ABSTRACT
Wolbachia are a genus of insect endosymbiotic bacteria which includes strains wMel and wAlbB that are being utilized as a biocontrol tool to reduce the incidence of Aedes aegypti-transmitted viral diseases like dengue. However, the precise mechanisms underpinning the antiviral activity of these Wolbachia strains are not well defined. Here, we generated a panel of Ae. aegypti-derived cell lines infected with antiviral strains wMel and wAlbB or the non-antiviral Wolbachia strain wPip to understand host cell morphological changes specifically induced by antiviral strains. Antiviral strains were frequently found to be entirely wrapped by the host endoplasmic reticulum (ER) membrane, while wPip bacteria clustered separately in the host cell cytoplasm. ER-derived lipid droplets (LDs) increased in volume in wMel- and wAlbB-infected cell lines and mosquito tissues compared to cells infected with wPip or Wolbachia-free controls. Inhibition of fatty acid synthase (required for triacylglycerol biosynthesis) reduced LD formation and significantly restored ER-associated dengue virus replication in cells occupied by wMel. Together, this suggests that antiviral Wolbachia strains may specifically alter the lipid composition of the ER to preclude the establishment of dengue virus (DENV) replication complexes. Defining Wolbachia’s antiviral mechanisms will support the application and longevity of this effective biocontrol tool that is already being used at scale.
IMPORTANCE
Aedes aegypti transmits a range of important human pathogenic viruses like dengue. However, infection of Ae. aegypti with the insect endosymbiotic bacterium, Wolbachia, reduces the risk of mosquito to human viral transmission. Wolbachia is being utilized at field sites across more than 13 countries to reduce the incidence of viruses like dengue, but it is not well understood how Wolbachia induces its antiviral effects. To examine this at the subcellular level, we compared how different strains of Wolbachia with varying antiviral strengths associate with and modify host cell structures. Strongly antiviral strains were found to specifically associate with the host endoplasmic reticulum and induce striking impacts on host cell lipid droplets. Inhibiting Wolbachia-induced lipid redistribution partially restored dengue virus replication demonstrating this is a contributing role for Wolbachia's antiviral activity. These findings provide new insights into how antiviral Wolbachia strains associate with and modify Ae. aegypti host cells.
KEYWORDS: Wolbachia, Aedes aegypti, dengue, flavivirus, antiviral, arbovirus
INTRODUCTION
Arthropod-borne viruses (arboviruses) including dengue (DENV), Zika (ZIKV), chikungunya (CHIKV), and yellow fever virus (YFV) are primarily transmitted by female Aedes aegypti and, to a lesser extent, by female Aedes albopictus (1–3). The global spread of these viruses has dramatically increased in recent decades. This is largely due to factors associated with the geographic distribution of Aedes spp. such as climate change (4–7), globalization (8, 9), urbanization (9, 10), resistance to insecticides (11), and the lack of effective vector control strategies (12).
A promising biological vector control strategy that has emerged in the last decade employs antiviral strains of Wolbachia to restrict mosquito-to-human transmission of Ae. aegypti-borne viruses (13–17). Wolbachia is an intracellular gram-negative endosymbiotic bacterium that infects a wide range of invertebrates, including arthropods and nematodes, but it is not naturally found in Ae. aegypti mosquitoes (18, 19). Wolbachia strains can be isolated from native hosts such as Drosophila melanogaster (wMel) (14, 20) and Ae. albopictus (wAlbB) (15, 21, 22) and then introduced into heterologous arthropods including Ae. aegypti. Strains such as wMel and wAlbB induce an antiviral state in Ae. aegypti reducing the rate of infection, dissemination, and transmission of +RNA viruses such as DENV (14, 20, 23). Wolbachia’s exceptional ability to infect and alter the host germ line to facilitate their vertical transmission through the maternal lineage underpins the application of this bacterium as a biocontrol tool (14). Additionally, many Wolbachia strains are capable of inducing cytoplasmic incompatibility (CI), a condition that gives Wolbachia-carrying females a reproductive advantage (24, 25). These strains can be introgressed into wild-type Ae. aegypti populations to reduce their transmission potential. This approach has been shown to dramatically and significantly reduce the incidence of dengue in communities (26–31).
DENV is a member of the Flaviviridae family with a single positive-strand RNA genome that is surrounded by the virally encoded capsid protein in a host-derived lipid bilayer (3, 32). The viral genome is released into the host cell cytosol after entering susceptible cells via endocytosis and is translated by host machinery at the rough endoplasmic reticulum (33, 34). DENV induces drastic rearrangements of endoplasmic reticulum (ER) membranes to form viral replication complexes that are critically required to coordinate the multiple steps of viral replication, genome translation, and virion assembly (33–39). Wolbachia’s antiviral impacts are believed to be systemic in mosquitoes (40), but also cell autonomous whereby Wolbachia-infected cells do not protect surrounding Wolbachia-free cells from viral infection (41). At the subcellular level, viral restriction is believed to occur early before viral replication is effectively initiated (42), and low levels of progeny virus produced in the presence of Wolbachia have reduced infectivity (43). How Wolbachia induces these antiviral effects is not well understood, but a variety of hypotheses have been examined. These include competition between Wolbachia and viruses for nutrients and physical space within host cells (13, 44–48) and altered expression of host pro- or antiviral genes (49–52), including priming of immune pathways (23, 53, 54). Implicating the mechanisms that drive the Wolbachia-induced antiviral state has been difficult due to technical limitations such as failure to genetically modify Wolbachia and to grow it axenically. However, new experimental models may help to advance our understanding of Wolbachia-induced host phenotypes. Specifically, we recently found that Wolbachia strain wPip (derived from Culex quinquefasciatus) does not inhibit flavivirus replication, dissemination, or transmission in Ae. aegypti (23, 55). We hypothesized that pairwise comparisons between Ae. aegypti infected with Wolbachia strains that do or do not induce an antiviral state may facilitate the dissociation of Wolbachia’s antiviral effects from the general symbiont effects. Here, we generated a unique panel of Ae. aegypti-derived cell lines infected with wMel, wAlbB (the antiviral stains being utilized in Ae. aegypti biocontrol programs), or wPip to identify the subcellular changes specifically induced by antiviral Wolbachia strains. We identify intimate Wolbachia-ER interactions and triacylglyceride biosynthesis for LD formation as key host cell modifications that contribute to the Wolbachia-induced antiviral state.
RESULTS
Generation of a panel of Wolbachia-infected Ae. aegypti-derived cell lines
To examine the subcellular changes specifically induced by antiviral Wolbachia strains, we generated a panel of Wolbachia-infected cell lines using the immunocompetent Ae. aegypti-derived cell line (Aag2). Aag2 cells infected with antiviral strain wMel and the wMel-cured line (wMel.Tet-Aag2) have been described previously (56). wAlbB-Aag2 and wPip-Aag2 cell lines were produced by stable infection of wMel.Tet-Aag2 using Wolbachia isolated from previously generated cell lines (RML12-wAlbB) or Ae. aegypti eggs (Rockefeller-wPip). This was done to account for insect-specific flaviviruses known to infect Aag2 cells but not wMel-Aag2 or wMel.Tet-Aag2 (herein referred to as Wolbachia-free-Aag2) (56).
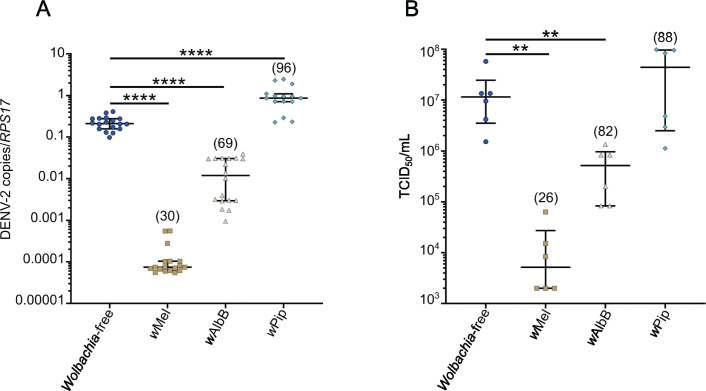
To confirm that wAlbB and wPip had antiviral and non-antiviral impacts in these cell lines, respectively, we evaluated replication of DENV serotype 2 (DENV-2) in Wolbachia-free, wMel-Aag2, wAlbB-Aag2, and wPip-Aag2 cells. Consistent with our previous findings in Ae. aegypti (23, 55), wPip did not restrict DENV-2 replication or production of infectious virus compared to the Wolbachia-free line (Fig. 1A and B). Notably, wPip was found to grow to a higher density than wMel and wAlbB (average of 96 Wolbachia per cell, compared to 30 and 69 for wMel and wAlbB, respectively) (Fig. 1A, in parentheses). By contrast, DENV-2 RNA copies and infectious virus were significantly reduced by wMel’s potent antiviral activity (approximately 3 log10 reduction compared to Wolbachia-free). wAlbB also significantly inhibited DENV-2 replication and infectious virus in Aag2 cell lines, but the antiviral activity was weaker than wMel (approximately 1 log10 reduction compared to Wolbachia-free). This confirms that the diverse antiviral phenotypes of Wolbachia strains previously demonstrated in Ae. aegypti mosquitoes (20, 22, 23) can be recapitulated in Aag2 cell lines.
Fig 1.
wPip does not restrict DENV-2 replication in Aag2 cells. Ae. aegypti-derived cell lines (Aag2) stably infected with wMel, wAlbB, and wPip strains were infected with DENV-2 at MOI 1 and compared to their matched Wolbachia-free line. Infectious virus and cellular RNA were collected from each cell line 7 days post-infection for analysis by tissue culture infectious dose 50 (TCID50) and qRT-PCR. The numbers in parentheses represent the average number of Wolbachia per cell (Wolbachia 16S rRNA/RPS17), determined in parallel wells for each independent experiment prior to DENV-2 inoculation. (A) Data are the median number of virus genome copies relative to the RPS17 mosquito housekeeping gene ±IQR. (B) Data are the median infectious viral titres ± IQR determined by TCID50. Data are derived from at least 3 (A) or 2 (B) independent experiments performed with triplicate biological replicates each. Statistical analyses were performed using Mann-Whitney test, where **P < 0.005, ****P < 0.0001.
Antiviral Wolbachia strains associate with host ER membranes
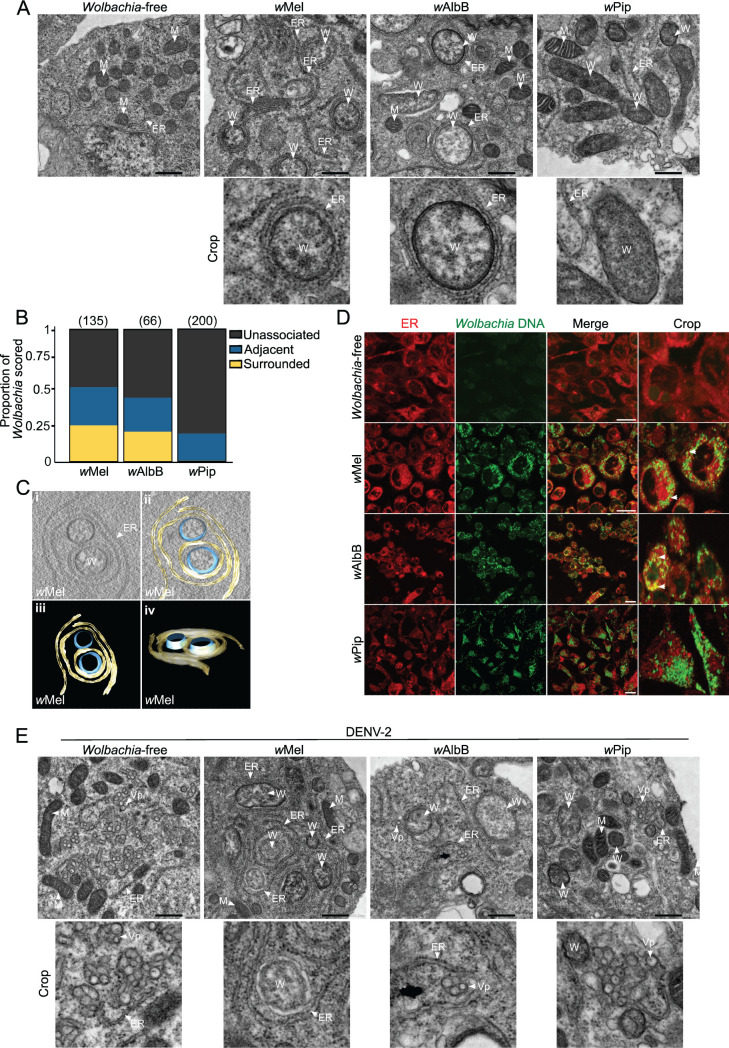
Wolbachia and viruses are both obligate intracellular residents of eukaryotic cells that rely on a variety of host structures and processes to complete their life cycles. Interestingly, Wolbachia titers and intracellular distribution have been shown to be influenced by the host’s genetic background (46, 57, 58). To gain insights into how Wolbachia strains may associate with Ae. aegypti cells to induce an antiviral phenotype, we assessed our panel of Aag2 cell lines using transmission electron microscopy (TEM). Micrographs of the three Wolbachia-infected mosquito cell lines show each cell line is heavily infected with their respective Wolbachia strain (top row, Fig. 2A; Fig. S1). Wolbachia appeared similar in size and shape to mitochondria but could be differentiated by a lower electron density and an absence of cristae. We observed that antiviral strains, wMel and wAlbB, were frequently associated with host membranes that appeared to be the host endoplasmic reticulum (ER), while the non-antiviral strain wPip was not (bottom row, Fig. 2A; Fig. S1e through p). By scoring the closeness of interaction between Wolbachia and the ER, a striking difference was evident between antiviral and non-antiviral strains: approximately 50% of wMel and wAlbB bacteria were in close contact with ER membranes in Aag2 cell lines. Of those, 25% were clearly surrounded by the membranes (Fig. 2B and C). This tight association was observed in our reconstructed tomograms of wMel where the bacterium was found to be surrounded by the ER double membrane and, in some instances, was wrapped multiple times (Fig. 2C—see Video S1 for the reconstructed tomogram). Meanwhile, over 75% of wPip bacteria were not located near any intracellular membranes, and only 25% were situated adjacent to any part of the ER (Fig. 2B; Fig. S1m through p). To further confirm that the membrane structures were, indeed, ER, we carried out live cell imaging using an ER tracker dye which binds to the sulfonylurea receptors of ATP-sensitive K+ channels present on ER membranes, and SYTO11 which preferentially stains Wolbachia DNA over eukaryote DNA (57, 59). Confocal fluorescence imaging confirmed the antiviral strains wMel and wAlbB exhibited substantial colocalization with ER membranes (white arrowheads, Fig. 2D), while wPip did not, instead forming a massive cluster of cytosolic Wolbachia (Fig. 2D).
Fig 2.
Antiviral Wolbachia strains associate with host endoplasmic reticulum membranes. (A) TEM micrographs of Aag2 cell lines stably infected with wMel, wAlbB, and wPip show their intracellular distribution and association with ER membranes. Scale bar = 500 nm. (B) Wolbachia-ER interactions were manually scored as “unassociated,” “adjacent,” or “surrounded” from two independent experiments, where “unassociated" Wolbachia did not contact any host membrane, “adjacent” Wolbachia were seen to have a contact point with a host membrane but were not surrounded, and “surrounded” Wolbachia were entirely encompassed by a host membrane. Wolbachia were counted from a minimum of 28 cells in total per line. The number of Wolbachia bacteria scored is in parentheses. (C) TEM tomography shows wMel strain surrounded by ER membranes. See Video S1 for the reconstructed tomogram. (D) Live imaging of all Aag2 cell lines stained with SYTO 11 (Wolbachia DNA—green) and ER tracker (red). Scale bar = 15 µm. (E) TEM of all Aag2 cell lines infected with DENV-2 at MOI 2 for 48 h. Scale bar = 500 nm. ER, endoplasmic reticulum; W, Wolbachia; M, mitochondria; Vp, vesicle packets.
DENV and other flaviviruses significantly remodel ER membranes to form replication complexes (32, 33, 35, 36, 60). To determine whether the distinct ER interaction of antiviral Wolbachia strains precludes DENV-2 association with the host ER, we infected each Aag2 cell line with DENV-2 and performed TEM analyses. We observed numerous viral replication sites containing many viral vesicle packets (Vp) in the cytosol of the Wolbachia-free line (white arrowheads, Fig. 2E), consistent with the high viral titers determined in this cell line (Fig. 1). We found no evidence of virus replication in any of the wMel-Aag2 cell line micrographs examined, and few and smaller Vp were observed in the wAlbB-Aag2 cell line (white arrowheads, Fig. 2E). Further consistent with our data in Fig. 1, wPip showed numerous Vp in the cytosol, similar to the Wolbachia-free control (white arrowheads, Fig. 2E). Together this data set provides the first insight into how antiviral Wolbachia strains differentially associate with Ae. aegypti ER membranes and prevent formation of typical DENV-2 replication complexes.
Antiviral Wolbachia strains increase lipid droplet formation in Aag2 cells
It has previously been reported that the ER-derived organelle, lipid droplets (LDs) are upregulated following DENV infection, contributing to the cellular antiviral response in both mammalian and Ae. aegypti-derived cell lines (61, 62). Since the ER provides most of the constituent molecules for LDs (63), we next hypothesized that the association of antiviral Wolbachia strains with host ER membranes could further manipulate other intracellular lipid sources.
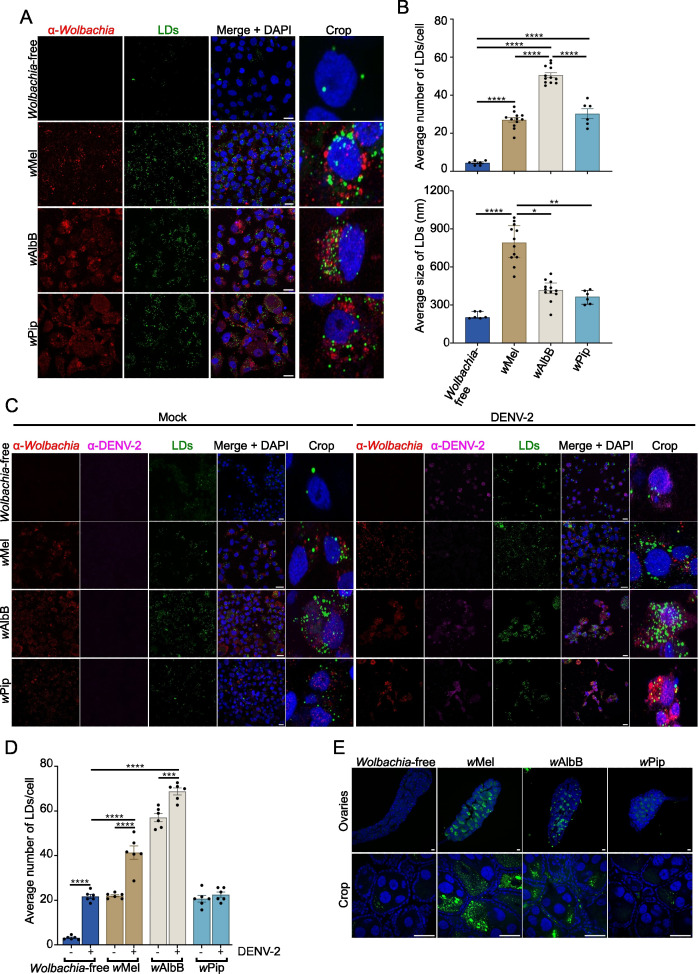
To examine intracellular LDs, we stained all Aag2 cell lines with BODIPY 493/503 and analyzed the number and size of LDs per cell. Notably, compared to the Wolbachia-free line, all Wolbachia-infected lines induced LD accumulation (Fig. 3A and B). Interestingly, wMel induced the accumulation of a similar number of LD’s per cell as wPip (27 and 30.2 per cell, respectively), while wAlbB induced nearly twice the amount (50.5 per cell). However, wMel-induced LDs were significantly larger than those induced by wAlbB and wPip (791.5, 417.5, and 365.5 nm, respectively) (Fig. 3B). This implies that antiviral Wolbachia strains might boost the total volume of LDs per cell.
Fig 3.
Antiviral Wolbachia strains increase lipid droplet formation in mosquito cells. (A–D) Aag2 cells stably infected with Wolbachia strains wMel, wAlbB, or wPip, or Wolbachia-free cells were mock-infected or DENV-2 infected at MOI 1 for 24 h. (A and C) All cells were stained with BODIPY (493/503) to visualize LDs (green) and DAPI to visualize the cell nuclei (blue). Wolbachia was detected with an α-WSP antibody (red) and 4G2 hybridoma fluid against flavivirus group E antigen for DENV-2 staining (magenta). Scale bars = 15 µm. (B) LD numbers and sizes were analyzed using Fiji software. Data are the mean LD number ± SEM (top) or median LD size ± IQR (bottom) from at least 6 fields of view representing at least 80 cells in total where each data point represents the mean LD number/size from a single field of view. Data are from one experiment, representative of two independent experiments, where each experiment included triplicate biological replicates. Statistical analyses were performed by one-way ANOVA with Tukey’s correction for multiple comparisons (top graph) or Kruskal-Wallis test with Dunn’s correction for multiple comparisons (bottom graph), *P < 0.05, **P < 0.005, ****P < 0.0001. (D) LD numbers were analyzed using Fiji software. Data are the mean LD number ± SEM from at least 6 fields of view representing at least 80 cells in total where each data point represents the mean LD number from a single field of view. Data are from one experiment, representative of two independent experiments, where each experiment included triplicate biological replicates. Statistical analyses were performed by two-way ANOVA with Tukey’s correction for multiple comparisons, *P < 0.05, **P < 0.005, ***P < 0.0005, ****P < 0.0001. (E) Ovaries were dissected from female mosquitoes 5–7 days post-emergence and stained with BODIPY (green) and DAPI (blue). Scale bars = 50 µm. All slides were imaged as 3-dimensional z-stacks and 2D images generated by Maximum Intensity Projection (MIP) using Fiji software. Images are from a single representative experiment performed on ovaries from at least three mosquitoes.
Lipid biosynthesis and degradation play a role in several stages of DENV infection, including viral replication, assembly, and energy supply (35, 64–66). Similarly, Wolbachia proliferation is tightly associated with changes in the host lipidome (67–69). Next, we investigated whether DENV-2 infection affected LD accumulation in the presence of each Wolbachia strain. In line with previous findings (61), DENV-2 induced the accumulation of LDs in Wolbachia-free cells (Fig. 3C and D). Interestingly, DENV-2 infection caused a further accumulation of LDs in wMel-Aag2 and wAlbB-Aag2 but not in wPip-Aag2 (Fig. 3D). Furthermore, the LD accumulation observed in both cell lines with antiviral Wolbachia strains after DENV-2 infection was significantly higher than the LD accumulation in Wolbachia-free cells infected with DENV-2 (Fig. 3D). By contrast, we measured similar levels of LDs in wPip- and Wolbachia-free Aag2 cells infected with DENV-2 (22.5 and 21.7 per cell, respectively) (Fig. 3D), indicating an association between high LD numbers and restriction of DENV-2 replication.
To investigate LD accumulation induced by different Wolbachia strains in vivo, we dissected female Ae. aegypti infected with wMel, wAlbB or wPip. We selected ovarian tissue since this tissue is rich in Wolbachia for all strains of interest (Fig. S2) (23). Ovaries were stained with BODIPY 493/503 for LDs and DAPI to demarcate nuclei. Supporting our in vitro findings, LDs were most prevalent in the presence of antiviral Wolbachia strains compared to the Wolbachia-free control (Fig. 3E). Taken together, our findings demonstrate that changes in intracellular lipid storage are key features of mosquito cells infected with antiviral Wolbachia strains.
Intracellular redistribution of lipids contributes to DENV-2 restriction in wMel-Aag2 cells
We next hypothesized that Wolbachia-induced LD formation may be required to induce an antiviral state. We utilized a series of inhibitors that restrict enzymes required for LD synthesis in mammalian cells. The enzyme fatty acid synthase (FAS) catalyzes the second step of the de novo triacylglycerol synthesis pathway which contributes molecules to the neutral core of LDs and membrane synthesis, while diacylglycerol acyltransferase (DGAT) 1 and DGAT 2 catalyze the final step in this pathway, converting diacylglycerol to triacylglycerol (70, 71). Inhibitors of these enzymes were applied to wMel- and Wolbachia-free-Aag2 cells to test if they were effective against Ae. aegypti orthologs. The DGAT 1 inhibitor (T863) and DGAT 2 inhibitor (PF-06424439) did not reduce LD formation in these cells. However, the fatty acid synthase (FAS) inhibitor C75 did effectively reduce LD accumulation (Fig. S3A and B).
We, therefore, compared DENV-2 replication and infectious virus production in Wolbachia-free and wMel-Aag2 cell lines treated with C75. We chose to focus on wMel since this strain induces a more potent antiviral effect in Aag2 cells (Fig. 1) and is the primary release strain used in field trials to date (27, 72, 73).
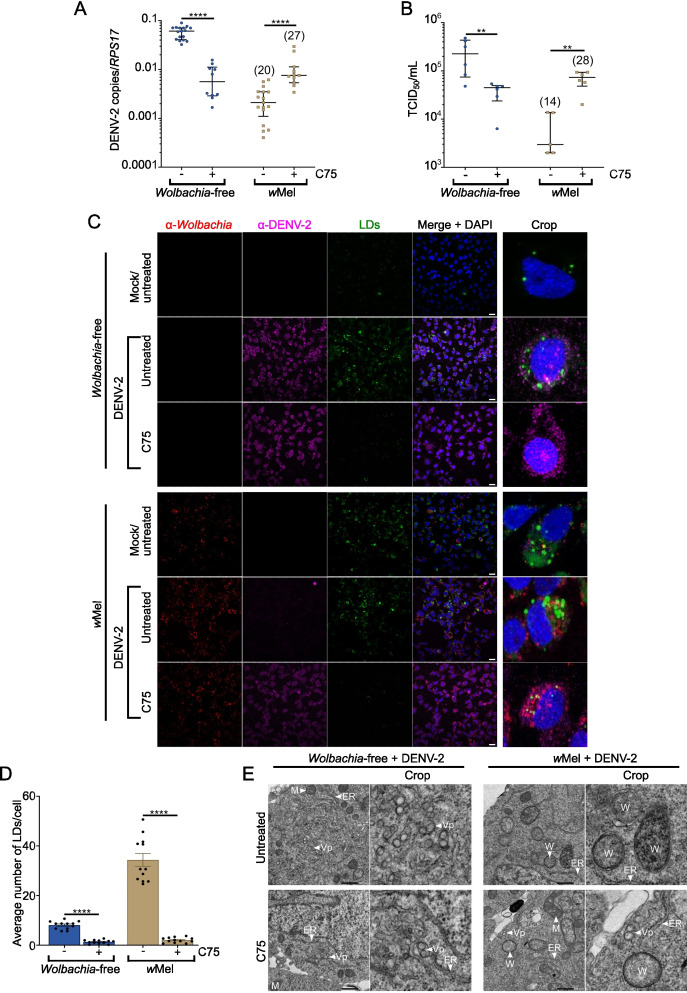
While C75 treatment reduced LD numbers in both cell lines DENV-2 replication was significantly reduced only in Wolbachia-free cells (by ~1 log10), consistent with previous reports of LDs playing a key role in DENV replication (66) (Fig. 4A ,B, C and D). By contrast, C75 treatment of the wMel-Aag2 cell line partially restored DENV-2 replication, with intracellular DENV-2 and infectious virus ~1 log10 higher than in untreated wMel-Aag2 cells. Notably, C75 treatment of wMel-Aag2 cell lines did not reduce Wolbachia density (Fig. 4A and B, in parentheses), indicating that FAS and wMel-induced LDs are not directly required for active Wolbachia replication and maintenance. However, it is possible that prolonged periods of LD depletion could affect Wolbachia survival.
Fig 4.
Disrupting Wolbachia-induced lipid droplets facilitates viral replication in wMel-Aag2 cell line. Wolbachia-free and wMel-Aag2 cell lines were mock-infected or DENV-2 infected at MOI 1 for 24 h. Prior to DENV-2 infection cells were pre-treated with C75 for 24 h, and C75 was maintained during the time of infection. (A–B) Cell-associated RNA and virus supernatant were collected from each cell line 24 h post-DENV-2-infection for analysis by qRT-PCR (A) and TCID50 (B). Data are the median number of viral genome copies ± IQR relative to the RPS17 mosquito housekeeping gene (A) or the median TCID50/mL ± IQR (B). The numbers in parentheses represent the average number of Wolbachia per cell (Wolbachia 16S rRNA/RPS17), determined in parallel wells for each independent experiment prior to DENV-2 inoculation. Data are representative of at least three (A) or two independent experiments (B) with triplicate biological replicates each. Statistical analyses were performed using Mann-Whitney test, where **P < 0.001 and ****P < 0.0001. (C) Wolbachia-free (top panel) and wMel-Aag2 cell lines (bottom panel) were stained with BODIPY (green) and DAPI (blue). Wolbachia was detected with an α-WSP antibody (red) and 4G2 hybridoma fluid against flavivirus group E antigen for DENV-2 staining (magenta). Slides were imaged as 3-dimensional z-stacks and 2D images generated by MIP using Fiji software. Scale bars = 15 µm. (D) LD numbers were quantified 24 h post-DENV-2 infection at MOI 1. LD numbers were analyzed using Fiji software. Data are the mean LD number ± SEM from at least 6 fields of view representing at least 80 cells in total, where each data point represents the mean LD number from a single field of view. Statistical analyses were performed by an unpaired two-tailed student’s t-test, where ****P < 0.0001. (E) TEM of Wolbachia-free and wMel-Aag2 cell lines mock-infected or DENV-2 infected at MOI 1 for 24 h and treated with C75 as previously described. Scale bar = 500 nm. ER, endoplasmic reticulum; W, Wolbachia; M, mitochondria; Vp, vesicle packets.
Finally, TEM micrographs of both Wolbachia-free and wMel-Aag2 cell lines treated with C75 were examined to verify whether DENV-2 replication could now be supported in cells near wMel bacterium. We observed a reduction in visible cytosolic Vps in the Wolbachia-free cell line, while, for the first time, we were able to find evidence of virus replication in cells already occupied by wMel (Fig. 4E). Together, these data show that blocking the de novo lipogenesis of triacylglycerol significantly compromises wMel-induced LD accumulation, and consequently, its antiviral activity in Ae. aegypti cells.
DISCUSSION
Wolbachia has been implemented as a biocontrol tool in cities in Oceania, Asia, and Latin America. Several epidemiological studies have now demonstrated the efficacy of Wolbachia-introgression in reducing the burden of mosquito-borne diseases in communities (26–31), and the Vector Control Advisory Group to the World Health Organization has endorsed its public health value (74). To best support the longevity of this method, and to predict the emergence of Wolbachia-resistant viruses, it is critical that we understand the mechanisms that underpin the antiviral activity of Wolbachia.
Given that Wolbachia strains are classified into major phylogenetic lineages known as supergroups (75), we can strategically compare host modifications induced by the antiviral strain wAlbB and the non-antiviral strain wPip because they both belong to supergroup B. Additionally, both strains are adapted to mosquito host species (75, 76). wMel, which belongs to supergroup A (75), has a well-characterized antiviral activity and comparisons with wAlbB enable us to determine whether antiviral strains from different supergroups employ similar strategies to induce the antiviral state in Ae. aegypti. In our cell culture models, we observed that supergroup B strains, wAlbB and wPip, grew to substantially higher densities (>80 Wolbachia/cell) than the distantly related wMel (<30 Wolbachia/cell). This is consistent with somatic tissues in mosquitoes where it has been reported that wAlbB and wPip generally reside at higher levels than wMel (20, 23, 55). Thus, our in vitro findings support previous research and show that Wolbachia density does not necessarily determine a strain’s antiviral activity.
Recent investigations have demonstrated that wMel resides nearby ER and Golgi organelles in its native host, D. melanogaster (46, 47, 57). Here, we remarkably observed that only antiviral Wolbachia strains associate closely with the ER network in Aag2 cell lines. Non-antiviral strain wPip instead formed a massive cytoplasmatic cluster with no apparent interaction with other organelles. TEM analyses revealed that wMel and wAlbB were individually surrounded by ER membranes at a high frequency. This may be strategic by these strains; for example, the wMel genome lacks pathways for metabolizing some membrane components, relying on its host for many of the materials required for their membrane formation (77–79). Since wPip does not demand this close association for its active replication, perhaps it has alternate ways of acquiring nutrients or has a more complete intrinsic metabolic capacity. Comparative genomic studies between related strains wAlbB and wPip may provide insight into this. Interestingly, based on our TEM micrographs, even in Aag2 cell lines highly infected with wAlbB or wPip strains (>80 Wolbachia/cell), we did not see any morphological evidence of increased ER activity linked to ER stress, such as enlarged tubules and cisternae, nor subcellular redistribution of this organelle as previously described for wMel in D. melanogaster (46, 47).
To our knowledge, there have not been any reports describing the association of Wolbachia strains with Ae. aegypti organelles using sectioned mosquitoes or dissected tissues. Such an examination will be important to confirm the in vivo relevance of the findings described here. Notably, wMel and wAlbB are known to reside at varying density throughout a variety of Ae. aegypti tissues. This includes tissues known to be important for transmission of DENV like the salivary glands (80). Thus, intracellular modification of organelles in these tissues could conceivably affect the potential for these tissues to support viral replication.
DENV and other flaviviruses considerably remodel ER membranes for their own replication (32, 33, 35, 36, 60). While it is interesting that only the antiviral Wolbachia strains were found to be substantially associated with this organelle, our TEM analyses did not indicate any spatial competition between antiviral Wolbachia strains and DENV-2 for ER membranes. Indeed, ER membranes are prolific in eukaryotic cells and we observed large regions of Aag2 cells that had ER membranes free from Wolbachia, with space to hypothetically support formation of viral replication complexes. Instead, we hypothesize that antiviral Wolbachia strains may biochemically alter the composition of the ER membranes in Aag2 cells by eliciting the formation of LDs. LDs may then have further direct or indirect roles in mediating viral restriction.
LDs are thought to have conflicting roles in viral infection: many reports have demonstrated that enveloped viruses usurp LDs and alter lipidomic profiles of host cells to enhance their viral life cycles, with lipids accounting for 20%–30% of the weight of the virion (61, 66, 81–85). DENV infection, for example, increases LD formation (61), recruiting FAS to the virus replication site (64), and its capsid protein accumulates on the surface of LDs, facilitating viral replication by providing a platform for nucleocapsid formation during encapsidation (66). By contrast, LDs have been shown to have a critical role in supporting innate immune signaling pathways, potentially acting as a platform to augment and coordinate the cell’s antiviral response (61, 62, 86, 87). Since recent studies have suggested that Wolbachia’s antiviral activity toward DENV is not driven by innate immune priming (23, 53), it remains unclear what role Wolbachia-induced LDs may have in viral restriction.
In this study, we noticed distinct LD profiles in Aag2 single-infected with Wolbachia strains. Antiviral Wolbachia strains induced a higher volume of LDs per cell than wPip. Additionally, we found that wMel- and wAlbB-Aag2 cell lines responded to DENV-2 infection by further accumulating LDs, which did not occur in wPip-Aag2 cell line. Based on these findings, it appears that DENV-2 replication in Aag2 cell lines requires a preferred LD accumulation threshold that, if crossed, e.g., in the presence of antiviral Wolbachia strains, no longer supports DENV-2 replication. Previously, Aag2 cells infected with wMelPop (a supergroup A pathogenic antiviral Wolbachia strain from D. melanogaster) were shown to have LDs enriched in esterified cholesterol. DENV-2 replication was partially rescued in these cells by dispersing localized cholesterol using the drug 2-hydroxypropyl-β-cyclodextrin. However, this phenotype was not reproducible in Ae. albopictus-derived cells, Aa23, infected with another supergroup A antiviral Wolbachia strain, wAu (88, 89). Therefore, assessment of all antiviral strains using a common approach will be important in the future to determine whether some antiviral strains utilize different antiviral mechanisms in Ae. aegypti.
Meanwhile, Manokaran and colleagues discovered that acyl-carnitines (intermediate molecules that transport activated fatty acids, FA-CoA, from the cytoplasm to the mitochondria) are downregulated in the presence of wMel, re-directing lipid sources from β-oxidation, and negatively affecting ATP production, which is required for efficient DENV-1 and Zika virus replication (48). All these findings support our hypothesis that antiviral Wolbachia strains affect essential lipid classes and limit their availability to sustain DENV replication.
C75 inhibition of LD formation has previously been shown to impact DENV-2 RNA replication in the absence of Wolbachia. This is thought to be because C75 treatment would disrupt localization of the capsid protein at host LDs (66, 90). Here, C75 treatment of wMel-Aag2 cells abrogated LD formation and instead supported an increase in intracellular and infectious titers of DENV-2, to levels comparable with C75-treated Wolbachia-free-Aag2 cells. This supports a model whereby excess LD’s induced by wMel have a strong antiviral role, while moderate levels of LDs have a pro-viral role (as seen in Wolbachia-free Aag2 cells), and LD abrogation leads to incomplete viral restriction (as we observed in both wMel- and Wolbachia-free Aag2 cells treated with C75). While C75 effectively reduced LD numbers in Wolbachia-free and wMel-Aag2 cells, DGAT1 and 2 inhibitors did not. This is most likely due to a lack of conservation of these enzymes, or the inhibitor binding sites between mammalian and Ae. aegypti orthologs.
It is important to acknowledge that the C75 target, FAS, does not directly drive formation of LDs, and instead catalyses synthesis of triacylglycerols. Excess free fatty acids are converted into neutral lipids and stored in cytosolic LDs. Primarily, FAS synthesizes palmitate from acetyl-CoA and malonyl-CoA in the presence of NADPH (71). Thus, although FAS inhibition was associated with reduced LD formation, we cannot exclude the possibility that other FAS-mediated changes in fatty acid homeostasis are responsible for the partial restoration of DENV-2 replication in wMel-Aag2 cells.
One impact of reducing LD formation is the accumulation of lipids at ER membranes, especially precursors of triacylglycerols and sterol esters (such as cholesterol), which constitute most of the structural core of LDs (91, 92). Within the several classes of lipids, sphingolipids and sterols are essential in determining membrane flexibility and stability (93, 94). Therefore, an adequate membrane-lipid composition in the ER is critical for membrane rearrangement and assembly and function of +RNA virus replication complexes (32, 33, 35, 36, 60). Thus, the reduction of LD formation following C75 treatment might facilitate DENV-2 acquisition of lipid classes in the ER that may not normally be available in the presence of antiviral Wolbachia strains. Moreover, dysregulation of LDs could modify not only the composition of the ER membranes but also the composition of the LDs, both of which are required for DENV replication. In reality, Wolbachia’s redistribution of lipids within cells may mask intracellular competition for lipids. That is, differential localization of specific lipid classes that prevent effective viral replication on ER membranes may not have been detected in past lipidomic studies if the overall prevalence of each lipid class remains the same (48, 69). The mechanism by which antiviral Wolbachia strains remove lipids from ER membranes and store them as LDs must therefore be further examined.
These results provide the first detailed analyses of how antiviral Wolbachia strains associate with and modify Ae. aegypti cell organelles to restrict viral replication. We identify two striking phenotypes induced only by antiviral strains, including intimate association with host ER membranes, and induction of bigger and/or more numerous LDs. Our data support a role for Wolbachia-induced lipid redistribution in restriction of DENV-2 replication.
MATERIALS AND METHODS
Cell line generation and maintenance wMel-Aedes aegypti-derived (Aag2) cell line and the Wolbachia-free wMel.Tet-Aag2 cell lines have been described previously (56). Briefly, wMel.Tet-Aag2 was generated by treating wMel-Aag2 cell line with 10 µg/mL tetracycline for three successive passages, to cure the line of wMel infection. This Wolbachia-free-Aag2 cell line was then further used to generate wAlbB-Aag2 and wPip-Aag2 cell lines. wAlbB strain was purified from the Ae. albopictus-derived cell line, RML-12, stably infected with wAlbB and infection of Wolbachia-free-Aag2 cells was done using the shell vial technique as described previously (58).
To generate wPip-Aag2, wPip was extracted from infected Ae. aegypti eggs (95). In an Eppendorf tube, 1,000–2,000 2- to 5-day-old eggs were rinsed 3–5 times in 1 mL distilled water. The eggs were passed through a 100 µm mesh sieve and placed in a new Eppendorf tube containing 1 mL 80% (vol/vol) ethanol. Eggs were sterilized by 3–5 washes in 80% (vol/vol) ethanol then rinsed three times in distilled water, sieved, and transferred to a new Eppendorf tube containing 500 µL SPG buffer (218 mM sucrose, 3.8 mM KH2PO4, 7.2 mM K2HPO4, 4.9 mM l- glutamate at pH 7.4). Eggs were rinsed three times in SPG buffer then homogenized in 300–500 µL SPG buffer using a plastic pestle. The homogenate was spun to remove debris, and the supernatant was collected in a fresh tube. This was repeated three to five times until the supernatant became clear. A pool of supernatant samples was created. Under sterile conditions, the wPip-containing supernatant was then passed through a 5-µm syringe filter followed by a 2.7-µm syringe filter. The filtrate was then centrifuged at 12,000 g to pellet Wolbachia. Extracted wPip pellet was finally resuspended in ~300 µL of sterile SPG buffer. Wolbachia-free-Aag2 cell line was then infected with the purified wPip using the shell vial technique (58).
All Aag2 cell lines (hereafter referred as Wolbachia-free-Aag2, wMel-Aag2, wAlbB-Aag2, and wPip-Aag2 cell lines) were routinely cultured at 26°C in maintenance media consisting in 1:1 Schneider’s Drosophila (Gibco)/Mitsuhashi and Maramorosch [CaCl2 0.151 g/L, MgCl2 0.047 g/L, KCl 0.2 g/L, NaCl 7 g/L, NaH2PO4 0.174 g/L, d(+)-glucose 4 g/L, yeast extract 5 g/L, lactalbumin hydrolysate 6.5 g/L, and NaHCO3 0.12 g/L at pH 6.9] medium supplemented with 10% fetal bovine serum (FBS—Gibco).
C6/36 cell lines of Aedes albopictus origin were supplied by the American Type Culture Collection (ATCC) were routinely cultured at 28°C with 5% CO2 in RPMI 1640 medium (Gibco) with GlutaMAX Supplement containing 10% FBS.
Wolbachia density
Wolbachia density was routinely monitored in each cell line. To determine the average number of Wolbachia per cell, we used qPCR to measure the relative abundance of the conserved Wolbachia 16S rRNA gene to that of the single-copy mosquito house-keeping gene RPS17 gene. Wolbachia-free cells and wMel-, wAlbB-, and wPip-Aag2 cell lines were lysed with squash extraction buffer [10 mM Tris Buffer, 1 mM ethylenediamine tetraacetic acid (EDTA), 50 mM NaCl in ultrapure water, 1:50 Proteinase K]. Cell lysates were then incubated at 56°C for 5 min, followed by 98°C for 5 min. Samples were diluted 1:10 in water, and qPCR was performed with 3 µL of each diluted cell lysate using the LightCycler 480 Probes Master mix (Roche) according to the manufacturer’s instructions. Probes and primers used are as described: For Wolbachia detection, 16S rRNA F (5′-GAGTGAAGAAGGCCTTTGGG-3′), 16S rRNA R (5′-CACGGAGTTAGCCAGGACTTC-3′), and 16S rRNA Cy5 probe (5′-LC640CTGTGAGTACCGTCATTATCTTCCTCACT-IowaBlackRQ-3′) were used. To detect the housekeeping gene, RPS17 F (5′-TCCGTGGTATCTCCATCAAGCT-3′), RPS17 R (5′-CACTTCCGGCACGTAGTTGTC-3′), and RPS17 FAM probe (5′FAM- CAGGAGGAGGAACGTGAGCGCAG-BHQ1-3′) were utilized. PCR cycling conditions were 95°C for 5 min, 45 cycles of 95°C for 10 s, 60°C for 15 s, 72°C for 1 s, followed by cooling at 40°C for 10 s. Wolbachia densities were quantified using the delta CT method (2CT(reference)/ 2CT(target)) (96).
DENV-2
Dengue virus serotype 2 (DENV-2) strain 92T (isolated from human serum collected from a patient from Townsville, Queensland/Australia, in 1992) (13) was prepared by inoculation of C6/36 cell lines with a multiplicity of infection (MOI) of 0.1 and collection of culture supernatant 13–14 days post-infection. Infectious titers were determined by TCID50.
DENV-2 infection of cell lines
Aag2 cell lines were seeded to reach 90% confluency in 24, 12, or 6 well-plates and incubated at 26°C for 48 h prior to viral infection. Unless otherwise stated, cells were infected with DENV-2 at MOI 1 in maintenance medium without FBS for 2 h at 26°C. Virus inoculum was then removed, and the cells were washed once with warm PBS. Maintenance media containing 2% FBS were added, and cells were then incubated at 26°C for the indicated time.
Quantification of DENV-2 RNA
To quantify DENV-2 genomic copies, total RNA from virus-infected cells was extracted using the RNeasy kit (Qiagen) or Isolate II RNA mini kit (Bioline). DENV-2 RNA was amplified by qRT-PCR (LightCycler Multiplex RNA Virus Master, Roche), using primers to the conserved 3′UTR: Forward 5′-AAGGACTAGAGGTTAGAGGAGACCC; Reverse 5′- CGTTCTGTGCCTGGAATGATG; Probe 5′-HEX- AACAGCATATTGACGCTGGGAGAGACCAGA-BHQ13’ (23); DENV-2 RNA copies were quantified relative to Ae. aegypti house-keeping gene RPS17 (primers and probe sequences as above) using the delta CT method (2CT(reference)/ 2CT(target)) (96). Reactions were run on a LightCycler 480 (Roche), and data analysis was carried out with the LightCycler 480 software. qRT-PCR conditions: 50°C for 10 min, 95°C for 30 s, followed by 45 cycles of 95°C for 10 s, 55°C for 5 s, and 72°C for 10 s.
Tissue culture infectious dose 50 ELISAs to determine viral titres
Infectious virus levels were determined by Tissue Culture Infectious Dose 50 (TCID50) as previously described (97). Briefly, serial dilutions (10-fold) of virus were inoculated onto C6/36 cell lines in flat-bottom 96 well plates containing maintenance medium (RPMI-1640, 2% FBS) and incubated for 13–14 days at 28°C. After the incubation period, media were aspirated, and cells were fixed with acetone fixative buffer (20% acetone/0.02% BSA in PBS) at 4°C for 24 h. The fixative was removed, and the plate was left to completely air-dry. Once dried, virus concentrations were immediately quantified by ELISA or plates were kept at –20°C until required.
ELISAs were performed using monoclonal antibody 4G2 to detect DENV E protein (98). Briefly, plates were blocked with 2% casein in TNE buffer (10 mM Tris; 0.2 M NaCl; 1 mM EDTA; 0.05% Tween20) for 1 h at room temperature. 4G2 (produced as hybridoma supernatant) (99) was diluted 1:200 in blocking solution and incubated on fixed cells for 1 h at 37°C. Plates were washed four times with PBST (0.05% Tween20). Anti-mouse secondary antibody was diluted 1:2,000 in blocking solution and incubated for 1 h at 37°C. Plates were washed six times with PBST and then incubated with 3,3′5,5′-tetramethylbenzidine (TMB) for up to 5 min. The reaction was stopped by the addition of 0.1 M HCl. The absorbance of each well was measured at 450 nm using a BioTek Gen5 microplate reader (Agilent, Santa Clara, CA, USA). The TCID50/mL was determined using the Reed-Muench calculation (100).
Cell fixation and processing for transmission electron microscopy
Aag2 cell lines were seeded in a 6-well cell culture plate in triplicate at a concentration of 3 × 106 cells/well for 48 h at 26°C. Unless otherwise stated, cells were mock-infected or infected for 2 h at 26°C with DENV-2 at MOI 2 in maintenance media without FBS. Virus inoculum was then removed, and cells were washed once with warm PBS. Maintenance media containing 2% FBS were added, and cells were incubated at 26°C for 2 days. For C75 treatment, cells were treated as described below and infected with DENV-2 at MOI 1. At 24 h post-infection, cells were washed once with warm PBS and fixed for 2 h at room temperature with 2% glutaraldehyde in 0.1 M sodium cacodylate buffer. Cells were gently washed three times for 10 min each with 0.1 M sodium cacodylate buffer.
Cells were post fixed in 1% osmium tetroxide in 0.1 M sodium cacodylate buffer for 30 min at room temperature which was then reduced with 1.5% potassium ferrocyanide in 0.1 M sodium cacodylate buffer for 30 min. Cells were washed three times for 10 min with ultrapure water. Staining with 2.5% uranyl acetate (aqueous) followed overnight at 4°C. Cells were once again washed 3 times for 10 minutes with ultrapure water. Following washing, cells were scraped and pelleted into 4% agarose in water. Pellets were dehydrated with increasing concentrations of ethanol (25%, 50%, 75%, 90%, 100%, 100%) followed by acetone (100%, 100%). Each dehydration step was aided by a microwave regime of 40 s at 150 W (Pelco Biowave). Pellets were infiltrated with epon resin using a microwave regime (25%, 50%, 75%, 100%, 100%; each step 3 min at 250 W under vacuum). Pellets were transfered to fresh 100% epon and left at room temperature overnight and finally embedded in a silicone mold for polymerization for 48 h at 60°C.
Transmission electron microscopy
Seventy nanometer of sections were cut on a ultramicrotome (Leica UC7) and collected on copper mesh grids. Transmission electron microscopy (TEM) imaging was conducted on a Jeol JEM1400-Plus at 80 kV. A combination of single snapshots and image montages was acquired; montages were automatically stitched by Jeol acquisition software (TEM Centre).
Electron tomography
Two hundred nanometer of sections were collected on copper mesh grids for electron tomography. Ten nanometer gold fiducials were added to both sides of the section. Single-axis tomography was performed on a Jeol JEM1400-Plus at 120 kV using Jeol Recorder software. Tilt series were recorded with tilt angles from +65° to −65° with varying tilt increments based on a Saxton scheme. IMOD software (101) was used to reconstruct tilt series (etomo package), manual segmentation, and visualization.
Immunofluorescence analyses
Live experiments
Cells were plated on 18 mm × 18 mm coverslips in a 6-well cell culture plate 48 h previously coated with gelatin (0.2%, vol/vol) (62, 86). To stain the ER, the cell culture medium was aspirated, and cells were washed with PBS once and incubated for 30 min at 26°C with 1 µM live ER-tracker red dye (Molecular Probes) diluted in the appropriate Aag2 cell culture medium. The ER-tracker solution was replaced by a 1/20,000 solution of SYTO-11 (Molecular Probes) DNA dye for 10 min at 26°C diluted in the appropriate Aag2 cell culture medium, washed twice with PBS, and cells were maintained in Aag2 culture medium during the confocal microscopy observations.
LDs staining and quantification
Cells were plated on 12 mm × 12 mm coverslips in a 12-well cell culture plate for 48 h previously coated with gelatin (0.2%, vol/vol) (62, 86). All cells were maintained in Aag2 cell culture medium with 10% FBS. Twenty-four hours post-DENV-2-infection, mock-infected and infected cells were fixed with 4% paraformaldehyde in PBS for 15 min, washed twice with PBS, permeabilized with 0.5% Saponin in PBS for 10 min, and washed three times with PBS. Cells were blocked with 5% BSA for 1 h, before antibody staining with polyclonal anti-WSP (98) and 4G2 hybridoma fluid against flavivirus group E antigen for DENV-2 staining. Cells were then incubated with Alexa Fluor 555 or 647 secondary antibodies (1:2,000) for 1 h. LDs were stained by incubating cells with BODIPY (493/503 4,4-difluoro 1,3,5,7,8 pentamethyl 4-bora3a,4a-diaza-s-indacene—Molecular Probes) at 1 ng/mL for 1 h, and nuclei were stained with DAPI (Sigma-Aldrich, 1 µg/mL) for 5 min. All incubations were at room temperature. Samples were then washed with PBS and mounted with Vectashield Antifade Mounting Medium (Vector Laboratories). The slides were imaged as 3-dimensional z-stacks and 2D images generated by Maximum Intensity Projection (MIP). For each condition, at least 6 fields of view were imaged at 63× magnification from different locations across each coverslip. LDs from at least 80 cells per biological replicate with a minimum of 2 biological replicates per experiment being analyzed for both LD number and average LD size. LD numbers and diameters were analyzed using quantitative data from the single raw CZI images (from Zen Blue) in ImageJ using the particle analysis tool.
For imaging of mosquito tissues, ovaries were dissected from non-blood-fed female mosquitoes 5–7 days post-emergence. Mosquito lines Rockefeller, Rockefeller-wMel, Rockefeller-wAlbB, and Rockefeller-wPip have been described previously (23). Ovaries were dissected from 4 to 5 mosquitoes per line, in PBS, then fixed on poly-lysine coated slides in cold 4% paraformaldehyde/PBS for 15 min. Slides were rinsed three times in PBS and then permeabilized in 0.1% Triton X-100 in PBS for 10 min. Samples were blocked with 1% BSA for 30 min and then stained with BODIPY (493/503) at 1 ng/mL for 1 h. Nuclei were stained with DAPI (Sigma-Aldrich, 1 µg/mL) for 5 min at room temperature. Samples were washed with PBS and mounted with Vectashield Antifade Mounting Medium (Vector Laboratories). All images were acquired using a Zeiss LSM 800 confocal microscope and processed using Zeiss analysis software (Zen Blue Edition version 10.1.19043, Jena, Germany) and FIJI analysis software. The slides were imaged as 3-dimensional z-stacks and 2D images generated by MIP.
Inhibition of FAS by C75 treatment
Unless otherwise stated, Wolbachia-free and wMel-Aag2 lines were seeded and infected as described above. Prior to DENV-2 infection cells were pre-treated with 1µM fatty acid synthase (FAS) inhibitor C75 (Abcam) or DMSO diluent only for 24 h. Cells were mock-infected or DENV-2-infected at MOI 1 for 24 h, and C75 was maintained during the time of infection in Aag2 cell culture medium with 10% FBS. LD quantification, viral replication, and TEM imaging were performed as described above.
Statistical analyses
Statistical analyses of parametric data were performed using one-way ANOVA or two-way ANOVA tests with a Tukey’s multiple comparison correction, or unpaired two-tailed Student’s t test, and data were expressed as mean ± Standard Error of the Mean (±SEM). Statistical analyses of non-parametric data were performed using a Kruskal-Wallis test with Dunn’s correction for multiple comparisons, or a two-tailed Mann-Whitney test, and data were expressed as median ± Interquartile Range (± IQR). All statistical analyses were performed using Prism 9.3.1 (GraphPad Software), with P < 0.05 considered to be significant.
ACKNOWLEDGMENTS
This research was funded by the Australian Research Council (DP220102997) and the National Health and Medical Research Council (APP1182432). The funders had no role in study design, data collection, and interpretation, or the decision to submit the work for publication.
We thank Professor Roy Hall and Jody Hobson-Peters (University of Queensland) for provision of the 4G2 antibody.
The authors would like to acknowledge the Ramaciotti Centre for Cryo Electron Microscopy, a node of Microscopy Australia. The authors acknowledge Monash Micro Imaging, Monash University, for the provision of instrumentation, training, and technical support. We would like to thank the La Trobe University Bioimaging Facility for their assistance with image acquisition, data collection, and image analysis essential to this work. Finally, we thank Katrina Ibay for technical support.
R.K.L. performed the majority of the experiments; E.A.M. assisted in the design of LD experiments, LD imaging, and image analyses. R.T. and G.R. assisted with the T.E.M. acquisition. E.A.M. and R.T. equally collaborated to this work. J.T.B. originally established wPip-Aag2 and wAlbB-Aag2 mosquito cell lines, and H.F. kindly provided the mosquito lines used in this study. J.M., K.J.H., and C.P.S. assisted in experimental direction. J.E.F. was responsible for the overall study design, alongside R.K.L. J.E.F. J.M.M. and C.P.S. acquired the funding for the project. J.E.F. and R.K.L. wrote the manuscript; all authors contributed to editing of the manuscript.
Contributor Information
Johanna E. Fraser, Email: johanna.fraser@monash.edu.
Nisha K. Duggal, Virginia Tech, Blacksburg, Virginia, USA
Rushika Perera, Colorado State University, Fort Collins, Colorado, USA.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/mbio.02495-23.
Supplemental text, figures, movie legend, and references.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Ogunlade ST, Meehan MT, Adekunle AI, Rojas DP, Adegboye OA, McBryde ES. 2021. A review: Aedes-borne arboviral infections, controls and Wolbachia-based strategies. Vaccines 9:32. doi: 10.3390/vaccines9010032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Weaver SC, Barrett ADT. 2004. Transmission cycles, host range, evolution and emergence of arboviral disease. Nat Rev Microbiol 2:789–801. doi: 10.1038/nrmicro1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mukhopadhyay S, Kuhn RJ, Rossmann MG. 2005. A structural perspective of the flavivirus life cycle. Nat Rev Microbiol 3:13–22. doi: 10.1038/nrmicro1067 [DOI] [PubMed] [Google Scholar]
- 4. Kraemer MUG, Reiner RC, Brady OJ, Messina JP, Gilbert M, Pigott DM, Yi D, Johnson K, Earl L, Marczak LB, et al. 2019. Past and future spread of the arbovirus vectors Aedes aegypti and Aedes albopictus. Nat Microbiol 4:854–863. doi: 10.1038/s41564-019-0376-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brady OJ, Golding N, Pigott DM, Kraemer MUG, Messina JP, Reiner Jr RC, Scott TW, Smith DL, Gething PW, Hay SI. 2014. Global temperature constraints on Aedes aegypti and Ae. albopictus persistence and competence for dengue virus transmission. Parasit Vectors 7:338. doi: 10.1186/1756-3305-7-338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ryan SJ, Carlson CJ, Mordecai EA, Johnson LR. 2019. Global expansion and redistribution of Aedes-borne virus transmission risk with climate change. PLOS Negl Trop Dis 13:e0007213. doi: 10.1371/journal.pntd.0007213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Goindin D, Delannay C, Ramdini C, Gustave J, Fouque F. 2015. Parity and longevity of Aedes aegypti according to temperatures in controlled conditions and consequences on dengue transmission risks. PLOS ONE 10:e0135489. doi: 10.1371/journal.pone.0135489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Morens DM, Folkers GK, Fauci AS. 2004. The challenge of emerging and re-emerging infectious diseases. Nature 430:242–249. doi: 10.1038/nature02759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kraemer MUG, Sinka ME, Duda KA, Mylne A, Shearer FM, Brady OJ, Messina JP, Barker CM, Moore CG, Carvalho RG, Coelho GE, Van Bortel W, Hendrickx G, Schaffner F, Wint GRW, Elyazar IRF, Teng H-J, Hay SI. 2015. The global compendium of Aedes aegypti and Ae. albopictus occurrence. Sci Data 2:150035. doi: 10.1038/sdata.2015.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu-Helmersson J, Quam M, Wilder-Smith A, Stenlund H, Ebi K, Massad E, Rocklöv J. 2016. Climate change and aedes vectors: 21st century projections for dengue transmission in Europe. EBioMedicine 7:267–277. doi: 10.1016/j.ebiom.2016.03.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Morales D, Ponce P, Cevallos V, Espinosa P, Vaca D, Quezada W. 2019. Resistance status of Aedes aegypti to deltamethrin, malathion, and temephos in ecuador. J Am Mosq Control Assoc 35:113–122. doi: 10.2987/19-6831.1 [DOI] [PubMed] [Google Scholar]
- 12. Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GRW, Simmons CP, Scott TW, Farrar JJ, Hay SI. 2013. The global distribution and burden of dengue. Nature 496:504–507. doi: 10.1038/nature12060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Moreira LA, Iturbe-Ormaetxe I, Jeffery JA, Lu G, Pyke AT, Hedges LM, Rocha BC, Hall-Mendelin S, Day A, Riegler M, Hugo LE, Johnson KN, Kay BH, McGraw EA, van den Hurk AF, Ryan PA, O’Neill SL. 2009. A Wolbachia symbiont in Aedes aegypti limits infection with dengue, chikungunya, and plasmodium. Cell 139:1268–1278. doi: 10.1016/j.cell.2009.11.042 [DOI] [PubMed] [Google Scholar]
- 14. Walker T, Johnson PH, Moreira LA, Iturbe-Ormaetxe I, Frentiu FD, McMeniman CJ, Leong YS, Dong Y, Axford J, Kriesner P, Lloyd AL, Ritchie SA, O’Neill SL, Hoffmann AA. 2011. The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature 476:450–453. doi: 10.1038/nature10355 [DOI] [PubMed] [Google Scholar]
- 15. Bian G, Xu Y, Lu P, Xie Y, Xi Z. 2010. The endosymbiotic bacterium Wolbachia induces resistance to dengue virus in Aedes aegypti. PLoS Pathog. 6:e1000833. doi: 10.1371/journal.ppat.1000833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Carrington LB, et al. 2018. “Field- and clinically derived estimates of Wolbachia-mediated blocking of dengue virus transmission potential in Aedes aegypti mosquitoes” Proceedings of the National Academy of Sciences, Vol. 115, p 361–366. doi: 10.1073/pnas.1715788115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Aliota MT, Walker EC, Uribe Yepes A, Velez ID, Christensen BM, Osorio JE. 2016. The wMel strain of Wolbachia reduces transmission of chikungunya virus in Aedes aegypti. PLOS Negl Trop Dis 10:e0004677. doi: 10.1371/journal.pntd.0004677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zug R, Hammerstein P. 2012. Still a host of hosts for Wolbachia: analysis of recent data suggests that 40% of terrestrial Arthropod species are infected. PLoS ONE 7:e38544. doi: 10.1371/journal.pone.0038544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ross PA, et al. 2020. An elusive endosymbiont: does Wolbachia occur naturally in Aedes aegypti?. Ecology and Evolution 10:1581–1591. doi: 10.1002/ece3.6012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ant TH, Herd CS, Geoghegan V, Hoffmann AA, Sinkins SP. 2018. The Wolbachia strain wAu provides highly efficient virus transmission blocking in Aedes aegypti. PLOS Pathog. 14:e1006815. doi: 10.1371/journal.ppat.1006815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xi Z, Khoo CCH, Dobson SL. 2005. Wolbachia establishment and invasion in an Aedes aegypti laboratory population. Science 310:326–328. doi: 10.1126/science.1117607 [DOI] [PubMed] [Google Scholar]
- 22. Flores HA, Taneja de Bruyne J, O’Donnell TB, Tuyet Nhu V, Thi Giang N, Thi Xuan Trang H, Thi Thuy Van H, Thi Long V, Thi Dui L, Le Anh Huy H, Thi Le Duyen H, Thi Van Thuy N, Thanh Phong N, Van Vinh Chau N, Thi Hue Kien D, Thuy Vi T, Wills B, O’Neill SL, Simmons CP, Carrington LB. 2020. Multiple Wolbachia strains provide comparative levels of protection against dengue virus infection in Aedes aegypti . PLOS Pathog. 16:e1008433. doi: 10.1371/journal.ppat.1008433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fraser JE, O’Donnell TB, Duyvestyn JM, O’Neill SL, Simmons CP, Flores HA. 2020. Novel phenotype of Wolbachia strain wPip in Aedes aegypti challenges assumptions on mechanisms of Wolbachia-mediated dengue virus inhibition. PLOS Pathog. 16:e1008410. doi: 10.1371/journal.ppat.1008410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sinkins SP, Braig HR, O’Neill SL. 1995. Wolbachia superinfections and the expression of cytoplasmic incompatibility . Proc. R. Soc. Lond. B 261:325–330. doi: 10.1098/rspb.1995.0154 [DOI] [PubMed] [Google Scholar]
- 25. Serbus LR, Casper-Lindley C, Landmann F, Sullivan W. 2008. The genetics and cell biology of Wolbachia -host interactions. Annu Rev Genet 42:683–707. doi: 10.1146/annurev.genet.41.110306.130354 [DOI] [PubMed] [Google Scholar]
- 26. Indriani C, Tantowijoyo W, Rancès E, Andari B, Prabowo E, Yusdi D, Ansari MR, Wardana DS, Supriyati E, Nurhayati I, Ernesia I, Setyawan S, Fitriana I, Arguni E, Amelia Y, Ahmad RA, Jewell NP, Dufault SM, Ryan PA, Green BR, McAdam TF, O’Neill SL, Tanamas SK, Simmons CP, Anders KL, Utarini A. 2020. Reduced dengue incidence following deployments of Wolbachia-infected Aedes aegypti in Yogyakarta, Indonesia: a quasi-experimental trial using controlled interrupted time series analysis. Gates Open Res 4:50. doi: 10.12688/gatesopenres.13122.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Utarini A, Indriani C, Ahmad RA, Tantowijoyo W, Arguni E, Ansari MR, Supriyati E, Wardana DS, Meitika Y, Ernesia I, Nurhayati I, Prabowo E, Andari B, Green BR, Hodgson L, Cutcher Z, Rancès E, Ryan PA, O’Neill SL, Dufault SM, Tanamas SK, Jewell NP, Anders KL, Simmons CP, AWED Study Group . 2021. Efficacy of Wolbachia-infected mosquito deployments for the control of dengue. N Engl J Med 384:2177–2186. doi: 10.1056/NEJMoa2030243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nazni WA, Hoffmann AA, NoorAfizah A, Cheong YL, Mancini MV, Golding N, Kamarul GMR, Arif MAK, Thohir H, NurSyamimi H, ZatilAqmar MZ, NurRuqqayah M, NorSyazwani A, Faiz A, Irfan F-RMN, Rubaaini S, Nuradila N, Nizam NMN, Irwan SM, Endersby-Harshman NM, White VL, Ant TH, Herd CS, Hasnor AH, AbuBakar R, Hapsah DM, Khadijah K, Kamilan D, Lee SC, Paid YM, Fadzilah K, Topek O, Gill BS, Lee HL, Sinkins SP. 2019. Establishment of Wolbachia strain wAlbB in malaysian populations of Aedes aegypti for dengue control. Curr Biol 29:4241–4248. doi: 10.1016/j.cub.2019.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. O’Neill SL, Ryan PA, Turley AP, Wilson G, Retzki K, Iturbe-Ormaetxe I, Dong Y, Kenny N, Paton CJ, Ritchie SA, Brown-Kenyon J, Stanford D, Wittmeier N, Anders KL, Simmons CP. 2019. Scaled deployment of Wolbachia to protect the community from dengue and other aedes transmitted arboviruses. Gates Open Res 2:36. doi: 10.12688/gatesopenres.12844.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ryan PA, Turley AP, Wilson G, Hurst TP, Retzki K, Brown-Kenyon J, Hodgson L, Kenny N, Cook H, Montgomery BL, Paton CJ, Ritchie SA, Hoffmann AA, Jewell NP, Tanamas SK, Anders KL, Simmons CP, O’Neill SL. 2019. Establishment of wMel Wolbachia in Aedes aegypti mosquitoes and reduction of local dengue transmission in cairns and surrounding locations in northern Queensland, Australia. Gates Open Res 3:1547. doi: 10.12688/gatesopenres.13061.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pinto SB, Riback TIS, Sylvestre G, Costa G, Peixoto J, Dias FBS, Tanamas SK, Simmons CP, Dufault SM, Ryan PA, O’Neill SL, Muzzi FC, Kutcher S, Montgomery J, Green BR, Smithyman R, Eppinghaus A, Saraceni V, Durovni B, Anders KL, Moreira LA. 2021. Effectiveness of Wolbachia-infected mosquito deployments in reducing the incidence of dengue and other aedes-borne diseases in Niteroi, Brazil: a quasi-experimental study. PLoS Negl Trop Dis 15:e0009556. doi: 10.1371/journal.pntd.0009556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Harapan H, Michie A, Sasmono RT, Imrie A. 2020. Dengue: a minireview. Viruses 12:829. doi: 10.3390/v12080829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Guzman MG, Gubler DJ, Izquierdo A, Martinez E, Halstead SB. 2016. Dengue infection. Nat Rev Dis Primers 2:16055. doi: 10.1038/nrdp.2016.55 [DOI] [PubMed] [Google Scholar]
- 34. van den Elsen K, Quek JP, Luo D. 2021. Molecular insights into the flavivirus replication complex. Viruses 13:956. doi: 10.3390/v13060956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Welsch S, Miller S, Romero-Brey I, Merz A, Bleck CKE, Walther P, Fuller SD, Antony C, Krijnse-Locker J, Bartenschlager R. 2009. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host & Microbe 5:365–375. doi: 10.1016/j.chom.2009.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Eyre NS, Johnson SM, Eltahla AA, Aloi M, Aloia AL, McDevitt CA, Bull RA, Beard MR. 2017. Genome-wide mutagenesis of dengue virus reveals plasticity of the Ns1 protein and enables generation of infectious tagged reporter viruses. J Virol 91:e01455-17. doi: 10.1128/JVI.01455-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lescar J, et al. 2018. In advances in experimental medicine and biology:115–129.
- 38. Mackenzie JM, Jones MK, Young PR. 1996. Improved membrane preservation of flavivirus-infected cells with cryosectioning. J Virol Methods 56:67–75. doi: 10.1016/0166-0934(95)01916-2 [DOI] [PubMed] [Google Scholar]
- 39. Westaway EG, Mackenzie JM, Kenney MT, Jones MK, Khromykh AA. 1997. Ultrastructure of Kunjin virus-infected cells: colocalization of Ns1 and Ns3 with double-stranded RNA, and of Ns2B with Ns3, in virus-induced membrane structures. J Virol 71:6650–6661. doi: 10.1128/JVI.71.9.6650-6661.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Amuzu HE, McGraw EA. 2016. Wolbachia-based dengue virus inhibition is not tissue-specific in Aedes aegypti. PLOS Negl Trop Dis 10:e0005145. doi: 10.1371/journal.pntd.0005145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nainu F, Trenerry A, Johnson KN. 2019. Wolbachia-mediated antiviral protection is cell-autonomous. J Gen Virol 100:1587–1592. doi: 10.1099/jgv.0.001342 [DOI] [PubMed] [Google Scholar]
- 42. Thomas S, Verma J, Woolfit M, O’Neill SL. 2018. Wolbachia-mediated virus blocking in mosquito cells is dependent on Xrn1-mediated viral RNA degradation and influenced by viral replication rate. PLOS Pathog. 14:e1006879. doi: 10.1371/journal.ppat.1006879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bhattacharya T, Newton ILG, Hardy RW. 2020. Viral RNA is a target for Wolbachia-mediated pathogen blocking. PLOS Pathog. 16:e1008513. doi: 10.1371/journal.ppat.1008513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Caragata EP, Rancès E, Hedges LM, Gofton AW, Johnson KN, O’Neill SL, McGraw EA. 2013. Dietary cholesterol modulates pathogen blocking by Wolbachia. PLoS Pathog. 9:e1003459. doi: 10.1371/journal.ppat.1003459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Caragata EP, Rancès E, O’Neill SL, McGraw EA. 2014. Competition for amino acids between Wolbachia and the mosquito host, Aedes aegypti. Microb Ecol 67:205–218. doi: 10.1007/s00248-013-0339-4 [DOI] [PubMed] [Google Scholar]
- 46. Cho K-O, Kim G-W, Lee O-K. 2011. Wolbachia bacteria reside in host golgi-related Vesicles whose position is regulated by polarity proteins. PLoS ONE 6:e22703. doi: 10.1371/journal.pone.0022703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. White PM, et al. 2017. Reliance of Wolbachia on high rates of host proteolysis revealed by a genome-wide Rnai screen of Drosophila cells. Genetics 205:1473–1488. doi: 10.1534/genetics.116.198903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Manokaran G, Flores HA, Dickson CT, Narayana VK, Kanojia K, Dayalan S, Tull D, McConville MJ, Mackenzie JM, Simmons CP. 2020. Modulation of acyl-carnitines, the broad mechanism behind Wolbachia -mediated inhibition of medically important flaviviruses in Aedes aegypti . Proc. Natl. Acad. Sci. U.S.A 117:24475–24483. doi: 10.1073/pnas.1914814117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang G, Hussain M, O’Neill SL, Asgari S. 2013. Wolbachia uses a host microRNA to regulate transcripts of a methyltransferase, contributing to dengue virus inhibition in Aedes aegypti . Proc. Natl. Acad. Sci. U.S.A 110:10276–10281. doi: 10.1073/pnas.1303603110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mayoral JG, Etebari K, Hussain M, Khromykh AA, Asgari S. 2014. Wolbachia infection modifies the profile, shuttling and structure of micrornas in a mosquito cell line. PLoS ONE 9:e96107. doi: 10.1371/journal.pone.0096107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lindsey ARI, Bhattacharya T, Hardy RW, Newton ILG, Hurst GDD, Dubilier N. 2021. Wolbachia and virus alter the host transcriptome at the interface of nucleotide metabolism pathways. mBio 12. doi: 10.1128/mBio.03472-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bhattacharya T, Yan L, Crawford JM, Zaher H, Newton ILG, Hardy RW. 2022. Differential viral RNA methylation contributes to pathogen blocking in Wolbachia-colonized arthropods. PLoS Pathog 18:e1010393. doi: 10.1371/journal.ppat.1010393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rancès E, Ye YH, Woolfit M, McGraw EA, O’Neill SL. 2012. The relative importance of innate immune priming in Wolbachia-mediated dengue interference. PLoS Pathog 8:e1002548. doi: 10.1371/journal.ppat.1002548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pan X, Zhou G, Wu J, Bian G, Lu P, Raikhel AS, Xi Z. 2012. Wolbachia induces reactive oxygen species (ROS)-dependent activation of the toll pathway to control dengue virus in the mosquito Aedes aegypti. Proc Natl Acad Sci U S A 109:E23–31. doi: 10.1073/pnas.1116932108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fraser JE, De Bruyne JT, Iturbe-Ormaetxe I, Stepnell J, Burns RL, Flores HA, O’Neill SL. 2017. Novel Wolbachia-transinfected Aedes aegypti mosquitoes possess diverse fitness and vector competence phenotypes. PLOS Pathog. 13:e1006751. doi: 10.1371/journal.ppat.1006751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. McLean BJ, Dainty KR, Flores HA, O’Neill SL. 2019. Differential suppression of persistent insect specific viruses in Trans-infected wMel and wMelPop-CLA Aedes-derived mosquito lines. Virology 527:141–145. doi: 10.1016/j.virol.2018.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fattouh N, Cazevieille C, Landmann F. 2019. Wolbachia endosymbionts subvert the endoplasmic reticulum to acquire host membranes without triggering ER stress. PLOS Negl Trop Dis 13:e0007218. doi: 10.1371/journal.pntd.0007218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. McMeniman CJ, Lane AM, Fong AWC, Voronin DA, Iturbe-Ormaetxe I, Yamada R, McGraw EA, O’Neill SL. 2008. Host adaptation of a Wolbachia strain after long-term serial passage in mosquito cell lines. Appl Environ Microbiol 74:6963–6969. doi: 10.1128/AEM.01038-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Casper-Lindley C, Kimura S, Saxton DS, Essaw Y, Simpson I, Tan V, Sullivan W. 2011. Rapid fluorescence-based screening for Wolbachia endosymbionts in drosophila germ line and somatic tissues. Appl Environ Microbiol 77:4788–4794. doi: 10.1128/AEM.00215-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Guzman MG, Harris E. 2015. Dengue. The Lancet 385:453–465. doi: 10.1016/S0140-6736(14)60572-9 [DOI] [PubMed] [Google Scholar]
- 61. Barletta ABF, Alves LR, Silva MCLN, Sim S, Dimopoulos G, Liechocki S, Maya-Monteiro CM, Sorgine MHF. 2016. Emerging role of lipid droplets in Aedes aegypti immune response against bacteria and dengue virus. Sci Rep 6:19928. doi: 10.1038/srep19928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Monson EA, Crosse KM, Duan M, Chen W, O’Shea RD, Wakim LM, Carr JM, Whelan DR, Helbig KJ. 2021. Intracellular lipid droplet accumulation occurs early following viral infection and is required for an efficient interferon response. Nat Commun 12:4303. doi: 10.1038/s41467-021-24632-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Olzmann JA, Carvalho P. 2019. Dynamics and functions of lipid droplets. Nat Rev Mol Cell Biol 20:137–155. doi: 10.1038/s41580-018-0085-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tang W-C, Lin R-J, Liao C-L, Lin Y-L. 2014. Rab18 facilitates Dengue virus infection by targeting fatty acid synthase to sites of viral replication. J Virol 88:6793–6804. doi: 10.1128/JVI.00045-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Junjhon J, Pennington JG, Edwards TJ, Perera R, Lanman J, Kuhn RJ. 2014. Ultrastructural characterization and three-dimensional architecture of replication sites in dengue virus-infected mosquito cells. J Virol 88:4687–4697. doi: 10.1128/JVI.00118-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Samsa MM, Mondotte JA, Iglesias NG, Assunção-Miranda I, Barbosa-Lima G, Da Poian AT, Bozza PT, Gamarnik AV. 2009. Dengue virus capsid protein USURPS lipid droplets for viral particle formation. PLoS Pathog. 5:e1000632. doi: 10.1371/journal.ppat.1000632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Molloy JC, Sommer U, Viant MR, Sinkins SP. 2016. Wolbachia modulates lipid metabolism in Aedes albopictus mosquito cells. Appl Environ Microbiol 82:3109–3120. doi: 10.1128/AEM.00275-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ponton F, Wilson K, Holmes A, Raubenheimer D, Robinson KL, Simpson SJ. 2015. Macronutrients mediate the functional relationship between Drosophila and Wolbachia. Proc Biol Sci 282:20142029. doi: 10.1098/rspb.2014.2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Koh C, Islam MN, Ye YH, Chotiwan N, Graham B, Belisle JT, Kouremenos KA, Dayalan S, Tull DL, Klatt S, Perera R, McGraw EA. 2020. Dengue virus dominates lipid metabolism modulations in Wolbachia-coinfected Aedes aegypti. Commun Biol 3:518. doi: 10.1038/s42003-020-01254-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Heier C, Kühnlein RP. 2018. Triacylglycerol metabolism in drosophila melanogaster Genetics 210:1163–1184. doi: 10.1534/genetics.118.301583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Chirala SS, Wakil SJ. 2004. Structure and function of animal fatty acid synthase. Lipids 39:1045–1053. doi: 10.1007/s11745-004-1329-9 [DOI] [PubMed] [Google Scholar]
- 72. O’Neill SL, Ryan PA, Turley AP, Wilson G, Retzki K, Iturbe-Ormaetxe I, Dong Y, Kenny N, Paton CJ, Ritchie SA, Brown-Kenyon J, Stanford D, Wittmeier N, Jewell NP, Tanamas SK, Anders KL, Simmons CP. 2019. Scaled deployment of Wolbachia to protect the community from dengue and other Aedes transmitted Arboviruses. Gates Open Res 2:36. doi: 10.12688/gatesopenres.12844.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Pinto SB, Riback TIS, Sylvestre G, Costa G, Peixoto J, Dias FBS, Tanamas SK, Simmons CP, Dufault SM, Ryan PA, O’Neill SL, Muzzi FC, Kutcher S, Montgomery J, Green BR, Smithyman R, Eppinghaus A, Saraceni V, Durovni B, Anders KL, Moreira LA. 2021. Effectiveness of Wolbachia-infected mosquito deployments in reducing the incidence of dengue and other aedes-borne diseases in niterói, Brazil: a quasi-experimental study. PLOS Negl Trop Dis 15:e0009556. doi: 10.1371/journal.pntd.0009556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. World Health Organization . 2021. Thirteenth meeting of the WHO vector control advisory group. Available from: https://www.who.int/publications/i/item/9789240021792
- 75. Baldo L, Dunning Hotopp JC, Jolley KA, Bordenstein SR, Biber SA, Choudhury RR, Hayashi C, Maiden MCJ, Tettelin H, Werren JH. 2006. Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl Environ Microbiol 72:7098–7110. doi: 10.1128/AEM.00731-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Landmann F. 2019. The Wolbachia endosymbionts. Microbiol Spectr 7. doi: 10.1128/microbiolspec.BAI-0018-2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wu M, Sun LV, Vamathevan J, Riegler M, Deboy R, Brownlie JC, McGraw EA, Martin W, Esser C, Ahmadinejad N, Wiegand C, Madupu R, Beanan MJ, Brinkac LM, Daugherty SC, Durkin AS, Kolonay JF, Nelson WC, Mohamoud Y, Lee P, Berry K, Young MB, Utterback T, Weidman J, Nierman WC, Paulsen IT, Nelson KE, Tettelin H, O’Neill SL, Eisen JA. 2004. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 2:E69. doi: 10.1371/journal.pbio.0020069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ant TH, Mancini MV, McNamara CJ, Rainey SM, Sinkins SP. 2023. Wolbachia-virus interactions and arbovirus control through population replacement in mosquitoes. Pathog Glob Health 117:245–258. doi: 10.1080/20477724.2022.2117939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Lin M, Rikihisa Y. 2003. Ehrlichia chaffeensis and anaplasma phagocytophilum lack genes for lipid A biosynthesis and incorporate cholesterol for their survival. Infect Immun 71:5324–5331. doi: 10.1128/IAI.71.9.5324-5331.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Fraser JE, O’Donnell TB, Duyvestyn JM, O’Neill SL, Simmons CP, Flores HA. 2020. Novel phenotype of Wolbachia strain wPip in Aedes aegypti challenges assumptions on mechanisms of Wolbachia-mediated dengue virus inhibition. PLoS Pathog 16:e1008410. doi: 10.1371/journal.ppat.1008410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Monson EA, Trenerry AM, Laws JL, Mackenzie JM, Helbig KJ. 2021. Lipid droplets and lipid mediators in viral infection and immunity. FEMS Microbiol Rev 45:fuaa066. doi: 10.1093/femsre/fuaa066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Heaton NS, Perera R, Berger KL, Khadka S, LaCount DJ, Kuhn RJ, Randall G. 2010. Dengue virus nonstructural protein 3 redistributes fatty acid synthase to sites of viral replication and increases cellular fatty acid synthesis. Proc. Natl. Acad. Sci. U.S.A 107:17345–17350. doi: 10.1073/pnas.1010811107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Perera R, Riley C, Isaac G, Hopf-Jannasch AS, Moore RJ, Weitz KW, Pasa-Tolic L, Metz TO, Adamec J, Kuhn RJ. 2012. Dengue virus infection perturbs lipid homeostasis in infected mosquito cells. PLoS Pathog. 8:e1002584. doi: 10.1371/journal.ppat.1002584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Hitakarun A, Williamson MK, Yimpring N, Sornjai W, Wikan N, Arthur CJ, Pompon J, Davidson AD, Smith DR. 2022. Cell type variability in the incorporation of lipids in the dengue virus Virion. Viruses 14:2566. doi: 10.3390/v14112566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Carro AC, Damonte EB. 2013. Requirement of cholesterol in the viral envelope for dengue virus infection. Virus Res 174:78–87. doi: 10.1016/j.virusres.2013.03.005 [DOI] [PubMed] [Google Scholar]
- 86. Monson EA, Crosse KM, Das M, Helbig KJ. 2018. Lipid droplet density alters the early innate immune response to viral infection. PLOS ONE 13:e0190597. doi: 10.1371/journal.pone.0190597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Monson EA, Whelan DR, Helbig KJ. 2021. Lipid droplet motility increases following viral immune stimulation. Int J Mol Sci 22:4418. doi: 10.3390/ijms22094418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Geoghegan V, Stainton K, Rainey SM, Ant TH, Dowle AA, Larson T, Hester S, Charles PD, Thomas B, Sinkins SP. 2017. Perturbed cholesterol and vesicular trafficking associated with dengue blocking in Wolbachia-infected Aedes aegypti cells. Nat Commun 8:526. doi: 10.1038/s41467-017-00610-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Rainey SM, Geoghegan V, Lefteri DA, Ant TH, Martinez J, McNamara CJ, Kamel W, de Laurent ZR, Castello A, Sinkins SP. 2023. Differences in proteome perturbations caused by the Wolbachia strain wAu suggest multiple mechanisms of Wolbachia-mediated antiviral activity. Sci Rep 13:11737. doi: 10.1038/s41598-023-38127-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Heaton NS, Randall G. 2010. Dengue virus-induced autophagy regulates lipid metabolism. Cell Host & Microbe 8:422–432. doi: 10.1016/j.chom.2010.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Thiele C, Penno A. 2016. Lipid droplets. Encyclopedia of Cell Biology:273–278. doi: 10.1016/B978-0-12-394447-4.20023 [DOI] [Google Scholar]
- 92. Gao M, Huang X, Song B-L, Yang H. 2019. The biogenesis of lipid droplets: lipids take center stage. Progress in Lipid Research 75:100989. doi: 10.1016/j.plipres.2019.100989 [DOI] [PubMed] [Google Scholar]
- 93. Zaitseva E, Yang S-T, Melikov K, Pourmal S, Chernomordik LV. 2010. Dengue virus ensures its fusion in late endosomes using compartment-specific lipids. PLoS Pathog. 6:e1001131. doi: 10.1371/journal.ppat.1001131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Kielian M, Chanel-Vos C, Liao M. 2010. Alphavirus entry and membrane fusion. Viruses 2:796–825. doi: 10.3390/v2040796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Fraser JE, De Bruyne JT, Iturbe-Ormaetxe I, Stepnell J, Burns RL, Flores HA, O’Neill SL. 2017. Novel Wolbachia-transinfected Aedes aegypti mosquitoes possess diverse fitness and vector competence phenotypes. PLoS Pathog 13:e1006751. doi: 10.1371/journal.ppat.1006751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 97. O’Brien CA, Hobson-Peters J, Yam AWY, Colmant AMG, McLean BJ, Prow NA, Watterson D, Hall-Mendelin S, Warrilow D, Ng M-L, Khromykh AA, Hall RA. 2015. Viral RNA intermediates as targets for detection and discovery of novel and emerging mosquito-borne viruses. PLOS Negl Trop Dis 9:e0003629. doi: 10.1371/journal.pntd.0003629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Lee E, Hien Nguyen T, Yen Nguyen T, Nam Vu S, Duong Tran N, Trung Nghia L, Mai Vien Q, Dong Nguyen T, Kriiger Loterio R, Iturbe-Ormaetxe I, Flores HA, O’Neill SL, Anh Dang D, Simmons CP, Fraser JE. 2022. Transient Introgression of Wolbachia into Aedes aegypti populations does not elicit an antibody response to Wolbachia surface protein in community members. Pathogens 11:535. doi: 10.3390/pathogens11050535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Gentry MK, Henchal EA, McCown JM, Brandt WE, Dalrymple JM. 1982. Identification of distinct antigenic determinants on dengue-2 virus using monoclonal antibodies. Am J Trop Med Hyg 31:548–555. doi: 10.4269/ajtmh.1982.31.548 [DOI] [PubMed] [Google Scholar]
- 100. Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am J Epidemiol 27:493–497. doi: 10.1093/oxfordjournals.aje.a118408 [DOI] [Google Scholar]
- 101. Kremer JR, Mastronarde DN, McIntosh JR. 1996. Computer visualization of three-dimensional image data using IMOD. J Struct Biol 116:71–76. doi: 10.1006/jsbi.1996.0013 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental text, figures, movie legend, and references.