.1074/jbc.M113.505495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Sineshchekov, O. A., Govorunova, E. G., Jung, K.-H., Zauner, S., Maier, U.-G., Spudich, J. L.. Rhodopsin-mediated photoreception in cryptophyte flagellates. Biophys. J. 89, 4310–4319 (2005). https://doi.org/10.1529/biophysj.105.070920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Wietek, J., Beltramo, R., Scanziani, M., Hegemann, P., Oertner, T. G., Wiegert, J. S.. An improved chloride-conducting channelrhodopsin for light-induced inhibition of neuronal activity in vivo. Sci. Rep. 5, 14807 (2015). https://doi.org/10.1038/srep14807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Berndt, A., Lee, S. Y., Wietek, J., Ramakrishnan, C., Steinberg, E. E., Rashid, A. J., et al. . Structural foundations of optogenetics: Determinants of channelrhodopsin ion selectivity. Proc. Natl. Acad. Sci. U.S.A. 113, 822–829 (2016). https://doi.org/10.1073/pnas.1523341113 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The evidence data generated and/or analyzed during the current study are available from the corresponding author on reasonable request.