Abstract
The human pathogen Chlamydia trachomatis is an obligate intracellular bacterium with a unique developmental cycle. Within the host cell cytoplasm, it resides within a membrane-bound compartment, the inclusion. A distinguishing characteristic of the C. trachomatis life cycle is the fusion of the chlamydia-containing inclusions with each other in the host cell cytoplasm. We report that fusion of inclusions does not occur at 32°C in multiple mammalian cell lines and with three different serovars of C. trachomatis. The inhibition of fusion was inclusion specific; the fusion with sphingolipid-containing secretory vesicles and the interaction with early endosomes were unaffected by incubation at 32°C. The inhibition of fusion of the inclusions was not primarily the result of delayed maturation of the inclusion, as infectious progeny was produced in host cells incubated at 32°C, and the unfused inclusions remained competent to fuse up to 48 h postinfection. The ability to reverse the inhibition of fusion by shifting the infected cells from 32 to 37°C allowed the measurement of the rate and the time of fusion of the inclusions after entry of the bacteria. Most significantly, we demonstrate that fusion of inclusions with each other requires bacterial protein synthesis and that the required bacterial protein(s) is present, but inactive or not secreted, at 32°C.
Chlamydia trachomatis, a gram-negative bacterium and an obligate intracellular parasite, is a major human pathogen worldwide. It is the leading cause of sexually transmitted disease in the Western world and the main cause of noncongenital blindness in developing nations (28). During its life cycle, C. trachomatis alternates between two distinct morphological forms: the replicative, intracellular reticulate body (RB) and the infectious but metabolically inactive elementary body (EB). Following binding to an as-yet-unidentified receptor, the bacteria are internalized, enveloped within membrane-bound compartments (inclusions), and transported to a perinuclear location. When multiple bacteria enter a host cell, the C. trachomatis-containing inclusions fuse with each other, resulting in a single large inclusion. Within the inclusion, EBs differentiate into RBs 6 to 8 h postinfection (hpi); the RBs subsequently undergo binary fission to yield approximately 500 to 1,000 progeny. Between 24 and 72 hpi, the RBs differentiate back into EBs and are released into the extracellular space to start a new round of infection (22).
C. trachomatis presents a unique setting for the study of the mechanism and regulation of vesicular fusion events. Appropriate trafficking of this organism within the host cell is of the utmost importance for their survival. When chlamydiae are transported to a phagolysosome, as is often the case in polymorphonuclear leukocytes, the bacteria are rapidly destroyed (38). In epithelial cells, however, the inclusion does not acquire lysosomal markers (8, 14, 35, 36) but does fuse with trans-Golgi network (TGN)-derived sphingolipid-containing secretory vesicles (11, 12, 31). The inclusion also becomes surrounded with transferrin (Tf)-containing endosomes very early after infection (36), although no fusion with these endosomes has been detected (11). Other host cell trafficking events are not affected by infection with Chlamydia psittaci (6) or C. trachomatis (31, 35a).
While much has been learned recently about the fusion of the C. trachomatis inclusion with TGN-derived vesicles (reviewed in reference 10), less is known about the role, regulation, and mechanism of the unique homotypic fusion of C. trachomatis inclusions. In electron microscopy studies, fusion has been observed to occur within several hours after entry into the host cell (15), but when fusion was quantitated more carefully, host cells could still be seen to contain multiple inclusions as late as 15 hpi, and fusion was not complete until 25 hpi (21). The exact time at which fusion initiated and the rate at which the inclusions fused with each other were not determined. Interpretation of these studies was further limited by the asynchronous nature of the chlamydial life cycle. Others have demonstrated that inclusions of one serovar can fuse with vesicles containing newly entered bacteria of a different serovar up to 24 hpi (25). Interestingly, the individual bacteria of the two serovars remained segregated within the inclusion without any evidence of fusion.
The cellular requirements for the fusion of inclusions vary for different C. trachomatis serovars. In the presence of the microfilament-disrupting drug cytochalasin D, perinuclear transport and fusion of the inclusions of the L2 serovar of lymphogranuloma venereum (LGV) was greatly delayed but the fusion of serovar E-containing inclusions was unaffected (29). In coinfection experiments, Ridderhof and Barnes found a decrease in the number of inclusions containing both serovar E and serovar F after treatment with cytochalasin D, indicating a microfilament-dependent step in the fusion of these inclusions (25).
In summary, many aspects of the fusion of inclusions remain unknown. The rate at which the inclusions fuse with each other, the time after entry of the bacteria at which fusion is initiated, the relationship of the fusion of inclusions with each other and with sphingolipid-containing secretory vesicles, and the possible involvement of bacterial proteins in the fusion process have not previously been described. A better understanding of the fusion events will lead to further insights into the regulation of the interaction of this membrane-bound compartment with the host cell and thus the mechanism by which the bacteria survive within the host. In this study, we exploited the unexpected observation that fusion of C. trachomatis-containing inclusions does not occur at 32°C to further investigate these questions. We show that the acquisition of sphingomyelin from the TGN and the colocalization of the inclusions with Tf-containing early endosomes are unaffected at this temperature. In a series of temperature shift experiments that allow synchronization of events, the time course of fusion was more precisely defined. Finally, we provide evidence that fusion of inclusions requires the synthesis of one or more bacterial proteins that are present, but inactive or not secreted, at 32°C.
MATERIALS AND METHODS
Cell culture.
HeLa cells (ATCC CCL2) were maintained in DMEM-5 (Dulbecco’s modified Eagle’s medium H-16 supplemented with 5% fetal bovine serum [FBS; GIBCO BRL, Bethesda, Md.]). L-929 cells (ATCC CCL1) were maintained in RPMI 1640 supplemented with 10% FBS. McCoy cells (ATCC CRL1696) were maintained in minimal essential medium supplemented with 10% FBS. CHO-K1 cells (a kind gift of Kentaro Hanada, National Institute of Infectious Diseases, Tokyo, Japan) were maintained in Ham’s F-12 medium supplemented with 10% FBS. HeLa, L-929, and McCoy cells were incubated at 37°C, in an atmosphere of 5% CO2, and propagated every 3 days. CHO-K1 cells were incubated at 32°C, in an atmosphere of 5% CO2, and propagated every 3 to 5 days. All media were purchased from the UCSF Cell Culture Facility (San Francisco, Calif.).
Growth of C. trachomatis.
The LGV L2 and mouse pneumonitis (MoPn) strains were propagated as previously described (36). Serovar E was kindly supplied by Priscilla Wyrick (University of North Carolina, Chapel Hill). Unless otherwise indicated, all experiments were carried out with the LGV L2 serovar. Infections of L2 and MoPn were carried out by adding bacteria, suspended in 200 μl of medium appropriate for the cell line being infected, to host cells grown on 12-mm glass coverslips at a multiplicity of infection sufficient to infect approximately 80% of the host cells per monolayer. For one set of experiments (see Fig. 2), 0.1 μl of EBs resulted in 80% infection. Host cells were incubated at 37°C for 1 h, after which the inoculum was removed, and the host cells were washed with medium. One milliliter of medium was added, and the host cells were incubated at either 32 or 37°C. For some experiments, heparin (0.5 mg/ml) was included in the medium to prevent reinfection (39). Infections with serovar E were performed identically, except that the host cells and bacteria were centrifuged at room temperature for 5 min at 1,900 × g prior to incubation at 37°C. Unless stated otherwise, the bacteria and host cells were visualized by fixation with cold methanol for 10 min, followed by staining with a fluorescein isothiocyanate (FITC)-conjugated anti-Chlamydia antibody and Evans blue as the counterstain (Meridian Diagnostic, Cincinnati, Ohio).
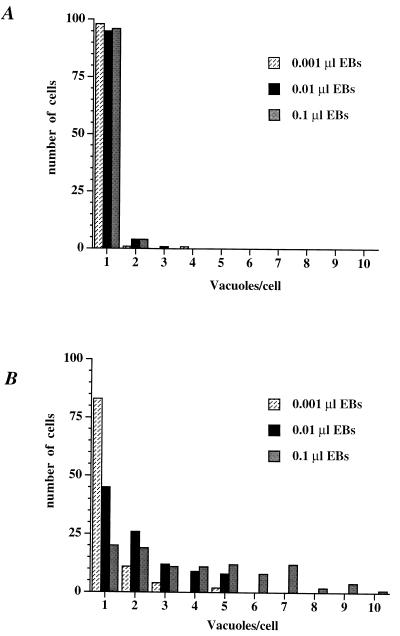
FIG. 2.
The number of C. trachomatis-containing inclusions per host cell increases as the inoculum size increases. L-929 cells were infected with C. trachomatis with the indicated inoculum for 1 h at 37°C. The host cells were subsequently incubated at 37°C (A) or 32°C (B) for 26 h. The chlamydiae and the host cells were visualized as described in the legend of Fig. 1. The number of inclusions per host cell was determined for 100 infected cells for each inoculum size at each temperature. The number of infected cells containing 1 to 10 inclusions is illustrated. The experiment was carried out twice with similar results.
Labeling of host cells with fluorescent ceramide or Tf.
HeLa cells were infected with L2 LGV for 1 h at 37°C and incubated at 32°C for 26 h. To label host cells with C6-NBD-ceramide [6-((N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)hexanoyl)sphingosine; Molecular Probes, Eugene, Oreg.], infected cells were washed with cold phosphate-buffered saline and C6-NBD-ceramide–bovine serum albumin complex was added to the infected host cells for 30 min at 4°C (23). The host cells were washed with cold phosphate-buffered saline and minimal essential medium containing 5% defatted bovine serum albumin (Sigma Chemical Co., St. Louis, Mo.) was added. After incubation at 32°C for 2 h, the samples were mounted on microscope slides and viewed on a Bio-Rad 600 confocal microscope.
To label C. trachomatis-infected HeLa cells with FITC-Tf, the infected cells were first incubated in the presence of serum-free DMEM-5 for 1 h at 32°C. This was replaced with serum-free DMEM-5 containing 30 μg of FITC-Tf (Molecular Probes) per ml, and the infected cells were incubated at 32°C for 1 h. The infected cells were fixed according to the pH shift protocol of Bacallao and Stelzer (1) with 4% paraformaldehyde in 80 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)]-KOH (pH 6.5), 5 mM EGTA, and 2 mM MgCl2 for 5 min, followed by fixation with 4% paraformaldehyde in 100 mM sodium borate (pH 11) for 10 min, and stained for C. trachomatis by utilizing the L2-I45 monoclonal antibody (a kind gift of Harlan Caldwell, NIH Rocky Mountain Laboratory) directly conjugated to Texas red according to the manufacturer’s instructions (Molecular Probes). The samples were mounted with 1.0% p-phenylenediamine in 90% glycerol (pH 8.2) and viewed by confocal microscopy with the Bio-Rad 600 confocal microscope.
Quantification of viable progeny.
HeLa cells grown in 12-well dishes were infected for 1 h with LGV L2. The inoculum was aspirated, and the infected host cells were incubated at 37°C in an atmosphere of 5% CO2 for 24 to 72 h. The infected host cells were removed with a cell scraper, and the well was washed once with DMEM-5. The host cells were pelleted in a microcentrifuge for 10 min at 17,000 × g. The host cells were washed once with medium, pelleted again, and lysed by drawing them sequentially through a 22-gauge and a 27-gauge needle. Two different aliquots of the lysate were diluted in DMEM-5 and used to infect HeLa cells plated on coverslips for 1 h. The fraction of these HeLa cells that became infected was determined 18 to 24 h later to determine the relative amount of infectious progeny. All experiments were performed in triplicate, and a minimum of 300 infected cells were counted per sample.
Inhibition of protein synthesis.
To inhibit bacterial protein synthesis, 50 μg of chloramphenicol (CAM; Sigma) per ml was added to the culture medium at the indicated time. Drug removal was accomplished by washing the HeLa cells three times with DMEM-5. This process was repeated twice at 1-h intervals to fully remove the CAM.
Statistical analysis.
All experiments were performed in triplicate, and data are shown as means ± 1 standard deviation from a representative of two to four independent experiments. Statistical analysis was performed by using the two-tailed Student t test with unequal variance.
RESULTS
Host cells contain multiple chlamydial inclusions at 32°C.
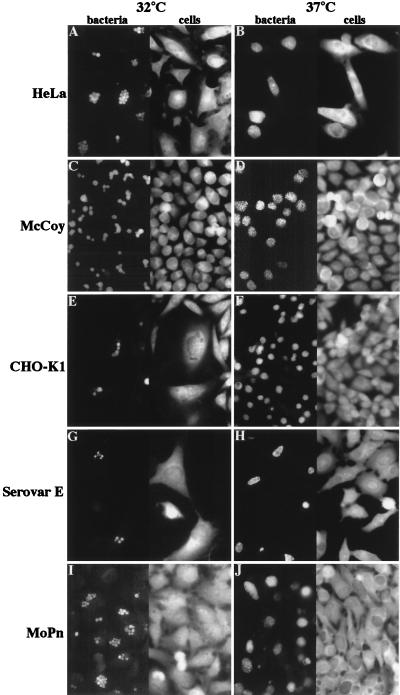
The LGV L2 serovar of C. trachomatis is routinely propagated in cell culture at 37°C. Surprisingly, when HeLa cells were infected with the L2 serovar of LGV for 1 h at 37°C and incubated at 32°C for 26 h, multiple inclusions were observed in many infected cells (Fig. 1A). In contrast, HeLa cells infected under identical conditions but incubated at 37°C predominantly contained single inclusions (Fig. 1B). To ascertain that this observation was not cell line specific, a mouse fibroblast cell line (McCoy) and a hamster ovarian epithelial cell line (CHO-K1) were infected at 37°C for 1 h and incubated for 26 h at 32 or 37°C. Figure 1 illustrates that these cell lines also contained multiple inclusions after incubation at 32°C but not at 37°C. The inclusions detected after incubation at 32°C appeared uniform in size, in contrast to the heterogeneously sized inclusions observed in infected cells grown at 37°C. Additionally, all inclusions in the host cells incubated at 32°C were positioned adjacent to the nucleus, comparable with the position of the inclusions in infected cells grown at 37°C. The number of inclusions seen within the host cells grown at 32°C varied from 1 to more than 10. The range in number of inclusions was most pronounced for HeLa cells; other cell lines usually showed a more moderate number of inclusions, but nonetheless contained multiple inclusions, all of approximately identical size.
FIG. 1.
The lack of fusion of C. trachomatis-containing inclusions at 32°C is not cell line or serovar specific. Three different cell lines, HeLa (A and B), McCoy (C and D), and CHO-K1 (E and F), were infected with the LGV L2 serovar of C. trachomatis for 1 h at 37°C and incubated at either 32°C (A, C, and E), or 37°C (B, D, and F) for 26 h. To test if inhibition of fusion was observed for multiple serovars, HeLa cells were infected for 1 h at 37°C with serovar E (G and H) and the MoPn strain (I and J) as described in Materials and Methods. The HeLa cells were subsequently incubated at 32°C (G and I) or 37°C (H and J) for 26 h. After fixation, the bacteria were visualized with FITC-conjugated anti-Chlamydia antibodies, and the host cells were visualized with Evan’s blue. The left-hand side of each panel shows the anti-Chlamydia staining, and the right-hand side of the panel shows the Evan’s blue staining of the same field of C. trachomatis-infected cells.
As inclusion formation differs even within Chlamydia species, the ability of two other C. trachomatis serovars to form multiple inclusions at 32°C was determined. HeLa cells were infected with either the MoPn or E serovar followed by incubation at either 32 or 37°C. Both serovars behaved identically to LGV: many infected cells contained multiple inclusions when incubated 32°C (Fig. 1G and I) but only a single large inclusion when incubated at 37°C (Fig. 1H and J). The formation of multiple inclusions at 32°C is thus not limited to the LGV serovar. Of note, serovar E is often propagated at 35°C, and a single inclusion is usually observed at this temperature (9).
Formation of multiple inclusions results from an inhibition of fusion of the inclusions.
The finding of multiple inclusions in C. trachomatis-infected cells grown at 32°C raises the possibility that they arise from a combination of multiple entry events followed by division of the inclusions, as is the case for the C. psittaci guinea pig inclusion conjunctivitis strain (26). If the inclusions divide, formation of multiple inclusions should be independent of the number of bacteria that enter a host cell; if the multiple inclusions derive from multiple entry events, the percentage of host cells containing more than one inclusion should increase with increasing inoculum size. To differentiate between these two possibilities, L cells were infected at 37°C over a 100-fold range of inocula and then incubated at 32 or 37°C for 26 h. As illustrated in Fig. 2, the number of host cells containing multiple inclusions at 32°C was dependent on the size of the inoculum; in contrast, at 37°C, multiple inclusions were not observed. Hence, the multiple inclusions formed at 32°C do not derive from division of an inclusion but instead result from multiple entry events.
Inhibition of fusion of inclusions does not prevent acquisition of sphingomyelin from the TGN or association with endosomes.
The lack of fusion of inclusions observed at 32°C could result from a general inhibition of interactions of the inclusion with host cell compartments or could be specific to homotypic fusion of inclusions. The only host cell compartment with which the C. trachomatis inclusion has been demonstrated to fuse is sphingolipid-containing TGN-derived secretory vesicles (11, 12, 31). In addition, we have demonstrated that there is a close interaction of the inclusion with Tf-containing endosomes (36). Therefore, we assessed the effect of temperature on the interaction of the inclusion with these two compartments.
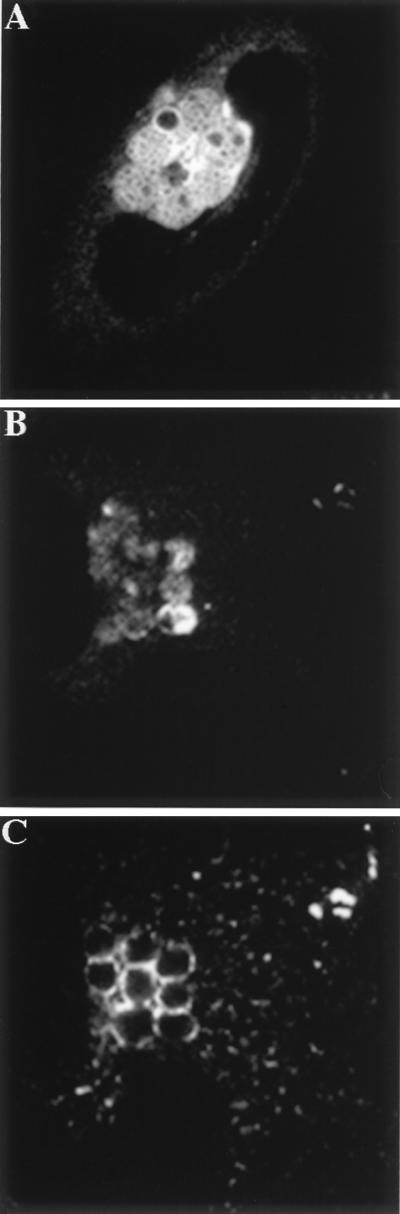
To determine if unfused inclusions could fuse with sphingolipid-containing vesicles at 32°C, HeLa cells were infected with LGV for 1 h at 37°C followed by incubation at 32°C for 26 h. Infected cells were labeled with the fluorescent Golgi complex marker C6-NBD-ceramide and observed by confocal microscopy. The fluorescent lipid was clearly detected within the bacterial envelope (Fig. 3A), indicating that fusion of inclusions with sphingolipid-containing secretory vesicles still occurred under conditions that otherwise inhibited fusion of inclusions.
FIG. 3.
Unfused inclusions retain the ability to fuse with sphingolipid-containing secretory vesicles and associate with Tf-containing endosomes at 32°C. HeLa cells were infected with C. trachomatis for 1 h at 37°C and incubated at 32°C for 26 h. (A) The host cells were labeled with C6-NBD-ceramide for 30 min at 4°C and subsequently incubated at 32°C for 2 h to allow the ceramide to be processed and transported to the inclusion and the plasma membrane. The samples were subsequently mounted and viewed by confocal microscopy. (B and C) The infected cells were labeled with FITC-Tf for 1 h, and then fixed and stained for chlamydiae. The samples were then viewed by confocal microscopy. Panel B shows an infected HeLa cell stained with an antibody to C. trachomatis, and panel C shows the distribution of FITC-Tf in the same cell.
The interaction of the chlamydial inclusion with early endosomes at 32°C was also investigated. HeLa cells were infected at 37°C for 1 h and incubated at 32°C for 26 h. Infected cells were labeled with FITC-Tf at 32°C for 1 h, fixed, and stained for chlamydiae. As shown in Fig. 3B and C, the unfused inclusions were surrounded by FITC-Tf-containing endosomes. Together these results indicate that incubation at 32°C specifically inhibits fusion of inclusions with each other but does not affect their interactions with secretory or endocytic vesicles.
Fusion of inclusions is not observed even at late times in the chlamydial developmental cycle.
In the experiments described so far, fusion of inclusions at the nonpermissive temperature was monitored only up to 26 hpi. Therefore, the lack of fusion of the inclusions could be due to a retardation of inclusion maturation or chlamydial development. Indeed, the inclusions observed after a 26-h incubation at 32°C were smaller than those seen at 37°C.
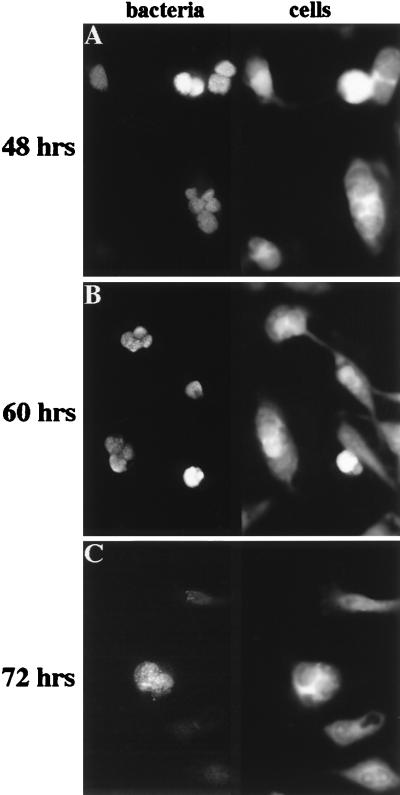
To determine if the inclusions fused at later time points in the developmental cycle, HeLa cells were infected at 37°C for 1 h and incubated at either 32 or 37°C for 48, 60, and 72 h in the presence of heparin, which prevents reinfection with progeny released during the course of the experiment (39). Heparin itself does not inhibit fusion when added after infection (data not shown). At 48 and 60 hpi, a large fraction of the infected host cells grown at 32°C contained multiple inclusions (Fig. 4A and B). The inclusions seen at these time points were also of approximately identical size. Even as late as 72 hpi, multiple inclusions could be detected in HeLa cells incubated at 32°C (Fig. 4C), although at this time point, no host cell was found to contain more than three inclusions. A possible explanation for this observation is that many infected host cells have already lysed at late time points; infected cells containing a high number of inclusions may rupture earlier, lowering the average number of inclusions per infected cell at these late time points, even in the absence of fusion of the inclusions. Alternatively, some fusion may occur very late in the developmental cycle.
FIG. 4.
C. trachomatis-containing inclusions remain unfused at late time points in the developmental cycle at 32°C. HeLa cells were infected with C. trachomatis for 1 h at 37°C and incubated at 32°C for 48 h (A), 60 h (B), or 72 h (C) in the presence of 0.5 mg of heparin per ml to prevent reinfection at later times during the experiment. C. trachomatis and host cells were visualized as described in the legend to Fig. 1.
To further investigate the growth and proliferation of the bacteria at 32°C, the amount of infectious progeny recovered from infected cells incubated at 32°C and 37°C was compared. While the onset of production of viable progeny was delayed in host cells incubated at 32°C compared to that in cells incubated at 37°C and the total amount of progeny appeared to decrease by approximately 50%, the bacteria were nonetheless capable of proliferating at 32°C (data not shown). These findings suggest that the C. trachomatis intracellular life cycle can proceed to completion at 32°C, albeit more slowly. The formation of inclusions demonstrates that the EBs can transform to RBs and proliferate at 32°C. Furthermore, the recovery of viable progeny indicates that the bacteria can transform from RBs back to EBs. Thus, the inhibition of fusion at lower temperatures is not due to a failure of the bacteria to grow or to complete the intracellular developmental cycle. Rather, the fusion process itself appears to be a temperature-sensitive step in the chlamydial life cycle.
Inclusions remain fusogenic for at least 48 hpi.
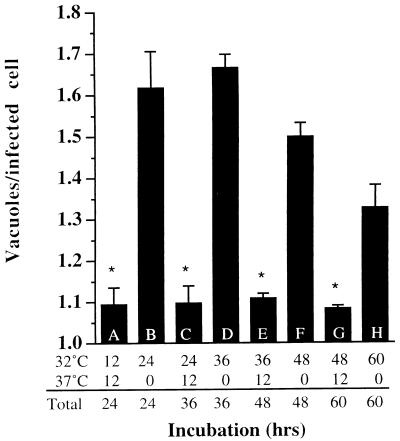
It was shown previously that up to 24 hpi, superinfection of C. trachomatis serovar F-infected cells with serovar E resulted in fusion of the different serovar-containing inclusions (25). The ability to induce fusion by shifting the cells from low to high temperature allowed us to test whether two preexisting inclusions of the same serovar could fuse at late times postinfection. Duplicate samples of HeLa cells were infected with C. trachomatis for 1 h and incubated at 32°C in the presence of heparin to prevent reinfection with bacteria released during the course of the experiment. After incubation for 12, 24, 36, and 48 h at 32°C, one set was transferred to 37°C for 12 h to allow fusion to occur, whereas an identical set was incubated at 32°C for an additional 12 h. The average number of inclusions per infected cell at each time point is shown in Fig. 5. At 12, 24, 36, and 48 hpi, shifting the infected cells to 37°C for 12 h decreased the average number of inclusions per cell compared to the control sample that had been kept at 32°C for an identical duration of infection (compare data for samples A and B, C and D, E and F, and G and H) with a certainty of a value of P < 0.005 for each pair (two-tailed Student t test). The inclusions thus remain competent to fuse late in the developmental cycle. As shown in Fig. 4, the number of inclusions per cell observed after 60 h incubation at 32°C had decreased slightly compared to that at 24 h (P < 0.02, two-tailed Student t test).
FIG. 5.
Inclusions retain the ability to fuse late in infection. HeLa cells were infected with C. trachomatis LGV L2 for 1 h at 37°C and incubated at 32°C in the presence of 0.5 mg of heparin per ml (to inhibit reinfection [39]) for the time indicated. Samples A, C, E, and G were transferred to 37°C after the indicated time for 12 h. Samples B, D, F, and H were incubated at 32°C continuously for the indicated time. After the total incubation time shown, the host cells were fixed and stained for C. trachomatis, as described in the legend to Fig. 1. Each value represents the mean of three determinations (200 cells counted for three slides each), and error bars represent 1 standard deviation. This experiment was carried out twice with similar results. Asterisks represent significant differences (P < 0.02) for the values of samples A and C compared to B, samples C and E compared to D, samples E and G compared to F, and sample G compared to H.
Fusion of inclusions is a gradual process that occurs between 10 and 16 hpi.
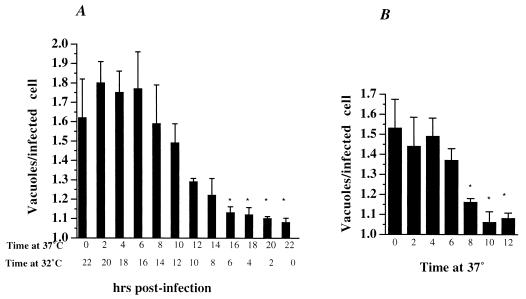
While fusion can be detected early after entry by electron microscopy, at least one group has reported observing host cells that contain multiple inclusions 15 hpi (21). In that study, a majority of the inclusions had fused by 25 hpi. The ability to manipulate the time of inclusion fusion by a temperature shift allowed us to more precisely define the time at which the inclusions fuse. HeLa cells were infected for 1 h and incubated at 37°C for 2 to 22 h, followed by further incubation at 32°C. When the total infection time reached 22 h, the host cells were fixed and stained for chlamydiae, and the distribution of the number of inclusions per infected host cell was determined. As shown in Fig. 6A, the average number of inclusions per infected host cell was first observed to decrease after 10 h at 37°C but did not approach a statistically significant difference (P < 0.05 compared to 0 h at 37°C) or an average of one per infected cell until 16 hpi. These results indicate that a majority of fusion of inclusions occurs between 10 and 16 hpi, a finding which confirms data in other published reports (15, 21).
FIG. 6.
Fusion of vacuoles is not initiated until 10 hpi and is a gradual process. (A) HeLa cells were infected with C. trachomatis for 1 h at 37°C and then incubated at 37°C for the time indicated. To block further fusion of the inclusions but still allow the bacteria to proliferate, the infected cells were transferred to 32°C at the times indicated. After a total infection time of 22 h, the host cells were fixed and stained for C. trachomatis as described in the legend to Fig. 1. (B) HeLa cells were infected with C. trachomatis for 1 h at 37°C and incubated at 32°C for 20 h. The infected cells were shifted to 37°C to initiate fusion of the inclusions and subsequently fixed and stained as before at the indicated time. Each value represents the mean of three determinations (200 cells counted for three slides each), and error bars represent 1 standard deviation. This experiment was carried out twice with similar results. Asterisks represent significant differences (P < 0.05) compared to the zero time value.
Previous investigations have not determined whether the late onset of fusion of inclusions is a result of the slow fusion of inclusions or a rapid process that is not initiated until later in the chlamydial developmental cycle. To distinguish between these two possibilities, the rate of fusion of mature inclusions was determined. HeLa cells were infected with C. trachomatis for 1 h and incubated at 32°C for 24 h. One sample was fixed at this time point to determine the average number of inclusions per host cell at this time. The remaining samples were shifted to 37°C for 2 to 12 h, after which they were fixed, and the average number of inclusions per infected host was determined. As shown in Fig. 6B, fusion of preexisting inclusions appears to be a gradual process. Not until 8 h after the temperature shift did the decrease in average inclusion per cell reach statistical significance (P < 0.05 compared to 0 h at 37°C), which corresponded to the levels normally seen for infected cells incubated continuously at 37°C.
Fusion of inclusions requires synthesis of one or more chlamydial proteins that is inactive at 32°C.
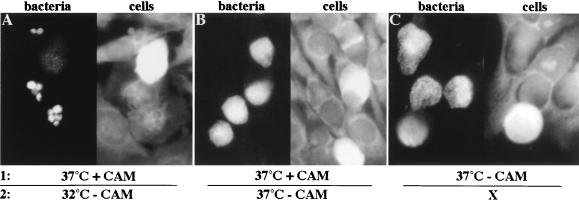
All known membrane fusion events in the host cell are tightly regulated to avoid a spurious joining of two membranes (for a review, see reference 27). Fusion of inclusions could be dependent on host cell factors or could be controlled by the bacteria themselves. To determine if bacterial protein synthesis was required for fusion of the inclusions, HeLa cells were infected with C. trachomatis for 1 h and incubated at 37°C for 3 h to allow the bacteria to aggregate in the peri-Golgi region. Bacterial protein synthesis was subsequently inhibited by the addition of CAM, and the infected cells were placed at 37°C for 20 h. To determine the amount of fusion that had occurred when bacterial protein synthesis was inhibited, the CAM was removed, and the infected host cells were incubated at 32°C for 26 h. This allowed the bacteria to grow in the absence of any further fusion of inclusions. As shown in Fig. 7A, many infected host cells still contained multiple inclusions after incubation at 32°C. Thus, no fusion of the inclusions had occurred during the 20-h incubation at 37°C. When infected host cells were incubated at 37°C after the removal of CAM, they contained only single inclusions (Fig. 7B). HeLa cells infected and grown at 37°C in the absence of CAM for 20 h likewise harbored only single inclusions (Fig. 7C). Infected host cells treated with CAM had no detectable bacterial growth after 20 h at 37°C (data not shown). Although quantitation of the fusion of inclusions was not possible due to the poor recovery of the bacteria after drug removal (32), these findings nonetheless indicate that bacterial protein synthesis is required for the fusion of chlamydial inclusions.
FIG. 7.
Fusion of inclusions requires bacterial protein synthesis. HeLa cells were infected with C. trachomatis at 37°C for 1 h. After 3 h, 50 μg of CAM per ml (A and B) or an equivalent amount of ethanol (C) was added to the samples, and they were incubated at 37°C for an additional 20 h. One sample was fixed and stained for chlamydiae to determine the amount of fusion of inclusions that had occurred in the absence of CAM (C). In the remaining samples, the drug was removed, and the cells were incubated at 32°C (A) or 37°C (B) for 26 h. The host cells and the bacteria were visualized as in Fig. 1.
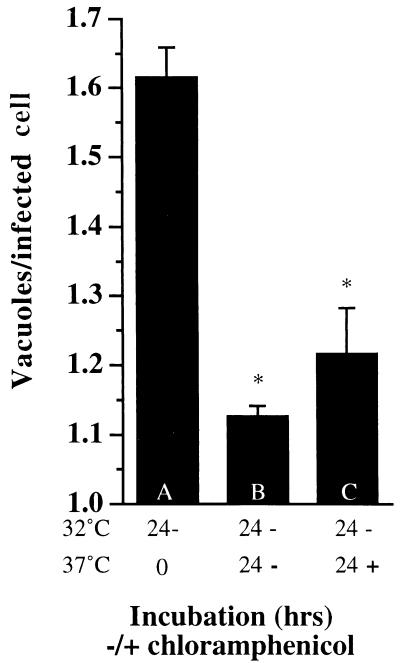
The lack of fusion at 32°C could be the result of the absence of the required chlamydial protein or the consequence of its inability to function or be localized correctly at 32°C. To differentiate between these two possibilities, HeLa cells were infected at 37°C for 1 h and incubated at 32°C for 24 h. A sample was fixed and stained to determine the average number of inclusions per infected cell after this time (Fig. 8, sample A). The remaining two samples were subsequently incubated at 37°C in the absence (sample B) or presence (sample C) of CAM to inhibit further bacterial protein synthesis. After 24 h at 37°C, the host cells were fixed and stained for chlamydiae, and the average number of inclusions per infected host cell was determined. If the protein(s) required for the fusion of inclusions is inactive but present at 32°C, fusion should be observed upon shifting of the infected cells to 37°C in the presence of CAM. Alternatively, if the required protein(s) is not synthesized at 32°C, it will likewise not be produced in the presence of CAM, and fusion should not occur even at 37°C when CAM is added. Therefore, there will be no change in the average number of inclusions compared to the infected cells fixed at the time of the temperature shift. As Fig. 8 demonstrates, the average number of inclusions per infected cell in the sample treated with CAM upon shift to 37°C does decrease greatly (sample C, P < 0.002 compared with sample A), falling almost to the level of infected cells incubated at 37°C without addition of CAM (sample B, P < 0.002 compared with sample A). The difference in the average number of vacuoles between samples B and C did not reach statistical significance (P > 0.14). Thus, even without ongoing bacterial protein synthesis, fusion between preexisting, mature inclusions can occur. The protein(s) responsible for the fusion must have been present on the surface of the inclusion at 32°C, because bacterial protein synthesis, but not fusion of the inclusions, was inhibited during the incubation at 37°C. Alternatively, the protein is produced and functional at 32°C but is not secreted properly until the infected cells are incubated at 37°C. The finding that restoration of fusion of the inclusions is not fully complete may reflect a short half-life of the protein(s) or a lower level of expression at 32°C that is insufficient to promote complete fusion.
FIG. 8.
Preexisting inclusions do not require bacterial protein synthesis for fusion. HeLa cells were infected with C. trachomatis for 1 h at 37°C and incubated at 32°C for 24 h. Sample A was fixed and stained for C. trachomatis at this time. Samples B and C were further incubated at 37°C for 24 h in the absence (B) or presence (C) of CAM. The bacteria and host cells were visualized as described in the legend to Fig. 1. Each value represents the mean of three determinations (200 cells counted for three slides each), and error bars represent 1 standard deviation. Asterisks represent significant differences (P < 0.002) compared to the value for sample A.
DISCUSSION
In this study, we further investigated the fusion of C. trachomatis-containing inclusions with each other. Our unexpected observation that this fusion event is inhibited at 32°C allowed us to probe several properties, such as the rate and the involvement of bacterial proteins, of this homotypic fusion event. Since all infections were initiated at 37°C, the formation of multiple inclusions at 32°C is not the result of a change in the entry mechanism at low temperatures but is instead due to an inhibition of a step in the fusion process. Although we did not determine the minimal temperature at which fusion occurs, several strains of C. trachomatis (including serovar E) are propagated at 35°C, and the inclusions fuse at that temperature (9). Thus, a temperature shift of as little as 3°C can affect the fusion of inclusions. While this is surprising, there is precedence for very small temperature shifts altering the function of a protein (for example, see reference 17), although usually the protein is inactive at the higher temperature. Effects of low-temperature incubation on membrane transport have also been reported. At 30°C, the redistribution of Golgi markers into the endoplasmic reticulum in response to brefeldin A is inhibited (30). It was not reported if this process was affected at 32°C as well.
We demonstrate that the presence of multiple inclusions at 32°C is likely to be the result of an inability of the inclusions to fuse, as increasing the inoculum size also increased the number of inclusions per infected cell. This contrasts with the formation of the multilobed inclusions characteristic of the C. psittaci guinea pig inclusion conjunctivitis strain, an event which is independent of the inoculum size (26). The number of inclusions fluctuated greatly from host cell to host cell, consistent with the notion that the number of bacteria that enter a host cell varies greatly within a population of host cells. Therefore, it is likely that the difference in inclusion sizes observed in infected cells grown at 37°C is the result of entry of different numbers of bacteria into a single host cell, followed by fusion to form inclusions of different sizes.
Careful measurements of the kinetics of the fusion of inclusions indicated that the fusion of inclusions is a gradual process that is not initiated until 10 hpi and is not fully completed until 16 to 20 hpi. This finding is in contrast to other reports that showed evidence of fusion of the inclusions shortly after infection (15). Those experiments utilized very large inocula in order to visualize the bacteria, which could have led to an increase in the fusion rate of the inclusions due to the high concentration of bacteria inside the cells. Our data complement those of Matsumoto and colleagues (21), who showed that at 15 hpi, many host cells contained multiple inclusions, but that at 25 hpi, these had fused to form single inclusions.
Two lines of evidence suggest that the bacteria at least partly mediate the fusion of inclusions with each other. We observed very little fusion prior to the conversion of EBs to RBs (6 to 8 hpi), the time at which most bacterial protein synthesis is initiated. Additionally, inhibition of bacterial protein synthesis after the aggregation of the inclusions prevented fusion of the inclusions. Thus, four critical steps in the C. trachomatis life cycle require bacterial protein synthesis: the aggregation of inclusions in the perinuclear region (20), the fusion of inclusions, the inhibition of lysosomal fusion (32), and the acquisition of sphingolipids (32). The bacteria clearly play an active role in their intracellular transport within the host cell and in the interaction of the vacuole with other membrane-bound compartments within the host cell. How the bacteria mediate all these functions is still unclear, but the recent discovery in the C. trachomatis and C. psittaci genomes of orthologs encoding putative homologs of the type III secretion system provides a potential mechanism for the secretion of bacterial proteins into the inclusion and the host cell (16, 34). Possibly, bacterial proteins directly interact with the host cell to mediate these functions, or host cell factors are recruited to the inclusion by bacterial proteins, which then further regulate the fusion and transport of the vacuole. Indeed, both dynein (5) and members of the annexin family (19) have been found to colocalize with the vacuole.
The role of fusion of the inclusions in the C. trachomatis life cycle remains a matter of speculation. The bacteria may have a growth advantage when the inclusion becomes more spacious. In addition, fusion of the inclusions may facilitate genetic recombination between individual RBs within the inclusion. Genetic exchange between extracellular EBs is unlikely, given their lack of close proximity to each other, their rigid outer membrane, and their lack of detectable metabolic activity. Indeed, polymorphisms within the omp1 gene, the gene encoding the major outer membrane protein of Chlamydia, have been described for different C. trachomatis serovars that could be explained by recombination within one or between two different serovars (3, 7, 13, 18, 24, 33, 37). In order for exchange of genetic material to occur, a host cell has to become superinfected with two different serovars of C. trachomatis. This has been observed both in a tissue culture model (25) and in human settings (2). As the outer membrane protein is a major target for the human immune response, alteration of its epitopes through recombination may play an important role in the interaction and survival of the bacterium within its eukaryotic host (4).
Our finding that the bacteria synthesize a protein that promotes this fusion points to an evolutionarily conserved function. In the future, a comparison of the genomes of Chlamydia species that include or exclude fusion of the inclusions during their developmental cycle may be informative. The finding that C. trachomatis inclusions fail to fuse with each other at 32°C will be a useful tool for dissecting this fusion event. Further studies investigating the role of host cell factors such as microfilaments, microtubules, Ca2+, and annexins in the fusion process are currently under way.
ACKNOWLEDGMENTS
We thank Keith Mostov, Frances Brodsky, Richard Stephens, and members of the Engel lab for suggestions and support. We also thank Priscilla Wyrick for supplying serovar E, Kentaro Hanada for sharing the CHO-K1 cells, and Harlan Caldwell for the L2-I45 antibody.
This work was supported by National Institutes of Health grants RO1 AI24436, R21 AI38271, and KO4 01348.
REFERENCES
- 1.Bacallao R, Stelzer E H. Preservation of biological specimens for observation in a confocal fluorescence microscope and operational principles of confocal fluorescence microscopy. Methods Cell Biol. 1989;31:437–452. doi: 10.1016/s0091-679x(08)61621-0. [DOI] [PubMed] [Google Scholar]
- 2.Barnes R C, Rompalo A M, Stamm W E. Comparison of Chlamydia trachomatis serovars causing rectal and cervical infections. J Infect Dis. 1987;156:953–958. doi: 10.1093/infdis/156.6.953. [DOI] [PubMed] [Google Scholar]
- 3.Brunham R, Yang C, Maclean I, Kimani J, Maitha G, Plummer F. Chlamydia trachomatis from individuals in a sexually transmitted disease core group exhibit frequent sequence variation in the major outer membrane protein (omp1) gene. J Clin Investig. 1994;94:458–463. doi: 10.1172/JCI117347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brunham R C, Plummer F A, Stephens R S. Bacterial antigenic variation, host immune response, and pathogen-host coevolution. Infect Immun. 1993;61:2273–2276. doi: 10.1128/iai.61.6.2273-2276.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clausen J D, Christiansen G, Holst H U, Birkelund S. Chlamydia trachomatis utilizes the host cell microtubule network during early events of infection. Mol Microbiol. 1997;25:441–449. doi: 10.1046/j.1365-2958.1997.4591832.x. [DOI] [PubMed] [Google Scholar]
- 6.Eissenberg L G, Wyrick P B. Inhibition of phagolysosome fusion is localized to Chlamydia psittaci-laden vacuoles. Infect Immun. 1981;32:889–896. doi: 10.1128/iai.32.2.889-896.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fitch W M, Peterson E M, de la Maza L M. Phylogenetic analysis of the outer membrane-protein genes of Chlamydiae, and its implication for vaccine development. Mol Biol Evol. 1993;10:892–913. doi: 10.1093/oxfordjournals.molbev.a040048. [DOI] [PubMed] [Google Scholar]
- 8.Friis R R. Interaction of L cells and Chlamydia psittaci: entry of the parasite and host responses to its development. J Bacteriol. 1972;110:706–721. doi: 10.1128/jb.110.2.706-721.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gordon F H, Quan A L. Occurrence of glycogen in inclusions of the psittacosis-lymphogranuloma venereum-trachoma agent. J Infect Dis. 1965;120:186–196. doi: 10.1093/infdis/115.2.186. [DOI] [PubMed] [Google Scholar]
- 10.Hackstadt T, Fischer E R, Scidmore M A, Rockey D D, Heinzen R A. Origins and functions of the chlamydial inclusion. Trends Microbiol. 1997;5:288–293. doi: 10.1016/S0966-842X(97)01061-5. [DOI] [PubMed] [Google Scholar]
- 11.Hackstadt T, Rockey D, Heinzen R, Scidmore M. Chlamydia trachomatis interrupts an exocytic pathway to acquire endogenously synthesized sphingomyelin in transit from the Golgi apparatus to the plasma membrane. EMBO J. 1996;15:964–977. [PMC free article] [PubMed] [Google Scholar]
- 12.Hackstadt T, Scidmore M A, Rockey D D. Lipid metabolism in Chlamydia trachomatis-infected cells: directed trafficking of Golgi-derived sphingolipids to the chlamydial inclusion. Proc Natl Acad Sci USA. 1995;92:4877–4881. doi: 10.1073/pnas.92.11.4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayes L J, Yearsley P, Treharne J D, Ballard R A, Fehler G H, Ward M E. Evidence for naturally occurring recombination in the gene encoding the major outer membrane protein of lymphogranuloma venereum isolates of Chlamydia trachomatis. Infect Immun. 1994;62:5659–5663. doi: 10.1128/iai.62.12.5659-5663.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heinzen R A, Scidmore M A, Rockey D D, Hackstadt T. Differential interaction with endocytic and exocytic pathways distinguish parasitophorous vacuoles of Coxiella burnetii and Chlamydia trachomatis. Infect Immun. 1996;64:796–809. doi: 10.1128/iai.64.3.796-809.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hodinka R L, Davis C H, Choong J, Wyrick P B. Ultrastructural study of endocytosis of Chlamydia trachomatis by McCoy cells. Infect Immun. 1988;56:1456–1463. doi: 10.1128/iai.56.6.1456-1463.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsia R C, Pannekoek Y, Ingerowski E, Bavoil P M. Type III secretion genes identify a putative virulence locus of Chlamydia. Mol Microbiol. 1997;25:351–359. doi: 10.1046/j.1365-2958.1997.4701834.x. [DOI] [PubMed] [Google Scholar]
- 17.Iiri T, Herzmark P, Nakamoto J M, van Dop C, Bourne H R. Rapid GDP release from Gs alpha in patients with gain and loss of endocrine function. Nature. 1994;371:164–168. doi: 10.1038/371164a0. [DOI] [PubMed] [Google Scholar]
- 18.Lampe M F, Suchland R J, Stamm W E. Nucleotide sequence of the variable domains within the major outer membrane protein gene from serovariants of Chlamydia trachomatis. Infect Immun. 1993;61:213–219. doi: 10.1128/iai.61.1.213-219.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Majeed M, Ernst J D, Magnusson K-E, Kihlström E, Stendahl O. Selective translocation of annexins during intracellular redistribution of Chlamydia trachomatis in HeLa and McCoy cells. Infect Immun. 1994;62:126–134. doi: 10.1128/iai.62.1.126-134.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Majeed M, Gustafsson M, Kihlström E, Stendahl O. Roles of Ca2+ and F-actin in intracellular aggregation of Chlamydia trachomatis in eucaryotic cells. Infect Immun. 1993;61:1406–1414. doi: 10.1128/iai.61.4.1406-1414.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsumoto A, Bessho H, Uehira K, Suda T. Morphological studies of the association of mitochondria with chlamydial inclusions and the fusion of chlamydial inclusions. J Electron Microsc. 1991;40:356–363. [PubMed] [Google Scholar]
- 22.Moulder J W. Interaction of chlamydiae and host cells in vitro. Microbiol Rev. 1991;55:143–190. doi: 10.1128/mr.55.1.143-190.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pagano R E. A fluorescent derivative of ceramide: Physical properties and use in studying the Golgi apparatus of animal cells. Methods Cell Biol. 1989;29:75–85. doi: 10.1016/s0091-679x(08)60188-0. [DOI] [PubMed] [Google Scholar]
- 24.Poole E, Lamont I. Chlamydia trachomatis serovar differentiation by direct sequence analysis of the variable segment 4 region of the major outer membrane protein gene. Infect Immun. 1992;60:1089–1094. doi: 10.1128/iai.60.3.1089-1094.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ridderhof J C, Barnes R C. Fusion of inclusions following superinfection of HeLa cells by two serovars of Chlamydia trachomatis. Infect Immun. 1989;57:3189–3193. doi: 10.1128/iai.57.10.3189-3193.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rockey D D, Fischer E R, Hackstadt T. Temporal analysis of the developing Chlamydia psittaci inclusion by use of fluorescence and electron microscopy. Infect Immun. 1996;64:4269–4278. doi: 10.1128/iai.64.10.4269-4278.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rothman J. Mechanism of intracellular protein transport. Nature. 1994;372:55–63. doi: 10.1038/372055a0. [DOI] [PubMed] [Google Scholar]
- 28.Schachter J, Dawson C R. The epidemiology of trachoma predicts more blindness in the future. Scand J Infect Dis. 1990;69:55–62. [PubMed] [Google Scholar]
- 29.Schramm N, Wyrick P B. Cytoskeletal requirements in Chlamydia trachomatis infection of host cells. Infect Immun. 1995;63:324–332. doi: 10.1128/iai.63.1.324-332.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sciaky N, Presley J, Smith C, Zaal K, Cole N, Moriera J, Terasaki M, Siggia E, Lippincott-Schwartz J. Golgi tubule traffic and the effects of brefeldin A visualized in living cells. J Cell Biol. 1997;139:1137–1150. doi: 10.1083/jcb.139.5.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scidmore M A, Fischer E R, Hackstadt T. Sphingolipids and glycoproteins are differentially trafficked to the Chlamydia trachomatis inclusion. J Cell Biol. 1996;134:363–374. doi: 10.1083/jcb.134.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scidmore M A, Rockey D D, Fischer E R, Heinzen R A, Hackstadt T. Vesicular interactions of the Chlamydia trachomatis inclusion are determined by chlamydial early protein synthesis rather than route of entry. Infect Immun. 1996;64:5366–5372. doi: 10.1128/iai.64.12.5366-5372.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stephens R S, Sanchez-Pescador R, Wagar E A, Inouye C, Urdea M S. Diversity of Chlamydia trachomatis major outer membrane protein genes. J Bacteriol. 1987;169:3879–3885. doi: 10.1128/jb.169.9.3879-3885.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stephens R S, Kalman S, Fenner C, Davis R. Chlamydia Genome Project. 1997. [Google Scholar]
- 35.Taraska T, Ward D M, Ajioka R S, Wyrick P B, Davis-Kaplan S R, Davis C H, Kaplan J. The late chlamydial inclusion membrane is not derived from the endocytic pathway and is relatively deficient in host proteins. Infect Immun. 1996;64:3713–3727. doi: 10.1128/iai.64.9.3713-3727.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35a.van Ooij, C., and T. Engel. Unpublished observations.
- 36.van Ooij C, Apodaca G, Engel J. Characterization of the Chlamydia trachomatis vacuole and its interaction with the host endocytic pathway in HeLa cells. Infect Immun. 1997;65:758–766. doi: 10.1128/iai.65.2.758-766.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang C L, Maclean I, Brunham R C. DNA sequence polymorphism of the Chlamydia trachomatis omp1 gene. J Infect Dis. 1993;168:1225–1230. doi: 10.1093/infdis/168.5.1225. [DOI] [PubMed] [Google Scholar]
- 38.Yong E C, Chi E Y, Chen W-J, Kuo C-C. Degradation of Chlamydia trachomatis in human polymorphonuclear leukocytes: an ultrastructural study of peroxidase-positive phagolysosomes. Infect Immun. 1986;53:427–431. doi: 10.1128/iai.53.2.427-431.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang J P, Stephens R S. Mechanism of Chlamydia trachomatis attachment to eukaryotic host cells. Cell. 1992;69:861–869. doi: 10.1016/0092-8674(92)90296-o. [DOI] [PubMed] [Google Scholar]