Abstract
Introduction
Acquired MET gene amplification, MET exon 14 skip mutations, or MET fusions can emerge as resistance mechanisms to tyrosine kinase inhibitors (TKIs) in patients with lung cancer. The efficacy and safety of combining MET TKIs (such as crizotinib, capmatinib, or tepotinib) with parent TKIs to target acquired MET resistance are not well characterized.
Methods
Multi-institutional retrospective chart review identified 83 patients with metastatic oncogene-driven NSCLC that were separated into the following two pairwise matched cohorts: (1) MET cohort (n = 41)—patients with acquired MET resistance continuing their parent TKI with a MET TKI added or (2) Chemotherapy cohort (n = 42)—patients without any actionable resistance continuing their parent TKI with a platinum-pemetrexed added. Clinicopathologic features, radiographic response (by means of Response Evaluation Criteria in Solid Tumors version 1.1), survival outcomes, adverse events (AEs) (by means of Common Terminology Criteria for Adverse Events version 5.0), and genomic data were collected. Survival outcomes were assessed using Kaplan-Meier methods. Multivariate modeling adjusted for lines of therapy, brain metastases, TP53 mutations, and oligometastatic disease.
Results
Within the MET cohort, median age was 56 years (range: 36–83 y). Most patients were never smokers (28 of 41, 68.3%). Baseline brain metastases were common (21 of 41, 51%). The most common oncogenes in the MET cohort were EGFR (30 of 41, 73.2%), ALK (seven of 41, 17.1%), and ROS1 (two of 41, 4.9%). Co-occurring TP53 mutations (32 of 41, 78%) were frequent. Acquired MET alterations included MET gene amplification (37 of 41, 90%), MET exon 14 mutations (two of 41, 5%), and MET gene fusions (two of 41, 5%). After multivariate adjustment, the objective response rate (ORR) was higher in the MET cohort versus the chemotherapy cohort (ORR: 69.2% versus 20%, p < 0.001). Within the MET cohort, MET gene copy number (≥10 versus 6–10) did not affect radiographic response (54.5% versus 68.4%, p = 0.698). There was no difference in ORR on the basis of MET TKI used (F [2, 36] = 0.021, p = 0.978). There was no difference in progression-free survival (5 versus 6 mo; hazard ratio = 0.64; 95% confidence interval: 0.34–1.23, p = 0.18) or overall survival (13 versus 11 mo; hazard ratio = 0.75; 95% confidence interval: 0.42–1.35, p = 0.34) between the MET and chemotherapy cohorts. In the MET cohort, dose reductions for MET TKI-related toxicities were common (17 of 41, 41.4%) but less frequent for parent TKIs (two of 41, 5%). Grade 3 AEs were not significant between crizotinib, capmatinib, and tepotinib (p = 0.3). The discontinuation rate of MET TKIs was 17% with no significant differences between MET TKIs (p = 0.315). Among pre- and post-treatment biopsies (n = 17) in the MET cohort, the most common next-generation sequencing findings were loss of MET gene amplification (15 of 17, 88.2%), MET on-target mutations (seven of 17, 41.2%), new Ras-Raf-MAPK alterations (three of 17, 17.6%), and EGFR gene amplification (two of 17, 11.7%).
Conclusions
The efficacy and safety of combining MET TKIs (crizotinib, capmatinib, or tepotinib) with parent TKIs for acquired MET resistance are efficacious. Radiographic response and AEs did not differ significantly on the basis of the underlying MET TKI used. Loss of MET gene amplification, development of MET on-target mutations, Ras-Raf-MAPK alterations, and EGFR gene amplification were molecular patterns found on progression with dual parent and MET TKI combinations.
Keywords: NSCLC, Tyrosine kinase inhibitor, Acquired resistance, MET amplification, MET exon 14 skipping
Introduction
The use of effective tyrosine kinase inhibitors (TKIs) for patients with oncogene-driven metastatic NSCLC (mNSCLC) has improved clinical outcomes.1 MET alterations have emerged as an important target. These can occur as de novo oncogenic drivers that are highly responsive to MET TKIs.2, 3, 4, 5 Outside of a primary oncogenic driver, MET alterations represent an important class of acquired resistance.6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16 Acquired MET resistance alterations are heterogeneous and include MET gene amplification, MET exon 14 skip mutations (METex14), and MET gene fusions.3,4,9 The clinical practice of combining MET TKIs with the parent TKI to target acquired MET resistance has been reported in small case series.17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28 Nevertheless, a systematic analysis of the overall frequency, efficacy, and safety of this approach across different oncogenic subtypes of NSCLC and a detailed comparison to the alternative strategy of using platinum-pemetrexed chemotherapy have not been described before. Here, we performed a detailed multi-institutional analysis of the efficacy and safety of combining MET TKIs with parent TKIs among patients with mNSCLC with a range of original driver oncogenes and MET acquired resistances.
Materials and Methods
Patients and Clinical Data Collection
The study was conducted in accordance with the Declaration of Helsinki and good clinical practice guidelines following approved institutional review board policies. Three U.S. academic oncology centers (University of Colorado, Vanderbilt University, and the University of Pennsylvania) were involved. All patients with mNSCLC (American Joint Committee on Cancer eighth edition) from January 2013 to January 2023 harboring EGFR, ALK, ROS1, RET, NTRK, BRAF V600E, or HER2 oncogenes that were treated with a parent TKI were eligible (Supplementary Fig. 1). A parent TKI was defined as a TKI that resulted in documented clinical and radiographic benefit for at least 3 months. All patients had comprehensive genomic profiling both at diagnosis and immediately before receiving post-progression treatment. Eligible patients with progression on their parent TKI were considered within one of the following two cohorts: (1) MET cohort—patients with acquired MET resistance alterations who received a combination of their parent TKI with a MET TKI and (2) Chemotherapy cohort—patients without a MET resistance alteration and no other actionable resistance mechanisms who received a combination of their parent TKI with platinum-pemetrexed chemotherapy (Supplementary Fig. 2). All eligible patients were on Food and Drug Administration (FDA)-approved starting doses of their parent TKI before receiving post-progression treatment. Within the MET cohort, all patients started with FDA-approved doses of crizotinib, tepotinib, or capmatinib. Within the chemotherapy cohort, all patients received carboplatin area under the curve 5 and pemetrexed 500 mg/m2 dosed every 21 days.
Demographics, pathology, molecular testing, imaging reports, treatment history, and clinical outcomes were extracted from the electronic health record. Investigator-assessed radiographic responses (by means of Response Evaluation Criteria in Solid Tumors version 1.1) were performed using the patient’s first on-treatment scan obtained within 8 to 12 weeks after starting treatment. Clinical outcome measures such as progression-free survival (PFS), time to next treatment (TTNT), and overall survival (OS) were collected (Supplementary Fig. 2). Adverse events (AEs) were graded according to Common Terminology Criteria for Adverse Events version 5.0. Exclusion criteria included the following: (1) nonmetastatic disease, (2) absence of molecular profiling at diagnosis, (3) absence of molecular profiling on progression of their parent TKI, (4) documented oncogenic MET alteration at diagnosis (e.g., METex14), (5) patients with documented acquired MET resistance who did not receive crizotinib, tepotinib, or capmatinib, and (6) incomplete follow-up.
Molecular Profiling Methods
Molecular testing was conducted on all patients in this study by Clinical Laboratory Improvement Amendments-certified laboratories using a mixture of commercially available or institution-specific next-generation sequencing (NGS) (Supplementary Table 1). Tissue samples at the University of Colorado (n = 46) were analyzed using both the Archer VariantPlex Solid Tumor and Archer FusionPlex sequencing panel (ArcherDx, Inc.). Bioinformatics analysis was carried out using version-controlled Archer Analysis (4.1.1.7). Fluorescence in situ hybridization (FISH) was performed on neutral formalin-fixed, paraffin-embedded tissue that was pretreated with proteinase K and hybridized with the MET DNA probe set (MetaSystems MET orange and chromosome 7 centromere green). Cell-free DNA (cfDNA) NGS testing was performed using the Guardant 360 assay (Guardant Health, Inc.), which was sequenced using the Illumina platform and hg19 as the reference genome.
We defined acquired MET resistance as the presence of new (previously undetected) MET gene amplification, METex14 skip mutations, or MET gene fusions found after radiographic progression on the parent TKI. The thresholds for calling high, intermediate, and low levels of MET gene amplification in this study were defined in advance (based on previously described methods29,30) and are found in Supplementary Table 2. In cases where both NGS and FISH were performed, priority was given to the MET-to-CEP7 ratio over the absolute gene copy number (GCN) in defining MET gene amplification.
Statistical Analyses
Descriptive statistics using Fisher exact test for categorical end points and paired t tests for continuous end points were used. Comparisons of radiographic response percentage, objective response rate (ORR), and disease control rate (DCR) were performed controlling for MET amplification (low, intermediate, high), specimen source (tissue alone, cfDNA alone, both), and method of detection (NGS, FISH, both). A Pearson product-moment correlation coefficient was computed to assess the relationship between radiographic response and PFS in both the MET and chemotherapy cohorts. Both univariate and multivariate Cox proportional hazard models were used for PFS, TTNT, and OS end points. Two-way (1:1) propensity score matching was also performed on the complete case data set using the nearest neighbors approach without replacement and a caliper of 0.15 to create balanced cohorts. Models were adjusted for prior lines of therapy, brain metastases, oligometastatic disease (defined as ≤3 metastatic sites), and TP53 mutations. Martingale residuals were used to select the best functional form of prior lines of therapy. Cox-Snell, Schoenfeld, and deviance residuals were further used to assess model fit. Unadjusted survival curves and corresponding medians were obtained with Kaplan-Meier methods. In each cohort, Cox proportional hazard models were used to assess the univariate associations between each survival outcome and the following variables: percentage change in tumor size, ORR, DCR, MET amplification (low, intermediate, high), specimen source (cfDNA, tissue, both), and method of detection (NGS, FISH, both). For safety analyses, multivariate logistic regression models, adjusting for age and lines of therapy, were used to compare the odds of grade greater than or equal to 3 AEs, dose reduction, dose hold, and treatment discontinuation across cohorts. The likelihood ratio test was used to calculate p values and confidence intervals (CIs). The CIs and corresponding p values were obtained using robust SEs. Statistical analyses were performed using GraphPad Prism software (version 9.51 for Windows, GraphPad Software, San Diego, CA) and R (version 4.2.2 for Windows, R Project, Vienna, Austria).
Results
Patient Characteristics
Across the three academic centers, 511 patients with oncogene-driven mNSCLC treated with parent TKIs were identified. Furthermore, 83 eligible patients with progression on parent TKIs were included for additional analyses (Table 1 and Supplementary Fig. 1) and analyzed across the two cohorts. The MET cohort included 41 patients with acquired MET resistance who received their parent TKI with a MET TKI. The chemotherapy cohort included 42 patients with no actionable resistance who received their parent TKI with platinum-pemetrexed chemotherapy. The TKIs included in this study are found in Supplementary Table 3.
Table 1.
Baseline Characteristics of the MET and Chemotherapy Cohorts Evaluated in This Study
| Characteristic | MET Cohort (Parent TKI + MET TKI) n = 41, n (%) | Chemotherapy Cohort (Parent TKI + Platinum–Pemetrexed) n = 42, n (%) | p Value |
|---|---|---|---|
| Median age at diagnosis, y (range) | 56.0 (36.0–83.0) | 61.0 (30.0–83.0) | 0.684 |
| Sex | |||
| Female | 21 (51.2) | 30 (71.4) | 0.073 |
| Male | 20 (48.8) | 12 (28.6) | 0.073 |
| Racea | |||
| Asian | 3 (7.31) | 7 (16.7) | 0.313 |
| Black | 2 (4.9) | 2 (4.8) | 1.00 |
| White | 35 (85.4) | 33 (78.5) | 0.57 |
| Pacific Islander | 1 (2.4) | 0 (0) | 0.494 |
| Smoking status | |||
| Never or light smokerb | 28 (68.3) | 34 (81.0) | 0.214 |
| Median pack-years | 0 (0–50.0) | 0 (0–40.0) | 0.292 |
| Nonsquamous | 38 (92.7) | 42 (100) | 0.116 |
| Oncogene | |||
| EGFR–typical mutationsc | 28 (68.3) | 28 (66.7) | 1.00 |
| EGFR–atypical mutationsc | 2 (4.9) | 4 (9.5) | 0.676 |
| ALK | 7 (17.1) | 7 (16.7) | 1.00 |
| ROS1 | 2 (4.9) | 2 (4.8) | 1.00 |
| BRAF V600E | 1 (2.4) | 1 (2.4) | 1.00 |
| HER2 exon 20 insertiond | 1 (2.4) | 0 (0.0) | 1.00 |
| Stage at diagnosis | |||
| Stages I–IIIBe | 2 (4.9) | 2 (4.8) | 1.00 |
| Stage IV | 39 (95.1) | 40 (95.2) | 1.00 |
| Metastatic profile | |||
| Brain metastases | 21 (51.2) | 17 (40.5) | 0.382 |
| Oligometastatic diseasef | 5 (12.2) | 5 (11.9) | 1.00 |
| Molecular profiling | |||
| Tissue NGS | 15 (36.6) | 16 (38.1) | 1.00 |
| ctDNA NGS | 4 (9.8) | 9 (21.4) | 0.227 |
| Both | 22 (53.7) | 17 (40.5) | 0.274 |
| MET alterations | |||
| MET gene amplification | 37 (90.2) | - | - |
| MET exon 14 skip | 2 (4.87) | - | - |
| MET fusion | 2 (4.87) | - | - |
| TP53 alterationsg | |||
| Present | 32 (78.0) | 27 (64.2) | 0.227 |
| Absent | 9 (21.9) | 15 (35.7) | 0.227 |
| Prior therapies | |||
| Prior lines of anticancer therapy, median (range) | 2.0 (1.0–9.0) | 1.0 (1.0–5.0) | 0.137 |
| Prior chemotherapy | 20 (48.8) | - | - |
| Prior TKIh | 15 (36.6) | 16 (38.1) | 1.00 |
ctDNA, circulating tumor DNA; HHS, Human Health Services; NGS, next-generation sequencing; TKI, tyrosine kinase inhibitor.
Racial categorizations are based on HHS data collection standards. Racial categories include: (1) White, (2) Black (or African American), (3) Asian, (4) Native Hawaiian or Other Pacific Islander, or (5) American Indian or Alaskan Native. The latter is not found because no patient met this category.
Light smoker defined as ≤10 pack years.
Typical EGFR mutations include sensitizing EGFR exon 19 deletions and L858R mutations. Atypical EGFR mutations include EGFR G709, L861Q, S768I, or exon 20 insertions.
This patient had a HER2 exon 20 insertion and received platinum doublet chemotherapy, trastuzumab deruxtecan, and then afatinib in the third-line setting. Although the patient was on afatinib, she developed MET amplification as a mechanism of resistance.
All these patients had metastatic relapse. On relapse, all molecular profiling was performed on the metastatic sample before the initiation of systemic therapy.
Oligometastatic disease is defined as less than or equal to three metastatic sites at diagnosis.
Refers to TP53 mutations identified at diagnosis.
In some cases, patients may have received a sequence of TKIs before MET alterations were identified (MET cohort) or before chemotherapy was initiated (chemotherapy cohort). For example, an ALK-positive patient may have received crizotinib, alectinib, and lorlatinib before a MET alteration was identified. For the purposes of this analysis, only the last TKI received before receiving post-progression treatments was considered.
The median age at diagnosis of mNSCLC for the MET cohort and chemotherapy cohort was 56 years (range: 36–83 y) and 61 years (range: 30–83 y), respectively. Most patients in both cohorts were never smokers (68.3% versus 81%). Adenocarcinoma was the dominant NSCLC sub-type (92% and 100%). All patients had central nervous system (CNS) imaging at baseline. Brain metastases at diagnosis were common in both cohorts (51.2% and 40.5%). Within the MET cohort, 48.8% of the patients received prior chemotherapy. All patients in the chemotherapy cohort were, by definition, chemotherapy naive. The percentage of patients within the MET and chemotherapy cohorts who received more than one TKI was 36.6% and 38.8%, respectively.
Genomic Characteristics
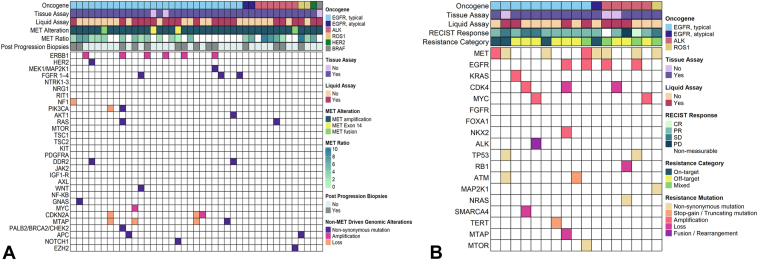
The distribution of driver oncogenes in both cohorts is found in Table 1. Sensitizing EGFR mutations were the dominant driver oncogene (68.3% and 66.7%) in both cohorts. Co-occurring TP53 mutations were common (78% and 52.4%) in both cohorts. The genomic alterations in the MET cohort before receiving a MET TKI are found in Figure 1A. All somatic alterations reported here were not present at diagnosis. Among 41 patients in the MET cohort, we found 37 cases with MET gene amplification. The distribution of high, intermediate, and low MET gene amplification was 54.0% (20 of 37), 27.0% (10 of 37), and 2.7% (one of 37), respectively. In 16.2% (six of 37) of the cases, MET gene amplification was called in the NGS report, but the GCN was not explicitly quantified. In two cases, METex14 skip resistance mutations were identified as novel resistance mechanisms to osimertinib (Supplementary Fig. 3). In addition, we identified two MET gene fusions as acquired resistance mechanisms. One was a novel CDC42EP3-MET fusion found on progression on osimertinib (Fig. 2D) and the other was an ST7-MET fusion found on progression on alectinib. In all cases, the METex14 skip mutations and MET gene fusions were not present at diagnosis and only detected on progression with the parent TKI.
Figure 1.
Genomic data of patients who received dual parent and MET TKI combinations. (A) A heatmap revealing genomic data from 41 samples before receiving dual parent and MET TKI combinations. (B) A heatmap revealing genomic mechanisms of resistance among 17 samples obtained after progression on dual parent and MET TKI combinations. TKI, tyrosine kinase inhibitor.
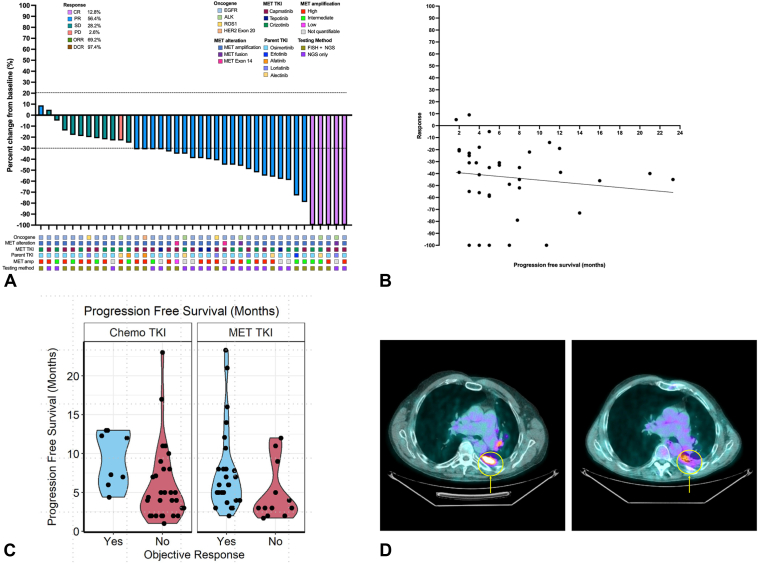
Figure 2.
Radiographic responses to dual parent TKI and MET TKI combinations for acquired MET resistance. (A) A waterfall plot of investigator-assessed radiographic response for 41 patients who received MET TKIs for acquired MET resistance. Each bar represents an individual patient and is color coded to represent radiographic response (by means of RECIST 1.1). (B) A scatterplot revealing no correlation between radiographic response and PFS among patients who received dual parent TKI and MET TKI combinations. (C) A violin plot revealing the relationship between PFS radiographic response in the MET and chemotherapy cohorts. (D) Radiographic response in a patient who developed a novel CDC42EP3-MET fusion as a resistance mechanism to osimertinib. The PET/CT with progression on osimertinib is found (left) with FDG avidity within the pleura. A PET/CT obtained after 12 weeks on osimertinib plus crizotinib reveals an interval decrease in FDG avidity within the pleura (right). CT, computed tomography; FDG, fluorodeoxyglucose; PET, positron emission tomography; PFS, progression-free survival; RECIST, Response Evaluation Criteria in Solid Tumors; TKI, tyrosine kinase inhibitor.
Radiographic and Tumor Response
Waterfall plots of the MET and chemotherapy cohorts are found in Figure 2A and Supplementary Figure 4. The median time to the first scan was 10 weeks (range: 8–12 wk). Two patients in the MET cohort and two patients in the chemotherapy cohort were excluded because they had assessable, but not measurable, disease per Response Evaluation Criteria in Solid Tumors version 1.1. The median percentage change in target lesion sizes at the first on-treatment assessment was −39% versus −17% (R2 = 0.76, 95% CI: 28.3–40.9, p < 0.001). The ORR and DCR were significantly higher in the MET cohort (ORR: 69.2% versus 20%, DCR: 97.4% versus 80%, p < 0.001). The degree of MET gene amplification (intermediate versus high) did not influence the radiographic response rate (54.5% versus 68.4%, p = 0.698; Supplementary Table 4). The method of NGS testing (cfDNA versus tissue) to identify MET gene amplification did not affect the radiographic response (ORR: 60.0 versus 72.7%, DCR: 60 versus 77.3%, p = 0.524; Supplementary Table 4). A one-way analysis of variance revealed no significant difference in radiographic response based on underlying MET TKI used (F [2, 36] = 0.021, p = 0.978).
Two patients with acquired METex14 resistance mutations had partial responses (Supplementary Fig. 3). One patient with a novel CDC42EP3-MET fusion resistance had a partial response after crizotinib was added to osimertinib (Fig. 2D). The other patient with a ST7-MET resistance to alectinib had a complete response with the addition of capmatinib. Patterns of intracranial and extracranial progression were captured. The chemotherapy cohort had a significantly higher rate of intracranial progression relative to the MET cohort (41.4% versus 14.8%, p = 0.004). Within the MET cohort, we sought to determine whether this difference was based on the use of CNS-penetrant MET TKIs. We found no difference in CNS progression rates in patients who received crizotinib versus capmatinib or tepotinib (43.8% versus 38.5%, p = 1.000).
Clinical Outcomes
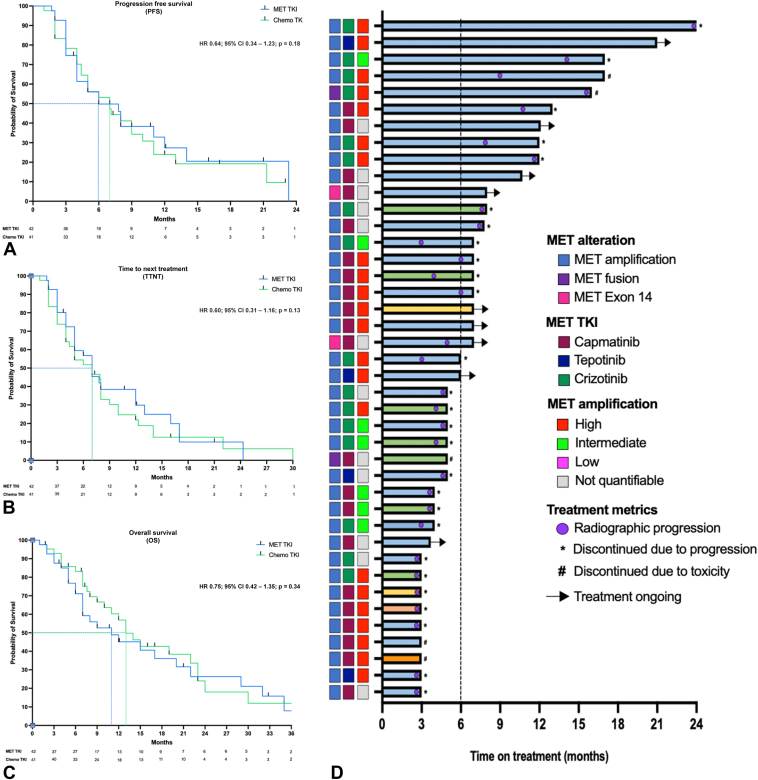
Median duration of follow-up was 12 months for both cohorts. Kaplan-Meier curves for PFS (Fig. 3A), TTNT (Fig. 3B), and OS (Fig. 3C), adjusted for brain metastases, oligometastatic disease (≤3 metastatic sites at diagnosis), TP53 mutations, and prior lines of therapy, are found in Figure 3. A Swimmer Plot showing time to next treatment is shown in Figure 3D. Between the MET and chemotherapy cohorts, there was no difference in PFS (5 versus 6 mo; hazard ratio [HR] = 0.64, 95% CI: 0.34–1.23, p = 0.18), TTNT (7 versus 7 mo; HR = 0.60, 95% CI: 0.31–1.16, p = 0.13), or OS (13 versus 11 mo; HR = 0.75, 95% CI: 0.42–1.35, p = 0.34). We separately performed two-way (1:1) propensity score matching on the complete case data set and found no difference in PFS (7.8 versus 4.4 mo; HR = 0.64, 95% CI: 0.34–1.23, p = 0.18), TTNT (7.8 versus 4.4 mo; HR = 0.60, 95% CI: 0.31–1.16, p = 0.13), and OS (11 versus 10 mo; HR = 0.75, 95% CI: 0.42–1.35, p = 0.34) between the MET and chemotherapy cohorts. To account for potential biases, multivariate models were repeated in select subpopulations which included (1) patients who received only one prior line of therapy and (2) patients with sensitizing EGFR mutations treated with osimertinib. Among all subset analyses, there was no difference in PFS, TTNT, or OS between the MET and chemotherapy cohorts (Supplementary Fig. 5A-F). After adjusting for brain metastases, oligometastatic disease, TP53 mutations, and prior lines of therapy, we found no association between depth of radiographic response and PFS (HR = 0.429, 95% CI: 0.191–0.962, p = 0.41) in the MET cohort (Fig. 2B and C). In the chemotherapy cohort, we did observe a positive correlation between depth of radiographic response and PFS (HR = 0.888, 95% CI: 0.303–2.60, p = 0.83) and OS (HR = 0.361, 95% CI: 0.119–1.10, p = 0.025) (Supplementary Fig. 6).
Figure 3.
Clinical outcome measures among the MET and chemotherapy cohorts in this study. (A) PFS between the MET and chemotherapy cohort. (B) TTNT between the MET and chemotherapy cohort. (C) OS between the MET and chemotherapy cohort. (D) A Swimmer plot of the MET cohort. Each bar represents an individual patient. Circles within the bars represent points of radiographic progression. The median time on treatment is indicated by the dotted line. OS, overall survival; PFS, progression-free survival; TTNT, time to next treatment.
Safety
The safety data within the MET and chemotherapy cohorts are found in Tables 2 and 3. Osimertinib was the most common parent TKI used in either cohort (68.2% and 76.2%). The MET TKIs used in this study included capmatinib (54%), crizotinib (37%), and tepotinib (9%). All TKI doses were initially administered at the FDA-approved starting dose levels. Anemia (57% versus 22%, p = 0.002) and neutropenia (24% versus 1%, p = 0.007) were significantly more common in the chemotherapy cohort. Peripheral edema (46% versus 17%, p = 0.004), dizziness (17% versus 2%, p = 0.029), and paronychia (15% versus 0%, p = 0.012) were significantly more common in the MET cohort. Dose reductions of the MET TKI because of treatment-attributable toxicities were common (41%) but rarely required for the parent TKI (5%). In one case, erlotinib (the parent TKI) was dose reduced to 100 mg because of grade 2 diarrhea and rash. In another case, a patient with BRAF V600E NSCLC with MET gene amplification who received encorafenib and binimetinib with capmatinib developed grade 4 pneumonitis and grade 3 hyponatremia. This was the only case of pneumonitis found in our entire data set, and both the parent and MET TKI were discontinued. The remainder of dose adjustments were due to MET TKI toxicities with peripheral edema (46%) being the most common treatment-related reason for dose modification. Diarrhea (any grade) was common when crizotinib was paired with the parent TKI (Table 3). In the MET cohort, the dose reduction rate was 41%, dose interruption rate was 34%, and the treatment discontinuation rate was 17% and not significantly different on the basis of the underlying MET TKI used (p = 0.315). After multivariate analysis, adjusting for age and lines of therapy, the odds of grade greater than or equal to 3 AEs were not significantly different among patients who received crizotinib, capmatinib, or tepotinib (p = 0.3). Grade 3 AEs were significantly higher in the chemotherapy cohort (45% versus 27%, p = 0.013), driven predominantly by anemia and thrombocytopenia. Nevertheless, there was no difference in rates of dose reduction (p = 0.63), dose hold (p = 0.27), or treatment discontinuation (p = 0.43) between the MET and chemotherapy cohorts. Number of prior systemic therapies was the largest predictor of grade greater than or equal to 3 AEs in either cohort. Receiving greater than or equal to two lines of systemic therapy was associated with 4.44 times the odds of developing grade greater than or equal to 3 AEs in either cohort (95% CI: 1.60–13.67, p = 0.004). Patient age alone was not significantly associated with increased odds of grade greater than or equal to 3 AEs in either cohort (p = 0.59).
Table 2.
Summary of Dose Adjustments and Adverse Event Profile Within the MET and Chemotherapy Cohorts
| Adverse Event and Dose Summary |
MET Cohort (n = 41) |
Chemotherapy Cohort (n = 42) |
p Value | ||
|---|---|---|---|---|---|
| Dose Adjustments | Parent TKI | MET TKI | Parent TKI | Chemotherapy | |
| Treatment discontinuation | 0 (0) | 7 (17) | 0 (0) | 12 (29) | 0.615 |
| Dose reduction | 1 (2) | 17 (41) | 0 (0) | 16 (38) | 0.658 |
| Dose interruption | 0 (0) | 14 (34) | 0 (0) | 10 (24) | 0.340 |
| Adverse event profile | Any grade | Grade ≥3 | Any grade | Grade ≥3 | |
| SAE | - | 11 (27) | - | 19 (45) | 0.013 |
| Adverse events occurring in ≥10% of patients | |||||
| Anemia | 9 (22) | 1 (2) | 24 (57) | 3 (7) | 0.002 |
| Neutropenia | 1 (2) | 1 (2) | 10 (24) | 7 (17) | 0.007 |
| Thrombocytopenia | 2 (5) | 0 (0) | 7 (17) | 0 (0) | 0.156 |
| Visual impairment | 5 (12) | 0 (0) | 1 (2) | 0 (0) | 0.109 |
| Diarrhea | 17 (41) | 1 (2) | 10 (24) | 1 (2) | 0.104 |
| Nausea | 14 (24) | 2 (5) | 16 (38) | 1 (2) | 0.820 |
| Constipation | 5 (12) | 0 (0) | 10 (24) | 0 (0) | 0.254 |
| Mucositis | 1 (2) | 0 (0) | 6 (14) | 0 (0) | 0.109 |
| Peripheral edema | 19 (46) | 3 (7) | 7 (17) | 0 (0) | 0.004 |
| Fatigue | 17 (41) | 3 (7) | 21 (50) | 1 (2) | 0.511 |
| Elevated AST or ALT | 4 (9) | 0 (0) | 4 (9) | 1 (2) | 0.99 |
| Hyponatremia | 3 (7) | 2 (5) | 5 (12) | 2 (5) | 0.713 |
| Hypoalbuminemia | 5 (12) | 0 (0) | 3 (7) | 0 (0) | 0.483 |
| Myalgia | 4 (9) | 0 (0) | 3 (7) | 0 (0) | 0.713 |
| Muscle weakness | 0 (0) | 0 (0) | 4 (9) | 1 (2) | 0.116 |
| Dizziness | 7 (17) | 0 (0) | 1 (2) | 0 (0) | 0.029 |
| Peripheral neuropathy | 4 (9) | 0 (0) | 7 (17) | 0 (0) | 0.519 |
| Kidney injury | 4 (9) | 0 (0) | 7 (17) | 5 (12) | 0.519 |
| Dyspnea | 3 (7) | 1 (2) | 5 (12) | 1 (2) | 0.713 |
| Cough | 1 (2) | 0 (0) | 5 (12) | 0 (0) | 0.201 |
| Maculopapular rash | 8 (20) | 0 (0) | 8 (20) | 1 (2) | 0.999 |
| Paronychia | 6 (15) | 0 (0) | 0 (0) | 0 (0) | 0.012 |
ALT, alanine transaminase; AST, aspartate transaminase; SAE, serious adverse event; TKI, tyrosine kinase inhibitor.
Bolded values indicate statistically significant differences (p < 0.05)
Table 3.
Adverse Events From Specific Parent TKI and MET TKI Combinations
| TKI Combinations | N = 42 (%) | Most Common AEs—Any Grade |
Grade ≥3 AEs |
Dose Adjustment and Attributiona |
Discontinuation Rate |
|||
|---|---|---|---|---|---|---|---|---|
| AE | n (%)b | AE | n (%)b | TKI | n (%) | n (%) | ||
| EGFR | ||||||||
| Osimertinib + capmatinib | 14 (33) | Edema | 6/19 (31) | Fatigue | 3/17 (17) | Osimertinib | 0 (0) | 2/14 (14) |
| Fatigue | 6/17 (35) | Edema | 2/19 (10) | Capmatinib | 8/14 (57) | |||
| Diarrhea | 5/17 (36) | |||||||
| Nausea | 5/14 (36) | |||||||
| Low albumin | 3/5 (60) | |||||||
| Kidney injury | 3/4 (75) | |||||||
| Osimertinib + crizotinib | 10 (24) | Diarrhea | 7/17 (41) | VTE | 1/3 (33) | Osimertinib | 0 (0) | 3/10 (30) |
| Nausea | 5/14 (36) | Anorexia | 1/5 (20) | Crizotinib | 5/10 (50) | |||
| Fatigue | 4/17 (23) | Nausea | 1/14 (7) | |||||
| Paronychia | 3/6 (50) | Diarrhea | 1/17 (6) | |||||
| Rash | 3/8 (38) | |||||||
| Osimertinib + tepotinib | 4 (10) | Edema | 3/19 (16) | VTE | 1/3 (33) | Osimertinib | 0 (0) | 0 (0) |
| Dizziness | 3/17 (17) | Anemia | 1/9 (11) | Tepotinib | 0 (0) | |||
| Fatigue | 1/17 (6) | |||||||
| VTE | 1/3 (33) | |||||||
| Afatinibc + capmatinib | 1 (2) | Diarrhea | 1/17 (6) | - | 0 (0) | Afatinib | 0 (0) | 0 (0) |
| Nausea | 1/14 (7) | Capmatinib | 0 (0) | |||||
| Fatigue | 1/17 (6) | |||||||
| VTE | 1/3 (33) | |||||||
| Dacomitinib + crizotinib | 1 (2) | AST or ALT elevation | 1/4 (25) | - | 0 (0) | Dacomitinib | 0 (0) | 0 (0) |
| Paronychia | 1/6 (17) | Crizotinib | 1/1 (100) | |||||
| Diarrhea | 1/17 (6) | |||||||
| Edema | 1/19 (5) | |||||||
| Erlotinib + crizotinib | 1 (2) | Diarrhea | 1/17 (6) | - | 0 (0) | Erlotinib | 1/1 (100) | 0 (0) |
| Nausea | 1/14 (7) | Crizotinib | 1/1 (100) | |||||
| Rash | 1/8 (12) | |||||||
| Low albumin | 1/5 (20) | |||||||
| ALK and ROS1 | ||||||||
| Alectinib + capmatinib | 1 (2) | Elevated bilirubin | 1/2 (50) | - | 0 (0) | Alectinib | 0 (0) | 0 (0) |
| Anemia | 1/9 (11) | Capmatinib | 0 (0) | |||||
| Alectinib + crizotinib | 3 (7) | AST or ALT elevation | 3/4 (75) | VTE | 1/3 (33) | Alectinib | 0 (0) | 0 (0) |
| Fatigue | 2/17 (12) | Nausea | 1/14 (7) | Crizotinib | 3/3 (100) | |||
| Edema | 2/19 (10) | Hyponatremia | 1/3 (33) | |||||
| Nausea | 2/14 (14) | Dyspnea | 1/3 (33) | |||||
| Rash | 2/8 (25) | |||||||
| VTE | 1/3 (33) | |||||||
| Lorlatinib + capmatinib | 5 (12) | Edema | 3/19 (16) | Edema | 1/19 (5) | Lorlatinib | 0 (0) | 1/5 (20) |
| Proteinuria | 1/1 (100) | Proteinuria | 1/1 (100) | Capmatinib | 1/5 (20) | |||
| Low albumin | 1/5 (20) | |||||||
| Fatigue | 1/17 (13) | |||||||
| Nausea | 1/14 (7) | |||||||
| BRAF V600E TKIs | ||||||||
| Encorafenib + binimetinib + capmatinib | 1 (2) | Pneumonitis | 1/1 (100) | Pneumonitis | 1/1 (100) | Encorafenib + binimetinib | 1/1 (100) | 1/1/ (100) |
| Hyponatremia | 1/3 (33) | Hyponatremia | 1/3 (33) | Capmatinib | 1/1 (100) | |||
| Kidney injury | 1/4 (25) | |||||||
| Anemia | 1/9 (11) | |||||||
AE, adverse event; ALT, alanine transaminase; AST, aspartate transaminase; PO, orally; TKI, tyrosine kinase inhibitor; VTE, venous thromboembolism.
Dose adjustment includes any dose reductions or dose hold for either parent or MET TKI. The attribution of which TKI contributed to a dose adjustment is also revealed.
Percentages are derived from dividing the specific AEs identified from a specific parent plus MET TKI combination from the total AEs identified in the MET cohort.
This was for a patient with a HER2 exon 20 insertion mutation who received carboplatin plus pemetrexed, trastuzumab deruxtecan, and then afatinib in the third-line setting. This patient was on afatinib 40 mg PO daily when MET gene amplification was discovered.
Mechanisms of Resistance to MET TKI Combinations
Of the 25 patients in the MET cohort with radiographic progression, 17 patients (68.0%) had post-progression biopsies (Fig. 1B). The distribution of parent driver oncogenes within these 17 patients was EGFR (58.8%; 10 of 17), ALK (35.3%; six of 17), and ROS1 (5.88%; one of 17). All 17 patients had MET gene amplification as the mechanism of resistance to their parent TKI. Among these 17 patients, 82.3% (14 of 17) had a tissue biopsy and 41.2% (seven of 17) had cfDNA collected on progression with the parent and MET TKI combination. Loss of MET gene amplification was found in 88.2% (15 of 17) cases (Fig. 1B and Supplementary Fig. 4). MET on-target resistance mutations occurred in 41.2% of the patients (seven of 17) with the following distribution: MET D1228N (four of seven, 57%), MET Y1230C (two of seven, 28.6%), MET L1195V, MET D1164Y, and MET Y1003N. On-target resistances to the parent TKI (e.g., EGFR C797S, ALK G1202R, or ROS1 G2032R) were not found. A new STRN-ALK gene fusion was observed as mechanism of resistance to osimertinib and capmatinib. EGFR gene amplification (previously undetected and identified on tissue NGS) was found in two patients with ALK NSCLC after treatment with alectinib plus capmatinib and lorlatinib plus capmatinib, respectively. Alterations within the Ras-Raf-MAPK pathway were observed in 17.6% (three of 17) cases. These included new NRAS G12D and MAP2K1 G128N mutations, along with KRAS gene amplification (3+, defined by using Guardant cfDNA assay).
Discussion
To our knowledge, our study is the largest series addressing the frequency, efficacy, and safety of dual parent and MET TKI combinations for acquired MET resistance across a variety of oncogenic drivers in NSCLC. Our study has several novel findings. We report that MET alterations are a shared resistance mechanism to a variety of oncogenic driver mutations. Although the efficacy and safety of osimertinib with MET TKIs have been previously reported,31, 32, 33 our data broaden the scope to include the efficacy and safety of combining a variety of MET TKIs with parent TKIs where acquired MET resistance (through MET gene amplification, METex14 skip mutations, or MET gene fusions) is clearly defined.
We report that dual parent TKI and MET TKI combinations for MET resistance yield an ORR of 69.2%, which is consistent with prior trial data.31, 32, 33 The INSIGHT-2 (NCT03940703) and TATTON (NCT02143466) trials address the efficacy and safety of combining osimertinib with either tepotinib or savolitinib, respectively. Both trials defined MET amplification as MET GCN greater than or equal to 5 or MET/CEP7 greater than or equal to 2 by FISH. The ORRs of the INSIGHT-2 trial and TATTON trial were 54.5% and 67%, respectively.31,32 Our data build on these findings by revealing an ORR of 69.2% across a variety of MET TKIs and across a variety of oncogenic driver alterations.33 Interestingly, our data reveal that high MET gene amplification (defined in our series as MET GCN ≥ 10 and/or MET/CEP7 ≥ 3) does not correlate with depth of radiographic response. Prior studies reveal that depth of radiographic response does correlate with PFS and OS among TKI-naive ALK NSCLC.34 Our data suggest that this relationship may not hold in the setting of acquired MET resistance. Nonetheless, the higher ORR in the MET cohort has implications for therapy selection, especially when patients have progression involving high-risk anatomical structures and need urgent cytoreduction. Our data suggest that if a MET resistance alteration is identified, dual parent TKI and MET TKI combinations can generate such a response.
We found no difference in PFS, TTNT, or OS between the MET and chemotherapy cohorts after adjusting for known drivers of poor outcomes and potential biases through multivariate analyses. In our chemotherapy cohort, we only included patients who continued their parent TKI with platinum-pemetrexed chemotherapy, rather than switching from TKI to chemotherapy. The efficacy of continuing TKIs with chemotherapy post-progression remains under debate35, 36, 37 and is being evaluated prospectively through the ongoing COMPEL (NCT04765059) and Lung-MAP (NCT05364645) clinical trials. Nonetheless, it is common practice across many academic centers in the United States. By only including a cohort of patients who continued their parent TKI with platinum-pemetrexed chemotherapy, we attempted to mitigate the concern about lack of CNS protection that could result from discontinuation of the parent TKI.
Our data reveal that patients with acquired MET resistance who receive dual parent and MET TKI combinations have clinical outcomes that are at least comparable to patients with no actionable resistance who continue their parent TKI with platinum-pemetrexed chemotherapy. An important, inherent, and unavoidable bias of the study is that the two cohorts compared are biologically different. Prior studies have suggested that MET signaling (either through increased cell surface expression or through gene amplification) may confer a worse prognosis.38,39 This is relevant when drawing conclusions between the two cohorts in our study. Nonetheless, our findings overlap with the INSIGHT trial (NCT01982955), which compared the efficacy of tepotinib and gefitinib in patients with EGFR mNSCLC with acquired MET resistance.40 MET resistance was defined as MET overexpression (immunohistochemistry 2–3+) or MET gene amplification (defined as MET gene copy ≥5 or MET/CEP7 ≥2). In contrast with our series, gefitinib (parent TKI) was not continued in the platinum doublet chemotherapy arm. In a subgroup analysis, there was no significant difference in mPFS between the tepotinib and gefitinib arm (4.9 mo; 90% CI: 3.9–6.9) and the platinum doublet chemotherapy arm (4.4 mo; 90% CI: 4.2–6.8 mo) among patients with EGFR mNSCLC with acquired MET resistance. Low accrual led to termination of this trial. The phase 3 GEOMETRY-E study (NCT04816214) recruited patients with EGFR-mutant NSCLC with MET amplification and randomized patients (2:1) to receive osimertinib plus capmatinib versus platinum doublet chemotherapy. This trial was terminated by the sponsor. The ongoing SACHI trial (NCT05015608) is prospectively comparing osimertinib and savolitinib versus platinum-pemetrexed chemotherapy for patients with EGFR mNSCLC with MET gene amplification who have progressed on osimertinib. To date, there are no prospective trial data that directly compare the efficacy of dual parent and MET TKI combinations to parent TKI with platinum-pemetrexed for patients with acquired MET resistance.
The safety of dual parent TKI and MET TKI combinations for acquired MET resistance is best characterized for EGFR mNSCLC. Prospectively, the INSIGHT-2 (NCT03940703), TATTON (NCT02143466), and SAVANNAH (NCT03778229) clinical trials explored combining osimertinib with tepotinib (INSIGHT-2) or savolitinib (TATTON, SAVANNAH) for EGFR-mutant NSCLC. Nausea, peripheral edema, paronychia, and diarrhea were the most common AEs,31, 32, 33 consistent with our data. A small phase 1 study of 27 patients receiving crizotinib with erlotinib found that the maximum tolerated dose for crizotinib and erlotinib was 150 mg and 100 mg daily, both of which are lower than the approved dose for either agent.41 We found that MET-associated peripheral edema was the most common reason for dose modification and that parent TKI toxicities rarely led to dose modifications. The type of MET TKI used (crizotinib, capmatinib, tepotinib) did not affect the discontinuation rate or development of grade greater than or equal to 3 AEs. We found that the number of prior lines of therapy was the single greatest predictor of grade greater than or equal to 3 AEs with dual parent and MET TKI combinations. Our data highlight the importance of testing for acquired MET alterations after progression on first-line TKIs, as the likelihood of tolerability of dual parent and MET TKI combinations may decrease with each subsequent line of therapy.
Resistances to dual parent and MET TKI combinations are not well described. There were 17 patients (68.0%) in the MET cohort who had molecular profiling on progression. Loss of MET gene amplification was found in 88.2% patients with progression on dual parent and MET TKI combinations. On-target MET mutations identified in this series include MET D1228N, MET Y1230C, MET L1195V, MET D1164Y, and MET Y1003N, all of which have been reported after osimertinib with salvolitinib.28 Detecting mechanisms of resistance after parent TKI and MET TKI therapy has practical relevance because there are type II MET TKIs in development (such as foretinib) that do have preclinical efficacy against secondary MET mutations at residues D1228 or Y1230, which raises the interesting possibility of switching MET TKIs on the basis of detected on-target MET mutations.42 Interestingly, on-target gatekeeper, solvent front, and ATP-binding mutations to the parent TKI (e.g., EGFR C797S, ALK G1202R, ROS1 G2032R) were not found in this series. EGFR and Ras-Raf-MAPK bypass signaling were found on progression with dual parent and MET TKI combinations, consistent with prior reports.27,28,32,43, 44, 45 EGFR bypass signaling is of specific interest given the availability of EGFR-MET bispecific antibodies (such as amivantamab), raising the possibility of using these agents if EGFR gene amplification is detected. These approaches should be explored in prospective clinical trials. Our data suggest that cancer cells treated with dual parent TKI and MET TKI combinations lose dependency on MET gene amplification and evolve resistance through either on-target MET mutations or through bypass signaling involving the EGFR or Ras-Raf-MAPK pathways.
There are several limitations to our analysis. Given the varied frequency of scan assessments, we used the first on-treatment scan (obtained within 8–12 wk) to determine ORR which may underestimate the best radiographic response. Not all patients had paired tissue and plasma genomic profiling, limiting our ability to draw conclusions on the sensitivity or specificity of either approach to detect acquired MET resistance. Patients were identified over a time period that spanned a decade. During these 10 years, the practice patterns regarding the use of NGS testing evolved, and thus, there was variability in the NGS testing methodologies. In addition, there was variability in the availability of MET TKIs (such as capmatinib and tepotinib), which limits our ability to draw strong conclusions on any potential efficacy or safety differences between crizotinib, capmatinib, or tepotinib. Most patients in our series had acquired MET gene amplification, and it remains an open question whether there may be unique and subtle differences in efficacy when dual parent and MET TKI combinations are used for some of the more rare resistance subtypes such as acquired METEx14 or MET gene fusions. Finally, although we controlled for many competing variables, the time horizon within our data yields inherent biases and our data need to be replicated in prospective clinical trials.
In conclusion, our series reveal that MET alterations are resistance mechanisms found across multiple oncogenic drivers in lung cancer. MET resistances are actionable, and use of a MET TKI can generate significant radiographic responses. Nevertheless, radiographic response alone does not predict clinical durability. Dual parent and MET TKI combinations are tolerable, which can play into shared decision-making with patients when considering alternative approaches with chemotherapy. Mechanisms of resistance to dual parent and MET TKI combinations are complex, with loss of MET gene amplification found in most patients. Further research on how cancer cells use MET signaling to escape evolutionary pressures from TKIs is needed. Finally, consensus on the diagnostic tests (or combination of tests) that best predict which patients with MET resistance may derive long-term benefit from MET TKIs is needed.
CRediT Authorship Contribution Statement
Tejas Patil: Conceptualization, Data curation, Formal analysis, Methodology, Project administration, Supervision, Validation, Visualization, Roles/Writing—original draft, and Writing—review and editing.
Alyse Staley: Data curation, Formal analysis, Methodology, Visualization, Roles/Writing—original draft.
Yunan Nie: Data curation, Methodology.
Mandy Sakamoto: Data curation, Methodology.
Margaret Stalker: Data curation, Methodology.
James M. Jurica: Data curation, Methodology.
Kenna Hornbeck: Data curation, Methodology.
Amanda Cass: Data curation, Methodology.
Kurt D. Davies: Data curation, Methodology, Formal analysis.
Hala Nijmeh: Data curation, Visualization.
Mary Haag: Data curation, Visualization.
Benjamin A. Yoder: Writing—review and editing.
Paul A. Bunn: Writing—review and editing.
Erin L. Schenk: Writing—review and editing.
Dara L. Aisner: Visualization, Writing—review and editing.
Wade T. Iams: Project administration, Supervision, Validation, Writing—review and editing.
Melina E. Marmarelis: Project administration, Supervision, Validation, Writing—review and editing.
D. Ross Camidge: Supervision, Roles/Writing—original draft, Writing—review and editing.
Disclosure
Dr. Patil reports personal fees from Astrazeneca, personal fees from Bicara, personal fees from Bristol-Myers-Squibb, grants and personal fees from EMD Soreno, grants and personal fees from Janssen, personal fees from Mirati Therapeutics, personal fees from Pfizer, personal fees from Sanofi, personal fees from Regeneron, personal fees from Roche/ Genentech, personal fees from Takeda, grants and personal fees from Gilead, other from Elevation Oncology, outside the submitted work; Dr. Camidge reports personal fees from Abbvie, personal fees from Anheart, personal fees from Appolomics, personal fees from AstraZeneca, personal fees from Beigene, personal fees from Dizal, personal fees from Elevation Oncology, personal fees from Eli Lily, personal fees from EMD Soreno, personal fees from Hengrui, personal fees from Hummingbird, personal fees from Imagene, personal fees from Immunocore, personal fees from Janssen, personal fees from Medtronic, personal fees from Mersana, personal fees from Mirati, personal fees from Nalo Therapeutics, personal fees from Onkure, personal fees from Prelude, personal fees from Regeneron, personal fees from Roche, personal fees from Sanofi, personal fees from Seattle Genetics, personal fees from Takeda, personal fees from Theseus, personal fees from Valence , personal fees from Xcovery, outside the submitted work; Dr. Schenk reports personal fees from Actinium Pharmaceuticals, personal fees from AstraZeneca, personal fees from BioAtla, personal fees from BeiGeneius, personal fees from Bionest Partners, personal fees from ClearView Healthcare Partners, personal fees from G1 Therapeutics, personal fees from Harpoon, personal fees from Janssen, personal fees from MECC Global Meetings, personal fees from Regeneron, personal fees from Takeda, personal fees and other from Thetis, outside the submitted work; Dr. Bunn reports personal fees from Ascentage, personal fees from Amgen, personal fees from Astrazeneca, personal fees from Bristol Myers Squibb, personal fees from CStone, personal fees from Daiichi, personal fees from Genentech, personal fees from Merck , personal fees from Verastem, outside the submitted work; Dr. Iams reports personal fees from Astrazeneca, personal fees from Amgen, personal fees from Bristol Myers Squibb, personal fees from Clinical Care Options, personal fees from Chardan, personal fees from Cello Health, personal fees from Curio Science, personal fees from Genentech, personal fees from GI Therapeutics, personal fees from Janssen, personal fees from Jazz Pharmaceuticals, personal fees from Mirati Therapeutics, personal fees from NovoCure, personal fees from Outcome Heights, personal fees from Sanofi, personal fees from Takeda, outside the submitted work; Dr. Marmarelis reports grants and personal fees from Astrazeneca, personal fees from Bayer, personal fees from Bristol Myers Squibb, personal fees from Eli Lily, personal fees from Genentech, personal fees from Ikena, personal fees from Janssen, personal fees from Takeda, outside the submitted work; Dr. Aisner reports personal fees from AbbVie, personal fees from Bristol Myers Squibb, personal fees from Inivata, outside the submitted work; the other authors have nothing to disclose.
Acknowledgments
This research was supported by NIH/NCATS Colorado CTSA (UL1 TR002535) and the Hamoui Foundation and Lungevity Foundation (2022-23). This work was also supported by the CCSG grant (P30CA46934) at the University of Colorado Cancer Center supporting the use of Genomics, Biostatistics, Bioinformatics, Molecular Correlates Laboratory, and Pathology Shared Resources.
Data Availability Statement
The data generated in this study are available on request from the corresponding author.
Footnotes
Cite this article as: Patil T, Staley A, Nie Y, et al. The efficacy and safety of treating acquired MET resistance through combinations of parent and MET tyrosine kinase inhibitors in patients with metastatic oncogene-driven NSCLC. JTO Clin Res Rep. 2024;5:100637.
Note: To access the supplementary material accompanying this article, visit the online version of the JTO Clinical and Research Reports at www.jtocrr.org and at https://doi.org/10.1016/j.jtocrr.2024.100637.
Supplementary Data
References
- 1.Howlader N., Forjaz G., Mooradian M.J., et al. The effect of advances in lung-cancer treatment on population mortality. N Engl J Med. 2020;383:640–649. doi: 10.1056/NEJMoa1916623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sakamoto M., Patil T. MET alterations in advanced non-small cell lung cancer. Lung Cancer. 2023;178:254–268. doi: 10.1016/j.lungcan.2023.02.018. [DOI] [PubMed] [Google Scholar]
- 3.Remon J., Hendriks L.E., Mountzios G., et al. MET alterations in NSCLC—current perspectives and future challenges. J Thorac Oncol. 2023;18:419–435. doi: 10.1016/j.jtho.2022.10.015. [DOI] [PubMed] [Google Scholar]
- 4.Paik P.K., Felip E., Veillon R., et al. Tepotinib in non-small cell lung cancer with MET exon 14 skipping mutations. N Engl J Med. 2020;383:931–943. doi: 10.1056/NEJMoa2004407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wolf J., Seto T., Han J.Y., et al. Capmatinib in MET exon 14-mutated or MET-amplified non-small-cell lung cancer. N Engl J Med. 2020;383:944–957. doi: 10.1056/NEJMoa2002787. [DOI] [PubMed] [Google Scholar]
- 6.Choudhury N.J., Marra A., Sui J.S.Y., et al. Molecular biomarkers of disease outcomes and mechanisms of acquired resistance to first-line osimertinib in advanced EGFR-mutant lung cancers. J Thorac Oncol. 2023;18:463–475. doi: 10.1016/j.jtho.2022.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engelman J.A., Zejnullahu K., Mitsudomi T., et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 8.Bean J., Brennan C., Shih J.-Y., et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci. 2007;104:20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drilon A., Cappuzzo F., Ou S.H.I., Camidge D.R. Targeting MET in lung cancer: will expectations finally be MET? J Thorac Oncol. 2017;12:15–21. doi: 10.1016/j.jtho.2016.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Capuzzo F., Jänne P.A., Skokan M., et al. MET increased gene copy number and primary resistance to gefitinib therapy in non-small-cell lung cancer patients. Ann Oncol. 2009;20:298–304. doi: 10.1093/annonc/mdn635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stewart E.L., Tan S.Z., Liu G., Tsao M.-S. Known and putative mechanisms of resistance to EGFR targeted therapies in NSCLC patients with EGFR mutations - a review. Transl Lung Cancer Res. 2015;4:67–81. doi: 10.3978/j.issn.2218-6751.2014.11.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dagogo-Jack I., Yoda S., Lennerz J.K., et al. MET alterations are a recurring and actionable resistance mechanism in ALK-positive lung cancer. Clin Can Res. 2020;11:2535–2545. doi: 10.1158/1078-0432.CCR-19-3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCoach C.E., Le A.T., Gowan K., et al. Resistance mechanisms to targeted therapies in ROS1+ and ALK+ non-small cell lung cancer. Clin Can Res. 2018;24:3334–3347. doi: 10.1158/1078-0432.CCR-17-2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tyler L.C., Le A.T., Chen N., et al. MET gene amplification is a mechanism of resistance to entrectinib in ROS1+ NSCLC. Thorac Cancer. 2022;13:3032–3041. doi: 10.1111/1759-7714.14656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takumi Y., Arai S., Suzuki C., et al. MET kinase inhibitor reverses resistance to entrectinib induced by hepatocyte growth factor in tumors with NTRK1 or ROS1 rearrangements. Cancer Med. 2023;12:5809–5820. doi: 10.1002/cam4.5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin J.J., Liu S.V., McCoach C.E., et al. Mechanisms of resistance to selective RET tyrosine kinase inhibitors in RET fusion-positive non-small-cell lung cancer. Ann Oncol. 2020;31:1725–1733. doi: 10.1016/j.annonc.2020.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takamori S., Seto T., Yamaguchi M., et al. Case report: success of tepotinib therapy in overcoming resistance to osimertinib in a patient with EGFR-mutant lung adenocarcinoma with a potential acquired MET exon 14 skipping mutation. Front Oncol. 2022;12 doi: 10.3389/fonc.2022.965741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davies K.D., Ng T.L., Estrada-Bernal A., et al. Dramatic response to crizotinib in a patient with lung cancer positive for an HLA-DRB1-MET gene fusion. JCO Precis Oncol. 2017;1:1–6. doi: 10.1200/PO.17.00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nelson A.W., Schrock A.B., Pavlick D.C., Ali S.M., Atkinson E.C., Chachoua A. Novel SPECC1L-MET fusion detected in circulating tumor DNA in a patient with lung adenocarcinoma following treatment with erlotinib and osimertinib. J Thorac Oncol. 2019;14:e27–e29. doi: 10.1016/j.jtho.2018.10.160. [DOI] [PubMed] [Google Scholar]
- 20.Riedel R., Fassunke J., Scheel A.H., et al. Brief Report: MET fusions in non-small cell lung cancer: clinicopathologic features and response to MET inhibition. J Thorac Oncol. 2024;19:160–165. doi: 10.1016/j.jtho.2023.06.020. [DOI] [PubMed] [Google Scholar]
- 21.York E.R., Varella-Garcia M., Bang T.J., Aisner D.L., Camidge D.R. Tolerable and effective combination of full-dose crizotinib and osimertinib targeting MET amplification sequentially emerging after T790M positivity in EGFR-mutant non-small cell lung cancer. J Thorac Oncol. 2017;12:e85–e88. doi: 10.1016/j.jtho.2017.02.020. [DOI] [PubMed] [Google Scholar]
- 22.Deng L., Kiedrowski L.A., Ravera E., Cheng H., Halmos B. Response to dual crizotinib and osimertinib treatment in a lung cancer patient with MET amplification detected by liquid biopsy who acquired secondary resistance to EGFR tyrosine kinase inhibition. J Thorac Oncol. 2018;13:e169–e172. doi: 10.1016/j.jtho.2018.04.007. [DOI] [PubMed] [Google Scholar]
- 23.Aubanel M., Swalduz A., Avrillon V., et al. Combining EGFR and MET inhibition with crizotinib in EGFR-mutated lung adenocarcinoma harboring MET amplification: a brief report. Clin Lung Cancer. 2020;21:e601–e606. doi: 10.1016/j.cllc.2020.05.015. [DOI] [PubMed] [Google Scholar]
- 24.Gautschi O., Menon R., Bertrand M., Murer C., Diebold J. Capmatinib and osimertinib combination therapy for EGFR-mutant lung adenocarcinoma. J Thorac Oncol. 2020;15:e13–e15. doi: 10.1016/j.jtho.2019.07.027. [DOI] [PubMed] [Google Scholar]
- 25.Rosen E.Y., Johnson M.L., Clifford S.E., et al. Overcoming MET-dependent resistance to selective RET inhibition in patients with RET fusion-positive lung cancer by combining selpercatinib with crizotinib. Clin Can Res. 2021;27:34–42. doi: 10.1158/1078-0432.CCR-20-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chou Y.T., Lin C.C., Lee C.T., Pavlick D.C., Su P.L. Durable response of dabrafenib, trametinib, and capmatinib in an NSCLC patient with co-existing BRAF-KIAA1549 fusion and MET amplification: a case report. Front Oncol. 2022;12 doi: 10.3389/fonc.2022.838798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang K., Du R., Roy-Chowdhuri S., et al. Brief report: clinical response, toxicity, and resistance mechanisms to osimertinib plus MET inhibitors in patients with EGFR-mutant MET amplified NSCLC. JTO Clin Res Rep. 2023;4 doi: 10.1016/j.jtocrr.2023.100533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dagogo-Jack I., Kierdowski L.A., Heist R.S., et al. Efficacy and tolerability of ALK/MET combinations in patients with ALK-rearranged lung cancer with acquired MET amplification: a retrospective analysis. JTO Clin Res Rep. 2023;4 doi: 10.1016/j.jtocrr.2023.100534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noonan S.A., Berry L., Lu X., et al. Identifying the appropriate FISH criteria for defining MET copy number-driven lung adenocarcinoma through oncogene overlap analysis. J Thorac Oncol. 2016;11:1293–1304. doi: 10.1016/j.jtho.2016.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsui D.C., Drusbosky L.M., Wienke S., et al. Oncogene overlap analysis of circulating cell-free tumor DNA to explore the appropriate criteria for defining MET copy number–driven lung cancer. Clin Lung Cancer. 2022;23:630–663. doi: 10.1016/j.cllc.2022.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mazieres J., Kim T.M., Lim B.K., et al. LBA52 tepotinib + osimertinib for EGFRm NSCLC with MET amplification (METamp) after progression on first-line (1L) osimertinib: initial results from the Insight 2 study. Ann Oncol. 2022;33(suppl 7):S808–S869. [Google Scholar]
- 32.Sequist L.V., Han J.Y., Anh M.J., et al. Osimertinib plus savolitinib in patients with EGFR mutation-positive, MET-amplified, non-small-cell lung cancer after progression on EGFR tyrosine kinase inhibitors: interim results from a multicentre, open-label, phase 1b study. Lancet Oncol. 2020;21:373–386. doi: 10.1016/S1470-2045(19)30785-5. [DOI] [PubMed] [Google Scholar]
- 33.Hartmeier R.J., Markovets A.A., Anh M.J., et al. Osimertinib + savolitinib to overcome acquired MET-mediated resistance in epidermal growth factor receptor-mutated, MET-amplified non-small cell lung cancer: TATTON. Cancer Discov. 2023;13:1–16. doi: 10.1158/2159-8290.CD-22-0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCoach C.E., Blumenthal G.M., Zhang L., et al. Exploratory analysis of the association of depth of response and survival in patients with metastatic non-small-cell lung cancer treated with a targeted therapy or immunotherapy. Ann Oncol. 2017;28:2707–2714. doi: 10.1093/annonc/mdx414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soria JC, Wu YL, Nakagawa K, et al. Gefitinib plus chemotherapy versus placebo plus chemotherapy in EGFR-mutation-positive non-small-cell lung cancer after progression on first-line gefitinib (IMPRESS): a phase 3 randomised trial. Lancet Oncol 215;16:990–998. [DOI] [PubMed]
- 36.Lin J.J., Schoenfeld A.J., Zhu V.W., et al. Efficacy of platinum/pemetrexed combination chemotherapy in ALK-positive NSCLC refractory to second-generation ALK inhibitors. J Thorac Oncol. 2020;15:258–265. doi: 10.1016/j.jtho.2019.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patil T., Tsui D.C., Nicklawsky A., et al. Effect of continuing osimertinib with chemotherapy in the post-progression setting on progression-free survival among patients with metastatic epidermal growth factor receptor (EGFR) positive non-small cell lung cancer. J Clin Oncol. 2021;39(suppl 15) 9124–9124. [Google Scholar]
- 38.Tsuta K., Kozu Y., Mimae T., et al. c-MET/Phospho-MET protein expression and MET gene copy number in non-small cell lung carcinomas. J Thorac Oncol. 2012;7:331–339. doi: 10.1097/JTO.0b013e318241655f. [DOI] [PubMed] [Google Scholar]
- 39.Finocchiaro G., Toschi L., Gianoncelli L., Baretti M., Santoro A. Prognostic and predictive value of MET deregulation in non-small cell lung cancer. Ann Transl Med. 2015;3:83. doi: 10.3978/j.issn.2305-5839.2015.03.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu Y.L., Cheng Y., Zhou J., et al. Tepotinib plus gefitinib in patients with EGFR-mutant non-small-cell lung cancer with MET overexpression or MET amplification and acquired resistance to previous EGFR inhibitor (Insight study): an open-label, phase 1b/2, multicentre, randomised trial. Lancet Respir Med. 2020;8:1132–1143. doi: 10.1016/S2213-2600(20)30154-5. [DOI] [PubMed] [Google Scholar]
- 41.Ou S.H.I., Govindan R., Eaton K.D., et al. Phase I results from a study of crizotinib in combination with erlotinib in patients with advanced nonsquamous non–small cell lung cancer. J Thorac Oncol. 2017;12:145–151. doi: 10.1016/j.jtho.2016.09.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fujino T., Suda K., Koga T., et al. Foretinib can overcome common on-target resistance mutations after capmatinib/tepotinib treatment in NSCLCs with MET exon 14 skipping mutation. J Hematol Oncol. 2022;15:79. doi: 10.1186/s13045-022-01299-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rotow J.K., Gui P., Wu W., et al. Co-occurring alterations in the Ras-MAPK pathway limit response to MET inhibitor treatment in MET exon 14 skipping mutation-positive lung cancer. Clin Can Res. 2020;26:439–449. doi: 10.1158/1078-0432.CCR-19-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suzawa K., Offin M., Lu D., et al. Activation of KRAS mediates resistance to targeted therapy in MET Exon 14-mutant non-small cell lung cancer. Clin Can Res. 2019;25:1248–1260. doi: 10.1158/1078-0432.CCR-18-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peters T.L., Patil T., Le A.T., et al. Evolution of MET and NRAS gene amplification as acquired resistance mechanisms in EGFR mutant NSCLC. NPJ Precis Oncol. 2021;5:91. doi: 10.1038/s41698-021-00231-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are available on request from the corresponding author.