Abstract
Analysis of expressed sequence tags (ESTs) constitutes a useful approach for gene identification that, in the case of human pathogens, might result in the identification of new targets for chemotherapy and vaccine development. As part of the Trypanosoma cruzi genome project, we have partially sequenced the 5′ ends of 1,949 clones to generate ESTs. The clones were randomly selected from a normalized CL Brener epimastigote cDNA library. A total of 14.6% of the clones were homologous to previously identified T. cruzi genes, while 18.4% had significant matches to genes from other organisms in the database. A total of 67% of the ESTs had no matches in the database, and thus, some of them might be T. cruzi-specific genes. Functional groups of those sequences with matches in the database were constructed according to their putative biological functions. The two largest categories were protein synthesis (23.3%) and cell surface molecules (10.8%). The information reported in this paper should be useful for researchers in the field to analyze genes and proteins of their own interest.
Partial cDNA sequencing to generate expressed sequence tags (ESTs) is being used at present for the fast and efficient obtainment of a detailed profile of genes expressed in various tissues, cell types, or developmental stages (1). Genome projects have taken advantage of EST studies because ESTs represent a particular type of sequence-tagged sites useful for the physical mapping of genomes (24). ESTs can serve the same purpose as sequence-tagged sites, with the additional bonus of pointing directly to expressed genes.
One of the most interesting applications of the EST database (dbEST) is gene discovery (6). A significant development with important implications in this field has been the enormous growth of the dbEST (5). Novel genes can be found by querying the dbEST with a protein or DNA sequence. Among a number of recent examples of findings made by following this approach, a new member of the human Ly-6 family was detected (10) and 66 human ESTs were identified and mapped based on their resemblance to 66 Drosophila genes (3).
In 1994, the Special Programme for Research and Training in Tropical Diseases of the World Health Organization launched an initiative to analyze the genomes of the parasites Filaria, Schistosoma, Leishmania, Trypanosoma brucei, and Trypanosoma cruzi. Five networks were established, with the aims of (i) gaining significant knowledge on the molecular biology of these parasites; (ii) identifying new genes and their products which could be used to design new drugs, to speed up vaccine development, and to improve diagnosis; and (iii) sharing material and expertise and providing an information system that is accessible globally to researchers in the field (32).
T. cruzi is the agent of the American trypanosomiasis, Chagas’ disease, for which there is neither a definitive chemotherapeutic treatment nor a vaccine being tested at present. This parasite has a complex life cycle in the Triatomine insect vector (epimastigote and metacyclic trypomastigote parasite stages) and in the mammalian host (the bloodstream trypomastigote and the intracellular amastigote stages). Thus, the expression of a number of stage-specific genes might be related to the different environments and requirements of each parasite stage. Given these facts, and as part of the T. cruzi genome project (32), we have started a project on gene discovery through EST sequencing. A total of 1,949 ESTs were sequenced from a normalized epimastigote cDNA library of the parasite clone (CL Brener) selected for this genome project (31). Their analysis revealed that the putative functions of about 18.4% of the ESTs might be deduced by sequence comparison with genes from other organisms, while about 67% have no sequence homologies in the databases and thus might represent some T. cruzi-specific sequences.
MATERIALS AND METHODS
cDNA library.
Poly(A)+ RNA isolated from CL Brener epimastigotes was used to construct a directional cDNA library in the plasmid vector pT7T318D with a modified polylinker, which consists of the restriction sites for SfiI, EcoRI, SnaBI, BamHI, PacI, NotI, and HindIII placed between the T7 and T3 promoters (7). This reduced polylinker was necessary for the efficiency of the subsequent normalization procedure. Normalization was done by partial reassociation kinetics and hydroxyapatite chromatography, whereby the excess of abundant cDNA clones was removed (7). Further details of the construction and characterization of the normalized library will be described elsewhere. Around 23,040 clones were randomly picked and plated in 384-well microplates in the laboratory of Ulf Pettersson (Uppsala, Sweden).
Nucleotide sequencing.
Aliquots (1 to 2 μl) of each clone from 384-well microplates were grown overnight at 37°C in 3 ml of 2xTY containing 100 μg of ampicillin per ml (26). The template DNA for the sequencing reaction was prepared from 1.5 ml of culture by an alkaline lysis method with minor modifications (26), followed by a polyethylene glycol 8000 precipitation. The amount of isolated DNA template was estimated on a 1.0% agarose gel by comparison to serial dilutions of pBluescript II KS(+) (Stratagene). Sequencing reactions were performed in a Genius thermal cycler (Techne) by using a Dye Terminator Cycle Sequencing Ready Reaction Kit with AmpliTaq DNA polymerase (FS enzyme) (Applied Biosystems) according to the protocols supplied by the manufacturer and were analyzed in an ABI prism 377 sequencer (Applied Biosystems). Single-pass sequencing was performed on each template with T7 primer, and sequences longer than 100 bases were further analyzed. The ESTs were edited to remove vector sequences from 5′ ends and to remove unreliable data from the 3′ ends by using the program Factura (Perkin-Elmer).
Sequence analysis.
The sequences were compared against the National Center for Biotechnology Information (NCBI) nonredundant protein database by using the program BLASTx (2) on the BLAST network service at NCBI. Sequences that did not match sequences in the protein databases were further analyzed by searching for similarities at the nucleotide level by using the BLASTn program against the nonredundant nucleotide sequence database.
Nucleotide sequence accession numbers.
EST sequence data has been deposited in the dbEST with the following accession numbers: AA867894 to AA867980, AA882519 to AA883010, AA890742 to AA891021, AA908031 to AA908158, AA926379 to AA926628, AA952317 to AA952754, AA958023 to AA958272, and AA960728 to AA960749.
RESULTS AND DISCUSSION
A normalized cDNA library was used to reduce considerably the number of high- and intermediate-abundance sequences and to maximize the chances of finding new genes through random sequencing (28). A total of 1,994 clones were randomly selected, and the 5′ ends of the inserts were sequenced. After deletion of vector sequences and unreliable data, an average length of 420 bases per clone was obtained and used for database searches. Sequence similarities identified by the BLAST programs were considered statistically significant with a Poisson P value of ≤10−5. Among the 1,994 sequences, 31 contained no insert and 14 exhibited homology with rRNA and were excluded from further analysis.
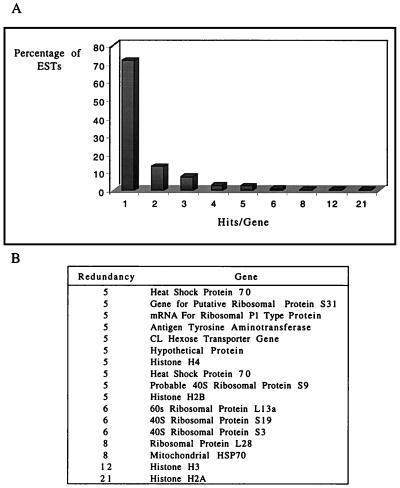
We first estimated the redundancy of our data on the basis of the redundancy of homology with sequences in the databases. A total of 644 ESTs were identified by homology with 398 different genes in the databases, representing a calculated level of redundancy of 27.9%. As shown in Fig. 1, data were classified according to the number of matches (hits) per gene. Among the 644 ESTs, 357 appeared more than once (redundant EST group), representing 111 putative genes, and 287 appeared only once. The most frequently represented genes in the library were those encoding histone H2A (accession no. gnl|PID|e290647) and histone H3 (gi|442456), which appeared 21 and 12 times, respectively (Fig. 1B). In contrast to the case for other organisms, histone transcripts in trypanosomatids are polyadenylated (19). Since the clones were picked from a normalized library, the redundancy of a cDNA clone should not be thought to represent the expression level of the gene.
FIG. 1.
Level of redundancy of ESTs that matched sequences in the NCBI nonredundant databases. (A) Percentage of ESTs with the indicated number of matches to the same gene. (B) Genes with five or more hits. The analysis was performed on a total of 644 ESTs.
On the basis of database searches, the 1,949 EST sequences were classified into four groups, as shown in Table 1. About 18.7 and 14.3% matched sequences from trypanosomatids and from other organisms, respectively. About 67% did not have a database match and thus might represent T. cruzi-specific genes. The percentage of ESTs with matches was somewhat higher (33%) than that obtained in other EST studies of protozoan parasites (11, 16, 20).
TABLE 1.
Database match categories of ESTs sequenced in T. cruzi
| EST category | No. of ESTs | % of ESTs |
|---|---|---|
| Total | 1,949 | 100 |
| Database matches to: | ||
| Total | 644 | 33 |
| T. cruzi | 285 | 14.6 |
| Other trypanosomatids | 80 | 4.1 |
| Other organisms | 279 | 14.3 |
| No database matcha | 1,305 | 67 |
ESTs without significant matches (P > 10−5) to database sequences.
Further analyses of our data were performed by taking into account only nonredundant ESTs. That is, when more than one EST showed homology to a gene annotated in the databases, only one EST was considered in the analysis.
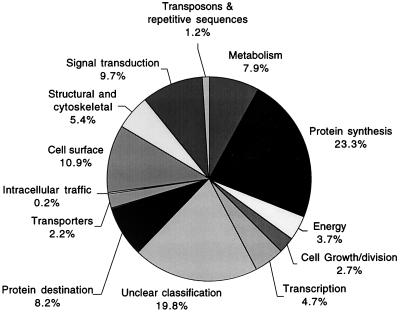
ESTs with predicted or known functions were classified into putative cellular roles (4). The proportion of ESTs in each role category is shown in Fig. 2. Of the 398 nonredundant ESTs analyzed, the largest number (23.3%) was related to protein synthesis; other categories include sequences related to metabolism (7.9%), protein destination (8.2%), transcription (4.7%), and energy (3.7%). Interestingly sequences related to cell surface proteins accounted for 10.9% of the analyzed ESTs (the second-largest category of known functions). It is well known that T. cruzi has a large number of surface proteins belonging to at least two main families: the mucin gene family and the superfamily of surface antigens.
FIG. 2.
Functional classification of T. cruzi ESTs, showing the proportions of predicted genes according to their putative biological functions. A total of 398 nonredundant ESTs having a P value of ≤10−5 were classified into 13 categories.
The mucin gene family, for which a minimum of 484 genes has been estimated (15), is composed of two groups of genes, as defined by their central domains. One group contains genes having a variable number of tandem repeats, whereas genes in the second group have nonrepetitive sequences (14). Six ESTs matched members of the mucin gene family; one matched members belonging to the former group (TENS0234), whereas the other five ESTs matched different members belonging to the second group of genes (TENS0206, TENS0592, TENS1868, TENS0163, and TENS1740).
The superfamily of surface antigens is composed of hundreds of members that can be grouped into four families (groups I to IV) based on their similarities (9, 13).
Several ESTs showed significant matches to members belonging to group II, which comprises the so-called GP85 surface glycoproteins (TENS0211, TENS0203, TENS0196, TENS0182, TENS0142, TENS0215, TENS1365, TENS0190, TENS0229, TENS1292, and TENS0222). Interestingly, the top-ranking sequences of the BLAST searches corresponding to the last two ESTs matched the sequences coding for amastigote surface protein-2 and -1, respectively, which have recently been described as the first trans-sialidase (TS) superfamily members preferentially expressed in the amastigote stage (21, 27). In contrast, members of group I (which contains some members that express TS activity), group III, and group IV were hit by only one EST each (TENS0149, TENS0779, and TENS1235, respectively).
The results reported above show that several ESTs have significant matches to trypomastigote- and amastigote-expressed members of the TS superfamily. Although these molecules are stage-specific proteins not present at detectable levels in the epimastigote stage, this result might be expected for trypanosomatids. Unlike transcriptional gene regulation in other organisms, gene regulation in these parasites takes place mainly by posttranscriptional mechanisms (23), even for the expression of stage-specific proteins (29). Thus, it is possible that a low level of trypomastigote- and amastigote-specific mature mRNAs coding for these proteins is present at the epimastigote stage, even though the encoded proteins are absent. Another possibility is that these cDNAs are derived from contaminating metacyclic trypomastigote forms (estimated to be at about 1%) present in the epimastigote culture.
We next organized the EST data set according to matches to the NCBI nonredundant databases. Table 2 lists all significant matches to non-T. cruzi entries in GenBank sorted according to matches to the “other trypanosomatids” and “other organisms” categories. In cases where several entries from various species had significant scores, only the top-ranking score is given. A complete (including matches to T. cruzi) and updated listing of matches to known sequences present in GenBank can be found at our laboratory home page (http://www.iib.unsam.edu.ar/genomelab/tcruzi/5ests.html). A detailed analysis of the putative genes identified is not within the scope of this work and will certainly be done by interested researchers in the field. However, a number of interesting matches with sequences from other organisms were observed. Among them are several proteins identified in other trypanomatids, including several metabolic enzymes (TENS1285, TENS1439, TENS1345, and TENS1204); a homolog to a recently described TRACK (receptor for activated C kinase) in T. brucei rhodesiense (TENS1408); a cyclophilin A (TENS0472); a nucleic acid-binding protein (homolog to the universal minicircle binding protein) (TENS1943); and a homolog to GP63-3 (TENS1942), a metalloprotease originally found in Leishmania and recently described for T. brucei rhodesiense (17). This protein seems to play an important role in the invasion (30) and survival (12) of the leishmanial parasites within the macrophage and has not been detected previously in T. cruzi. This result emphasizes the efficacy of the EST approach, which has allowed us to identify a gene potentially important in the host-parasite interplay.
TABLE 2.
T. cruzi EST matches to known sequences from trypanosomatids (not T. cruzi) and other organisms in NCBI databasesa
| EST (TENS no.)b | Putative identificationc | Accession no. | BLASTd |
|---|---|---|---|
| Other trypanosomatids | |||
| 1273 | 40S ribosomal protein L14 | sp|P55842| | X |
| 0051 | 40S ribosomal protein S12 | sp|Q03253| | X |
| 0057 | 40S ribosomal protein S14 | sp|P19800| | X |
| 1630 | 60S ribosomal protein L18 | sp|P50885| | X |
| 1451 | 60S ribosomal protein L30 | sp|P49153| | X |
| 1271 | Activated protein kinase C receptor homolog mRNA | gb|U72205| | N |
| 1408 | Activated protein kinase C receptor homolog TRACK | gi|2952301| | X |
| 0472 | Cyclophilin A | gi|1532210| | X |
| 1314 | Cytochrome c oxidase polypeptide I | sp|P04371| | X |
| 1285 | Fructose-bisphosphate aldolase | pir||A54500 | | X |
| 1354 | GP63-3 surface protease homolog | gi|2196917 | | X |
| 1942 | GP63-3 surface protease homolog | gi|2196917 | X |
| 0362 | H+-transporting ATPase (EC 3.6.1.35) | pir||A45598| | X |
| 1233 | Hypothetical protein 2 | pir||A05123 | | X |
| 1614 | Intergenic region from the EF-1alpha upstream-associated gene-1 to the EF-1alpha gene | gb|U52680| | N |
| 1421 | Kinetoplastid membrane protein 11 | gnl|PID|e225864 | X |
| 0020 | mRNA for S12-like ribosomal protein | emb|Z15031| | N |
| 1636 | mRNA, clone Q14R1 | emb|Z86119| | X |
| 1943 | Nucleic acid-binding protein | gi|1841864 | X |
| 0506 | ORF 1 | gnl|PID|e37082 | X |
| 1439 | Phosphoglycerate kinase | sp|P41760| | X |
| 1204 | Phosphoglycerate kinase, glycosomal | sp|P41762| | X |
| 0072 | Probable 40S ribosomal protein S9 | sp|P17959| | X |
| 1291 | Putative serine/threonine protein kinase | sp|Q08942| | X |
| 0021 | Ribosomal protein L27a | gb|U96757| | N |
| 1345 | Thioredoxin peroxidase | sp|Q26695| | X |
| Other organisms | |||
| 1260 | 1,5-Heptosyltransferase I (Rfac) and Flax genes, complete Cds | gb|U40862| | N |
| 0451 | 10-kDa heat shock protein, mitochondrial (Hsp10) | pir||S47532| | X |
| 1352 | 14-3-3-Like protein | gi|1773328| | X |
| 1290 | 2-Oxoglutarate dehydrogenase E1 component precursor | sp|P20967| | X |
| 1838 | 3,2-trans-Enoyl-coenzyme A isomerase | sp|P42125| | X |
| 1264 | 31.1-kDa protein In Dcm-Seru intergenic region | sp|P31658| | X |
| 1485 | 40S ribosomal protein | sp|Q06559| | X |
| 0047 | 40S ribosomal protein S10 | sp|Q07254| | X |
| 1750 | 40S ribosomal protein S13 | sp|Q05761| | X |
| 0904 | 40S ribosomal protein S15 | sp|P20342| | X |
| 0046 | 40S ribosomal protein S16 | sp|P46294| | X |
| 0084 | 40S ribosomal protein S17 | sp|O01692| | X |
| 0037 | 40S ribosomal protein S19 | sp|P40978| | X |
| 0012 | 40S ribosomal protein S2 | sp|P25444| | X |
| 1725 | 40S ribosomal protein S23 | sp|P39028| | X |
| 0063 | 40S ribosomal protein S25 | sp|P46301| | X |
| 0079 | 40S ribosomal protein S26e | sp|P21772| | X |
| 0053 | 40S ribosomal protein S3 | sp|Q06559| | X |
| 0045 | 40S ribosomal protein S4 | sp|P47961| | X |
| 0038 | 40S ribosomal protein S6 | sp|P02365| | X |
| 0056 | 40S ribosomal protein Sa | sp|P38981| | X |
| 0077 | 50S ribosomal protein L13 | sp|O06260| | X |
| 0949 | 55.2-kDa protein in Hxt8 5′ region | sp|P39976| | X |
| 0028 | 60S ribosomal protein L10 | sp|Q09127| | X |
| 0027 | 60S ribosomal protein L11 | sp|P42922| | X |
| 0075 | 60S ribosomal protein L12 | sp|P30050| | X |
| 0954 | 60S ribosomal protein L13a | sp|P35427| | X |
| 1482 | 60S ribosomal protein L17 | sp|P24049| | X |
| 1794 | 60S ribosomal protein L18a | sp|P41093| | X |
| 0054 | 60S ribosomal protein L2 | sp|P29766| | X |
| 1589 | 60S ribosomal protein L21 | sp|Q43291| | X |
| 1003 | 60S ribosomal protein L22 | sp|P13732| | X |
| 1923 | 60S ribosomal protein L24 | sp|P38663| | X |
| 0049 | 60S ribosomal protein L26 | sp|P47832| | X |
| 0003 | 60S ribosomal protein L26-B | sp|P53221| | X |
| 0008 | 60S ribosomal protein L3 | sp|P35684| | N |
| 0043 | 60S ribosomal protein L31 | sp|P46290| | X |
| 1875 | 60S ribosomal protein L32 | sp|Q94460| | X |
| 0081 | 60S ribosomal protein L35 | sp|P42766| | X |
| 0085 | 60S ribosomal protein L37a | sp|P32046| | X |
| 0953 | 60S ribosomal protein L5 | sp|Q26481| | X |
| 0061 | 60S ribosomal protein L7 | sp|P11874| | X |
| 1925 | 60S ribosomal protein L7b | sp|P25457| | X |
| 0033 | 60S ribosomal protein L9 | sp|P49209| | X |
| 1917 | Acidic ribosomal protein P1 | gi|2865615| | X |
| 1468 | Actin-interacting protein 2 | sp|P46681| | X |
| 1830 | Acyl carrier protein | sp|P53665| | X |
| 1801 | Adenosylhomocysteinase | pir||A45569| | X |
| 1946 | ADP-ribosylation factor 1 | sp|P35676| | X |
| 0459 | Af-9 Protein | sp|P42568| | X |
| 1326 | Alpha NAC/1.9.2. protein | gi|1142653| | X |
| 1289 | Alpha proteasome | gnl|PID|e321980 | X |
| 1374 | Alpha-adaptin | gnl|PID|d1022258 | X |
| 1381 | Alpha-enolase/tau-crystallin | gi|213085| | X |
| 1520 | Alpha-gliadin storage protein pseudogene | gb|U51305| | N |
| 1944 | TBP-interacting protein (TIP 49) | gnl|PID| d1029109 | X |
| 1301 | Alternative oxidase | dbj||AB003176_1 | X |
| 1358 | Arg kinase | prf||2020435A | X |
| 1329 | ATP synthase delta′ chain, mitochondrial precursor | sp|Q41000| | X |
| 1582 | ATP synthase F1 subunit alpha | gi|2258360| | X |
| 1242 | ATP-dependent RNA helicase, DEAD family (Dead) | gi|2648271| | X |
| 1300 | B0025.2 gene product | gi|1938574| | X |
| 0281 | BAC-146N21 chromosome X contains iduronate-2-sulfatase gene | gb|AC002315| | N |
| 0265 | BBC1 protein | gnl|PID|d1024629 | X |
| 1303 | Bop1 | gi|1679772| | X |
| 1322 | C25a1.6 | gnl|PID|e275630 | X |
| 1635 | CAGH26 mRNA | gb|U80739| | N |
| 0644 | Calmodulin | gi|167676| | X |
| 0259 | Caltractin | gb|U03270| | X |
| 1184 | Cctalpha chaperonin subunit | gi|2231589| | X |
| 1281 | Cell binding factor 2 | sp|Q46105| | X |
| 0416 | Chaperonin containing T complex polypeptide 1, beta subunit; CCT-beta | gi|2559012| | X |
| 1227 | Chromosome 21q22.2 PAC clone P169K17, complete sequence | gb|AF015720| | N |
| 1599 | Cnjb | gi|161752| | X |
| 1331 | Contains similarity to enoly-coenzyme A hydratases | gi|2854202| | X |
| 1592 | Contains similarity to human spliceosome-associated protein | gi|2384908| | X |
| 1862 | Cyclophylin | gnl|PID|e267528 | X |
| 1294 | Cytochrome b5 | gi|2062405| | X |
| 1856 | Cytochrome P450-like TBP | gnl|PID|d1011583 | X |
| 1304 | Cytoplasmic malate dehydrogenase | gi|2286153| | X |
| 1272 | Deoxyhypusine synthase mRNA | gb|U40579| | N |
| 1435 | Dihydrolipoamide acetyltransferase component (E2) of pyruvate dehydrogenase complex (Pdc-E2) | sp|P08461| | X |
| 1279 | Dihyroorotate dehydrogenase | sp|P28272| | X |
| 1851 | DNA polymerase delta small subunit | gnl|PID|e243837 | X |
| 1376 | DNA-directed DNA polymerase | pir||A55874| | X |
| 1338 | Dnaj protein | sp|P35515| | X |
| 1293 | Drome Pelota protein | sp|P48612| | X |
| 1406 | Dynein beta chain, flagellar outer arm | sp|Q39565| | X |
| 1274 | Enolase 1 | P51555 | X |
| 1320 | Enoyl-coenzyme A Hydratase, mitochondrial precursor | sp|P14604| | X |
| 0438 | Estb = esterase II | gb|S79600| | N |
| 0501 | Eukaryotic translation initiation factor 1a | sp|P38912| | X |
| 1633 | Excision repair protein Ercc-6 | sp|Q03468| | X |
| 1602 | F21b7.26 | gi|2809257| | X |
| 1313 | F421: this 421-aa ORF is 31% identical (3 gaps) to 91 residues of an approximately 864-aa protein, LOX3_SOYBN SW: P09186 | gi|1787042| | X |
| 1699 | F44g4.1 | gnl|PID|e236517 | X |
| 0581 | Fast tropomyosin isoform | gi|2660868| | X |
| 1284 | G10 protein homolog | sp|P34313| | |
| 0002 | Gene for putative ribosomal protein S31 | emb|X14247| | N |
| 0351 | Genes for ORF1, ORF2, ORF3, ORF4, and Srb, partial and complete Cds | dbj|D64116| | N |
| 1356 | Glucosamine-6-phosphate isomerase | sp|P44538| | X |
| 1722 | Glycine cleavage system H protein precursor | sp|P23434| | X |
| 1308 | GTP-binding protein Ypt3 | sp|P17610| | X |
| 1400 | Guanine nucleotide-binding protein alpha subunit | sp|P43151| | X |
| 1327 | H protein subunit of glycine decarboxylase mRNA, complete Cds | gb|AF022731| | X |
| 1687 | Heat shock protein 10 | gi|2623879| | X |
| 1493 | Heat shock protein 75 | gi|2865466| | X |
| 0670 | Heat shock protein HSLV | sp|P31059| | X |
| 1437 | Helicase | gi|780410| | X |
| 0088 | Histone H3 | sp|P40285| | X |
| 0094 | Histone H4 | gnl|PID|e324304 | X |
| 1192 | Hit family protein 1 | sp|Q04344| | X |
| 0448 | Homologous to acyl-coenzyme A dehydrogenase | gi|436861| | X |
| 0421 | Hydroproline-rich protein mRNA | gb|J03625| | X |
| 1380 | Hypothetical 20.8-kDa protein in Fgf-Vubi intergenic region | sp|P21286| | X |
| 1341 | Hypothetical 22.6-kDa protein F46c5.8 in chromosome Ii | sp|P52879| | X |
| 1328 | Hypothetical 23.5-kDa protein in Rfa2-Stb1 intergenic region | sp|P42844| | X |
| 1910 | Hypothetical 24.9-kDa protein in Sura-Hepa intergenic region | sp|P39219| | X |
| 1330 | Hypothetical 31.9-kDa protein in Gog5-Clg1 intergenic region | sp|P53081| | X |
| 1302 | Hypothetical 39.3-kDa protein in Gcn4-Wbp1 intergenic region | sp|P40004| | X |
| 1364 | Hypothetical 41.9-kDa protein in Sds3-Ths1 intergenic region | sp|P40506| | X |
| 1177 | Hypothetical 44.5-kDa protein in Pgpb-Pyrf intergenic region precursor | sp|P45576| | X |
| 1824 | Hypothetical 47.3-kDa protein in Ompx-Moeb | sp|P38821| | X |
| 1585 | Hypothetical 54.2-kDa protein in Cdc12-Orc6 intergenic region | sp|P38821| | X |
| 0386 | Hypothetical 90.8-kDa protein T05h10.7 in chromosome Ii | sp|Q10003| | X |
| 1298 | Hypothetical protein | gnl|PID|e326877 | X |
| 1385 | Hypothetical protein | pir||S57550 | X |
| 1323 | Hypothetical protein | gnl|PID|e339926 | X |
| 1618 | Hypothetical protein | gnl|PID|e276614 | X |
| 1360 | Hypothetical protein | gnl|PID|d1018647 | X |
| 1185 | Hypothetical protein and to PIR:C48583 stress-inducible protein ST11 | gi|1213541| | X |
| 1186 | Hypothetical protein YDR531w | pir||S69586 | X |
| 1812 | Hypothetical protein YPL235w | pir||S61029 | X |
| 1476 | Initiation factor 5a (Eif-5a) (Eif-4d) | sp|P56332| | X |
| 1741 | Insulinase | pir||SNHUIN | X |
| 1369 | Isocitrate dehydrogenase | gi|1277203| | X |
| 1431 | JC8.C | gnl|PID|e1247056 | X |
| 1580 | KIAA0107-like protein | gi|2982297| | X |
| 1805 | Kiaa0305 | gnl|PID|d1021601 | X |
| 1317 | L1231-38 | gi|2194152| | X |
| 1315 | L1231-6d | gi|2194149| | X |
| 1609 | L1439-18 | gi|2266918| | X |
| 1407 | L4 protein (aa 1–256) | gi|4396(X17204) | X |
| 0069 | Large ribosomal subunit protein L13 | sp|P38014| | X |
| 1392 | Male sterility 2-like protein | gnl|PID|e258459 | X |
| 1395 | Meiotic spindle formation Protein Mei-1 | sp|P34808| | X |
| 0287 | Mel-13a transcript | gb|U35309| | N |
| 1889 | Membrane-associated diazepam-binding inhibitor | prf||1911410A | X |
| 1692 | Mex-1 | gi|1899062| | X |
| 1399 | Mitochondrial trifunctional enzyme beta subunit precursor | sp|Q60587| | X |
| 1375 | Mitotic centromere-associated kinesin | prf||2103270A | X |
| 1515 | mRNA for ribosomal protein L12 | emb|X53504| | N |
| 0018 | mRNA for ribosomal protein S17 | emb|X07257| | N |
| 1443 | mRNA for surface antigen P2 | emb|X56810| | N |
| 1336 | No definition line found | gi|2384956| | X |
| 1900 | No definition line found | gi|2570931| | X |
| 1391 | Novel serine/threonine protein kinase | gnl|PID|d1006875 | X |
| 1335 | N-terminal acetyltransferase complex Ard1 subunit homolog | sp|Q05885| | X |
| 1941 | NUC-1 negative regulatory protein PREG | sp|Q06712| | X |
| 1505 | Nucleoside diphosphate kinase | sp|P27950| | X |
| 0667 | Peptidase T (aminotripeptidase) (tripeptidase) | sp|P29745| | X |
| 1311 | Peptidylprolyl Isomerase | pir||S50141| | X |
| 1905 | Peroxisome targeting signal 2 receptor | gi|1907315| | X |
| 1373 | Phosphoglucomutase isoform 1 (glucose phosphomutase) | sp|P00949| | X |
| 1347 | Phosphoinositide-specific phospholipase C | prf||2123392A | X |
| 1945 | Phosphorylation regulatory protein HP-10 | pir||A61382 | X |
| 1353 | Phosphotyrosyl phosphatase activator | gi|974837| | X |
| 1762 | Potential Caax prenyl protease 1 (prenyl protein-specific endoprotease 1) | sp|Q10071| | X |
| 0055 | Probable 60S ribosomal protein L35 | sp|P49180| | X |
| 1370 | Probable cell division control protein P55cdc | pir||A56021 | X |
| 1382 | Probable membrane protein | pir||S51473 | X |
| 1412 | Probable reductase protein | pir||A32950 | X |
| 1844 | Proteasome iota chain (macropain iota chain) | sp|P34062| | X |
| 1377 | Proteasome subunit P112 | gnl|PID|d1008506 | X |
| 1581 | Protein kinase isolog | gi|2347199| | X |
| 1359 | Protein transport protein Sec61 alpha subunit | sp|P79088| | X |
| 1393 | Putative dimethyladenosine transferase | gi|2529685| | X |
| 1390 | Putative mevalonate kinase | sp|Q09780| | X |
| 1371 | Putative protein | gnl|PID|1253348 | X |
| 0016 | Putative ribosomal protein L7A | gi|2529665| | X |
| 1250 | Pyruvate dehydrogenase E1 component, beta subunit precursor | sp|Q09171| | X |
| 1947 | RAS homolog GTPase rab28 isoform S | sp|P51157| | X |
| 1948 | RAS-related protein RAB-2 | sp|Q05975| | X |
| 0394 | RAS-related protein Rab-23 (Rab-15) | sp|P35288| | N |
| 1240 | Red-1 | gnl|PID|e209012 | X |
| 0062 | Rer1 protein | sp|P25560| | X |
| 1612 | Ribonucleoprotein La | pir||A53781 | X |
| 0026 | Ribosomal protein | gnl|PID|d1019682 | X |
| 0010 | Ribosomal protein (Rp112) | gb|L04280| | N |
| 0022 | Ribosomal protein 15a (40S subunit) | emb|Z21673| | N |
| 1882 | Ribosomal protein L10, cytosolic | pir||JN0273 | X |
| 0065 | Ribosomal protein L13.E, fruit fly | pir||S42877 | X |
| 0078 | Ribosomal protein L15.E | sp|P30736| | X |
| 0004 | Ribosomal protein L3 | sp|P39023| | X |
| 1207 | Ribosomal protein S11 homolog | pir||A48583 | X |
| 1526 | Ribosomal protein S30 | gnl|PID|e1173009 | X |
| 1332 | SC2 = synaptic glycoprotein | pir||I56573 | X |
| 1318 | Serine/threonine protein phosphatase 2b catalytic subunit, beta isoform | sp|P20651| | X |
| 1297 | Seryl-tRNA synthetase | pir||S71293 | X |
| 1758 | Similar to acetyltransferases | gi|1825778| | X |
| 1256 | Similar to mammalian ZFP36 proteins in zinc finger regions | gi|1255428| | X |
| 1819 | Similar to pig tubulin-tyrosine ligase | gnl|PID|d1012156 | X |
| 1387 | Similar to Saccharomyces cerevisiae BCS1 Protein, SWISS-PROT Accession no. P32839 | dbj||D89136_1 | X |
| 1720 | Similar to S. cerevisiae unknown, EMBL Accession no. Z68195 | gnl|PID|d1014559 | X |
| 0319 | Spermidine synthase mRNA | gnl|PID|e267359 | X |
| 1253 | Succinate dehydrogenase | gnl|PID|e341165 | X |
| 1191 | Succinyl coenzyme A synthetase alpha Subunit mRNA | gb|U23408| | N |
| 1193 | Succinyl-Coa ligase (Gdp-forming) | sp|P13086| | X |
| 1278 | Sulfated surface glycoprotein SSG185 | prf||1604369 | X |
| 1397 | Symbiosis-related protein | gi|2072023| | X |
| 1684 | T-complex protein 1, alpha subunit | sp|O15891| | X |
| 1309 | Thermostable carboxypeptidase 1 | sp|P42663| | X |
| 1288 | Thyroid receptor-interacting protein 12 | sp|Q14669| | X |
| 1949 | Translation initiation factor 5A | gnl|PID|e266087 | X |
| 1416 | Triacylglycerol lipase | sp|P21811| | X |
| 1368 | Ubiquinol--cytochrome C reductase | pir||A44033 | X |
| 1389 | UDP-glucose 4-epimerase (Gale-2) | gi|2648515| | X |
| 1405 | Unknown | gnl|PID|e223630 | X |
| 1436 | Vacuolar aminopeptidase I precursor | gi|699234| | X |
| 1307 | Wd40 repeat protein 2 | sp|P54686| | X |
| 1182 | Weak similarity to SP:YAD5_CLOAB (P33746) hypothetical protein and to PIR:C48583 stress-inducible protein STI1 | gi|1213541 | X |
| 1325 | White | gi|2182784 | X |
| 0449 | Yeast probable phosphatidylinositol-4-phosphate 5-kinase | sp|P34756| | X |
| 1324 | ZK795.D | gnl|PID|e1188511 | X |
All significant similarities (P ≤ 10−5) of nonredundant ESTs against non-T. cruzi entries in NCBI nonredundant databases are listed, together with the accession numbers and the program used for the search. Matches are sorted according to the “Other trypanosomatids” and “Other organisms” categories. A complete (including matches to T. cruzi) and more detailed table is available at http://www.iib.unsam.edu.ar/genomelab/tcruzi/5ests.html.
EST names in the dbEST are the four-digit numbers given here preceded by TENS.
ORF, open reading frame; aa, amino acids.
N, BLASTn; X, BLASTx.
Other ESTs matched known proteins in other organisms, including TATA-binding protein-interacting protein 49 (TENS1944), serine/threonine protein kinase (TENS1391), serine/threonine protein phosphatase 2b catalytic subunit (calcineurin) (TENS1318), phosphorylation-regulatory protein HP-10 (TENS1945), meiotic spindle formation proteins (TENS1395, and TENS1293), mitotic centromere-associated kinesin (TENS1375), α and p112 proteosome subunits (TENS1289 and TENS1377), DNAJ protein (TENS1338), ADP-ribosylation factor (TENS1946), a probable cell division control protein (TENS1370), several RAS-related proteins (TENS1644, -1947, -1948, and -0394), translation initiation factor 5A (TENS1949), a negative regulatory factor of a transcriptional activator (TENS1941), enolases (TENS1381 and -1274), and a phosphoinositide-specific phospholipase C (TENS1347). Interestingly this last EST showed significant matches to phosphatidylinositol-specific phospholipases C from different organisms and did not show any significant match either to an already-reported T. cruzi glycosylphosphatidylinositol-specific phospholipase C (PID|e329378) or to glycosylphosphatidylinositol-specific phospholipases from other trypanosomatids, suggesting the presence of at least two different enzymes in T. cruzi. Some of the sequences mentioned above have also been identified in a recently published paper (8).
Several ESTs had strong matches with hypothetical, probable, or putative proteins (Table 2), many of them derived from genome sequencing projects for different organisms (mouse, human, Drosophila, yeast, and Arabidopsis, etc.). Although statistically significant similarities do not necessarily mean that these putative proteins actually exist, some of the highly significant matches might indicate that they are indeed real proteins conserved during evolution. Obviously, further sequence analysis and biochemical work are needed to distinguish among these and other possible alternatives.
Until the budget for the complete sequencing of the T. cruzi genome is available, a reasonable accomplishment will be the identification of a large proportion of the gene content in T. cruzi. This might be done by EST or genomic sequencing (18) in the near future. The next step in the short run would be the analysis of the data and the development of new approaches both for the identification of targets for chemotherapy and for vaccine development. Given the difficulties in the treatment of parasitic diseases and the frequent appearance of mutants resistant to chemotherapeutic agents among some protozoa such as Plasmodium and Leishmania (22, 25), gene discovery might be a cost-efficient way to contribute to the eradication of these diseases, which mostly affect developing countries.
ACKNOWLEDGMENTS
We are indebted to Diego Rey Serantes and Judith Eva Princ for their valuable help in DNA purification and sequencing, to Lena Åslund for providing cDNAs ordered on microplates, and to J. J. Cazzulo for reading the manuscript.
This work was supported by grants from the World Bank/UNDP/WHO Special Program for Research and Training in Tropical Diseases (TDR); the Swedish Agency for Research Cooperation with Developing Countries (SAREC); the Consejo Nacional de Investigaciones Científicas y Técnicas, Argentina; and the Ministerio de Cultura y Educación, Argentina. The research of A.C.C.F. was supported in part by an International Research Scholars Grant from the Howard Hughes Medical Institute. A.C.C.F. and D.O.S. are members of the Research Career of the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), Argentina. R.E.V. is a fellow from the Universidad Nacional de General San Martín.
REFERENCES
- 1.Adams M D, Kelley J M, Gocayne J D, Dubnick M, Polymeropoulos M H, Xiao H, Merril C R, Wu A, Olde B, Moreno R F, Kerlavage A R, McCombie W R, Venter J C. Complementary DNA sequencing: expressed sequence tags and human genome project. Science. 1991;252:1651–1656. doi: 10.1126/science.2047873. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Banfi S, Borsani G, Rossi E, Bernard L, Guffanti A, Rubboli F, Marchitiello A, Giglio S, Coluccia E, Zollo M, Zuffardi O, Ballabio A. Identification and mapping of human cDNAs homologous to Drosophila mutant genes through EST database searching. Nat Genet. 1996;13:167–174. doi: 10.1038/ng0696-167. [DOI] [PubMed] [Google Scholar]
- 4.Bevan M, Bancroft I, Bent E, Love K, Goodman H, Dean C, Bergkamp R, Dirkse W, Van Staveren M, Stiekema W, Drost L, Ridley P, Hudson S A, Patel K, Murphy G, Piffanelli P, Wedler H, Wedler E, Wambutt R, Weitzenegger T, Pohl T M, Terryn N, Gielen J, Villarroel R, Chalwatzis N. Analysis of 1.9 Mb of contiguous sequence from chromosome 4 of Arabidopsis thaliana. Nature. 1998;391:485–488. doi: 10.1038/35140. [DOI] [PubMed] [Google Scholar]
- 5.Boguski M S, Lowe T M, Tolstoshev C M. dbEST—database for “expressed sequence tags.”. Nat Genet. 1993;4:332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- 6.Boguski M S, Tolstoshev C M, Bassett D E., Jr Gene discovery in dbEST. Science. 1994;265:1993–1994. doi: 10.1126/science.8091218. [DOI] [PubMed] [Google Scholar]
- 7.Bonaldo M F, Lennon G, Soares M B. Normalization and subtraction: two approaches to facilitate gene discovery. Genome Res. 1996;6:791–806. doi: 10.1101/gr.6.9.791. [DOI] [PubMed] [Google Scholar]
- 8.Brandão A, Urmenyi T, Rondinelli E, Gonzalez A, de Miranda A B, Degrave W. Identification of transcribed sequences (ESTs) in the Trypanosoma cruzigenome project. Mem Inst Oswaldo Cruz. 1997;92:863–866. doi: 10.1590/s0074-02761997000600024. [DOI] [PubMed] [Google Scholar]
- 9.Campetella O E, Sánchez D O, Cazzulo J J, Frasch A C C. A superfamily of Trypanosoma cruzisurface antigens. Parasitol Today. 1992;8:378–381. doi: 10.1016/0169-4758(92)90175-2. [DOI] [PubMed] [Google Scholar]
- 10.Capone M C, Gorman D M, Ching E P, Zlotnik A. Identification through bioinformatics of cDNAs encoding human thymic shared Ag-1/stem cell Ag-2. A new member of the human Ly-6 family. J Immunol. 1996;157:969–973. [PubMed] [Google Scholar]
- 11.Chakrabarti D, Reddy G R, Dame J B, Almira E C, Laipis P J, Ferl R J, Yang T P, Rowe T C, Schuster S M. Analysis of expressed sequence tags from Plasmodium falciparum. Mol Biochem Parasitol. 1994;66:97–104. doi: 10.1016/0166-6851(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 12.Chaudhuri G, Chaudhuri M, Pan A, Chang K-P. Surface acid proteinase (gp63) of Leishmania mexicana. J Biol Chem. 1989;264:7483–7489. [PubMed] [Google Scholar]
- 13.Cross G A, Takle G B. The surface trans-sialidase family of Trypanosoma cruzi. Annu Rev Microbiol. 1993;47:385–411. doi: 10.1146/annurev.mi.47.100193.002125. [DOI] [PubMed] [Google Scholar]
- 14.Di Noia J M, Sánchez D O, Frasch A C C. The protozoan Trypanosoma cruzihas a family of genes resembling the mucin genes of mammalian cells. J Biol Chem. 1995;270:24146–24149. doi: 10.1074/jbc.270.41.24146. [DOI] [PubMed] [Google Scholar]
- 15.Di Noia J M, D’Orso I, Åslund L, Sánchez D O, Frasch A C C. The Trypanosoma cruzimucin family is transcribed from hundreds of genes having hypervariable regions. J Biol Chem. 1998;273:10843–10850. doi: 10.1074/jbc.273.18.10843. [DOI] [PubMed] [Google Scholar]
- 16.El-Sayed N M, Alarcon C M, Beck J C, Sheffield V C, Donelson J E. cDNA expressed sequence tags of Trypanosoma brucei rhodesienseprovide new insights into the biology of the parasite. Mol Biochem Parasitol. 1995;73:75–90. doi: 10.1016/0166-6851(95)00098-l. [DOI] [PubMed] [Google Scholar]
- 17.El-Sayed N M, Donelson J E. African trypanosomes have differentially expressed genes encoding homologues of the Leishmania GP63 surface protease. J Biol Chem. 1997;272:26742–26748. doi: 10.1074/jbc.272.42.26742. [DOI] [PubMed] [Google Scholar]
- 18.El-Sayed N M, Donelson J E. A survey of the Trypanosoma brucei rhodesiensegenome using shotgun sequencing. Mol Biochem Parasitol. 1997;84:167–178. doi: 10.1016/s0166-6851(96)02792-2. [DOI] [PubMed] [Google Scholar]
- 19.Galanti N, Galindo M, Sabaj V, Espinosa I, Toro G C. Histone genes in trypanosomatids. Parasitol Today. 1998;14:64–70. doi: 10.1016/s0169-4758(97)01162-9. [DOI] [PubMed] [Google Scholar]
- 20.Levick M P, Blackwell J M, Connor V, Coulson R M, Miles A, Smith H E, Wan K L, Ajioka J W. An expressed sequence tag analysis of a full-length, spliced-leader cDNA library from Leishmania majorpromastigotes. Mol Biochem Parasitol. 1996;76:345–348. doi: 10.1016/0166-6851(95)02569-3. [DOI] [PubMed] [Google Scholar]
- 21.Low H P, Tarleton R L. Molecular cloning of the gene encoding the 83 kDa amastigote surface protein and its identification as a member of the Trypanosoma cruzisialidase superfamily. Mol Biochem Parasitol. 1997;88:137–149. doi: 10.1016/s0166-6851(97)00088-1. [DOI] [PubMed] [Google Scholar]
- 22.McKie J H, Douglas K T, Chan C, Roser S A, Yates R, Read M, Hyde J E, Dascombe M J, Yuthavong Y, Sirawaraporn W. Rational drug design approach for overcoming drug resistance: application to pyrimethamine resistance in malaria. J Med Chem. 1998;41:1367–1370. doi: 10.1021/jm970845u. [DOI] [PubMed] [Google Scholar]
- 23.Nilsen T W. Unusual strategies of gene expression and control in parasites. Science. 1994;264:1868–1869. doi: 10.1126/science.7912006. [DOI] [PubMed] [Google Scholar]
- 24.Olson M, Hood L, Cantor C, Botstein D. A common language for physical mapping of the human genome. Science. 1989;245:1434–1435. doi: 10.1126/science.2781285. [DOI] [PubMed] [Google Scholar]
- 25.Ouellette M, Papadopoulou B. Mechanisms of drug resistance in Leishmania. Parasitol Today. 1993;9:150–153. doi: 10.1016/0169-4758(93)90135-3. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 27.Santos M A, Garg N, Tarleton R L. The identification and molecular characterization of Trypanosoma cruziamastigote surface protein-1, a member of the trans-sialidase gene super-family. Mol Biochem Parasitol. 1997;86:1–11. [PubMed] [Google Scholar]
- 28.Soares M B, Bonaldo M F, Jelene P, Su L, Lawton L, Efstratiadis A. Construction and characterization of a normalized cDNA library. Proc Natl Acad Sci USA. 1994;91:9228–9232. doi: 10.1073/pnas.91.20.9228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teixeira S M, Russell D G, Kirchhoff L V, Donelson J E. A differentially expressed gene family encoding “amastin,” a surface protein of Trypanosoma cruziamastigotes. J Biol Chem. 1994;269:20509–20516. [PubMed] [Google Scholar]
- 30.Wilson M E, Hardin K K. The major concanavalin A-binding surface glycoprotein of Leishmania donovani chagasipromastigotes is involved in attachment to human macrophages. J Immunol. 1988;141:265–272. [PubMed] [Google Scholar]
- 31.Zingales B, Pereira M E, Oliveira R P, Almeida K A, Umezawa E S, Souto R P, Vargas N, Cano M I, da Silveira J F, Nehme N S, Morel C M, Brener Z, Macedo A. Trypanosoma cruzigenome project: biological characteristics and molecular typing of clone CL Brener. Acta Trop. 1997;68:159–173. doi: 10.1016/s0001-706x(97)00088-0. [DOI] [PubMed] [Google Scholar]
- 32.Zingales B, Rondinelli E, Degrave W, Franco da Silveira J, Levin M, Le Paslier D, Modabber F, Dobrokhotov B, Swindle J, Kelly J M, Åslund L, Hoheisel J D, Ruiz A M, Cazzulo J J, Pettersson U, Frasch A C C. The Trypanosoma cruzigenome initiative. Parasitol Today. 1997;13:16–22. [Google Scholar]