Summary
Direct acting antivirals (DAAs) represent critical tools for combating severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern (VOCs) that have escaped vaccine-elicited spike-based immunity and future coronaviruses with pandemic potential. Here, we used bioluminescence imaging to evaluate therapeutic efficacy of DAAs that target SARS-CoV-2 RNA-dependent RNA polymerase (favipiravir, molnupiravir) or main protease (nirmatrelvir) against Delta or Omicron VOCs in K18-hACE2 mice. Nirmatrelvir displayed the best efficacy followed by molnupiravir and favipiravir in suppressing viral loads in the lung. Unlike neutralizing antibody treatment, DAA monotherapy regimens did not eradicate SARS-CoV-2 in mice, but combining molnupiravir with nirmatrelvir exhibited superior additive efficacy and led to virus clearance. Furthermore, combining molnupiravir with caspase-1/4 inhibitor mitigated inflammation and lung pathology whereas combining molnupiravir with COVID-19 convalescent plasma demonstrated synergy, rapid virus clearance, and 100% survival. Thus, our study provides insights into in vivo treatment efficacies of DAAs and other effective combinations to bolster COVID-19 therapeutic arsenal.
Subject areas: Virology, Optical imaging
Graphical abstract
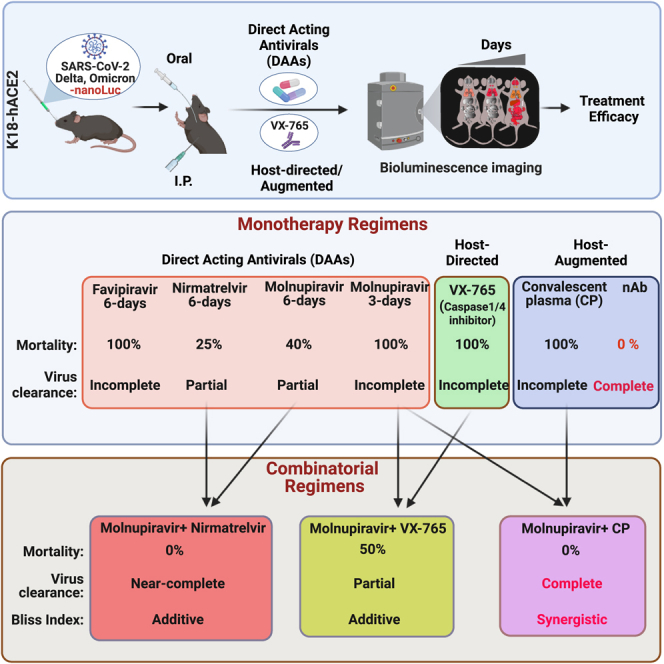
Highlights
-
•
Monotherapy with approved drugs is inefficient in clearing SARS-CoV-2 in mice
-
•
Combining molnupiravir with nirmatrelvir clears SARS-CoV-2 infection in mice
-
•
Combining molnupiravir with caspase-1/4 inhibitor mitigates inflammation and pathology
-
•
Combining molnupiravir with convalescent plasma yields rapid virus clearance
Virology; Optical imaging
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is an enveloped positive-strand RNA virus of the Coronaviridae family that is the causative agent of the coronavirus disease-19 (COVID-19) pandemic and has caused over 6.24 million deaths worldwide to date.1 Immunity provided by neutralizing antibodies (nAbs) elicited by several vaccines and treatment using nAb cocktails have significantly reduced the mortality rate caused by the COVID-19 pandemic.1,2,3,4 However, mutations incurred in SARS-CoV-2 variants of concern (VOCs; including Alpha, Beta, Gamma, Delta, and Omicron) has led to evasion of spike-based immunity, and several waves of pandemic.5 We are experiencing accelerated evolution of virus and enhanced immune escape as well as a notable decrease in vaccine protection resulting in requirement for additional bivalent booster doses especially in the elderly population.6,7 Although COVID-19 is no longer considered a global health emergency by the World Health Organization (WHO),8 long-term COVID and post-acute sequelae associated with SARS-CoV-2 infections remain a concern. Moreover, new variants, like EG.5.1, HV.1, and JN.1 that have resulted in an increased rate of COVID-19-related hospitalizations, have the potential to develop into new VOCs. It is important to note that Sarbecovirus subgenus consists of many zoonotic viruses that can potentially spread into human populations. Several members of this family have caused epidemics with much greater mortality rate, including SARS-CoV-1 (2002/3) and Middle Eastern respiratory syndrome virus (MERS) (2012). The lessons learned from previous and current coronavirus outbreaks demand that we continue to develop multipronged countermeasures consisting of vaccines, therapeutic antibodies, and antiviral drugs as well as host-directed and host-augmented strategies to blunt the impact of current and future outbreaks in the human population.9
Several direct acting antivirals (DAAs) repurposed from former antiviral programs have been approved or received emergency use authorization for COVID-19 treatment.10 They are essential to combating evolving VOCs, especially in the elderly, immunocompromised, and diseased population where vaccination fails to produce the desired protective response, as well as in vaccine-hesitant groups. Unlike nAbs, whose efficacy is compromised against VOCs, DAAs target conserved viral enzymes that play an essential role in virus replication and are expected to be overall effective despite presence of nAb-escape mutations in spike proteins of VOCs. However, equivalent efficacy against VOCs cannot be guaranteed and must be ascertained through experimentation. Two classes of antiviral drugs that target viral RNA-dependent RNA polymerase (RdRp) (molnupiravir, favipiravir) and main viral protease (nirmatrelvir) are currently available in the clinic and are approved by the Food and Drug Administration (FDA) or are available for off-label use to treat high-risk adults with SARS-CoV-2. Molnupiravir (MK-4482, EIDD-2801, beta-D-N4-hydroxycytidine, NHC) is an orally available ribonucleoside analog.11 After oral administration, molnupiravir, isopropylester prodrug of NHC (EIDD-1931), is phosphorylated intracellularly to the active NHC triphosphate form. During virus replication, NHC triphosphate is incorporated into viral RNA by the viral RNA polymerase and then promotes misincorporation of guanosine or adenosine resulting in mutagenesis. The virus is ultimately rendered non-infectious and unable to replicate after accumulating deleterious errors throughout its genome which is also known as “error catastrophe.” Favipiravir (6-fluoro-3-hydroxypyrazine-2-carboxamine; available for off-label use)12,13 demonstrates broad spectrum antiviral activity against a variety of RNA viruses. It is also a prodrug that is metabolized intracellularly to the active ribonucleoside 5′-triphosphate form that acts as a nucleotide analog to selectively inhibit RdRp and functions by inducing lethal mutagenesis or chain termination if two molecules of favipiravir are incorporated together. Several studies have reported in vitro inhibitory activity of favipiravir against SARS-CoV-2.13,14 Nirmatrelvir was originally developed in response to SARS-CoV-1 and targets the highly conserved main protease/3-chymotrypsin-like protease (Mpro, 3CLPRO) of SARS-CoV-1, SARS-CoV-2, and Betacoronaviruses.15,16 Mpro is involved in the processing of the viral polyprotein into functional proteins for the virus to form replication complexes. Nirmatrelvir-mediated inhibition of Mpro leads to the formation of non-functional viral proteins, preventing the virus from replicating and spreading. Several studies have shown that both molnupiravir and nirmatrelvir can effectively suppress acute virus replication, or associated disease in mice, Syrian hamster, ferrets, and macaques infected with SARS-CoV-2 ancestral strain.17,18,19,20 Moreover, results from in vitro studies or in hamsters suggest that both the drugs retain activity against multiple VOCs including Delta and Omicron variants.21 However, DAA monotherapy can be a double-edged sword that promote accelerated virus evolution, eventual drug resistance, and rebound after cessation of treatment especially in immunocompromised individuals.22,23,24 Combination antiviral drug treatments are common for chronic RNA virus infections like HIV-1 to combat drug resistance and increase the utility duration of DAAs. However, there is currently no approved antiviral combination therapy for SARS-CoV-2. Therefore, safe and effective (additive or synergistic) combinations of DAAs that ideally target different steps of virus replication or host-dependent pathways may be required to effectively eradicate the virus.25
As SARS-CoV-2 evolves, independent preclinical evidence for efficacy of antiviral drugs or their combinations in vivo are needed and must be generated due to accrued mutations, differences in replication rate, titers, and pathogenesis of VOCs. In this regard, we have developed a non-invasive bioluminescence imaging (BLI)-guided infection model to monitor virus replication and dissemination using SARS-CoV-2 nanoLuc luciferase (nLuc) reporters.26,27 A BLI-guided analysis can reduce the number of animals used in experiments and conform to the 3Rs (refine, reduce, replace) of animal research. Furthermore, BLI allows monitoring the impact of antiviral regimens on virus replication and spread from lung to distal organs, such as the brain and gastrointestinal (GI) tract, while also capturing the ability of the tested interventions to distribute into the affected tissues which may differ due to pharmacokinetic differences and the activity of drug transporters such p-glycoprotein.26 In this study we applied BLI to visualize SARS-CoV-2 VOC (Delta and Omicron) spread and persistence for evaluating efficacy of therapeutic drugs in K18-hACE2 transgenic mice. These mice are highly susceptible to human-tropic SARS-CoV-2 variants, (except Omicron and its sub lineages) and succumb to infection-induced mortality characterized by acute respiratory disorder syndrome (ARDS) due to infection in the lung followed by lethal encephalitis.28 We evaluated the anti-SARS-CoV-2 activities of favipiravir, molnupiravir, nirmatrelvir, or a combination of molnupiravir with nirmatrelvir, in combating the highly transmissible Delta and Omicron VOCs. While monotherapy regimens of favipiravir, molnupiravir, or nirmatrelvir at the tested dosage reduced virus replication in lungs, combination therapy with molnupiravir and nirmatrelvir was significantly more efficacious and demonstrated an additive in vivo efficacy against Delta and Omicron VOC in our preclinical model analyses. Inflammasome activation and pyroptosis-induced cell death mechanisms play a critical role in exacerbating COVID-19.29,30 Hence, we also evaluated the efficacy of caspase-1/4 inhibitor (VX-765) in combination with short-term molnupiravir dosing under therapeutic settings. This combination was also additive and significantly improved survival over monotherapy in addition to mitigating SARS-CoV-2-induced imbalanced inflammatory response and lung pathology. We also explored the potential of combining antiviral drugs with COVID-19 convalescent plasma (CCP) therapy that is an available treatment option in many of low- and middle-income countries (LMICs) where resources are limited. Our analyses demonstrate that CCP with Fc-effector functions effectively synergized with short-term molnupiravir treatment and rapidly cleared established infection mice. Thus, our study provides valuable insights into treatment efficacy of approved drugs, and their combinations with host-directed (inflammasome inhibitors) or host-augmented (CCPs) measures as part of the COVID-19 therapeutic arsenal.
Results
Comparative efficacy of favipiravir, molnupiravir, or nirmatrelvir monotherapy against Delta (B.1.617.2) VOC in K18-hACE2 mice
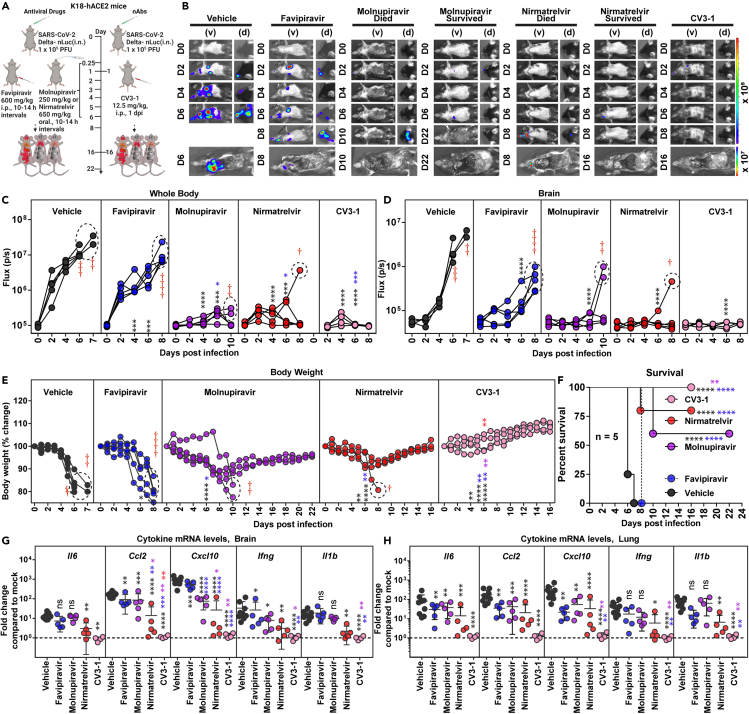
We tested the therapeutic efficacy of off-label use/FDA-approved DAAs favipiravir31 (600 mg/kg intraperitoneal, i.p.), molnupiravir (250 mg/kg; oral), or nirmatrelvir (650 mg/kg, oral) in K18-hACE2 mice challenged with reporter Delta VOC expressing nLuc (Delta-nLuc) (intranasal, i.n.; 1 × 105 plaque-forming units, PFUs). Delta VOC is known to replicate to higher levels than other VOCs in the lungs.32 Pharmacokinetic analyses demonstrated a considerable decrease in serum drug concentrations at 24 h after oral or i.p. dosing (Figure S1). Therefore, we administered the drugs 6 h post infection (hpi) followed by two doses per 10–14 h intervals until 6 days post infection (dpi) to test for effective suppression of virus replication (1,200 mg/kg favipiravir; 500 mg/kg molnupiravir or 1,300 mg/kg nirmatrelvir for ∼18–20 g mice). A similar regimen (twice daily) is also used in patients. The primary dosage we used in mice for evaluation was above the animal equivalent doses33 for each drug used in the clinic (average human weight: ∼60 kg, favipiravir, 656 mg/kg31; molnupiravir, 328 mg/kg34; nirmatrelvir, 123 mg/kg35 per day for 5 days; see STAR methods for details). Vehicle-treated mice were used as controls (Figure 1A). Virus infection was monitored with BLI every two days, and morbidity assessed using daily body weight measurements. BLI and subsequent quantification of nLuc flux (photons/sec) revealed that Delta-nLuc was able to spread from the nose to establish infection in the lungs of vehicle-treated mice by 2 dpi (Figures 1B–1D). The virus subsequently expanded in the lungs before spreading to the brain between 5 and 7 dpi, resulting in a concomitant loss in body weight and 100% mortality by 6–7 dpi (Figures 1E and 1F). Analyses of inflammatory cytokine mRNA expression (Il6, Ccl2, Cxcl10, Ifng, Il1b) in the brain and lung revealed 10- to 1,000-fold induction in vehicle-treated mice compared to uninfected control (Figures 1G and 1H). A single-dose treatment of a well-characterized nAb CV3-1 (12.5 mg/kg, i.p., 1 dpi) served as a positive control to compare the degree of virologic control engendered by the drug regimen. As expected from our previous studies,26 CV3-1 nAb treatment completely controlled Delta VOC replication as indicated by near absence of nLuc signals in all tissues (non-invasively and after necropsy), and body weight loss with 100% survival as well as baseline levels of inflammatory cytokines and nucleocapsid (N) mRNA expression.
Figure 1.
In Vivo efficacy of favipiravir, molnupiravir, and nirmatrelvir monotherapy regimen in K18-hACE2 mice against SARS-CoV-2 Delta VOC
(A) Experimental design for monitoring therapeutic efficacy of indicated drugs and neutralizing antibody (nAb) CV3-1 in K18-hACE2 mice challenged intranasally (i.n.) with 1 × 105 PFU of reporter SARS-CoV-2-Delta-nLuc VOC. favipiravir (600 mg/kg body weight, b.w.) and nAb CV3-1 (12.5 mg/kg b.w.) were administered intraperitoneally (i.p.) while molnupiravir (250 mg/kg) and nirmatrelvir (650 mg/kg) given via oral gavage (oral) under a therapeutic regimen starting 0.25 dpi. All drugs except nAb (single dose) were given twice a day (BID) for 6 days. Vehicle-treated (Vehicle) mice (n = 5–10) were used as control.
(B) Representative BLI images of SARS-CoV-2-Delta-nLuc-infected mice under indicated treatment regimens in ventral (v) and dorsal (d) positions. Mice that succumb and survive challenge are shown separately for clarity.
(C and D) Temporal quantification of nLuc signal as flux (photons/sec) computed non-invasively.
(E) Temporal changes in mouse body weight with initial body weight set to 100% for an experiment shown in A.
(F) Kaplan-Meier survival curves of mice (n = 5 per group) statistically compared by log rank (Mantel-Cox) test for experiment as in A.
(G and H) Fold changes in indicated cytokine mRNA expression in brain and lung tissues of mice under specified treatment regimens after necropsy upon death or at 14 or 22 dpi in the surviving animals. Data were normalized to Gapdh mRNA in the same sample and that in non-infected mice after necropsy.
Scale bars in (B) denote radiance (photons/s/cm2/steradian). Grouped data in (C-E) and (G and H) were analyzed by two-way ANOVA followed by Tukey’s multiple comparison tests. Statistical significance for group comparisons to Vehicle are shown in black, with favipiravir are shown in blue, with molnupiravir are shown in purple, with nirmatrelvir are shown as red and with CV3-1 are shown as pink. ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001; ∗∗∗∗, p < 0.0001; ns not significant; mean values ±SD are depicted. Points enclosed in dotted circles and red cross signs are used to indicate mice that succumbed to infection. b.w.: body weight.
Favipiravir therapy significantly reduced virus replication in the lungs (N mRNA expression, viral loads) and gut (nLuc flux). However, the inhibition at this dosage used was not enough to curb body weight loss and eventual delayed spread of virus to the brain and nose as indicated by post-necropsy analyses (Figures 1A–1E and S2A–S2F). Consequently, all the mice in the cohort succumbed to Delta VOC-induced mortality with a one-day delay in death (Figure 1F). Despite failing to achieve 100% survival, favipiravir treatment significantly reduced the mRNA expression of specific inflammatory cytokines (Ccl2, Cxcl10, 3- to 31-fold) in brain and (Il6, Ccl2, Cxcl10, 2–15 -fold) lung tissues (Figures 1G and 1H; Table S1). Comparatively, molnupiravir or nirmatrelvir monotherapy significantly reduced Delta VOC-induced morbidity (body weight loss) and virus replication in target tissues (lungs, nose, brain, and gut) of surviving mice resulting in 60% and 75% survival, respectively, in addition to delaying death by 1–4 days in mice that succumbed to infection. BLI revealed that death of the mice in molnupiravir- and nirmatrelvir-treated cohort was likely due to virus neuroinvasion (Figures 1E, 1F, and S2A–S2D). Post-necropsy analyses for tissue viral loads (nLuc flux, N mRNA expression, viral titers) in the nose, lung, brain, and gut also showed that molnupiravir or nirmatrelvir monotherapy significantly reduced tissue viral loads (Figures S2A–S2D). Accordingly, we also observed significant reductions in all inflammatory cytokine mRNA expression in target tissues of molnupiravir- or nirmatrelvir-treated surviving mice (Figures 1G and 1H). There have been reports documenting persistence of SARS-CoV-2 infection in the gut after respiratory clearance in pediatric and adult patients.34,35 However, as expected for orally delivered DAA, molnupiravir or nirmatrelvir treatment showed near-baseline levels of nLuc signals especially in surviving mice compared to mice that succumbed to infection in all cohorts where we observed high level of SARS-CoV-2 infection in gut tissues (Figures S2E and S2F). Notably, our analyses revealed that virus loads in tissue homogenates on an average from mice under molnupiravir or nirmatrelvir monotherapy estimated on Vero E6 target cells (nLuc activity) were ∼92,000- and ∼4,000-fold above baseline values, respectively. Hence, although DAA monotherapy at the dosage used provided partial protection against Delta VOC, it did not achieve virus clearance to the same extent as nAb CV3-1 which displayed near-baseline values (1.13-fold above background). Therefore, our analyses indicated a superior in vivo efficacy for nAb therapy compared to drug monotherapy regimens at the administered dosage (Table S1).
Therapy using combination of molnupiravir with nirmatrelvir demonstrates additive efficacy against Delta (B.1.617.2) VOC in K18-hACE2 mice
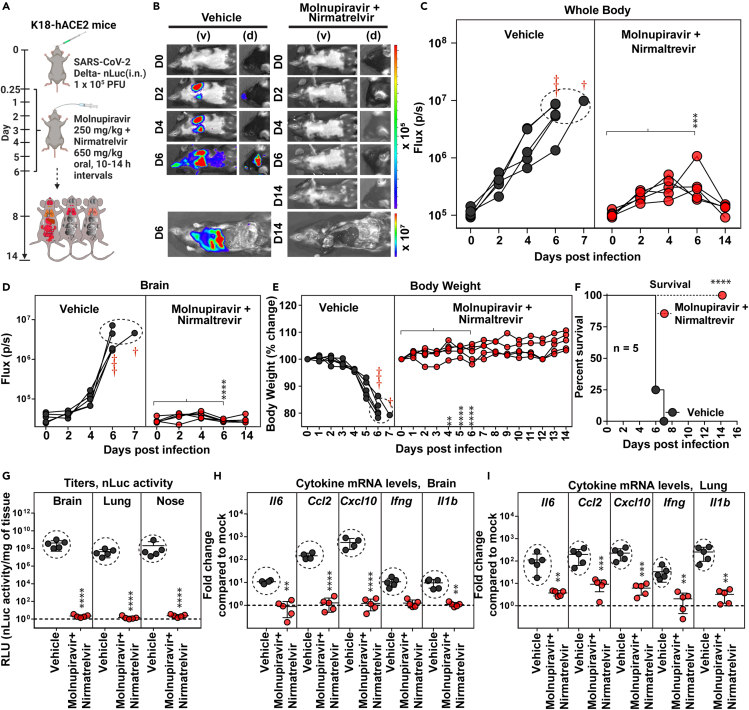
We next investigated if combination of DAAs molnupiravir (250 mg/kg) and nirmatrelvir (650 mg/kg) (every 10–14 h, for 6 days, starting 6 hpi) that target two distinct SARS-CoV-2 enzymes, polymerase and the protease, respectively, would demonstrate enhanced in vivo efficacy against Delta VOC (Figure 2A). Indeed, the combination treatment significantly reduced virus replication below detection limits of BLI and prevented neuroinvasion suggesting successful elimination of virus-infected cells (Figures 2B–2D). These results were in accordance with reduction in body weight loss phenotype and 100% survival in mice receiving combination drug therapy as compared with vehicle-treated cohorts (Figures 2E and 2F; Table S1). Moreover, post-necropsy analyses of viral loads in the nose, lung, brain, and gut also indicated virologic control (N mRNA expression, nLuc activity) as well as significant reduction (5- to 450-fold) in inflammatory cytokine mRNA expression (Figures 2G–2I and S3A–S2E; Table S1). To assess lung pathology, we used Keratin 8 (Krt8) mRNA expression, a marker for persistence of danger associated transitional progenitor (DATP) cells derived from alveolar epithelial cells during healing/repair after lung injury.36,37 While monotherapy reduced lung pathology as measured by Krt8 expression especially in surviving mice, CV3-1 nAb and combination therapy were significantly superior in reducing expression of lung injury/repair marker Krt8 (Figures S4A and S4B). Thus, our results demonstrate that simultaneous inhibition of viral protease and polymerase activity is effective in clearing established pool of SARS-CoV-2-infected cells.
Figure 2.
Treatment efficacy of molnupiravir and nirmatrelvir combination in K18-hACE2 mice lethally challenged with SARS-CoV-2 Delta VOC
(A) Experimental design to test therapeutic efficacy of molnupiravir (250 mg/kg b.w.) with nirmatrelvir (650 mg/kg b.w.) in K18-hACE2 mice challenged with SARS-CoV-2-Delta-nLuc (1 × 105 PFU, i.n.). The drugs were administered twice daily (BID) for 6 days beginning at 0.25 hpi. Vehicle-treated (Vehicle) mice (n = 5) were used as control.
(B) Representative BLI images of SARS-CoV-2-Delta-nLuc-infected mice in ventral (v) and dorsal (d) positions.
(C and D) Temporal quantification of nLuc signal as flux (photons/sec) computed non-invasively.
(E) Temporal changes in mouse body weight with initial body weight set to 100% for an experiment shown in A.
(F) Kaplan-Meier survival curves of mice (n = 5 per group) statistically compared by log rank (Mantel-Cox) test for experiment as in A.
(G) Viral loads (nLuc activity/mg) in indicated organs from mice under specified treatment regimens evaluated using Vero E6 cells as targets when mice died from infection or at 14 dpi for surviving mice.
(H and I) Fold changes in indicated cytokine mRNA expression in brain and lung tissues of mice under specified treatment regimens after necropsy upon death or at 14 dpi in the surviving animals. Data were normalized to Gapdh mRNA in the same sample and that in non-infected mice after necropsy.
Scale bars in (B) denote radiance (photons/s/cm2/steradian). Grouped data in (C–E), and (G–I) were analyzed by two-way ANOVA followed by Tukey’s multiple comparison tests. ∗, p < 0.05; ∗∗, p < 0.01; ns not significant; Mean values ±SD are depicted. Red cross signs and dots within indicated circles denote the mice that have succumbed to infection at specified times post infection. b.w.: body weight.
We next carried out a short-term (two-day treatment, 6 hpi) dose response study in Delta VOC-challenged K18-hACE2 mice to determine whether the drug combinations demonstrated synergistic or additive effects (Figure S5). Analyses of virus burden using BLI (organ flux), N mRNA expression, and titers (nLuc activity) in the lung at 3 dpi showed that the combined treatment with molnupiravir and nirmatrelvir significantly reduced virus replication as compared with drug monotherapy at the three drug concentrations (Figures S5B–S5E). The Bliss index, calculated using normalized viral burden as a response parameter, ranged from 0.01 to 1.68 across the three drug concentrations (Figure S5F). These data suggested that the combination therapy exhibited additive (Bliss index: −10 to 10, additive drug interaction) in vivo efficacy. The assessment of the Bliss index for the 6-day drug treatment regimens at human equivalent dosages, determined using nine parameters (viral loads: titer, inflammation: Ccl2 and Cxcl10 in both brain and lungs; mortality and delay in death, see method details) to compute the total disease burden, yielded a mean value of 0.9 (Figure S4D). Thus, taken together, our data suggested an additive effect for the two-drug combination regimen against SARS-CoV-2 Delta VOC in mice.
Comparative efficacy of favipiravir, molnupiravir, nirmatrelvir, and the combination of molnupiravir with nirmatrelvir against Omicron (B.1.1.529) VOC in K18-hACE2 mice
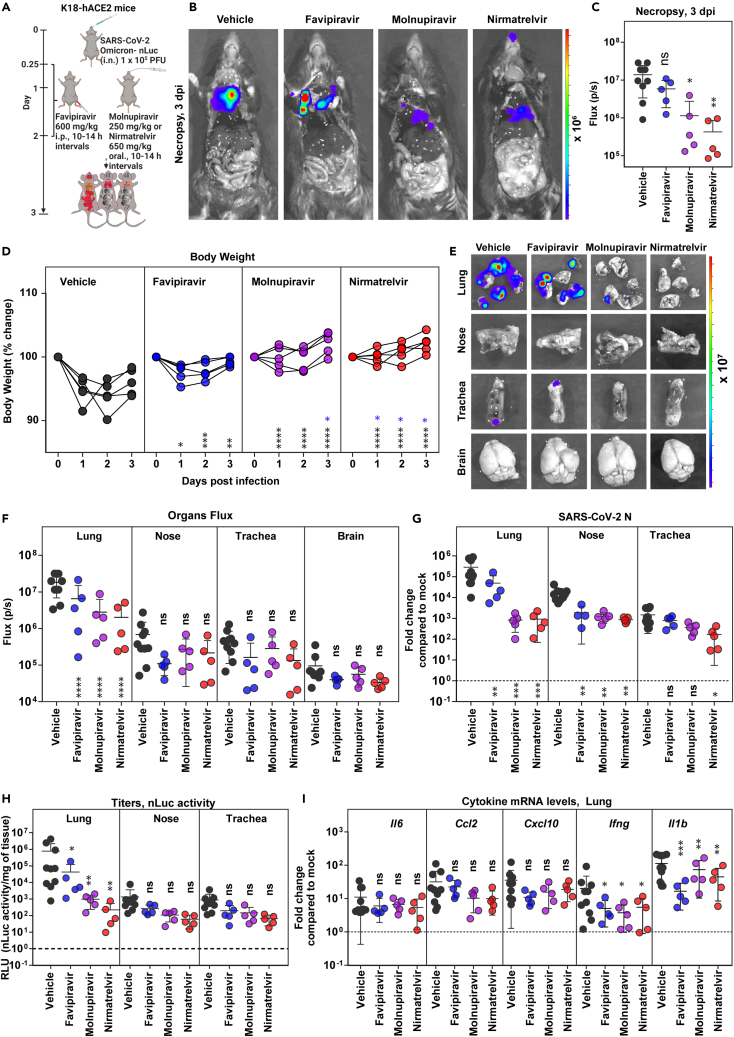
While Omicron VOC demonstrates high transmissibility, replication is attenuated in lungs of K18-hACE2 mice due to inefficient usage of TMPRSS2.38 In addition, Omicron VOC infection is limited to the nose, trachea, and lungs and does not spread consistently to the brain in K18-hACE2 mice. This is manifested by non-lethal infection and only a transient loss in body weight with recovery after 3 dpi. In vivo efficacy studies of DAAs against Omicron VOC were therefore terminated at 3 dpi by treating mice at the same dosage as for Delta VOC challenges but from 0.25 to 2 dpi (every 10–14 h) (Figure 3A). We were able to monitor replication of Omicron-nLuc VOC after necropsy using whole-body BLI and compared the effect of drug treatments on viral loads in the nose, trachea, and lungs. As was the case with Delta VOC, monotherapy regimens were able to significantly reduce nLuc signals (2- to 10-fold) in the lung but did not eliminate virus-infected cells at the experimental endpoint and the dosage used (Figures 3B and 3C; Table S1). Among the three drugs evaluated, favipiravir was less efficient than molnupiravir or nirmatrelvir in preventing body weight loss and reducing virus loads in the lungs (N mRNA expression and nLuc activity) compared to vehicle-treated cohorts of animals (Figures 3D–3H; Table S1). Notably, the mRNA expression of Ifng and Il1b in the lung and N mRNA in the nose of all DAA-treated cohorts were significantly lower than those in vehicle-treated mice (Figure 3I). Our data suggest that monotherapy with DAAs is effective against Omicron VOC in significantly reducing morbidity.
Figure 3.
Efficacy of favipiravir, molnupiravir, and nirmatrelvir monotherapy in K18-hACE2 mice against SARS-CoV-2 Omicron VOC
(A) Experimental design to test therapeutic efficacy of favipiravir (600 mg/kg b.w., i.p.), molnupiravir (250 mg/kg b.w. oral), and nirmatrelvir (650 mg/kg b.w., oral) against SARS-CoV-2-Omicron-nLuc (1 × 105 PFU, i.n.) in K18-hACE2 mice. The drugs were administered twice daily (BID) starting 6 hpi (0.25 dpi) for 2 days. Vehicle-treated (Vehicle) mice (n = 5–10) mice were used as control.
(B) Representative BLI images of SARS-CoV-2-Omicron-nLuc-infected mice after necropsy (3 dpi).
(C) Quantification of nLuc signal as flux (photons/sec) computed after necropsy at 3 dpi.
(D) Temporal changes in mouse body weight with initial body weight set to 100% for an experiment shown in A.
(E and F) Ex vivo imaging of indicated organs and quantification of nLuc signal as flux (photons/sec) after necropsy for an experiment as in A. (G) Fold changes in Nucleocapsid (N) mRNA expression in lung, nasal cavity, and trachea after necropsy at 3 dpi. Data were normalized to Gapdh mRNA in the same sample and that in non-infected mice.
(H) Viral loads (nLuc activity/mg) from indicated tissues at 3 dpi using Vero E6 cells as targets.
(I) Fold changes in indicated cytokine mRNA expression in lung tissues after normalization with Gapdh mRNA in the same sample and that in non-infected mice after necropsy.
Scale bars in (B) and (E) denote radiance (photons/s/cm2/steradian). The data in (C) was analyzed by one-way ANOVA followed by Kruskal-Wallis’s test and grouped data in (D), (F–I) were analyzed by two-way ANOVA followed by Tukey’s multiple comparison tests. Statistical significance for group comparisons to Vehicle are shown in black, with favipiravir are shown in blue, with molnupiravir are shown in purple, with nirmatrelvir are shown as red. ∗, p < 0.05; ∗∗, p < 0.01; ns not significant; Mean values ±SD are depicted. b.w.: body weight.
See also Table S1.
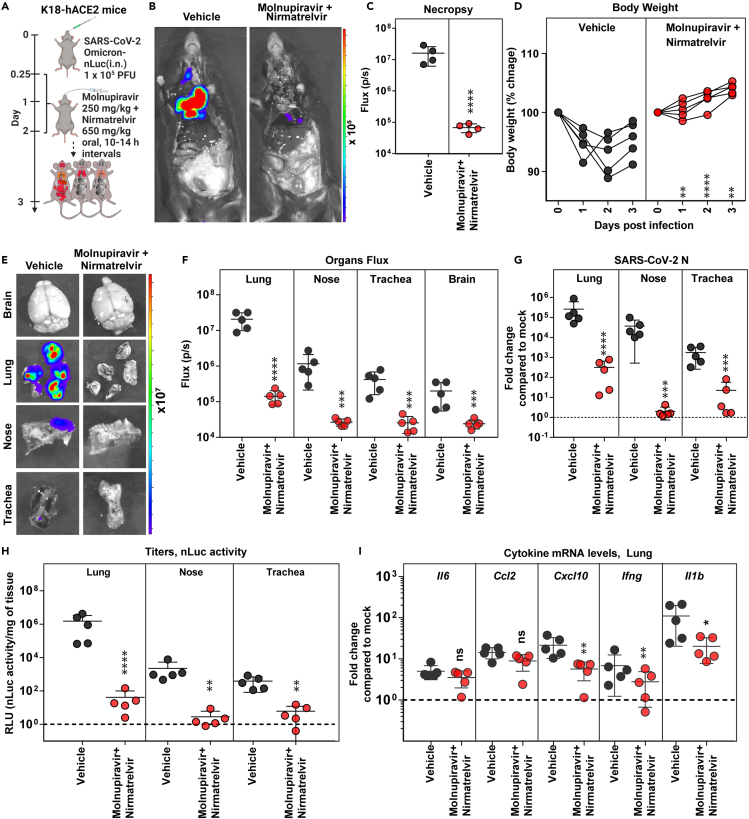
We next tested if the combination therapy of molnupiravir (250 mg/kg) and nirmatrelvir (650 mg/kg) (every 10–14 h, for 2 days, starting 6 hpi) can augment the antiviral effect against Omicron VOC like Delta VOC (Figure 4A). Indeed, the combination therapy was highly effective in controlling Omicron VOC replication as ascertained by absence of body weight loss phenotype as well as nLuc signals (BLI) after necropsy (Figures 4B–4D). Post-necropsy imaging of organs (lung, nose, trachea, and brain), viral load analyses (BLI, N mRNA, and titers), and inflammatory cytokine mRNA expression (lung: Cxcl10, Ifng, and Il1b) also revealed significant reduction in viral loads (70- to 18285-fold) and inflammation (2- to 6 -fold) compared to vehicle-treated cohorts (Figures 4E–4I; Table S1). This was corroborated by significantly reduced Krt8 mRNA expression suggesting reduced lung pathology upon treatment with combination of drugs compared to vehicle and monotherapy regimens (Figure S4C). A Bliss index of −6.7 (−10 to 10 range) calculated based on disease burden (5 parameters, see method details) also suggested an additive effect for the DAA combinations. Thus, the dual therapy of molnupiravir and nirmatrelvir was more effective in controlling replication and spread of the highly immune-evasive Omicron VOC than the DAA monotherapy regimen evaluated in the study.
Figure 4.
Therapeutic efficacy of molnupiravir with nirmatrelvir in K18-hACE2 mice against SARS-CoV-2 Omicron VOC
(A) Experimental design for testing efficacy of molnupiravir (250 mg/kg b.w., oral) with nirmatrelvir (600 mg/kg, b.w. oral) in K18-hACE2 mice challenged with SARS-CoV-2-Omicron-nLuc (1 × 105 PFU; i.n.). The drugs were administered twice daily (BID) starting 6 hpi (0.25 dpi) for 2 days. Vehicle-treated (Vehicle) mice (n = 5) were used as control.
(B and C) Representative BLI images and temporal quantification of nLuc signal as flux (photons/sec) after necropsy at 3 dpi.
(D) Temporal changes in mouse body weight with initial body weight set to 100% for an experiment shown in A.
(E and F) Ex vivo imaging of indicated organs and quantification of nLuc signal as flux (photons/sec) after necropsy for an experiment shown in A. (G) Fold changes in nucleocapsid (N) mRNA expression in lung, nasal cavity, and trachea tissues and treatment regimens at 3 dpi. Data were normalized to Gapdh mRNA in the same sample and that in non-infected mice after necropsy.
(H) Estimated viral loads (nLuc activity/mg) from indicated tissues and treatment regimens using Vero E6 cells as targets at 3 dpi after necropsy.
(I) Fold changes in indicated cytokine mRNA expression for specified treatment regimens in the lung. Data were normalized to Gapdh mRNA in the same sample and that in non-infected mice.
Scale bars in (B) and (E) denote radiance (photons/s/cm2/steradian). The data in (C) were analyzed by t test followed by non-parametric Mann-Whitney U tests. Grouped data in (D), (F-I) were analyzed by two-way ANOVA followed by Tukey’s multiple comparison tests. ∗, p < 0.05; ∗∗, p < 0.01; ns; not significant. Mean values ±SD are depicted. b.w.: body weight.
See also Table S1.
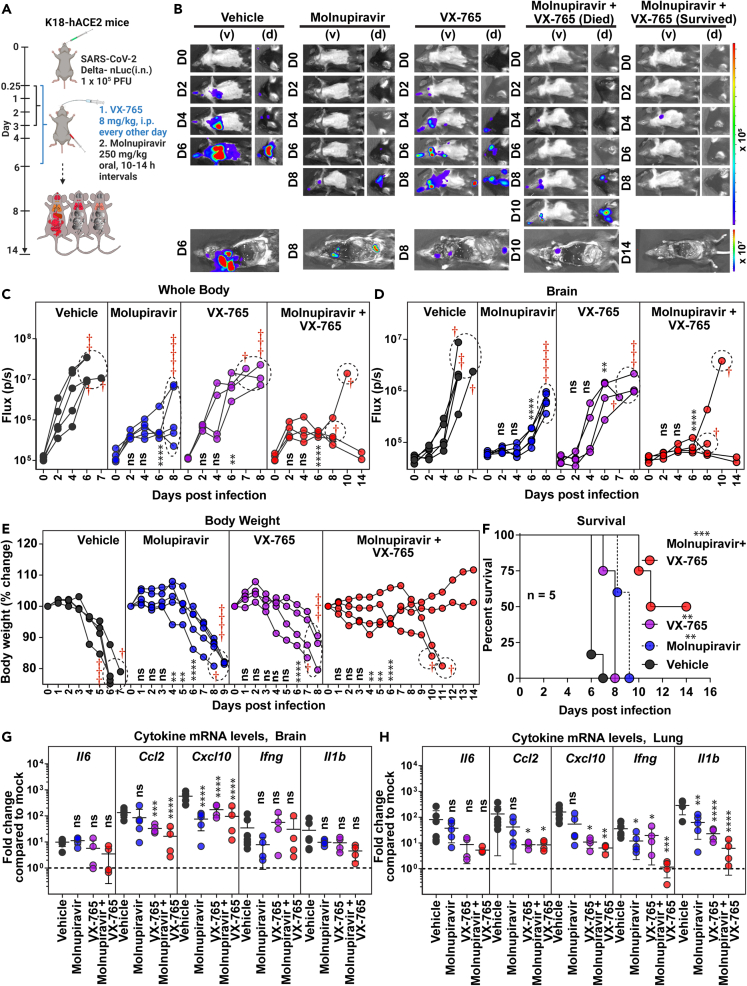
Caspase 1/4 inhibitor additively improves molnupiravir therapeutic efficacy and extends survival against Delta VOC challenge
SARS-CoV-2 infection elicits an imbalanced hyperinflammatory response primarily through activation of NLRP3-driven inflammasome pathway.39 Mitigating SARS-CoV-2-induced inflammation alleviates lung pathology, whereas inhibiting inflammasome pathway reverses chronic lung pathology.40,41,42 We therefore explored if host-directed agents like VX-765 that specifically inhibit executor inflammatory caspases 1 and 4 (i.p. 8 mg/kg, 0.25–6 dpi), which control cell death activated by inflammasomes,43 can synergize with molnupiravir treatment regimen. To assess synergy, we chose a short-term 3-day molnupiravir treatment regimen (250 mg/kg, oral BID, 0.25–3 dpi) which was less efficient in inhibiting Delta VOC replication based on aforementioned 6-day administration experiments (Figure 5A). Indeed, temporal BLI combined with nLuc signal quantification and body weight measurements revealed that, although 3-day molnupiravir treatment significantly reduced Delta VOC replication in the lung, it only delayed neuroinvasion and subsequent death (Figures 5B–5D). Interestingly, we also observed reduced virus replication in the lung, delayed neuroinvasion (nLuc flux quantitation; 6 dpi), and a slower weight loss rate (6 dpi) in VX-765-treated mice compared to vehicle-treated cohorts (Figure 5E). These data suggest that reducing inflammation can also reduce SARS-CoV-2 replication and spread (Figures S6A–S6D). Despite the reduction in inflammatory cytokine mRNA expression and virus replication, all the mice in the VX-765-treated cohort succumbed to Delta VOC-induced death (Figure 5F). Moreover, we did not observe any significant reduction in lung pathology/repair marker as assessed by Krt8 mRNA expression in VX-765 or short-term molnupiravir-treated mice (Figure S4B). Consequently, we explored if VX-765 and molnupiravir can be combined for enhanced efficacy against lethal Delta VOC challenge. BLI revealed that the 3-day molnupiravir dosing, when combined with VX-765 treatment, led to virologic control in 50% of the mice and a 3–4-day delay in neuroinvasion in the rest (Figures 5B–5D). Accordingly, half of the mice receiving the combination treatment displayed significant delay in body weight loss before succumbing to infection by 10–11 dpi while the other half survived (Figures 5E and 5F). Post-necropsy analyses (BLI, RNA analyses) corroborated the virologic control in 50% of the surviving mice in addition to significant overall reduction in inflammatory cytokine as well as Krt8 mRNA expression (lung injury) in the lung (Figures 5G, 5H, S6A–S6F, and S4B; Table S1) compared to individual drug- and vehicle-treated mice. In addition, a Bliss index of 0.001 also suggested an additive in vivo therapeutic efficacy for molnupiravir combination with VX-765 against Delta VOC (Figure S4D). Thus, inhibition of the inflammasome pathway has the potential to augment monotherapy drug regimens and can have significant impact on preserving lung function.
Figure 5.
Caspase-1/4 inhibitor can augment molnupiravir therapeutic efficacy in K18-hACE2 mice against Delta VOC
(A) Experimental design for testing efficacy of molnupiravir therapy (250 mg/kg b.w., oral) with caspase 1/4 inhibitor (VX-765; 8 mg/kg b.w., i.p.) in K18-hACE2 mice challenged with SARS-CoV-2-Delta-nLuc (1 × 105 PFU, i.n.). Molnupiravir was administered twice daily (BID) starting 6 hpi (0.25 dpi) for 3 days, and VX-765 was injected once every other day starting 6 hpi till 6 dpi vehicle-treated (Vehicle) mice (n = 5) mice were used as control.
(B) Representative BLI images of SARS-CoV-2-Delta-nLuc-infected mice under indicated treatment regimens in ventral (v) and dorsal (d) positions. Mice that succumb and survive challenge are shown separately for clarity.
(C and D) Temporal quantification of nLuc signal as flux (photons/sec) computed non-invasively.
(E) Temporal changes in mouse body weight with initial body weight set to 100% for an experiment shown in A.
(F) Kaplan-Meier survival curves of mice (n = 5 per group) statistically compared by log rank (Mantel-Cox) test for experiment as in A.
(G and H) Fold changes in indicated cytokine mRNA expression in lung and brain under specified drug regimens. RNAs were extracted when the mice succumbed to infection or for surviving mice at 14 dpi. Data were normalized to Gapdh mRNA expression in the same sample and that in non-infected mice after necropsy.
Scale bars in (B) denote radiance (photons/s/cm2/steradian). Grouped data in (C–E), and data in (G and H) were analyzed by two-way ANOVA followed by Tukey’s multiple comparison tests. Statistical significance for group comparisons to Vehicle are shown in black, with molnupiravir are shown in blue, with VX-765 are shown in purple, with molnupiravir+ VX-765 are shown as red. ∗, p < 0.05; ∗∗, p < 0.01; ns not significant; Mean values ±SD are depicted. Dotted circles with red cross signs at indicated time points are used to indicate mice that succumbed to infection. b.w.: body weight.
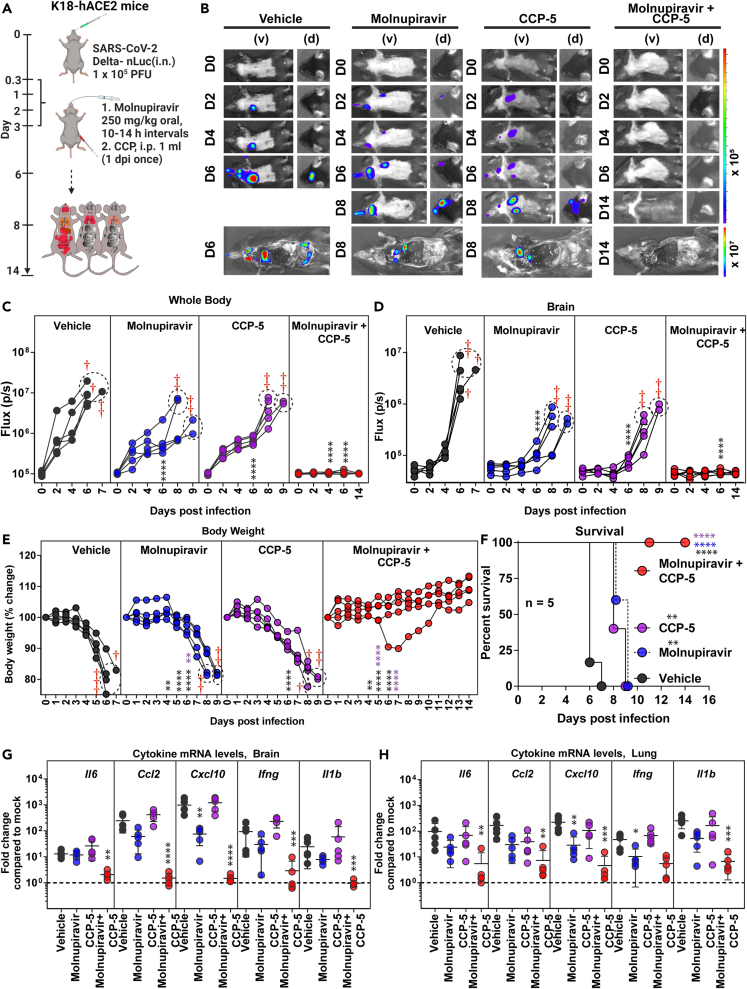
CCP synergizes with short-term molnupiravir treatment regimen for effective virologic control of Delta VOC
We next investigated if CCP can augment the short 3-day monotherapy with molnupiravir [250 mg/kg, oral, twice daily (BID), 0.25–3 dpi]. We chose an ancestral strain-elicited CCP (CCP-5, previously characterized in detail)27 with robust Fc-effector activity required to eliminate persistent infection, but with low neutralizing activity against Delta VOC to mimic conditions resulting from immune evasion displayed by circulating VOCs. CCP-5 was administered only once at 1 dpi (i.p.) either alone or in combination with molnupiravir (Figure 6A). Longitudinal BLI and nLuc flux quantification revealed that, although monotherapy using molnupiravir or CCP-5 reduced viral infection, delayed neuroinvasion, and extended survival by 2–3 days, they failed to protect mice against Delta VOC-induced mortality (Figures 6B–6F). Notably, combining short-term molnupiravir treatment with CCP-5 resulted in rapid virologic control (nLuc flux quantification) and significantly reduced morbidity as demonstrated by prevention of body weight loss and 100% survival (Figures 6E and 6F). These data were corroborated by post-necropsy analyses that showed elimination of nLuc signal as well as viral loads in all analyzed organs (Figures S7A–S7F). We also observed a significant reduction in inflammatory cytokine mRNA expression in both the brain and the lung, as well as less lung damage (Krt8 mRNA expression), in mice treated together with CCP-5 and molnupiravir (Figures 6G, 6H, and S4B; Table S1). Remarkably, a Bliss index of 24 (>10) implied that the treatment involving molnupiravir with CCP-5 exhibited synergy and was superior to other drug combinations assessed in our study (Figure S4D). Thus, combining a widely available resource such as CCP with a DAA has the potential to synergize with antiviral drug therapies and facilitate rapid virus clearance.
Figure 6.
COVID-19 convalescent plasma 5 (CCP-5) can augment molnupiravir therapeutic efficacy in K18-hACE2 mice against Delta VOC
(A) Experimental design for testing synergy of molnupiravir therapy (250 mg/kg b.w., oral) with CCP-5 (i.p., 1 mL, 1 dpi) in K18-hACE2 mice challenged with SARS-CoV-2-Delta-nLuc (i.n., 1 × 105 PFU). Vehicle-treated (Vehicle) mice (n = 5) mice were used as control.
(B) Representative BLI images of SARS-CoV-2-Delta-nLuc-infected mice under indicated treatment regimens in ventral (v) and dorsal (d) positions.
(C and D) Temporal quantification of nLuc signal as flux (photons/sec) computed non-invasively.
(E) Temporal changes in mouse body weight with initial body weight set to 100% for an experiment shown in A.
(F) Kaplan-Meier survival curves of mice (n = 5 per group) statistically compared by log rank (Mantel-Cox) test for experiment as in A.
(G and H) Fold changes in indicated cytokine mRNA expression in brain and lung tissues under specified treatment conditions. RNA was extracted from tissues of mice when they succumbed to infection or for surviving mice at 14 dpi. Data were normalized to Gapdh mRNA expression in the same sample and that in non-infected mice after necropsy.
Scale bars in (B) denote radiance (photons/s/cm2/steradian). Grouped data in (C-E), and data in (G-H) were analyzed by two-way ANOVA followed by Tukey’s multiple comparison tests. Statistical significance for group comparisons to Vehicle are shown in black, with molnupiravir are shown in blue, with CCP-5 are shown in purple, with molnupiravir combined with CCP-5 are shown as red. ∗, p < 0.05; ∗∗, p < 0.01; ns not significant; Mean values ±SD are depicted. Dotted circles with red cross signs at indicated time points are used to denote mice that succumbed to infection. b.w.: body weight.
Discussion
Antiviral interventions remain vital as SARS-CoV-2 VOCs may continue to emerge periodically with unpredictable clinical severity and ability to escape existing anti-spike-directed vaccine and infection-induced immunity. DAAs, molnupiravir/favipiravir and nirmatrelvir, that are approved or available for off-label use target SARS-CoV-2 polymerase and protease, respectively, work independently from an effect on spike protein to fill this gap.44,45 These DAAs were discovered for use against either influenza or SARS-CoV-1 and were repurposed for use against SARS-CoV-2. However, apart from improving the half-life through inhibition of cytochrome P450 3A (CYP3A) family of oxidizing enzymes by combining with ritonavir, an HIV protease inhibitor, accelerated improvement of DAAs like nirmatrelvir subsided after SARS-CoV-1 outbreak.46,47 While acknowledging that further development would have helped significantly in countering the current SARS-CoV-2 pandemic, this was expected due to lack of available funding and patients for human trials. In this context, preclinical animal models of infection play a pivotal role in advancing potential antiviral candidates toward near-final product stages. They are instrumental in identifying effective combinations, be it synergistic or additive, thereby informing therapeutic regimens and contributing significantly to pandemic preparedness.
Our BLI-guided analyses in K18-hACE2 model of SARS-CoV-2 infection revealed that nirmatrelvir had better in vivo efficacy than molnupiravir, while favipiravir was the least effective at the administered dose. Furthermore, molnupiravir monotherapy was more efficacious when administered over a six-day period than over a shorter three-day period (60% vs. 0% survival). These data support a longer course of monotherapy for effective virologic control. A longer duration may be especially important for treatment of patients with long-term COVID.48 However, none of the DAAs under monotherapy could match the in vivo efficacy and rapid virus clearance offered by the nAb CV3-1. The inability to eliminate the virus during monotherapy treatment may have consequences for virus evolution and emergence of drug-resistant VOCs especially for mutagenic DAAs like molnupiravir and favipiravir. Suboptimal concentrations have been shown to support emergence of SARS-CoV-2 mutants in hamster models.49 Furthermore, widespread use of molnupiravir has led to circulation of SARS-CoV-2 virus populations that carry molnupiravir-induced signature mutations (C to T and G to A) in the United Kingdom, the United States of America, and Australia.50 Recent evidence suggests that the random mutations induced by molnupiravir can contribute to accelerated evolution of SARS-CoV-2.22 Therefore, effective combinations that demonstrate enhanced virus clearance than DAA monotherapy are required to combat virus evolution and drug resistance and improve clinical outcomes.
Previous studies have examined several drug combinations targeting different steps of SARS-CoV-2 replication or host pathways in culture systems and organoids to identify effective combinations that display additive or synergistic viral control.25,48,51 In vivo analyses are required to ascertain if the identified drug combinations demonstrate enhanced antiviral activity as pharmacokinetics of individual drugs and their interactions with host and microbiota can impact overall efficacy.52 Here, we found that the combination of molnupiravir with nirmatrelvir demonstrated additive efficacy and showed promise in achieving reduction in virus loads that came close to nAb therapy for the highly pathogenic Delta VOC. The combination also showed additive efficacy against Omicron VOC than monotherapy regimens. Our data agree with previous studies in animal models that showed enhanced efficacy by combination DAA therapy.53,54,55 These findings highlight the importance of developing a panel of DAAs that can match the rapid virus clearance mediated by nAbs and, preferably, be used in combinations and for longer durations targeting distinct stages of the viral life cycle to remain ahead of the expected emergence of drug-resistance mutations as has been implemented for treatment of HIV with combinatorial antiretroviral therapy.
An exacerbated inflammatory response is the hallmark of SARS-CoV-2 infection, and reducing inflammation through use of corticosteroids like dexamethasone was beneficial in patients with severe COVID-19 that required respiratory support.56 We found that mitigating host inflammasome activation through use of caspase-1/4 inhibitor (VX-765) augmented recovery and survival (0% vs. 50%) of lethally challenged K18-hACE2 mice when combined with molnupiravir over individual treatment regimen. These data demonstrate the potential of combining DAAs with host-directed inhibition to augment virus clearance and recovery. Interestingly, reducing inflammation using VX-765 resulted in reduced viral loads suggesting that exacerbated inflammation, a hallmark of COVID-19, contributes to conditions beneficial for SARS-CoV-2 replication in vivo. Consequently, VX-765 treatment alone also reduced viral loads in addition to DAA treatment. Thus, our data suggest a model where individual DAA requires help from a second virus load-reducing measure. This can be accomplished either via combination therapy with a second DAA or with host-directed treatments such as VX-765.
The data from treatment with a combination of DAA and convalescent plasma further reinforced the importance of multipronged strategy. Notably, an nAb by itself operates like a combination therapy. The neutralizing activity targets the virus directly like a DAA while the Fc-effector function adds a second line of attack against both free virus particles (through complement, opsinophagocytosis, antibody-dependent cellular phagocytosis) and productively infected cells (antibody-dependent cellular cytotoxicity) through innate immune cell engagement. Although the efficacy of various clinically approved monoclonal nAb cocktails to neutralize SARS-CoV-2 VOCs has decreased due to escape mutations leading to the discontinuation of their therapeutic use, the polyclonal Fc-effector functions in plasma have shown resilience.27,57 This resilience is attributed to the ability of spike-binding antibodies to bind via conserved or non-neutralizing epitopes.57,58,59 Remarkably, combining molnupiravir with ancestral strain-elicited convalescent plasma 5, with robust Fc-effector activity,27 demonstrated synergy and was superior to other combinations evaluated in the study. Fc-effector functions provided through CCP offered a second mechanism to reduce viral loads to synergistically augment DAA monotherapy. Vice versa, the diminished neutralizing ability of polyclonal Abs in CCP-5 was complemented by direct acting molnupiravir. Hence, our study presents a model for augmenting the effectiveness of presently approved drugs and proposes the necessity of a combinatorial therapeutic approach. This could involve the addition of a second DAA or combinations with inflammasome inhibitors, convalescent plasma, or neutralizing mAbs to enhance efficacy against COVID-19.
Limitations of the study
We used K18-hACE2 mice as our model for in vivo efficacy analyses as they are highly sensitive to SARS-CoV-2 infection. By setting a high threshold for evaluating in vivo efficacy, this model allows identification of antiviral interventions with better translational potential. However, K18-hACE2 mice succumb due to virus neuroinvasion. While virus neuroinvasion remains debatable in humans, more recent studies have demonstrated that SARS-CoV-2 can infect astrocytes in the human brain.60,61,62 In addition, acute and long-term neurological dysfunction are one of the most frequent extrapulmonary complications in 30% of COVID-19 patients, and there is strong evidence of brain-related abnormalities even in patients with mild COVID-19 disease.63,64 The inability of the monotherapy regimens to clear the virus at the dosage used is maybe due to extreme susceptibility of K18-hACE2 mice to SARS-CoV-2 that support high levels of virus replication. It remains possible that longer duration (more than 6 days) as has been suggested for long COVID patients or higher concentrations of drugs during monotherapy may display improved efficacy and clear SARS-CoV-2 infection.48 Our study revealed the utility of a CCP to complement the action of molnupiravir to achieve virologic control in mice. However, CCPs are complex compared to purified nAbs and necessitate careful evaluation through a clinical trial before a combination with drugs like molnupiravir/nirmatrelvir is implemented. Alternatively, a combination of well-studied and safe spike-binding mAb with robust Fc-effector and broad neutralizing activity together with DAAs can also be tested for improved resilience to evolving SARS-CoV-2 VOCs. This strategy will harness the rapid virus clearance mediated by mAbs and considerably mitigate lung pathology that remains a major concern even after the virus is cleared in the aged population.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Fc block anti mouse-CD16/CD32 (93) | BioLegend Inc | Cat# 101302; RRID: AB_312801 |
| Anti-Cytokeratin 8 Antibody, clone TROMA-1 | Millipore Sigma | Cat# MABT329; RRID: AB_2891089 |
| Goat anti-Rat IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor™ 568 | Invitrogen | Cat# A-11077; RRID: AB_2534121 |
| Mouse anti-SARS-CoV-2 Nucleocapsid Antibody (clone A20087H) | BioLegend | Cat# 946102; RRID: AB_2892515 |
| CV3-1 (IgG1) | Finzi Lab, Université de Montréal Ullah et al., 202126 |
|
| Deposited data | ||
| Raw data used to generate all graphs shown in Figures | This manuscript | Mendeley Data, V2, https://doi.org/10.17632/n9ztbbbstm.2 |
| Bacterial and virus strains | ||
| SARS-CoV-2, Isolate hCoV-19/USA/PHC658/2021 (Delta Variant) | Yale New Haven Hospital/Craig Wilen Lab | GenBank Accession # MZ468047 |
| SARS-CoV-2, Isolate hCoV-19/USA/MD-HP01542/2021 (BA.1 Omicron variant) | Yale New Haven Hospital/Craig Wilen Lab | GenBank Accession # ON425981 |
| SARS-CoV-2, Isolate hCoV-19/USA/PHC658/2021 (Delta Variant) Expressing nanoluc luciferase |
Mothes/Kumar Lab | N/A |
| SARS-CoV-2, Isolate hCoV-19/USA/MD-HP01542/2021 (Omicron variant) Expressing nanoluc luciferase | Mothes/Kumar Lab | N/A |
| Biological samples | ||
| CCP-5 (Age: 30, Male, O Rh-, 30 dpi) | Ullah et al., 202327 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Molnupiravir (Mr: 329.31 Da) | TCG Lifesciences Pvt. Ltd. | #DNDI0003947753 CAS 2492423-29-5 |
| Nirmatrelvir (Mr: 587.69 Da) | TCG Lifesciences Pvt. Ltd. | #DNDI0003970297 CAS 2628280-40-8 |
| Favipiravir (Mr:157.1 Da) | TCG Lifesciences Pvt. Ltd. | #DNDI0003948820 CAS 259793-96-9 |
| VX-765, Caspase 1/4 inhibitor | InvivoGen | Cat# Inh-vx765i-1 CAS 273404-37-8 |
| Gibco™ RPMI 1640 medium | Thermo Fisher Scientific | Cat# 11875093 |
| Gibco™ Dulbecco’s modified Eagle’s medium (DMEM) | Thermo Fisher Scientific | Cat# 11965118 |
| Gibco™ MEM Non-essential amino acid (NEAA) solution | Thermo Fisher Scientific | Cat# 11140050 |
| Gibco™ Penicillin-streptomycin solution (10,000 U/ml) | Thermo Fisher Scientific | Cat# 15140122 |
| Gibco™ Dulbecco’s Phosphate Buffered Saline (DPBS) | Thermo Fisher Scientific | Cat# 14190144 |
| Gibco™ L-Glutamine (200mM) | Thermo Fisher Scientific | Cat# 25030081 |
| Gibco™ 0.05% Trypsin-EDTA, phenol red | Thermo Fisher Scientific | Cat# 25300054 |
| Fetal bovine serum | Atlanta Biologicals | Cat# S11550 |
| eBioscienceTM RBC Lysis Buffer (1X) | Invitrogen | Cat# 00-4333-57 |
| Bovine Serum Albumin (BSA) | Sigma-Aldrich | Cat# A9647-100G CAS: 9048-46-8 |
| Accutase | BioLegend Inc | Cat# 423201 |
| 0.05% Trypsin-EDTA (1X) | Life Technologies | Cat# 25300-054 |
| Sodium pyruvate (100 mM) | Life technologies | Cat# 11360-070 |
| 2-Mercaptoethanol | Sigma-Aldrich | Cat# M3148 |
| Polyethylenimine (PEI) “MAX” (Mr:40,000) | Polysciences Inc. | Cat# 24765 |
| L-Glutamine (200mM) | Life technologies | Cat# 25030-081 |
| Tris-buffered saline (TBS) | Thermo Fisher Scientific | Cat# BP24711 |
| Passive lysis buffer | Promega | Cat# E1941 |
| Paraformaldehyde (16% PFA) | Electron Microscopy Sciences | Cat# 19200 CAS: 30525-89-4 |
| Dimethyl sulfoxide (DMSO) | Sigma-Aldrich | Cat# D2650-5X5ML CAS: 67-68-5 |
| Sodium azide | Sigma-Aldrich | Cat# S-8032 EC No: 247-852-1 |
| Glycine | American Bioanalytical | Cat# AB00730-01000 CAS: 56-40-6 |
| The PEG-it Virus precipitation solution (5X) | System Bioscience | Cat# LV810A-1 |
| Avicel® Pharma Grade | FMC, Honko et al., 202065 | Cat# RC-581 NF |
| Puromycin dihydrochloride | Millipore Sigma | Cat# P8833 |
| Hydrochloric acid | Biolab | Cat# 351285–212 |
| Sodium acetate | Sigma Aldrich | Cat# S2889-250g |
| Carbonate, 0.5M buffer soln., pH 9.6 250mL | Fisher Scientific | Cat# AAJ62610AK |
| Sodium bicarbonate | Sigma Aldrich | Cat# S6014 |
| Sodium carbonate | Sigma Aldrich | Cat# S2127 |
| Tween 20 | Fisher Scientific | Cat# BP337-500 |
| PEG400 | Sigma Aldrich | CAS 25322-68-3 |
| Kolliphor-EL | Sigma Aldrich | CAS 61791-12-6 |
| Methyl cellulose | Sigma Aldrich | CAS 9004-67-5 |
| Tween 80 | Sigma Aldrich | CAS 9005-65-6 |
| Isopropyl Alcohol | Applied Biosystems | Cat# AB07015-01000 |
| Isoflurane, USP | Covetrus | Cat# 029405 |
| Critical commercial assays | ||
| Nano-Glo Luciferase Assay System (nanoluc substrate) | Promega | Cat# N1120 |
| KAPA SYBR FAST qPCR Master Mix (2X) Kit | KAPA Biosystems | Cat# KK4600 and KK4601 |
| Ambion DNase I (RNase-free) | ThermoFisher Scientific | Cat# AM2222 |
| RNeasy Mini Kit (50) | Qiagen | Cat# 74104 |
| MagMax mirVana Total RNA Isolation Kit | Fisher Scientific | Cat# A27828 |
| iScript advanced cDNA kit | Bio Rad | Cat# 1725038 |
| iQ Multiplex Powermix | Bio Rad | Cat# 1725848 |
| iScript™ cDNA Synthesis Kit | Bio Rad | Cat# 95047-100 |
| PrimeScript Reverse Transcriptase | Takara Bio | Cat# 2680A |
| PrimeStar GXL DNA Polymerase | Takara Bio | Cat# R050A |
| Mix-n-Stain CF 488A Antibody Labeling Kit (50–100 μg) | Millipore-Sigma | Cat# MX488AS100 |
| Experimental models: Cell lines | ||
| Vero-E6 (female, Chlorocebus sabaeus) | ATCC | Cat# CRL-1586; RRID: CVCL_0574 |
| Vero E6-ACE2-TMPRSS2 (VeroAT) (female, Chlorocebus sabaeus) | BEI resources | Cat# NR-54970 |
| HEK293 (female, Homo sapiens) | ATCC | Cat# CRL-1573; RRID: CVCL_0045 |
| HEK293T (female, Homo sapiens) | ATCC | Cat# CRL-3216; RRID: CVCL_0063 |
| Experimental models: Organisms/strains | ||
| C57BL/6J (B6) (males and females); 6–12 weeks old | The Jackson Laboratory | The Jackson Laboratory Stock No: 000664; RRID: IMSR_JAX:000664 |
| B6.Cg-Tg(K18-ACE2)2Prlmn/J (males and females); 6–12 weeks old | The Jackson Laboratory | Stock No: 034860; RRID:IMSR_JAX:034860 |
| Oligonucleotides | ||
| SARS-CoV-2 N F: 5′-ATGCTGCAATCGTGCTACAA-3′ | Yale School of Medicine, W. M. Keck Foundation, Oligo Synthesis Resource | N/A |
| SARS-CoV-2 N R: 5′-GACTGCCGCCTCTGCTC-3′ | Yale School of Medicine, W. M. Keck Foundation, Oligo Synthesis Resource | N/A |
| FAM-GAPDH | Bio Rad | Cat# 12001950 |
| HEX-IL6 | Bio Rad | Cat# 10031228 |
| TEX615-CCL2 | Bio Rad | Cat# 10031234 |
| Cy5-CXCL10 | Bio Rad | Cat# 10031231 |
| Cy5.5-IFNg | Bio Rad | Cat# 10031237 |
| HEX-IL1B | Bio Rad | Cat# 10031228 |
| Transgene Forward: GAC CCC TGA GGG TTT CAT ATA G | Yale School of Medicine, W. M. Keck Foundation, Oligo Synthesis Resource | #53437, Genotyping primers for K18-hACE2 mice. The Jackson Laboratory |
| Common: CAC CAA CAC AGT TTC CCA AC | Yale School of Medicine, W. M. Keck Foundation, Oligo Synthesis Resource | #53438, Genotyping primers for K18-hACE2 mice. The Jackson Laboratory |
| Wildtype forward: AAG TTG GAG AAG ATG CTG AAA GA | Yale School of Medicine, W. M. Keck Foundation, Oligo Synthesis Resource | #53439, Genotyping primers for K18-hACE2 mice. The Jackson Laboratory |
| Software and algorithms | ||
| Adobe Photoshop CC | Adobe Systems Inc | RRID:SCR_014199 |
| Adobe Illustrator CC | Adobe Systems Inc | RRID:SCR_010279 |
| BioRender (schematics in figures) | BioRender.com | RRID:SCR_018361 |
| CFX MaestroTM Software (qPCR analyses) | Bio-rad Inc | RRID:SCR_018064 |
| Graphpad Prism v9.0.1 | GraphPad Software | https://www.graphpad.com/RRID:SCR_002798 |
| Living Image software | Perkin Elmer |
http://www.perkinelmer.com/catalog/category/id/living%20image%20software RRID:SCR_014247 |
| Other | ||
| TriStar LB 941 Multimode Microplate Reader and Luminometer | BERTHOLD TECHNOLOGIES GmbH & Co. KG | Mothes Lab |
| King Fisher Apex | ThermoFisher Scientific | Cat# 5400930 |
| C1000 Touch thermal cycler | Bio-Rad | RRID:SCR_019688 |
| CFX Connect™ Real-Time PCR Detection System | Bio-Rad | RRID:SCR_018064 |
| Nanodrop Spectrophotometer ND-1000 | ThermoFisher Scientific | RRID:SCR_016517 |
| 27G × ½’’ insulin syringe with needle | TERUMO | Cat# SS∗05M2713 |
| 31G insulin syringe | BD Biosciences | Cat# 328468 |
| Acrodisc 25 mm Syringe Filter w/0.45 μm HT Tuffryn Membrane | PALL Life Sciences | Cat# 4184 |
| Zirconium Beads | MP Biomedicals | Cat# 116913100 |
| BeadBug 6 Homogenizer | Benchmark Scientific, TEquipment Inc | Cat# Z742682 |
| GloMax Explorer Luminometer | Promega Inc | Cat# GM3510 |
| 96-well white plates for luciferase assays | Costar | Cat# 3917 |
| Cryotubes | Thermo Scientific Nunc | Cat# 340711 |
| Polystyrene Round-bottom Tube | FALCON | Cat# 352058 |
| Optical Flat 8-Cap Strips for 0.2 mL tube stripes/plates | Bio-Rad | Cat# TCS0803 |
| Individual PCR tubes 8-tube Strip, clear | Bio-Rad | Cat# TLS0801 |
| ThermalGrid Rigid Strip PCR tubes | Denville Scientific INC | Cat# C18064 |
| 96 well U bottom plate | FALCON | Cat# 353077 |
| XIC-3 animal isolation chamber | PerkinElmer | Cat# 123997 |
| Perkin Elmer IVIS Spectrum In-Vivo Imaging System | PerkinElmer | Yale University ABSL-3 facility; RRID:SCR_018621 |
| RAS-4 Rodent Anesthesia System | PerkinElmer | Cat# CLS146737 |
| Millex-GV Filter, 0.22uM | Fisher Scientific | Cat# SLGV013SL |
| Legend 21R Micro centrifuge | Thermo Scientific | Cat# 75002446 |
| Sorvall X4 Pro Refrigerated Centrifuge SRVLX4R PRO-MD, 120V TX-1000CC |
Thermo Scientific | Cat# 75016052 |
Resource availability
Lead contact
Requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Pradeep Uchil (pradeep.uchil@yale.edu).
Materials availability
All unique reagents generated in this study are available from the lead contact with a completed Materials Transfer Agreement.
Data and code availability
-
•
Raw data used to generate all the graphs shown in the manuscript have been deposited at Mendeley Data, V2, https://doi.org/10.17632/n9ztbbbstm.2 and are publicly available as of the 29 Nov 2023. DOIs are listed in the key resources table.
-
•
This paper does not report the original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from lead contact upon request.
Experiment model and details
Cell and viruses
Vero E6 (CRL-1586, American Type Culture Collection (ATCC), were cultured at 37°C in RPMI supplemented with 10% fetal bovine serum (FBS), 10 mM HEPES pH 7.3, 1 mM sodium pyruvate, 1× non-essential amino acids, and 100 U/ml of penicillin–streptomycin. SARS-CoV-2 B.1.617.2 (Delta) and Omicron VOC was isolated from a patient in Yale New Haven Hospital. All viruses were sequence confirmed using Next Genome Sequencing (Yale Keck Facility). We generated nanoluc luciferase (nLuc) expressing reporter viruses for Omicron and Delta VOC for non-invasive BLI imaging of infected mice using circular polymerization extension reaction (CPER) as described previously.66 Briefly, viral RNA was converted into cDNA using PrimeScript RT kit (Takara Bio) using a mix of random and oligo dT primers. The cDNA was then used as the template to amplify 11 overlapping fragments that were ∼2–3 kb long using specific primers and PrimeStar GXL polymerase (Takara Bio). A nLuc-P2A cassette was inserted at the N-terminus of ORF8 (nLuc-P2A-Orf8). The gel-purified overlapping fragments were circularized by CPER using a Hepatitis Delta virus ribozyme-spacer-CMV promoter cassette that overlapped with the 3′ and 5′end of the genome. CPER reactions were transfected into a T-25 flask of HEK293 cells using polyethyleneimine and cultured under BSL3 conditions. The following day the HEK293 cells were resuspended and co-cultured with Vero E6 ACE2/TMPRSS2 (VeroAT) cells for 5–9 days until CPE was clearly visible and nLuc activity was detected in the culture supernatants. Delta-nLuc and Omicron-nLuc was propagated in VeroAT cells by infecting them in T150 cm2 flasks at a MOI of 0.1. The culture supernatants were collected after 18–24 h when cytopathic effects were clearly visible. The cell debris was removed by centrifugation and filtered through 0.45-micron filter to generate virus stocks. Viruses were concentrated by adding one volume of cold (4°C) 4x PEG-it Virus Precipitation Solution (40% (w/v) PEG-8000 and 1.2 M NaCl; System Biosciences) to three volumes of virus-containing supernatant. The solution was mixed by inverting the tubes several times and then incubated at 4°C overnight. The precipitated virus was harvested by centrifugation at 1,500 × g for 60 min at 4°C. The concentrated virus was then resuspended in PBS then aliquoted for storage at −80°C. All work with infectious SARS-CoV-2 was performed in Institutional Biosafety Committee approved BSL3 and ABSL3 facilities at Yale University School of Medicine.
Ethics statement
CCP-5 was obtained from individual who was infected during the first wave of the pandemic, after at least fourteen days of resolution of COVID-19 symptoms.67 The participant consented to the study (Héma-Québec, CER #2020–004). Research adhered to the standards indicated by the Declaration of Helsinki. All participants were adults and provided informed written consent prior to enrollment in accordance with Institutional Review Board approval.
Plasma samples
Recovered COVID-19 patient who had received a COVID-19 diagnosis by the Québec Provincial Health Authority and met the donor selection criteria for plasma donation in use at Héma-Québec were recruited. The participant was allowed to donate plasma at least 14 days after complete resolution of COVID-19 symptoms. A volume of 500 mL–750 mL of plasma was collected by plasmapheresis (TRIMA Accel, Terumo BCT). Disease severity (date of symptoms onset, end of symptoms, type, and intensity of symptoms, need for hospitalization/ICU) was documented using a questionnaire administered at the time of recruitment. For additional details of CCP-5 (sex, age, blood group of the convalescent donor and day of collection post infection, please refer to key resources table.
Mouse experiments
All animals were maintained in the (SPF-free) barrier facility of the Yale University Animal Resource Center (YARC) within a 14:10 light: dark cycle. Breeding population of mice and infected animals are maintained in separate rooms. All SARS-CoV-2-infected animals were housed in animal room under BSL3 containment. Cages, animal waste, bedding, and animal carcasses were disposed and decontaminated following the guidelines of Environmental Health Services at Yale. All replication competent virus-infected animals were handled under ABSL3 conditions with personnel’s donning pressurized air purified respirators (PAPR), double gloves, shoe covers, sleeve covers and disposable gowns. All experiments described here were approved by Institutional Animal Care and Use Committees (IACUC) as well as SOPs approved by Institutional Environmental Health and Biosafety committee. hACE2 transgenic B6 mice (heterozygous) were obtained from Jackson Laboratory. 6–8-week-old male and female mice were used for all the experiments. The heterozygous mice were crossed and genotyped to select heterozygous mice for experiments by using the primer sets recommended by Jackson Laboratory. Each cohort size was n = 4–8 to allow statistical testing and conducted as 2–3 biological replicates (n = 2–3 per replicate) to allow parallel evaluation of different cohorts. The number of animals (n = 4–8 per cohort) needed to achieve statistically significant results were calculated based on a priori power analysis. We calculated power and sample sizes required based on data from pilot experiments and previous studies.26,27,68,69 Animals with sex- and age-matched littermates were included randomly in the experiments. No animals were excluded due to illness after the experiments. At the time of experimentation, care was taken to include equal numbers of male and female mice whenever possible to ensure that sex of the animals does not constitute a biological variable during analysis.
Method details
SARS-CoV-2 infection and treatment conditions
For all in vivo experiments, 6 to 8 weeks male and female mice were intranasally challenged with 1 × 105 PFU SARS-CoV-2-nLuc Delta or Omicron-nLuc VOCs in 25–30 μL volume under anesthesia (0.5–5% isoflurane delivered using precision Dräger vaporizer with oxygen flow rate of 1 L/min). The starting body weight was set to 100%. For survival experiments, mice were monitored every 8–12 h starting six days after virus challenge. Lethargic and moribund mice or mice that had lost more than 20% of their body weight were sacrificed and considered to have succumbed to infection for Kaplan-Meier survival plots. Mice were considered to have recovered if they gained back all the lost weight.
Favipiravir was prepared at concentration of 50 mg/mL in 0.3% (w/v) sodium bicarbonate solution made in water. The drug was dissolved for 1h at room temperature by shaking in a thermomixer. Favipiravir solution was prepared fresh on the day of administration for dosing. Favipiravir administration was initiated at 6 hpi via intraperitoneal injection (i.p., 26 gauge) (600 mg/kg body weight) followed by twice daily administrations with morning dose at 8 a.m. and evening dose at 6 p.m. (total dose of 1200 mg/kg every 10–14 h intervals) till 6 dpi. In humans, Favipiravir is administered twice daily at a dosage of 1600 mg bringing the total dose to 3200 mg per day or 53.3 mg/kg for an average human weighing 60 kg.31 Therefore, animal equivalent dose was 656 mg/kg daily for 20 g mice which can be calculated by factoring 53.3 with 12.3 [correction factor/Km ratio which is estimated by dividing the average body weight (kg) of species to its body surface area (m2)] .33
Molnupiravir was dissolved in solution (Vehicle) of 10% PEG400 and 2.5% Kolliphor-EL in sterile milli Q water by shaking in thermomixer for 15 min and then by vortexing for 5 min. The solution was kept in thermomixer till it was ready for dosing. molnupiravir solution was prepared fresh daily. molnupiravir (250 mg/kg body weight) was made in a volume of 150 μL and administered orally using an oral gavage needle (20 gauge) starting at 6 hpi. On the following days, 250 mg/kg body weight of molnupiravir was administered two times daily (morning dose at 8 a.m., evening dose at 6 p.m., 10–14 h interval) until 3 dpi or 6 dpi (total dose of 500 mg/kg per 10–14 h). The COVID-19 Treatment Guidelines Panel (the Panel) recommends using molnupiravir 800 mg orally (PO) twice daily for 5 days which translates to 26.66 mg/kg for 60 kg average human.70 Therefore, animal equivalent dose conversion was calculated by factoring 26.66 with 12.3 to obtain 328 mg/kg daily for 20 g mice.33
Nirmatrelvir was prepared fresh daily by dissolving the drug in 0.5% (w/v) aqueous methyl cellulose in 2% Tween 80 prepared in MilliQ water. The drug was dissolved by shaking in thermomixer for 2 h at 37°C nirmatrelvir was dosed at 650 mg/kg body weight in a volume of 250 μL orally using an oral gavage needle (20 gauge) starting at 6 h post infection. On the following days, 650 mg/kg body weight of nirmatrelvir was administered two times daily (morning dose at 8 a.m., evening dose at 6 p.m., 10–14 h interval; total dose 1300 mg/kg) till 6 dpi. The COVID-19 Treatment Guidelines Panel recommends using nirmatrelvir 300 mg twice daily for 5 days which translates to 10 mg/kg of nirmatrelvir for 60 kg average human.71 Therefore, animal equivalent dose conversion was calculated by factoring 10 with 12.3 to obtain 123 mg/kg daily for 20 g mice.33
To test efficacy of nAb against Delta VOC, nAb CV3-1 was administered once (12.5 mg/kg, i.p.) at 1 dpi using a 26-gauge needle. Caspase-1/4 inhibitor (VX-765) (InvivoGen, in vivo grade) was injected intraperitoneally at 8 mg/kg body weight starting at 6 hpi followed by every other day till 6 dpi. VX-765 was administered either alone (VX-765 cohort) or in combination with molnupiravir (250 mg/kg body weight). 1 mL of COVID-19 convalescent plasma-5 (CCP-5)27 was administered intraperitoneally once at 1 dpi either alone (CCP-5 cohort) or in combination with molnupiravir (250 mg/kg body weight, oral). molnupiravir was administered till 3 dpi as mentioned above for all experiments designed to test synergy with VX-765 or CCP-5.
Dose response analysis
To delineate the interaction dynamics in DAA combination therapy, we conducted dose response analyses for molnupiravir, nirmatrelvir, and their combinations. Mice were infected with Delta-nLuc VOC as above and DAA treatment started at 0.25 hpi. DAAs were prepared at doses of 80 mg/kg, 40 mg/kg, and 20 mg/kg body weight. These drug solutions were made in a volume of 150 μL and administered orally as described above. Subsequently, the same amount of drug doses was administered twice daily, with a morning dose at 8 a.m. and an evening dose at 6 p.m., ensuring a 10–14 h interval between doses. This treatment regimen was continued until 2 dpi, resulting in a total cumulative dose of 160 mg/kg, 80 mg/kg, or 40 mg/kg, respectively, within the 10–14 h interval. The drug treatment was stopped 24 h before necropsy and the mice sacrificed at 3 dpi. BLI of whole body and individual lung lobe was carried out after necropsy to monitor virus replication. In addition, lung lobes were homogenized for viral load estimations using real-time PCR analyses for N mRNA expression and viral titers by infecting VeroAT cells followed by estimating nLuc activity.
Bliss index score
To determine if the combinatorial regimens were antagonistic, additive or synergistic, Bliss index scores were computed using multiparametric analyses to calculate overall disease burden. For experiments involving Delta VOC infection a total of nine parameters were included to calculate total disease burden. They were viral loads [N mRNA expression, titers (nLuc activity)] and inflammation [Ccl2 and Cxcl10 mRNA expression] in both the brain and lung tissues, lung injury (Krt8 mRNA expression), mortality, and extent of delay in death. For each parameter, data were normalized by setting the values obtained for vehicle treated animals to 100. Total disease burden was calculated by summing the normalized values of each parameter (viral load, inflammation, lung injury, mortality) and subtracting the normalized value for delayed death. In our experiments, we recorded a maximum delay in death of up to 4.5 days (from 6.5 days in vehicle-treated mice to 11 days in drug-treated animals). Therefore, 4.5 days was set as 100, and for each day of delayed death, a value of 22.2 was subtracted to calculate the overall disease burden. In vehicle-treated animals %Delay in death was 0. The total summed value was divided by 9 for nine parameters analyzed to normalize the data.
For Omicron VOC experiments, five parameters were used for computing total disease burden: viral loads in the lung (measured by N mRNA and nLuc activity), Ccl2, Cxcl10 and Krt8 mRNA expression in the lung. The summed value was divided by 5 for five parameters analyzed to normalize the data.
For short-term drug-dose response experiments, two parameters were used: viral loads in the lung (measured by N mRNA and nLuc activity). The summed value was divided by 2 for two parameters analyzed to normalize the data.
Bliss index scores were calculated as done previously72 using the formula below:
A Bliss index score of less than −10 was considered antagonistic, an index between −10 and +10 was considered an additive effect, and an index more than 10 was considered synergistic.
Pharmacokinetics of antiviral drugs
Favipiravir (600 mg/kg body weight, i.p) and nirmatrelvir (650 mg/kg body weight, oral) was administered in K18-hACE2 transgenic as described above. The whole blood samples (150μL per time point) were collected at 0, 4 and 24 h post administration. The samples were then allowed to clot by leaving undisturbed at room temperature for 4 h. Neat clear serum was collected by sedimenting the samples at 14,000 x g for 20 min at room temperature. At the 24 h timepoint mice were sacrificed and tissue samples (brain, nose, lung, trachea, and gut) were collected after necropsy. Tissue samples were accurately weighed and were homogenized in 800 μL of 1x Dulbecco’s PBS in 2 mL tube containing 1.5 mm Zirconium beads in a BeadBug 6 homogenizer (Benchmark Scientific, TEquipment Inc). The homogenized tissues were then sedimented at 13,000 rpm for 20 min at 4°C and the clear supernatant was frozen and stored at −80°C until further analysis.
Mass spectrometry
Favipiravir was extracted from mouse serum and tissue homogenates using protein precipitation as sample preparation technique. 200 μL of internal standard (IS) solution (500 ng/mL of 13C5-ribavirin in acetonitrile) was added to an aliquot of 25 μL of sample (serum or tissue homogenate). The sample was vortexed for approximately 5 s and let stand for a period of 10 min, then centrifuged at 16,000 x g for 10 min and the supernatant was transferred to an injection vial for analysis The analysis was performed using a Thermo Scientific TSQ Quantiva triple quadrupole mass spectrometer interfaced with a Thermo Scientific Ultimate 3000XRS UHPLC system using a heated electrospray ion source (HESI). MS detection was performed in negative ion mode, using selected reaction monitoring (SRM). The precursor-ion reactions for favipiravir and IS were set to 156.1 → 113.1 and 248.1 → 111.1, respectively. Isocratic elution was used with an Agilent Poroshell HILIC-Z column (100∗2.1 mm I.D., 2.7μm) and guard cartridge operating operating at 45°C. The mobile phase conditions consisted of acetonitrile and 20 mM ammonium formate in type 1 water pH 3 at a ratio of 92:8, respectively. Data acquisition and analysis were performed using Thermo Scientific Xcalibur 4.2.47. The analytical range was set from 10.0 to 50,000 ng/mL and the sample concentrations were interpolated from the standard curve.
Nirmatrelvir was extracted from mouse serum and tissue homogenates using protein precipitation as sample preparation technique. 180 μL of internal standard (IS) solution (100 ng/mL of EMD 281014 in methanol) was added to an aliquot of 20 μL of sample (serum or tissue homogenate). The sample was vortexed for approximately 5 s and let stand for a period of 10 min, then centrifuged at 16,000 x g for 10 min. One hundred and fifty microliters of the supernatant were transferred to a new tube and eight hundred and fifty microliters of 10 mM ammonium formate pH 3 buffer was added. The sample was vortexed for approximately 5 s and transferred to an injection vial for analysis. The analysis was performed using a Thermo Scientific TSQ Quantiva triple quadrupole mass spectrometer interfaced with a Thermo Scientific Ultimate 3000XRS UHPLC system using a heated electrospray ion source (HESI). MS detection was performed in positive ion mode, using selected reaction monitoring (SRM). The precursor-ion reactions for nirmatrelvir and IS were set to 500.3 → 319.1 and 377.2 → 209.1, respectively. A gradient mobile phase was used with a Thermo Scientific Accucore RP-MS analytical column (100 × 2.1 mm I.D., 2.6 μm) operating at 40°C. The initial mobile phase conditions consisted of acetonitrile and 10 mM ammonium formate in type 1 water pH 3.0 at a ratio of 10:90, respectively, and this ratio was maintained for 1 min. From 1 to 2.5 min a linear gradient was applied up to a ratio of 95:5 and maintained for 1.5 min. At 4.1 min, the mobile phase composition was reverted to the original conditions and the column was allowed to equilibrate for 5.9 min for a total run time of 10.0 min. Data acquisition and analysis were performed using ThermoScientific Xcalibur 4.2.47. The analytical range was set from 5.00 to 50,000 ng/mL and the sample concentrations were interpolated from the standard curve.
Molnupiravir in-life phase study was performed at Charles River Den Bosch, ‘s-Hertogenbosch, The Netherlands. Female BALB/c mice were dosed on a single occasion by oral gavage with molnupiravir formulated as a solution in 10% PEG400 and 2.5% Kolliphor-EL in Elix water. molnupiravir was administrated at doses of 3, 15 and 75 mg/kg to 3 groups of 6 animals. Blood samples (∼30 μL using K2EDTA-coated hematocrit capillaries) were collected from animals in all dose groups at 0.5, 1, 2, 6, 12 and 24 h post dose by sampling from the jugular vein. Blood samples were centrifuged within 1 h after blood sampling at approximately 3000g for 10 min at 2°C–8°C. Immediately after centrifugation, the plasma was transferred to a labeled polypropylene tube and stored in an ultra-low freezer set to maintain −80°C until shipment on dry ice to the bioanalytical laboratory. Plasma samples were then analyzed for molnupiravir metabolite EIDD-1931 with a standard HPLC-MS/MS method using an internal standard (Research Grade Assay 1).
Bioluminescence imaging (BLI) of SARS-CoV-2 infection
All standard operating procedures and protocols for In Vivo Imaging System (IVIS) imaging of SARS-CoV-2-infected animals under ABSL3 conditions were approved by IACUC, IBSCYU and YARC. All the imaging was carried out using IVIS Spectrum (PerkinElmer) in XIC-3 animal isolation chamber (PerkinElmer) that provided biological isolation of anesthetized mice or individual organs during the imaging procedure. All mice were anesthetized via isoflurane inhalation (3–5% isoflurane, oxygen flow rate of 1.5 L/min) prior and during BLI using the XGI-8 Gas Anesthesia System. Prior to imaging, 100 μL of nanoluc luciferase (nLuc) substrate, furimazine (NanoGlo, Promega, Madison, WI) diluted 1:40 in endotoxin-free PBS was retroorbitally administered to mice under anesthesia. The mice were then placed into XIC-3 animal isolation chamber (PerkinElmer) pre-saturated with isothesia and oxygen mix. The mice were imaged in both dorsal and ventral position at indicated days post infection. The animals were then imaged again after euthanasia and necropsy by spreading additional 200 μL of substrate on to exposed intact organs. Infected areas identified by carrying out whole-body imaging after necropsy were isolated, washed in PBS to remove residual blood and placed onto a clear plastic plate. Additional droplets of furimazine in PBS (1:40) were added to organs and soaked in substrate for 1–2 min before BLI.
Images were acquired and analyzed with Living Image v4.7.3 in vivo software package (PerkinElmer Inc). Image acquisition exposures were set to auto, with imaging parameter preferences set in order of exposure time, binning, and f/stop, respectively. Images were acquired with luminescent f/stop of 2, photographic f/stop of 8. Binning was set to medium. Comparative images were compiled and batch-processed using the image browser with collective luminescent scales. Photon flux was measured as luminescent radiance (p/sec/cm2/steradian). During luminescent threshold selection for image display, luminescent signals were regarded as background when minimum threshold setting resulted in displayed radiance above non-tissue-containing or known uninfected regions.
Plaque forming assay
Titers of virus stocks was determined by standard plaque assay. Briefly, the 4 x 105 Vero-E6 cells were seeded on 12-well plate. 24 h later, the cells were infected with 200 μL of serially diluted virus stock followed by overlaying with 1 mL of pre-warmed 0.6% Avicel (RC-581 FMC BioPolymer) made in complete RPMI medium. Plaques were resolved at 48 h post infection by fixing in 10% paraformaldehyde for 15 min followed by staining for 1 h with 0.2% crystal violet made in 20% ethanol. Plates were rinsed in water to visualize plaques.
Measurement of viral burden
Indicated organs (nasal cavity, brain, lungs) from infected or uninfected mice were collected, weighed, and homogenized in 1 mL of serum free RPMI media containing penicillin-streptomycin and homogenized in 2 mL tube containing 1.5 mm Zirconium beads with BeadBug 6 homogenizer (Benchmark Scientific, TEquipment Inc). Virus titers were measured using two highly correlative methods. Frist, the total RNA was extracted from homogenized tissues using RNeasy plus Mini kit (Qiagen Cat # 74136), reverse transcribed with iScript advanced cDNA kit (Bio-Rad Cat #1725036) followed by an SYBR Green Real-time PCR assay for determining copies of SARS-CoV-2 N gene RNA using primers SARS-CoV-2 N F: 5’-ATGCTGCAATCGTGCTACAA-3′ and SARS-CoV-2 N R: 5′-GACTGCCGCCTCTGCTC-3’. All the real-time PCR assays based on SYBR Green have a built-in melt-curve analyses to ensure estimation of only specific PCR products and not false-positives.
Second, we used nLuc activity as a shorter surrogate for plaque assay. Serially diluted clarified tissue homogenates were used to infect Vero-E6 cell culture monolayer. Infected cells were washed with PBS and then lysed using 1X Passive lysis buffer. The lysates transferred into a 96-well solid white plate (Costar Inc) and nLuc activity was measured using Tristar multiwell Luminometer (Berthold Technology, Bad Wildbad, Germany) for 2.5 s by adding 20 μL of Nano-Glo substrate in nanoluc assay buffer (Promega Inc, WI, USA). Uninfected monolayer of Vero-E6 cells treated identically served as controls to determine basal luciferase activity to obtain normalized relative light units. The data were processed and plotted using GraphPad Prism 8 v8.4.3.
Analyses of signature inflammatory cytokines mRNA expression
Brain and lung samples were collected from mice at the time of necropsy. Approximately, 20 mg of tissue was suspended in 500 μL of RLT lysis buffer, and RNA was extracted using RNeasy plus Mini kit (Qiagen Cat# 74136), reverse transcribed with iScript advanced cDNA kit (Bio-Rad Cat# 1725036). To determine mRNA copy numbers of signature inflammatory cytokines, multiplex qPCR was conducted using iQ Multiplex Powermix (Bio Rad Cat# 1725848) and PrimePCR Probe Assay mouse primers FAM-GAPDH, HEX-IL6, TEX615-CCL2, Cy5-CXCL10, Cy5.5-IFNgamma and HEX-IL1B. The reaction plate was analyzed using CFX96 touch real-time PCR detection system. Scan mode was set to all channels. The PCR conditions were 95°C 2 min, 40 cycles of 95°C for 10 s and 60°C for 45 s, followed by a melting curve analysis to ensure that each primer pair resulted in amplification of a single PCR product. mRNA copy numbers of Il6, Ccl2, Cxcl10, Ifng and Il1b in the cDNA samples of infected mice were normalized to Gapdh mRNA with the formula ΔCt(target gene) = Ct(target gene)-Ct(Gapdh). The fold increase was determined using 2−ΔΔCt method comparing treated mice to uninfected controls.
Cryo-immunohistology of lung tissue
Lung tissues were isolated after necropsy and fixed in 1X PBS containing freshly prepared 4% PFA for 12 h at RT or 4°C. They were then washed with PBS, cryoprotected with 10, 20 and 30% ascending sucrose series, snap-frozen in O.C.T.TM compound (Tissue-Tek) inside cryomolds and stored at −80°C. 10 μm thick frozen sections were then cut and mounted on Superfrost plus slides. The sections were allowed to dry at 37°C for 1 h and stored at −20°C. For staining, the slides were equilibrated at RT, washed with 1X PBS to remove excess O.C.T and permeabilized with 0.2% Triton X-100 and treated with Fc receptor blocker (Innovex Biosciences) before staining with antibodies to SARS-CoV-2 nucleocapsid directly conjugated to Alexa FluorTM 488 or antibodies to mouse Krt8 followed by detection using secondary antibody conjugated to Alexa FluorTM 568 in PBS containing 2% BSA and 0.1% fetal bovine serum. Stained sections were treated with TrueVIEW Autofluorescence Quenching Kit (Vector Laboratories) and mounted in VECTASHIELD Vibrance Antifade Mounting Medium. Images were acquired and stitched using EVOS M7000 imaging system. The images were processed using Nikon Elements AR version 4.5 software (Nikon Instruments Inc, Americas) and figures assembled with Photoshop CC and Illustrator CC (Adobe Systems, San Jose, CA, USA).
Quantification and statistical analysis
Data were analyzed and plotted using GraphPad Prism software (La Jolla, CA, USA). Statistical significance for pairwise comparisons were derived by applying non-parametric Mann-Whitney test (two-tailed).
To obtain statistical significance for survival curves, grouped data were compared by log rank (Mantel-Cox) test. To obtain statistical significance for grouped data we employed two-way ANOVA followed by Tukey’s multiple comparison tests. p values lower than 0.05 were considered statistically significant. p values were indicated as ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001; ∗∗∗∗, p < 0.0001.
Acknowledgments
This project was funded through DNDi under the support by the Wellcome Trust Grant ref. 222489/Z/21/Z through the “COVID-19 Therapeutics Accelerator” and subcontracted to P.D.U., NIH grant R01AI163395 to W.M., a CIHR operating Pandemic and Health Emergencies Research grant #177958 to W.M. and A.F, CIHR operating grant #487578 to P.D.U. and A.F., and NIH R24OD026440-03S1 (COVID-19 Administrative Supplement) to P.K. and P.D.U. A.F. is the recipient of Canada Research Chair on Retroviral Entry no. RCHS0235 950-232424.
Author contributions
Conceptualization, P.D.U., E.C., F.E., I.S., C.M., and L.F.; methodology, P.D.U., I.U., F.E., I.S., and E.C.; investigation, I.U., Z.G., and P.D.U.; writing – original draft, P.D.U. and I.U.; writing – review & editing, P.D.U., A.F., W.M., P.K., R.B., P.B., M.C., I.U., E.C., F.E., I.S., C.M., and L.F.; funding acquisition, E.C., C.M., L.F., P.D.U., W.M., and A.F.; resources, W.M., P.K., A.F., and R.B.; supervision, P.D.U., E.C., L.F. C.M., and P.K.
Declaration of interests
The authors declare no competing interests.
Published: January 30, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.109049.
Contributor Information
Priti Kumar, Email: priti.kumar@yale.edu.
Eric Chatelain, Email: echatelain@dndi.org.
Pradeep D. Uchil, Email: pradeep.uchil@yale.edu.
Supplemental information
References
- 1.Larkin H.D. Global COVID-19 Death Toll May Be Triple the Reported Deaths. JAMA. 2022;327:1438. doi: 10.1001/jama.2022.4767. [DOI] [PubMed] [Google Scholar]
- 2.Marcelin J.R., Pettifor A., Janes H., Brown E.R., Kublin J.G., Stephenson K.E. COVID-19 Vaccines and SARS-CoV-2 Transmission in the Era of New Variants: A Review and Perspective. Open Forum Infect. Dis. 2022;9:ofac124. doi: 10.1093/ofid/ofac124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suthar A.B., Wang J., Seffren V., Wiegand R.E., Griffing S., Zell E. Public health impact of covid-19 vaccines in the US: observational study. BMJ. 2022;377 doi: 10.1136/bmj-2021-069317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dye C. The benefits of large scale covid-19 vaccination. BMJ. 2022;377:o867. doi: 10.1136/bmj.o867. [DOI] [PubMed] [Google Scholar]
- 5.Alter G., Yu J., Liu J., Chandrashekar A., Borducchi E.N., Tostanoski L.H., McMahan K., Jacob-Dolan C., Martinez D.R., Chang A., et al. Immunogenicity of Ad26.COV2.S vaccine against SARS-CoV-2 variants in humans. Nature. 2021;596:268–272. doi: 10.1038/s41586-021-03681-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agency, U.H.S. COVID-19 Vaccine Surveillance Report Week 19.
- 7.Tartof S.Y., Slezak J.M., Puzniak L., Hong V., Xie F., Ackerson B.K., Valluri S.R., Jodar L., McLaughlin J.M. Durability of BNT162b2 vaccine against hospital and emergency department admissions due to the omicron and delta variants in a large health system in the USA: a test-negative case-control study. Lancet Respir. Med. 2022;10:689–699. doi: 10.1016/S2213-2600(22)00101-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lenharo M. WHO declares end to COVID-19's emergency phase. Nature. 2023 doi: 10.1038/d41586-023-01559-z. [DOI] [PubMed] [Google Scholar]
- 9.Gandhi S., Klein J., Robertson A.J., Peña-Hernández M.A., Lin M.J., Roychoudhury P., Lu P., Fournier J., Ferguson D., Mohamed Bakhash S.A.K., et al. De novo emergence of a remdesivir resistance mutation during treatment of persistent SARS-CoV-2 infection in an immunocompromised patient: a case report. Nat. Commun. 2022;13:1547. doi: 10.1038/s41467-022-29104-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abdalhamid B., Bilder C.R., McCutchen E.L., Hinrichs S.H., Koepsell S.A., Iwen P.C. Assessment of Specimen Pooling to Conserve SARS CoV-2 Testing Resources. Am. J. Clin. Pathol. 2020;153:715–718. doi: 10.1093/ajcp/aqaa064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Syed Y.Y. Molnupiravir: First Approval. Drugs. 2022;82:455–460. doi: 10.1007/s40265-022-01684-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furuta Y., Gowen B.B., Takahashi K., Shiraki K., Smee D.F., Barnard D.L. Favipiravir (T-705), a novel viral RNA polymerase inhibitor. Antivir. Res. 2013;100:446–454. doi: 10.1016/j.antiviral.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kato H., Takayama-Ito M., Satoh M., Kawahara M., Kitaura S., Yoshikawa T., Fukushi S., Nakajima N., Komeno T., Furuta Y., Saijo M. Favipiravir treatment prolongs the survival in a lethal mouse model intracerebrally inoculated with Jamestown Canyon virus. PLoS Neglected Trop. Dis. 2021;15 doi: 10.1371/journal.pntd.0009553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boretti A. Favipiravir use for SARS CoV-2 infection. Pharmacol. Rep. 2020;72:1542–1552. doi: 10.1007/s43440-020-00175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Owen D.R., Allerton C.M.N., Anderson A.S., Aschenbrenner L., Avery M., Berritt S., Boras B., Cardin R.D., Carlo A., Coffman K.J., et al. An oral SARS-CoV-2 M(pro) inhibitor clinical candidate for the treatment of COVID-19. Science. 2021;374:1586–1593. doi: 10.1126/science.abl4784. [DOI] [PubMed] [Google Scholar]
- 16.Joyce R.P., Hu V.W., Wang J. The history, mechanism, and perspectives of nirmatrelvir (PF-07321332): an orally bioavailable main protease inhibitor used in combination with ritonavir to reduce COVID-19-related hospitalizations. Med. Chem. Res. 2022;31:1637–1646. doi: 10.1007/s00044-022-02951-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeong J.H., Chokkakula S., Min S.C., Kim B.K., Choi W.-S., Oh S., Yun Y.S., Kang D.H., Lee O.J., Kim E.G., et al. Combination therapy with nirmatrelvir and molnupiravir improves the survival of SARS-CoV-2 infected mice. Antivir. Res. 2022;208 doi: 10.1016/j.antiviral.2022.105430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abdelnabi R., Foo C.S., Jochmans D., Vangeel L., De Jonghe S., Augustijns P., Mols R., Weynand B., Wattanakul T., Hoglund R.M., et al. The oral protease inhibitor (PF-07321332) protects Syrian hamsters against infection with SARS-CoV-2 variants of concern. Nat. Commun. 2022;13:719. doi: 10.1038/s41467-022-28354-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosenke K., Lewis M., Feldmann F., Bohrnsen E., Schwarz B., Okumura A., Bohler W.F., Callison J., Shaia C., Bosio C. Combined molnupiravir and nirmatrelvir treatment improves the inhibitory effect on SARS-CoV-2 in Rhesus Macaques. bioRxiv. 2022 doi: 10.1101/2022.09.03.506479. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosenke K., Okumura A., Lewis M.C., Feldmann F., Meade-White K., Bohler W.F., Griffin A., Rosenke R., Shaia C., Jarvis M.A., Feldmann H. Molnupiravir inhibits SARS-CoV-2 variants including Omicron in the hamster model. JCI insight. 2022;7 doi: 10.1172/jci.insight.160108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vangeel L., Chiu W., De Jonghe S., Maes P., Slechten B., Raymenants J., André E., Leyssen P., Neyts J., Jochmans D. Remdesivir, Molnupiravir and Nirmatrelvir remain active against SARS-CoV-2 Omicron and other variants of concern. Antivir. Res. 2022;198 doi: 10.1016/j.antiviral.2022.105252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanderson T., Hisner R., Donovan-Banfield I., Hartman H., Løchen A., Peacock T.P., Ruis C. A molnupiravir-associated mutational signature in global SARS-CoV-2 genomes. Nature. 2023;623:594–600. doi: 10.1038/s41586-023-06649-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tian H., Yang C., Song T., Zhou K., Wen L., Tian Y., Tang L., Xu W., Zhang X. Efficacy and safety of paxlovid (nirmatrelvir/ritonavir) in the treatment of COVID-19: An updated meta-analysis and trial sequential analysis. Rev. Med. Virol. 2023;33:e2473. doi: 10.1002/rmv.2473. [DOI] [PubMed] [Google Scholar]
- 24.Focosi D., McConnell S., Shoham S., Casadevall A., Maggi F., Antonelli G. Nirmatrelvir and COVID-19: development, pharmacokinetics, clinical efficacy, resistance, relapse, and pharmacoeconomics. Int. J. Antimicrob. Agents. 2023;61 doi: 10.1016/j.ijantimicag.2022.106708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wagoner J., Herring S., Hsiang T.Y., Ianevski A., Biering S.B., Xu S., Hoffmann M., Pöhlmann S., Gale M., Jr., Aittokallio T., et al. Combinations of Host- and Virus-Targeting Antiviral Drugs Confer Synergistic Suppression of SARS-CoV-2. Microbiol. Spectr. 2022;10 doi: 10.1128/spectrum.03331-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ullah I., Prévost J., Ladinsky M.S., Stone H., Lu M., Anand S.P., Beaudoin-Bussières G., Symmes K., Benlarbi M., Ding S., et al. Live imaging of SARS-CoV-2 infection in mice reveals that neutralizing antibodies require Fc function for optimal efficacy. Immunity. 2021;54:2143–2158.e15. doi: 10.1016/j.immuni.2021.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ullah I., Beaudoin-Bussières G., Symmes K., Cloutier M., Ducas E., Tauzin A., Laumaea A., Grunst M.W., Dionne K., Richard J., et al. The Fc-effector function of COVID-19 convalescent plasma contributes to SARS-CoV-2 treatment efficacy in mice. Cell Rep. Med. 2023;4 doi: 10.1016/j.xcrm.2022.100893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tarrés-Freixas F., Trinité B., Pons-Grífols A., Romero-Durana M., Riveira-Muñoz E., Ávila-Nieto C., Pérez M., Garcia-Vidal E., Perez-Zsolt D., Muñoz-Basagoiti J., et al. Heterogeneous infectivity and pathogenesis of SARS-CoV-2 variants beta, delta and omicron in transgenic K18-hACE2 and wildtype mice. Front. Microbiol. 2022;13:1382. doi: 10.3389/fmicb.2022.840757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodrigues T.S., de Sá K.S.G., Ishimoto A.Y., Becerra A., Oliveira S., Almeida L., Gonçalves A.V., Perucello D.B., Andrade W.A., Castro R., et al. Inflammasomes are activated in response to SARS-CoV-2 infection and are associated with COVID-19 severity in patients. J. Exp. Med. 2021;218 doi: 10.1084/jem.20201707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Rivero Vaccari J.C., Dietrich W.D., Keane R.W., de Rivero Vaccari J.P. The inflammasome in times of COVID-19. Front. Immunol. 2020;11:2474. doi: 10.3389/fimmu.2020.583373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agrawal S., Goel A.D., Gupta N. Emerging prophylaxis strategies against COVID-19. Monaldi Arch. Chest Dis. 2020;90 doi: 10.4081/monaldi.2020.1289. [DOI] [PubMed] [Google Scholar]
- 32.da Silva Santos Y., Gamon T.H.M., de Azevedo M.S.P., Telezynski B.L., de Souza E.E., de Oliveira D.B.L., Dombrowski J.G., Rosa-Fernandes L., Palmisano G., de Moura Carvalho L.J., et al. Virulence Profiles of Wild-Type, P. 1 and Delta SARS-CoV-2 Variants in K18-hACE2 Transgenic Mice. Viruses. 2023;15:999. doi: 10.3390/v15040999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nair A.B., Jacob S. A simple practice guide for dose conversion between animals and human. J. Basic Clin. Pharm. 2016;7:27–31. doi: 10.4103/0976-0105.177703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lehmann M., Allers K., Heldt C., Meinhardt J., Schmidt F., Rodriguez-Sillke Y., Kunkel D., Schumann M., Böttcher C., Stahl-Hennig C., et al. Human small intestinal infection by SARS-CoV-2 is characterized by a mucosal infiltration with activated CD8+ T cells. Mucosal Immunol. 2021;14:1381–1392. doi: 10.1038/s41385-021-00437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arostegui D., Castro K., Schwarz S., Vaidy K., Rabinowitz S., Wallach T. Persistent SARS-CoV-2 nucleocapsid protein presence in the intestinal epithelium of a pediatric patient 3 months after acute infection. JPGN Rep. 2022;3:e152. doi: 10.1097/PG9.0000000000000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi J., Park J.E., Tsagkogeorga G., Yanagita M., Koo B.K., Han N., Lee J.H. Inflammatory Signals Induce AT2 Cell-Derived Damage-Associated Transient Progenitors that Mediate Alveolar Regeneration. Cell Stem Cell. 2020;27:366–382.e7. doi: 10.1016/j.stem.2020.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strunz M., Simon L.M., Ansari M., Kathiriya J.J., Angelidis I., Mayr C.H., Tsidiridis G., Lange M., Mattner L.F., Yee M., et al. Alveolar regeneration through a Krt8+ transitional stem cell state that persists in human lung fibrosis. Nat. Commun. 2020;11:3559. doi: 10.1038/s41467-020-17358-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Halfmann P.J., Iida S., Iwatsuki-Horimoto K., Maemura T., Kiso M., Scheaffer S.M., Darling T.L., Joshi A., Loeber S., Singh G., et al. SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters. Nature. 2022;603:687–692. doi: 10.1038/s41586-022-04441-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vora S.M., Lieberman J., Wu H. Inflammasome activation at the crux of severe COVID-19. Nat. Rev. Immunol. 2021;21:694–703. doi: 10.1038/s41577-021-00588-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eltobgy M.M., Zani A., Kenney A.D., Estfanous S., Kim E., Badr A., Carafice C., Daily K., Whitham O., Pietrzak M., et al. Caspase-4/11 exacerbates disease severity in SARS-CoV-2 infection by promoting inflammation and immunothrombosis. Proc. Natl. Acad. Sci. USA. 2022;119 doi: 10.1073/pnas.2202012119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sefik E., Qu R., Junqueira C., Kaffe E., Mirza H., Zhao J., Brewer J.R., Han A., Steach H.R., Israelow B., et al. Inflammasome activation in infected macrophages drives COVID-19 pathology. bioRxiv. 2022 doi: 10.1101/2021.09.27.461948. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Junqueira C., Crespo Â., Ranjbar S., de Lacerda L.B., Lewandrowski M., Ingber J., Parry B., Ravid S., Clark S., Schrimpf M.R., et al. FcγR-mediated SARS-CoV-2 infection of monocytes activates inflammation. Nature. 2022;606:576–584. doi: 10.1038/s41586-022-04702-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Doitsh G., Galloway N.L.K., Geng X., Yang Z., Monroe K.M., Zepeda O., Hunt P.W., Hatano H., Sowinski S., Muñoz-Arias I., Greene W.C. Cell death by pyroptosis drives CD4 T-cell depletion in HIV-1 infection. Nature. 2014;505:509–514. doi: 10.1038/nature12940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Islam T., Hasan M., Rahman M.S., Islam M.R. Comparative evaluation of authorized drugs for treating Covid-19 patients. Health Sci. Rep. 2022;5 doi: 10.1002/hsr2.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fan H., Lou F., Fan J., Li M., Tong Y. The emergence of powerful oral anti-COVID-19 drugs in the post-vaccine era. Lancet. Microbe. 2022;3:e91. doi: 10.1016/S2666-5247(21)00278-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hammond J., Leister-Tebbe H., Gardner A., Abreu P., Bao W., Wisemandle W., Baniecki M., Hendrick V.M., Damle B., Simón-Campos A., et al. Oral Nirmatrelvir for High-Risk, Nonhospitalized Adults with Covid-19. N. Engl. J. Med. 2022;386:1397–1408. doi: 10.1056/NEJMoa2118542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hashemian S.M.R., Sheida A., Taghizadieh M., Memar M.Y., Hamblin M.R., Bannazadeh Baghi H., Sadri Nahand J., Asemi Z., Mirzaei H. Paxlovid (Nirmatrelvir/Ritonavir): A new approach to Covid-19 therapy? Biomed. Pharmacother. 2023;162 doi: 10.1016/j.biopha.2023.114367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cohen A.K., Jaudon T.W., Schurman E.M., Kava L., Vogel J.M., Haas-Godsil J., Lewis D., Crausman S., Leslie K., Bligh S.C., et al. Impact of extended-course oral nirmatrelvir/ritonavir (Paxlovid) in established Long COVID: Case series and research considerations. Res. Sq. 2023 doi: 10.21203/rs.3.rs-3359429/v1. [DOI] [Google Scholar]
- 49.Illingworth C., Guerra-Assuncao J.A., Gregg S., Charles O.J., Pang J., Roy S., Abdelnabi R., Neyts J., Breuer J. Genetic consequences of effective and suboptimal dosing with mutagenic drugs in a hamster model of SARS-CoV-2 infection. bioRxiv. 2023 doi: 10.1101/2023.02.20.529243. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Callaway E. COVID drug drives viral mutations - and now some want to halt its use. Nature. 2023;614:399. doi: 10.1038/d41586-023-00347-z. [DOI] [PubMed] [Google Scholar]
- 51.Bobrowski T., Chen L., Eastman R.T., Itkin Z., Shinn P., Chen C.Z., Guo H., Zheng W., Michael S., Simeonov A., et al. Synergistic and Antagonistic Drug Combinations against SARS-CoV-2. Mol. Ther. 2021;29:873–885. doi: 10.1016/j.ymthe.2020.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu Y., Shrestha N., Préat V., Beloqui A. An overview of in vitro, ex vivo and in vivo models for studying the transport of drugs across intestinal barriers. Adv. Drug Deliv. Rev. 2021;175 doi: 10.1016/j.addr.2021.05.005. [DOI] [PubMed] [Google Scholar]
- 53.Abdelnabi R., Foo C.S., Kaptein S.J.F., Zhang X., Do T.N.D., Langendries L., Vangeel L., Breuer J., Pang J., Williams R., et al. The combined treatment of Molnupiravir and Favipiravir results in a potentiation of antiviral efficacy in a SARS-CoV-2 hamster infection model. EBioMedicine. 2021;72 doi: 10.1016/j.ebiom.2021.103595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jeong J.H., Chokkakula S., Min S.C., Kim B.K., Choi W.S., Oh S., Yun Y.S., Kang D.H., Lee O.J., Kim E.G., et al. Combination therapy with nirmatrelvir and molnupiravir improves the survival of SARS-CoV-2 infected mice. Antivir. Res. 2022;208 doi: 10.1016/j.antiviral.2022.105430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rosenke K., Lewis M.C., Feldmann F., Bohrnsen E., Schwarz B., Okumura A., Bohler W.F., Callison J., Shaia C., Bosio C.M., et al. Combined molnupiravir-nirmatrelvir treatment improves the inhibitory effect on SARS-CoV-2 in macaques. JCI Insight. 2023;8 doi: 10.1172/jci.insight.166485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.The RECOVERY Collaborative Group Dexamethasone in hospitalized patients with Covid-19. N. Engl. J. Med. Overseas. Ed. 2021;384:693–704. doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Richardson S.I., Manamela N.P., Motsoeneng B.M., Kaldine H., Ayres F., Makhado Z., Mennen M., Skelem S., Williams N., Sullivan N.J., et al. SARS-CoV-2 Beta and Delta variants trigger Fc effector function with increased cross-reactivity. Cell Rep. Med. 2022;3 doi: 10.1016/j.xcrm.2022.100510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grunst M.W., Uchil P.D. Fc effector cross-reactivity: A hidden arsenal against SARS-CoV-2's evasive maneuvering. Cell Rep. Med. 2022;3 doi: 10.1016/j.xcrm.2022.100540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yamin R., Jones A.T., Hoffmann H.H., Schäfer A., Kao K.S., Francis R.L., Sheahan T.P., Baric R.S., Rice C.M., Ravetch J.V., Bournazos S. Fc-engineered antibody therapeutics with improved anti-SARS-CoV-2 efficacy. Nature. 2021;599:465–470. doi: 10.1038/s41586-021-04017-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stein S.R., Ramelli S.C., Grazioli A., Chung J.Y., Singh M., Yinda C.K., Winkler C.W., Sun J., Dickey J.M., Ylaya K., et al. SARS-CoV-2 infection and persistence in the human body and brain at autopsy. Nature. 2022;612:758–763. doi: 10.1038/s41586-022-05542-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Etter M.M., Martins T.A., Kulsvehagen L., Pössnecker E., Duchemin W., Hogan S., Sanabria-Diaz G., Müller J., Chiappini A., Rychen J., et al. Severe Neuro-COVID is associated with peripheral immune signatures, autoimmunity and neurodegeneration: a prospective cross-sectional study. Nat. Commun. 2022;13:6777. doi: 10.1038/s41467-022-34068-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Crunfli F., Carregari V.C., Veras F.P., Silva L.S., Nogueira M.H., Antunes A.S.L.M., Vendramini P.H., Valença A.G.F., Brandão-Teles C., Zuccoli G.D.S., et al. Morphological, cellular, and molecular basis of brain infection in COVID-19 patients. Proc. Natl. Acad. Sci. USA. 2022;119 doi: 10.1073/pnas.2200960119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Douaud G., Lee S., Alfaro-Almagro F., Arthofer C., Wang C., McCarthy P., Lange F., Andersson J.L.R., Griffanti L., Duff E., et al. SARS-CoV-2 is associated with changes in brain structure in UK Biobank. Nature. 2022;604:697–707. doi: 10.1038/s41586-022-04569-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mao L., Jin H., Wang M., Hu Y., Chen S., He Q., Chang J., Hong C., Zhou Y., Wang D., et al. Neurologic Manifestations of Hospitalized Patients With Coronavirus Disease 2019 in Wuhan, China. JAMA Neurol. 2020;77:683–690. doi: 10.1001/jamaneurol.2020.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Honko A.N., Storm N., Bean D.J., Henao Vasquez J., Downs S.N., Griffiths A. Rapid Quantification and Neutralization Assays for Novel Coronavirus SARS-CoV-2 Using Avicel RC-591 Semi-Solid Overlay. Preprints.org. 2020 doi: 10.20944/preprints202005.0264.v1. [DOI] [Google Scholar]
- 66.Amarilla A.A., Sng J.D.J., Parry R., Deerain J.M., Potter J.R., Setoh Y.X., Rawle D.J., Le T.T., Modhiran N., Wang X., et al. A versatile reverse genetics platform for SARS-CoV-2 and other positive-strand RNA viruses. Nat. Commun. 2021;12:3431. doi: 10.1038/s41467-021-23779-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Perreault J., Tremblay T., Fournier M.J., Drouin M., Beaudoin-Bussières G., Prévost J., Lewin A., Bégin P., Finzi A., Bazin R. Waning of SARS-CoV-2 RBD antibodies in longitudinal convalescent plasma samples within 4 months after symptom onset. Blood. 2020;136:2588–2591. doi: 10.1182/blood.2020008367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen Y., Prévost J., Ullah I., Romero H., Lisi V., Tolbert W.D., Grover J.R., Ding S., Gong S.Y., Beaudoin-Bussières G., et al. Molecular basis for antiviral activity of two pediatric neutralizing antibodies targeting SARS-CoV-2 Spike RBD. iScience. 2023;26 doi: 10.1016/j.isci.2022.105783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xiao A.T., Tong Y.X., Zhang S. Profile of RT-PCR for SARS-CoV-2: a preliminary study from 56 COVID-19 patients. Clin. Infect. Dis. 2020;71:2249–2251. doi: 10.1093/cid/ciaa460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Panel COVID-19 Treatment Guidelines-Molnupiravir. https://www.covid19treatmentguidelines.nih.gov/therapies/antivirals-including-antibody-products/molnupiravir/
- 71.Panel Treament guidelines for Paxlovid. https://www.covid19treatmentguidelines.nih.gov/therapies/antivirals-including-antibody-products/ritonavir-boosted-nirmatrelvir--paxlovid-/
- 72.Ianevski A., Giri A.K., Aittokallio T. SynergyFinder 2.0: visual analytics of multi-drug combination synergies. Nucleic Acids Res. 2020;48:W488–W493. doi: 10.1093/nar/gkaa216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Raw data used to generate all the graphs shown in the manuscript have been deposited at Mendeley Data, V2, https://doi.org/10.17632/n9ztbbbstm.2 and are publicly available as of the 29 Nov 2023. DOIs are listed in the key resources table.
-
•
This paper does not report the original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from lead contact upon request.