Abstract
Background
During the regression of hypertrophic scars, autophagy and apoptosis are the main ways of cell death. Recent investigations demonstrated effective inhibition of resveratrol on hypertrophic scar fibroblasts (HSFs). But its therapeutic value is limited by chemical instability and hydrophobicity, as well as the mechanism of its role in regulation of autophagy and apoptosis remains unknown.
Aim of the study
We prepared a mesoporous silica nanoparticle laden with resveratrol (MSN@Res) which can effectively improve the solubility and stability of resveratrol. The purpose of this study was to investigate whether MSN@Res regulate autophagy and apoptosis of HSFs via inhibition of ROS/p38/HIF-1α/p53 signaling axis, as to reveal its pharmacological action and target.
Materials and methods
Network pharmacology, molecular docking, and in vitro assays were carried out in this study. An in vitro model of fibroblasts cultivated in hypoxic and ischemic situations was established to simulate the scar in the proliferative phase.
Results
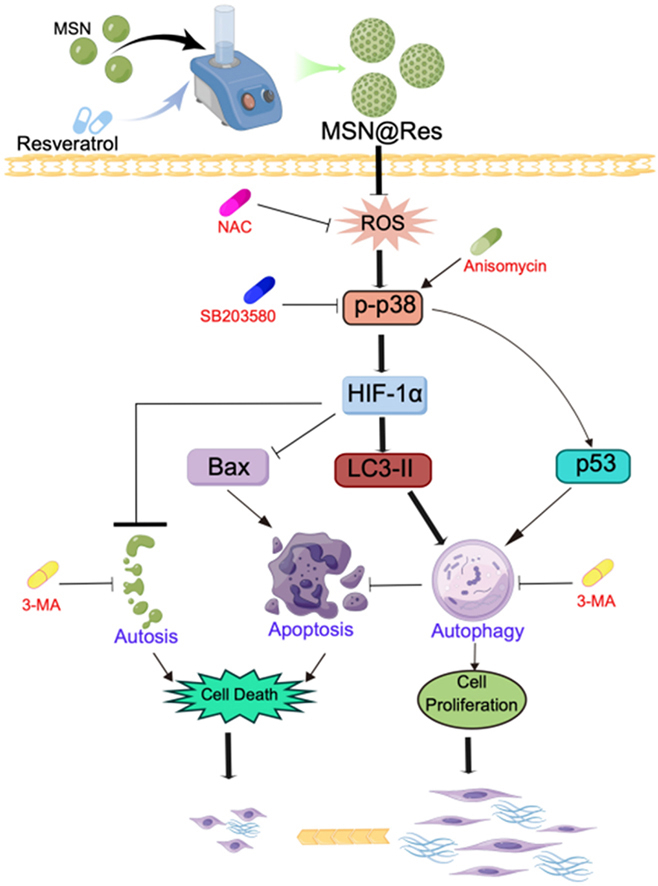
MSN@Res surpresses HSFs by reducing physiological autophagy and inducing apoptosis, autosis may be another cell death involed in this process. According to the network pharmacological analysis and molecular docking, the mechanism by which MSN@Res alleviates hypertrophic scar may be closely related to the MAPK signaling pathway. MSN@Res significantly downregulate the expression of HIF-1α and p53 through the inhibition of ROS induced p38-MAPK phosphorylation with corresponding changes in the expression of autophagy and apoptosis related protein.
Conclusion
MSN@Res is a novel drug delivery system with excellent chemical stability and drug release performance. It can inhibit protective autophagy of fibroblasts in hypoxic environment, and induce the apoptosis and autosis via the ROS -mediated p38-MAPK/HIF-1α/p53 signaling axis.
Keywords: Network pharmacology, MAPK signaling pathway, HIF-1α, p53, Autophagy, Apoptosis, Autosis, Nanoparticles, Hypertrophic scar fibroblast, Resveratrol
Graphical abstract
A Schematic diagram for surpression of Hypertrophic Scar Fibroblasts induced by MSNs@Res.
1. Introduction
Without treatment, hypertrophic scar (HS) could regress spontaneously, and become softer and flattered several years later. This phenomenon is thought to be related to autophagy and apoptosis in hypoxic environments. Apoptosis is regarded as the main way of cell death during the regression and maturation of HS [1]. Autophagy is a form of conservative catabolism whereby substances from the cytoplasm are transported to lysosomes for degradation by autophagosomes, and the products are utilized for cell survival [2]. Overautophagy, also referred to as autophagic cell death or autosis, can result in cell death. Physiological autophagy promotes fibroblast proliferation and scar formation. Conversely, Overautophagy facilitates scar regression and maturation [3]. Hypoxia plays an important role in autophagy inducement [4], and the inhibition of physiological autophagy could trigger cell apoptosis [5]. The transition of autophagy and apoptosis of fibroblast depends on the dominant expression of HIF-1α or p53 [6].
It has been reported that resveratrol (Res), a naturally occurring plant polyphenol, exerts numerous beneficial effects on the human body, including anti-oxidation [7], anti-inflammation [8], and anti-cancer [9]. Reactive oxygen species (ROS) are important in the early stages of wound healing and play a critical role in fibroblast activation and fibrosis [10]. Previous studies have indicated that resveratrol has potentia benefits on skin fibrosis, which may be associated with its powerful antioxidant properties [11,12]. However, the precise molecular mechanism remains unclear and requires further investgation, specifically regarding the involvement of fibroblast autophagy and apoptosis in the inhibition of cell vitality and collagen deposition induced by resveratrol. While network pharmacology as a new system biology tool that explores the mechanisms of complex diseases and medications within the context of comprehensive biological networks shows light on the research of plant compounds and complex diseases [13].
To improve the solubility, stability and bioavailability of Res, enhance its permeability and therapeutic efficacy, and reduce its toxicity, the drug has been loaded into synthetic nanodelivery systems [14]. Mesoporous silica nanoparticles (MSN) are also widely used in drug delivery and biomedicine due to their large surface area and pore volume. It has been reported that encapsulating Res in colloidal MSN with high loading capacity (20 % w/w) and excellent encapsulation efficiency (100 %) can enhance its solubility by 95 % and improve in vitro release kinetics, leading to stronger anti-inflammatory and anti-tumor activities than free Res [15]. Thus we prepared a MSN laden with resveratrol (MSN@Res), the purpose of this study was to investigate its physicochemical property and biocompatibility, as well as the potential pharmacological effect and mechanism on fibroblast autophagy and apoptosis upon the network pharmacology and experimental verification.
2. Methods and materials
2.1. Targets investigation and network construction for Res and HS
The targets of Res were obtained from Traditional Chinese Medicine Systems Pharmacology (TCMSP) [16], SwissTargetPrediction [17], STITCH [18], and Similarity ensemble approach (SEA) [19] databases. The HS related targets were obtained from Genecards (http://www.genecards.org/) [20], Online Mendelian Inheritance in Man (OMIM) (https://omim.org/) [21], Malacard and DisGeNET databases. The Uniprot database (https://www.uniprot.org/) was used to standardize the target information by including the species information “homo sapiens”. The overlapping part called the intersecting targets of Res and HS was screened by the Venn (http://bioinformatics.psb.ugent.be/webtools/Venn/). Cytoscape3.8.0 was adopted for constructing the drug-targets-disease network [22]. Additionally, the STRING database (http://string-db.org) [23] was used to build the protein–protein interactions (PPI) network, and the results were visualized with Cytoscape 3.8.0. The DAVID database (https://david.ncifcrf.gov)was employed for Gene Ontology (GO) and Kyoto Encyclopaedia of Genes and Genome (KEGG) enrichment analysis [24]. p-Value <0.05 was considered as a potential pathway for all GO terms and KEGG pathways. Finally, the top10 core targets and the main signaling pathways were obtained.
2.2. Molecular docking
The 3D structure of Res and protein of core targets were obtained from PubChem (https://pubchem.ncbi.nlm.nih.gov/) and Protein Data Bank (http://www.pdb.org/) respectively, the unwanted molecules included water, ions, and ligands were removed [25], the Res and core targets protein were docking on Autodock to analyze the lowest binding energy, the bonds, and interaction forces between the functional groups of molecular ligands and interactive amino acids of proteins, and the docking results were visualized on the Pymol software [26].
2.3. Preparation and characterization of MSN@Res
MSN@Res was prepared as reported previously [27]. Firstly, 6 mL of ammonia solution was added to a mixture of 150 mL anhydrous ethanol and 3 mL deionized water at room temperature for 7 h. Then, 3 mL of tetraethyl orthosilicate (TEOS) was added to the mixture and stirred for 15 h. Secondly, the mixed solution was centrifuged at 10,000 rpm for 15 min, which was washed twice with ethanol, and the resulting solid silica spheres were suspended in 40 mL of deionized water. Thirdly, 1.5 g of CTAB, 53.4 μL of TEA, and 64.5 mL of water were vigorously stirred at 80 °C to which the SiO2 suspension was added, and then the mixture was cooled to 50 °C and then 9 mL of saturated Na2CO3 solution was added to etch the silica sphere for 2 h. Fouth, The hollow silica spheres were obtained by centrifugationat 15,000 rpm for 15 min which were dispersed in 100 mL of HCl/ethanol solution (v/v = 1:10) at 60 °C for 2 h to remove CTAB. After that, mesoporous silica nanoparticles (MSN) were obtained. For the encapsulation of Res in MSN, 40 mg of Res was placed into 10 mL of ethanol and sonicated for 2 min, 40 mg of MSN was then added and sonicated for a further 5 min. The mixture was oscillated at room temperature in the dark for 24 h to obtain MSN@Res. After centrifugation, the MSN@Res precipitation was slowly evaporated until all ethanol was removed and a dry powder was observed, which was collected, stored covered in aluminum foil to protect it from photo degradation and administered in solution for further study. The amount of Res adsorbed on the MSN nanoparticles was determined by subtracting the amount of Res remaining in the supernatant (WSup) after incubation from the initial addition amount of Res (WRes)and the Res loading was calculated as: (WRes-WSup)/WResx100 %.
1 mL of MSN@Res was transferred to a dialysis bag (MW: 100Da), then it was placed in 5 mL PBS solution and incubated at 37 °C. The supernatant was collected at different time points (1, 3, 6, 12, 24, 48, 72, 96, 120, 144 h) and replenished with the same volume of PBS solution (PH: 7.4). The collected supernatant was tested for Res content using a UV spectrophotometer to calculate the cumulative release rate (%): release amount/total amount × 100 %.
The size distribution of MSN@Res was assessed using a Zeta-Sizer Nano ZS (Malvern, UK), and its morphological property was characterized using transmission electron microscopy (TEM) (H-800, Hitachi) at 200 kV (×100,000),UV spectrophotometry was performed to confirm the absorption peaks of Res , MSN, and MSN@Res, as well characterizations of which were determined by FTIR spectrometry and X-ray diffractograms.
2.4. Cell culture and treatment
The skin samples were obtained from five HS patients (three female, two male, age from 26 to 42years), all of whom had pathological diagnoses. The dermal portion of scar tissues were chopped into small pieces (0.4 cm × 0.4 cm × 0.4 cm), which were then cultured at 37 °C in 5 % CO2 for 6–8 h. After that, DMEM (C1199500BT, Gibco) containing10 % FBS (42F7180K, Gibco)was added to the flasks for culture. The samples collection was approved by the ethics committee of the first affiliated hospital of Xinjiang Medical University (Urumqi, China). Written informed consents were obtained from the patients (20170214-71). Besides Res and MSN@Res, autophagy inhibitors 3-MA (HY19312, MCE), caspase inhibitor z-VAD (HY166588, MCE), p38 inhibitor SB203580(HY10256, MCE)and activator anisomycin(HY18982, MCE)were also used to treat HSFs in following assays.
2.5. Cell viability assay
The HSFs (2.5 × 103 cells/well) were incubated in 96-well plates under normoxic conditions (21 % O2/10 % FBS) overnight, then treated with varying dose (25–400 μg/mL) of MSN@Res under 5 % O2/5 % FBS at 37 °C to simulate moderate hypoxia and malnutrition for 24 and 48 h. The cells treated with MSN, Res, and MSN@Res for 24, 48 and 72 h were harvested and washed with PBS, then the cell viability was determined using a CCK8 kit (C0038, China Biotech). For live/dead staining, cells were stained using calcein acetoxymethyl ester and propidium iodide (Calcein/PI). Finally, images were captured using a fluorescence microscope (DS-Fi3, Nikon).
2.6. Flow cytometry analysis of Cell apoptosis
HSFs (2 × 105 cells/plate) were cultured in 60 mm plate overnight and treated with MSN, Res, and MSN@Res for 48 h. The cells were harvested and stained with Annexin V-FITC and PI (640914, Biolegend) in the dark. After being washed, the frequency of apoptotic cells was analyzed by flow cytometry (CytoFLEX, Beckman).
2.7. Mitochondrial membrane potential
HSFs were treated as described in 2.5. Then stained with JC-1(C2006, China Biotech) and DAPI for 30 min at 37 °C in the dark respectively. After being washed, the cells were analyzed under a fluorescence microscope.
2.8. Measurement of concentrations of Cytosol Ca2+
HSFs (2 × 104 cells/well) were seeded on a 24-well plate and treated as described in 2.5. The adherent cells were detached with trypsin-EDTA, aftert cenrifugation, the collected cells were resuspended using 5 μM Fluo-4 (China Biotech, AMS1060) and incubated at 37 °C in a CO2 incubator for 40 min, the stained cells were washed with PBS and fluorescent intensity was analyzed using a flow cytometer (CytoFLEX, Beckman).
2.9. Analysis of reactive oxygen Species (ROS)
HSFs were treated as described in 2.5 and after being washed, the cells were cultured in serum-free DMEM in the presence of 10 μM DCFH-DA (S0033S, China Biotech) at 37 °C for 30 min in the dark. Then the cells were harvested for fluorescence analysis.
2.10. Visualization of MDC-labeled vacuoles
MDC is specific for autolysosomes, which are concentrated in autophagic vacuole membrane structures distributed in the cytoplasm [28]. HSFs (2 × 105 cells/well) were cultured in 6-well culture plates for 24 h, then treated as described in 2.5, the cells were incubated with 0.05 mM MDC(C3018S, Beyotime) for 20 min at 37 °C, and fluorescence changes were observed by fluorescence microscopy (Nikon, DS-Fi3).
2.11. Immunofluorescence of LC3-II and tunel assay
HSFs were harvested after pre-incubation with SB203580(10 μM) or H2O2 (300 μM) for 2 h, followed by the treatment of MSN@Res for 48 h, with or without the co-incubation of anisomycin (25 μg/mL). After that, the cells were fixed with 4 % paraformaldehyde for 20 min, Triton X-100 was then incubated for 10 min at room temperature. Following that, the primary LC3 antibody (14600-1-AP, PTG; dilution ratio: 1:400)was diluted with 5 % BSA and incubated overnight at 4 °C. The cells were then incubated with CY3-labeled goat anti-rabbit secondary antibody (AS-1109, Aspen; dilution ratio: 1:100) at 37 °C in the dark for 40 min. Cell apoptosis was detected using a TUNEL detection kit (G1504, Servicebio). Finally, the cells were harvested for fluorescence analysis.
2.12. Western blot analysis
After the same treatment as described in 2.10, HSFs were harvested and lyzed in RIPA buffer (AS1004, ASPEN) containing inhibitors of protease and phosphatase (AS1008, ASPEN), followed by centrifugation (12000 rpm × 5min). The BCA protein kit (AS1086, ASPEN) was used to assess the protein contents in the lysate. The cell lysate samples (50μg/lane) were separated by SDS-PAGE gels (AS1012, ASPEN) and transferred to PVDF membranes (IPVH00010, Millipore). After being blocked with 5 % bovine serum, the membranes were incubated with primary antibodies at 4 °C overnight. The membrane was then incubated with the secondary antibody at room temperature for 1 h after being washed with TBST. The proteins were detected using an ECL chemiluminescence kit (AS1059, ASPEN). The primary antibodies included p-p38 (28796-1-AP, Proteintech), p38 (14064-1-AP, Proteintech), p-JNK(#4668, CST), JNK(#9252, CST), p-ERK (28733-1-AP, Proteintech), ERK (11257-1-AP, Proteintech), Bax (#2772, CST), Cleaved caspase3 (19677-1-AP, Proteintech), HIF-1α (20960-1-AP, Proteintech), p53 (10442-1-AP, Proteintech), Beclin-1 (AF5128, affinity), LC3-II (ab48394, abcam), β-Actin (TDY051, 1:10000).
2.13. Transmission electron Microscopy (TEM)
Cells were harvested and fixed with 2.5 % glutaraldehyde at 4 °C overnight. After 2 h of fixing in 1 % osmic acid/0.1 M phosphate buffer PBS (PH7.4), the cells were dehydrated with a series of graded alcohols. After being embedded in epoxy resin, the samples were examined by transmission electron microscopy (H-800, Hitachi).
2.14. Statistical analyses
GraphPad Prism software version 9.5.1 was used for statistical analysis. The mean and SD were used to express the data. A one-way analysis of variance was used to assess differences between two or more groups. P < 0.05 was accepted as significant.
3. Results
3.1. Common targets of Res and HS
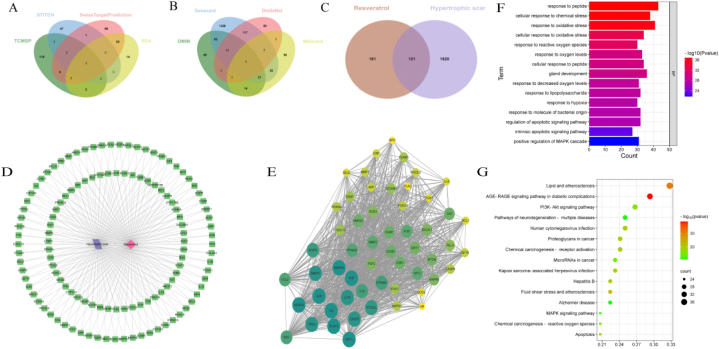
We initially investigated the potential mechanisms of Res that may contribute to the treatment of HS. The keyword “Resveratrol” was input into the Swiss Target Prediction, STITCH, TCMSP and SEA databases, and 282 potential drug targets were obtained after the repetition was removed (Fig. 1A). “Hypertrophic Scar”were entered into the DisGeNET, OMIM, Malacard and GeneCards databases, and 1741 targets were obtained (Fig. 1B). Then a total of 121 common targets of Res and HS were obtained through a Venn diagram (Fig. 1C).
Fig. 1.
Network pharmacology predicts the anti-Hypertrophic Scar mechanism of Resveratrol. (A) Putative targets of Resveratrol. (B) Hypertrophic Scar-related targets. (C) Co-targets of Resveratrol and Hypertrophic Scar. (D) Interaction between Resveratrol, Hypertrophic Scar and co-targets. (E) Ranking of co-targets for Resveratrol and Hypertrophic Scar, the proteins were ranked by their degree values, and the deeper 10 core nodes in color represent greater importance. (F),(G) GO and KEGG enrichment analysis: (F) Enrichment results of biological processes. (G) Enrichment results of KEGG pathways. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The 121 common targets were processed by the STRING database to obtain a PPI network (Fig. 1D). The results of the first 55 targets interacting with each other were imported into Cytoscape 3.8.0 to obtain a PPI network sorted by degree value from large to small (Fig. 1E). The top 10 core targets were AKT1,MAPK3,STAT3,JUN,VEGFR,TNF,IL6,IL1B,TP53 and CASP3.
3.2. GO and KEGG enrichment analysis
GO analysis revealed that these biological process (BP) were primarily concentrated on multiple processes, such as response to peptide, response to oxidative stress, response to hypoxia, regulation of apoptotic signaling pathway and positive regulation of MAPK cascade (Fig. 1F).The interacting targets in the PPI network were input into the DAVID database to conduct KEGG pathway enrichment. The top 15 KEGG pathways mainly included Lipid and atherosclerosis, PI3K-Akt signaling pathway, Apoptosis, Chemical carcinogenesis-reactive oxygen species (ROS) and MAPK signaling pathway (Fig. 1G).
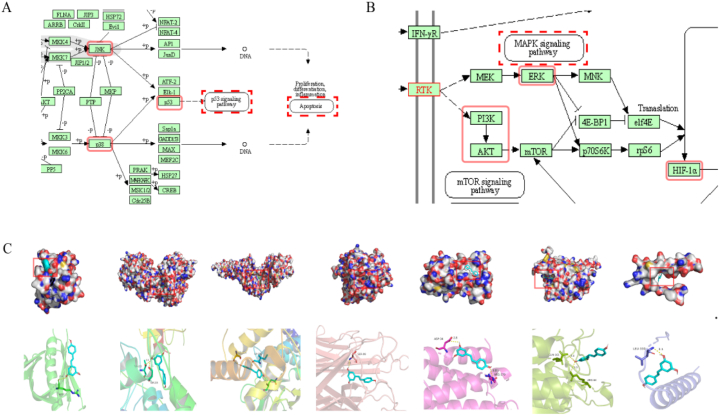
AKT1, MAPK3, STAT3, TNF, IL6, CASP3 and TP53, which ranked top 10 core targets, were enriched in the MAPK and HIF-1 signaling pathway. Further results came out by searching “MAPK signaling pathway” and “HIF-1 signaling pathway” in KEGG Mapper: the p53 and apoptosis pathway are the downstream of p38/JNK-MAPK pathway, while HIF-1α pathway is a downstream of ERK-MAPK and PI3K-Akt pathway (Fig. 2A and B). Based on these results and previous studies [6,[29], [30], [31]], we predict that ROS-MAPK - HIF-1α/p53 axis may be involved in the regulation of cell apoptosis and autophagy induced by Res.
Fig. 2.
Molecular docking analyses of the binding modes of resveratrol with the active sites of core genes. (A),(B) KEGG Mapper analysis: (A) Multiple coretargets were enriched in the MAPK signaling pathway.(B) Multiple coretargets were enriched in the HIF-1 signaling pathway. (C) Molecular docking 3D mode. From left to right, the molecular docking of resveratrol with AKT1, IL6, SATA3, TNF, MAPK3, CASP3 and TP53 are shown.
3.3. Molecular docking
Molecular docking was carried out to analyze whether Res could play a role in inhibition of HS by combining with the core targets. Given that a lower docking score represents a stronger binding affinity, and a score<−5 indicates strong binding activity [32]. The result showed that Res had a strong affinity with the chosen proteins AKT1, MAPK3, STAT3, TNF, IL6, CASP3 and TP53. The docking scores were−5.6, −7.4, −6.5, −6.5, −5.9, −5.5, and−6.1 kcal/mol, respectively. Among them, the docking of MAPK3 had the lowest binding score −7.4 kcal/mol. As shown in (Fig. 2C), the active sites of MAPK3 including THR-223 and SER-225 interplayed with Res by several shorter hydrogen bonds.
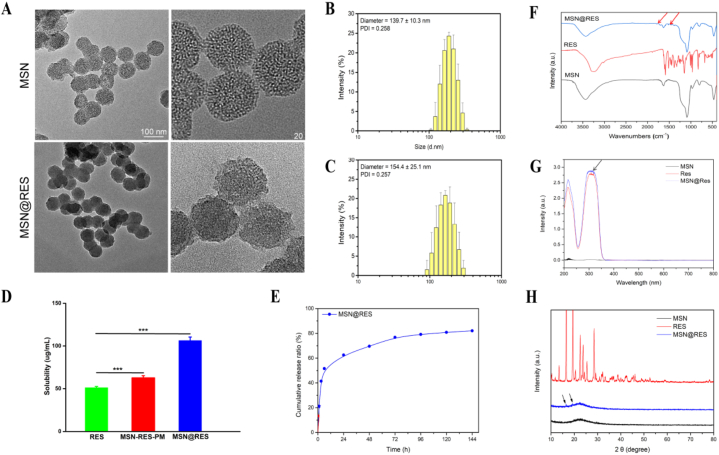
3.4. Characterization of MSN@Res nanoparticles
TEM analysis was performed to characterize the shape, size, and homogeneity of MSN and MSN@Res, the results showed that these nanoparticles have the mesoporous property which increases the surface area and drug loading of MSN (Fig. 3A). The polydispersity index (PDI) of the nanoparticles was 0.25, indicating the modified MSN still had good dispersion and stability. According to the Nano ZS zetasizer, the MSN and MSN@Res had a uniform particle size distribution with an average particle size of 139.7 ± 10.3 nm and 154.4 ± 25.1 nm, respectively (Fig. 3B and C).
Fig. 3.
Characterization of MSN@Res nanoparticles (A) TEM image of MSN and MSN@Res. (B) (C) Size distribution of MSN and MSN@Res analyzed by DLS. (D) Saturated aqueous solubility of pure Res, MSN-Res-PM and MSN@Res (n = 3, ∗∗∗P < 0.001). (E) The drug release curve of MSN@Res in vitro. (F) FTIR spectroscopy of MSN@Res, Res and MSN. (G) UV spectrophotometry of Res, MSN and MSN@Res. (H) Wide angle X-ray diffraction (XRD) pattern of MSN@Res, Res and MSN.
FTIR spectroscopy (Fig. 3F) indicated that the vibration peaks of Res were observed between 1750 cm−1 and 1250 cm−1, most of the characteristic peaks of Res disappeared when it was loaded in MSN nanoparticles, while samll peaks (red arrows) were also visible at a very low intensity in MSN@Res which were absence in MSN, this was also supported by the UVspectrum (Fig. 3G), which showed a small peak (black arrows) at 300 nm, these results demonstrated that Res was encapsulated successfully. Fig. 3D reveal that Res solubility was significantly enhanced approximately 2.1 fold in MSN@Res (106.3 ± 4.11 μg/mL) compared to Res (51.2 ± 1.5 μg/mL). Moreover, the solubility was also significantly higher in MSN@Res than in its physical mixture MSN-Res-PM (62.7 ± 2.58 μg/mL).The enhanced solubility of Res is attributed to the confinement of Res in amorphous form confirmed by XRD studies. As shown in (Fig. 3H), Res shows sharp diffraction peaks between 10◦and 30◦indicative of its crystalline nature in its free powdered form. These peaks (black arrows) were also visible at a very low intensity in MSN@Res, while such diffraction peaks were absent in MSN, suggesting incomplete encapsulation and the amorphous nature of Res within nanoparticles. The encapsulation of MSN@Res was 90.38 %, and the drug loading was 45.19 %. In addition, MSN@Res showed a linear release trend within 6 h, with an accumulated release of about 51.6 %, and then slowly released for up to 6 days (Fig. 3E).
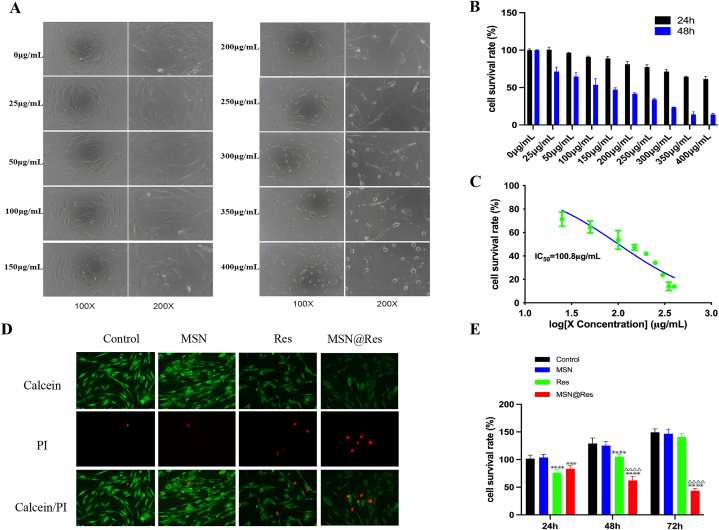
3.5. MSN@Res decreases the viability of hypertrophic scar fibroblasts
To examine the direct effect of MSN@Res on cells viability, HSFs were treated with MSN@Res at different concentrations (0–400 μg/mL) up to 24 and 48 h, then their viabilities were determined by CCK-8 assays. As shown in (Fig. 4A and B), concentrations ranging from 0 to 300 μg/mL were not significantly toxic to HSFs for 24 h; while treatment with MSN@Res from 25 to 400 μg/mL significantly decreased the viability of HSFs in a dose-dependent manner for 48 h. The half maximal inhibitory concentration (IC50) of MSN@Res for HSFs with different duration were 503.5 μg/mL (24 h), 100.8 μg/mL (48 h), respectively (Fig. 4C). Hence, MSN@Res treatment decreased the viability of HSFs in a dose and time-dependent manner. Futher results demonstrated that Res exhibited a diminishing trend of inhibition, which may be associated with its poor stability, while MSN@Res had a better suppressive effect on HSFs than Res at 48 and 72 h, which suggest that the sustained release of Res from MSN@Res almost entirely inhibited the proliferation of the HSFs (Fig. 4E). The live/dead assay also confirmed these results. (Fig. 4D).
Fig. 4.
MSN@Res reduces the viability of HSFs. (A) The cell morphology after treatment with different concentrations (0–400 μg/mL) of MSN@Res for 48 h. (B) After treatment of MSN@Res for 24 and 48 h, the cell viability of HSFs were determined by CCK-8 assay (n = 3). (C) The IC50 value of MSN@Res treatment on HSFs for 48 h.(D)The Live/dead assay after treatment with Res, MSN or MSN@Res for 48 h was confirmed by Calcein/PI staining.(E) CCK-8 assay was used to detect the viability of HSFs after treatment with Res, MSN or MSN@Res for 24 h, 48 h, and 72 h (n = 6,Res or MSN@Res vs Control,∗∗∗P < 0.001, ****P < 0.0001; MSN@Res vs Res,△△△△P < 0.0001).
3.6. MSN@Res increases the apoptosis of hypertrophic scar fibroblasts
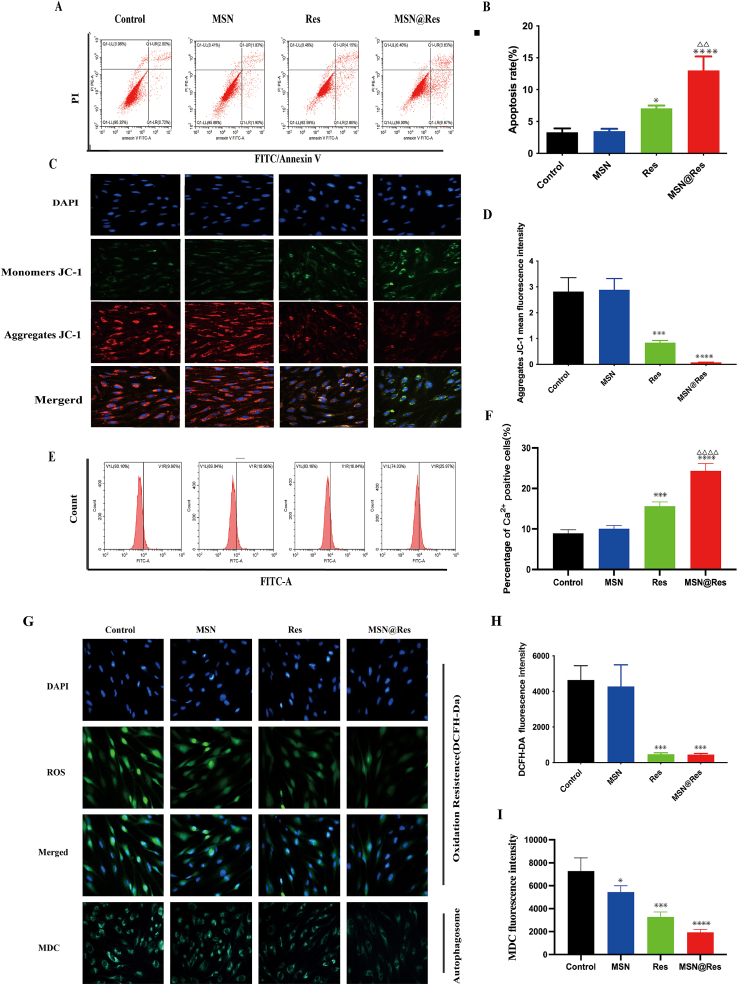
The effect of MSN@Res on fibroblast apoptosis was tested. HSFs were treated with Res, MSN or MSN@Res for 48 h, the frequency of apoptotic cells was determined by flow cytometry, treatment with Res or MSN@Res increased the percentages of apoptotic HSFs, and MSN@Res had a better induction effect than Res(Fig. 5A and B). To further confirm that MSN@Res acted through the mitochondrial apoptotic pathway, the mitochondrial membrane potential (MMP) of individual groups of cells was measured by fluorescence after staining with JC-1. As shown in (Fig. 5C and D), red fluorescence intensity (aggregates JC-1) represents MMP, compared with the cells treated with vehicle alone, both Res and MSN@Res decreased the MMP, without significant difference between these two groups. To investigate the changes in intracellular calcium levels, HSFs were stained with Fluo-4M to conduct the flow cytometry analysis, in comparison with vehicle-treated cells, both Res and MSN@Res increased the intracellular calcium levels, and MSN@Res exhibits a more significant effect (Fig. 5E and F). These results are consistent with previous studies, which suggest that a MMP loss leads to increased permeability, resulting in a elevation of intracellular calcium [33].
Fig. 5.
The apoptosis, autophagy and ROS generation of HSFs treated with MSN@Res, MSN or Res. (A)and(B) Flow cytometric analysis of the frequency of apoptotic HSFs at 48 h post treatment (n = 3). (C)and(D) HSFs were treated with MSN@Res or Res for 48 h, and the mitochondrial membrane potential (MMP) was analyzed by fluorescent imaging after staining the cells with JC-1 (n = 3). (E)and(F) Flow cytometric analysis with Fluo-4M was used to investigate the intracellular calcium levels (n = 3). (G)and(H) The levels of intracellular ROS were analyzed by fluorescence after staining the cells with DCFH-DA (n = 3, Scale bar = 400 μm). (G)and(I) The formation of autolysosomes after staining with MDC were examined by fluorescence microscopy (n = 3). (Data shown as mean ± SD, compared with control, *P < 0.05,∗∗∗P < 0.001,****P < 0.0001; compared with Res,△△P < 0.05,△△△△P < 0.0001).
3.7. MSN@Res reduces the reactive oxygen species and autolysosomes production of hypertrophic scar fibroblasts
To understand the pharmacological action of MSN@Res, the effect of Res, MSNs or MSN@Res on ROS production was tested by fluorescent microscopy, as shown in (Fig. 5G and H), Compared with the cells treated with vehicle alone, Res and MSN@Res both decreased the ROS levels in HSFs cells.
In order to ascertain how MSN@Res affect autophagy on HSFs cells, the formation of autolysosomes after staining with MDC was examined. As shown in (Fig. 5G–I), the accumulation of MDC in MSN@Res and Res treated cells decreased, indicating MSN@Res and Res decreased the number of autolysosomes compared to untreated cells, without significant difference between these two groups.
High levels of ROS can induce oxidative stress and cell apoptosis, while our results are inconsistent with previous research [33], suggesting that MSN@Res may induce apoptosis by reducing oxidative stress and inhibiting cell autophagy, rather than by promoting the ROS generation and oxidative stress.
3.8. MSN@Res surpresses protective autophagy contributes to Cell apoptosis of hypertrophic scar fibroblasts via inhibition of p38-MAPK signaling pathways
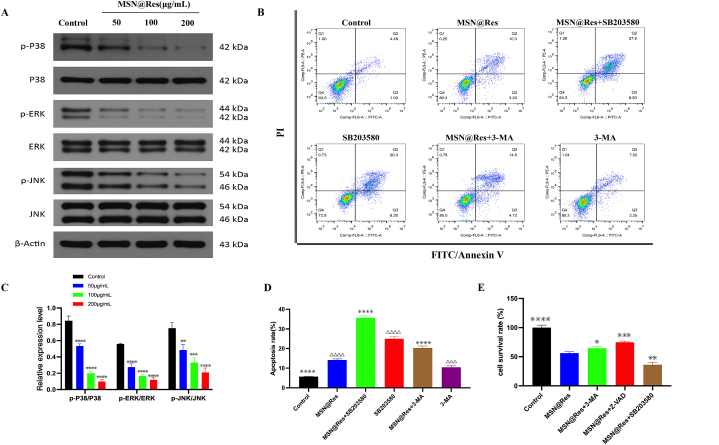
To reveal the potential mechanisms about the MSN@Res repressed autophagy and induced apoptosis, we investigated the effects of MSN@Res on MAPK signaling. Firstly, HSFs were treated with MSN@Res at different concentrations (50,100, 200 μg/mL) for 48 h, Western blot analysis revealed that treatment with MSN@Res effectively inhibited phosphorylation of ERK, JNK and p38 in a dose-dependent manner (Fig. 6A–C). Subsequently, HSFs were pretreated with the p38 inhibitor SB203580, apoptosis inhibitor Z-VAD or the autophagy inhibitor 3-MA,compared to the MSN@Res alone-treated group, SB203580 and 3-MA pretreatment significantly increased MSN@Res induced apoptosis (Fig. 6B–D). Furthermore, CCK8 assay was used for the detection of cell viability, as shown in (Fig. 6E), when compared with MSN@Res alone treatment, the cell growth inhibition induced by MSN@Res were significantly decreased or enhanced after pretreated with Z-VAD or SB203580 respectively. Interestingly, the 3-MA pretreatment also partially reversed the inhibition of cell viability induced by MSN@Res. These results indicated that the physiological autophagy surpression induced by MSN@Res contributes to cell apoptosis and other forms of cell death via inhibition of p38-MAPK signaling, results in cell viability inhibition of HSFs. While 3-MA can promote apoptosis of HSFs by inhibiting physiological autophagy, it may also inhibit other forms of cell death, such as autosis [3], the effect of this death form on cell viability was greater than that of apoptosis.
Fig. 6.
Effects of p38-MAPK signaling and autophagy on MSN@Res-induced apoptosis in HSFs cells. (A)and(C) Western blot analysis of different concentrations of MSN@Res on the phosphorylation of ERK, p38, and JNK (n = 3) (compared with control, ∗∗P < 0.01,∗∗∗P < 0.001,****P < 0.0001). (B)and(D) Flow cytometry analysis of pretreated with p38 inhibitor SB203580 or autophagy inhibitor 3-MA on MSN@Res-induced cell apoptosis (n = 3,compared with control,△△△P < 0.001,△△△△P < 0.0001;compared with MSN@Res,****P < 0.0001). (E) CCK8 assay analysis of pretreated with SB203580, 3-MA or apoptosis inhibitor Z-VAD on MSN@Res-induced cell viability inhibition (n = 6,compared with MSN@Res,*P < 0.05,∗∗P < 0.01,∗∗∗P < 0.001 ****P < 0.0001).
3.9. MSN@Res regulates autophagy and apoptosis of hypertrophic scar fibroblasts via inhibition of p38/HIF-1α/p53 signaling axis
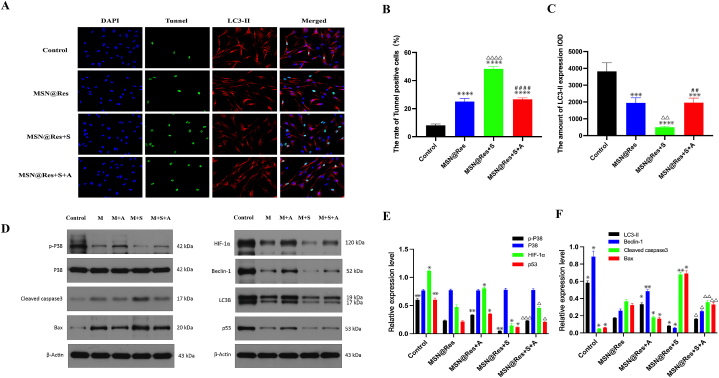
To further illuminate the mechanism of MSN@Res induced transition from autophagy to apoptosis, HSFs were pretreated with p38 inhibitor SB203580 or activator anisomycin, followed by the treatment of MSN@Res, with or without the co-incubation of anisomycin. The immunofluorescence results revealed that MSN@Res reduced the autophagy marker LC3-II expression and increased the Tunnel-positive cells, and the SB203580 pretreatment could enhanced MSN@Res -induced apoptosis and autophagy inhibition, which was completely reversed by the addition of anisomycin (Fig. 7A,B,C). Western blotting results demonstrated that MSN@Res upregulated the expression of cleaved caspase3 and Bax, while decreasing phospho-p38, HIF-1α, p53, LC3-II and Beclin-1 protein expression, and the upregulation of apoptosis-related proteins induced by MSN@Res was enhanced by the SB203580 pretreatment, which also enchanced the downregulation of HIF-1α, p53 and autophagy-related proteins (LC3-II and Beclin-1) induced by MSN@Res. Furthermore, anisomycin abolished the up or downregulation of these related proteins expression induced by MSN@Res(Fig. 7D,E,F). The Western blotting results were consistent with those of fluorescence, indicating that MSN@Res induced transition from fibroblast (protective) autophagy to apoptosis via inhibition of p38/HIF-1α/p53 signaling axis under hypoxic conditions. It is noteworthy that the effect of MSN@Res on p53 protein expression is inconsistent with previous research results.
Fig. 7.
The inactivation of p38-MAPK/HIF-1α/p53 signaling axis is triggered by MSN@Res and its role in regulation of fibroblast autophagy and apoptosis.
(A),(B)and(C) The HSFs were pretreated with 10 μM SB203580 for 2 h prior to 100 μM MSNs@Res treatment for 48 h with or without 25 μg/mL anisomycin, and then the transition from autophagy to apoptosis in HSFs was determined by immunofluorescence analysis, where as green fluorescence represents Tunnel-positive cells, indicating apoptosis, and red fluorescence represents LC3-II-positive cells, indicating autophagy (n = 3, compared with control,△△P< 0.01,△△△△P < 0.0001;compared with MSN@Res, ∗∗∗P < 0.0001, ****P < 0.0001;compared with MSN@Res+S, ##P < 0.01,####P < 0.0001). (D),(E)and(F) Western blot analysis of HIF-1α, p53, autophagy and apoptosis-related proteins expression. (n = 3, compared with MSN@Res , **P < 0.01, *P < 0.05; compared with MSN@Res + S , △P < 0.05, △△P < 0.01,△△△P < 0.001) Note: M: MSN@Res, A: Anisomycin, S: SB203580. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.10. MSN@Res suppresses oxidative stress-mediated p38/HIF-1α/p53 signaling activation of hypertrophic scar fibroblasts
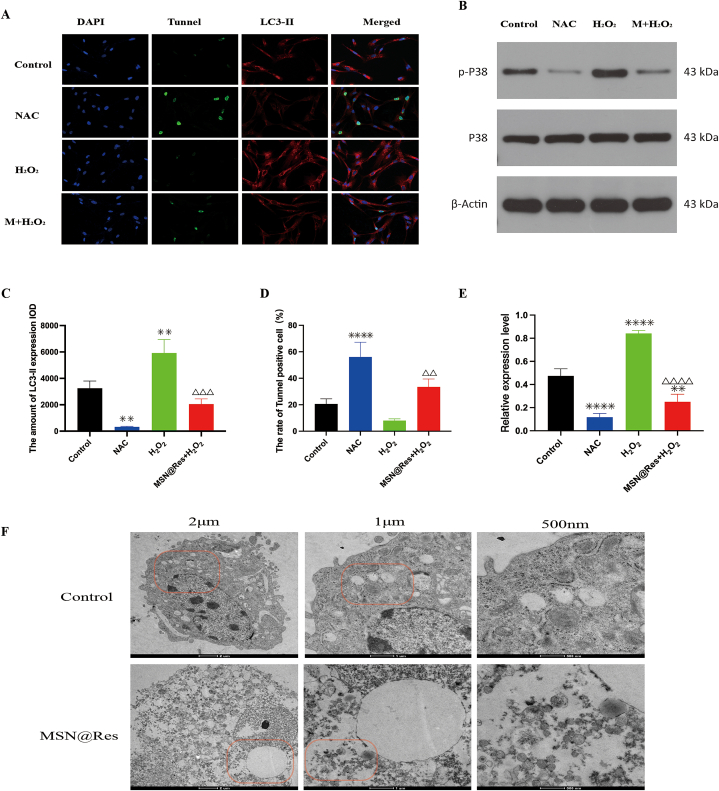
Studies have shown that oxidative stress is one of the initiators of fibroblast activation [10]. H2O2 is an important component of ROS. Our fluorescence results revealed that MSN@Res reduced ROS production, we need to further varify the role of ROS on MSN@Res-induced transition of fibroblasts from autophagy to apoptosis and its association between the p38-MAPK/HIF-1α/p53 signaling axis. Immunofluorescence results revealed that NAC, an antioxidant, incresed Tunnel-positive cells and reduced LC3-II expression significantly, while LC3-II expression was markedly enhanced by incubation with H2O2, which was completely reversed by the addition of MSN@Res with a corresponding increase of Tunnel-positive cells (Fig. 8A–C,D). Western blotting was used to measure the expression of total and phosphorylated p38-MAPK, as shown in (Fig. 8B–E), phospho-p38 was downregulated in the NAC group compared to the control group, while phospho-p38 expression was markedly enhanced by incubation with H2O2, which was completely reversed by the addition of MSNs@Res. The Western blotting results were consistent with those of fluorescence, indicating that MSN@Res suppresses oxidative stress-mediated p38-MAPK/HIF-1α/p53 signaling activation, thus induced transition of fibroblasts from autophagy to apoptosis, leading to cell viability inhibition. In addition, the transmission electron microscopy showed autosis was also detected in MSN@Res treated fibroblasts, which is another form of cell death induced by autophagy [3](Fig. 8F).
Fig. 8.
Roles of ROS in autophagy and apoptosis regulated by MSN@Res and its association between the p38-MAPK/HIF-1α/p53 signaling axis. (A),(C)and(D) The HSFs were treated with 100 μM NAC or 300 μM H2O2 alone for 2 h, or pretreated with 300 μM H2O2 for 2 h prior to 100 μM MSN@Res treatment for 48 h, and then the autophagy and apoptosis in HSFs was determined by immunofluorescence analysis, where as green fluorescence represents Tunnel-positive cells, indicating apoptosis, and red fluorescence represents LC3-II-positive cells, indicating autophagy (n = 3, compared with control, ∗∗P < 0.01, ****P < 0.0001; compared with H2O2,△△P < 0.01, △△△P < 0.001 ). (B)and(E) Western blot analysis of phospho-p38 protein expression. (n = 3, compared with Control, ∗∗P < 0.01,****P < 0.0001; compared with H2O2 , △△△△P < 0.0001). (F) TEM analysis of cell ultrastructural characteristics in control and MSN@Res treated cells, the formation of autophagosomes was observed in control treated cells under hypoxia/malnutrition condition, while vacuolated cytoplasm and ruptured cell membrane were detected in MSN@Res treated cells, showing a typical morphology of autosis. Note: M: MSN@Res. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
Resveratrol (Res) is a natural antioxidant polyphenolic compound rich in various plants such as grapes, peanuts, and berries [34]. Res has been demonstrated to not only prevent and inhibit the emergence and development of various tumors [[35], [36], [37]], but also to inhibit hypertrophic scar fibroblasts proliferation and collagen synthesis via regulation of autophagy and apoptosis [38,39]. However, its application is limited by factors, such as water solubility and low bioavailability [40]. Recent researches have shown that these problems could be solved by nanotechnology-driven drug delivery systems, such as poly (lactic-co-glycolic acid) nanoparticles loaded with Res and functionalized nanostructured liposomes. These systems have great potential to improve drug concentration and bioavailability in tumor lesions [41,42]. As a result of the tunable structure, low toxicity, controllable drug release and good biocompatibility after mesoporous modification [43], mesoporous silica nanoparticles (MSN) have been widely used in drug delivery systems, such as transferrin-protease B-cleavable peptide-modified MSN@Res (Tf-Res-MSN), which have high targeting specificity for MCF-7 cells and excellent drug release [44]. Additionally, it has been reported that a variety of hydrogels containing free Res or Res-loaded nanoparticles are used to treat skin injuries, promoting angiogenesis and cell proliferation, reducing inflammation, and accelerating wound healing [[45], [46], [47]].
Inspired by the previous investigations, we successfully prepared MSN loaded with Res and characterized its physicochemical properties. The results demonstrated that the modified MSN particles had a slightly increased diameter and a more blurred mesoporous structure, indicating that the drug was successfully loaded while retaining excellent dispersibility and stability. The hollow spherical MSN particles had a porous structure on the surface, which served as the structural basis for their excellent drug carrier. Although Res released linearly from the nanoparticles for the first 6 h, with a cumulative release of up to 51.6 %, it then slowed drastically 24 h later and released slowly at a near-constant rate for up to 6 days, indicating that MSN@Res had good drug release controllability, this may be related to the large number of silanol groups on the MSN surface, which are easy to functionalize [48]. This study revealed that MSN@Res exhibited a more potent inhibitory impact on HSFs proliferation in hypoxic conditions compared to free Res. The improved effect can be attributed to the consistent release of Res at a stable concentration from the nanoparticles, as well as the potential improvement of Res's amorphization through the encapsulation process. These characteristics contribute to the strengthened chemical stability of Res [48].
Clinical observations have provided empirical evidence that, in the absence of therapy, hypertrophic scars have a tendency to progressively grow softer, flatter, and in some cases, may even fade over years. The primary mechanism of cell death implicated in this regression phase is fibroblast apoptosis. Scar lesions are characterized with hypoxic and ischemic microenvironments, in which autophagy is a compensatory process for fibroblasts to maintain normal metabolism. Recent researches indicate that physiological autophagy might facilitate fibroblast proliferation and scar formation [49], while excessive autophagy may result in autosis, which participates in the regression and maturation of hypertrophic scars in combination with apoptosis [3]. Moreover, it has been observed that the expression of HIF-1α and p53 exhibit a seesaw-like switch, which is intricately associated with the transition between fibroblast autophagy and apoptosis, as well as the regression and hyperplasia of hypertrophic scars [50]. Oxidative stress serves as a catalyst for the process of “fibroblast activation”, which is the main feature of hypertrophic scars, characterized by persistent fibroblast activation and accumulation of extracellular matrix. In a rat model of type 2 diabetic cardiomyopathy, metformin can improve myocardial fibrosis by the reduction of ROS levels, as well as the downregulation of left ventricular p53 and collagen expression, indicating a significant correlation between the ROS-p53-collagen axis and the development of myocardial fibrosis [29]. p38-MAPK is known to be active during the fibrosis process of hypertrophic scar and is essential for collagen production of dermal fibroblasts [30]. Resveratrol may inhibit the proliferation of rheumatoid arthritis-related fibroblast-like synoviocyte by reducing ROS accumulation and suppressing phosphorylation of p38 and JNK-MAPK [31].
Network pharmacology predictions revealed that 121 common genes were mainly enriched in hypoxia and oxidative stress response, regulation of apoptosis, and positive regulation of MAPK cascade reaction. Based on these results, we speculate that MSN@Res may modulate fibroblast autophagy and apoptosis through the attenuation of oxidative stress and inhibition of the MAPK cascade pathway. Consequently, this modulation induced a seesaw-like switch between HIF-1α and p53, ultimately facilitating the regression of hypertrophic scar within a hypoxic and ischemic environment. According to previous studies, depolarization of the mitochondrial membrane potential increases membrane permeability, resulting in the release of calcium ions that are stored in mitochondria or endoplasmic reticulum into the cytoplasm. As a result, mitochondria produce a significant amount of ROS, which further facilitates a continuous elevation of intracellular calcium ions, triggering cellular apoptosis ultimately [51]. Nevertheless, we have discovered that MSN@Res has the potential to mitigate ROS levels in fibroblasts, possibly attributed to its robust antioxidant properties. The activation of the oxidative stress response is not part of the process by which it triggers cell apoptosis. Additionally, resveratrol has been shown to increase HSF autophagy by up-regulating miR-4654, which inhibits HS formation [51]. This is contradictory with our findings, and it may be related to the different cell culture conditions. Resveratrol has previously been reported to suppress HS by inducing autosis of HSFs in normoxic conditions [39], whereas MSN@Res may reduce cell viability by inhibiting physiological autophagy of HSFs in hypoxic and ischemic environments in our research, which are more similar to the actual internal environment of hypertrophic scar lesions.
Although p38-MAPK activation might stimulate dermal fibroblast collagen production and the formation of hypertrophic scars, the precise upstream and downstream pathways have not been identified [30]. We noticed that MSN@Res may trigger cell apoptosis by blocking p38-MAPK phosphorylation, indicating that p38-MAPK is involved in MSN@Res-induced fibroblast apoptosis and activity inhibition. In addition, MSN@Res-induced suppression of fibroblast activity has been linked to several kinds of cell death, including autosis and apoptosis. Interestingly, inhibiting autophagy does not reverse the suppression of MSN@Res on cell viability, implying that autophagy inhibitors may not only enhance cell apoptosis by decreasing physiological autophagy but may also suppress other forms of cell death than apoptosis, such as autosis [3].
The process of autophagy relies on the expression of HIF-1α, and the inhibition of its expression may surppress autophagy and cell survival with a inducement of cell apoptosis [52,53]. We discovered that the inhibitory effect of MSN@Res on the expression of HIF-1α and p53 proteins in fibroblast either diminished or strengthened in accordance with the activation or inhibition of p38-MAPK phosphorylation. The seesaw-like transition observed in the expression of apoptosis markers Cleaved caspase3, Bax, and autophagy markers LC3I/II, Beclin-1 was consistent with findings from previous research [50]. The immunofluorescence results also revealed that the transition between cell autophagy and apoptosis is consistent with the expression of marker proteins, indicating that MSN@Res can regulate this transition by downregulating HIF-1 and p53 protein expression via the inhibition of p38-MAPK phosphorylation. It is worth noting that previously described seesaw-like transition of HIF-1 and p53 proteins has not been observed. p53 is a tumor suppressor protein, as well as a regulatory factor for cell cycle progression and apoptosis, which has been extensively studied. Early research reported that knocking down p53 in HCT-116 cells increased mice survival by promoting autophagy [54], whereas re-transfection with wild-type p53 inhibited protective autophagy [55]. Conversely, some researches suggested that p53 can trigger autophagy by upregulating AMPK [56]. Recent studies revealed that silencing p53 weakens autophagy and increases the cell death of colon cancer induced by the novel ERK inhibitor CC90003, indicating that p53 is intimately related to protective cell autophagy [57]. Similarly, we found that MSN@Res can downregulate p53 expression in fibroblasts by inhibiting p38-MAPK phosphorylation, which is accompanied by a reduction in physiological autophagy and a rise in apoptosis, implying that p53's role in cell physiological autophagy may be more important than its effect on cell apoptosis. In addition to cell apoptosis, MSN@Res suppresses fibroblast physiological autophagy and induces autosis, which may be its primary mechanism to promote the HS regression.
The growth inhibition of A549 and NCI–H292 lung cancer cells for Bruceine D, as well as the induction of apoptosis and autophagy, may be related to the activation of the ERK and JNK-MAPK signaling pathways caused by ROS generation, while NAC can almost reverse these effects [58]. Consequently, our findings indicate that a reduction in oxidative stress not only hinders p38-MAPK phosphorylation but also inhibits physiological autophagy in fibroblast along with apoptosis promotion. Conversely, an increase in oxidative stress reverses these processes, which is consistent with the prevailing consensus.
5. Conclusion
In summary, we have prepared Res loaded MSN nanoparticles to explore the effect on solubility and drug release, as well as in-vitro anti-fibroblasts activity using in-vitro assays. It was found that when Res was encapsulated in MSNs the solubility increased, and MSN@Res had excellent stability and drug release performance. Additionally, MSN@Res showed enhanced dose-dependent cytotoxicity compared with Res, which may also be related to inhibition of physiological autophagy and inducement of apoptosis and autosis in fibroblasts under hypoxic environment through the downregulation of ROS/p38-MAPK/HIF-1α/p53 axis. This study provide novel insights into MSN as a drug delivery carrier for hydrophobic drugs, while also adding to the established body of evidence regarding the usefulness of Res as an anti-hypertrophic scar drug. Although in vivo bioavailability assays had not been conducted in this study, testing the effects of surface functionalization on solubility, in vitro release, cytotoxicity will be involved in our future study, and the development of dual sustained-release functional hydrogels loaded with MSN@Res and its in vivo bioavailability assays will be the focus in the future.
Funding
1.Regional Foundation of National Natural Science Foundation of China (No. 81760345); 2.Youth Program of Natural Science Foundation of Hunan Province (No. 2021JJ40487).
Data availability statement
Data will be made available on request.
Ethics declarations
All participants/patients (or their proxies/legal guardians) provided informed consent to participate in the study.
CRediT authorship contribution statement
Jun Zuo: Writing – review & editing, Writing – original draft, Investigation, Data curation. Shaolin Ma: Writing – review & editing, Supervision, Project administration, Investigation, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
None.
Contributor Information
Jun Zuo, Email: zjzjwfwf123@163.com.
Shaolin Ma, Email: mashaolin9@163.com.
References
- 1.Lynam E.C., Xie Y., Dawson R., et al. Severe hypoxia and malnutrition collectively contribute to scar fibroblast inhibition and cell apoptosis. Wound Repair Regen. 2015;23(5):664–671. doi: 10.1111/wrr.12343. [DOI] [PubMed] [Google Scholar]
- 2.Chen H., Xu K., Sun C., et al. Inhibition of ANGPT2 activates autophagy during hypertrophic scar formation via PI3K/AKT/mTOR pathway. An. Bras. Dermatol. 2023;98(1):26–35. doi: 10.1016/j.abd.2021.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang J., Song F., Wang X.Q. Role of autosis of fibroblasts in Hyper tropic Scar regression. J. Shanghai Jiao Tong Univ. (Sci.) 2022;42(1):44–50. [Google Scholar]
- 4.Qureshi-Baig K., Kuhn D., Viry E., et al. Hypoxia-induced autophagy drives colorectal cancer initiation and progression by activating the PRKC/PKC-EZR (ezrin) pathway. Autophagy. 2020;16(8):1436–1452. doi: 10.1080/15548627.2019.1687213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levine B., Kroemer G. Biological Functions of autophagy genes: a disease perspective. Cell. 2019;176(1–2):11–42. doi: 10.1016/j.cell.2018.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giordano A., Avantaggiati M.L. p300 and CBP: partners for life and death. J. Cell. Physiol. 1999;181(2):218–230. doi: 10.1002/(SICI)1097-4652(199911)181:2<218::AID-JCP4>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 7.Xia N., Daiber A., Förstermann U., et al. Antioxidant effects of resveratrol in the cardiovascular system. Br. J. Pharmacol. 2017;174(12):1633–1646. doi: 10.1111/bph.13492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meng T., Xiao D., Muhammed A., et al. Anti-inflammatory action and mechanisms of resveratrol. Molecules. 2021;26(1):229. doi: 10.3390/molecules26010229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rauf A., Imran M., Butt M.S., et al. Resveratrol as an anti-cancer agent: a review. Crit. Rev. Food Sci. Nutr. 2018;58(9):1428–1447. doi: 10.1080/10408398.2016.1263597. [DOI] [PubMed] [Google Scholar]
- 10.Akasaka E., Kleiser S., Sengle G., et al. Diversity of mechanisms underlying latent TGF-β activation in recessive dystrophic epidermolysis bullosa. J. Invest. Dermatol. 2021;141(6):1450–1460.e9. doi: 10.1016/j.jid.2020.10.024. [DOI] [PubMed] [Google Scholar]
- 11.Ikeda K., Torigoe T., Matsumoto Y., et al. Resveratrol inhibits fibrogenesis and induces apoptosis in keloid fibroblasts. Wound Repair Regen. 2013;21(4):616–623. doi: 10.1111/wrr.12062. [DOI] [PubMed] [Google Scholar]
- 12.He W., Zhang J., Gan T.Y., et al. Advanced glycation end products induce endothelial-to-mesenchymal transition via downregulating Sirt 1 and upregulating TGF-β in human endothelial cells. BioMed Res. Int. 2015;2015 doi: 10.1155/2015/684242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou W., Zhang H., Wang X., et al. Network pharmacology to unveil the mechanism of Moluodan in the treatment of chronic atrophic gastritis. Phytomedicine. 2022;95 doi: 10.1016/j.phymed.2021.153837. [DOI] [PubMed] [Google Scholar]
- 14.Huang Y., Zhao Y., Liu F., et al. Nano traditional Chinese medicine: current progresses and future challenges. Curr. Drug Targets. 2015;16(13):1548–1562. doi: 10.2174/1389450116666150309122334. [DOI] [PubMed] [Google Scholar]
- 15.Li C., Wang Z., Lei H., etal Recent progress in nanotechnology-based drug carriers for resveratrol delivery. Drug Deliv. 2023;30(1) doi: 10.1080/10717544.2023.2174206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ru J., Li P., Wang J., et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J. Cheminf. 2014;6:13. doi: 10.1186/1758-2946-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gfeller D., Grosdidier A., Wirth M., et al. SwissTargetPrediction: a web server for target prediction of bioactive small molecules. Nucleic Acids Res. 2014;42(Web Server issue):W32–W38. doi: 10.1093/nar/gku293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuhn M., von Mering C., Campillos M., et al. STITCH: interaction networks of chemicals and proteins. Nucleic Acids Res. 2008;36(Database issue):D684–D688. doi: 10.1093/nar/gkm795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keiser M.J., Roth B.L., Armbruster B.N., et al. Relating protein pharmacology by ligand chemistry. Nat. Biotechnol. 2007;25(2):197–206. doi: 10.1038/nbt1284. [DOI] [PubMed] [Google Scholar]
- 20.Stelzer G., Rosen N., Plaschkes I., et al. The GeneCards suite: from gene data mining to disease Genome sequence analyses. Curr Protoc Bioinformatics. 2016;54:1. doi: 10.1002/cpbi.5. 30.1-1.30.33. [DOI] [PubMed] [Google Scholar]
- 21.Amberger J.S., Bocchini C.A., Schiettecatte F., et al. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43(Database issue):D789–D798. doi: 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shannon P., Markiel A., Ozier O., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szklarczyk D., Gable A.L., Nastou K.C., et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021 Oct 11;49(18) doi: 10.1093/nar/gkab835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan H., Li Y., Yang B., et al. Exploring the mechanism of action of Yiyi Fuzi Baijiang powder in colorectal cancer based on network pharmacology and molecular docking studies. Biotechnol. Genet. Eng. Rev. 2023:1–21. doi: 10.1080/02648725.2023.2167765. [DOI] [PubMed] [Google Scholar]
- 25.Formagio A.S.N., Vilegas W., Volobuff C.R.F., et al. Exploration of essential oil from Psychotria poeppigiana as an anti-hyperalgesic and anti-acetylcholinesterase agent: chemical composition, biological activity and molecular docking. J. Ethnopharmacol. 2022;296 doi: 10.1016/j.jep.2022.115220. [DOI] [PubMed] [Google Scholar]
- 26.Tang Z., Lu L., Zhou X., et al. A new cytotoxic polycyclic polyprenylated acylphloroglucinol from Garcinia nujiangensis screened by the LC-PDA and LC-MS. Nat. Prod. Res. 2020;34(17):2448–2455. doi: 10.1080/14786419.2018.1539983. [DOI] [PubMed] [Google Scholar]
- 27.Xie X., Nie H., Zhou Y., et al. Eliminating blood oncogenic exosomes into the small intestine with aptamer-functionalized nanoparticles. Nat. Commun. 2019;10(1):5476. doi: 10.1038/s41467-019-13316-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meschini S., Condello M., Calcabrini A., et al. The plant alkaloid voacamine induces apoptosis-independent autophagic cell death on both sensitive and multidrug resistant human osteosarcoma cells. Autophagy. 2008;4(8):1020–1033. doi: 10.4161/auto.6952. [DOI] [PubMed] [Google Scholar]
- 29.Al-Ani B., Alzamil N.M., Hewett P.W., et al. Metformin ameliorates ROS-p53-collagen axis of fibrosis and dyslipidemia in type 2 diabetes mellitus-induced left ventricular injury. Arch. Physiol. Biochem. 2023;129(3):734–740. doi: 10.1080/13813455.2020.1869265. [DOI] [PubMed] [Google Scholar]
- 30.Chai C.Y., Song J., Tan Z., et al. Adipose tissue-derived stem cells inhibit hypertrophic scar(HS) fibrosis via p38/MAPK pathway. J. Cell. Biochem. 2019;120(3):4057–4064. doi: 10.1002/jcb.27689. [DOI] [PubMed] [Google Scholar]
- 31.Yang G., Chang C.C., Yang Y., et al. Resveratrol alleviates rheumatoid arthritis via reducing ROS and inflammation, inhibiting MAPK signaling pathways, and suppressing angiogenesis. J. Agric. Food Chem. 2018;66(49):12953–12960. doi: 10.1021/acs.jafc.8b05047. [DOI] [PubMed] [Google Scholar]
- 32.Liu X., Wang J., Dou P., et al. The ameliorative effects of arctiin and arctigenin on the oxidative injury of lung induced by silica via TLR-4/NLRP3/TGF-β signaling pathway. Oxid. Med. Cell. Longev. 2021;2021 doi: 10.1155/2021/5598980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bae H., Park S., Ham J., et al. ER-mitochondria calcium flux by β-sitosterol promotes cell death in ovarian cancer. Antioxidants. 2021;10(10):1583. doi: 10.3390/antiox10101583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xue Y.Q., Di J.M., Luo Y., etal Resveratrol oligomers for the prevention and treatment of cancers. Oxid. Med. Cell. Longev. 2014;2014 doi: 10.1155/2014/765832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.AminiP, NodooshanSJ, AshrafizadehM, et al. Resveratrol induces apoptosis and attenuates proliferation of MCF-7 cells in combination with radiation and hyperthermia. Curr. Mol. Med. 2021;21(2):142–150. doi: 10.2174/1566524020666200521080953. [DOI] [PubMed] [Google Scholar]
- 36.VernousfaderaniEK AkhtariN., RezaeiS RezaeeY., et al. Resveratrol and colorectal cancer: a molecular approach to clinical researches. Curr. Top. Med. Chem. 2021;21(29):2634–2646. doi: 10.2174/1568026621666211105093658. [DOI] [PubMed] [Google Scholar]
- 37.ZaffaroniN BerettaGL. Resveratrol and prostatecancer:thepower of phytochemicals. Curr. Med. Chem. 2021;28(24):4845–4862. doi: 10.2174/0929867328666201228124038. [DOI] [PubMed] [Google Scholar]
- 38.Zeng G., Zhong F., Li J., et al. Resveratrol-mediated reduction of collagen by inhibiting proliferation and producing apoptosis in human hypertrophic scar fibroblasts. Biosci. Biotechnol. Biochem. 2013;77(12):2389–2396. doi: 10.1271/bbb.130502. [DOI] [PubMed] [Google Scholar]
- 39.Pang K., Li B., Tang Z., et al. Resveratrol inhibits hypertrophic scars formation by activating autophagy via the miR-4654/Rheb axis. Mol. Med. Rep. 2020;22(4):3440–3452. doi: 10.3892/mmr.2020.11407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng Y., Jia R., Li J., etal Curcumin- and resveratrol-co-loaded nanoparticles in synergistic treatment of hepatocellular carcinoma. J. Nanobiotechnol. 2022;20(1):339. doi: 10.1186/s12951-022-01554-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bhatt P., Fnu G., Bhatia D., et al. Nanodelivery of resveratrol-loaded PLGA nanoparticles for age-related macular degeneration. AAPS PharmSciTech. 2020;21:291. doi: 10.1208/s12249-020-01836-4. [DOI] [PubMed] [Google Scholar]
- 42.Poonia N., Narang J.K., Lather V., et al. Resveratrol loaded functionalized nanostructured lipid carriers for breast cancer targeting: systematic development, characterization and pharma- cokinetic evaluation. Colloids Surf. B Biointerfaces. 2019;181:756–766. doi: 10.1016/j.colsurfb.2019.06.004. [DOI] [PubMed] [Google Scholar]
- 43.Zhu X., Wen Y., Zhao Y., et al. Functionalized chitosan-modified defect-related luminescent mesoporous silica nanoparticles as new inhibitors for hIAPP aggregation. Nanotechnology. 2019;30 doi: 10.1088/1361-6528/ab13ef. [DOI] [PubMed] [Google Scholar]
- 44.Li D., Song C., Zhang J., Zhao X. Targeted delivery and apoptosis induction activity of peptide-transferrin targeted mesoporous silica encapsulated resveratrol in MCF-7 cells. J. Pharm. Pharmacol. 2023;75(1):49–56. doi: 10.1093/jpp/rgac028. [DOI] [PubMed] [Google Scholar]
- 45.Zhao C.C., Zhu L., Wu Z., et al. Resveratrol-loaded peptide-hydrogels inhibit scar formation in wound healing through suppressing inflammation. Regen Biomater. 2020;7(1):99–107. doi: 10.1093/rb/rbz041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miao L., Daozhou L., Ying C., etal A resveratrol-loaded nanostructured lipid carrier hydrogel to enhance the anti-UV irradiation and anti-oxidant efficacy. Colloids Surf. B Biointerfaces. 2021;204 doi: 10.1016/j.colsurfb.2021.111786. [DOI] [PubMed] [Google Scholar]
- 47.Amanat S., Taymouri S., Varshosaz J., etal Carboxymethyl cellulose-based wafer enriched with resveratrol-loaded nanoparticles for enhanced wound healing. Drug Deliv Transl Res. 2020;10(5):1241–1254. doi: 10.1007/s13346-020-00711-w. [DOI] [PubMed] [Google Scholar]
- 48.Lin M., Yao W., Xiao Y., et al. Resveratrol-modified mesoporous silica nanoparticle for tumor-targeted therapy of gastric cancer. Bioengineered. 2021;12(1):6343–6353. doi: 10.1080/21655979.2021.1971507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cao C., Wang W., Lu L., et al. Inactivation of Beclin-1-dependent autophagy promotes ursolic acid-induced apoptosis in hypertrophic scar fibroblasts. Exp. Dermatol. 2018;27(1):58–63. doi: 10.1111/exd.13410. [DOI] [PubMed] [Google Scholar]
- 50.Li M., Su Y., Gao X., et al. Transition of autophagy and apoptosis in fibroblasts depends on dominant expression of HIF-1α or p53. J. Zhejiang Univ. - Sci. B. 2022;23(3):204–217. doi: 10.1631/jzus.B2100187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim S.H., Kim K.Y., Yu S.N., et al. Monensin induces PC-3 prostate cancer cell apoptosis via ROS production and Ca2+ homeostasis disruption[J] Anticancer Res. 2016;36(11):5835–5843. doi: 10.21873/anticanres.11168. [DOI] [PubMed] [Google Scholar]
- 52.Zhang Y., Liu D., Hu H., et al. HIF-1α/BNIP3 signaling pathway-induced-autophagy plays protective role during myocardial ischemia-reperfusion injury. Biomed. Pharmacother. 2019;120 doi: 10.1016/j.biopha.2019.109464. [DOI] [PubMed] [Google Scholar]
- 53.Lu N., Li X., Tan R., et al. HIF-1α/Beclin1-Mediated autophagy is involved in neuroprotection induced by hypoxic preconditioning. J. Mol. Neurosci. 2018;66(2):238–250. doi: 10.1007/s12031-018-1162-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tasdemir E., Maiuri M.C., Galluzzi L., et al. Regulation of autophagy by cytoplasmic p53. Nat. Cell Biol. 2008;10:676–687. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morselli E., Tasdemir E., Maiuri M.C., et al. Mutant p53 protein localized in the cytoplasm inhibits autophagy. Cell Cycle. 2008;7:3056–3061. doi: 10.4161/cc.7.19.6751. [DOI] [PubMed] [Google Scholar]
- 56.Jing K., Song K.S., Shin S., et al. Docosahexaenoic acid induces autophagy through p53/AMPK/mTOR signaling and promotes apoptosis in human cancer cells harboring wild-type p53. Autophagy. 2011;7:1348–1358. doi: 10.4161/auto.7.11.16658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mi W., Wang C., Luo G., et al. Targeting ERK induced cell death and p53/ROS-dependent protective autophagy in colorectal cancer. Cell Death Dis. 2021;7(1):375. doi: 10.1038/s41420-021-00677-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fan J., Ren D., Wang J., et al. Bruceine D induces lung cancer cell apoptosis and autophagy via the ROS/MAPK signaling pathway in vitro and in vivo. Cell Death Dis. 2020;11(2):126. doi: 10.1038/s41419-020-2317-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.