Abstract
The success of colloidal semiconductor nanocrystals (NCs) in science and optoelectronics is inextricable from their surfaces. The functionalization of lead halide perovskite NCs1–5 poses a formidable challenge because of their structural lability, unlike the well-established covalent ligand capping of conventional semiconductor NCs6,7. We posited that the vast and facile molecular engineering of phospholipids as zwitterionic surfactants can deliver highly customized surface chemistries for metal halide NCs. Molecular dynamics simulations implied that ligand–NC surface affinity is primarily governed by the structure of the zwitterionic head group, particularly by the geometric fitness of the anionic and cationic moieties into the surface lattice sites, as corroborated by the nuclear magnetic resonance and Fourier-transform infrared spectroscopy data. Lattice-matched primary-ammonium phospholipids enhance the structural and colloidal integrity of hybrid organic–inorganic lead halide perovskites (FAPbBr3 and MAPbBr3 (FA, formamidinium; MA, methylammonium)) and lead-free metal halide NCs. The molecular structure of the organic ligand tail governs the long-term colloidal stability and compatibility with solvents of diverse polarity, from hydrocarbons to acetone and alcohols. These NCs exhibit photoluminescence quantum yield of more than 96% in solution and solids and minimal photoluminescence intermittency at the single particle level with an average ON fraction as high as 94%, as well as bright and high-purity (about 95%) single-photon emission.
Subject terms: Materials chemistry, Surface chemistry, Inorganic chemistry, Nanoscale materials
Phospholipids enhance the structural and colloidal integrity of hybrid organic–inorganic lead halide perovskites and lead-free metal halide nanocrystals, which then exhibit enhanced robustness and optical properties.
Main
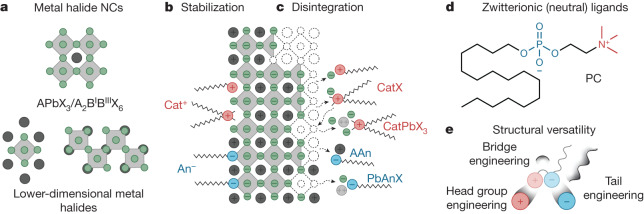
Lead halide perovskite (LHP) nanocrystals (NCs), of the general formula APbX3 (A = Cs, MA, FA (FA, formamidinium; MA, methylammonium); X = Cl, Br, I), receive immense scientific and practical interest as narrow-band emitters for displays or as quantum light sources8–13, whereas other metal halides (Fig. 1a) are pursued as broadband emitters for solid-state lighting, scintillation and thermometry14–16. An imminent challenge impeding the progress in the chemistry and applications of many metal chloride, bromide and iodide NCs is that strongly binding ligands readily outcompete the relatively low internal lattice energy17. For instance, typical anionic and cationic surfactants that attach to halide NC surfaces in an ionic manner, displacing surface ions with the ligand head groups (Fig. 1b)4,18–20, also readily engage in adverse solubilization equilibria with the ions constituting the inorganic NC cores (Fig. 1c). This problem culminates with hybrid organic–inorganic perovskite compositions—MAPbX3 and FAPbX3 NCs. We proposed that zwitterionic, hence charge-neutral, capping molecules offer a general mitigation strategy for avoiding these adverse ionic metathesis reactions with NC cores, as motivated by our recent work on CsPbBr3 NCs with several commercial long-chain zwitterions (phosphocholines (PCs) (Fig. 1d), γ-amino acids and sulfobetaines)21–23. Herein, we sought to design and implement a library of phospholipid capping ligands (Fig. 1e) to broaden the compositional scope of metal halides available in the form of finely engineered NCs. Notably, only through this approach were we able to produce surface-robust, highly emissive MAPbX3 and FAPbX3 NCs. The zwitterionic group engineering was computationally guided by assessing ligand–surface binding with molecular dynamics (MD) simulations. Solid-state nuclear magnetic resonance (NMR) and Fourier-transform infrared (FTIR) spectroscopy then corroborated the atomistic models. Our design of chemically pure phospholipids with various head, bridge and tail groups leverages facile synthesis methods developed over the past few decades24–30. The utility of the entire library of ligands was assayed through post-synthetic anchoring to the NC surfaces. The vast molecular engineering of phospholipids endowed the compatibility of ligand-capped NCs with solvents of diverse polarity, ranging from hydrocarbons to alcohols, and enhanced luminescent properties in thin films and at the single particle level.
Fig. 1. Surface chemistry of soft metal halide NCs.
a, Examples of ionic metal halides. b,c, Atomistic depiction of surface stabilization (b) and disintegration (c) of APbX3 perovskite by common long-chain cationic (Cat+) and anionic (An−) ligands due to excess ligand quantity and low internal crystal energy. d, Zwitterionic molecules offer stronger (multidentate) binding and reduced reactivity owing to their neutral charge. e, Structural engineering of the head, bridge and tail groups unlocks their broad utility for stabilizing diverse metal halide NCs and dispersing them in various media.
Determining the binding mode of zwitterionic ligands
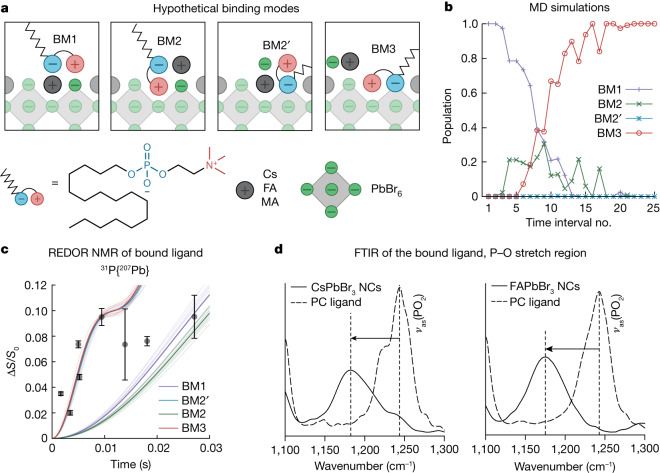
Empirically, we have seen the efficacy of commercial long-chain zwitterions as ligands for CsPbX3 NCs21–23,31, mainly CsPbBr3. A champion ligand was natural lecithin (a physiological mixture of glycerophospholipids with quaternary and primary ammonium moieties), affording robust CsPbBr3 colloids down to small NC sizes of 3 nm. Further progress towards customizable phospholipid ligands for the entire family of LHP NCs and beyond requires a concerted computational and experimental effort to rationalize the binding of phospholipids at the molecular level. We first set out to determine whether both charged groups of the ligand participate in anchoring to the NC surface. We used a combination of classical MD simulations, FTIR spectroscopy and rotational-echo double-resonance (REDOR) NMR spectroscopy. The surface of the NCs was modelled with a slab of bulk cubic FAPbBr3, terminated with FABr-rich (100) crystallographic planes (Supplementary Note 1). One alkyl PC ligand was placed at 0.5 nm above the surface, and the system was solvated with toluene. During equilibration, the ligand quickly adsorbs onto the surface, assuming a conformation that is referred to as binding mode (BM) 1 (Fig. 2a). We anticipated that the ligand might also displace some of the native ions from the surface, similar to oleylammonium (OAm) binding to the surface of CsPbBr3 NCs20. Thus, three possibilities—displacement of FA (BM2), bromide (BM2′) or both FA and bromide (BM3)—are depicted in Fig. 2a. A replica-exchange32,33 MD simulation is then used to explore the basic BMs and to determine which of them has the lowest free energy at room temperature (Methods, Supplementary Notes 1 and 2 and Extended Data Fig. 1). The population of the starting BM1 diminishes as the simulation progresses, whereas BM3 dominates the ensemble in the late stages of the simulation (Fig. 2b). Similar results for CsPbBr3 surfaces indicate that A cation and the slight difference in the crystal structure do not play a substantial role (Extended Data Fig. 2). Furthermore, BM3 also prevails at other surface stoichiometries, with FABr and ligand concentrations of between 0 and 1 per binding site (Extended Data Fig. 3). Attachment of the PC ligand to FAPbBr3 and CsPbBr3 NCs through the phosphate group binding to the surface Pb atoms is attested with 31P–207Pb REDOR NMR (Fig. 2c; see Methods and Supplementary Figs. 7 and 8 for details) and by the FTIR spectroscopy (Fig. 2d, Supplementary Note 3 and Supplementary Figs. 9–12). Notably, MD simulations suggest that the relatively bulky trimethylammonium head group of the bound ligand is elevated by roughly 0.15 nm, compared with the surface FA cations (Fig. 3a and Supplementary Table 1). Therefore, we have investigated whether the surface can sustain an ever-increasing degree of FABr substitution for the PC ligand. The complete surface stability map as a function of ligand and FABr concentrations relative to the maximum possible surface coverage is presented in Extended Data Fig. 4. Although a stable surface is still observed at 50% FABr substitution, a small fraction of PC ligands, as well as FA and Br ions, do not participate in surface passivation (Fig. 3c and Extended Data Fig. 3c). Further increase of the [ligand]/[FABr] ratio leads to a complete rupture of the PbBr underlayer (Fig. 3e), suggesting that achieving greater than 50% surface coverage with the PC ligand is unlikely.
Fig. 2. Binding of zwitterionic ligands to the FAPbBr3 NC surface.
a, Schematics of different modes for binding of zwitterionic ligands, whose plausibility was assessed with replica-exchange MD simulations. b, Evolution of the BM populations, computed in a 50-ns-long replica-exchange MD simulation of a single PC ligand that was initially placed on the pristine FABr-rich (100) surface of FAPbBr3. BM3 prevails in the ensemble in the late stages of the simulation. c, Results of a REDOR NMR experiment for 31P–207Pb coupling supports the theoretical prediction of surface anchoring with the phosphate group. Theoretical REDOR curves were calculated using conformations obtained from the MD simulations. d, FTIR spectroscopy analysis of the (P–O) stretching region, wherein the P–O bond weakening on ligand binding (that is, P–O–Pb bridge formation) shifts the signal to lower frequencies. νas, asymmetric stretching vibration.
Extended Data Fig. 1. Classification of ligand binding modes.
a, A typical map of configurations observed in replica-exchange MD simulations of the PC ligand on the (100) FAPbBr3 surface. dN-slab and dP-slab define distances from the ligand head groups (nitrogen and phosphorus atoms, correspondingly) to the middle atomic plane of the slab. Four well-defined clusters of configurations correspond to the different binding modes of the ligand (Fig. 2). The diffuse region at large ligand-slab separations corresponds to free ligands. b–e, Snapshots of the observed binding modes - BM2 (b), BM3 (c), BM1 (d) and BM2′ (e).
Extended Data Fig. 2. Generality of the prediction that zwitterionic ligands tend to displace both A and X ions from the perovskite surfaces.
a–f, Evolution of binding mode populations in replica-exchange MD simulations of single PC (a,b,c) and PEA (d,e,f) ligand molecules on the FAPbBr3 surface (a,d), (001) CsPbBr3 surface (b,e), and (010) CsPbBr3 surface (c,f). Binding with a displacement of both A and X ions (BM3) is thermodynamically preferred in all studied systems. *Crystallographic orientations refer to the primitive unit cell of CsPbBr3 (see Supplementary Note 1 for more details).
Extended Data Fig. 3. FAPbBr3 surfaces at various [FABr] and [Lig] concentrations.
a–f, Evolution of binding mode populations in systems with [Lig] = 50% and with a varying amount of surface FABr – 100% (a,b), 50% (c,d), and 0% (e,f). g,h, Evolution of binding mode populations in systems with [Lig] = 100% and [FABr] = 0%. In all scenarios, BM3 was identified as a dominant binding mode. The population of BM3 is also systematically higher for PEA ligand compared to PC, indicating a better fit of the former to the FAPbBr3 surface. In addition, some new binding modes were discovered in systems where the rupture of the PbBr underlayer is observed — BM-B (ammonium in the PbBr layer), BM-B’ (phosphate in the PbBr layer), and BM-C (both ammonium and phosphate in the PbBr layer). However, these are marginal and are encountered only along the phase boundaries between the newly exposed FABr- and the original PbBr-terminated surfaces. i–p, Corresponding MD snapshots with ligands being color-coded according to their binding mode — BM1 (blue), BM2 (green), BM2’ (orange), BM3 (red), unbound (black), and other (gray).
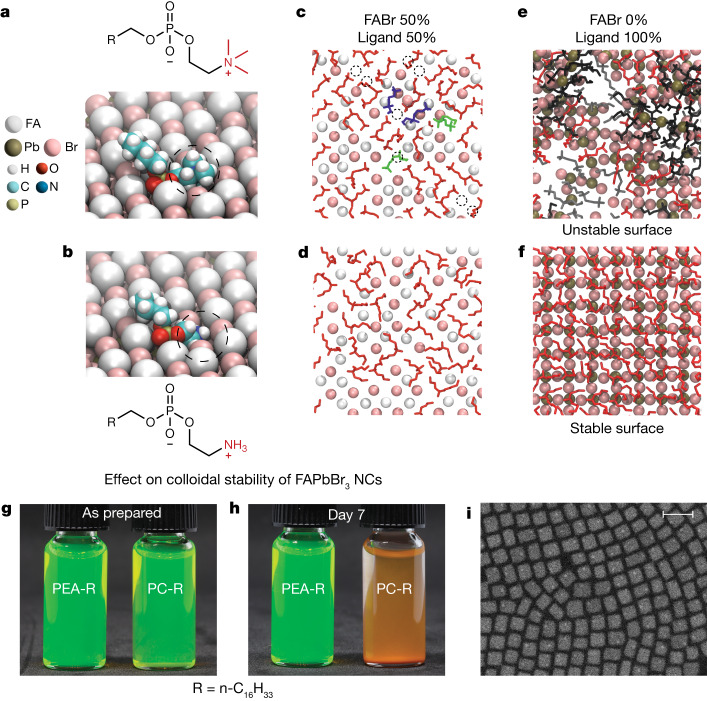
Fig. 3. Ligand head-group engineering.
a,b, Geometries of the PC (a) and PEA (b) head groups on the (100) surface of FAPbBr3, emphasizing the improved PEA fitness for the surface A-site. c–f, Snapshots from replica-exchange MD simulations of the FAPbBr3 surfaces in which 50% (c,d) or 100% (e,f) of FABr pairs were substituted with the ligands. Ligand molecules are colour-coded according to their BM—BM1 (blue), BM2 (green), BM2' (orange) and BM3 (red). Although stable surfaces are observed at 50% substitution for both ligands, PC (c) and PEA (d), a noticeable number of PC ligands and FA and Br ions desorb from a PC-capped surface, leaving behind vacancies in the top-most surface layer (black dashed circles in c). At 100% substitution, the PC-capped surface starts to rupture (e), whereas it remains stable in the case of the PEA ligand (f). g,h, Colloids of purified hexadecyl-PEA-capped and hexadecyl-PC-capped purified FAPbBr3 NCs (8.5 nm), as prepared (g) and after 7 days (h). i, Typical HAADF-STEM image of FAPbBr3 NCs capped with the alkyl-PEA ligand. Scale bar, 20 nm.
Extended Data Fig. 4. Stoichiometry-dependent stability of (100) FAPbBr3 surfaces.
a,b, Surface stability maps as a function of ligand and FABr concentrations computed for PC (a), and PEA (b) ligands. c–f, MD snapshots that illustrate three different regions observed on the stability maps. At high ligand+FABr concentrations, excess ions and ligands separate from the surface (blue area on the map) (c). As a result, the system acquires equilibrium stoichiometry corresponding to the green region on the map (d). At low ligand and/or FABr concentrations the surface becomes unstable again (a red region on the map) — segregation into two surfaces is observed for incomplete FABr passivation (f), whereas partial coverage solely with the ligand causes rupture of the PbBr underlayer (e). Ligand molecules in the snapshots are color-coded according to their binding mode — BM1 (blue), BM2 (green), BM2′ (orange), BM3 (red), unbound (black), and other (gray).
Tailoring the head-group affinity
Rethinking the cationic moiety was a major leap in the project. Compared with the PC, an analogous zwitterionic head group with primary ammonium moiety (instead of fully methylated), namely phosphoethanolamine (PEA), allows for an excellent geometric fit on A-site (Fig. 3a,b) and theoretically allows up to 100% surface coverage (Fig. 3d,f). The absence of structural degradation or ligand desorption in simulations (Fig. 3d,f and Extended Data Figs. 3 and 4) hints that PEA is better suited for passivating FAPbBr3 NC under otherwise identical conditions. We also note that primary ammonium ligands, such as OAm, were the first kind of ligand used for producing monodisperse CsPbBr3 and FAPbBr3 LHP NCs1,2 and were shown to strongly bind to the surface A-site pockets (Fig. 1b)20,34,35. At the same time, such cationic–ligand binding proved highly labile owing to the acid–base equilibrium with the neutral primary amine, seen as a rapid loss of ligands and, consequently, a loss of the NC integrity on repetitive purification of NCs34.
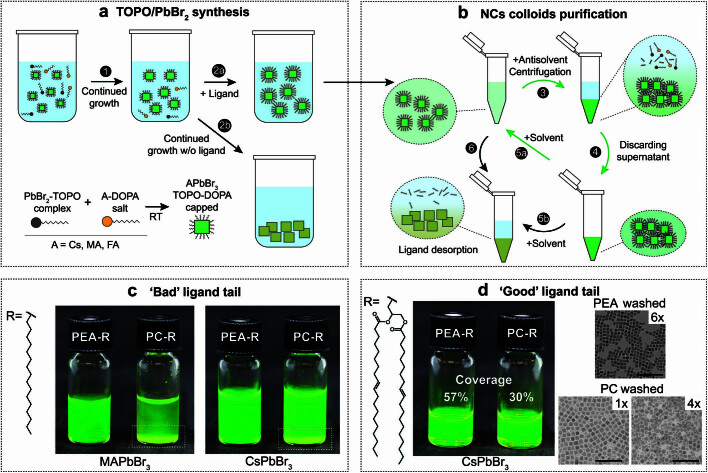
The computational design was put to test by synthesizing FAPbBr3, as well as CsPbBr3 and MAPbBr3, NCs, capped with both PC and PEA ligands. We used the trioctylphosphine oxide (TOPO)/PbBr2 room-temperature synthesis method to form these NCs31, followed by the post-synthetic displacement of weakly bound trialkylphosphine oxide and alkylphosphinic acid ligands with the ligand of choice (phospholipids), and subsequent isolation and purification of NCs (Methods and Extended Data Fig. 5a,b). PEA-ligand binding through the phosphate-group coordination to lead atoms along with the ammonium-group insertion on the surface A-site (BM3 in Fig. 2a) is confirmed by the FTIR and 31P–207Pb REDOR NMR spectroscopies (Supplementary Figs. 8–12). When comparing NCs capped with these ligands, initially stable hexadecyl-PC-capped FAPbBr3 NCs lose colloidal stability after several days, whereas monodisperse hexadecyl-PEA-capped NCs of the same size remain stable in colloids for months (Fig. 3g–i and Supplementary Table 13). Furthermore, MAPbBr3 and CsPbBr3 NCs exhibited an even stronger drop in colloidal stability with hexadecyl-PC ligand (compared with PEA-analogue; Extended Data Fig. 5c).
Extended Data Fig. 5. Synthesis of APbBr3 NCs and testing of ligands.
a, In TOPO-DOPA synthesis31, PbBr2-trioctylphosphine (TOPO) complex reacts with diisooctylphosphinic acid (DOPA) salt of the corresponding A cation at room temperature, yielding monodisperse NCs (1). TOPO and DOPA are known as “bad ligands” and can be readily exchanged for zwitterion ligands (2a). If no capping ligands are added promptly after NCs are formed, the NCs rapidly lose their colloidal and structural integrity (2b). b, To purify phospholipid-capped NCs (see also Supplementary Note 6), a suited antisolvent is added, followed by centrifugation (3), whereas a supernatant containing unreacted precursors and free ligand molecules is discarded (4). If the ligand is’ good’, the resulting NCs pellet is redispersed in a suited solvent, yielding a stable colloid (5a). With a ‘bad’ ligand, NCs redisperse incompletely, do not redisperse (5b), or lose their colloidal integrity upon storage (6). Such a purification cycle (steps 3, 4, and 5a) can be repeated several times. c,d, In this experiment, two head groups and two tails were compared. Combining a ‘bad’ head group and a ‘bad’ tail (c) leads to the worst colloidal stability, while combining a ‘bad’ head group and a ‘good’ tail might still yield long-term stable colloids (d). Similar to FAPbBr3, MAPbBr3, and CsPbBr3 (c) NCs with PC head-group and a ‘bad’ hexadecyl tail precipitate after three rounds of purification, with visible deposit highlighted with a white box on the photo. CsPbBr3 NC colloids with both PEA or PC head-groups and a ‘good’ glycerodioleyl tail (d, commercially available) yield stable colloids; however, with different ligand coverage, in agreement with MD prediction. Furthermore, PC-based surface ligation is more labile, with both “good” and “bad” tails: NCs tend to increase their mean particle size (inset TEM images, scale bars 50 nm) and acquire more irregular NC morphology with storage or several purification cycles.
Ligand tail engineering
The apparent colloidal stability is a combined effect of the binding group affinity to the NC surface and the structure of the ligand tail. For instance, single hydrocarbon tails (such as hexadecyl) tend to form crystalline domains and are inferior to more entropic tails comprising bent oleyl or branched hydrocarbons36 in instilling efficient steric repulsion. Thus, it was unsurprising to observe the efficacy of the ligand in which the PC-head group is paired with the dioleyl-glycerophosphate fragment and, likewise, the efficacy of natural lecithin comprising diverse long-chain fatty acid substituents21,37. Besides suited steric repulsion, the ligand shall yield long-term colloidal stability when the binding strength outcompetes the solvation-induced desorption of the ligand. For a given ‘good’ tail, transitioning from a PC to a PEA head group afforded a twofold increase in the estimated surface coverage (by NMR) and robust purification through multiple cycles of precipitation with a non-solvent, retaining uniform size and shape (Extended Data Fig. 5d).
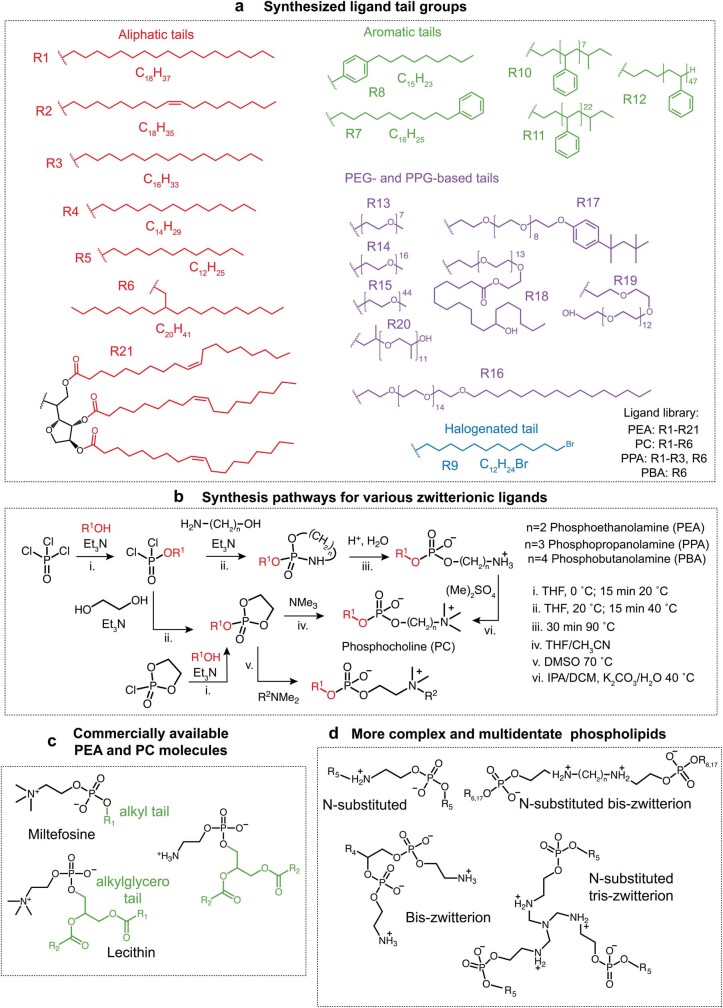
We thus set out to synthesize and test a library of PEA-based capping molecules, reasoning that anchoring tail groups (R) of aliphatic, aromatic, halogenated or polyether structures (Extended Data Fig. 6a) will render the resulting NCs dispersible in a broad range of common organic solvents. Synthesis of PC-terminated phospholipids with aliphatic chains has already been extensively studied with a multitude of optimized reactions30, owing to the prevalence of PC lipids in biological membranes38 and their broad use for engineering liposomes for medical applications (for example, drug or gene delivery)24,39,40 or as drugs on their own41,42. We first find that hexadecyl-PEA can be conveniently isolated by adopting the synthesis of hexadecyl-PC from hexadecanol by Eibl and Engel29, skipping the last methylation step. We then generalized this synthesis approach for various alcohols (ROH), converting them into preparative quantities of the respective R-PEA ligands beginning with POCl3 in three steps (Fig. 4a). The first two steps of gradual Cl exchange are one-pot reactions to form oxazaphospholane cycle, followed by acidic hydrolysis (Methods).
Extended Data Fig. 6. Survey of synthesized and commercial phospholipid ligands.
a, Tail groups of the synthesized PEA ligand library. b, Synthetic pathways for PPA, PEA, PC and N-alkyl substituted PEA ligands. b, Commercially available PC and PEA ligands. d, Multi-zwitterionic ligands.
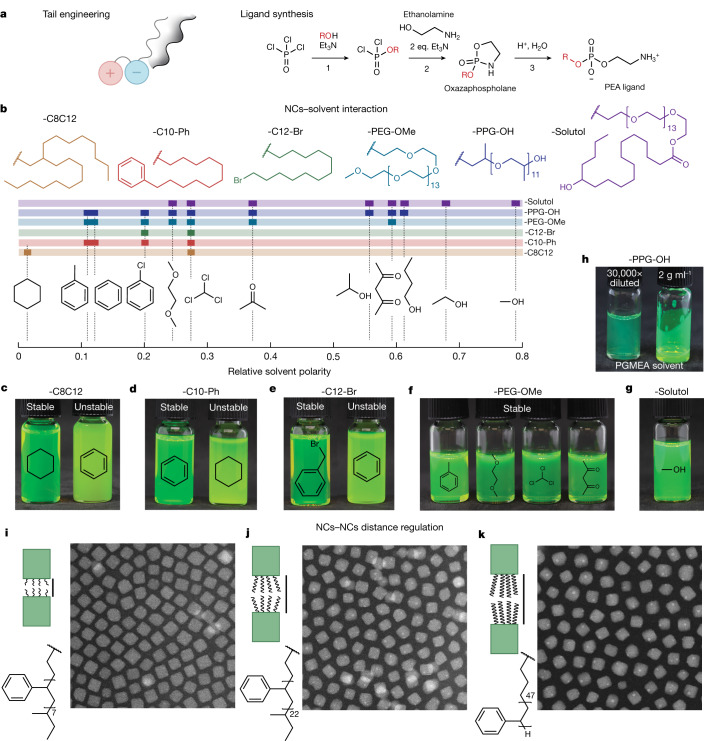
Fig. 4. Examples of functional tail engineering.
a, Synthesis scheme for PEA ligands tested in this work, with 21 ligand tails shown in Extended Data Fig. 6. b–g, Different tails enable matching of the solvent polarity (b) with highly specific dispersibility, shown for FAPbBr3 NCs (c–e) capped with aliphatic (c), aromatic (d), halogenated (e) tails and CsPbBr3 NCs (f,g) with polyether tails. h, PPG-PEA-capped CsPbBr3 NCs in PGMEA at a concentration of 2 g (CsPbBr3) per ml of dispersion. i–k, Engineering inter-NC distance in monolayers of CsPbBr3 NCs capped with PEA ligands with polystyrene tails by adjusting the ligand molecular weight: Mn = 900 Da (h), 1,200 Da (i) and 5,000 Da (j). Scale bars, 20 nm (i–k, main).
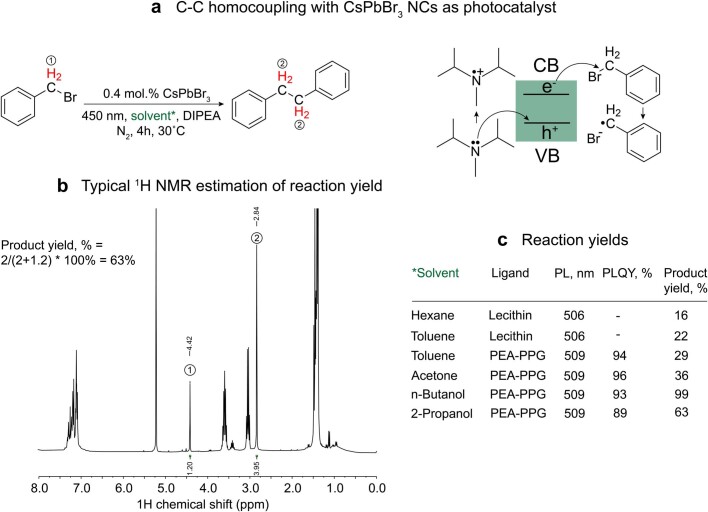
Each of the tested ligand tails attached to a single PEA head group (21 in total; Extended Data Fig. 6a) imparted long-term colloidal stability to LHP NCs in specific solvents, echoing the ‘like dissolves the like’ principle (Fig. 4b). For each ligand–solvent pair, the purification methodology needs to be adjusted, particularly the selection of the anti-solvent (Supplementary Note 6 and Supplementary Tables 11 and 12). A branched aliphatic tail (-C8C12) is most compatible with aliphatic hydrocarbon solvents, whereas phenyl- or halide-terminated tails render LHP NCs preferentially dispersible in, respectively, aromatic or halogenated solvents (Fig. 4c–e and Supplementary Fig. 19). Dispersing LHP NCs in common polar solvents, such as acetone, alcohols or acetylacetone, without compromising their morphology had thus far remained a formidable challenge. The matter is resolved in this study using PEA-ligands with poly(ethylene) glycol (PEG) and poly(propylene) glycol (PPG) tails (-PEG-OMe, -Solutol (-PEG-OH) and -PPG-OH), all affording long-term colloidal stability and retaining monodispersity, cuboid shape and high emissivity (Fig. 4f–g and Supplementary Figs. 20–22). For instance, PPG–PEA, unlike apolar aliphatic ligands (that is, lecithin), renders FAPbBr3 and CsPbBr3 NCs highly dispersible (up to 67% by weight, inorganic basis) in propylene glycol methyl ether acetate (PGMEA; Fig. 4h and Supplementary Fig. 23), an environmentally benign solvent of broad use in optoelectronics, particularly for formulating quantum dot inks in manufacturing displays43,44. Another exciting avenue lies in the ability to fine-tune the inter-NC separation in NC solids, as routinely accomplished for self-assembled NC superlattices10,45,46. We thus have devised stable colloids with polystyrene–PEA ligands synthesized from commercial OH-terminated polystyrenes of adjustable molecular weight (0.93–21 kDa). Rigid polystyrene tails increase the inter-NC spacing to at least 5 nm in NC monolayers (Fig. 4i–k), compared with 1.2 nm with C8C12-PEA capping. Inexpensive LHP NCs are of growing interest also as photocatalysts in organic synthesis, owing to reduced carrier trapping and optical tunability47; yet the studies have been stalled by the lability of LHP NCs in common polar, green solvents—alcohols and ethers. PPG–PEA–ligand is an enticing ligand choice for feasibility studies, as it imparts robustness to CsPbBr3 NC colloids in diverse organic solvents. We conducted reductive C–C bond coupling with benzyl bromide as a substrate, previously reported with CsPbBr3 perovskite NCs48, but not with common Ir-based photocatalysts, to our knowledge. A drastic increase in the product yield (in %) was reached on transitioning from toluene (29%) to n-butanol (99%), for a reaction run time of 4 h at 0.4 mol% of catalyst (Extended Data Fig. 7). Lecithin-capped quantum dots, dispersible in hexane and toluene only, reached a product yield of just 16% and 22%, respectively.
Extended Data Fig. 7. Photocatalysis with poly(propylene glycole) PEA capped CsPbBr3 NCs.
a, Photocatalytic C–C coupling reaction through activation of C–Br bond in benzyl bromide, catalyzed by CsPbBr3 NCs. b, 1H NMR of the final reaction mixture showing peaks from the starting material (around 4.4 ppm) and product (around 2.2 ppm) and how the reaction yield was calculated from integrating the two peaks. c, A table summarizing product yields in different solvents and with NCs capped with either commercial lecithin or poly(propylene glycole) (PPG)-PEA.
Light-emissivity of alkyl-PEA-capped APbBr3 NCs
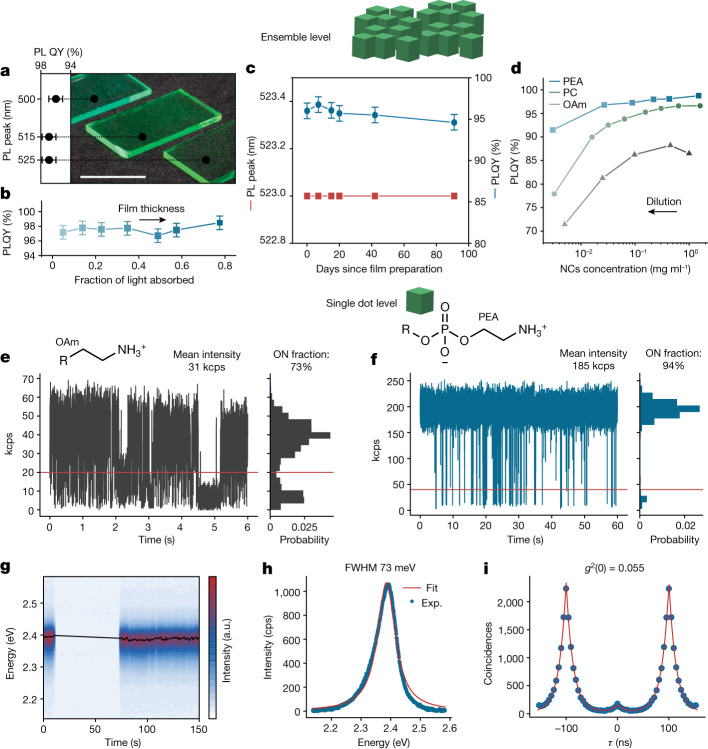
TOPO/PbBr2 synthesis31 coupled with subsequent PEA-ligand capping affords highly robust colloids of FAPbBr3 and MAPbBr3 quantum dots (NC sizes down to just a few nanometres), and thus brings closer their much awaited exploration at ensemble and single-particle levels and comparison with thoroughly studied CsPbBr3 NCs. For instance, C8C12-PEA-capped 6 nm FAPbBr3 NCs exhibit unaltered optical properties after ten rounds of purification (Supplementary Fig. 25). MAPbBr3 NCs (10 nm) have similar stability (Supplementary Fig. 26). Compact, spin-coated films of C8C12-PEA-capped FAPbBr3 NCs exhibit room-temperature photoluminescence (PL) quantum yields (QYs) of 96–97% (5.5–12 nm size range, 500–525 nm PL peak range; Fig. 5a and Supplementary Figs. 29 and 30). The measured PL QY value is retained when altering the optical density (film thickness; Fig. 5b) by roughly an order of magnitude, attesting to the inherent near-unity PL quantum efficiency. Both PL QY and PL peak wavelengths of colloids and films sustain at least 3 months of storage under ambient conditions without encapsulation (Fig. 5c).
Fig. 5. Light emission from C8C12-PEA-capped FAPbBr3 NCs on the ensemble and single dot levels.
a–d, Highlighted optical properties of FAPbBr3 NCs in an ensemble. Films of FAPbBr3 NCs of various sizes demonstrate equally high PL QY and tunable green emission (a). High PL QY is retained in thick films (b) and is not influenced by film storage in ambient conditions for at least 90 days (c). PL QY of greater than 90% is preserved on approximately 1,000-fold dilution of C8C12-PEA-capped NCs with octane, whereas a pronounced drop is observed for C8C12-PC- and OAm-capped NCs (d), indicative of dilution-induced surface degradation. e–i, Optical properties of FAPbBr3 NCs at the single-particle level. Single OAm-capped NCs exhibit pronounced blinking (e), while single PEA-capped NCs exhibit high brightness and suppressed PL blinking (f), good PL stability (g), narrow PL linewidth (full-width at half-maximum (FWHM)) (h) and high single-photon purity (i). PL QY measurement error in (d) is ±1%. a.u., arbitrary unit. Scale bar, 10 mm.
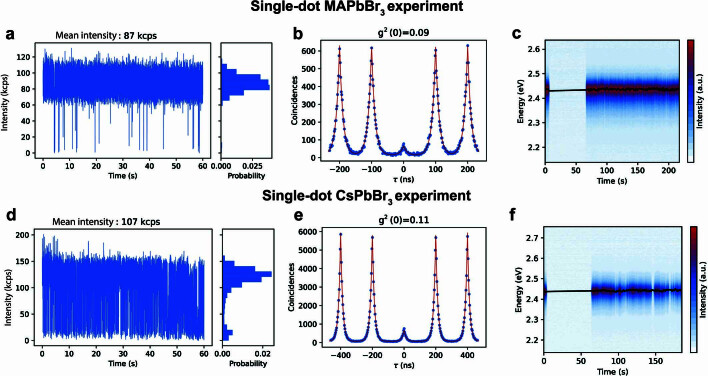
Not only do purified C8C12-PEA-capped NCs exhibit higher PL QY, compared with PC and OAm ligands, but they also retain their emissivity on strong dilution of the solution (up to 1,000-fold in Fig. 5d). Far more diluted samples (×104–105) are required for preparing samples for single-particle spectroscopy in conventional micro-PL set-ups. Detrimental processes—ligand desorption, which caused NC aggregation, and chemical reactivity towards trace water and oxygen or even the polymer used as a matrix—are drastically aggravated on extreme dilution. Structurally labile CsPbBr3 NCs were reported to shrink in size, alter their morphology and surface composition and photobleach49,50. Fluorescence blinking—stochastic switching between bright ON and dim OFF states—are universally observed in almost all quantum emitters at room temperature. Blinking is commonly attributed to the trapping of charges on surface defects on photoexcitation, and along with PL intensity of single NCs, serves as a diagnostics of the surface electronic state. Single-dot PL data (Fig. 5e–i) evidence a profound role played by the capping ligand in achieving spectrally stable PL, suppressed blinking (94% ‘ON’ state) and high single-photon purity, with values of the second-order correlation function g2(τ) at zero delay time (τ = 0), g2(0), down to 0.055 from single C8C12-PEA-capped FAPbBr3 NCs. We note that achieving nearly blinking-free emission from CdSe-based NCs required epitaxial overgrowth with minimally strained lattice-matched wider-band-gap material51–53. For comparison, the average outcome from single OAm-capped FAPbBr3 NCs, which are minimally purified to reduce ligand desorption, is 73% of the time in the bright state and overall brightness of only 20–30% of the PEA-coated counterparts. These substantial differences in favour of PEA-ligated NCs, as well as the narrower dispersion of PL characteristics, are echoed by statistics over a total of 78 quantum dots (Supplementary Fig. 30). Analogous improvements have been realized also for CsPbBr3 and MAPbBr3 NCs (Extended Data Fig. 8). We note that single PEA-capped FAPbBr3 NCs also greatly outperform NCs capped with commercial lecithin (Supplementary Figs. 31–33). Notably, single PEA-capped quantum dots retain high brightness (about 3 × 105 cps) and high ON fraction (about 90%) beyond 1 h of continuous operation (Supplementary Fig. 33b,c).
Extended Data Fig. 8. MAPbBr3 and CsPbBr3 single-dot emission at room temperature.
These NCs were capped with C8C12-PEA-ligands and purified three times, followed by dilution (×105) and spin-coating onto a glass substrate. a,d, Blinking traces of MAPbBr3 and CsPbBr3 single dots. b,e, Pronounced antibunching evidence high purity of single photon emission. c,f, Stable and narrow emission from a single MAPbBr3 NC (c) or CsPbBr3 NC (f).
Broader implications of phospholipid ligand capping
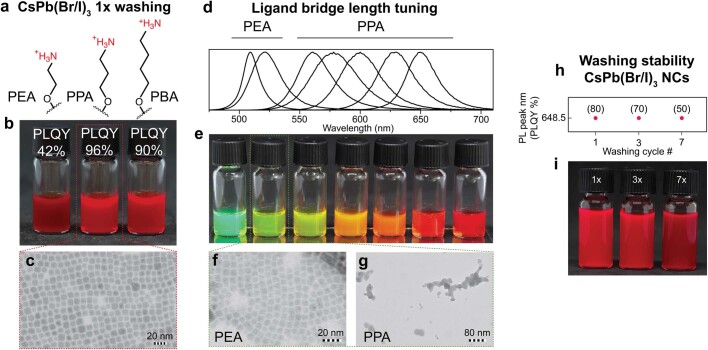
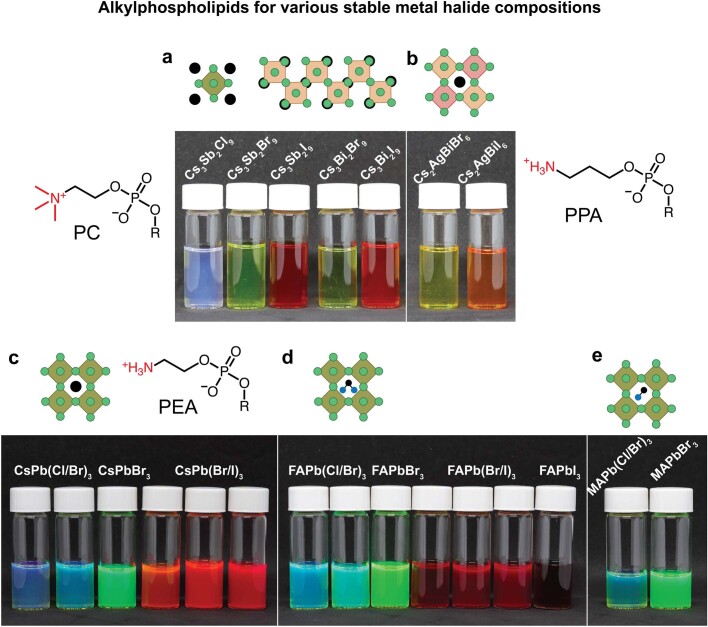
Thus far, we have comprehensively presented the design of phospholipid ligation for one compositional family of LHP NCs (APbBr3; A = Cs, FA, MA) and a single binding head group (PEA), validated computationally and through solid-state NMR, and then synthetically paired with 21 structurally different tail groups. A far broader deployment of phospholipids as capping ligands is anticipated from the feasibility studies. For instance, adjusting the spacing between the ammonium and phosphate groups aids in matching the larger lattice constant, illustrated for iodide-rich LHP NCs (Extended Data Fig. 9), which are stable with phosphopropanolamine (PPA, C3-bridge) or phosphobutanolamine (PBA, C4-bridge), but not PEA ligand (C2-bridge). Furthermore, we used well-documented, facile and high-yield reaction schemes, exemplified in Extended Data Fig. 6b, to further extend the relevant structural space of phospholipid capping ligands. In addition to the molecules with a single head group (Extended Data Fig. 6c), molecules comprising several zwitterionic fragments (Extended Data Fig. 6d) were also validated as efficient surfactants (Supplementary Fig. 35). The broad scope of NC core materials is seen in stable colloids obtained across the entire family of LHP NCs and for the main kinds of lead-free materials—double perovskite NCs and low-dimensional Sb- and Bi-based metal halide NCs (Extended Data Fig. 10). Notably, new capping ligands can be applied not only through the post-synthetic ligand exchange, but also in the direct synthesis of NCs (Supplementary Note 7). Future studies might extend to phospholipid-stabilized colloids, oxides and fluorides, as well as two-dimensional inorganic materials such as MXenes or transition metal dichalcogenides.
Extended Data Fig. 9. Head-group optimization by tuning the bridge length.
a–c, Lead iodide perovskites have a larger distance between A and X surface sites than bromides. Positive-to-negative moieties distance in the zwitterion thus has a pronounced effect on the ligand binding. Mixed halide CsPb(Br/I)3 NCs (without anti-solvent purification, synthesis details in Supplementary Table 7) capped with phosphoalkylamine ligands featuring different distances between ammonium and phosphate functionalities (a): PEA, PPA, and PBA. After the first purification step with anti-solvent (b), PEA-capped NCs drop in PLQY from 95% to 42%, while longer-bridge PPA and PBA ligands retain high PLQY and NCs shape (c). d–g, In general, PEA ligands better suit Br-rich compositions, while PPA ligands make for a better choice for I-rich compositions. h,i, Mixed halide CsPb(Br/I)3 NCs capped with 3-ammoniopropyl (2-octyl-1-dodecyl) phosphate (PPA-R6) display remarkable spectral stability during purification cycles with antisolvent (ethyl acetate:acetonitrile).
Extended Data Fig. 10. A compositional variety of metal halide NCs that can be stabilized with alkylphospholipid zwitterionic capping ligands.
a,b, Lead-free metal halides: low-dimensional (a) or double perovskites (b). c–e, Stable lead halide perovskite NCs with all three cations: Cs (c), FA (d) and MA (e), as well as of varying halide composition, can be prepared using zwitterionic ligands with phosphate and primary ammonium head-groups and varying bridge length (PEA, PPA, PBA) (additionally see Supplementary Figs. 15–17).
Methods
Computational model
The surface of perovskite NCs was modelled using a crystal slab terminated with (100) crystallographic planes, in line with the high-resolution scanning transmission electron microscopy images of FAPbBr3 and CsPbBr3 NCs capped with alkylphospholipid ligands (Supplementary Fig. 5). Both ABr- and PbBr-rich lattice terminations were considered. One of the slab surfaces was then passivated by placing a certain number of ligands (1, 32 or 64) at a distance of 0.5 nm from the surface (measured to the head group). The ligand tail was truncated at five carbon atoms to reduce computational cost. If needed, some ion pairs were removed from the surface to yield the desired system stoichiometry defined by two quantities—ligand concentration , and ABr concentration . Finally, the system was solvated with toluene. Further details of the slab model can be found in Supplementary Note 1. Interactions between ions comprising the slab were modelled by Coulomb and Lennard-Jones potentials, with parameters adopted from refs. 54–56. This classical perovskite model was tested in simulations of bulk CsPbBr3 and FAPbBr3 crystals with a fully anisotropic pressure coupling and yielded good agreement with experimental structural properties (Supplementary Fig. 6). The all-atom optimized potentials for liquid simulations (OPLS-AA) force field57–59 was used to model interactions between atoms in the organic part of the system. Ligand parameters were taken from the corresponding models of phospholipids60,61. Missing O–C–C–C and O–C–C–H dihedral parameters at the point of attachment of the tail to the head group were taken from the analogous dihedral parameters for ether/alcohol62.
Replica-exchange MD simulations
The complete simulation boxes were equilibrated for 20 ps in the constant number, volume, temperature (NVT) ensemble with positions of all ions and ligand head groups tightly restrained and then for 1 + 10 ns in the constant number, pressure, temperature (NPT) ensemble with released restraints. In all simulations, positions of lead ions in the middle layer of the slab were restrained to the origin of the z axis and to their crystallographic positions in the x and y directions by applying weak harmonic restraining potential with a force constant of k = 1,000 kJ mol−1 nm−2, to prevent floating of the slab across the simulation box.
The final pre-equilibrated structures were used as starting points for replica-exchange MD simulations33 in the NVT ensemble. A total of 120 replicas were exponentially distributed between 300 and 2,200 K, allowing the efficient crossing of potential energy barriers. Exchanges between neighbouring replicas were attempted every 1 ps, and each replica was simulated for 50 ns. The average exchange rate was ensured to be above 10% for all neighbouring replica pairs. Setting all atomic masses to 12 atomic units allowed us to increase the simulation time step to 1 fs without affecting the configurational phase space of the system. Special restraining potentials were used to prevent the crystal’s high-temperature melting and limit diffusive movements of the ions and ligand molecules. These are discussed in more detail in Supplementary Note 2.
All reported simulations were performed using GROMACS software package63. Electrostatic interactions were computed using the smooth particle-mesh Ewald method64.
Binding mode populations were computed in two steps. First, distances from the ligands’ nitrogen and phosphorus atoms to the middle atomic plane of the slab, and , were calculated over the whole replica-exchange MD run and plotted as a two-dimensional map. At most, four well-defined clusters were observed in the case of AX-terminated surfaces, which were assigned to four different BMs—BM1, BM2, BM2′ and BM3 (Extended Data Fig. 1). The broad tail that extends to long ligand-slab separations was attributed to unbound ligands. In the next step, ligands were classified according to their BM using the corresponding cut-offs on the N–slab and P–slab distances. Binding mode populations were computed as ensemble averages in 2 ns intervals and presented as time traces.
PEA, PPA and PBA ligand synthesis
The synthesis route starting with alcoholysis of phosphorous oxychloride was adopted from ref. 29. Solution of alcohol substrate (0.025 mol; 1 equiv) dissolved in dry tetrahydrofuran (THF) (25 ml) along with triethylamine (0.275 mol; 1.1 equiv; 3.83 ml) is added dropwise with vigorous stirring to a solution of phosphorous oxychloride (0.03 mol; 1.2 equiv; 2.78 ml) in THF (2.5 ml) on an ice water bath. The reaction mixture is subsequently kept at 20 °C for 15 min to complete the reaction. Next, ethanolamine (0.03 mol; 1.2 equiv; 1.81 ml) and triethylamine (0.06 mol; 2.4 equiv) in THF (37.5 ml) are added dropwise under vigorous stirring to the reaction mixture kept in a room-temperature water bath. Subsequently, the mixture is heated to 40 °C for 15 min to complete the ring closure. Finally, the reaction mixture is filtered to remove precipitated triethylamine hydrochloride, and the filtrate solution is dried. An oily residue, that is, alkyl-2-oxo-1,2,3-oxazaphospholane, is dissolved in a mixture of acetic acid (5.7 ml) and distilled water (2.6 ml) at 70 °C. After 30 min, ring scission at the P–N bond is complete, and the product is separated by beating with acetone (125–150 ml). After cooling to 10 °C, alkylphosphoethanolamine is collected and dried overnight under a vacuum at 40–50 °C. For PPA or PBA ligands, 3-aminopropan-1-ol and 4-aminobutanol-1-ol were used instead of 2-aminoethan-1-ol. For PBA, the hydrolysis step was conducted at 90 °C.
PC ligand synthesis
The synthesis procedure, beginning with the alcoholysis of 2-chloro-2-oxo-1,3,2-dioxaphospholane (COP), was adopted from ref. 65. A solution of COP (0.7 mol; 1equiv; 10 g) in dry THF (30 ml) was added dropwise to a mixture of alcohol substrate (0.7 mol; 1 equiv) and triethylamine (0.7 mol; 1 equiv) in dry THF (140 ml) at 0 °C under vigorous stirring. After the addition, stirring was continued for 1 h at room temperature. After filtering, the filtrate solution was concentrated by at least a factor of two or by evaporation and dry acetonitrile (150 ml) was added, and the reaction mixture was placed into a glass pressure bottle. At −20 °C, 2 M trimethylamine in THF (0.14 mol; 2 equiv; 70 ml) was added and the reaction was carried out at 70 °C for 12 h. After 12 h, the reaction mixture was cooled to −20 °C to precipitate the product. The product was then filtered off and dried under a vacuum overnight.
Further synthetic conditions and characterization of all obtained ligands can be found in Supplementary Figs. 13 and 14 and Supplementary Note 4.
‘Ligand-exchange’ synthesis of CsPbBr3, FAPbBr3 and MAPbBr3 NCs capped with zwitterionic ligands
NCs were synthesized according to the modified TOPO–diisooctylphosphinic acid (DOPA) procedure described elsewhere31. Pb precursor was prepared from PbBr2 (0.2 mmol, 73.4 mg) and TOPO (90%) (1.1 mmol, 429.6 mg) dissolved in n-octane (2.5 ml) at 120 °C on a hotplate in the air. Pb precursor is then diluted with various quantities of hexane to obtain NCs of different sizes. Cs precursor was obtained by reacting Cs2CO3 (100 mg) with DOPA (1 ml) in octane (2 ml) at 120 °C, followed by dilution with hexane (27 ml). FA precursor was prepared from formamidine acetate, 64 mg (0.61 mmol), DOPA (3 ml) and OA (2 ml), dissolved in n-octane (5 ml) at 120 °C in a 40 ml vial on a hotplate in the air. MA precursor was prepared by mixing MA in THF (2 M, 0.3 ml) along with DOPA (3 ml), OA (2 ml) and n-octane (5 ml). To synthesize NCs, A precursor is swiftly injected into the Pb precursor on stirring at room temperature and left for a defined amount of time to nucleate and grow NCs, followed by the addition of the zwitterionic ligands. Synthesis details, specific to NC size and composition, and further characterization can be found in Supplementary Note 5, Supplementary Tables 4–10 and Supplementary Figs. 15–17.
Purification of zwitterion-capped NCs
When NCs remain dispersed in n-hexane after the ligand exchange, 2–3 equiv. of an anti-solvent (list of anti-solvents is given in Supplementary Table 11) are added to precipitate NCs, followed by centrifugation. The colourless supernatant is discarded, and the precipitate is redispersed in a desired solvent. When the ligand-capped NCs are incompatible with hexane (for example, polystyrene, polyethyglycol-based ligands), they precipitate after ligand exchange. NCs are further collected by centrifugation and redispersed in suitable solvent, completing the first washing cycle. Further, 3–4 equiv of n-hexane (or other suitable anti-solvent) are added to precipitate NCs again for the next washing cycle. The washing cycle can be repeated as many times as required. After three cycles, impurities of TOPO, DOPA and OA are absent, as evidenced by 31P NMR (Supplementary Fig. 25). Further details can be found in Supplementary Note 6 and Supplementary Figs. 25 and 26.
Ligand coverage estimation by 31P NMR
To estimate the ligand coverage, stable and purified NC colloids in toluene capped with PEA or PC ligands were dissolved in dimethyl sulfoxide-d6 (DMSO-d6), destroying the NCs and freeing the bound ligands. A known amount of phosphor-containing standard (for example, tetrabutylphosphonium bromide) was added to the toluene-DMSO-d6 sample, and the 31P NMR one-dimensional spectrum was measured. Integration of the P signal was readily recalculated to the ligand concentration, knowing the average NC size. The NC concentration was estimated from the extinction coefficient34, and absorption was measured from the sample before destruction with DMSO.
FTIR spectroscopy
Ligand BM in PEA- and PC-capped perovskite NCs was assessed with FTIR spectroscopy in conjunction with ab initio molecular dynamics simulations (details are in Supplementary Note 3). FTIR spectra of solid-state samples, that is, ligands and dry NC powders, were obtained in an N2-filled glovebox by means of a Thermo Fisher Nicolet iS5 FTIR spectrometer with a deuterated triglycine sulfate detector, a KBr beam splitter and an iD5 attenuated total reflectance unit comprising a diamond crystal.
Solid-state NMR spectroscopy
31P–207Pb REDOR experiments were performed on a Bruker narrow-bore 16.4 T (600 MHz) and 9.4 T (400 MHz) NMR spectrometers equipped with a Bruker Avance III HD console. All experiments were performed on a 2.5 mm HXY probe configured in a 1H–31P–207Pb mode. A magic-angle spinning frequency of 20 kHz was used for all experiments. The π/2 pulse was optimized to 3 µs for 1H, to 6.1 µs for for 31P and to 6.5 µs for 207Pb. Chemical shifts were externally referenced to tetramethylsilane (1H), phosphoric acid (31P) and tetramethyl lead (207Pb). 31P spectra were acquired with a π-pulse excitation and 1H decoupling (SPINAL64) during acquisition. A recycle delay of 1 s was used. 207Pb spectra were acquired using a π/2–π Hahnecho sequence. The echo delay was set to one rotor cycle (50 µs). A recycle delay of 0.5 s was used.
1H–31P(207Pb) cross-polarization (cp) REDOR experiments were performed on a Bruker wide-bore 14.1 T NMR spectrometer equipped with a Bruker Avance III HD console. All experiments were performed on a 3.2 mm HXY DNP probe configured in a 1H–31P–207Pb mode. A magic-angle spinning frequency of 9 and 11 kHz was used for all experiments. The 1H π/2 pulse was optimized to 2.7 µs. The 31P π/2 pulse was optimized to 7 µs. A saturation pulse train with 16 π/2 pulses on 1H and 31P was applied. A ramp pulse (2 ms) was used for 1H–31P contact. 1H–31P cp spectra with varying recoupling times were acquired with (S) and without (S0) dephasing pulses on 207Pb. The duration of dipolar recoupling was incremented linearly. Dephasing was induced by 207Pb π pulses (15.2 µs). A recycle delay of 3 s was used. The recoupling curves were obtained by plotting (S0 − S)/S0 versus the recoupling time.
The recoupling curves are determined as
where and are the integrals of the NMR spectra with and without dephasing pulses at varying recoupling times (expressed as number of rotations ). Using uncertainty propagation, the corresponding error is
The errors and are determined from the noise level of the spectrum and are identical as number of scans and noise levels are identical.
Computation of theoretical REDOR curves
Theoretical REDOR curves (BM1–BM3) were generated by our Python implementation of an approach reported elsewhere20. The source code is available at https://gitlab.ethz.ch/kovalenkolab/redor. This approach simulates REDOR curves for a multi-spin system (more than two spins), including heteronuclear coupling, while assuming that homonuclear coupling can be neglected. This assumption holds for the initial slope of the REDOR curve (short dephasing times)66. The geometries for the theoretical curves were either based on the crystal structure of FAPbBr3 or chosen from the replica-exchange MD simulation of a single PC ligand on the FABr-terminated perovskite surface. Ten randomly selected structures were used for each BM in the latter case. Because BM2′ was not observed in this simulation, a separate 10 ns plain MD simulation with the ligand initially placed in BM2′ was performed. We note that BM2′ was found to be metastable across the entire simulation. The theoretical REDOR curve on Fig. 2c was scaled by coefficient 0.3 to fit the initial data slope to account for dynamics and inefficient recoupling due to the potential broadening of 207Pb signal of Pb atoms bound to phosphate.
Electron microscopy
Transmission electron microscopy images were collected using a Hitachi HT7700 microscope operated at 100 kV. High-angle annular dark field scanning transmission electron microscopy (HAADF-STEM) images were obtained using an FEI Titan Themis aberration-corrected microscope operated at 300 kV and with a probe-corrected cubed Thermo Fisher Scientific Themis Z Microscope operating at 300 kV with a probe semi-convergence angle of 20 rad. Images were processed using Image J.
Optical spectroscopy
Room-temperature PL spectra of purified quantum dots were recorded with a Fluorolog iHR 320 Horiba Jobin Yvon with an excitation at 350 nm. Absorption spectra were recorded with a Jasco V670 spectrometer. PL QY was measured using the Quantaurus-QY spectrometer C11347-11 from Hamamatsu Photonics; for samples with absorbance higher than 0.4, a self-absorption correction procedure was used as implemented in the software (U6039-05 for Quantaurus QY). Single-dot spectroscopy and related sample preparation were conducted under a nitrogen atmosphere. First, the NC solutions were diluted by a factor of 30,000–60,000 with dry and filtered n-octane (99+% extra dry, Acros Organics). A sparse NC film was obtained by spin-coating 100 µl of the diluted solution at 150 reps for 60 s onto a clean cover glass (thickness 170 ± 5 μm; diameter 25 mm; Thorlabs). Micro-PL measurements were carried out with a home-build set-up under irradiation with a pulsed 405 nm laser (10 MHz, pulses less than 50 ps, greater than 100 W cm−2, PicoQuant). The laser is focused (1/e2 = 1 μm) by an oil immersion objective (numerical aperture = 1.3) onto the sample, and the same objective collects the emitted light. The collected light is passed through a dichroic mirror to filter out the residual light from the excitation laser and sent either to a Hanbury Brown–Twiss experiment or to a monochromator and electron-multiplying charge-coupled device camera (1 s binning, Princeton Instruments). The Hanbury Brown–Twiss experiment consists of a 50/50 beam splitter, two avalanche photodiodes (temporal resolution = 250 ps, Excelitas) and photon-counting electronics (PicoQuant), enabling the acquisition of time-tagged time-resolved (TTTR) fluorescence data. Single-dot measurements were carried out in the weak excitation regime at a fluence of 0.8–1.3 μJ cm−2 (less than one exciton per pulse). The spectra of the NCs were obtained by averaging the first five frames of the spectra series; they were then fitted with a Lorentzian peak to find the peak centre and full-width at half-maximum. The second-order correlation function (g2(τ)) was calculated from the TTTR data with the pycorrelate package using the algorithm by Laurence et al.67 and fitted by a biexponential function (shared lifetimes, no constant offset) to predict the g2(0). The blinking traces were obtained by binning the TTTR data into 1 ms bins, and the fraction of time spent in the ON state was determined by thresholding after visual inspection of intensity histograms and traces (Supplementary Fig. 30).
Photocatalysis with CsPbBr3 NCs
Benzyl bromide (69.5 µl) and a photocatalyst (CsPbBr3 NCs, 1.35 mg) were combined with the solvent of choice (1 ml) and N,N-diisopropylethylamine (306 µl) in a 4 ml vial that is then sealed with Parafilm. The vial is placed in the temperature-controlled photoredox device PhotoRedOx TC from HepatoChem (HCK1006-01-025) equipped with a 450 nm blue light-emitting diode (30 W, 250 V) for 4 h at 30 °C. After this time, the reaction mixture was transferred to a round-bottom flask with the help of dichloromethane and all solvent was evaporated. The dry leftover was dissolved in 0.5–0.6 ml of CDCl3 for NMR (300 Hz) to analyse the product yield.
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at 10.1038/s41586-023-06932-6.
Supplementary information
Supplementary Notes 1–7, Figs. 1–36, Tables 1–13 and references—see Contents page for details.
Acknowledgements
This work was financially supported by the Air Force Office of Scientific Research and the Office of Naval Research (award no. FA8655-21-1-7013), by the Swiss National Science Foundation (National Centre of Competence in Research, NCCR Catalysis, grant no. 180544), by the European Research Council through the European Union’s Horizon 2020 programme (ERC Consolidator Grant SCALE-HALO, agreement no. 819740) and by the Swiss Innovation Agency (Innosuisse, grant no. 32908.1 IP-EE).
Extended data figures and tables
Author contributions
V.M. and M.V.K. conceived the project. V.M. and M.S. synthesized phospholipids and, together with C.J.K. and J.A., tested them as capping ligands and evaluated ensemble optical properties. A.S. and A.B. conducted MD simulations of NC–ligand interfaces. S.C.B. acquired and interpreted FTIR spectra. M.A., L.G.F., V.M. and A.S. acquired and analysed REDOR spectra. N.J.S., S.B. and I.C. acquired high-resolution electron microscopy images. L.G.F. and G.R. conducted single-particle PL studies. Y.S. and D.N.D. performed photocatalytic studies. V.M., A.S. and M.V.K. wrote the manuscript with the contribution of all co-authors. M.V.K. supervised the work. All authors discussed the results and commented on the manuscript.
Peer review
Peer review information
Nature thanks the anonymous reviewers for their contribution to the peer review of this work.
Funding
Open access funding provided by Swiss Federal Institute of Technology Zurich.
Data availability
The data that support the findings of this study are available on Zenodo public depository and from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
is available for this paper at 10.1038/s41586-023-06932-6.
Supplementary information
The online version contains supplementary material available at 10.1038/s41586-023-06932-6.
References
- 1.Protesescu L, et al. Nanocrystals of cesium lead halide perovskites (CsPbX3, x = Cl, Br, and I): novel optoelectronic materials showing bright emission with wide color gamut. Nano Lett. 2015;15:3692–3696. doi: 10.1021/nl5048779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Protesescu L, et al. Monodisperse formamidinium lead bromide nanocrystals with bright and stable green photoluminescence. J. Am. Chem. Soc. 2016;138:14202–14205. doi: 10.1021/jacs.6b08900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akkerman QA, Raino G, Kovalenko MV, Manna L. Genesis, challenges and opportunities for colloidal lead halide perovskite nanocrystals. Nat. Mater. 2018;17:394–405. doi: 10.1038/s41563-018-0018-4. [DOI] [PubMed] [Google Scholar]
- 4.Almeida G, Infante I, Manna L. Resurfacing halide perovskite nanocrystals. Science. 2019;364:833–834. doi: 10.1126/science.aax5825. [DOI] [PubMed] [Google Scholar]
- 5.Dey A, et al. State of the art and prospects for halide perovskite nanocrystals. ACS Nano. 2021;15:10775–10981. doi: 10.1021/acsnano.0c08903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boles MA, Ling D, Hyeon T, Talapin DV. The surface science of nanocrystals. Nat. Mater. 2016;15:141–153. doi: 10.1038/nmat4526. [DOI] [PubMed] [Google Scholar]
- 7.Owen J. The coordination chemistry of nanocrystal surfaces. Science. 2015;347:615–616. doi: 10.1126/science.1259924. [DOI] [PubMed] [Google Scholar]
- 8.Tamarat P, et al. The ground exciton state of formamidinium lead bromide perovskite nanocrystals is a singlet dark state. Nat. Mater. 2019;18:717–724. doi: 10.1038/s41563-019-0364-x. [DOI] [PubMed] [Google Scholar]
- 9.Park YS, Guo SJ, Makarov NS, Klimov VI. Room temperature single-photon emission from individual perovskite quantum dots. ACS Nano. 2015;9:10386–10393. doi: 10.1021/acsnano.5b04584. [DOI] [PubMed] [Google Scholar]
- 10.Cherniukh I, et al. Perovskite-type superlattices from lead halide perovskite nanocubes. Nature. 2021;593:535–542. doi: 10.1038/s41586-021-03492-5. [DOI] [PubMed] [Google Scholar]
- 11.Kovalenko MV, Protesescu L, Bodnarchuk MI. Properties and potential optoelectronic applications of lead halide perovskite nanocrystals. Science. 2017;358:745–750. doi: 10.1126/science.aam7093. [DOI] [PubMed] [Google Scholar]
- 12.Kwon JI, et al. Ultrahigh-resolution full-color perovskite nanocrystal patterning for ultrathin skin-attachable displays. Sci. Adv. 2022;8:eadd0697. doi: 10.1126/sciadv.add0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaplan AEK, et al. Hong–Ou–Mandel interference in colloidal CsPbBr3 perovskite nanocrystals. Nat. Photonics. 2023;17:775–780. doi: 10.1038/s41566-023-01225-w. [DOI] [Google Scholar]
- 14.Luo JJ, et al. Efficient and stable emission of warm-white light from lead-free halide double perovskites. Nature. 2018;563:541–545. doi: 10.1038/s41586-018-0691-0. [DOI] [PubMed] [Google Scholar]
- 15.Lian LY, et al. Efficient and reabsorption-free radioluminescence in Cs3Cu2I5 nanocrystals with self-trapped excitons. Adv. Sci. 2020;7:2000195. doi: 10.1002/advs.202000195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yakunin S, et al. High-resolution remote thermometry and thermography using luminescent low-dimensional tin-halide perovskites. Nat. Mater. 2019;18:846–852. doi: 10.1038/s41563-019-0416-2. [DOI] [PubMed] [Google Scholar]
- 17.Schutt, K., Berry, J. J., Wheeler, L. M. & Rosales, B. A. Leveraging low-energy structural thermodynamics in halide perovskites. ACS Energy Lett.8, 1705–1715 (2023).
- 18.Bodnarchuk MI, et al. Rationalizing and controlling the surface structure and electronic passivation of cesium lead halide nanocrystals. ACS Energy Lett. 2019;4:63–74. doi: 10.1021/acsenergylett.8b01669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imran M, et al. Simultaneous cationic and anionic ligand exchange for colloidally stable CsPbBr3 nanocrystals. ACS Energy Lett. 2019;4:819–824. doi: 10.1021/acsenergylett.9b00140. [DOI] [Google Scholar]
- 20.Chen Y, et al. Surface termination of CsPbBr3 perovskite quantum dots determined by solid-state NMR spectroscopy. J. Am. Chem. Soc. 2020;142:6117–6127. doi: 10.1021/jacs.9b13396. [DOI] [PubMed] [Google Scholar]
- 21.Krieg F, et al. Stable ultraconcentrated and ultradilute colloids of CsPbX3 (X = Cl, Br) nanocrystals using natural lecithin as a capping ligand. J. Am. Chem. Soc. 2019;141:19839–19849. doi: 10.1021/jacs.9b09969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krieg F, et al. Colloidal CsPbX3 (X = Cl, Br, I) nanocrystals 2.0: zwitterionic capping ligands for improved durability and stability. ACS Energy Lett. 2018;3:641–646. doi: 10.1021/acsenergylett.8b00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krieg F, et al. Monodisperse long-chain sulfobetaine-capped CsPbBr3 nanocrystals and their superfluorescent assemblies. ACS Cent. Sci. 2021;7:135–144. doi: 10.1021/acscentsci.0c01153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu S, et al. Membrane-destabilizing ionizable phospholipids for organ-selective mRNA delivery and CRISPR-Cas gene editing. Nat. Mater. 2021;20:701–710. doi: 10.1038/s41563-020-00886-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zumbuehl A. Artificial phospholipids and their vesicles. Langmuir. 2019;35:10223–10232. doi: 10.1021/acs.langmuir.8b02601. [DOI] [PubMed] [Google Scholar]
- 26.Harayama T, Riezman H. Understanding the diversity of membrane lipid composition. Nat. Rev. Mol. Cell Bio. 2018;19:281–296. doi: 10.1038/nrm.2017.138. [DOI] [PubMed] [Google Scholar]
- 27.Fedotenko IA, Holme MN, Tanasescu R, Zaffalon PL, Zumbuehl A. Putting the ‘P’ into phospholipids. Chimia. 2011;65:859–862. doi: 10.2533/chimia.2011.859. [DOI] [PubMed] [Google Scholar]
- 28.Magolda RL, Johnson PR. A new efficient and versatile synthesis of alkyl prosphorylcholines. Tetrahedron Lett. 1985;26:1167–1170. doi: 10.1016/S0040-4039(00)98424-7. [DOI] [Google Scholar]
- 29.Eibl H, Engel J. Synthesis of hexadecylphosphocholine (miltefosine) Prog. Exp. Tumor Res. 1992;34:1–5. doi: 10.1159/000420825. [DOI] [PubMed] [Google Scholar]
- 30.Fiore M. The synthesis of mono-alkyl phosphates and their derivatives: an overview of their nature, preparation and use, including synthesis under plausible prebiotic conditions. Org. Biomol. Chem. 2018;16:3068–3086. doi: 10.1039/C8OB00469B. [DOI] [PubMed] [Google Scholar]
- 31.Akkerman QA, et al. Controlling the nucleation and growth kinetics of lead halide perovskite quantum dots. Science. 2022;377:1406–1412. doi: 10.1126/science.abq3616. [DOI] [PubMed] [Google Scholar]
- 32.Swendsen RH, Wang JS. Replica Monte Carlo simulation of spin glasses. Phys. Rev. Lett. 1986;57:2607–2609. doi: 10.1103/PhysRevLett.57.2607. [DOI] [PubMed] [Google Scholar]
- 33.Sugita Y, Okamoto Y. Replica-exchange molecular dynamics method for protein folding. Chem. Phys. Lett. 1999;314:141–151. doi: 10.1016/S0009-2614(99)01123-9. [DOI] [Google Scholar]
- 34.De Roo J, et al. Highly dynamic ligand binding and light absorption coefficient of cesium lead bromide perovskite nanocrystals. ACS Nano. 2016;10:2071–2081. doi: 10.1021/acsnano.5b06295. [DOI] [PubMed] [Google Scholar]
- 35.Stelmakh A, Aebli M, Baumketner A, Kovalenko MV. On the mechanism of alkylammonium ligands binding to the surface of CsPbBr3 nanocrystals. Chem. Mater. 2021;33:5962–5973. doi: 10.1021/acs.chemmater.1c01081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang Y, et al. Entropic ligands for nanocrystals: from unexpected solution properties to outstanding processability. Nano Lett. 2016;16:2133–2138. doi: 10.1021/acs.nanolett.6b00730. [DOI] [PubMed] [Google Scholar]
- 37.Mir WJ, et al. Lecithin capping ligands enable ultrastable perovskite-phase CsPbI3 quantum dots for Rec. 2020 bright-red light-emitting diodes. J. Am. Chem. Soc. 2022;144:13302–13310. doi: 10.1021/jacs.2c04637. [DOI] [PubMed] [Google Scholar]
- 38.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Bio. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh RP, Gangadharappa HV, Mruthunjaya K. Phospholipids: unique carriers for drug delivery systems. J. Drug Deliv. Sci. Tec. 2017;39:166–179. doi: 10.1016/j.jddst.2017.03.027. [DOI] [Google Scholar]
- 40.Zhang YB, Sun CZ, Wang C, Jankovic KE, Dong YZ. Lipids and lipid derivatives for RNA delivery. Chem. Rev. 2021;121:12181–12277. doi: 10.1021/acs.chemrev.1c00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dorlo TPC, Balasegaram M, Beijnen JH, de Vries PJ. Miltefosine: a review of its pharmacology and therapeutic efficacy in the treatment of leishmaniasis. J. Antimicrob. Chemoth. 2012;67:2576–2597. doi: 10.1093/jac/dks275. [DOI] [PubMed] [Google Scholar]
- 42.Eibl H, Unger C. Hexadecylphosphocholine – a new and selective antitumor drug. Cancer. Treat. Rev. 1990;17:233–242. doi: 10.1016/0305-7372(90)90053-I. [DOI] [PubMed] [Google Scholar]
- 43.Hahm D, et al. Direct patterning of colloidal quantum dots with adaptable dual-ligand surface. Nat. Nanotechnol. 2022;17:952–958. doi: 10.1038/s41565-022-01182-5. [DOI] [PubMed] [Google Scholar]
- 44.Hahm D, et al. Surface engineered colloidal quantum dots for complete green process. ACS Appl. Mater. Interfaces. 2020;12:10563–10570. doi: 10.1021/acsami.9b23265. [DOI] [PubMed] [Google Scholar]
- 45.Ye XC, et al. Structural diversity in binary superlattices self-assembled from polymer-grafted nanocrystals. Nat. Commun. 2015;6:10052. doi: 10.1038/ncomms10052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cherniukh I, et al. Shape-directed co-assembly of lead halide perovskite nanocubes with dielectric nanodisks into binary nanocrystal superlattices. ACS Nano. 2021;15:16488–16500. doi: 10.1021/acsnano.1c06047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu XL, et al. Lead halide perovskites for photocatalytic organic synthesis. Nat. Commun. 2019;10:2843. doi: 10.1038/s41467-019-10634-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rosa-Pardo I, et al. The synergy between the CsPbBr3 nanoparticle surface and the organic ligand becomes manifest in a demanding carbon-carbon coupling reaction. Chem. Commun. 2020;56:5026–5029. doi: 10.1039/D0CC01339K. [DOI] [PubMed] [Google Scholar]
- 49.Raino G, et al. Ultra-narrow room-temperature emission from single CsPbBr3 perovskite quantum dots. Nat. Commun. 2022;13:2587. doi: 10.1038/s41467-022-30016-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rainò G, et al. Underestimated effect of a polymer matrix on the light emission of single CsPbBr3 nanocrystals. Nano Lett. 2019;19:3648–3653. doi: 10.1021/acs.nanolett.9b00689. [DOI] [PubMed] [Google Scholar]
- 51.Chen O, et al. Compact high-quality CdSe-CdS core-shell nanocrystals with narrow emission linewidths and suppressed blinking. Nat. Mater. 2013;12:445–451. doi: 10.1038/nmat3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi JJ, et al. All-optical fluorescence blinking control in quantum dots with ultrafast mid-infrared pulses. Nat. Nanotechnol. 2021;16:1355–1361. doi: 10.1038/s41565-021-01016-w. [DOI] [PubMed] [Google Scholar]
- 53.Chen Y, et al. ‘Giant’ multishell CdSe nanocrystal quantum dots with suppressed blinking. J. Am. Chem. Soc. 2008;130:5026–5027. doi: 10.1021/ja711379k. [DOI] [PubMed] [Google Scholar]
- 54.de Araujo AS, Sonoda MT, Piro OE, Castellano EE. Development of new Cd2+ and Pb2+ Lennard-Jones parameters for liquid simulations. J. Phys. Chem. B. 2007;111:2219–2224. doi: 10.1021/jp064835t. [DOI] [PubMed] [Google Scholar]
- 55.Lybrand TP, Ghosh I, Mccammon JA. Hydration of chloride and bromide anions – determination of relative free-energy by computer-simulation. J. Am. Chem. Soc. 1985;107:7793–7794. doi: 10.1021/ja00311a112. [DOI] [Google Scholar]
- 56.Aqvist J. Ion water interaction potentials derived from free-energy perturbation simulations. J. Phys. Chem. 1990;94:8021–8024. doi: 10.1021/j100384a009. [DOI] [Google Scholar]
- 57.Jorgensen WL, Tirado-Rives J. Potential energy functions for atomic-level simulations of water and organic and biomolecular systems. Proc. Natl Acad. Sci. USA. 2005;102:6665–6670. doi: 10.1073/pnas.0408037102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dodda LS, Vilseck JZ, Tirado-Rives J, Jorgensen WL. 1.14*CM1A-LBCC: localized bond-charge corrected CM1A charges for condensed-phase simulations. J. Phys. Chem. B. 2017;121:3864–3870. doi: 10.1021/acs.jpcb.7b00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dodda LS, Cabeza de Vaca I, Tirado-Rives J, Jorgensen WL. LigParGen web server: an automatic OPLS-AA parameter generator for organic ligands. Nucleic Acids Res. 2017;45:331–336. doi: 10.1093/nar/gkx312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maciejewski A, Pasenkiewicz-Gierula M, Cramariuc O, Vattulainen I, Rog T. Refined OPLS all-atom force field for saturated phosphatidylcholine bilayers at full hydration. J. Phys. Chem. B. 2014;118:4571–4581. doi: 10.1021/jp5016627. [DOI] [PubMed] [Google Scholar]
- 61.Kulig W, Pasenkiewicz-Gierula M, Rog T. Cis and trans unsaturated phosphatidylcholine bilayers: a molecular dynamics simulation study. Chem. Phys. Lipids. 2016;195:12–20. doi: 10.1016/j.chemphyslip.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 62.Jorgensen WL, Maxwell DS, Tirado-Rives J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 1996;118:11225–11236. doi: 10.1021/ja9621760. [DOI] [Google Scholar]
- 63.Van Der Spoel D, et al. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 64.Essmann U, et al. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. doi: 10.1063/1.470117. [DOI] [Google Scholar]
- 65.Kang EC, Kataoka S, Kato K. Synthesis and properties of alkyl phosphorylcholine amphiphiles with a linear and an asymmetrically branched alkyl chain. B. Chem. Soc. Jpn. 2005;78:1558–1564. doi: 10.1246/bcsj.78.1558. [DOI] [Google Scholar]
- 66.Goetz JM, Schaefer J. REDOR dephasing by multiple spins in the presence of molecular motion. J. Magn. Reson. 1997;127:147–154. doi: 10.1006/jmre.1997.1198. [DOI] [PubMed] [Google Scholar]
- 67.Laurence TA, Fore S, Huser T. Fast, flexible algorithm for calculating photon correlations. Opt. Lett. 2006;31:829–831. doi: 10.1364/OL.31.000829. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Notes 1–7, Figs. 1–36, Tables 1–13 and references—see Contents page for details.
Data Availability Statement
The data that support the findings of this study are available on Zenodo public depository and from the corresponding author on reasonable request.