BLURB
Integrator broadly drives promoter-proximal termination of RNAPII through phosphatase and RNA endonuclease activities. Here, we summarize recent work illuminating the molecular underpinnings of Integrator activity. Further, we describe the critical role Integrator plays in human health and disease, through its regulation of gene expression and noncoding RNA synthesis.
SUMMARY
Integrator is a metazoan-specific protein complex capable of inducing termination at all RNAPII-transcribed loci. Integrator recognizes paused, promoter-proximal RNAPII and drives premature termination using dual enzymatic activities: an endonuclease that cleaves nascent RNA and a protein phosphatase that removes stimulatory phosphorylation associated with RNAPII pause release and productive elongation. Recent breakthroughs in structural biology have revealed the overall architecture of Integrator and provided insights into how multiple Integrator modules are coordinated to elicit termination effectively. Further, functional genomics and biochemical studies have unraveled how Integrator-mediated termination impacts protein-coding and noncoding loci. Here, we review the current knowledge about the assembly and activity of Integrator and describe the role of Integrator in gene regulation, highlighting the importance of this complex for human health.
INTRODUCTION
In Metazoans, the regulated pausing of RNA polymerase II (RNAPII) and its controlled release into productive elongation are major points of gene regulation1,2. After synthesizing 20–50 nt of RNA, RNAPII is bound by the elongation factor SPT5 and the NELF complex, which promote the stable pausing of RNAPII3–7. During pausing, RNAPII remains active and engaged on the DNA template while awaiting further signals for productive elongation. Recruitment of the kinase P-TEFb allows for the release of paused RNAPII into the gene body, in large part because phosphorylation of SPT5 and the carboxy-terminal domain (CTD) of the largest subunit of RNAPII triggers the dissociation of NELF and binding of elongation factors that stimulate transcription elongation1–3,8.
As RNAPII transcribes across the gene body, the CTD is further phosphorylated, and elongation continues processively until RNAPII reaches the polyadenylation sequence (PAS) at the gene 3′ end9. The PAS sequence designates the appropriate location for pre-mRNA cleavage by the Cleavage and Polyadenylation (CPA) machinery, which is coupled with the dephosphorylation of SPT5 and the RNAPII CTD10. Together, dephosphorylation of the elongation complex and RNA cleavage by the CPA machinery slow elongation and facilitate transcription termination, wherein RNAPII releases both the nascent RNA and DNA template.
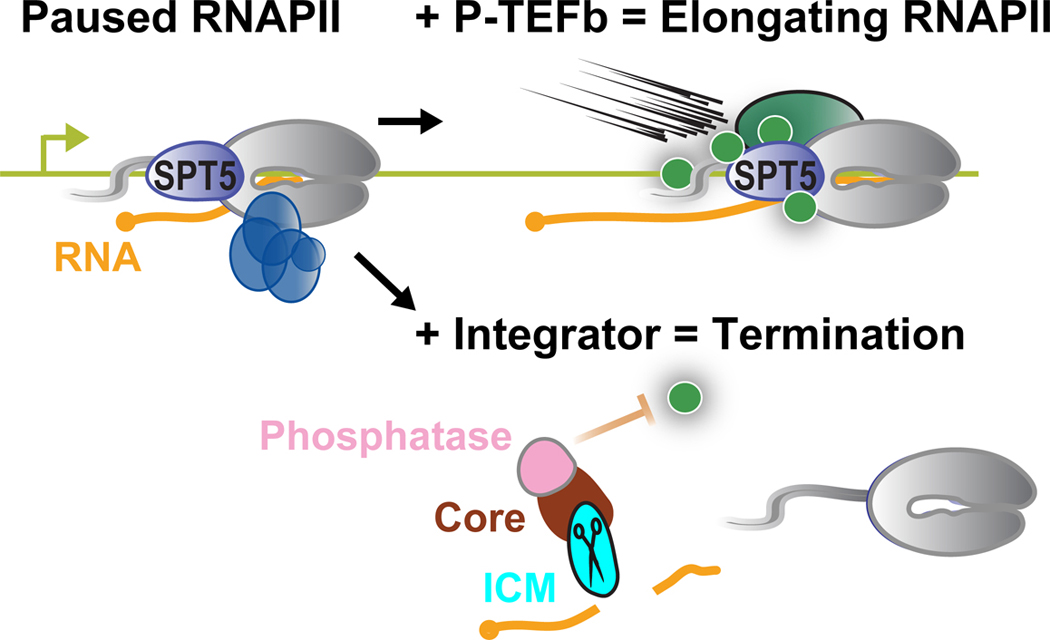
Importantly, not all promoter paused RNAPII is destined to transcribe a full-length RNA, and increasing evidence supports a model where pause release is balanced with an alternate fate of promoter-proximally paused RNAPII, namely premature termination (Figure 1). Accordingly, interest in premature termination as a gene regulatory strategy has grown substantially, as has the appreciation that much of this is carried out by Integrator, a termination complex that is highly implicated in both development and disease11,12. Here, we describe recent progress towards understanding premature termination driven by Integrator and highlight the conceptual and functional similarities with the CPA machinery. More details on canonical transcription termination at gene 3′ ends are provided in an accompanying review by Passmore and colleagues.
Figure 1. Integrator contains modules with both endonuclease and phosphatase activities.
Shown are schematics depicting the balance between pause release by P-TEFb vs. Integrator-mediated termination, depicting the cleavage activity of the Integrator endonuclease, and the phosphatase activity of Integrator-associated PP2A.
Discovery of Integrator and overall architecture of the complex
Integrator was initially purified as a complex associated with the RNAPII CTD13, comprised of 12 subunits that were numbered by descending size (INTS1-INTS12)13. Integrator was found to participate in 3′-end formation of the U-rich small nuclear RNAs (snRNAs) that form central components of the spliceosome13, implicating this novel complex in RNA processing and transcription termination.
Two additional Integrator subunits, INTS13 and INTS14, were subsequently identified through a genome-wide RNAi screen for factors required for snRNA biogenesis14, and validated as subunits of Integrator using immunoprecipitation and mass spectrometry15,16. More recently, multiple studies have provided evidence for the existence of INTS15 using systems biology and biochemical approaches17–20.
Several additional factors have been found to play central roles within Integrator. Most notably, subunits of the PP2A phosphatase demonstrate a biochemically stable association with Integrator and cryo-EM structures of the Integrator-PP2A complex reveal intimate interactions of the PP2A-A scaffold subunit and PP2A-C enzymatic subunit with multiple surfaces on Integrator21–23. Incorporating a phosphatase within Integrator has profound implications since the P-TEFb-mediated phosphorylation of the paused elongation complex is critical for RNAPII progression into productive elongation24. Indeed, the Integrator-associated PP2A phosphatase was recently shown to antagonize transcriptional kinases to suppress pause release and transcription elongation (Figure 1)21–23. Beyond PP2A, mass spectrometry studies have identified a collection of weak interactions with proteins involved in various cellular processes15,16,25.
Upon the identification of Integrator, primary sequence inspection of its subunits yielded little insight into the function of the complex, with the key exception of INTS9 and INTS11, which are members of β-CASP/metallo-β-lactamase (MβL) family of DNA/RNA endonucleases (Figure 2)26,27. These two Integrator subunits are paralogous to the cleavage and polyadenylation specificity factors CPSF100 and CPSF73, respectively28. These observations provided critical clues that Integrator could cleave nascent RNA. The parallels between Integrator and the CPA machinery extend further, as INTS9 and INTS11 interact in a manner reminiscent of the CSPF100/73 heterodimer29, and INTS11, like CPSF73, possesses catalytic activity, whereas INTS9, like CPSF100, lacks several critical amino acids thought to be required for activity. Notably, cleavage of nascent RNA by Integrator or the CPA machinery releases an RNA with a protective 5’ cap and leaves RNAPII associated with a short, uncapped (5′-monophosphate) RNA (Figure 1). This cleavage event can facilitate termination of the elongating RNAPII because the uncapped RNA 5’-end provides an entry point for exonucleases such as XRN2 and/or helicases that destabilize the elongation complex30–32. However, the direct connections between Integrator endonuclease activity and transcription termination remain to be fully elucidated.
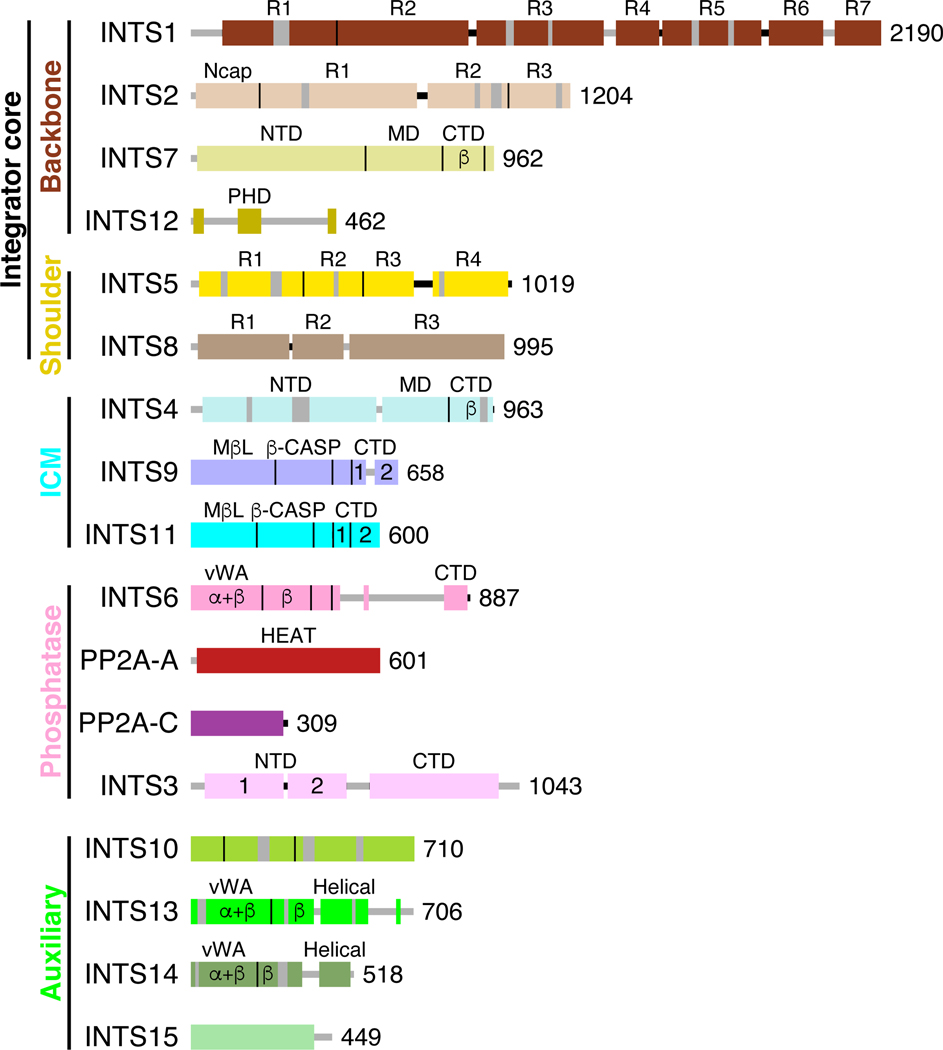
Figure 2. Domain organizations of Integrator and PP2A subunits.
Domains are indicated as boxes while vertical lines indicate boundaries between neighboring domains. The size of each subunit is provided as number of amino acids in the human ortholog. Flexible segments in the subunits are shown in gray. Abbreviations are: N-terminal domain (NTD); Middle domain (MD); C-terminal domain (CTD); N-terminal cap (Ncap); Plant Homeodomain Finger (PHD); Metallo-β-Lactamase (MβL); Metallo-β-Lactamase-associated CPSF73, Artemis, SNM, and PSO (β-CASP); von Willebrand factor type A (vWA); Huntington, Elongation factor 3, Protein Phosphatase A, Tor1 (HEAT). Domains containing α+β or only β secondary structure elements are labeled. Domains in INTS9 and INTS11 contain α+β elements. All other domains contain only α helices.
STRUCTURAL CHARACTERIZATION OF INTEGRATOR AND INTERACTIONS WITH PAUSED RNAPII
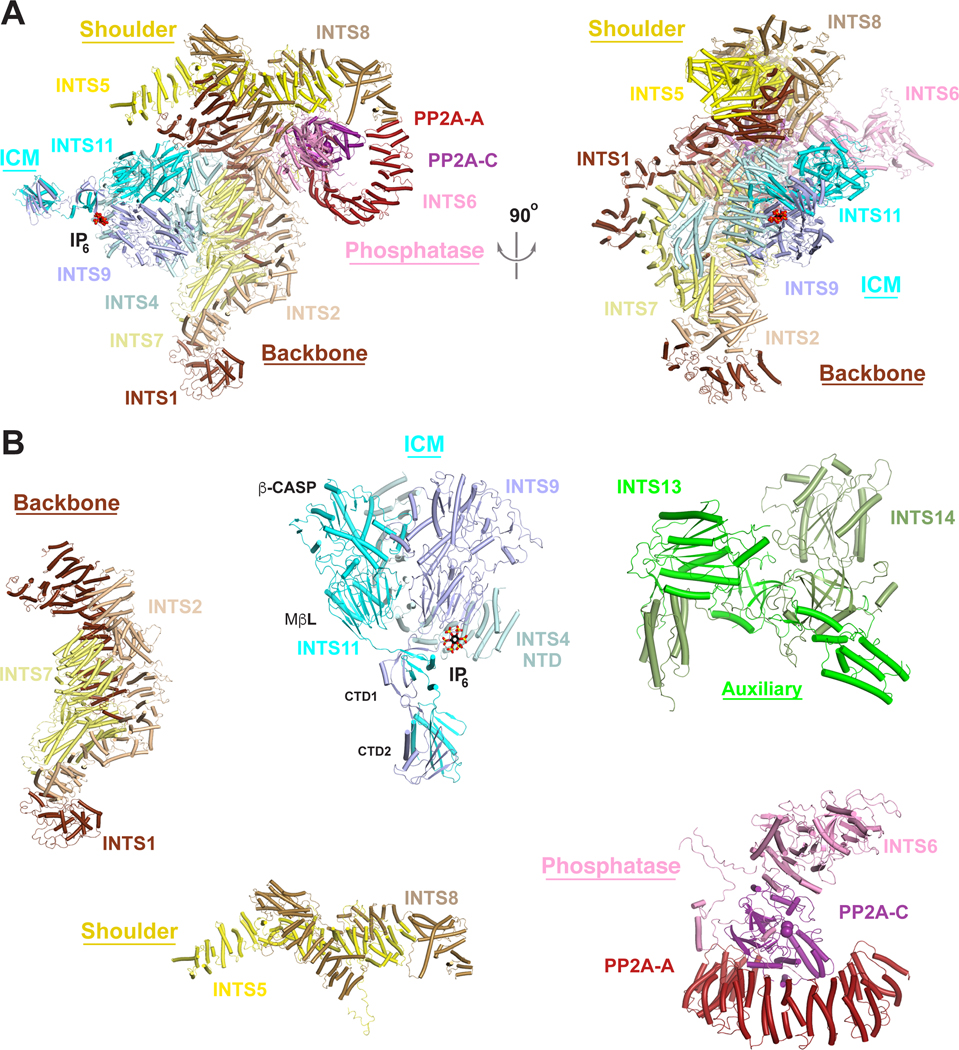
Recent breakthroughs have yielded structures of Integrator associated with PP2A-A and PP2A-C23, as well as Integrator-PP2A bound to paused RNAPII33,34, which have provided insight into the overall physical organization of the complex and its mechanism of activation. These and other recent structural studies35–41 demonstrate that Integrator is assembled from a ‘core’ constructed of backbone and shoulder modules (Figures 2 and 3A) which are bound by discrete endonuclease, phosphatase, and auxiliary modules. Notably, while the entire Integrator complex was included in the sample for structural studies, the auxiliary module was not observed, nor were INTS3 or INTS12, likely because they are flexibly tethered in this state of Integrator. Likewise, many segments of individual subunits are not present in the atomic model due to flexibility. Nevertheless, the general architecture and, most notably, the modularity of the complex is apparent.
Figure 3. The overall architecture of Integrator in an inactive state.
(A). (Left) Schematic of the structure of Integrator-PP2A complex in an inactive state33, with subunits colored as in Fig. 2. IP6 observed in the structure of Drosophila ICM36 is shown in a sphere model. The metal ions in the active site of INTS11 and manganese ions in the active site of PP2A-C are shown as spheres. (Right) Structure shown at left after 90° rotation around the vertical axis. (B). Structures of the individual Integrator modules. Module structures are derived from the structure shown in panel 3A with the exception of the INTS13/INTS14 structure, which is from39. Structure figures were produced with PyMOL (www.pymol.org).
The Integrator core: backbone and shoulder modules
The Integrator backbone module consists of INTS1, INTS2, and INTS7 (Figures 2 and 3B). Not surprisingly, INTS1 makes extensive contacts with other members of Integrator, consistent with it being the largest subunit of the complex. There are direct contacts between the C-terminal domains of INTS1 and INTS2 (Figure 3B). The INTS7 N-terminal domain (NTD) adopts a crescent-shaped structure and interacts with INTS1 and INTS2, while the INTS7 middle domain (MD) interacts with INTS2 Ncap and helical repeat R1 (Figure 3B). Although not seen in any Integrator structures, INTS12 likely associates with the Integrator backbone module34. Biochemical, yeast two-hybrid, and cellular studies demonstrate that INTS12 utilizes a ‘microdomain’ to interact with the N-terminal region of INTS142.
The Integrator shoulder module contains the heterodimer of INTS5 and INTS837, with INTS5 wrapping around INTS8, generating a relatively inflexible structure (Figure 3B). The shoulder module has intimate contacts with the C-terminal repeats of INTS1 and INTS2 and is arranged perpendicular to the backbone module, with the two modules forming a cruciform shape. Altogether, the backbone and shoulder modules function as a scaffold for interactions with the other modules.
The Integrator phosphatase module
Early purifications of Integrator revealed an association with PP2A15,16,25 while independent identification of factors associated with PP2A yielded Integrator subunits43,44. The critical role of PP2A-mediated dephosphorylation of the transcription machinery for Integrator function was recently demonstrated, in both human cells and Drosophila, thus revealing the importance and conservation of the Integrator-PP2A interaction21. The phosphatase module consists of PP2A-C, PP2A-A, and INTS6 (Figure 3B)22,23. This assembly contrasts all previously described PP2A complexes, which include a PP2A-B regulatory subunit45. Given the established role of B regulatory subunits in guiding the recognition of PP2A substrates, we and others have proposed that Integrator serves a similar regulatory purpose, directing PP2A activity towards RNAPII and elongation factors21–23. Notably, while INTS3 was not observed in the structure of Integrator, it binds INTS635 and crosslinking-mass spectrometry supports its proximity to the phosphatase module34. INTS3, however, is not uniformly associated with Integrator, and is also in the sensor of single-stranded DNA (SOSS) complex, which is important for DNA double-strand break repair38.
The stable association of the phosphatase module is strongly dependent on interactions with the shoulder module (Figure 3A). PP2A-C and INTS6 associate with INTS2, INTS5, and INTS8 to form a critical interface between the phosphatase module and the Integrator core. Indeed, excluding INTS5 or INTS8 from recombinant Integrator complexes results in loss of the phosphatase module, whereas removal of INTS11 has no effect23. Further, a highly conserved WFEFLL motif within INTS8 directly contacts PP2A-A. Yeast two-hybrid studies show that INTS8 and PP2A-A can interact in the absence of other Integrator subunits, and their association is dependent on the WFEFLL motif21. Moreover, mutation of WFEFL residues causes a loss of PP2A from both human and fly Integrator21, resulting in a dramatic increase in phosphorylation of the RNAPII elongation complex and pause release.
The Integrator cleavage module (ICM)
As suggested by homology with the CPSF100/73 heterodimer within the canonical CPA machinery, dimerization of INTS9/11 proteins is required for endonuclease activity29. Further, genetic and biochemical studies demonstrated a requirement for INTS4 for Integrator-mediated RNA cleavage46,47. Subsequent structural studies have shed light on the molecular basis of ICM assembly and architecture36,37,40. The ICM is located on one side of the Integrator core (Figure 3A), making direct contacts with the backbone module through INTS4. In comparison, INTS9 and INTS11 have no reported contacts with other Integrator subunits, suggesting that INTS4 anchors the ICM to the Integrator core. The catalytic segment of INTS11 (MβL and β-CASP domains) forms a pseudo-dimer with the equivalent segment of INTS9, which is likely stabilized by the NTD of INTS4 acting as a scaffold (Figure 3B). The organization of this pseudo-dimer is similar to that of CPSF100/73 observed in the active human U7 snRNP48. The INTS11 CTD1 and CTD2 have tight interactions with the equivalent regions of INTS9 (Figures 2 and 3B), and the CTD1 of INTS11 is crucial for recruiting INTS437. However, in the structure of Integrator-PP2A complex, INTS11 is in an inactive, closed state and there is no room to accommodate the RNA substrate.
The overall structures of the isolated human and Drosophila ICM36,37 are essentially the same as the human ICM within the Integrator complex23, suggesting that there are no conformational changes in ICM when incorporated into Integrator. Unexpectedly, an inositol hexakisphosphate (IP6) molecule was found in the structure of Drosophila ICM (Figure 2B)36, and EM density consistent with IP6 is present in human ICM37. The binding site is located at the interface of all three subunits of ICM, with IP6 having ionic interactions with several highly conserved residues (Figure 3B) in an electrostatically positive pocket. Although mutations of residues interacting with IP6 do not abolish Integrator assembly, they disrupt Integrator function in Drosophila and human cells36. The binding site is 55 Å away from the active site of INTS11, suggesting an allosteric regulation of activity.
The Integrator auxiliary module
The least understood Integrator module consists of INTS10, INTS13, INTS14, and INTS15. Loss of these subunits gives rise to only modest levels of snRNA misprocessing and minimal changes to the transcriptome, suggesting that this module is not critical to Integrator’s broad termination function14,20,39,49. The structure of the INTS13-INTS14 complex shows that the two molecules are highly intertwined (Figure 3B), with an extensive interface39. The domain organizations and structures of INTS13 and INTS14 are similar to Ku70 and Ku80, which are required for DNA double-strand break repair (Figure 2)50. However, INTS13-INTS14 is expected to have a distinct nucleic-acid binding mode, as the DNA bound to Ku70-Ku80 clashes with INTS13-INTS14. Accordingly, INTS13-INTS14 is suggested to bind RNA rather than DNA39, although the functional consequence of this interaction remains unclear. Curiously, the domain organization of INTS6 is also similar to that of INTS13 and INTS14 (Figure 2), and its β-barrel domain shares a similarity with Ku70-Ku8033. The auxiliary module also has interactions with the ICM34,37,39,51, through a segment in the C-terminal region of INTS13 39. Although INTS10 primarily contacts INTS14 in this module14,39, recent studies have revealed interactions between INTS10 and INTS15 (CG5274 in Drosophila and C7orf26 in human)19, unveiling INTS15 as another subunit of the auxiliary module (Figure 2).
The function of the auxiliary module is enigmatic. INTS13 has been reported to associate with EGR1 and NAB2 to promote enhancer activation, but it is unclear if this function requires the entire auxiliary module49. Notably, the association of the INTS13 C-terminus with the ICM suggests a potential role in regulating Integrator cleavage activity, which is supported by mutations in this domain51.
Integrator adopts an active conformation when associated with paused RNAPII
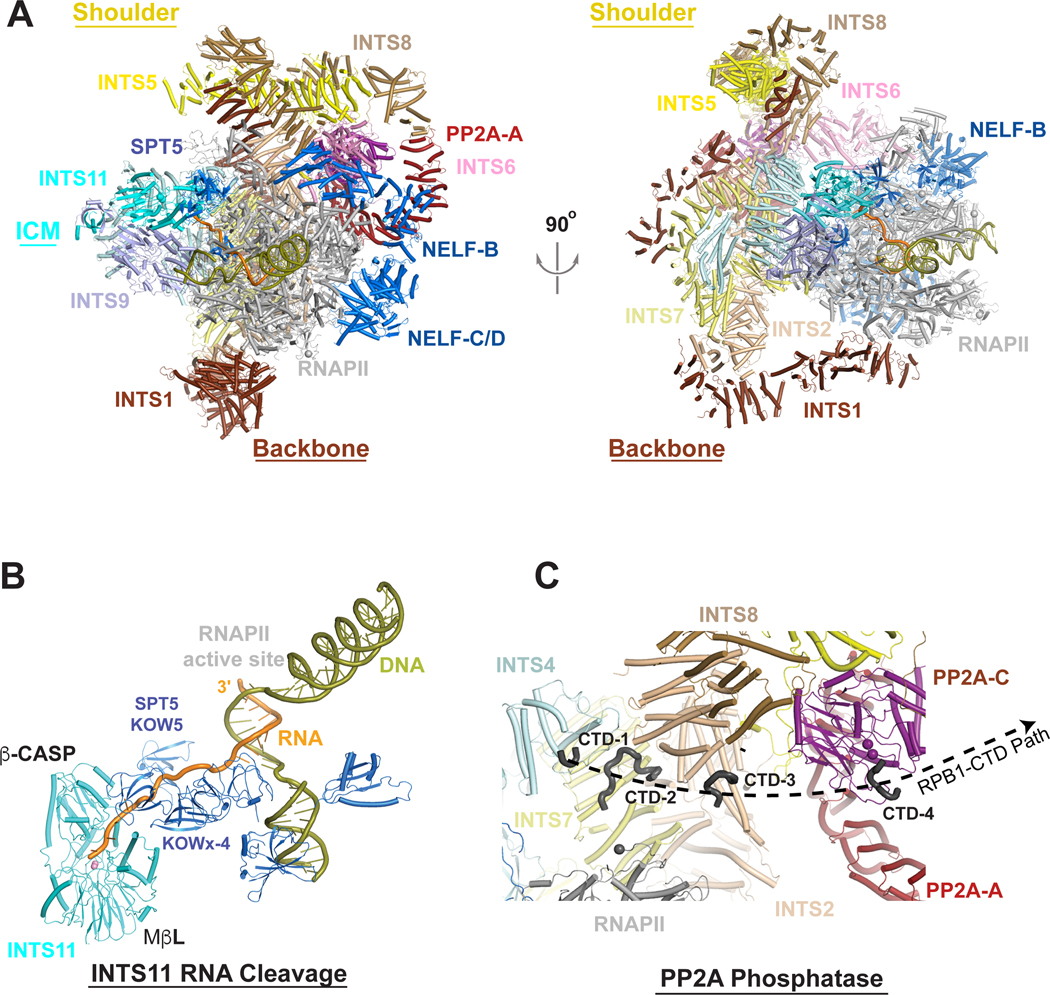
Structures of the isolated ICM or Integrator-PP2A capture INTS11 in an inactive conformation that would not accommodate RNA23,36,37. However, the active conformation of INTS11 was observed in structures of Integrator-PP2A associated with a paused elongation complex (PEC) which includes SPT5 and NELF (Figures 4A and 4B)33,34. In this complex, the PEC is embraced by ‘arms’ from Integrator, and the N-terminal repeats of INTS1 become ordered, enabling Integrator to contact the RPB2 subunit of RNAPII ~70 Å away from the body of Integrator (Figure 4A).
Figure 4. Overall architecture of Integrator in an active state.
(A). (Left) Schematic of the structure of Integrator-PP2A-RNAPII PEC complex in an active state 34. Integrator subunits are colored according to Fig. 2, with RNAPII in gray, NELF and SPT5 in marine. (Right) Structure shown at left after 90° rotation around the vertical axis. (B). Nascent RNA in the active site of INTS1133. The RNA is shown in orange, and DNA in olive. The nascent RNA exits RNAPII, and SPT5 helps to direct it to the active site of INTS11. (C) Zoom-in of CTD peptides associated with Integrator subunits with a numbered projections towards the active site of PP2A-C. The black sphere represents residue 1487 of RPB1 and is the last amino acid modeled in the structure. INTS1, INTS6, and INST11 are omitted for clarity.
There is a change in the position of ICM in the complex with PEC compared to Integrator-PP2A alone, which facilitates the interaction with SPT5 and the activation of INTS1134. The catalytic segment of INTS11 is in direct contact with the KOWx-4 domains of SPT5, the only connection between INTS11 and the PEC (Figures 4A and 4B). This contact likely helps to bring INTS11 into an active conformation, with a 17° rotation of its β-CASP domain that opens the active site. This active state of INTS11 is similar to that of active CPSF73 observed in the histone 3′-end processing machinery48. In addition, the SPT5 KOWx-4 domains located near the RNA exit site of RNAPII appear to direct the RNA towards the INTS11 active site (Figure 4B). The structures indicate a distance of ~22 nucleotides between the active sites of the polymerase and INTS11, which agrees with cell-based measurements of Integrator-mediated RNA cleavage52,53.
The interactions observed between Integrator-PP2A and the PEC provide a compelling explanation for the enrichment of Integrator with paused, promoter-proximal RNAPII21,52–57. First, the three-helix bundle in the N-terminal region of INTS6 (Figure 2) interacts with NELF-B (Figure 4A), which is uniquely present in paused RNAPII33,34. Second, the structure of the PEC complex indicates that Integrator would sterically clash with transcription initiation factors and Mediator, implying that Integrator would not associate with a pre-initiation complex34,58. Third, several Integrator binding sites on RNAPII are occluded upon association of SPT6 and PAF1 during the conversion of the paused RNAPII to a productive elongation complex, suggesting that association of Integrator with RNAPII during productive elongation would require significant structural rearrangements or would exhibit a lower binding affinity8,34. Finally, the mode of RNAPII CTD interaction and catalytic activity of the Integrator phosphatase module is most consistent with action on a paused polymerase33,34. The CTD repeats interact with several Integrator subunits (Figure 4C), and in the presence of NELF and DSIF, Integrator exhibits no preference for CTD phosphorylation status33. Critically, CTD interactions with Integrator appear to form a path radiating to the active site of PP2A-C33, suggesting that while Integrator can interact with RNAPII harboring a phosphorylated CTD, PP2A activity will lead to dephosphorylation (Figure 4C). Accordingly, Integrator-PP2A removes phosphates from the RNAPII CTD as well as SPT521. The consequence of this phosphatase activity is to prevent the transition of paused RNAPII to productive RNA synthesis, and to reduce RNAPII elongation rate. Notably, slower elongation could facilitate RNA cleavage by the ICM, analogous to the role of PNUTS-PP1 phosphatase within the CPA machinery10. Importantly, pausing is a general feature of all RNAPII transcription, with evidence of paused elongation complexes at mRNAs, upstream antisense RNAs (uaRNAs), long noncoding RNAs (lncRNAs) and enhancer RNAs (eRNAs)59,60. Thus, Integrator can broadly associate with PECs at coding and noncoding loci by recognizing specific features of paused RNAPII.
INTEGRATOR LOCALIZATION, SPECIFICITY AND FUNCTION
Despite a widespread convergence of data indicating that Integrator is a termination complex acting on RNAPII paused in early elongation, many questions remain about the specificity of Integrator activity and the impact of termination on transcription levels.
Integrator is globally enriched near transcription start sites
Given the intimate contacts of Integrator subunits with paused RNAPII it is not surprising that all Integrator subunits analyzed to date by ChIP-seq display enrichment just downstream of TSSs21,52–57, and Integrator occupancy closely tracks with levels of promoter-associated RNAPII. Consistently, most Integrator-mediated termination occurs on RNAPII very early in elongation21,52,53,55. Despite this promoter enrichment, Integrator subunits can remain associated with RNAPII as it enters the gene body, and Integrator has been implicated in termination at some canonical mRNA 3′ ends54,61. Although questions remain about the prevalence of Integrator within gene bodies and which factor(s) might stabilize Integrator-RNAPII interactions once NELF dissociates, intriguing data indicate a role for Integrator in mRNA 3′-end formation under stress conditions54,61. Specifically, cellular challenges such as osmotic stress or viral infection can cause failures in mRNA cleavage and 3′-end formation, resulting in RNAPII elongation >10kb beyond the typical site of transcription termination62,63. This Downstream of Gene (DoG) transcription64 represents a fundamental defect in the termination process and can allow RNAPII readthrough into neighboring genes, raising the specter of transcriptional interference65. Notably, the mRNA genes that generate DoGs during stress partially overlap with genes that show evidence of readthrough past the 3′-end when INTS11 is depleted. Further, hyperosmotic stress was found to reduce Integrator association with RNAPII61, suggesting that a subset of protein-coding genes deploy Integrator as a backup to the CPA machinery under conditions of stress or immune challenge. However, in normal cellular conditions, INTS11 depletion affects canonical 3′-end formation at a limited number of transcripts54,66. Long-term depletion of INTS11 was found to alter the expression of CPSF7354 suggesting an interesting level of feedback among 3′-end processing machineries that could confound long-term depletion studies. Indeed, a recent study used a fast-acting degron to deplete INTS11 in mouse ES cells found no significant role for Integrator in canonical mRNA 3′-end formation under normal growth conditions67.
Specificity of Integrator activity
Early data suggested that Integrator functioned uniquely at snRNA genes, with Integrator directly recruited to these promoters by interactions with the transcription factor snRNA activating protein complex (SNAPc) and with RNA cleavage directed by the ‘3′ box motif’ (Baillat and Wagner, 2015; Baillat et al., 2005). However, since this time, Integrator has been found to act at nearly every species of noncoding RNA (ncRNA), including lncRNA68,69, PIWI-interacting RNAs70, telomerase RNA71 uaRNAs66,72 and eRNAs66,73. Moreover, Integrator targets RNAPII at mRNA TSSs, regulating protein-coding gene activity52,55–57,74. This broad spectrum of targets makes it difficult to envision models involving selective promoter recruitment by TFs, and indeed Integrator occupancy broadly correlates with RNAPII levels rather than specific TF motifs or protein factors56,57. This widespread association of Integrator with paused RNAPII raises questions about regulation of the INTS11 endonuclease. Given data from snRNAs, an appealing model was that motifs in RNA modulate INTS11 activity. However, in contrast to the CPA machinery, no Integrator subunit contains a sequence-specific RNA binding domain, and sequences resembling the 3′ box were not observed near most Integrator target genes66,75. Thus, it remains an open and intriguing question how Integrator-mediated cleavage might be controlled.
Integrator’s functional roles
Below, we highlight current models and remaining questions about the consequences of Integrator-mediated termination, using enhancers and protein-coding genes as examples.
Integrator at enhancers
RNAs generated at enhancers are typically short (<300 nt) and display heterogeneous 3′ ends, some of which are generated by Integrator activity. Integrator efficiently terminates paused RNAPII at enhancers, driving a rapid turnover of early elongation complexes and promoting the synthesis of short RNA species52,60,73. Integrator loss delays eRNA 3′-end formation, with cleavage and termination carried out farther downstream by alternative termination complexes such as the CPA machinery. Accordingly, depletion of Integrator subunits results in the formation of eRNAs that are longer, yet less abundant49,52,66,67,73. In the absence of a clear model for eRNA function76 however, the consequences of Integrator activity at enhancers remain unclear. One study reported that Integrator facilitates enhancer-promoter looping at several stimulus-dependent genes73 but this remains to be investigated more broadly. We propose that Integrator-mediated recycling of RNAPII at enhancers could maintain RNAPII dynamically engaged at the locus so that it is rapidly available for transfer to the promoter during gene activation. Moreover, if early termination by Integrator enables rapid re-initiation of transcription at enhancers, this could promote the synthesis of a short-lived ‘cloud’ of eRNAs around the enhancer that serve as binding surfaces for transcription factors or co-activators77. Conversely, increased production of extended eRNAs with longer retention times on chromatin in the absence of Integrator might promote RNA-protein interactions. Future studies of eRNA function and a more detailed analysis of Integrator action at enhancers are thus warranted.
Premature termination at protein-coding genes
Long term depletion of Integrator (e.g., using 48–96 h RNAi treatment) in mammalian or Drosophila cells consistently reveals up- and down-regulation of hundreds of protein coding genes52,53,56,78,79. The differential effects of Integrator loss on gene activity have suggested that Integrator could be repressive, stimulatory, or inconsequential for mRNA expression depending on the gene and the context. Fundamentally, either repressive or stimulatory effects could be envisioned for Integrator, depending on the status of the RNAPII complexes that are targeted for termination. For example, if Integrator terminates transiently paused elongation complexes that would otherwise produce a mature RNA, then Integrator activity would be repressive for transcription21,52,66. Indeed, models wherein premature termination attenuates gene activity are well established in bacteria, yeast and metazoan systems80. If instead, Integrator terminates inactive RNAPII that has stalled and is obstructing the DNA template, then Integrator would serve an activating role53,57,74. Critically, these models are not mutually exclusive, and either scenario could dominate depending on the cellular conditions. For example, under normal growth conditions, Integrator might primarily serve to attenuate expression of stress-responsive genes, but upon activation of stress- or DNA-damaging pathways, Integrator could become critical for removal of stalled RNAPII and gene induction.
Genes within stress- and signal-responsive pathways are recurrently affected by Integrator loss across cell types and species, with a particular enrichment of immediate early genes such as Jun and Fos21,22,52,53,55–57. These findings suggest a common set of targets or pathways, despite a lack of evidence for gene-specific Integrator recruitment12,21,57.
Recent work using rapid, degron-mediated depletion of INTS11 in mouse embryonic stem cells sheds light on the function and specificity of Integrator. Acute degradation of INTS11 causes universal increases in RNAPII complexes released from promoter regions into genes67, suggesting that Integrator broadly limits RNAPII release into elongation. However, loss of INTS11 did not significantly increase the expression of most genes, due to elongation defects in RNAPII. Investigation of these defects revealed that rapid loss of INTS11 did not dissociate other Integrator subunits from RNAPII, with evidence that the phosphatase module remained active on elongating polymerase. Consequently, phosphorylation of RNAPII and SPT5 was impaired, impacting the rate and processivity of elongation. As a result, only short mRNA genes were upregulated, along with a repertoire of inherently short ncRNAs. Of note, rapidly inducible and stress-responsive factors, including Jun and Fos, tend to be encoded by short transcripts with short or no introns81. These results suggest that the consistent activation of specific immediate-early genes encoding TFs, kinases and signaling regulators reflects the length of these genes rather than specific activities of Integrator.
INTEGRATOR IMPORTANCE IN PHYSIOLOGY AND DISEASE
The broad presence of Integrator would suggest fundamental importance to cellular function and organismal development. Indeed, depletion of Integrator subunits disrupts an array of cellular processes and differentiation pathways71,82–86. Consistently, homozygous loss of most Integrator subunits causes lethality in multiple model organisms46,82,87–89. The one exception to this trend is INTS6, which was initially identified as DICE1 (Deleted in Cancer 1) based on its frequent deletion in cancers90. However, the lack of essentiality of INTS6 likely reflects that it is the only Integrator subunit with a paralog within the human genome (INTS6-like).
Studies have revealed particular importance for Integrator in developing and differentiating neuronal cell types. In mice, the Integrator core has been found to interact with Cohesin subunit Nipbl and ZFP609 and modulate the expression of genes important for neuronal migration during development79. In Drosophila, depletion of backbone or shoulder module subunits leads to increased type II neuroblasts, and thus these subunits are required to prevent de-differentiation of intermediate neural progenitor cells91. In humans, mutation of INTS1 or INTS8 has been found to cause severe neurodevelopmental defects, including profound intellectual disability, epilepsy, and structural brain abnormalities92,93. More recently, BRAT1 has been found to interact with the INTS9/11 heterodimer and mutations in BRAT1 are associated with numerous neurodevelopmental disorders94.
Deleterious human Integrator mutations have also been informative on how Integrator modules interact with each other. In the case of INTS8, patients presenting neurological dysfunction are hypomorphic and predominantly express a form containing a three amino acid deletion of a conserved EVL motif near the C-terminal region92. The INTS8-ΔEVL was found to associate with the rest of the complex poorly, and recent structural findings indicate that the EVL motif lies within a region of INTS8 that is likely critical to maintain tight interaction between the shoulder and phosphatase modules23. Similarly, two distinct mutations near the C-terminus of INTS13 that are causative of a specific ciliopathy disease51 are predicted to disrupt the cleavage-module binding motif34,39 underscoring the importance of ICM interactions with the auxiliary module51. These examples highlight the value of characterizing diseased states caused by disrupted Integrator interactions.
FUTURE PERSPECTIVES
While the first 15 years of Integrator research have provided significant insight into its function, many questions remain unanswered. From a structural perspective, it is not yet known if Integrator only exists as a full complex or whether individual modules are separable. Importantly, whereas long term depletion of Integrator subunits often destabilizes the entire complex, short term degradation strategies are now allowing more surgical removal of specific subunits and modules. Such approaches will elucidate whether the two catalytic activities of Integrator are independent or coordinated. Additionally, a clear understanding of where the auxiliary module associates with the rest of the complex and how it contributes to Integrator function is still lacking. Finally, provocative biochemical and structural connections between Integrator and DNA damage sensing machinery35,38,95 have been observed, but whether these interactions represent a novel function for Integrator or repurposing of its established termination activity is unknown.
The selectivity and regulation of Integrator activity also remains to be defined, with broad Integrator occupancy raising the question of how cells govern the balance between premature termination and pause release. We propose that mechanisms exist to deactivate Integrator or evict it from the paused elongation complex to enable gene induction. Possible candidates for this are factors that recruit P-TEFb, which could destabilize Integrator association with RNAPII by phosphorylation of SPT5 and the CTD, and dissociation of NELF. However, how P-TEFb activity might be coordinated to specifically overcome the Integrator-associated PP2A phosphatase function remains an active area of research. Further, how Integrator determines where to cleave nascent RNA is not clear. While sequence elements appear to govern snRNA 3′-end formation, these sequences are not found at other Integrator targets75. We therefore propose that INTS11-mediated cleavage activity is modulated by protein factors associated with the paused RNAPII, potentially SPT5 or NELF.
Finally, while mutations within Integrator subunits can have dire consequences on human development and health, the specific gene sets most sensitive to these mutations are only beginning to be understood67. Moreover, it isn’t clear why specific tissue types or developmental stages are asymmetrically impacted by reduced Integrator integrity. Regardless of these unknowns, our understanding of Integrator constituency and function in gene expression has undergone a remarkable evolution, and progress is expected to continue at a rapid pace.
Acknowledgments
We thank Patrick Cramer and Yanhui Xu for sharing atomic structures before their release from the PDB. This research is supported by NIH grants R01GM134539 (to K.A. and E.J.W.) and R35GM118093 (to L.T.).
Footnotes
Declaration of Interests
K.A. is on the SAB of CAMP4 Therapeutics, received research funding from Novartis and is a member of the Advisory Board of Molecular Cell.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Adelman K, and Lis JT (2012). Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet 13, 720–731. 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Core L, and Adelman K. (2019). Promoter-proximal pausing of RNA polymerase II: a nexus of gene regulation. Genes Dev 33, 960–982. 10.1101/gad.325142.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vos SM, Farnung L, Urlaub H, and Cramer P. (2018). Structure of paused transcription complex Pol II-DSIF-NELF. Nature 560, 601–606. 10.1038/s41586018-0442-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Missra A, and Gilmour DS (2010). Interactions between DSIF (DRB sensitivity inducing factor), NELF (negative elongation factor), and the Drosophila RNA polymerase II transcription elongation complex. Proc Natl Acad Sci U S A 107, 11301–11306. 10.1073/pnas.1000681107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujinaga K, Irwin D, Huang Y, Taube R, Kurosu T, and Peterlin BM (2004). Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Mol Cell Biol 24, 787–795. 10.1128/MCB.24.2.787-795.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamaguchi Y., Inukai N., Narita T., Wada T., and Handa H. (2002). Evidence that negative elongation factor represses transcription elongation through binding to a DRB sensitivity-inducing factor/RNA polymerase II complex and RNA. Mol Cell Biol 22, 2918–2927. 10.1128/MCB.22.9.2918-2927.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henriques T, Gilchrist DA, Nechaev S, Bern M, Muse GW, Burkholder A, Fargo DC, and Adelman K. (2013). Stable pausing by RNA polymerase II provides an opportunity to target and integrate regulatory signals. Mol Cell 52, 517–528. 10.1016/j.molcel.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vos SM, Farnung L, Boehning M, Wigge C, Linden A, Urlaub H, and Cramer P. (2018). Structure of activated transcription complex Pol II-DSIF-PAF-SPT6. Nature 560, 607–612. 10.1038/s41586-018-0440-4. [DOI] [PubMed] [Google Scholar]
- 9.Proudfoot NJ (2011). Ending the message: poly(A) signals then and now. Genes Dev 25, 1770–1782. 10.1101/gad.17268411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cortazar MA, Sheridan RM, Erickson B, Fong N, Glover-Cutter K, Brannan K, and Bentley DL (2019). Control of RNA Pol II Speed by PNUTS-PP1 and Spt5 Dephosphorylation Facilitates Termination by a “Sitting Duck Torpedo” Mechanism. Mol Cell 76, 896–908 e894. 10.1016/j.molcel.2019.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rienzo M, and Casamassimi A. (2016). Integrator complex and transcription regulation: Recent findings and pathophysiology. Biochim Biophys Acta 1859, 1269–1280. 10.1016/j.bbagrm.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 12.Kirstein N, Gomes Dos Santos H, Blumenthal E, and Shiekhattar R. (2021). The Integrator complex at the crossroad of coding and noncoding RNA. Curr Opin Cell Biol 70, 37–43. 10.1016/j.ceb.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baillat D, Hakimi MA, Naar AM, Shilatifard A, Cooch N, and Shiekhattar R. (2005). Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell 123, 265–276. [DOI] [PubMed] [Google Scholar]
- 14.Chen J, Ezzeddine N, Waltenspiel B, Albrecht TR, Warren WD, Marzluff WF, and Wagner EJ (2012). An RNAi screen identifies additional members of the Drosophila Integrator complex and a requirement for cyclin C/Cdk8 in snRNA 3’-end formation. RNA 18, 2148–2156. 10.1261/rna.035725.112rna.035725.112 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malovannaya A, Lanz RB, Jung SY, Bulynko Y, Le NT, Chan DW, Ding C, Shi Y, Yucer N, Krenciute G, et al. (2011). Analysis of the human endogenous coregulator complexome. Cell 145, 787–799. S0092-8674(11)00532–0 [pii] 10.1016/j.cell.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malovannaya A., Li Y., Bulynko Y., Jung SY., Wang Y., Lanz RB., O’Malley BW., and Qin J. (2010). Streamlined analysis schema for high-throughput identification of endogenous protein complexes. Proc Natl Acad Sci U S A 107, 2431–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drew K, Wallingford JB, and Marcotte EM (2021). hu.MAP 2.0: integration of over 15,000 proteomic experiments builds a global compendium of human multiprotein assemblies. Mol Syst Biol 17, e10016. 10.15252/msb.202010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Funk L, Su K-C, Ly J, Feldman D, Singh A, Moodie B, Blainey PC, and Cheeseman IM (2021). The phenotypic landscape of essential human genes. Cell 185, 1–20. 10.1016/j.cell.2022.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan J, Kwon JJ, Talamas JA, Borah AA, Vazquez F, Boehm JS, Tsherniak A, Zitnik M, McFarland JM, and Hahn WC (2022). Sparse dictionary learning recovers pleiotropy from human cell fitness screens. Cell Syst 13, 286–303 e210. 10.1016/j.cels.2021.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Replogle JM, Saunders RA, Pogson AN, Hussmann JA, Lenail A, Guna A, Mascibroda L, Wagner EJ, Adelman K, Lithwick-Yanai G, et al. (2022). Mapping information-rich genotype-phenotype landscapes with genome-scale Perturb-seq. Cell 185, 2559–2575 e2528. 10.1016/j.cell.2022.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang KL, Jee D, Stein CB, Elrod ND, Henriques T, Mascibroda LG, Baillat D, Russell WK, Adelman K, and Wagner EJ (2020). Integrator Recruits Protein Phosphatase 2A to Prevent Pause Release and Facilitate Transcription Termination. Mol Cell 80, 345–358 e349. 10.1016/j.molcel.2020.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vervoort SJ, Welsh SA, Devlin JR, Barbieri E, Knight DA, Offley S, Bjelosevic S, Costacurta M, Todorovski I, Kearney CJ, et al. (2021). The PP2A-Integrator-CDK9 axis fine-tunes transcription and can be targeted therapeutically in cancer. Cell 184, 3143–3162 e3132. 10.1016/j.cell.2021.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng H, Qi Y, Hu S, Cao X, Xu C, Yin Z, Chen X, Li Y, Liu W, Li J, et al. (2020). Identification of Integrator-PP2A complex (INTAC), an RNA polymerase II phosphatase. Science 370. 10.1126/science.abb5872. [DOI] [PubMed] [Google Scholar]
- 24.Peterlin BM, and Price DH (2006). Controlling the elongation phase of transcription with P-TEFb. Mol Cell 23, 297–305. 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 25.Baillat D, Russell WK, and Wagner EJ (2016). CRISPR-Cas9 mediated genetic engineering for the purification of the endogenous integrator complex from mammalian cells. Protein Expr Purif 128, 101–108. 10.1016/j.pep.2016.08.011. [DOI] [PubMed] [Google Scholar]
- 26.Callebaut I, Moshous D, Mornon JP, and de Villartay JP (2002). Metallo-beta-lactamase fold within nucleic acids processing enzymes: the beta-CASP family. Nucleic Acids Res 30, 3592–3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dominski Z., Yang XC., Purdy M., Wagner EJ., and Marzluff WF. (2005). A CPSF-73 homologue is required for cell cycle progression but not cell growth and interacts with a protein having features of CPSF-100. Mol Cell Biol 25, 1489–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mandel CR, Kaneko S, Zhang H, Gebauer D, Vethantham V, Manley JL, and Tong L. (2006). Polyadenylation factor CPSF-73 is the pre-mRNA 3’-end-processing endonuclease. Nature 444, 953–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Albrecht TR, and Wagner EJ (2012). snRNA 3’ end formation requires heterodimeric association of integrator subunits. Mol Cell Biol 32, 1112–1123. MCB.06511–11 [pii] 10.1128/MCB.06511-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.West S, Gromak N, and Proudfoot NJ (2004). Human 5’ --> 3’ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature 432, 522–525. 10.1038/nature03035. [DOI] [PubMed] [Google Scholar]
- 31.Kim M, Krogan NJ, Vasiljeva L, Rando OJ, Nedea E, Greenblatt JF, and Buratowski S. (2004). The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature 432, 517–522. 10.1038/nature03041. [DOI] [PubMed] [Google Scholar]
- 32.Eaton JD, Francis L, Davidson L, and West S. (2020). A unified allosteric/torpedo mechanism for transcriptional termination on human protein-coding genes. Genes Dev 34, 132–145. 10.1101/gad.332833.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng H, Jin Q, Qi Y, Liu W, Ren Y, Wang X, Chen F, Cheng J, Chen X, and Xu Y. (2022). Structural basis of INTAC-regulated transcription. BioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fianu I, Chen Y, Dienemann C, Dybkov O, Linden A, Urlaub H, and Cramer P. (2021). Structural basis of Integrator-mediated transcription regulation. Science 374, 883–887. 10.1126/science.abk0154. [DOI] [PubMed] [Google Scholar]
- 35.Jia Y, Cheng Z, Bharath SR, Sun Q, Su N, Huang J, and Song H. (2021). Crystal structure of the INTS3/INTS6 complex reveals the functional importance of INTS3 dimerization in DSB repair. Cell Discov 7, 66. 10.1038/s41421-021-00283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin MH, Jensen MK, Elrod ND, Huang KL, Welle KA, Wagner EJ, and Tong L. (2022). Inositol hexakisphosphate is required for Integrator function. Nat Commun 13, 5742. 10.1038/s41467-022-33506-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pfleiderer MM, and Galej WP (2021). Structure of the catalytic core of the Integrator complex. Mol Cell 81, 1246–1259 e1248. 10.1016/j.molcel.2021.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ren W, Chen H, Sun Q, Tang X, Lim SC, Huang J, and Song H. (2014). Structural basis of SOSS1 complex assembly and recognition of ssDNA. Cell Rep 6, 982–991. 10.1016/j.celrep.2014.02.020. [DOI] [PubMed] [Google Scholar]
- 39.Sabath K., Staubli ML., Marti S., Leitner A., Moes M., and Jona S. (2020). INTS10-INTS13-INTS14 form a functional module of Integrator that binds nucleic acids and the cleavage module. Nat Commun 11, 3422. 10.1038/s41467-020-17232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu Y, Albrecht TR, Baillat D, Wagner EJ, and Tong L. (2017). Molecular basis for the interaction between Integrator subunits IntS9 and IntS11 and its functional importance. Proc Natl Acad Sci U S A 114, 4394–4399. 10.1073/pnas.1616605114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J, Ma X, Banerjee S, Baruah S, Schnicker NJ, Roh E, Ma W, Liu K, Bode AM, and Dong Z. (2021). Structural basis for multifunctional roles of human Ints3 C-terminal domain. J Biol Chem 296, 100112. 10.1074/jbc.RA120.016393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen J, Waltenspiel B, Warren WD, and Wagner EJ (2013). Functional analysis of the integrator subunit 12 identifies a microdomain that mediates activation of the Drosophila integrator complex. J Biol Chem 288, 4867–4877. 10.1074/jbc.M112.425892M112.425892 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yadav L, Tamene F, Goos H, van Drogen A, Katainen R, Aebersold R, Gstaiger M, and Varjosalo M. (2017). Systematic Analysis of Human Protein Phosphatase Interactions and Dynamics. Cell Syst 4, 430–444 e435. 10.1016/j.cels.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 44.Herzog F, Kahraman A, Boehringer D, Mak R, Bracher A, Walzthoeni T, Leitner A, Beck M, Hartl FU, Ban N, et al. (2012). Structural probing of a protein phosphatase 2A network by chemical cross-linking and mass spectrometry. Science 337, 1348–1352. 10.1126/science.1221483. [DOI] [PubMed] [Google Scholar]
- 45.Shi Y. (2009). Serine/threonine phosphatases: mechanism through structure. Cell 139, 468–484. 10.1016/j.cell.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 46.Ezzeddine N., Chen J., Waltenspiel B., Burch B., Albrecht T., Zhuo M., Warren WD., Marzluff WF., and Wagner EJ. (2011). A subset of Drosophila integrator proteins is essential for efficient U7 snRNA and spliceosomal snRNA 3’-end formation. Mol Cell Biol 31, 328–341. MCB.00943–10 [pii] 10.1128/MCB.00943-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Albrecht TR, Shevtsov SP, Wu Y, Mascibroda LG, Peart NJ, Huang KL, Sawyer IA, Tong L, Dundr M, and Wagner EJ (2018). Integrator subunit 4 is a ‘Symplekin-like’ scaffold that associates with INTS9/11 to form the Integrator cleavage module. Nucleic Acids Res 46, 4241–4255. 10.1093/nar/gky100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun Y, Zhang Y, Aik WS, Yang XC, Marzluff WF, Walz T, Dominski Z, and Tong L. (2020). Structure of an active human histone pre-mRNA 3’-end processing machinery. Science 367, 700–703. 10.1126/science.aaz7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barbieri E, Trizzino M, Welsh SA, Owens TA, Calabretta B, Carroll M, Sarma K, and Gardini A. (2018). Targeted Enhancer Activation by a Subunit of the Integrator Complex. Mol Cell 71, 103–116 e107. 10.1016/j.molcel.2018.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walker JR, Corpina RA, and Goldberg J. (2001). Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature 412, 607–614. 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 51.Mascibroda LG, Shboul M, Elrod ND, Colleaux L, Hamamy H, Huang KL, Peart N, Singh MK, Lee H, Merriman B, et al. (2022). INTS13 variants causing a recessive developmental ciliopathy disrupt assembly of the Integrator complex. Nat Commun 13, 6054. 10.1038/s41467-022-33547-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Elrod ND, Henriques T, Huang KL, Tatomer DC, Wilusz JE, Wagner EJ, and Adelman K. (2019). The Integrator Complex Attenuates Promoter-Proximal Transcription at Protein-Coding Genes. Mol Cell 76, 738–752 e737. 10.1016/j.molcel.2019.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beckedorff F, Blumenthal E, daSilva LF, Aoi Y, Cingaram PR, Yue J, Zhang A, Dokaneheifard S, Valencia MG, Gaidosh G, et al. (2020). The Human Integrator Complex Facilitates Transcriptional Elongation by Endonucleolytic Cleavage of Nascent Transcripts. Cell Rep 32, 107917. 10.1016/j.celrep.2020.107917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dasilva LF, Blumenthal E, Beckedorff F, Cingaram PR, Gomes Dos Santos H, Edupuganti RR, Zhang A, Dokaneheifard S, Aoi Y, Yue J, et al. (2021). Integrator enforces the fidelity of transcriptional termination at protein-coding genes. Sci Adv 7, eabe3393. 10.1126/sciadv.abe3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Skaar JR, Ferris AL, Wu X, Saraf A, Khanna KK, Florens L, Washburn MP, Hughes SH, and Pagano M. (2015). The Integrator complex controls the termination of transcription at diverse classes of gene targets. Cell Res 25, 288–305. 10.1038/cr.2015.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stadelmayer B., Micas G., Gamot A., Martin P., Malirat N., Koval S., Raffel R., Sobhian B., Severac D., Rialle S., et al. (2014). Integrator complex regulates NELF-mediated RNA polymerase II pause/release and processivity at coding genes. Nat Commun 5, 5531. 10.1038/ncomms6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gardini A, Baillat D, Cesaroni M, Hu D, Marinis JM, Wagner EJ, Lazar MA, Shilatifard A, and Shiekhattar R. (2014). Integrator Regulates Transcriptional Initiation and Pause Release following Activation. Mol Cell 56, 128–139. 10.1016/j.molcel.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen X, Yin X, Li J, Wu Z, Qi Y, Wang X, Liu W, and Xu Y. (2021). Structures of the human Mediator and Mediator-bound preinitiation complex. Science 372. 10.1126/science.abg0635. [DOI] [PubMed] [Google Scholar]
- 59.Bunch H, Lawney BP, Burkholder A, Ma D, Zheng X, Motola S, Fargo DC, Levine SS, Wang YE, and Hu G. (2016). RNA polymerase II promoter-proximal pausing in mammalian long non-coding genes. Genomics 108, 64–77. 10.1016/j.ygeno.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Henriques T, Scruggs BS, Inouye MO, Muse GW, Williams LH, Burkholder AB, Lavender CA, Fargo DC, and Adelman K. (2018). Widespread transcriptional pausing and elongation control at enhancers. Genes Dev 32, 26–41. 10.1101/gad.309351.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rosa-Mercado NA, Zimmer JT, Apostolidi M, Rinehart J, Simon MD, and Steitz JA (2021). Hyperosmotic stress alters the RNA polymerase II interactome and induces readthrough transcription despite widespread transcriptional repression. Mol Cell 81, 502–513 e504. 10.1016/j.molcel.2020.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vilborg A, Passarelli MC, Yario TA, Tycowski KT, and Steitz JA (2015). Widespread Inducible Transcription Downstream of Human Genes. Mol Cell 59, 449–461. 10.1016/j.molcel.2015.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nemeroff ME, Barabino SM, Li Y, Keller W, and Krug RM (1998). Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3’end formation of cellular pre-mRNAs. Mol Cell 1, 991–1000. 10.1016/s1097-2765(00)80099-4. [DOI] [PubMed] [Google Scholar]
- 64.Rosa-Mercado NA, and Steitz JA (2022). Who let the DoGs out? - biogenesis of stress-induced readthrough transcripts. Trends Biochem Sci 47, 206–217. 10.1016/j.tibs.2021.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martens JA, Laprade L, and Winston F. (2004). Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature 429, 571–574. 10.1038/nature02538. [DOI] [PubMed] [Google Scholar]
- 66.Lykke-Andersen S., Zumer K., Molska ES., Rouviere JO., Wu G., Demel C., Schwalb B., Schmid M., Cramer P., and Jensen TH. (2021). Integrator is a genome-wide attenuator of non-productive transcription. Mol Cell 81, 514–529 e516. 10.1016/j.molcel.2020.12.014. [DOI] [PubMed] [Google Scholar]
- 67.Stein CB, Field AR, Mimoso CA, Zhao C, Huang KL, Wagner EJ, and Adelman K. (2022). Integrator endonuclease drives promoter-proximal termination at all RNA polymerase II-transcribed loci. Mol Cell. 10.1016/j.molcel.2022.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nojima T, Tellier M, Foxwell J, Ribeiro de Almeida C, Tan-Wong SM, Dhir S, Dujardin G, Dhir A, Murphy S, and Proudfoot NJ (2018). Deregulated Expression of Mammalian lncRNA through Loss of SPT6 Induces R-Loop Formation, Replication Stress, and Cellular Senescence. Mol Cell 72, 970–984 e977. 10.1016/j.molcel.2018.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barra J, Gaidosh GS, Blumenthal E, Beckedorff F, Tayari MM, Kirstein N, Karakach TK, Jensen TH, Impens F, Gevaert K, et al. (2020). Integrator restrains paraspeckles assembly by promoting isoform switching of the lncRNA NEAT1. Sci Adv 6, eaaz9072. 10.1126/sciadv.aaz9072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Beltran T, Pahita E, Ghosh S, Lenhard B, and Sarkies P. (2021). Integrator is recruited to promoter-proximally paused RNA Pol II to generate Caenorhabditis elegans piRNA precursors. EMBO J 40, e105564. 10.15252/embj.2020105564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rubtsova MP, Vasilkova DP, Moshareva MA, Malyavko AN, Meerson MB, Zatsepin TS, Naraykina YV, Beletsky AV, Ravin NV, and Dontsova OA (2019). Integrator is a key component of human telomerase RNA biogenesis. Sci Rep 9, 1701. 10.1038/s41598-018-38297-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu X, Guo Z, Han J, Peng B, Zhang B, Li H, Hu X, David CJ, and Chen M. (2022). The PAF1 complex promotes 3’ processing of pervasive transcripts. Cell Rep 38, 110519. 10.1016/j.celrep.2022.110519. [DOI] [PubMed] [Google Scholar]
- 73.Lai F, Gardini A, Zhang A, and Shiekhattar R. (2015). Integrator mediates the biogenesis of enhancer RNAs. Nature 525, 399–403. 10.1038/nature14906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yue J, Lai F, Beckedorff F, Zhang A, Pastori C, and Shiekhattar R. (2017). Integrator orchestrates RAS/ERK1/2 signaling transcriptional programs. Genes Dev 31, 1809–1820. 10.1101/gad.301697.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Baillat D, and Wagner EJ (2015). Integrator: surprisingly diverse functions in gene expression. Trends Biochem Sci 40, 257–264. 10.1016/j.tibs.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Field A, and Adelman K. (2020). Evaluating Enhancer Function and Transcription. Annu Rev Biochem 89, 213–234. 10.1146/annurev-biochem011420-095916. [DOI] [PubMed] [Google Scholar]
- 77.Sigova AA, Abraham BJ, Ji X, Molinie B, Hannett NM, Guo YE, Jangi M, Giallourakis CC, Sharp PA, and Young RA (2015). Transcription factor trapping by RNA in gene regulatory elements. Science 350, 978–981. 10.1126/science.aad3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tatomer DC., Elrod ND., Liang D., Xiao MS., Jiang JZ., Jonathan M., Huang KL., Wagner EJ., Cherry S., and Wilusz JE. (2019). The Integrator complex cleaves nascent mRNAs to attenuate transcription. Genes Dev 33, 1525–1538. 10.1101/gad.330167.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.van den Berg DL, Azzarelli R, Oishi K, Martynoga B, Urban N, Dekkers DH, Demmers JA, and Guillemot F. (2017). Nipbl Interacts with Zfp609 and the Integrator Complex to Regulate Cortical Neuron Migration. Neuron 93, 348–361. 10.1016/j.neuron.2016.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kamieniarz-Gdula K, and Proudfoot NJ (2019). Transcriptional Control by Premature Termination: A Forgotten Mechanism. Trends Genet 35, 553–564. 10.1016/j.tig.2019.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lopes I, Altab G, Raina P, and de Magalhaes JP (2021). Gene Size Matters: An Analysis of Gene Length in the Human Genome. Front Genet 12, 559998. 10.3389/fgene.2021.559998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tao S, Cai Y, and Sampath K. (2009). The Integrator subunits function in hematopoiesis by modulating Smad/BMP signaling. Development 136, 2757–2765. [DOI] [PubMed] [Google Scholar]
- 83.Takata H, Nishijima H, Maeshima K, and Shibahara K. (2012). The integrator complex is required for integrity of Cajal bodies. J Cell Sci 125, 166–175. 10.1242/jcs.090837. [DOI] [PubMed] [Google Scholar]
- 84.Otani Y, Nakatsu Y, Sakoda H, Fukushima T, Fujishiro M, Kushiyama A, Okubo H, Tsuchiya Y, Ohno H, Takahashi S, et al. (2013). Integrator complex plays an essential role in adipose differentiation. Biochem Biophys Res Commun 434, 197–202. 10.1016/j.bbrc.2013.03.029. [DOI] [PubMed] [Google Scholar]
- 85.Jodoin JN, Shboul M, Albrecht TR, Lee E, Wagner EJ, Reversade B, and Lee LA (2013). The snRNA-processing complex, Integrator, is required for ciliogenesis and dynein recruitment to the nuclear envelope via distinct mechanisms. Biol Open 2, 1390–1396. 10.1242/bio.20136981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kapp LD, Abrams EW, Marlow FL, and Mullins MC (2013). The integrator complex subunit 6 (ints6) confines the dorsal organizer in vertebrate embryogenesis. PLoS Genet 9, e1003822. 10.1371/journal.pgen.1003822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Han SM., Lee TH., Mun JY., Ki MJ., Kritikou EA., Lee SJ., Han SS., Hengartner MO., and Koo HS. (2006). Deleted in cancer 1 (DICE1) is an essential protein controlling the topology of the inner mitochondrial membrane in C. elegans. Development 133, 3597–3606. [DOI] [PubMed] [Google Scholar]
- 88.Hata T, and Nakayama M. (2007). Targeted disruption of the murine large nuclear KIAA1440/Ints1 protein causes growth arrest in early blastocyst stage embryos and eventual apoptotic cell death. Biochim Biophys Acta 1773, 1039–1051. [DOI] [PubMed] [Google Scholar]
- 89.Rutkowski RJ, and Warren WD (2009). Phenotypic analysis of deflated/Ints7 function in Drosophila development. Dev Dyn 238, 1131–1139. [DOI] [PubMed] [Google Scholar]
- 90.Wieland I, Arden KC, Michels D, Klein-Hitpass L, Bohm M, Viars CS, and Weidle UH (1999). Isolation of DICE1: a gene frequently affected by LOH and downregulated in lung carcinomas. Oncogene 18, 4530–4537. 10.1038/sj.onc.1202806. [DOI] [PubMed] [Google Scholar]
- 91.Zhang Y, Koe CT, Tan YS, Ho J, Tan P, Yu F, Sung WK, and Wang H. (2019). The Integrator Complex Prevents Dedifferentiation of Intermediate Neural Progenitors back into Neural Stem Cells. Cell Rep 27, 987–996 e983. 10.1016/j.celrep.2019.03.089. [DOI] [PubMed] [Google Scholar]
- 92.Oegema R, Baillat D, Schot R, van Unen LM, Brooks A, Kia SK, Hoogeboom AJM, Xia Z, Li W, Cesaroni M, et al. (2017). Human mutations in integrator complex subunits link transcriptome integrity to brain development. PLoS Genet 13, e1006809. 10.1371/journal.pgen.1006809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Krall M, Htun S, Schnur RE, Brooks AS, Baker L, de Alba Campomanes A, Lamont RE, Gripp KW, Care 4 Rare Canada, C., Schneidman-Duhovny, D., et al. (2019). Biallelic sequence variants in INTS1 in patients with developmental delays, cataracts, and craniofacial anomalies. Eur J Hum Genet 27, 582–593. 10.1038/s41431-018-0298-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cihlarova Z, Kubovciak J, Sobol M, Krejcikova K, Sachova J, Kolar M, Stanek D, Barinka C, Yoon G, Caldecott KW, and Hanzlikova H. (2022). BRAT1 links Integrator and defective RNA processing with neurodegeneration. Nat Commun 13, 5026. 10.1038/s41467-022-32763-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Huang J, Gong Z, Ghosal G, and Chen J. (2009). SOSS complexes participate in the maintenance of genomic stability. Mol Cell 35, 384–393. 10.1016/j.molcel.2009.06.011S1097-2765(09)00402-X [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]