Summary
The response to infection is generally heterogeneous and diverse, with some individuals remaining asymptomatic while others present with severe disease or a diverse range of symptoms. Here, we address the role of host genetics on immune phenotypes and clinical outcomes following viral infection by studying genetically diverse mice from the Collaborative Cross (CC), allowing for use of a small animal model with controlled genetic diversity while maintaining genetic replicates. We demonstrate variation by deeply profiling a broad range of innate and adaptive immune cell phenotypes at steady-state in 63 genetically distinct CC mouse strains and link baseline immune signatures with virologic and clinical disease outcomes following infection of mice with herpes simplex virus 2 (HSV-2) or severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). This work serves as a resource for CC strain selection based on steady-state immune phenotypes or disease presentation upon viral infection, and further, points to possible pre-infection immune correlates of survival and early viral clearance upon infection.
Subject areas: Immunology, Virology
Graphical abstract

Highlights
-
•
The diverse response to infection is difficult to capture in a single inbred mouse model
-
•
Immune cells from 63 distinct Collaborative Cross strains were extensively phenotyped
-
•
We link baseline immune signatures with virologic and disease outcomes upon infection
-
•
This work is a resource for CC strain selection based on steady-state immune phenotypes
Immunology; Virology
Introduction
The past century has seen a dramatic advancement in our understanding of the immune system, in large part aided by study of immunity in mouse models. Mouse model systems are controllable and tunable, as laboratory mice can be selected based on age and sex and are exposed to a standardized environment including diet, antibiotics and other drugs, microbiome, and potential pathogens. Furthermore, mice are highly tractable; for example, genes can be knocked out or transgenes inserted, mice can be infected with pathogens in a highly controlled manner with longitudinal sampling, mice can be treated with investigational compounds or injected with tumors, and all types of tissues can be removed upon euthanasia so that immune cells in distinct tissue compartments can be assessed. Finally, the use of standard laboratory inbred strains of mice has allowed us to reduce variability within experiments by using genetically constrained, virtually identical mice. While this makes experiments more reproducible and lessens variability between research subjects, it minimizes the ability to examine host genetic contributions to immunity, and further, it potentially limits the applicability of the findings from such studies to improving health within the human population, the ultimate goal of biomedical research. It is widely recognized that the human immune system is characterized by massive inter-individual variation, which is likely to be due to a combination of differences between individuals in environmental factors, sex, and age, as well as host genetic differences, the latter heavily influenced by microbial infections.1,2
To improve modeling of the human immune system, we and others have made use of the Collaborative Cross (CC), first conceived of over 20 years ago. The CC is a panel of recombinant inbred (RI) mouse strains that are derived from eight founder strains (C57BL/6J, A/J, 129S1/SvImJ, NOD/ShiLtJ, NZO/HlLtJ, CAST/EiJ, PWK/PhJ, and WSB/EiJ) representing the three major Mus musculus subspecies. Through funnel breeding of these founder strains, genetic aspects of all eight founder strains are incorporated within each CC strain, followed by at least 20 generations of inbreeding,3 which has yielded over 60 inbred strains of CC mice that can be purchased. Thus, the CC provides a reproducible model of genetic variation more akin to humans, while still retaining the benefits of a small animal model system, including a controllable environment, replicate research subjects that are genetically identical, and a tractable model system.
Studies of the CC have already proved useful for immunology and infectious disease research; we and others have demonstrated immense diversity in immune phenotypes across F1 crosses of CC strains or among limited sets of CC strains.4,5,6,7 Further, differences in clinical disease symptoms following various infections have been noted across CC strains, including influenza A virus, Ebola, SARS-CoV, West Nile virus (WNV), Rift Valley fever virus, Mycobacterium tuberculosis, and Pseudomonas aeruginosa.8,9,10,11,12,13,14,15,16,17,18,19,20 This highlights that host genetics certainly play a role in host responses to infection, and yet studies involving only a single strain of inbred mice, such as C57BL/6 mice, restricts our ability to learn about immunity and disease in diverse genetic backgrounds, and is more akin to studying immunity and the host response to infection in a single individual. Expanding these studies to a larger reference population can allow for discovery of improved mouse models that better mimic human disease responses, and identification of mouse strains that can be used by the research community at large as improved disease models. For example, we previously used the CC to identify a new mouse model of chronic WNV infection that can be used to elucidate the immunological mechanisms underlying chronic flaviviral infection and disease in humans.11
It is clear that the CC is a useful resource for studying immunology and host-pathogen interactions,21 and yet without considerable resources available to perform a screen, it remains difficult to select strains of interest depending on the research question. Here, we demonstrate wide variation within innate and adaptive immune cell phenotypes at steady-state across 63 CC strains, as well as virologic and survival outcomes following infection of mice from these strains with herpes simplex virus 2 (HSV-2) or mouse-adapted severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). We provide data on these immune cell populations and clinical outcomes following virus infection as a resource to aid in CC strain selection for futures studies using this model. Additionally, we demonstrate the utility of these data and use of the CC in identifying immune correlates of survival and early viral clearance upon infection.
Results
Collaborative Cross recombinant inbred (CC-RI) mouse strains exhibit diversity in lymphocyte splenic abundance and phenotypes at steady-state
We performed a screen of genetically diverse mice from the CC for immune phenotypes within the spleen from 63 recombinant inbred CC strains (Table 1). Whole spleens from 2 male and 2 female mice from each of these strains were shipped on ice overnight from the University of North Carolina at Chapel Hill to the Fred Hutchinson Cancer Center in Seattle, and spleens were prepared for flow cytometric analysis of immune cells using three distinct panels to examine T cells, B cells, and various innate immune cells (Tables 2, 3, and 4; Figures S1–S3). Additionally, spleens from C57BL/6J and BALB/cJ mice were included in the analysis as a reference population, as these strains are commonly used as models in immunology research. As shown in Figure 1, and as we have previously shown for T cells using F1 crosses of CC strains,4 there is great diversity among the recombinant inbred CC (CC-RI) strains in any lymphocyte population examined. Further, this diversity encompasses a much greater range than the differences between C57BL6/J and BALB/cJ mice alone, with CC strains presenting with extreme phenotypes at both the low and high ends of the spectrum. In particular, the number of CD8 T cells, CD4 conventional T cells (Foxp3−; Tconv), or Foxp3+ regulatory T cells (Treg) vary by approximately 10-fold across the 63 CC strains examined (Figures 1A and 1B). There is also a range within the number of splenic B cells across CC strains, although C57BL/6J is at the high end of this range (Figure 1C). Similarly, when the frequency of each of these subsets of cells in the spleen was enumerated, we found a wide range across CC-RI strains, as expected for a genetically diverse population more similar to studying humans: the frequency of CD8 T cells among live lymphocytes varied from 1.4% to 19% across CC-RI strains, with C57BL/6J at 5.9% and BALB/cJ at 5.6%; the frequency of CD4 Tconv cells among live lymphocytes varied from 3.7% to 23.7% across CC-RI strains, with C57BL/6J at 10% and BALB/cJ at 15.4%; and the frequency of Treg among total CD4 T cells varied from 5.9% to 33.2% across CC-RI strains, with C57BL/6J at 9.3% and BALB/cJ at 11.7% (Figure 1D). The frequency of splenic B cells among live lymphocytes varied from 39.7% to 78.6% across CC-RI strains, with C57BL/6J at 60.9% and BALB/cJ at 50.7% (Figure 1D). Data from each of the mice analyzed from each CC-RI strain can be found in Tables S1 and S2 and serve as a resource for immunologists looking to select CC-RI strains off the shelf based on baseline adaptive immune cell phenotypic characteristics.
Table 1.
CC strains used for baseline immunophenotyping
| CC001/Unc | CC018/Unc | CC037/TauUnc | CC060/Unc |
| CC002/Unc | CC019/TauUnca | CC038/GeniUnca | CC061/GeniUnc |
| CC003/Unca | CC021/Unc | CC039/Unca | CC062/Unc |
| CC004/TauUnca | CC023/GeniUnca | CC040/TauUnc | CC065/Unc |
| CC005/TauUnca | CC024/GeniUnc | CC041/TauUnca | CC068/TauUnc |
| CC006/TauUnca | CC025/GeniUnca | CC042/GeniUnc | CC071/TauUnca |
| CC007/Unca | CC026/GeniUnc | CC043/GeniUnc | CC072/TauUnc |
| CC008/GeniUnca | CC027/GeniUnca | CC044/Unca | CC074/Unc |
| CC009/Unca | CC028/GeniUnc | CC045/GeniUnc | CC075/Unca |
| CC010/GeniUnca | CC029/Unc | CC046/Unc | CC078/Unca |
| CC011/Unca | CC030/GeniUnca | CC049/TauUnca | CC079/TauUnca |
| CC012/GeniUnc | CC031/GeniUnc | CC051/TauUnc | CC080/TauUnc |
| CC013/GeniUnc | CC032/GeniUnca | CC053/Unc | CC081/Unc |
| CC015/Unca | CC033/GeniUnc | CC057/Unca | CC083/Unc |
| CC016/GeniUnc | CC035/Unca | CC058/Unc | CC084/TauJUnca |
| CC017/Unc | CC036/Unca | CC059/TauUnc |
Italicized strains were used in the HSV-2 infection screen.
Strains used in the MA SARS-CoV-2 infection screen.
Table 2.
T cell panel used for baseline immunophenotyping
| Channel | Fluorochrome | Antibody | Clone | Dilution | Company | Catalog # |
|---|---|---|---|---|---|---|
| B515 | FITC | Foxp3 | FJK-16s | 1:800 | Invitrogen | 11-5773-80 |
| B710 | PerCP-Cy5.5 | Neuropilin-1 | 3E12 | 1:40 | Biolegend | 145207 |
| R660 | APC | CTLA-4 | UC10-4B9 | 1:100 | Biolegend | 106310 |
| R780 | APC-eFluor780 | CD25 | PC61.5 | 1:100 | Invitrogen | 47-0251-82 |
| G575 | PE | CD122 | 5H4 | 1:400 | Invitrogen | 12-1221-82 |
| G610 | PE-CF594 | CD49d | R1-2 | 1:800 | BD | 564395 |
| G660 | PE-Cy5 | ICOS | 7E.17G9 | 1:250 | Invitrogen | 15-9942-82 |
| G780 | PE-Cy7 | GITR | DTA-1 | 1:500 | Invitrogen | 25-5874-82 |
| V450 | Pacific Blue | Granzyme B | GB11 | 1:100 | Biolegend | 515408 |
| V510 | Aqua | Live/dead | 1:1000 | Invitrogen | L34966 | |
| V610 | BV605 | CD4 | RM4-5 | 1:400 | Biolegend | 100548 |
| V655 | BV650 | CD8α | 53–6.7 | 1:800 | Biolegend | 100742 |
| V710 | BV711 | CD44 | IM7 | 1:400 | Biolegend | 103057 |
| V780 | SB780 | PD-1 | J43 | 1:80 | Invitrogen | 78-9985-82 |
| U395 | BUV395 | CD3 | 145-2C11 | 1:1600 | BD | 563565 |
| U730 | BUV737 | CD62L | MEL-14 | 1:200 | BD | 612833 |
Table 3.
B cell panel used for baseline immunophenotyping
| Channel | Fluorochrome | Antibody | Clone | Dilution | Company | Catalog # |
|---|---|---|---|---|---|---|
| B515 | FITC | CD23 | B3B4 | 1:50 | BD | 553138 |
| R660 | APC | CD40 | HM40-3 | 1:160 | Invitrogen | 17-0402-82 |
| G575 | PE | CD21/CD35 | 7E9 | 1:320 | Biolegend | 123409 |
| G780 | PE-Cy7 | IgM | R6-60.2 | 1:20 | BD | 552867 |
| V450 | Pacific Blue | IgD | 11-26c.2a | 1:100 | Biolegend | 405712 |
| V510 | Aqua | Live/dead | 1:1000 | Invitrogen | L34966 | |
| V780 | BV786 | CD45R/B220 | RA3-6B2 | 1:400 | BD | 563894 |
| U395 | BUV395 | CD3 | 145-2C11 | 1:1600 | BD | 563565 |
| U660 | BUV661 | CD19 | 1D3 | 1:50 | BD | 612971 |
Table 4.
Innate immune cell panel used for baseline immunophenotyping
| Channel | Fluorochrome | Antibody | Clone | Dilution | Company | Catalog # |
|---|---|---|---|---|---|---|
| B515 | BB515 | CD11b | M1/70 | 1:200 | BD | 564455 |
| B710 | PerCP-Cy5.5 | Ly-6C | HK1.4 | 1:150 | Invitrogen | 45-5932-80 |
| R660 | APC | CD40 | HM40-3 | 1:160 | Invitrogen | 17-0402-82 |
| R780 | APC-eFluor780 | CD8α | 53–6.7 | 1:100 | Invitrogen | 47-0081-80 |
| G575 | PE | CD86 | PO3.1 | 1:160 | Invitrogen | 12-0861-81 |
| G610 | PE/Dazzle594 | F4/80 | BM8 | 1:200 | Biolegend | 123145 |
| G660 | PE-Cy5 | CD80 | 16-10A1 | 1:200 | Invitrogen | 15-0801-81 |
| V450 | eFluor450 | MHC Class II | M5/114.15.2 | 1:800 | Invitrogen | 48-5321-80 |
| V510 | Aqua | Live/dead | 1:1000 | Invitrogen | L34966 | |
| V610 | BV605 | CD4 | RM4-5 | 1:400 | Biolegend | 100548 |
| V710 | BV711 | NKp46 | 29A1.4 | 1:100 | Biolegend | 137621 |
| U396 | BUV395 | Ly-6G | 1A8 | 1:50 | BD | 563978 |
| U500 | BUV496 | CD11c | N418 | 1:133 | BD | 750450 |
| U570 | BUV563 | CD3 | 145-2C11 | 1:200 | BD | 749277 |
| U660 | BUV661 | CD19 | 1D3 | 1:50 | BD | 612971 |
Figure 1.
Heterogeneity in abundance of T and B lymphocytes in the spleen of Collaborative Cross mouse strains
The spleens from 2 male and 2 female 10-week-old mice from each of 63 CC recombinant inbred mouse strains, as well as C57BL/6J and BALB/cJ mice were prepared for flow cytometric analysis of T and B cell populations using antibodies as described in Tables 2 and 3, respectively.
(A) The number of CD8+ T cells, CD4+Foxp3− conventional T cells, or Foxp3+ Treg are quantified and rank-ordered, with each black histogram bar representing the number from an individual CC strain.
(B) Heatmaps showing the mean number of CD8 T cells, CD4 Foxp3− conventional T cells (Tconv) or Foxp3+ Treg across the strains examined.
(C) The number of CD3−CD19+ B cells are quantified and rank-ordered.
(D) The fraction of CD8 T cells (as a % of live lymphocytes), CD4 conventional T cells (T conv; as a % of live lymphocytes), Treg (as a % of total CD4 T cells), or B cells (as a % of live lymphocytes) is depicted.
In addition to examining baseline splenic T and B cell numbers and frequencies, we more deeply characterized these cell subsets using many phenotypic and activation markers (Tables 2, 3, S1, and S2; Figure S4). While there is heterogeneity across the CC-RI strains for any immune phenotype measured, we herein display a limited set of immune cell phenotypes to demonstrate the extensive heterogeneity seen across the CC-RI strains (Figures 2 and S4). For example, the frequency of CD8 T cells expressing CD44 ranged from 11.5% to 79.7% across CC strains, and the frequency of CD8 T cells with a CD49dloCD122+CD44+ virtual memory T cell (TVM) phenotype ranged from 1.7% to 39.1%, whereas for common inbred lab strains of C57BL/6J and BALB/c, the frequency is less than 10% (Figure 2A). Thus, study of unconventional memory CD8 T cells could benefit from use of CC strains with extreme phenotypes of either very high or very low abundance TVM, for example to better understand the signals that govern or restrict their development. Furthermore, the frequency of CD8 T cells with a central memory (TCM) or effector memory (TEM) phenotype also varies dramatically across the CC (8.7%–73.1% for TCM and 2.0%–21.7% for TEM; Figure 2A). There is also a wide range of phenotypes in conventional, Foxp3− CD4 T cells, with 8.0%–56.5% expressing CD44 across CC strains, and 0.9%–11.2% for CD25 (Figure 2B). For Foxp3+ Treg, there is similarly a high degree of heterogeneity in the frequency of Tregs expressing activation or suppression markers CTLA-4, ICOS, PD-1, and Nrp1 across CC strains (Figure 2C), thereby suggesting that not only the abundance but also the potential functional and phenotypic properties vary across genetically distinct strains of mice. Thus, CC strains could be utilized as models to investigate Treg biology and mechanisms of immunosuppression. Finally, we also assessed various B cell populations, including mature B cells, follicular B cells, marginal zone B cells, and transitional B cells, and identified a wide range of abundance of each of these cell types across CC strains that far exceeds the differences seen in the two common laboratory inbred strains examined (C57BL/6J and BALB/cJ; Figures 2D and S4D). In sum, we demonstrate that use of the Collaborative Cross mouse population models the diversity in lymphocyte populations and phenotypes observed across humans, and our data provides a resource for those seeking mouse models with specific immunologic signatures or characteristics beyond what can be achieved by examination of a single common inbred laboratory strain of mice.
Figure 2.
The frequency of T and B cell subsets and activation states varies within different recombinant inbred strains of the Collaborative Cross
Spleens from 2 male and 2 female mice from each of 63 CC recombinant inbred mouse strains, as well as C57BL/6J and BALB/cJ mice were prepared for flow cytometric analysis of T and B cell subsets using antibodies described in Tables 2 and 3, respectively. The percent CD44+ of CD8 T cells, % CD49dloCD44+CD122+ virtual memory (TVM) of CD8 T cells, % CD44+CD62L + central memory (TCM) of CD8 T cells, % CD44+CD62L- (TEM) of CD8 T cells, and % CD25+ of CD8 T cells is shown in (A). The percent CD44+ of CD4 Tconv cells or % CD25+ of CD4 Tconv cells is shown in (B). The percent of Foxp3+ Treg that are CTLA-4+, ICOS+, PD-1+, or Nrp1+ are shown in (C). The percent of mature, IgD+ B cells, the % of IgD+CD23+ follicular B cells, the % of IgD+CD23−CD21/35hi marginal zone (MZ) B cells, and the % of IgD− transitional B cells as a fraction of live lymphocytes is shown in (D).
Baseline abundance and phenotype of innate immune cells varies within CC-RI strains
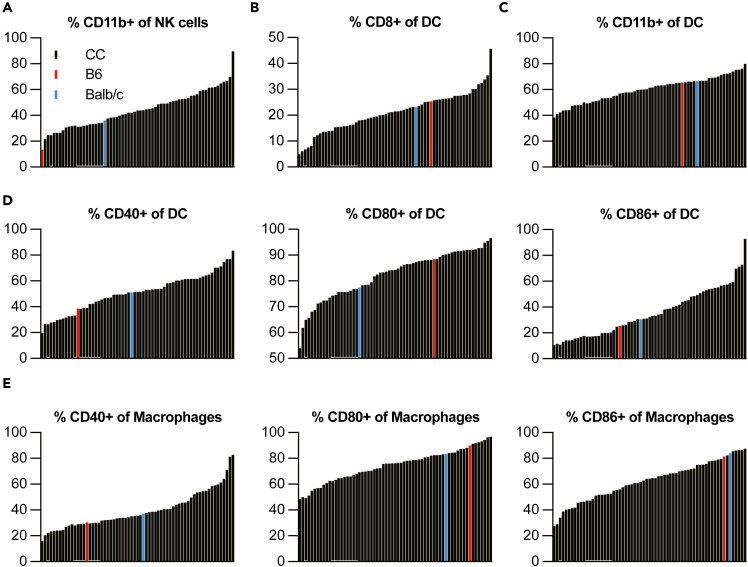
In addition to characterizing lymphocytes, we also profiled innate immune cells within the spleen of 63 CC mouse strains (Table 1) at steady state using a flow cytometry panel with markers for neutrophils, natural killer (NK) cells, monocytes, macrophages, and dendritic cells (DC; Tables 4 and S3; Figure S3). We first examined the number and frequency of each of these categories of cells across the CC strains as well as in C57BL/6J and BALB/cJ mice (Figure 3). Like our findings with lymphocytes (Figure 1), there is a great deal of diversity in abundance of each of these types of innate immune cells across the CC, with generally a 10-fold or more difference in the cell number and a wide range of frequency as well (Figure 3). When we characterize activation or phenotypic markers within these innate immune cell subsets, we identified additional heterogeneity (Figure 4). For example, for NK cells, we found that the fraction of cells expressing maturation marker CD11b ranged from 21.8 to 89.6% at steady state (Figure 4A). For splenic DC, we observed a very large difference in the frequency of CD8a+ and CD11b+ DCs across CC strains of 4.9%–45.7% and 38.3%–79.9%, respectively (Figures 4B and 4C). There are also sizable differences in the fraction of activated DC and macrophages in the spleen, as detected by percent of cells expressing CD40, CD80, or CD86 at baseline (Figures 4D and 4E). Thus, it is possible that antigen presentation varies significantly across these genetically distinct mouse strains, and thus the CC could be useful for studying innate immune signaling and consequences.
Figure 3.
The number and frequency of neutrophils, NK cells, and antigen-presenting cell subsets in the spleen varies with host genetics
The spleens from 2 male and 2 female mice from each of 63 CC recombinant inbred mouse strains, as well as C57BL/6J and BALB/cJ mice were prepared for flow cytometric analysis of innate immune cell populations using antibodies as described in Table 4.
(A) Number of splenic neutrophils (CD3−CD19-Ly-6G + CD11b+) or % neutrophils of total live lymphocytes are shown rank ordered by mouse strain, with each black histogram bar representing the number from an individual CC strain.
(B) Number of splenic NK cells (CD3−CD19-Ly-6G-NKp46+) or % NK cells of total live lymphocytes are shown rank ordered by mouse strain, with each black histogram bar representing the number from an individual CC strain.
(C) Number of splenic monocytes (CD3−CD19-Ly-6G-Ly-6C-hiCD11b+) or % monocytes of total live lymphocytes are shown rank ordered by mouse strain, with each black histogram bar representing the number from an individual CC strain.
(D) Number of splenic macrophages (CD3−CD19-Ly-6G-F4/80+) or % macrophages of total live lymphocytes are shown rank ordered by mouse strain, with each black histogram bar representing the number from an individual CC strain.
(E) Number of splenic dendritic cells (DC; CD3−CD19-Ly-6G-F4/80-CD11c+) or % DC of total live lymphocytes are shown rank ordered by mouse strain, with each black histogram bar representing the number from an individual CC strain.
(F) Heatmap showing the mean frequency of indicated populations of innate immune cells of total live lymphocytes in the spleen of CC strains.
Figure 4.
Variability in activation status of splenic innate immune cells across mouse strains within the Collaborative Cross
The spleens from 2 male and 2 female mice from each of 63 CC recombinant inbred mouse strains, as well as C57BL/6J and BALB/cJ mice were prepared for flow cytometric analysis of innate immune cell populations using antibodies as described in Table 4.
(A) The percent of NK cells that are CD11b+ are rank ordered by mouse strain, with each black histogram bar representing the number from an individual CC strain.
(B) The % of DC that are CD8+ are rank ordered by mouse strain, with each black histogram bar representing the number from an individual CC strain.
(C) The % of DC that are CD11b+ are rank ordered by mouse strain, with each black histogram bar representing the number from an individual CC strain.
(D) The % of DC that are CD40+, CD80+, or CD86+ are rank ordered by mouse strain, with each black histogram bar representing the number from an individual CC strain.
(E) The % of macrophages that are CD40+, CD80+, or CD86+ are rank ordered by mouse strain, with each black histogram bar representing the number from an individual CC strain.
Use of CC-RI strains to model vaginal HSV-2 infection outcomes
A second cohort of mice from 22 CC strains (Table 1) was infected vaginally with wild-type (WT) HSV-2. Whereas WT HSV-2 is generally lethal in naive, SPF C57BL/6 mice,22 our screen of 22 strains led to identification of 6 strains that had a degree of survival following infection (CC001, CC003, CC013, CC018, CC030, and CC065, from 20 to 60% survival; Figure 5A). Thus, these inbred mouse strains could prove useful for immunologic studies, including those of memory responses to WT HSV-2 infection, since lethality in the C57BL/6 model precludes such efforts.
Figure 5.
Distinct baseline splenic immune phenotypes correlate with survival upon vaginal HSV-2 infection in CC mouse strains
Five mice from each of 22 CC strains (as identified in Table 1) were infected intravaginally with HSV-2 and survival to day 30 was assessed.
(A) The percent survival is shown for the survival and no survival strains. The survival strains are CC001, CC003, CC013, CC018, CC030, and CC065. The no survival strains are CC012, CC015, CC016, CC019, CC023, CC025, CC029, CC035, CC040, CC041, CC043, CC059, CC061, CC074, CC075, and CC078.
Another cohort of these CC strains were used to evaluate baseline splenic T cell, B cell, and innate immune cell phenotypes by flow cytometry using the markers shown in Tables 2, 3, and 4. Pre-infection splenic immune phenotypes were examined for each of the 22 strains in these two groups and the statistically significant associations are shown for T cells are shown in (B). All associations tested for Treg are shown in the heatmap in (C), with an asterix indicating statistical significance. Statistically significant associations for B cells are shown in (D), and for innate immune cells in (E). Statistical significance was determined by two-tailed unpaired t test, with ∗ indicating p 0.05, ∗∗ indicating p 0.01, and ∗∗∗ indicating p 0.001.
Baseline immunophenotype signatures predict survival following HSV-2 infection in CC-RI strains
In addition to identification of improved mouse models, our discovery of CC mouse strains that survive HSV-2 infection affords us the ability to study baseline immune correlates of survival upon infection to identify elements of a successful anti-HSV-2 immune response. To this end, we looked for baseline splenic immune signatures that correlated with survival upon WT vaginal HSV-2 infection. We first looked for evidence that abundance of splenic T or B cells at baseline prior to infection could predict HSV-2 survival outcome and found no statistically significant difference in number of CD8 T cells, CD4 Tconv, or B cells, though an increased number of splenic Tregs at baseline was associated with survival upon infection with WT HSV-2 (Figure S5A). In addition, we looked at numbers of 18 different CD8 T cell types in the spleen (Figure S5B) and found that increased numbers of granzyme B+ CD8+ TVM, granzyme B+ CD8 T cells, and granzyme B+ TCM in the spleen at baseline correlated with survival upon subsequent HSV-2 infection (Figures 5B and S5B). Given that we have recently demonstrated that CD8 T cells can act in an innate-like fashion to provide local protection upon vaginal HSV-2 infection,22 we hypothesize that having an increased number of CD8 T cells armed with granzyme B at steady state could contribute to improved protection from and survival upon HSV-2 infection. We also compared the numbers of 8 different phenotypes of CD4 Tconv, none of which were statistically significantly different in strains that survived HSV-2 infection compared to those that did not (Figure S5C). Given that we observed a difference in baseline Treg numbers, we more deeply profiled Treg phenotypes through examination of 26 different Treg phenotypes (Figure 5C) and found 16 of these phenotypes associated with survival from HSV-2 infection (Figures 5C and S6), thereby suggesting that Tregs may play a protective role following vaginal HSV-2 infection. As an example, Treg-mediated regulation of an innate like TVM-driven response could be beneficial to restrict excess immunopathology from a rapid and exuberant response. Indeed, we and others have previously demonstrated that Tregs are essential to coordinate and restrict robust anti-pathogen immune responses in order to limit damage to host tissue,23,24,25,26,27,28,29,30,31,32,33,34 and so this could be at play as well with survival upon HSV-2 infection.
In addition to T cells, we also more deeply profiled the numbers of different B cell types in the spleen at baseline, including mature B cells, follicular B cells, marginal zone B cells, transitional B cells, and T1, T2, and T3 B cells (Figure S5D). Notably, the only significant association we found that correlated with survival upon vaginal HSV-2 infection was an increased number of T1 cells (Figures 5D and S5D). Finally, we also investigated abundance of 25 different categories of innate immune cells to probe for baseline splenic immunophenotypes that predicted survival upon HSV-2 infection (Figure S5E). We found that an increased number of CD86+ DC and CD86+ CD11b+ DC in the baseline spleen were each associated with survival upon HSV-2 infection (Figures 5E and S5E). In summary, we report that use of the CC as a genetic reference population is useful to describe and model the heterogeneity within the host immune system that is reported to be present in the more genetically diverse human population, as well as to model the different survival outcomes upon vaginal HSV-2 infection.
Steady-state immune correlates of early viral clearance upon SARS-CoV-2 MA10 infection
A third cohort of mice from 32 CC strains (Table 1) was used for infection studies using the mouse-adapted SARS-CoV-2 MA10 strain.35,36 Male and female mice from these 32 strains were infected intranasally with SARS-CoV-2 MA10, and cohorts of mice were euthanized at day 7 post-infection. At the time of euthanasia, lung viral loads were determined by viral plaque assay. We identified 18 strains that had early viral clearance by day 7 post-infection, while 14 strains had some degree of viral persistence (Figure 6A). To determine if baseline immune signatures could predict which mice would go on to have persistent virus present in lung tissue, we looked for differences in steady-state baseline splenic immune cell abundance as well as differences in phenotypic or functional aspects of different immune cell populations (immunophenotype categories as for the HSV-2 analysis shown in Figures S5 and 5C) in strains from these two groups—viral persistence or early viral clearance. Notably, we found a statistically significant difference in the number of granzyme B+ virtual memory CD8 T cells (TVM), with elevated numbers found prior to infection in the group that would go on to clear SARS-CoV-2 from the lung by day 7 post-infection (Figure 6B). TVM are unconventional memory-like CD8 T cells that are CD49dlo but express CD44 and CD122 and develop without exposure to foreign antigens. TVM are poised to rapidly produce effector molecules including IFNγ following exposure to proinflammatory cytokines,37,38 and indeed confer bystander-mediated protection against pathogens including Listeria monocytogenes and influenza virus.39,40,41 Thus, our data point to an additional role for TVM in contributing to timely viral clearance of SARS-CoV-2 from the lung.
Figure 6.
Lung viral persistence following SARS-CoV-2 infection correlates with reduced abundance of potentially cytolytic T cells and NK cells in the spleen
Viral loads at day 7 post intranasal SARS-CoV-2 MA10 infection were measured from 32 CC strains (shown in Table 1). Strains with virus in the lungs at day 7 post-infection were considered to have delayed viral clearance (CC010, CC011J, CC025, CC026, CC035J, CC036, CC037, CC038, CC039, CC041, CC044J, CC049, CC071, and CC079J), whereas strains with no measurable virus in the lungs at day 7 p.i. were considered to have viral clearance (CC001, CC003, CC004, CC005, CC006, CC007, CC008, CC009, CC015, CC018, CC019, CC023, CC030J, CC032J, CC057, CC075J, CC078, and CC084J). The day 7 viral loads from these 32 CC strains categorized into delayed and clearance groups are depicted in (A). A second cohort of these CC strains were used to evaluate baseline splenic T cell, B cell, and innate immune cell phenotypes by flow cytometry using the markers shown in Tables 2, 3, and 4. Pre-infection splenic immune phenotypes were examined for each of the 32 strains in these two groups and the statistically significant associations are shown in (B–D). Statistical significance was determined by two-tailed unpaired t test, with ∗ indicating p 0.05.
In addition to a difference in TVM, we also identified a difference in the number of splenic transitional-3 (T3) B cells based on day 7 post-infection viral clearance. Transitional B cells in mice are immature and IgD−, and can be further divided into T1, T2, and T3 cells based on expression of CD23 and IgM, with T3 cells being CD23+ and IgM−.42 We found a statistically significant increase in the number of splenic T3 B cells in mice that went on to clear virus by day 7 post-infection upon SARS-CoV-2 infection (Figure 6C). As opposed to T1 and T2 B cells, T3 B cells do not give rise to mature B cells and in fact are hyporesponsive upon stimulation through the B cell receptor,43 and so their function in anti-pathogen immunity is unclear.
Finally, we found a statistically significant increased number of CD11b+ NK cells in the spleen of CC strains that will successfully clear virus by day 7 post-SARS-CoV-2 infection (Figure 6D). CD11b has been used as a marker to define maturation states of NK cells, with CD11b+ NK cells having a stronger cytotoxic potential compared to CD11b− NK cells.44,45 Thus, it is possible that having a larger baseline population of these more mature and potentially cytotoxic NK cells prior to infection sets up the host for enhanced and rapid viral control during the innate immune response. In sum, we show an example whereby the CC is useful to more successfully model the variety of virologic outcomes following a viral infection in the human population. Further, use of improved mouse models of human disease may assist in identification of immune correlates of protection from severe and/or prolonged disease. Finally, we also use our data as a model for how the CC can be used for immunology and infectious disease research, and here demonstrate, without the need for prospectively collected longitudinal human samples, that pre-infection innate and adaptive immune signatures associate with survival or viral pathogenesis upon HSV-2 and SARS-CoV-2 infection.
Discussion
We and others have previously investigated immunological phenotypes within different cohorts of CC mice with the goal to better model the genetic diversity found in the human population. Rather than using knockout mice, in which a phenotype is either present or absent, the use of the CC allows for a range of immune phenotypes, more similar to what is observed in the human population. Collin et al. investigated 18 immunological traits in CC mice, focusing on subsets of NK cells, DCs, and T cells, which led to identification of genetic loci linked to some of these immune traits.6 In our study, we expand upon these 18 immunological traits to now profile hundreds of different immunophenotypes at steady state within the CC (Tables S1, S2, and, S3). Additionally, other groups have also explored post-infection immune responses using CC mice. Martin et al. used an LCMV model to characterize subsets of effector and memory T cells in 47 strains of CC mice, observing that the size of the memory CD8 T cell pool correlated with the size of the effector CD8 T cell pool formed after this systemic infection, with these subsets varying greatly between different strains.5 Jensen et al. used the CC mice to further test and identify a specific strain that would be useful for examining adaptive NK cells in a model of cytomegalovirus infection, identifying a novel mouse model.46 In our previous studies, we used F1 crosses of CC-RI strains to investigate immunity in the context of WNV infection.12 These RIX lines were heterozygous for the H-2b MHC haplotype, allowing for the use of WNV-specific T cells by using major histocompatibility complex (MHC) class I tetramer. While this provided additional antigen-specific phenotypic data, these crosses were individually bred for that screening project. Using the knowledge and experience gained from these previously mentioned studies, we were able to conduct our current study using CC-RI strains that are available for purchase, and we opted to use three distinct cohorts so that both pre- and post-infection data could be collected using two different virus infection models. Unlike previous studies, we used three high-parameter flow cytometry panels to enable a deep profiling of not only T cells, but also B cells, NK cells and other innate immune cells, and antigen-presenting cells. Thus, our data builds on previous resources to now profile even more cell types and markers in the near full panel of CC mice that are commercially available from the Systems Genetics Core Facility at UNC-Chapel Hill.
It is clear that the CC is a useful resource for studying immunology and host-pathogen interactions, yet without considerable resources available to perform a screen, it remains difficult to select strains of interest depending on the research question. We provide data on these immune cell populations and outcomes following mucosal HSV-2 and SARS-CoV-2 infection as a resource to aid in CC strain selection for futures studies using this model. Using extensive immunophenotyping panels in this population, we demonstrate wide variation within innate and adaptive immune cell phenotypes at steady-state across 63 CC strains, as well as virologic and survival outcomes following infection of mice from these strains with HSV-2 or mouse-adapted SARS-CoV-2. We demonstrate the utility of these data and use of the CC in identifying immune correlates of protection from death upon HSV-2 infection or from viral persistence following SARS-CoV-2. Future studies could involve smaller cohorts of CC strains selected to study specific outcomes that were observed in the screen, for example, an investigation of post-acute sequelae of SARS-CoV-2 (PASC) could be conducted using strains that were found to show viral persistence in this study. Finally, identification of pre-infection immune correlates of protection from viral persistence upon SARS-CoV-2 infection has the potential to identify individuals that may need additional risk mitigation and/or supportive care upon infection, and importantly, use of the CC mouse model enabled this discovery, whereas a very large and prospectively collected human cohort would have been required to perform a similar study in humans.
In summary, we hereby present a comprehensive characterization of different types of immune cells in the spleen of mice from 63 CC-RI strains. Further, we profile viral pathogenesis and survival outcomes following mouse-adapted SARS-CoV-2 or vaginal HSV-2 infection, respectively, in these CC-RI strains. This allowed us to demonstrate the utility of the CC in modeling different disease states observed in humans during the COVID-19 pandemic. Our study also serves to point to the utility of the CC in identification of baseline immune correlates of survival or protection from viral persistence upon virus infection.
Limitations of the study
While our data provide a powerful resource for the research community studying immunology and infectious disease, there are limitations to note from our study. First, having three cohorts of mice for each strain, one pre-infection, and two post-infection, afforded us the ability to correlate baseline phenotypes to infection outcomes such as survival or viral persistence. However, we do not have corresponding post-infection flow cytometry data to compare to baseline immunophenotypes in these cohorts. Such studies will be useful, for example to further elucidate elements of a successful immune response to HSV-2 and SARS-CoV-2, as well as other infections, across CC strains using a larger sample size. In addition, we utilize our post-infection outcome data to demonstrate the potential utility of the CC to define host immune factors that are linked to susceptibility or resistance to infection, but our data are thus far only correlative. Future studies will be required to mechanistically define immune parameters, either pre-infection or post-infection, that are required for resistance or susceptibility to particular clinical outcomes following infections. Furthermore, in our study of SARS-CoV-2-infected mice, the latest endpoint post-infection was day 7, but we found that not all mice had cleared virus by this time point. Future studies of CC strains presenting with viral persistence at later time points would be particularly useful in understanding the types of immune responses that lead to viral clearance without immune-mediated pathology and limited clinical symptoms and could possibly be useful in elucidating immune correlates of protection from PASC. Finally, while our studies using the CC allowed for examination of effects of host genetics on immunity and infection outcomes, there are other environmental factors that we did not account for or examine and yet that are known to impact immunity, including age, season, and microbiota or previous infection history.47,48,49
In addition to these technical limitations to our study, several limitations to the use of the CC model exist, many of which are related to the many genetic differences between the individual strains. For example, we have evaluated differences in survival to WT HSV-2 infection in mice, and identified six lines of the 22 screened that survive infection, unlike B6 mice. However, further studies must be performed to evaluate the immune mechanisms behind this survival, and we hypothesize that this may be due to several different factors based on distinct genetic differences in those six individual mouse strains. At this point, we cannot identify if survival comes from limiting initial viral replication, clearing the virus more rapidly, limiting tissue damage or disease dissemination—or likely, a complex combination of these factors. For these reasons, investigators may need to perform a “mini screen” of strains of interest in order to better select phenotypes for modeling and narrow down the strains for further studies. Finally, cost is also a limiting factor, as the purchase price of one CC mouse is approximately an order of magnitude higher than a standard inbred laboratory strain such as C57BL/6. Due to this high cost associated with use of CC mice, we hope that the dataset presented herein will be useful to focus in on particular CC strains when designing future studies.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Foxp3 FITC | Invitrogen | Cat# 11-5773-80; RRID:AB_465242 |
| Neuropilin-1 PerCP-Cy5.5 | Biolegend | Cat# 145207; RRID:AB_2562033 |
| CTLA-4 APC | Biolegend | Cat# 106310; RRID:AB_2087653 |
| CD25 APC-eFluor780 | Invitrogen | Cat# 47-0251-82; RRID:AB_1272179 |
| CD122 PE | Invitrogen | Cat# 12-1221-82; RRID:AB_465833 |
| CD49 d PE-CF594 | BD Biosciences | Cat# 564395; RRID:AB_2738788 |
| ICOS PE-Cy5 | Invitrogen | Cat# 15-9942-82; RRID:AB_468826 |
| GITR PE-Cy7 | Invitrogen | Cat# 25-5874-82; RRID:AB_10548516 |
| Granzyme B Pac Blue | Biolegend | Cat# 515408; RRID:AB_2562196 |
| CD4 BV605 | Biolegend | Cat# 100548; RRID:AB_2563054 |
| CD8α BV650 | Biolegend | Cat# 100742; RRID:AB_2563056 |
| CD44 BV711 | Biolegend | Cat# 103057; RRID:AB_2564214 |
| PD-1 SB780 | Invitrogen | Cat# 78-9985-82; RRID:AB_2734947 |
| CD3 BUV395 | BD Biosciences | Cat# 563565; RRID:AB_2738278 |
| CD62L BUV737 | BD Biosciences | Cat# 612833; RRID:AB_2870155 |
| CD23 FITC | BD Biosciences | Cat# 553138; RRID:AB_394653 |
| CD40 APC | Invitrogen | Cat# 17-0402-82; RRID:AB_10853008 |
| CD21/CD35 PE | Biolegend | Cat# 123409; RRID:AB_940413 |
| IgM PE-Cy7 | BD Biosciences | Cat# 552867; RRID:AB_394500 |
| IgD Pac Blue | Biolegend | Cat# 405712; RRID:AB_1937244 |
| CD45R/B220 BV786 | BD Biosciences | Cat# 563894; RRID:AB_2738472 |
| CD19 BUV661 | BD Biosciences | Cat# 612971; RRID:AB_2870243 |
| CD11b BB515 | BD Biosciences | Cat# 564454; RRID:AB_2665392 |
| Ly-6C PerCP-Cy5.5 | Invitrogen | Cat# 45-5932-80; RRID:AB_2723342 |
| CD40 APC | Invitrogen | Cat# 17-0402-82; RRID:AB_10853008 |
| CD8α APC-eFluor780 | Invitrogen | Cat# 47-0081-80; RRID:AB_1272221 |
| CD86 PE | Invitrogen | Cat# 12-0861-81; RRID:AB_465764 |
| F4/80 PE/Dazzle594 | Biolegend | Cat# 123145; RRID:AB_2564133 |
| CD80 PE-Cy5 | Invitrogen | Cat# 15-0801-81; RRID:AB_468773 |
| MHC Class II eFluor450 | Invitrogen | Cat# 48-5321-80; RRID:AB_1272241 |
| NKp46 BV711 | Biolegend | Cat# 137621; RRID:AB_2563289 |
| Ly-6G BUV395 | BD Biosciences | Cat# 563978; RRID:AB_2716852 |
| CD11c BUV496 | BD Biosciences | Cat# 750450; RRID:AB_2874611 |
| CD3 BUV563 | BD Biosciences | Cat# 749277; RRID:AB_2873652 |
| Bacterial and virus strains | ||
| WT HSV-2 | Lund lab, Fred Hutch Cancer Center | N/A |
| SARS-CoV-2 MA10 | Leist et al., 2020 | GenBank: MT952602 |
| Experimental models: Organisms/strains | ||
| CC001/Unc | RRID:MMRRC:066433-UNC | |
| CC002/Unc | RRID:MMRRC:066434-UNC | |
| CC003/Unc | RRID:MMRRC:066435-UNC | |
| CC004/TauUnc | RRID:MMRRC:066436-UNC | |
| CC005/TauUnc | RRID:MMRRC:066437-UNC | |
| CC006/TauUnc | RRID:MMRRC:066438-UNC | |
| CC007/Unc | RRID:MMRRC:066439-UNC | |
| CC008/GeniUnc | RRID:MMRRC:066440-UNC | |
| CC009/Unc | RRID:MMRRC:066441-UNC | |
| CC010/GeniUnc | RRID:MMRRC:066442-UNC | |
| CC011/Unc | RRID:MMRRC:066443-UNC | |
| CC012/GeniUnc | RRID:MMRRC:066444-UNC | |
| CC013/GeniUnc | RRID:MMRRC:066445-UNC | |
| CC015/Unc | RRID:MMRRC:065941-UNC | |
| CC016/GeniUnc | RRID:MMRRC:066446-UNC | |
| CC017/Unc | RRID:MMRRC:066447-UNC | |
| CC018/Unc | RRID:MMRRC:066492-UNC | |
| CC019/TauUnc | RRID:MMRRC:066448-UNC | |
| CC021/Unc | RRID:MMRRC:066449-UNC | |
| CC023/GeniUnc | RRID:MMRRC:066450-UNC | |
| CC024/GeniUnc | RRID:MMRRC:066451-UNC | |
| CC025/GeniUnc | RRID:MMRRC:066452-UNC | |
| CC026/GeniUnc | RRID:MMRRC:066453-UNC | |
| CC027/GeniUnc | RRID:MMRRC:066454-UNC | |
| CC028/GeniUnc | RRID:MMRRC:066493-UNC | |
| CC029/Unc | RRID:MMRRC:066455-UNC | |
| CC030/GeniUnc | RRID:MMRRC:066456-UNC | |
| CC031/GeniUnc | RRID:MMRRC:066457-UNC | |
| CC032/GeniUnc | RRID:MMRRC:066458-UNC | |
| CC033/GeniUnc | RRID:MMRRC:066459-UNC | |
| CC035/Unc | RRID:MMRRC:066460-UNC | |
| CC036/Unc | RRID:MMRRC:066461-UNC | |
| CC037/TauUnc | RRID:MMRRC:066462-UNC | |
| CC038/GeniUnc | RRID:MMRRC:066463-UNC | |
| CC039/Unc | RRID:MMRRC:066464-UNC | |
| CC040/TauUnc | RRID:MMRRC:066465-UNC | |
| CC041/TauUnc | RRID:MMRRC:066466-UNC | |
| CC042/GeniUnc | RRID:MMRRC:066467-UNC | |
| CC043/GeniUnc | RRID:MMRRC:066468-UNC | |
| CC044/Unc | RRID:MMRRC:066469-UNC | |
| CC045/GeniUnc | RRID:MMRRC:066470-UNC | |
| CC046/Unc | RRID:MMRRC:066471-UNC | |
| CC049/TauUnc | RRID:MMRRC:066472-UNC | |
| CC051/TauUnc | RRID:MMRRC:066473-UNC | |
| CC053/Unc | RRID:MMRRC:066474-UNC | |
| CC057/Unc | RRID:MMRRC:066476-UNC | |
| CC058/Unc | RRID:MMRRC:066477-UNC | |
| CC059/TauUnc | RRID:MMRRC:066478-UNC | |
| CC060/Unc | RRID:MMRRC:066479-UNC | |
| CC061/GeniUnc | RRID:MMRRC:066480-UNC | |
| CC062/Unc | RRID:MMRRC:066481-UNC | |
| CC065/Unc | RRID:MMRRC:066482-UNC | |
| CC068/TauUnc | RRID:MMRRC:066494-UNC | |
| CC071/TauUnc | RRID:MMRRC:066483-UNC | |
| CC072/TauUnc | RRID:MMRRC:066484-UNC | |
| CC074/Unc | RRID:MMRRC:066485-UNC | |
| CC075/Unc | RRID:MMRRC:066486-UNC | |
| CC078/TauUnc | RRID:MMRRC:066487-UNC | |
| CC079/TauUnc | RRID:MMRRC:066488-UNC | |
| CC080/TauUnc | RRID:MMRRC:066489-UNC | |
| CC081/Unc | RRID:MMRRC:066490-UNC | |
| CC083/UncJ | RRID:MMRRC:066495-UNC | |
| CC084/TauJUnc | RRID:MMRRC:066491-UNC | |
| C57BL/6J | Jackson Laboratory | Strain #000664; RRID:IMSR_JAX:000664 |
| BALB/cJ | Jackson Laboratory | Strain #000651; RRID:IMSR_JAX:000651 |
| Software and algorithms | ||
| FlowJo | BD | N/A |
| Prism | GraphPad | N/A |
| Illustrator | Adobe | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Jennifer Lund (jlund@fredhutch.org).
Materials availability
Any material generated in this paper may be available upon request to lead contact.
Collaborative cross mice are available to order through the Mutant Mouse Resource and Research Center (MMRRC), a NIH repository operated by the Systems Genetics Core Facility at UNC. More information on specific strain availability, lead times, and usage considerations can be found on the MMRRC website: https://www.mmrrc.org/news.php?post_id=94. The website maintains a list of all the Collaborative Cross strains, their corresponding RRIDs, and any strain-specific information. Ordering inquiries may be directed to MMRRC_CC@unc.edu.
Data and code availability
-
•
All data reported in this paper are shared within Tables S1, S2, and S3 and/or will be shared by the lead contact upon request.
-
•
This paper does not report any original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
Mice and infections
10 week old Male and female Collaborative Cross (CC) mice50 were obtained from the Systems Genetics Core Facility at the University of North Carolina at Chapel Hill. One cohort of 22 strains of CC mice (Table 1, obtained 2020–2021) was injected with depo provera 7 days prior to infection to synchronize mice in the diestrous phase of the estrous cycle for consistent infection, then inoculated vaginally with 104 PFU WT HSV-2 using a pipette. Mice were monitored for survival daily using a clinical scoring rubric as previously described.22 WT HSV-2 was grown and tittered on Vero cells. A second cohort of mice from 32 CC strains was infected with SARS-CoV-2 MA10. The mouse-adapted (MA) SARS-CoV-2 virus was used for in vivo infection of mice.35,36 Male and female CC mice were anesthetized using ketamine and xylazine prior to intranasal infection with SARS-CoV-2 MA10 (104 PFU). Weight loss, clinical signs of disease, and morbidity were monitored daily. At day 7 post-infection, randomly assigned animals were euthanized by an overdose of isoflurane and lung tissues were collected for assessment of viral loads. Lung viral titers were measured by viral plaque assay on Vero cells. A third cohort of mice (obtained 2021) were euthanized in the laboratory of M. Ferris at 6–8 weeks of age, with spleens in complete RPMI placed on wet ice, then shipped overnight to the Lund laboratory for baseline immunophenotyping studies. All animal experiments were approved by the University of North Carolina (UNC) and the Fred Hutchinson Cancer Center (FHCC) Institutional Animal Care and Use Committee. The Office of Laboratory Animal Welfare of the NIH Approved UNC and the FHCC, and this study was carried out in strict compliance with the Public Health Service (PHS) Policy on Humane Care and Use of Laboratory Animals.
Method details
Flow cytometry
Mouse spleens were minced with scissors in 0.5 mg/mL collagenase complete media, and then incubated at 37° on a nutating mixer for 45 min. The spleen tissues were then homogenized through a 70 um cell strainer and treated with ammonium chloride potassium (ACK) lysis buffer to remove red blood cells. The splenocytes were then washed and counted in trypan blue and resuspended in fluorescence-activated cell sorting (FACS) buffer (0.5% fetal bovine serum in PBS). Cells were plated at 1 × 106 cells/well, and AmCyan live/dead stain (Invitrogen) was used to identify live cells. Cells were then stained for surface markers for 15 min on ice, fixed and permeabilized (Foxp3 fixation/permeabilization kit, Ebioscience), and then stained intracellularly with antibodies for 30 min on ice. Flow cytometry was performed on a BD Fortessa machine using BD FACSDiva software. Analysis was performed using FlowJo software.
Directly conjugated antibodies were used in three separate staining panels. Antibodies were tested using cells from the 8 CC founder strains to confirm that antibody clones were compatible with the CC mice prior to being used for testing (with the exception of MHC Class II, which works only on strains with the correct haplotype). Tables 2, 3, and 4 contain antibody names, fluorochromes, and clones for each of the antibodies used.
Quantification and statistical analysis
All statistics were performed using Prism software (Graphpad). Unpaired t tests were used to compare the means between two groups. For comparison of more than 2 groups, one-way ANOVAs with Tukey’s multiple comparison test were used. Two-way ANOVAs were used for clinical scoring data analysis. Log rank tests were used to analyze survival curves. The error bars represent standard deviation SD and significance was defined if p < 0.05.
Acknowledgments
Funding for this study was provided by NIH grant U19 AI100625 (to R.S.B.), R01 AI157253 (to R.S.B.), P01 AI132130 (to M.T.F. and F.P.M.d.V.), and R21 AI152559 (to J.M.L.). We wish to thank our collaborators in the Systems Immunogenetics Group for helpful discussions and generation of mice.
Author contributions
J.B.G. and J.L.S. performed all flow cytometry experiments and all HSV-2 infection experiments. S.R.L. and A.S. performed all SARS-CoV-2 infection experiments. T.A.B., P.H., J.F., and G.D.S. collected spleens from CC mice and shipped to Seattle. M.T.F., F.P.M.d.V., R.S.B., and J.M.L. supervised and mentored staff and contributed to study design. J.B.G., J.L.S., and J.M.L. conceptualized the study and wrote the first draft of the manuscript. All the authors contributed to editing and approved the final draft.
Declaration of interests
S.R.L. and R.S.B. are listed on a patent for the SARS-CoV-2 MA10 virus (US 11225508 B1, “Mouse-adapted SARS-CoV-2 viruses and methods of use thereof”). Funding for this study was provided by NIH grant U19 AI100625 (to R.S.B.), R01 AI157253 (to R.S.B.), P01 AI132130 (to M.T.F. and F.P.M.d.V.), and R21 AI152559 (to J.M.L.).
Published: February 2, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.109103.
Supplemental information
References
- 1.Liston A., Humblet-Baron S., Duffy D., Goris A. Human immune diversity: from evolution to modernity. Nat. Immunol. 2021;22:1479–1489. doi: 10.1038/s41590-021-01058-1. [DOI] [PubMed] [Google Scholar]
- 2.Orrù V., Steri M., Sidore C., Marongiu M., Serra V., Olla S., Sole G., Lai S., Dei M., Mulas A., et al. Complex genetic signatures in immune cells underlie autoimmunity and inform therapy. Nat. Genet. 2020;52:1036–1045. doi: 10.1038/s41588-020-0684-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Churchill G.A., Airey D.C., Allayee H., Angel J.M., Attie A.D., Beatty J., Beavis W.D., Belknap J.K., Bennett B., Berrettini W., et al. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat. Genet. 2004;36:1133–1137. doi: 10.1038/ng1104-1133. [DOI] [PubMed] [Google Scholar]
- 4.Graham J.B., Swarts J.L., Mooney M., Choonoo G., Jeng S., Miller D.R., Ferris M.T., McWeeney S., Lund J.M. Extensive Homeostatic T Cell Phenotypic Variation within the Collaborative Cross. Cell Rep. 2017;21:2313–2325. doi: 10.1016/j.celrep.2017.10.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin M.D., Sompallae R., Winborn C.S., Harty J.T., Badovinac V.P. Diverse CD8 T Cell Responses to Viral Infection Revealed by the Collaborative Cross. Cell Rep. 2020;31 doi: 10.1016/j.celrep.2020.03.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collin R., Balmer L., Morahan G., Lesage S. Common Heritable Immunological Variations Revealed in Genetically Diverse Inbred Mouse Strains of the Collaborative Cross. J. Immunol. 2019;202:777–786. doi: 10.4049/jimmunol.1801247. [DOI] [PubMed] [Google Scholar]
- 7.Dupont M.S.J., Guillemot V., Campagne P., Serafini N., Marie S., Montagutelli X., Di Santo J.P., Vosshenrich C.A.J. Host genetic control of natural killer cell diversity revealed in the Collaborative Cross. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2018834118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elbahesh H., Schughart K. Genetically diverse CC-founder mouse strains replicate the human influenza gene expression signature. Sci. Rep. 2016;6 doi: 10.1038/srep26437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferris M.T., Aylor D.L., Bottomly D., Whitmore A.C., Aicher L.D., Bell T.A., Bradel-Tretheway B., Bryan J.T., Buus R.J., Gralinski L.E., et al. Modeling host genetic regulation of influenza pathogenesis in the collaborative cross. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graham J.B., Swarts J.L., Leist S.R., Schäfer A., Menachery V.D., Gralinski L.E., Jeng S., Miller D.R., Mooney M.A., McWeeney S.K., et al. Baseline T cell immune phenotypes predict virologic and disease control upon SARS-CoV infection in Collaborative Cross mice. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graham J.B., Swarts J.L., Wilkins C., Thomas S., Green R., Sekine A., Voss K.M., Ireton R.C., Mooney M., Choonoo G., et al. A Mouse Model of Chronic West Nile Virus Disease. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graham J.B., Thomas S., Swarts J., McMillan A.A., Ferris M.T., Suthar M.S., Treuting P.M., Ireton R., Gale M., Jr., Lund J.M. Genetic diversity in the collaborative cross model recapitulates human West Nile virus disease outcomes. mBio. 2015;6:e00493. doi: 10.1128/mBio.00493-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gralinski L.E., Ferris M.T., Aylor D.L., Whitmore A.C., Green R., Frieman M.B., Deming D., Menachery V.D., Miller D.R., Buus R.J., et al. Genome Wide Identification of SARS-CoV Susceptibility Loci Using the Collaborative Cross. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leist S.R., Pilzner C., van den Brand J.M.A., Dengler L., Geffers R., Kuiken T., Balling R., Kollmus H., Schughart K. Influenza H3N2 infection of the collaborative cross founder strains reveals highly divergent host responses and identifies a unique phenotype in CAST/EiJ mice. BMC Genom. 2016;17:143. doi: 10.1186/s12864-016-2483-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lorè N.I., Iraqi F.A., Bragonzi A. Host genetic diversity influences the severity of Pseudomonas aeruginosa pneumonia in the Collaborative Cross mice. BMC Genet. 2015;16:106. doi: 10.1186/s12863-015-0260-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rasmussen A.L., Okumura A., Ferris M.T., Green R., Feldmann F., Kelly S.M., Scott D.P., Safronetz D., Haddock E., LaCasse R., et al. Host genetic diversity enables Ebola hemorrhagic fever pathogenesis and resistance. Science. 2014;346:987–991. doi: 10.1126/science.1259595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith C.M., Proulx M.K., Olive A.J., Laddy D., Mishra B.B., Moss C., Gutierrez N.M., Bellerose M.M., Barreira-Silva P., Phuah J.Y., et al. Tuberculosis Susceptibility and Vaccine Protection Are Independently Controlled by Host Genotype. mBio. 2016;7 doi: 10.1128/mBio.01516-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith C.M., Baker R.E., Proulx M.K., Mishra B.B., Long J.E., Park S.W., Lee H.N., Kiritsy M.C., Bellerose M.M., Olive A.J., et al. Host-pathogen genetic interactions underlie tuberculosis susceptibility in genetically diverse mice. Elife. 2022;11 doi: 10.7554/eLife.74419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cartwright H.N., Barbeau D.J., Doyle J.D., Klein E., Heise M.T., Ferris M.T., McElroy A.K. Genetic diversity of collaborative cross mice enables identification of novel rift valley fever virus encephalitis model. PLoS Pathog. 2022;18 doi: 10.1371/journal.ppat.1010649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schäfer A., Leist S.R., Gralinski L.E., Martinez D.R., Winkler E.S., Okuda K., Hawkins P.E., Gully K.L., Graham R.L., Scobey D.T., et al. A Multitrait Locus Regulates Sarbecovirus Pathogenesis. mBio. 2022;13 doi: 10.1128/mbio.01454-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noll K.E., Ferris M.T., Heise M.T. The Collaborative Cross: A Systems Genetics Resource for Studying Host-Pathogen Interactions. Cell Host Microbe. 2019;25:484–498. doi: 10.1016/j.chom.2019.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arkatkar T., Davé V., Cruz Talavera I., Graham J.B., Swarts J.L., Hughes S.M., Bell T.A., Hock P., Farrington J., Shaw G.D., et al. Memory T cells possess an innate-like function in local protection from mucosal infection. J. Clin. Invest. 2023;133 doi: 10.1172/JCI162800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lund J.M., Hsing L., Pham T.T., Rudensky A.Y. Coordination of early protective immunity to viral infection by regulatory T cells. Science. 2008;320:1220–1224. doi: 10.1126/science.1155209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belkaid Y., Piccirillo C.A., Mendez S., Shevach E.M., Sacks D.L. CD4+CD25+ regulatory T cells control Leishmania major persistence and immunity. Nature. 2002;420:502–507. doi: 10.1038/nature01152. [DOI] [PubMed] [Google Scholar]
- 25.Belkaid Y., Rouse B.T. Natural regulatory T cells in infectious disease. Nat. Immunol. 2005;6:353–360. doi: 10.1038/ni1181. [DOI] [PubMed] [Google Scholar]
- 26.Bedoya F., Cheng G.S., Leibow A., Zakhary N., Weissler K., Garcia V., Aitken M., Kropf E., Garlick D.S., Wherry E.J., et al. Viral antigen induces differentiation of Foxp3+ natural regulatory T cells in influenza virus-infected mice. J. Immunol. 2013;190:6115–6125. doi: 10.4049/jimmunol.1203302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brincks E.L., Roberts A.D., Cookenham T., Sell S., Kohlmeier J.E., Blackman M.A., Woodland D.L. Antigen-specific memory regulatory CD4+Foxp3+ T cells control memory responses to influenza virus infection. J. Immunol. 2013;190:3438–3446. doi: 10.4049/jimmunol.1203140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fulton R.B., Meyerholz D.K., Varga S.M. Foxp3+ CD4 regulatory T cells limit pulmonary immunopathology by modulating the CD8 T cell response during respiratory syncytial virus infection. J. Immunol. 2010;185:2382–2392. doi: 10.4049/jimmunol.1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lanteri M.C., O'Brien K.M., Purtha W.E., Cameron M.J., Lund J.M., Owen R.E., Heitman J.W., Custer B., Hirschkorn D.F., Tobler L.H., et al. Tregs control the development of symptomatic West Nile virus infection in humans and mice. J. Clin. Invest. 2009;119:3266–3277. doi: 10.1172/JCI39387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee D.C.P., Harker J.A.E., Tregoning J.S., Atabani S.F., Johansson C., Schwarze J., Openshaw P.J.M. CD25+ natural regulatory T cells are critical in limiting innate and adaptive immunity and resolving disease following respiratory syncytial virus infection. J. Virol. 2010;84:8790–8798. doi: 10.1128/JVI.00796-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruckwardt T.J., Bonaparte K.L., Nason M.C., Graham B.S. Regulatory T cells promote early influx of CD8+ T cells in the lungs of respiratory syncytial virus-infected mice and diminish immunodominance disparities. J. Virol. 2009;83:3019–3028. doi: 10.1128/JVI.00036-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scott-Browne J.P., Shafiani S., Tucker-Heard G., Ishida-Tsubota K., Fontenot J.D., Rudensky A.Y., Bevan M.J., Urdahl K.B. Expansion and function of Foxp3-expressing T regulatory cells during tuberculosis. J. Exp. Med. 2007;204:2159–2169. doi: 10.1084/jem.20062105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suvas S., Azkur A.K., Kim B.S., Kumaraguru U., Rouse B.T. CD4+CD25+ regulatory T cells control the severity of viral immunoinflammatory lesions. J. Immunol. 2004;172:4123–4132. doi: 10.4049/jimmunol.172.7.4123. [DOI] [PubMed] [Google Scholar]
- 34.Veiga-Parga T., Suryawanshi A., Mulik S., Giménez F., Sharma S., Sparwasser T., Rouse B.T. On the role of regulatory T cells during viral-induced inflammatory lesions. J. Immunol. 2012;189:5924–5933. doi: 10.4049/jimmunol.1202322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dinnon K.H., 3rd, Leist S.R., Schäfer A., Edwards C.E., Martinez D.R., Montgomery S.A., West A., Yount B.L., Jr., Hou Y.J., Adams L.E., et al. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature. 2020;586:560–566. doi: 10.1038/s41586-020-2708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leist S.R., Dinnon K.H., 3rd, Schäfer A., Tse L.V., Okuda K., Hou Y.J., West A., Edwards C.E., Sanders W., Fritch E.J., et al. A Mouse-Adapted SARS-CoV-2 Induces Acute Lung Injury and Mortality in Standard Laboratory Mice. Cell. 2020;183:1070–1085.e12. doi: 10.1016/j.cell.2020.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akue A.D., Lee J.Y., Jameson S.C. Derivation and maintenance of virtual memory CD8 T cells. J. Immunol. 2012;188:2516–2523. doi: 10.4049/jimmunol.1102213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haluszczak C., Akue A.D., Hamilton S.E., Johnson L.D.S., Pujanauski L., Teodorovic L., Jameson S.C., Kedl R.M. The antigen-specific CD8+ T cell repertoire in unimmunized mice includes memory phenotype cells bearing markers of homeostatic expansion. J. Exp. Med. 2009;206:435–448. doi: 10.1084/jem.20081829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lanzer K.G., Cookenham T., Reiley W.W., Blackman M.A. Virtual memory cells make a major contribution to the response of aged influenza-naive mice to influenza virus infection. Immun. Ageing. 2018;15:17. doi: 10.1186/s12979-018-0122-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee J.Y., Hamilton S.E., Akue A.D., Hogquist K.A., Jameson S.C. Virtual memory CD8 T cells display unique functional properties. Proc. Natl. Acad. Sci. USA. 2013;110:13498–13503. doi: 10.1073/pnas.1307572110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sosinowski T., White J.T., Cross E.W., Haluszczak C., Marrack P., Gapin L., Kedl R.M. CD8α+ dendritic cell trans presentation of IL-15 to naive CD8+ T cells produces antigen-inexperienced T cells in the periphery with memory phenotype and function. J. Immunol. 2013;190:1936–1947. doi: 10.4049/jimmunol.1203149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Allman D., Lindsley R.C., DeMuth W., Rudd K., Shinton S.A., Hardy R.R. Resolution of three nonproliferative immature splenic B cell subsets reveals multiple selection points during peripheral B cell maturation. J. Immunol. 2001;167:6834–6840. doi: 10.4049/jimmunol.167.12.6834. [DOI] [PubMed] [Google Scholar]
- 43.Teague B.N., Pan Y., Mudd P.A., Nakken B., Zhang Q., Szodoray P., Kim-Howard X., Wilson P.C., Farris A.D. Cutting edge: Transitional T3 B cells do not give rise to mature B cells, have undergone selection, and are reduced in murine lupus. J. Immunol. 2007;178:7511–7515. doi: 10.4049/jimmunol.178.12.7511. [DOI] [PubMed] [Google Scholar]
- 44.Mujal A.M., Delconte R.B., Sun J.C. Natural Killer Cells: From Innate to Adaptive Features. Annu. Rev. Immunol. 2021;39:417–447. doi: 10.1146/annurev-immunol-101819-074948. [DOI] [PubMed] [Google Scholar]
- 45.Chiossone L., Chaix J., Fuseri N., Roth C., Vivier E., Walzer T. Maturation of mouse NK cells is a 4-stage developmental program. Blood. 2009;113:5488–5496. doi: 10.1182/blood-2008-10-187179. [DOI] [PubMed] [Google Scholar]
- 46.Jensen I.J., Martin M.D., Tripathy S.K., Badovinac V.P. Novel Mouse Model of Murine Cytomegalovirus-Induced Adaptive NK Cells. Immunohorizons. 2022;6:8–15. doi: 10.4049/immunohorizons.2100113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wyse C., O'Malley G., Coogan A.N., McConkey S., Smith D.J. Seasonal and daytime variation in multiple immune parameters in humans: Evidence from 329,261 participants of the UK Biobank cohort. iScience. 2021;24 doi: 10.1016/j.isci.2021.102255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hamilton S.E., Badovinac V.P., Beura L.K., Pierson M., Jameson S.C., Masopust D., Griffith T.S. New Insights into the Immune System Using Dirty Mice. J. Immunol. 2020;205:3–11. doi: 10.4049/jimmunol.2000171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mogilenko D.A., Shchukina I., Artyomov M.N. Immune ageing at single-cell resolution. Nat. Rev. Immunol. 2022;22:484–498. doi: 10.1038/s41577-021-00646-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Welsh C.E., Miller D.R., Manly K.F., Wang J., McMillan L., Morahan G., Mott R., Iraqi F.A., Threadgill D.W., de Villena F.P.M. Status and access to the Collaborative Cross population. Mamm. Genome. 2012;23:706–712. doi: 10.1007/s00335-012-9410-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper are shared within Tables S1, S2, and S3 and/or will be shared by the lead contact upon request.
-
•
This paper does not report any original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.