Abstract
The ura-blaster technique for the disruption of Candida albicans genes has been employed in a number of studies to identify possible genes encoding virulence factors of this fungal pathogen. In this study, the URA3-encoded orotidine 5′-monophosphate (OMP) decarboxylase enzyme activities of C. albicans strains with ura-blaster-mediated genetic disruptions were measured. All strains harboring genetic lesions via the ura-blaster construct showed reduced OMP decarboxylase activities compared to that of the wild type when assayed. The activity levels in different gene disruptions varied, suggesting a positional effect on the level of gene expression. Because the URA3 gene of C. albicans has previously been identified as a virulence factor for this microorganism, our results suggest that decreased virulence observed in strains constructed with the ura-blaster cassette cannot accurately be attributed, in all cases, to the targeted genetic disruption. Although revised methods for validating a URA3-disrupted gene as a target for antifungal drug development could be devised, it is clearly desirable to replace URA3 with a different selectable marker that does not influence virulence.
Fungal species have become an increasingly common cause of human infection, with Candida albicans emerging as the predominant fungal pathogen in both superficial and systemic infections (7). This polymorphic fungus is carried as a commensal by many individuals; however, in patients rendered immunocompromised by circumstances such as AIDS, chemotherapy, and organ transplantation, C. albicans readily becomes pathogenic.
The development of effective strategies for the treatment of C. albicans infections continues to be a major challenge. Effective antifungal agents may prove to be those which target virulence mechanisms important in the pathogenesis of this microorganism. This approach to drug development utilizes molecular biological strategies to identify fungal virulence genes for the determination of targets (23). Unfortunately, induction and analysis of specific genetic lesions required to identify these virulence factors are difficult tasks. Many conventional genetic techniques utilized in the study of bacterial pathogens cannot be applied to C. albicans due to its asexual life cycle and diploid genome (26). Fortunately, proven techniques used with the haploid yeast Saccharomyces cerevisiae are applicable, in certain cases, to the study of C. albicans.
A construct termed the ura-blaster was developed for the disruption of S. cerevisiae genes by Alani et al. (1). It consists of the URA3 gene of S. cerevisiae flanked by direct repeats of a Salmonella hisG sequence and portions of the target gene which can then be used for the disruption of a targeted gene of interest by integrative transformation with URA3 as a selectable marker. Once introduced into the genome, the hisG direct repeats may undergo mitotic recombination to eliminate the URA3 gene, leaving behind a single copy of the hisG repeat sequence at the site of the original integration in the target gene. This disruption technique became attractive in the study of C. albicans because strains could be constructed with targeted disruptions by using the C. albicans URA3 gene as the single selectable marker in a uracil auxotrophic background (ura3/ura3) (8). The final result of the ura-blaster genetic disruption method is an intact copy of the C. albicans URA3 gene located within the ura-blaster cassette at the position in the genome corresponding to one allele of the target gene.
The URA3 gene of C. albicans encodes orotidine 5′-monophosphate (OMP) decarboxylase, the enzyme that catalyzes the conversion of OMP to uridine 5′-monophosphate (UMP), the last step in the de novo pyrimidine biosynthetic pathway. Alterations within this pathway, such as disrupting URA3, result in significant decreases in the in vivo growth rate and pathogenicity of C. albicans (15). Therefore, CAI4, the strain used for the creation of ura-blaster genetic disruptions in C. albicans, is not virulent due to the absence of a functional URA3 gene. Any strain created to test the effect of a gene on virulence must have an intact, functional URA3 gene.
Strains constructed by the ura-blaster disruption technique have the URA3 gene of C. albicans inserted into the targeted gene of interest. These genetically engineered strains are then used to test for altered virulence in a murine model of infection to ascertain whether the targeted gene is a virulence factor. The attenuated virulence frequently observed when investigating these strains is generally attributed to the targeted genetic disruption via the ura-blaster. However, an alteration in URA3 activity within the disruption cassette may also contribute to a decrease in virulence. We undertook the analysis of URA3 gene expression within a variety of strains genetically disrupted by the ura-blaster technique to determine if the ura-blaster mode of disruption alters the activity of the URA3 gene product.
MATERIALS AND METHODS
Strains and media.
The C. albicans strains utilized in this study and their relevant genotypes are listed in Table 1. Some of these strains were acquired from other investigators including William A. Fonzi, Gerald R. Fink, and Judith Berman. For the maintenance of C. albicans strains, YPD medium (1% yeast extract, 2% Bacto Peptone, 2% dextrose) was used as a complex growth medium. For minimal medium, SD medium (0.67% Difco yeast nitrogen base without amino acids, 2% dextrose) lacking uridine supplementation was used. Two percent agar was added for the solidification of both YPD and SD-ura medium.
TABLE 1.
C. albicans strains used in this study
| Strain | Parent strain | Genotype | Reference or source |
|---|---|---|---|
| SC5314a | Clinical isolate | URA3/URA3 | 9a |
| CAF2-1a | SC5314 | Δura3::imm434/URA3 | 8 |
| CAI4a | CAF2-1 | Δura3::imm434/Δura3::imm434 | 8 |
| CAI4-2312L | CAI4 | Δura3::imm434/Δura3::imm434 + URA3b | 3 |
| CAI4-01 | CAI4 | ΔCamdr1::hisG-URA3-hisG/CaMDR1 | 3 |
| CAI4-116 | CAI4-01 | ΔCamdr1::hisG-URA3-hisG/ΔCamdr1::hisG | 3 |
| CJC01c | CAI4 | Δben38::hisG-URA3-hisG-URA3-hisG/BEN38 | Unpublished data |
| CJC024c | CJC01 | Δben38::hisG-URA3-hisG/Δben38::hisG | Unpublished data |
| CAI4-10 | CAI4 | Δrsr1::hisG-URA3-hisG/RSR1 | 29 |
| CAI4-5 | CAI4-10 | Δrsr1::hisG-URA3-hisG/Δrsr1::hisG | 29 |
| CWJ429 | CAI4 | ΔtopoI::hisG-URA3-hisG/TOPOI | 13 |
| CWJ430 | CAI4 | ΔtopoI::hisG-URA3-hisG/TOPOI | 13 |
| JKC19d | JKC17 | ΔcphI::hisG-URA3-hisG/ΔcphI::hisG | 18 |
| CACB8B-5 | CACB8B-6 | Δchs3-3::hisG-URA3-hisG-URA3-hisG/Δchs3-2::hisG | 5 |
| CACB8B-6 | CAI4 | Δchs3-3::hisG-URA3-hisG-URA3-hisG/CHS3 | 5 |
| CACB10B-8 | CAI4 | Δchs3-1::hisG-URA3-hisG/CHS3 | 5 |
| CACB10B-10 | CACB10B-8 | Δchs3-1::hisG-URA3-hisG/Δchs3-2::hisG | 5 |
| CAG1e | CAI4 | Δint1::hisG-URA3-hisG/INT1 | 9 |
| CAG3e | CAG1 | Δint1::hisG-URA3-hisG/Δint1::hisG | 9 |
| CAG4e | CAG3 | Δint1::hisG/Δint1::hisG | 9 |
| CAG5e | CAG4 | Δint1::hisG/Δint1::hisG + INT1-URA3f | 9 |
| RSM3c | CAI4 | Δcdr1::hisG-URA3-hisG-URA3-hisG/CDR1 | Unpublished data |
| RSM4c | CAI4 | Δcdr1::hisG-URA3-hisG/CDR1 | Unpublished data |
| RSM7c | RSM3 | Δcdr1::hisG-URA3-hisG/Δcdr1::hisG | Unpublished data |
| RSM11c | RSM4 | Δcdr1::hisG-URA3-hisG/Δcdr1::hisG | Unpublished data |
| RSM17c | CAI4-116 | ΔCamdr1::hisG/ΔCamdr1::hisG/Δcdr1::hisG-URA3-hisG/Δcdr1::hisG | Unpublished data |
a Received from William A. Fonzi.
b + URA3 indicates that URA3 was reintegrated into the chromosome at the URA3 locus.
c Received from Millennium Pharmaceuticals, Inc., Cambridge, Mass.
d Received from Gerald R. Fink.
e Received from Judith Berman.
f +INTI-URA3 indicates that INT1-URA3 was reintegrated into the chromosome at the INT1 locus.
Materials.
OMP, potassium phosphate (monobasic), and dithiothreitol (DTT) were obtained from Sigma Chemical Co., St. Louis, Mo. β-Mercaptoethanol and potassium phosphate (dibasic) were acquired from Mallinckrodt Speciality Chemicals Co., Paris, Ky. The protein assay reagent used in determining protein concentrations was acquired from Bio-Rad Laboratories, Hercules, Calif.
Assay of OMP decarboxylase enzyme activity.
Enzyme activity was monitored by using a modified version of assays described previously (22, 30). Cells from a 50-ml culture were collected at mid-log phase by centrifugation, washed once with 0.1 M potassium phosphate buffer (pH 7.5) containing 1 mM DTT, and resuspended to a final volume of 4 ml in the same buffer. The entire suspension was added to 3 g of glass beads (0.50-mm diameter; B. Braun Biotech International) and vortexed for 2 min at room temperature. The resulting extract was transferred to a clean tube, placed on ice, and cleared of particulate matter by centrifugation at 14,000 × g for 20 min at 4°C. The volume of the final cell lysate, which was stored at −80°C in 30% glycerol for later use, was approximately 2 ml. Protein concentrations within the lysates were determined by the method described by Bradford (4) with bovine serum albumin (BSA) as the standard. The conversion of OMP to UMP was measured spectrophotometrically at 23°C by observing the decrease in absorbance of the OMP substrate at 285 nm for a period of 25 min. The assay mixture in a total volume of 0.5 ml consisted of the cell lysate (150 μl), 0.1 M potassium phosphate buffer (pH 6.0), 1.0 μmol of β-mercaptoethanol, and 0.15 μmol of the OMP substrate. The concentration of OMP in the assay mixture was calculated by Beer’s law with a molar extinction coefficient of 1.65 × 103 cm−1 M−1 (22). A unit of enzyme activity was defined as the quantity of enzyme which catalyzed the conversion of 1 μmol of OMP to UMP per min. Specific activity was defined as units of enzyme activity per milligram of protein. Each assay with a particular cell extract was repeated at least three times, with specific activities reported as means ± standard deviations of the means.
Growth rate determination.
The doubling times of C. albicans strains were calculated in 2× YPD medium and SD-ura medium at 37°C. An overnight culture grown at 37°C was diluted 1 to 500 into 50 ml of either fresh 2× YPD or SD-ura medium. The culture was incubated with shaking at 37°C. The optical density at 600 nm (OD600) of each culture was determined every hour. The doubling times shown in Tables 2 and 3 are the averages of three independent experiments.
TABLE 2.
OMP decarboxylase activities of various C. albicans strains
| Straina | Phenotypeb | Sp actc | No. of copies of URA3 per genome | Doubling time 2× YPD (hr)d | Doubling time SD-ura (hr)d |
|---|---|---|---|---|---|
| SC5314 | Ura+ | 16.00 ± 0.47 | 2 | 1.06 ± 0.11 | |
| 17.23 ± 0.31 | |||||
| 17.53 ± 0.73 | |||||
| SC5314 (SD-ura medium) | Ura+ | 15.09 ± 0.42 | 2 | 1.38 ± 0.08 | |
| CAI4 | Ura− | 0.60 ± 0.39 | 0 | 0.89 ± 0.08 | |
| 0.40 ± 0.32 | |||||
| 0.71 ± 0.14 | |||||
| CAF2-1 | Ura+ | 19.61 ± 1.26 | 1 | 0.85 ± 0.09 | 1.41 ± 0.04 |
| CAI4-2312L | Ura+ | 17.61 ± 0.35 | 1 | 0.96 ± 0.12 |
a All strains were grown in YPD medium unless noted.
b For genotype, see Table 1.
c Reported as micromoles of OMP converted to UMP per minute per milligram of protein. Values are means ± standard deviations.
d Reported as the averages of three independent cultures ± standard deviations.
TABLE 3.
C. albicans strains constructed with the ura-blaster
| Straina | Sp actb | Fold reduction in enzyme activity compared to wild typec | Gene copy no.
|
Doubling time in 2× YPD medium (h)d | Doubling time in SD-ura medium (h)d | |
|---|---|---|---|---|---|---|
| URA3 | Target gene | |||||
| RSM3 | 12.87 ± 0.87 | 1.3 | 2 | 1 | 1.41 ± 0.10 | 1.46 ± 0.01 |
| RSM4 | 8.98 ± 0.44 | 1.9 | 1 | 1 | 1.39 ± 0.04 | |
| RSM11 (SD-ura) | 8.75 ± 1.96 | 1.9 | 1 | 0 | ||
| CAI4-01 | 8.28 ± 1.33 | 2.1 | 1 | 1 | 1.27 ± 0.05 | |
| RSM11 | 8.11 ± 0.37 | 2.1 | 1 | 0 | 1.38 ± 0.08 | |
| CWJ430 | 7.85 ± 0.19 | 2.2 | 1 | 1 | 1.23 ± 0.11 | |
| JKC19 | 7.53 ± 0.94 | 2.3 | 1 | 0 | 0.90 ± 0.06 | |
| CAI4-10 | 7.49 ± 0.98 | 2.3 | 1 | 1 | 1.34 ± 0.14 | |
| CACB8B-6 | 7.49 ± 0.73 | 2.3 | 2 | 1 | 1.35 ± 0.15 | 1.42 ± 0.05 |
| CACB8B-5 | 7.19 ± 0.13 | 2.4 | 2 | 1 | 1.22 ± 0.05 | |
| CACB10B-8 | 6.46 ± 0.96 | 2.6 | 1 | 1 | 1.25 ± 0.03 | |
| CAI4-116 | 6.22 ± 0.37 | 2.7 | 1 | 0 | 1.33 ± 0.11 | |
| CJC01 | 6.05 ± 0.97 | 2.8 | 2 | 1 | 1.21 ± 0.10 | |
| RSM7 | 5.53 ± 1.01 | 3.1 | 1 | 0 | 1.36 ± 0.11 | 1.70 ± 0.06 |
| CAG1 | 4.55 ± 0.41 | 3.7 | 1 | 1 | 1.23 ± 0.13 | |
| CJC024 | 3.75 ± 0.20 | 4.5 | 1 | 0 | 1.24 ± 0.08 | |
| CAG5 | 3.31 ± 0.38 | 5.1 | 1 | 1 | 1.24 ± 0.10 | |
| CAI4-5 | 3.24 ± 0.60 | 5.2 | 1 | 0 | 1.39 ± 0.07 | 2.11 ± 0.03 |
| CACB10B-10 | 3.16 ± 0.41 | 5.4 | 1 | 0 | 1.30 ± 0.12 | |
| CAG3 | 2.13 ± 0.10 | 8.0 | 1 | 0 | 1.30 ± 0.08 | |
| CWJ429 | 1.62 ± 0.76 | 10 | 1 | 1 | 1.15 ± 0.09 | |
| RSM17 | 1.23 ± 1.06 | 18 | 1 | 0 | 1.40 ± 0.06 | 1.87 ± 0.05 |
| CAG4 | 0.56 ± 0.11 | 30 | 0 | 0 | 1.27 ± 0.05 | |
a For genotype, see Table 1. Strains were grown in YPD medium unless otherwise noted.
b Reported as micromoles of OMP converted to UMP per minute per milligram of protein. Values are means ± standard deviations.
c For purposes of comparison, wild-type specific enzyme activity was 17.
d Reported as the averages of three independent cultures ± standard deviations.
RESULTS
OMP decarboxylase activities of C. albicans strains.
OMP decarboxylase enzyme activity was first assayed for control strains SC5314 (Ura+) and CAI4 (Ura−). SC5314, a clinical isolate, is the parent strain of CAF2-1 and the grandparent strain of CAI4. SC5314 was used as the relevant wild-type control in this work, since CAI4 is the beginning strain used in the construction of the disrupted strains assayed in this study. OMP decarboxylase assays of SC5314 and CAI4 were performed with three different cell extracts, each yielding results highly comparable to each other (Table 2). The enzyme activity of the C. albicans wild-type strain SC5314 was not significantly affected by the growth of the culture in SD-ura minimal medium. The specific activity of OMP decarboxylase was significantly decreased in the CAI4 strain which contained deletions of both genomic URA3 copies. There was no detectable conversion of OMP to UMP when the substrate was incubated in the presence of buffer alone or with boiled cell extract. The small amount of conversion of OMP to UMP observed in the CAI4 assays was probably due to enzymes of nucleotide anabolism other than OMP decarboxylase.
Also assayed were C. albicans CAF2-1 and CAI4-2312L (Table 2). CAF2-1, a strain heterozygous for the URA3 gene, and CAI4-2312L, a Δura3/Δura3 strain with one copy of URA3 reintegrated into the genome, both showed specific activities comparable to the SC5314-positive control strain.
Assay results of C. albicans strains constructed with ura-blaster-mediated genetic disruptions.
Twenty-three constructed C. albicans strains containing one or more genetic disruptions via the ura-blaster construct were assayed for OMP decarboxylase enzyme activity (Table 3). Compared to the wild-type SC5314 strain, the test strains assayed showed 2- to 18-fold less OMP decarboxylase enzyme activity. Repeated assays of various test strains with cell extracts prepared from several independent cultures yielded highly comparable values for specific enzyme activity (data not shown). To examine the possible effect of growth medium on these strains, the enzyme activity of RSM11 grown in SD-ura medium was also determined. YPD or SD-ura growth medium had no apparent effect on the specific activity obtained for test strain RSM11 (Table 3).
Growth rate data and germ tube formation of C. albicans strains.
In order to determine whether growth rate or germ tube formation was affected by the decrease in OMP decarboxylase enzyme activity within the C. albicans strains constructed with the ura-blaster, the doubling times in 2× YPD medium and the ability of each strain to form germ tubes in serum were examined. The doubling times of the control strains SC5314 (Ura+) and CAI4 (Ura−) did not vary significantly in 2× YPD medium at the P > 0.10 level (Table 2). In addition, with the exception of strain JKC19, the doubling times of the constructed strains were not significantly different from one another (P > 0.03) in 2× YPD and appeared to be somewhat lower in their growth rates than that of the CAI4 strain from which they were derived. The doubling times of several strains in SD-ura medium were also determined (Tables 2 and 3). The doubling times of strains RSM3 and CACB8B-6 were not statistically different from those of the Ura+ control strains SC5314 and CAF2-1 at the P > 0.10 level. However, the doubling times of strains RSM7, RSM17, and CAI4-5, which all display low levels of OMP decarboxylase enzyme activity, were significantly different from that of the wild-type SC5314 strain.
By a method described previously (12), the strains in the study were examined for their ability to form germ tubes. The control strains SC5314 (Ura+) and CAI4 (Ura−) formed germ tubes at rates indistinguishable from one another, indicating that the level of URA3 gene activity does not directly affect the ability of cells to form germ tubes (data not shown). With the exception of strains CAG1, CAG3, CAG4, CAG5, and CAI4-5, the strains constructed with the ura-blaster were able to form germ tubes at wild-type levels (data not shown). Strains CAG1, CAG3, CAG4, CAG5, and CAI4-5 contain disruptions in one or both copies of the INT1 or RSR1 gene, both of which are involved in germ tube formation (9, 29).
Relationship between OMP decarboxylase activity and virulence of C. albicans strains.
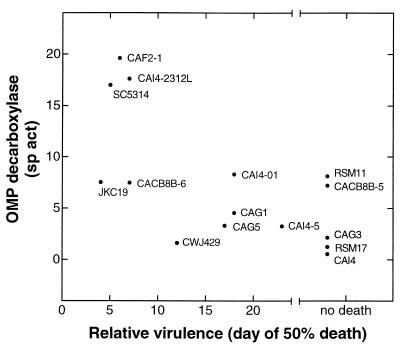
Virulence data for 14 of the strains assayed in this study are available (3, 5, 9, 18, 29). To compare results from these previous studies, the day of death of 50% of the mice within a group of immunocompetent mice injected with 106 C. albicans cells via the lateral vein was related to the OMP decarboxylase enzyme activity within the strain (Fig. 1).
FIG. 1.
Relationship between OMP decarboxylase specific activities in C. albicans strains and the known experimental virulence of the strains previously reported. Relative virulence is represented as the day of death for 50% of a group of immunocompetent mice injected with 106 C. albicans cells via the lateral tail vein. Mice injected with avirulent strains remain healthy and do not achieve 50% death after 24 to 30 days of observation. OMP decarboxylase specific activities are reported as units of enzyme activity per milligram of protein within the assayed cell lysate of each strain.
Strains SC5314, CAF2-1, and CAI4-2312L, which contain at least one functional copy of the URA3 gene in its native locus, are fully virulent, whereas CAI4, a strain lacking URA3, is avirulent and has only trace OMP decarboxylase activity. However, an examination of the C. albicans strains with ura-blaster-mediated genetic disruptions does not reveal a definitive relationship between enzyme activity and virulence. Most notably, two avirulent strains (CAG3 and RSM17) have markedly decreased OMP decarboxylase enzyme activities. Other avirulent strains (RSM11 and CACB8B-5), as well as two fully virulent strains (JKC19 and CACB8B-6), have intermediate levels of enzyme activity. Strains with intermediate virulence (CAI4-01, CAG1, CAG5, and CWJ429) vary from intermediate to low levels of enzyme activity.
Relationship between URA3 gene copy number and OMP decarboxylase specific activity.
Four C. albicans strains assayed in this study—RSM3, CACB8B-6, CACB8B-5, and CJC01—contain an alternative ura-blaster-mediated disruption in which one allele contains an additional copy of hisG and URA3 (see Table 1 for exact genotypes). The existence of tandem duplications has been noted previously in the process of disrupting Candida genes via the ura-blaster cassette (5, 10), possibly due to the formation of a circularized intermediate form of the linear ura-blaster cassette during transformation.
OMP decarboxylase specific activities and the numbers of URA3 copies were compared to determine whether these extra copies of URA3 within the disruption cassette at the targeted gene in strains RSM3, CACB8B-6, CACB8B-5, and CJC01 affected enzyme activity (Table 3). There is no definitive correlation between OMP decarboxylase activity and URA3 gene copy number within the genome of each strain. Strains CAF2-1 and CAI4-2312L show that wild-type enzyme activity is achieved with one functional copy of URA3 at its native locus. C. albicans strains containing one copy of URA3 in the targeted gene locus within the hisG-URA3-hisG disruption cassette have OMP decarboxylase activities ranging from 1.23 to 8.98. Compared to disrupted strains harboring one copy of the URA3 gene, an extra copy of URA3 present within the disrupted alleles of RSM3, CACB8B-6, CACB8B-5, and CJC01 did not notably increase OMP decarboxylase activity in all of the strains. RSM3, a Δcdr1/CDR1 strain with a hisG-URA3-hisG-URA3-hisG sequence present within the disrupted gene, did show the highest enzyme activity of any of the disrupted strains assayed. However, since the three other strains containing the hisG-URA3-hisG-URA3-hisG sequence did not have high activities as well, the observed OMP decarboxylase activity of RSM3 probably cannot be attributed to an extra copy of URA3 within the disrupted allele.
Relationship between OMP decarboxylase activity and total number of genetic disruptions.
C. albicans strains with one, two, or four ura-blaster-mediated genetic disruptions were assayed in this study (see Table 1 for exact genotypes). The OMP decarboxylase specific activities of the strains were compared to the numbers of genes disrupted with the ura-blaster cassette in order to determine if enzyme activity was affected by the number of times the ura-blaster cassette was used for disruption. No positive correlation between OMP decarboxylase specific activities and the numbers of genes disrupted was observed. Homozygous knockout strains do not, in general, show less activity than their heterozygous counterparts. For example, CACB8B-5, a Δchs3/Δchs3 strain, has an activity similar to that of CACB8B-6, a Δchs3/CHS3 strain. RSM17, a C. albicans strain in which the ura-blaster cassette has been used to disrupt two copies of the CDR1 gene and two copies of the CaMDR1 gene, has a very low OMP decarboxylase specific activity. However, the low enzyme activity of RSM17 cannot definitely be attributed to the fact that it contains four ura-blaster-mediated genetic disruptions.
DISCUSSION
We have shown that the OMP decarboxylase enzyme activities of C. albicans strains assayed in this study are significantly reduced and quite variable compared to the wild-type enzyme activity observed in Ura+ control strains. In each of these strains, the ura-blaster cassette, which contains either one or two tandem copies of the C. albicans URA3 gene, disrupts the targeted gene of interest in a Δura3/Δura3 background. It is of critical importance that the OMP decarboxylase enzyme, as encoded by the URA3 gene within the ura-blaster cassette, retains sufficient activity, since both uracil auxotrophy and mutations in the pyrimidine biosynthetic pathway have previously been shown to diminish virulence in C. albicans (15).
The ura-blaster disruption technique uses URA3, a gene known to affect the virulence of C. albicans, within a hisG-URA3-hisG disruption cassette, in order to identify other genes involved in pathogenesis. Our results indicate that it is necessary to closely examine the OMP decarboxylase activity of a ura-blaster-constructed strain before determining its virulence in order to determine if the gene of interest is actually related to C. albicans pathogenesis. Our results also indicate that the level of URA3 gene activity does not by itself influence the process of germ tube formation. However, reduced enzyme activity contributed to the lower growth rate of some of the tested strains in SD-ura medium. This lower rate of growth in some of the strains due to inadequate OMP decarboxylase activity may cause a decrease in virulence in vivo because a more slowly growing strain can be eliminated by the host’s defenses more effectively than a strain which is rapidly reproducing. However, when considering the growth rate data, it is important to note that two strains examined, RSM3 and CACB8B-6, have reduced levels of OMP decarboxylase enzyme activity but are able to grow in SD-ura medium at rates similar to those of the wild-type strain. These results suggest that a wild-type level of enzyme activity is not necessary to maintain a wild-type rate of growth in SD-ura medium.
One strain in this study, JKC19 (ΔcphI/ΔcphI), showed decreased OMP decarboxylase activity but retained wild-type virulence. Since the engineered disruption with the ura-blaster within this strain does not affect its virulence, it can be concluded that the targeted gene within JKC19, CPHI, is not a virulence factor. The reduced OMP decarboxylase activity of JKC19 does not affect its virulence, implying that C. albicans strains may not require wild-type enzyme activity to maintain a wild-type level of virulence. Therefore, some strains with intermediate levels of enzyme activity are able to maintain wild-type virulence (JKC19) or wild-type rates of growth in minimal medium (RSM3 and CACB8B-6). The OMP decarboxylase enzyme levels within these strains may represent a threshold or adequate level of enzyme activity necessary for these strains to behave like their wild-type parental strains. However, our results do not accurately define the adequate or sufficient amount of OMP decarboxylase enzyme activity required by C. albicans strains.
Strains RSM11 and CACB8B-5, which are both avirulent, have OMP decarboxylase specific activities similar to that of JKC19. Therefore, RSM11 and CACB8B-5 may have sufficient OMP decarboxylase activity, and the avirulence observed is due to the targeted genetic disruptions. RSM17 and CAG3 are avirulent strains which showed very low OMP decarboxylase activities compared to that of the wild type. In this situation, the reason for the apparent lack of virulence is difficult to assign. Avirulence could be the result of decrease in OMP decarboxylase activity, the targeted genetic disruption, or a combination of both factors. The same problem arises with several other strains in the study, such as CAI4-01 and CAG1, which display intermediate virulence in murine models and also have decreased OMP decarboxylase activities. Again, a distinction between the factors which could result in this decreased virulence cannot be determined.
The hisG-URA3-hisG disruption cassettes present within each strain are all derived from the same plasmid (8) and are presumably identical. Therefore, the reduction of OMP decarboxylase activity in the disrupted C. albicans strains is most likely due to altered URA3 expression within the ura-blaster construct. Several phenomena could result in this altered URA3 expression. After insertion into the targeted gene for disruption, the URA3 gene no longer resides at its native locus in the genome. Epigenetic gene regulation or different states of gene expression caused by differential effects of chromosome or chromatin packaging have been observed for many eukaryotes, including fungi (14, 28). An example of this effect is commonly referred to as position effect variegation (11). The S. cerevisiae URA3 gene, which shares 66.8% sequence identity with that of C. albicans (20), is known to be regulated at the level of transcription (2) and is susceptible to positional and orientation effects when manipulated (24). It is possible that the URA3 gene within the ura-blaster construct is not in close proximity to its proper regulatory elements, causing the observed decrease in OMP decarboxylase activity. Positional effects on URA3 expression and regulation may also explain the variability in enzyme activities observed within the constructed C. albicans strains.
The presence of flanking hisG direct repeats within the disruption cassette may also alter the URA3 gene in these constructed C. albicans strains. A proportion of the cells may have lost URA3 due to mitotic recombination between the hisG repeats, resulting in an overall decrease in OMP decarboxylase activity. Repeat-induced mutation (RIP) should also be noted when examining the ura-blaster construct. RIP is a phenomenon observed in some fungal species in which the presence of heterologous flanking repeated sequences increases the frequency of mutation of the gene being flanked (27). It is possible that the URA3 gene within the ura-blaster construct undergoes frequent mutation due to the presence of flanking hisG repeats from Salmonella, resulting in decreased OMP decarboxylase activity. Finally, it has not escaped our attention that the altered expression of URA3, as measured by us in C. albicans cell extracts, may not reflect expression during host infection when many environmental factors may modulate enzyme activity.
Since its application to the study of C. albicans, the ura-blaster technique for sequential gene disruption has been used extensively in searching for genes responsible for the virulence of this fungal pathogen (3, 5, 6, 13, 16, 18, 19, 21, 25, 31). The importance of C. albicans as a significant infectious microorganism in both healthy and immunocompromised individuals warrants the proper identification of genes necessary for its pathogenesis. Our results imply that the decreased in virulence of C. albicans strains constructed with the ura-blaster cassette cannot be definitively attributed, in all cases, to the targeted genetic disruption. The decrease in OMP decarboxylase activities within these strains may contribute to an attenuation in virulence. We conclude that the interpretation of virulence data obtained from this method of genetic disruption must take into account the decreased activity of the URA3 gene product.
There is currently a trend when examining C. albicans strains to reintroduce the wild-type gene back into a strain with both copies of the targeted gene disrupted. One strain employed in this study, CAG5, is representative of this technique. It contains both copies of the INT1 gene disrupted by a hisG sequence with one copy of INT1 and one copy of URA3 reintegrated into the genome. Strain CAG5 displays intermediate virulence and reduced OMP decarboxylase enzyme activity, whereas CAG3, the homozygous disruptant INT1 strain, is avirulent and has reduced enzyme activity. Restoration of one copy of the INT1 gene in CAG5 resulted in a partial recovery of virulence, indicating that INT1 could be a virulence factor. However, the problem of reduced URA3 gene activity still persists. It is possible that full, wild-type virulence was not restored in CAG5 due to inadequate OMP decarboxylase activity.
Alternative methods should most likely be employed in constructing a C. albicans strain harboring genetic disruptions to identify possible virulence factors. One revised ura-blaster disruption method would use a stable plasmid to reintroduce the URA3 gene encoding wild-type levels of OMP decarboxylase. Such a method has been reported elsewhere (17), but plasmid stability may not be optimized until an authentic C. albicans centromere is identified and incorporated into a plasmid. Another revised method would reintroduce the URA3 gene into its native locus, where it should be properly regulated and expressed. Preliminary data have shown that returning URA3 to its native location in the genome of a Ura− strain restores both wild-type OMP decarboxylase enzyme activity and wild-type virulence (data not shown). However, these additional genetic manipulations have the disadvantage of introducing mutations complicating the analysis of virulence studies. Perhaps the most desirable alternative to the construction of strains for virulence studies would be to use a selectable marker that encodes an activity not required for virulence or host survival.
ACKNOWLEDGMENTS
We thank the personnel of the Becker laboratory at the University of Tennessee for their support. We are especially grateful to Melinda Hauser for her input in numerous fruitful discussions regarding this project. We also thank Gerald Fink, William Fonzi, and Judith Berman for the strains used in this study.
REFERENCES
- 1.Alani E, Cao L, Kleckner N. A method for gene disruption that allows for repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bach M, Lacroute F, Botstein D. Evidence for transcriptional regulation of orotidine-5′-phosphate decarboxylase in yeast by hybridization of mRNA to the yeast structural gene cloned in Escherichia coli. Genetics. 1979;76:386–390. doi: 10.1073/pnas.76.1.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker J M, Henry L K, Jiang W, Koltin Y. Reduced virulence of Candida albicans mutants affected in multidrug resistance. Infect Immun. 1995;63:4515–4518. doi: 10.1128/iai.63.11.4515-4518.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford M M. A rapid and sensitive assay for the quantitation of microgram quantities of protein using the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Bulawa C E, Miller D W, Henry L K, Becker J M. Attenuated virulence of chitin-deficient mutants of C. albicans. Proc Natl Acad Sci USA. 1995;92:10570–10574. doi: 10.1073/pnas.92.23.10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diez-Orejas R, Molero G, Navarro-Garcia F, Pla J, Nombela C, Sanchez-Perez M. Reduced virulence of Candida albicans MKC1 mutants: a role for mitogen-activated protein kinase in pathogenesis. Infect Immun. 1997;65:833–837. doi: 10.1128/iai.65.2.833-837.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edwards E J J. Candida species. In: Mandell G L, Douglas R G, Bennett J E, editors. Principles and practice of infectious diseases. New York, N.Y: Churchill Livingstone; 1990. pp. 1435–1446. [Google Scholar]
- 8.Fonzi W A, Irwin M. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gale C A, Bendel C M, McClellan M, Hauser M, Becker J M, Berman J, Hostetter M K. Linkage of adhesion, filamentous growth and virulence in Candida albicans to a single gene, INT1. Science. 1998;279:1355–1358. doi: 10.1126/science.279.5355.1355. [DOI] [PubMed] [Google Scholar]
- 9a.Gillum A M, Tsay E Y, Kirsch D R. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet. 1984;198:179–182. doi: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
- 10.Gow N A R, Robbins P W, Lester J W, Brown A J P, Fonzi W A, Chapman T, Kinsman O S. A hyphal-specific chitin synthase gene (CHS2) is not essential for growth, dimorphism or virulence of Candida albicans. Proc Natl Acad Sci USA. 1994;91:6216–6220. doi: 10.1073/pnas.91.13.6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hendrich B D, Willard H F. Epigenetic regulation of gene expression: the effect of altered chromatin structure from yeast to mammals. Hum Mol Genet. 1995;4:1765–1777. doi: 10.1093/hmg/4.suppl_1.1765. [DOI] [PubMed] [Google Scholar]
- 12.Ibrahim A S, Mirbod F, Filler S G, Banno Y, Cole G T, Kitajima Y, Edwards J E, Nozawa Y, Ghannoum M A. Evidence implicating phospholipase as a virulence factor in Candida albicans. Infect Immun. 1995;63:1993–1998. doi: 10.1128/iai.63.5.1993-1998.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang W, Gerhold D, Kmiec E B, Hauser M, Becker J M, Koltin Y. The topoisomerase I gene from Candida albicans. Microbiology. 1997;143:377–386. doi: 10.1099/00221287-143-2-377. [DOI] [PubMed] [Google Scholar]
- 14.Kellum R, Schedl P. A position-effect assay for boundaries of higher order chromosomal domains. Cell. 1991;64:941–950. doi: 10.1016/0092-8674(91)90318-s. [DOI] [PubMed] [Google Scholar]
- 15.Kirsch D R, Whitney R R. Pathogenicity of Candida albicans auxotrophic mutants in experimental infections. Infect Immun. 1991;59:3297–3300. doi: 10.1128/iai.59.9.3297-3300.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leberer E, Harcus D, Broadbent I D, Clark K L, Dignard D, Ziegelbauer K, Schmidt A, Gow N A R, Brown A J P, Thomas D Y. Signal transduction through homologs of the Ste20p and Ste7p protein kinases can trigger hyphal formation in the pathogenic fungus Candida albicans. Proc Natl Acad Sci USA. 1996;93:13217–13222. doi: 10.1073/pnas.93.23.13217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leberer E, Ziegelbauer K, Schmidt A, Harcus D, Dignard D, Ash J, Johnson L, Thomas D Y. Virulence and hyphal formation of Candida albicans require the Ste20p-like protein kinase CaCla4p. Curr Biol. 1997;7:539–546. doi: 10.1016/s0960-9822(06)00252-1. [DOI] [PubMed] [Google Scholar]
- 18.Liu H, Köhler J, Fink G R. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science. 1994;266:1723–1726. doi: 10.1126/science.7992058. . (Erratum, 267:17, 1995.) [DOI] [PubMed] [Google Scholar]
- 19.Lo H, Köhler J, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink G R. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- 20.Losberger C, Ernst J F. Sequence and transcript analysis of the C. albicans URA3 gene encoding orotidine-5′-phosphate decarboxylase. Curr Genet. 1989;16:153–157. doi: 10.1007/BF00391471. [DOI] [PubMed] [Google Scholar]
- 21.Mio T, Yabe T, Sudoh M, Satoh Y, Nakajima T, Arisawa M, Yamada-Okabe H. Role of three chitin synthase genes in the growth of Candida albicans. J Bacteriol. 1996;178:2416–2419. doi: 10.1128/jb.178.8.2416-2419.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Myers K M, Sypherd P S, Fonzi W A. Use of URA3 as a reporter of gene expression in C. albicans. Curr Genet. 1995;27:243–248. doi: 10.1007/BF00326156. [DOI] [PubMed] [Google Scholar]
- 23.Perfect J R. Fungal virulence genes as targets for antifungal chemotherapy. Antimicrob Agent Chemother. 1996;40:1577–1583. doi: 10.1128/aac.40.7.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rose M, Grisafi P, Botstein D. Structure and function of the yeast URA3 gene: expression in Escherichia coli. Gene. 1984;29:113–124. doi: 10.1016/0378-1119(84)90172-0. [DOI] [PubMed] [Google Scholar]
- 25.Sarthy A V, McGonigal T, Coen M, Frost D J, Meulbroek J A, Goldman R C. Phenotype in Candida albicans of a disruption of the BGL2 gene encoding a 1,3-β-glucosyltransferase. Microbiology. 1997;143:367–376. doi: 10.1099/00221287-143-2-367. [DOI] [PubMed] [Google Scholar]
- 26.Scherer S, Magee P T. Genetics of Candida albicans. Microbiol Rev. 1991;54:226–241. doi: 10.1128/mr.54.3.226-241.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Selker E. Premeiotic instability of repeated sequences in Neurospora crassa. Annu Rev Genet. 1990;24:579–613. doi: 10.1146/annurev.ge.24.120190.003051. [DOI] [PubMed] [Google Scholar]
- 28.Tartof K D. Position effect variegation in yeast. Bioessays. 1994;16:713–714. doi: 10.1002/bies.950161004. [DOI] [PubMed] [Google Scholar]
- 29.Yaar L, Mevarech M, Koltin Y. A Candida albicans RAS-related gene (CaRSR1) is involved in budding, cell morphogenesis and hypha development. Microbiology. 1997;143:3033–3044. doi: 10.1099/00221287-143-9-3033. [DOI] [PubMed] [Google Scholar]
- 30.Yoshimoto A, Umezu K, Kobayashi K, Tomita K. Orotidylate decarboxylase (yeast) Methods Enzymol. 1978;54:74–79. doi: 10.1016/s0076-6879(78)51013-6. [DOI] [PubMed] [Google Scholar]
- 31.Zhao X, McElhaney-Feser G E, Sheridan M J, Broedel S E, Cihlar R L. Avirulence of Candida albicans FAS2 mutants in a mouse model of systemic candidiasis. Infect Immun. 1997;65:829–832. doi: 10.1128/iai.65.2.829-832.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]