Abstract
DNAs from bacteria and variety of nonvertebrate organisms, including nematodes, mollusks, yeasts, and insects, cause polyclonal activation of murine B lymphocytes. Similar studies have not been reported for bovine B cells, and to date no studies have reported mitogenic properties of protozoal DNA for any species. However, we and others have observed that protozoal parasite antigens can induce the proliferation of lymphocytes from nonexposed donors. Extending these studies, we now show that the mitogenic property of protozoal antigen preparations is in part attributable to parasite DNA and that Babesia bovis DNA is directly mitogenic for bovine B cells. DNase treatment of B. bovis extracts abrogated B. bovis-induced proliferation of peripheral blood mononuclear cells from nonexposed cattle. Like DNAs from other organisms that were mitogenic for murine B cells, B. bovis DNA is largely nonmethylated and induced a dose-dependent proliferation of bovine B cells, which was reduced upon methylation. Furthermore, B. bovis and E. coli DNAs enhanced immunoglobulin secretion by cultured B cells, inducing moderate increases in immunoglobulin G1 and stronger increases in immunoglobulin G2. Because certain nonmethylated CpG motifs present in bacterial DNA are known to stimulate proliferation of murine and human B cells, an 11-kb fragment of B. bovis DNA was analyzed for CG dinucleotide content and for the presence of known immunostimulatory sequences (ISS) centered on a CG motif. The frequency of CG dinucleotides was approximately one-half of the expected frequency, and several CpG hexameric sequences with known activity for murine B cells were identified. An oligodeoxynucleotide containing one of these ISS (AACGTT), which is present within the rhoptry-associated protein-1 (rap-1) open reading frame, was shown to stimulate B-cell proliferation. These ISS may be involved in host immune modulation during protozoal infection and may be useful as vaccine adjuvants.
The mitogenic properties of bacterial DNA include its ability to stimulate murine B cells to proliferate and secrete antibody (33) and its ability to activate macrophages to secrete cytokines (interleukin-6 [IL-6], IL-12, IL-18, alpha interferon [IFN-α], and tumor necrosis factor alpha [TNF-α]) involved in inflammation and promotion of a type-1 immune response (reviewed in reference 42). Additional effects of bacterial DNA include its induction of IL-1β and inducible nitric oxide synthase (iNOS) in IFN-γ-treated macrophages (52). Specific sequences present in microbial DNA, consisting of a nonmethylated CG core flanked by two 5′ purines and two 3′ pyrimidines, are largely responsible for its mitogenic properties. Mammalian DNA, which is predominantly methylated and has a suppressed frequency of CG dinucleotides (4), is not mitogenic. However, DNAs from a variety of nonvertebrate organisms, including insects (e.g., Drosophilia melanogaster), yeasts (e.g., Schizosaccharomyces pombe), nematodes (e.g., Caenorhabditis elegans), and mollusks (e.g., Mytilus edulis) have mitogenic properties for murine B cells that are similar to those of bacterial DNA, which correlated with the presence of nonmethylated CG dinucleotides (57, 58).
Protozoal parasites and crude antigenic fractions stimulate proliferation of peripheral blood mononuclear cells (PBMC) obtained from nonexposed donors, but the mitogenic components have not been completely defined. Examples include the stimulation of PBMC by Theileria parva (24, 40, 41), Plasmodium falciparum (15, 23, 29), Leishmania sp. (31), and Trypanosoma cruzi (44). We have similarly observed nonspecific proliferation of bovine PBMC in response to a membrane-enriched subcellular fraction prepared from Babesia bovis merozoites cultured in bovine erythrocytes (11, 12). The recent finding that DNAs from many types of nonvertebrate organisms are mitogenic for B cells led us to test the hypothesis that the stimulation of PBMC from naive donors by protozoal extracts is also in part attributable to DNA.
This study is the first to demonstrate the mitogenic properties of protozoal DNA for mammalian leukocytes. We show that B. bovis DNA is largely nonmethylated and stimulates B-cell proliferation and immunoglobulin G (IgG) secretion. Furthermore, B. bovis DNA contains CpG immunostimulatory sequences (ISS). We identify a potential mechanism by which protozoal parasites modulate host immune responses, and our results support the use of ISS as vaccine adjuvants to enhance type-1 immune responses in cattle.
MATERIALS AND METHODS
B-lymphocyte purification.
B cells were purified from bovine PBMC by negative selection by using a modified panning procedure (21, 50) or by positive selection with anti-bovine CD21-coated magnetic beads (62). For negative selection, macrophages were removed by the addition of 15 μl of a 4% carbonyl iron suspension in sterile phosphate-buffered saline (PBS) to 60 ml of blood collected in 2 ml of EDTA (0.5 M, pH 8.0) and incubated at 37°C for 30 min with gentle agitation. PBMC were isolated by Histopaque (Sigma Chemical Co., St. Louis, Mo.) density centrifugation, washed twice in Alsever’s solution (Sigma), and resuspended in panning solution (3% bovine serum albumin [BSA] fraction V [Sigma] in Hanks balanced salt solution, pH 7.4, with 0.9 mM Mg2+ and 1.25 mM Ca2+). After centrifugation at 250 × g for 10 min at 10°C, the cells were resuspended at a concentration of 107 cells per ml in panning solution, and 9 ml of cell suspension was placed in a T-75 flask (Corning, Cambridge, Mass.) and allowed to adhere at room temperature for 1 h, with gentle swirling after 30 min. The nonadherent cells were removed, and after careful rinsing with complete RPMI 1640 medium (11) the adherent, enriched B-cell population was collected by vigorous agitation. The cells were washed and resuspended in Hanks balanced salt solution, and CD3+ T cells were removed after incubation of 107 cells per ml with 15 μg of sodium azide-free bovine CD3-specific monoclonal antibody (MAb) MM1A per ml (kindly provided by William C. Davis, Washington State University, Pullman) for 30 min at 4°C, incubation with goat anti-mouse IgG-coated magnetic beads (Dynabead M-450; Dynal, Inc., Lake Success, N.Y.), and removal of bead-bound cells with a magnet according to the manufacturer’s protocol. The remaining cells were washed in complete RPMI 1640, and aliquots were removed for cell surface phenotype analysis.
Positive selection of B cells from PBMC was performed essentially as described previously (62). Briefly, B lymphocytes were isolated from PBMC by using anti-bovine CD21 MAb GB25A and goat anti-mouse IgG-coated magnetic beads (Dynal) according to the manufacturer’s instructions. MAb GB25A was purchased from the Washington State University Monoclonal Antibody Center. MAb GB25A was added at 0.25 μg per 106 PBMC, and magnetic beads were added at 4.5 × 105 beads per 106 PBMC.
Flow cytometric analysis of purified B cells.
Cell surface phenotype analysis was performed by using flow cytometry as described earlier (7) and MAb (15 μg/ml) specific for bovine CD2 (MUC 2A), CD3 (MM1A), CD4 (CACT 138A), CD8 α and β chains (CACT 80C and BAT 82A), γδ TcR1-N12 (CACT-61A), and CD14 (CAM36A). These MAbs were kindly provided by William C. Davis. A MAb (IL-A24) that stains a molecule present on dendritic cells and macrophages and monocytes (20) was obtained from the International Laboratory for Research on Animal Diseases, Nairobi, Kenya). Fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse immunoglobulin (a mixture of IgG, IgA, and IgM affinity-purified F(ab′)2 fragments; Cappel/Organon Teknika, Malvern, Pa.) was used as a secondary antibody, and background staining was indicated by staining with this antibody alone. FITC-labeled, affinity-purified (Fab′)2 goat anti-bovine IgG (Fab′)2-specific antibody (Jackson Immunoresearch Laboratories, Inc., Avondale, Pa.) was used (50 μg/ml) to label the B cells. After the negative selection panning procedure, there were no residual cells that expressed TcR1, CD3, CD4, CD8, CD14, or the molecule recognized by IL-A24, indicating that T cells, monocytes and macrophages, and dendritic cells were depleted. The majority (ca. 90%) of the panned cells expressed surface immunoglobulin (sIg), indicating the purity of the B-cell population, and in some experiments 2 to 10% of the cells expressed CD2 in the absence of other surface markers. After positive selection with anti-CD21-coated beads, approximately 94% of the cells expressed sIg, and less than 2% of the recovered B cells expressed TcR1, CD2, CD3, CD4, CD8, or CD14.
Preparation of DNA.
Calf thymus DNA and lyophilized Escherichia coli were purchased from Sigma. Lyophilized E. coli (1 g) was resuspended in 6 ml of TE buffer (100 mM NaCl, 10 mM Tris-HCl [pH 8.0], 25 mM EDTA [pH 8.0]) containing 10 mg of lysozyme per ml (Sigma) and incubated at 37°C for 1.5 h; an additional 4 ml of lysozyme in TE buffer was added for the last 30 min of incubation. The mixture was then diluted to 20 ml with TE buffer, proteinase K (Sigma) was added to a final concentration 0.5 mg/ml, and sodium dodecyl sulfate (SDS) was added to a final concentration of 0.6% (vol/vol). The mixture was incubated at 50°C for 1 h. The volume was then increased to 25 ml with TE buffer, proteinase K was added to a final concentration of 2 mg/ml, and SDS was added to a final concentration of 0.6% (vol/vol). The mixture was incubated at 50°C overnight with continuous shaking. DNA was extracted once with phenol, seven times with phenol-chloroform-isoamyl alcohol, and once with chloroform-isoamyl alcohol. E. coli DNA was precipitated with 3 M sodium acetate (pH 5.0) and absolute ethanol. The pellet was washed once with 70% ethanol and allowed to air dry. B. bovis DNA was prepared from the Texas or Mexico strains that were maintained in continuous culture in bovine erythrocytes (11). Merozoites were freed from erythrocytes as described earlier (11) and washed in PBS. DNA was prepared from merozoites that were resuspended in digestion buffer containing 100 mM NaCl, 10 mM Tris-HCl (pH 8.0), 25 mM EDTA (pH 8.0), 0.5% (vol/vol) sodium dodecyl sulfate, and 0.1 mg of proteinase K per ml and then incubated with shaking at 50°C overnight. The DNA was extracted once with phenol, twice with phenol-chloroform-isoamyl alcohol, and once with chloroform-isoamyl alcohol; it was then precipitated with 3 M ammonium acetate (pH 5.0) and absolute ethanol and washed with 70% ethanol, and the pellet was allowed to air dry. The E. coli and B. bovis DNA pellets were dissolved in PBS overnight at 37°C. The DNA concentrations were determined spectrophotometrically by optical density analysis. Immediately prior to assay, the DNAs were denatured (single stranded) by heating to 95°C for 5 min and then chilled on ice for 5 min before dilution in complete RPMI 1640 medium or DNase I buffer (33).
DNase treatment of B. bovis antigen and DNA.
B. bovis merozoite crude membrane (CM) antigen was prepared as described previously (11) by rupturing free merozoites with a French pressure cell at 1,500 lb/in2, pelleting the merozoite organelles and membranes by centrifugation at 145,000 × g for 1 h, and resuspending the pellet in PBS. CM antigen was incubated for 2 h at 37°C at a concentration of 1 mg of protein per ml of DNase I buffer containing 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, and 0.5 mg of BSA per ml with a final concentration of 1 mg of DNase I (Sigma) per ml. Purified DNA (150 to 200 μg/ml) was similarly treated with DNase for 2 h at 37°C and then stored at −20°C until use. Prior to use in proliferation assays, the DNA concentration was determined again by spectrophotometric analysis. Complete digestion of the DNA was confirmed by agarose gel electrophoresis. E. coli and B. bovis DNA contained <12 pg of endotoxin per ml when tested by the Limulus amebocyte lysate assay (Whittaker M. A. Bioproducts, Walkersville, Md.).
Methylation of E. coli and B. bovis DNA.
DNA was methylated essentially as described for Drosophila DNA (58). CpG methylase (SssI methylase) and HpaII restriction endonuclease were purchased from New England BioLabs (Beverly, Mass.), and methylation was performed according to the manufacturer’s instructions. DNA was treated with SssI methylase (1 to 2 U/μg of DNA) in NEB2 buffer (New England BioLabs) supplemented with S-adenosylmethionine at 37°C, with replenishment of S-adenosylmethionine every 4 h. Aliquots were removed periodically, and the extent of methylation was determined by the measuring the resistance of the treated DNA samples to cleavage by HpaII restriction endonuclease. Aliquots of untreated or methylated DNAs were digested with HpaII for 1 h at 37°C and electrophoresed on 1% agarose gels. Complete methylation was achieved after incubation with SssI methylase for 24 h. DNA was then extracted twice with phenol-chloroform-isoamyl alcohol and once with chloroform-isoamyl alcohol, precipitated with 3 M sodium acetate and absolute ethanol, washed with 70% ethanol, air dried, and dissolved in PBS. The concentration was determined by spectrophotometric analysis.
Oligodeoxynucleotides.
Oligodeoxynucleotides were designed by using published sequences (33, 55) and purchased from Operon Technologies (Alameda, Calif.). Phosphorothioate-modified oligodeoxynucleotides were purchased from Oligos Etc. (Wilsonville, Oreg.). Each oligonucleotide was further purified by precipitation with 3 M sodium acetate and absolute ethanol, air dried, and dissolved in PBS for use in B-cell proliferation assays. All oligonucleotides (20 to 40 μM concentrations) were negative for endotoxin contamination when tested by the Limulus amebocyte lysate assay, which has a sensitivity of 3.0 pg of endotoxin/ml (Whittaker).
Lymphocyte proliferation assays.
PBMC were obtained from three head of cattle (B1, B2, and C2) with no known exposure to B. bovis and from a cow (C15) that was immunized by three inoculations of cultured B. bovis merozoites (11). PBMC were cultured in triplicate wells at a density of 2 × 106 cells per ml in 100-μl volumes in 96-well round-bottomed plates (Costar, Cambridge, Mass.) in complete RPMI 1640 medium with 1 to 25 μg of untreated or DNase-treated B. bovis CM antigen per ml or 10 μg of concanavalin A (ConA; Sigma) per ml for 3 or 6 days (11). Purified B cells were cultured for 72 h in duplicate or triplicate at a density of 2 × 106 cells per ml in 100-μl volumes in complete RPMI 1640 medium with 1 to 25 μg of pokeweed mitogen (PWM; Sigma) per ml, 0.25 to 50 μg of DNA purified from E. coli (Sigma) per ml, 1 to 50 μg of calf thymus DNA (Sigma) per ml, or 1 to 100 μg of DNA purified from the Mexico or Texas strains of B. bovis parasites per ml. DNAs were tested alone or after treatment with DNase I. As a control, buffer containing DNase I, equal in amount to that present in 25 μg of B. bovis CM antigen per ml, was added to cultures of PBMC with ConA or to B cells with PWM to rule out any toxic effect of the DNase on cellular proliferation. In some experiments, polymyxin B sulfate (Sigma) was added at a final concentration of 10 μg/ml. Oligonucleotides were added to B-cell proliferation assays at a concentration of 0.15 to 40 μM. B cells were cultured for 48 or 72 h and either radiolabeled for the last 6 to 18 h of culture with 0.25 μCi of [3H]thymidine (New England Nuclear, Boston, Mass.) or, for assays with oligonucleotides, radiolabeled with 0.25 to 0.5 μCi of [3H]uridine (New England Nuclear) (33). The cells were then harvested and counted in a liquid scintillation counter. Results are presented as the mean counts per minute (cpm) of replicate cultures ± 1 standard deviation (SD) or as the stimulation index (SI), which was determined by dividing the mean cpm in replicate cultures of B lymphocytes with mitogen, DNA, or oligonucleotide by the mean cpm in replicate control cultures of B lymphocytes with medium. Proliferation was analyzed for statistical significance by Student’s one-tailed t test.
Mycoplasma detection assay.
Aliquots of B. bovis CM antigen, soluble, cytosolic (HSS) antigen, and purified B. bovis DNA were tested for Mycoplasma by PCR with a Mycoplasma primer set (Stratagene, La Jolla, Calif.) according to the manufacturer’s protocol. More than 40 samples collected over a period of several years were found to be negative for the species of Mycoplasma detected by this PCR assay. Positive and negative controls included in the assays verified the accuracy and sensitivity of the assay.
Analysis of IgG secreted by B cells stimulated with DNA.
Purified B cells were cultured in duplicate wells of 96-well round-bottomed plates at a density of 4 × 105 or 1.6 × 106 cells per ml in 250-μl volumes for 6 days in complete RPMI 1640 medium alone or with 10 μg of PWM, 25 μg of E. coli DNA, or 50 μg of B. bovis DNA per ml in the absence or presence of 20 U of recombinant human IL-2 (Boehringer Mannheim, Indianapolis, Ind.), 20 ng of recombinant bovine IL-4 (22), or 5 ng of recombinant bovine IFN-γ (kindly provided by Lorne Babiuk, VIDO, Sasketoon, Saskatchewan, Canada) per ml. The bovine IFN-γ had a biological activity of approximately 0.6 U/ng when assayed for neutralization of vesicular stomatitis virus (53). Supernatants were harvested after centrifugation of the plates at 250 × g for 10 min and were stored at −20°C. Prior to the assay, supernatants were again centrifuged (5,000 × g for 10 min) to pellet any cellular debris. Each duplicate sample was analyzed two or more times by sandwich capture enzyme-linked immunosorbent assay (ELISA) to detect IgM, IgG1, and IgG2 by using reagents specific for these immunoglobulin classes and subclasses as described earlier (21, 22). To detect IgM, a symmetrical sandwich capture ELISA was performed as described previously (22). Briefly, 96-well U-bottom immunoassay plates (Dynatech) were coated overnight at 4°C with 1 μg of goat anti-bovine IgM (Kirkegaard & Perry Laboratories, Gaithersburg, Md.) diluted in PBS per well. The plates were washed three times, blocked for 1 h at 37°C with 10% horse serum (GIBCO-BRL, Grand Island, N.Y.) diluted in PBS, and washed another three times; culture supernatants diluted 1:2 to 1:10 in PBS were then added to the wells in triplicate and incubated for 1 h at 37°C. To measure IgG1 and IgG2, plates were coated with 1 μg of goat anti-mouse IgG (Kirkegaard & Perry) in PBS per well overnight, washed three times in PBS, and blocked with 10% horse serum in PBS for 1 h at 37°C. Mouse anti-bovine IgG1 (1:500 dilution) or anti-bovine IgG2 (1:1,000 dilution) MAb purchased from Serotec, Ltd. (Oxford, United Kingdom) was added and incubated for 1 h at 37°C. Test supernatants diluted in 5% IgG-free normal horse serum (GIBCO-BRL) in PBS were added, and the plates were incubated for 1 h at 37°C. The plates were then washed, and alkaline phosphate-conjugated goat anti-bovine IgM (Kirkegaard & Perry) or alkaline phosphate-conjugated goat anti-bovine IgG (Kirkegaard & Perry), which had been previously adsorbed against mouse IgG, was added. The plates were incubated for 1 h at 37°C and then washed; substrate was then added for 1 h at 37°C. The reaction was developed by using a kit supplied by Kirkegaard & Perry according to the manufacturer’s instructions, and the optical density was determined with a Dynatech MR5000 ELISA plate reader at 405 nm. In each assay plate, immunoglobulin standards were included that consisted of purified bovine IgM (Sigma) or protein G affinity-purified bovine IgG1 and IgG2 (Jackson Immunoresearch, Westgrove, Pa.). Results are presented as the mean concentration in nanograms per milliliter ± 1 SD of IgM, IgG1, or IgG2 in duplicate cultures of B cells. The levels of secreted immunoglobulin were analyzed for statistical significance by Student’s one-tailed t test.
Analysis of B. bovis genomic DNA.
An 11-kb fragment of genomic DNA obtained from a λEMBL BamHI genomic library prepared from the MO7 biological clone of the Mexico strain of B. bovis, which contains a total of five open reading frames (ORF), including two encoding identical RAP-1 genes, was recently described (GenBank accession number AF027149 [55]). This 11-kb sequence was analyzed for the presence of CG dinucleotides and for specific AACGTT hexamers with the GCG (version 8.0) software package (17).
RESULTS
Nonspecific stimulation of lymphocyte proliferation by B. bovis merozoite antigen is DNase sensitive.
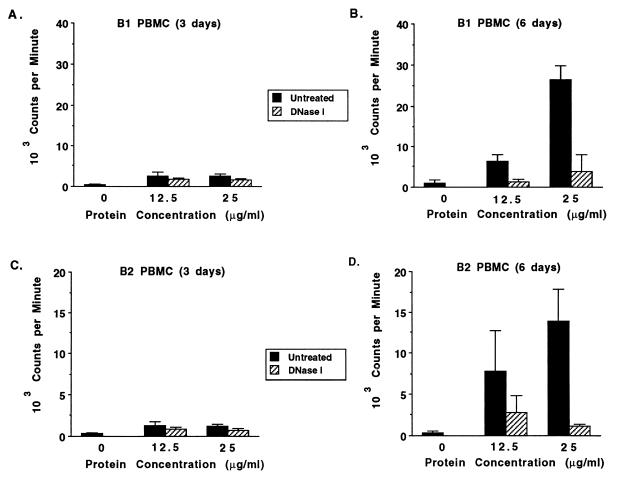
Over the course of our studies examining antigen-specific proliferation of B. bovis-immune and control PBMC, we observed nonspecific proliferation by PBMC from nonexposed cattle when cultured with the B. bovis merozoite antigen. Unlike the proliferative response to a T-cell mitogen, such as ConA, which is maximal at 2 to 3 days poststimulation (11), the response to B. bovis antigen by nonimmune PBMC peaked at 6 days poststimulation (data for two cows are shown in Fig. 1). The response by nonimmune PBMC was abrogated by treatment with DNase (Fig. 1B and D), whereas the proliferation of antigen-specific PBMC obtained from B. bovis-immune cow C15 (11) was only partially inhibited by DNase (data not shown). Nonspecific inhibition by either DNase or the buffer was ruled out, since the addition of equivalent amounts of these reagents to PBMC stimulated with mitogenic doses of ConA had no effect (data not shown). Similarly, the addition of DNase to mitogenic doses of PWM did not inhibit B-cell proliferation (data not shown).
FIG. 1.
Nonspecific stimulation by unfractionated B. bovis merozoite antigen is DNase sensitive. PBMC from two calves never exposed to babesial parasites were cultured for 3 (A and C) or 6 (B and D) days with medium alone (0) or 12.5 or 25 μg of B. bovis CM antigen per ml that was untreated (solid bars) or DNase I treated (striped bars), and proliferation was determined. Results are expressed as the mean cpm ± 1 SD of triplicate cultures.
Stimulation of B-lymphocyte proliferation by B. bovis and E. coli DNA.
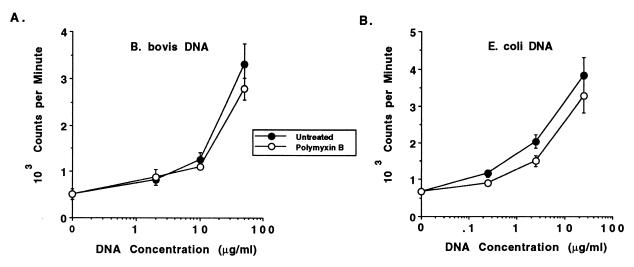
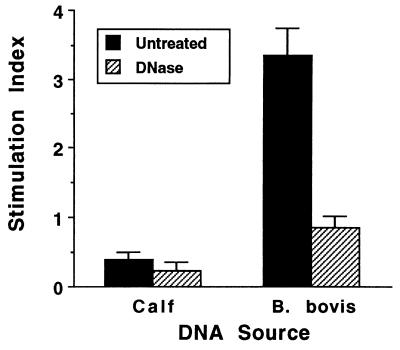
DNAs from a variety of different organisms, including bacteria, insects, yeasts, nematodes, and mollusks, have been shown to have mitogenic properties for murine B cells (33, 43, 57, 58). However, bacterial DNA was not mitogenic for human B cells, even though stimulation by selected phosphorothioate-modified oligonucleotides has been observed (5, 35), and to date there have been no reports of the capability of protozoal DNA to stimulate B lymphocytes from any species. To extend the studies with mitogenic nonvertebrate DNA from mice to an outbred species and to determine if DNA purified from B. bovis merozoites could directly activate B lymphocytes from the natural host of this protozoal parasite, bovine B lymphocytes were purified from PBMC of cattle not exposed to B. bovis and were assayed for stimulation with DNA purified from E. coli (positive control), calf thymus (negative control), or B. bovis. In eight experiments with B cells purified from PBMC by negative selection from donor cattle and DNA prepared from either the Mexico or the Texas strain of B. bovis, we detected a dose-dependent proliferative response to B. bovis DNA (a representative experiment is presented in Fig. 2). The SI values ranged from 3.0 to 6.5 when 25 to 50 μg of B. bovis DNA per ml was used. Treatment of B. bovis DNA with DNase I completely abrogated stimulation, as shown in Fig. 3, which represents the mean of three independent experiments. Calf thymus DNA did not stimulate bovine B cells at any of the concentrations from 0.2 to 50 μg/ml (data not shown and Fig. 3). E. coli DNA was generally more effective at inducing proliferation than was B. bovis DNA, since higher responses were typically observed with lower concentrations of E. coli DNA (Fig. 2). Significant endotoxin contamination of E. coli and B. bovis DNA was ruled out by the Limulus assay, which revealed the presence of <12 pg of endotoxin in 50 μg of DNA. This concentration of endotoxin is far below a mitogenic one, since in proliferation assays performed with bovine PBMC or B cells, LPS was not mitogenic at 100 ng/ml (data not shown). Furthermore, inclusion of 10 μg of the LPS inhibitor polymyxin B sulfate per ml with either E. coli or B. bovis DNA had no effect on the stimulation of B-cell proliferation (Fig. 2). At this concentration polymyxin B sulfate inhibits up to 1 μg of E. coli LPS per ml when tested for the induction of proliferation of bovine PBMC (data not presented). Potential contamination of B. bovis parasite cultures with Mycoplasma sp., which was recently described for numerous stocks of cultured malarial parasites (63), was also ruled out. More than 25 samples of B. bovis (Texas and Mexico strains) merozoite antigen preparations collected over a 1-year period and four samples of B. bovis DNA obtained from parasites cultured during this time were tested for the presence of Mycoplasma sp. All of the samples were negative by PCR with primers that detect several different species of Mycoplasma (reference 54 and data not shown).
FIG. 2.
Dose-dependent proliferation of B cells to DNA prepared from B. bovis or E. coli is not inhibited by polymyxin B. Negatively selected B cells were cultured for 3 days with medium or 2, 10, or 50 μg of B. bovis DNA per ml (A) or 0.25, 2.5, or 25 μg of E. coli DNA per ml (B) without (solid circles) or with (open circles) 10 μg of polymyxin B sulfate per ml. Results are presented as the mean cpm ± 1 SD of duplicate cultures.
FIG. 3.
Stimulation of B-cell proliferation by B. bovis DNA is abrogated by DNase treatment. Negatively selected B cells were cultured in duplicate wells for 3 days with medium or 50 μg of either calf thymus DNA or B. bovis DNA per ml that was untreated (solid bars) or treated (striped bars) with DNase I. Results are presented as the SI and represent the mean ± 1 SD of three independent experiments.
B. bovis DNA stimulates IgG production by bovine B cells.
To determine whether the ability of Babesia DNA to stimulate B-cell proliferation correlated with enhanced immunoglobulin secretion (33), we measured immunoglobulin production by bovine B cells cultured with either DNA alone or in the presence of IL-4 or IFN-γ, which in cattle have been shown to be factors involved in enhancing IgG1 and IgG2 production, respectively (21, 22). As a control, B cells were nonspecifically activated with PWM. In the presence of IL-4 and PWM, IgG1 production was upregulated 3.2-fold, whereas in the presence of IFN-γ and PWM, IgG2 production was upregulated 18.3-fold (Table 1, experiment 1). In contrast, E. coli DNA and B. bovis DNA both induced the production of moderate levels of IgG1 (approximately two- and threefold more than that produced by cells cultured with medium or PWM) and high levels of IgG2 (44- and 67-fold more than that produced by B cells cultured with PWM) in the absence of exogenous cytokine (Table 1, experiment 1). The addition of IL-4 or IFN-γ had no additive effect on respective IgG1 and IgG2 levels in the cultures (data not shown). In other experiments, the effect of B. bovis DNA on IgG1 was similar (1.7-fold increase), but the upregulation of IgG2 was only 2.5-fold (experiment 2). In the presence of IL-2 and IFN-γ, B. bovis DNA enhanced IgG2 production approximately eightfold (Table 1, experiment 3), but it had no effect on IgG1 production by B cells cocultured with IL-4 (data not shown). IgM production was not significantly enhanced by DNA.
TABLE 1.
B. bovis DNA induces IgG production by bovine B cells
| Expt and stimulusa | Cytokineb | Immunoglobulin class or subclass concn (ng/ml)c
|
||
|---|---|---|---|---|
| IgM | IgG1 | IgG2 | ||
| Expt 1 | ||||
| None | None | 2,589 ± 349 | 686 ± 305 | <10d |
| PWM | None | 2,757 ± 282 | 726 ± 392 | 240 ± 51 |
| PWM | IL-4 | 2,135 ± 177 | 2,053 ± 933 | 840 ± 51 |
| PWM | IFN-γ | 1,920 ± 205 | 779 ± 71 | 4,398 ± 548 |
| B. bovis DNA | None | 3,222 ± 412 | 2,071 ± 21∗ | 16,165 ± 1,035∗∗ |
| E. coli DNA | None | 3,603 ± 363 | 1,240 ± 152 | 10,640 ± 302∗∗ |
| Expt 2 | ||||
| None | None | 1,556 ± 120 | 1,828 ± 8 | 624 ± 164 |
| PWM | None | 2,662 ± 654 | 2,718 ± 550 | 1,738 ± 222 |
| B. bovis DNA | None | 1,798 ± 210 | 3,068 ± 348∗ | 1,568 ± 228∗ |
| Expt 3 | ||||
| None | IL-2 + IFN-γ | 11,753 ± 769 | 283 ± 99 | 158 ± 31 |
| PWM | IL-2 + IFN-γ | 13,210 ± 348 | 692 ± 26 | 2,603 ± 1,086 |
| B. bovis DNA | IL-2 + IFN-γ | 9,817 ± 618 | 155 ± 155 | 1,265 ± 108∗∗ |
B cells were negatively selected (experiments 1 and 3) or positively selected (experiment 2). For experiments 1 and 2, purified B cells were cultured for 6 days at a density of 4 × 105 cells/ml with 10 μg of PWM, 50 μg of B. bovis DNA, or 25 μg of E. coli DNA per ml. For experiment 3, purified B cells were cultured for 6 days at a density of 1.6 × 106 cells/ml with 10 μg of PWM or 50 μg of B. bovis DNA per ml.
In some assays, 5 ng of recombinant bovine IFN-γ, 20 ng of recombinant bovine IL-4, or 20 U of recombinant human IL-2 per ml were included in the cultures.
Supernatants were harvested, stored at −20°C, and assayed by ELISA for total IgM, IgG1, or IgG2. IgG1 or IgG2 levels were significantly greater in supernatants of B cells cultured with DNA than in supernatants of B cells cultured in medium alone (∗, P < 0.05; ∗∗, P < 0.005).
IgG2 was below the level of detection by ELISA.
Methylation of B. bovis DNA partially inhibits mitogenic activity.
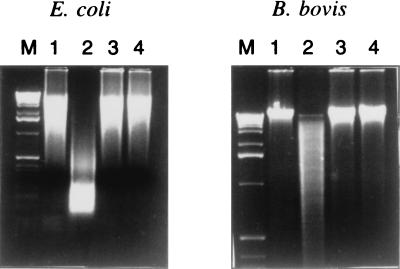
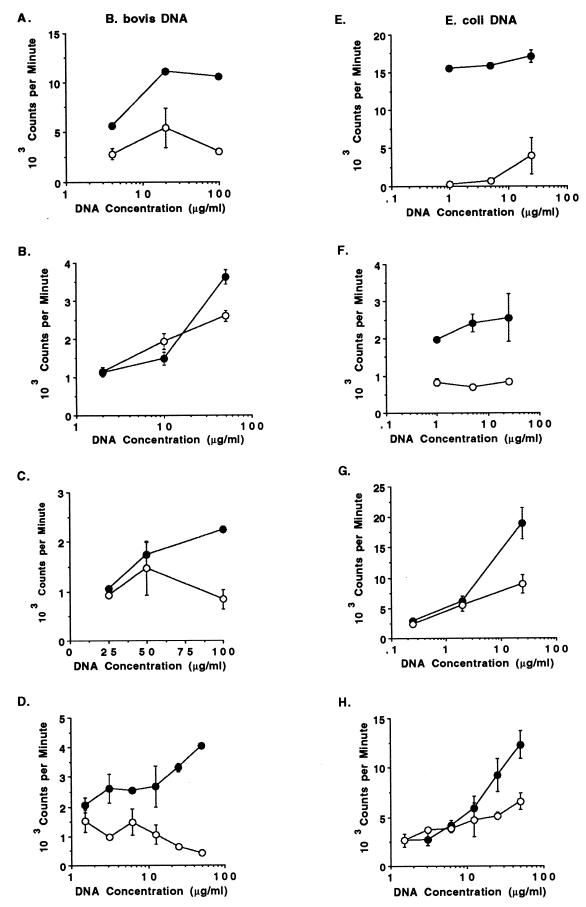
Experiments with E. coli and Drosophila DNA showed that the mitogenic effect on murine B cells was reversed by methylation with CpG methylase (33, 58), whereas the response to methylated yeast DNA was incompletely abrogated (57). Methylation of the cytosine residue of the CG dinucleotide renders the DNA resistant to cleavage with HpaII. To determine whether the mitogenicity of B. bovis DNA was due to the presence of nonmethylated CpG motifs, untreated DNA was compared with methylated DNA for its sensitivity to HpaII and for its ability to induce B-cell proliferation. Although B. bovis DNA appeared to be somewhat less sensitive than E. coli DNA to cleavage by HpaII (Fig. 4, lanes 1 and 2), both DNAs were completely resistant to HpaII cleavage after methylation (Fig. 4, lanes 3 and 4). These data suggest that B. bovis DNA may have fewer unmethylated CG dinucleotides than E. coli DNA, but they also verify that the CpG methylase treatment was successful. Proliferation of B cells stimulated by B. bovis and E. coli DNA was reduced, although it was not completely abrogated in every experiment, after methylation of the DNA (Fig. 5).
FIG. 4.
Resistance of methylated B. bovis DNA to cleavage by HpaII. DNAs prepared from E. coli or B. bovis were treated with SssI methylase and compared with untreated DNA for sensitivity to HpaII digestion. DNA was visualized after electrophoresis on ethidium bromide-stained agarose gels. Lanes: M, 1-kb markers; 1, untreated DNA; 2, untreated DNA incubated with HpaII; 3, methylated DNA; 4, methylated DNA incubated with HpaII.
FIG. 5.
Effect of methylating B. bovis and E. coli DNA with CpG methylase on stimulating B-cell proliferation. DNA was extracted from either B. bovis (A to D) or E. coli (E to H) and treated with SssI methylase as described in the text. Untreated (solid circles) and methylated (open circles) DNAs were then assayed for dose-dependent proliferation with 2 × 105 B cells purified by positive (panels A, D, and H) or negative (panels B, C, E, F, and G) selection and cultured for 3 days with 0.15 to 100 μg of B. bovis DNA or 0.15 to 50 μg of E. coli DNA per ml. Results are presented as the mean cpm ± 1 SD of duplicate or triplicate cultures.
Immunostimulatory CpG motifs in the B. bovis RAP-1 coding sequence stimulate B-cell proliferation.
An 11-kb segment of B. bovis DNA, which contains five ORF, including two tandem rhoptry-associated protein-1 (rap-1) genes (55), was analyzed for the presence of CG dinucleotides. This analysis revealed a relatively low abundance (0.033) of the CG dinucleotide, which is about one-half of the expected frequency of 1 per 16 (0.0625) dinucleotide pairs. A similar frequency of CG dinucleotides was present within the 1,695-bp ORF of the rap-1 gene. Examination of the RAP-1 coding region identified several known ISS, including AACGTT and GACGTT. To determine whether such nonmethylated CpG motifs present in B. bovis DNA were mitogenic for bovine B cells, one ISS consisting of nucleotides 1054 to 1065 (TAAAAACGTTAC; GenBank accession number 38218), which encodes amino acids 312 to 315 (KNVT) in the RAP-1 protein (56), was synthesized as a phosphodiester oligonucleotide and tested for induction of B-cell proliferation. This oligonucleotide was compared with an oligonucleotide (GGTCAACGTTGA) that was shown by others to be mitogenic for murine B cells and human natural killer (NK) cells (1, 32, 33). Control oligonucleotides were synthesized that contained either the CG in reverse orientation or a methylated cytosine residue in this position. In five experiments performed with four different sets of oligonucleotides, the oligonucleotide GGTCAACGTTGA was found to be mitogenic for bovine B cells, whereas the complementary oligonucleotides GGTCAAGCTTGA and GGTCAAQCTTGA (where Q represents 5-methylcytosine) had no or little activity (representative experiments are shown in Table 2, experiments 1 and 2). Similarly, the B. bovis-derived oligonucleotide TAAAAACGTTAC was clearly mitogenic for B cells, whereas control oligonucleotides with the CG in reverse orientation, a methylated cytosine at this position, or lacking the CG residues had little activity (Table 2, experiments 3 and 4). Phosphorothioate-modified oligonucleotides had activities comparable to those of phosphodiester oligonucleotides (Table 3).
TABLE 2.
Stimulation of bovine B-cell proliferation by B. bovis-derived phosphodiester oligodeoxynucleotides that contain the CpG motif
| Expt and oligodeoxynucleotidea | Proliferation (mean cpm ± SD)b | SIc |
|---|---|---|
| Expt 1 | ||
| 5′ GGTCAACGTTGA 3′ | 20,894 ± 2,799 | 34.5 |
| 5′ GGTCAAGCTTGA 3′ | 998 ± 81 | 1.6 |
| 5′ GGTCAAQGTTGA 3′ | 954 ± 48 | 1.6 |
| Expt 2 | ||
| 5′ GGTCAACGTTGA 3′ | 16,477 ± 964 | 27.8 |
| 5′ GGTCAAGCTTGA 3′ | 2,393 ± 144 | 4.0 |
| 5′ GGTCAAQGTTGA 3′ | 3,391 ± 86 | 5.7 |
| Expt 3 | ||
| 5′ GGTCAACGTTGA 3′ | 2,684 ± 357 | 11.6 |
| 5′ TAAAAACGTTAC 3′ | 4,267 ± 431 | 18.5 |
| 5′ TAAAAAGCTTAC 3′ | 810 ± 80 | 3.5 |
| 5′ TAAAAAQGTTAC 3′ | 684 ± 11 | 2.9 |
| 5′ TAAAAATTAC 3′ | 845 ± 95 | 3.7 |
| Expt 4 | ||
| 5′ TAAAAACGTTAC 3′ | 13,954 ± 288 | 86.6 |
| 5′ TAAAAAGCTTAC 3′ | 745 ± 205 | 4.6 |
Bovine B cells (2 × 105) purified by negative selection were cultured for 48 h with 20 μM (experiment 1) or 40 μM (experiments 2 to 4) concentrations of the indicated oligonucleotides and pulsed with 0.25 μCi of [3H]uridine for an additional 18 h before being harvested. The phosphodiester oligonucleotides are indicated in upper-case letters. The CG motif is underlined, and the methylated cytosines are indicated by the letter “Q.”
Results are expressed as the mean cpm ± 1 SD of duplicate or triplicate cultures.
The SI was derived by dividing the mean cpm of B-cell cultures with oligonucleotide by the mean cpm of B-cell cultures with medium.
TABLE 3.
Stimulation of negatively or positively selected B cells by B. bovis-derived phosphorothioate-modified oligodeoxynucleotides that contain the CpG motif
| Expt and oligonucleotide (or DNA)a | B-cell selection | Proliferation (mean cpm ± SD)b | SIc |
|---|---|---|---|
| Expt 1 | |||
| 5′ ggtcaacgttga 3′ | Positive | 17,821 ± 78 | 19.6 |
| 5′ ggtcaagcttga 3′ | Positive | 2,082 ± 58 | 2.3 |
| 5′ ggtcaaqgttga 3′ | Positive | 3,764 ± 1,496 | 4.1 |
| 5′ taaaaacgttac 3′ | Positive | 15,384 ± 1,266 | 16.9 |
| 5′ taaaaagcttac 3′ | Positive | 6,085 ± 1,970 | 6.7 |
| 5′ taaaaaqgttac 3′ | Positive | 2,561 ± 205 | 2.8 |
| 5′ taaaaattac 3′ | Positive | 2,794 ± 101 | 3.1 |
| E. coli DNA | Positive | 13,431 ± 20 | 14.8 |
| Expt 2 | |||
| 5′ taaaaacgttac 3′ | Negative | 18,598 ± 1,879 | 18.7 |
| 5′ taaaaagcttac 3′ | Negative | 2,471 ± 595 | 2.5 |
| 5′ taaaaaqgttac 3′ | Negative | 3,162 ± 1,388 | 3.2 |
| 5′ taaaaattac 3′ | Negative | 3,760 ± 277 | 3.8 |
Bovine B cells (2 × 105) purified by positive or negative selection were cultured for 64 h with 40 μM concentrations of the indicated oligonucleotides and pulsed with 0.25 μCi of [3H]uridine for an additional 6 h before being harvested. Phosphorothioate-modified oligonucleotides are indicated in lower-case letters. The “cg” motif is underlined, and the methylated cytosines are indicated by the letter “q”. E. coli DNA (10 μg/ml) cultured with 10 μg of polymyxin B per ml was included as a positive control.
Results are expressed as the mean cpm ± 1 SD of duplicate or triplicate cultures.
The SI was derived by dividing the mean cpm of B-cell cultures with oligonucleotide or DNA by the mean cpm of B-cell cultures with medium.
Negatively selected B cells can contain a small percentage (ca. 10% or less) of CD3− CD2+ cells. To verify that the proliferation described in these experiments was not due to contaminating non-B cells, B cells were also purified by positive selection with anti-CD21-coated magnetic beads. B cells purified in this manner contained fewer than 2% contaminating CD3+ or CD2+ cells. When compared with negatively selected B cells, positively selected B cells gave equivalent or better proliferative responses to bacterial DNA and CpG oligonucleotides (Table 3 and Fig. 5), ruling out the presence of contaminating cells as the responder population in our assays. Furthermore, all oligonucleotides were negative for endotoxin when tested by the Limulus amebocyte lysate assay.
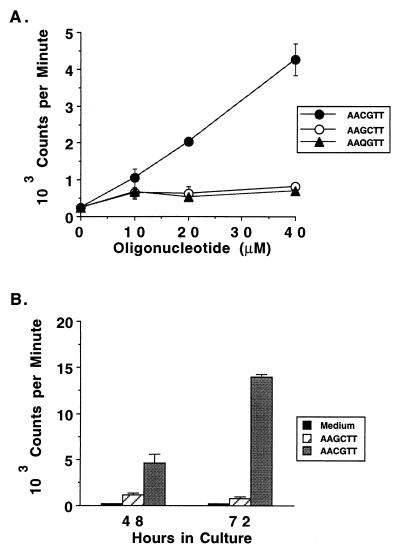
Although the background level of B-cell activation varied from experiment to experiment, proliferation was dose dependent (Fig. 6A) and maximal proliferation was induced with the highest concentration of oligonucleotide tested, which was either 20 μM (Table 3, experiment 1) or 40 μM (Table 3 [experiments 2 to 4], Table 4, and Fig. 6A). Studies with murine B cells have shown that optimal B-cell proliferation in response to CpG oligonucleotides (33) or Drosophila DNA (58) was observed at 24 to 48 h and waned by 72 h. However, when comparing bovine B cells stimulated for 48 or 72 h with the mitogenic B. bovis-derived oligonucleotide, maximal proliferation was achieved by 72 h (Fig. 6B).
FIG. 6.
Dose dependency and time course of the proliferative response of B cells to the CpG phosphodiester oligonucleotide derived from B. bovis. B cells purified by negative selection were tested for proliferation in a 3-day assay with medium or 10, 20, and 40 μM concentrations of either the B. bovis RAP-1-derived ISS oligonucleotide TAAAAACGTTAC (solid circles) or the control oligonucleotides TAAAAAGCTTAC (open circles) and TAAAAAQGTTAC (solid triangles) (A). In a different experiment, negatively selected B cells were cultured for either 48 or 72 h and radiolabeled for the last 6 h of culture with medium (solid bars) or 40 μM TAAAAACGTTAC (gray bars) or TAAAAAGCTTAC (striped bars). The CG dinucleotide is indicated with an underscore (B).
DISCUSSION
Bacterial DNA has multiple effects on different leukocyte subsets, acting both directly on B cells and macrophages and indirectly on NK cells and T cells, to enhance B-cell survival and immunoglobulin secretion in an antigen- and apparently T-cell-independent manner and to upregulate type-1 cytokine responses (42, 46). The present study reports, for the first time, that protozoal DNA has mitogenic properties for B cells that are qualitatively and quantitatively similar to those of bacterial DNA.
The stimulatory properties of protozoal parasites for lymphocytes from nonexposed donors have been described for the genera Theileria, Plasmodium, Leishmania, Trypanosoma, and Babesia (12, 15, 23, 29, 31, 40, 41, 44, and the present study). Factors that may contribute to the stimulation of PBMC from nonexposed donors include the expression of stimulatory cytokines by parasitized host cells, as shown for T. parva (9, 18). However, Babesia infects exclusively erythrocytes, ruling out cytokine production by the infected cell as the stimulus. Alternatively, it has been proposed that the response by naive donors is mediated by CD4+ T cells with a memory phenotype and directed against undefined cross-reactive antigens, as indicated for humans with no known exposure to P. falciparum (15, 29), Leishmania sp. (31), or T. cruzi (44). The kinetics of the PBMC response of nonexposed cattle to B. boris is like that of an antigen-driven response, and proliferation of murine spleen cells to Drosophila cells also peaked later than that of B cells to Drosophila DNA (58). However, the proliferative response to B. bovis antigen by PBMC from nonexposed cattle was largely abrogated by DNase treatment, indicating that at least for Babesia, proliferation is mediated by immunostimulatory DNA and not solely due to cross-reactive T-helper cell epitopes. In support of this, lymphocytes from three head of B. bovis-naive cattle cultured for 2 weeks with B. bovis antigen were predominantly γδ T cells and contained very few CD4+ αβ T cells (12), whereas the majority of B. bovis-stimulated lymphocytes from B. bovis-immune cattle were CD4+ T cells (10). Because bacterial and other nonvertebrate DNAs are not directly mitogenic for T cells (26, 57), the delayed proliferative response by PBMC to B. bovis DNA could reflect an indirect activation of T cells by cytokines, such as IL-12, that are produced by macrophages in response to DNA and known to stimulate T-cell proliferation (8, 61).
Most studies examining B-cell activation by microbial DNA have been performed with murine B cells, although the mitogenic activity of a variety of oligodeoxynucleotides for human B cells has been reported (5, 35). It is interesting that E. coli DNA did not stimulate human peripheral blood B-cell proliferation, even though oligonucleotides did (35). The present study is the first to demonstrate mitogenic properties of microbial DNA for bovine B cells and as such is an important extension of the mouse studies to a nonrodent and outbred species. E. coli and D. melanogaster DNA stimulated polyclonal murine B-cell activation (33, 57), and E. coli DNA enhanced IgM production in murine B cells stimulated with anti-μ (33). The studies performed with human B cells reported that oligonucleotides induced proliferation and secretion of both IgM and IgG (5, 35). E. coli and B. bovis DNA also stimulated bovine IgG production, with moderate effects on IgG1 and more dramatic effects on IgG2. The ability of B. bovis and E. coli DNA to stimulate IgG2 production by purified B cells in the absence of exogenous IFN-γ was probably not due to contaminating NK cells or T cells, since fewer than 1% CD3+ or CD2+ cells were detected in the purified B-cell preparations (data not shown).
The proliferation of B cells in response to B. bovis DNA was generally lower than the proliferation induced by E. coli DNA, in that equivalent levels of proliferation were achieved by two- to tenfold less E. coli DNA. This result could reflect the differences in CG dinucleotide content in B. bovis and E. coli DNAs. First, the overall CG content in B. bovis is relatively low. Second, in contrast to most bacterial, yeast, and Drosophila DNAs, which have the expected relative abundance of CG dinucleotides (13), CG dinucleotides were present at only one-half of the expected frequency in the 11-kb sequence of B. bovis DNA examined. Underrepresentation of CG dinucleotides in other protozoa, including P. falciparum, has also been reported (30). Third, although the results of HpaII digestion indicate that the majority of CpG residues in B. bovis DNA are nonmethylated, some methylated CpG residues may be present, which is likely since partial methylation of CpG residues was reported for P. falciparum DNA (45). Nevertheless, our results indicate the potential for the presence of nonmethylated CpG sequences capable of activating bovine B cells, since methylation of B. bovis DNA reduced its stimulatory activity in several experiments. Furthermore, identification of an ISS with demonstrated mitogenic activity for bovine B cells within the rap-1 ORF verifies the possibility that this and additional ISS within the genomic DNA of babesial parasites are stimulatory for B cells.
In several experiments with B. bovis DNA and in one experiment with E. coli DNA, the stimulation of B-cell proliferation was only partially reduced, even though methylation of the DNA with CpG methylase resulted in resistance to HpaII restriction endonuclease digestion. Sun et al. (57) similarly observed an incomplete abrogation of murine B-cell proliferation to methylated yeast DNA. It is possible that in some cases the DNA was incompletely methylated by SssI methylase treatment. It is also possible that residues in addition to nonmethylated CpG motifs present in B. bovis DNA can stimulate bovine B-cell proliferation. When synthetic oligonucleotides containing CpG, GpC, or methylated CpG sequences and oligonucleotides lacking CpG sequences were compared for B-cell mitogenic activity, the non-CpG oligonucleotides often induced some degree of B-cell proliferation above background levels, although the CpG oligonucleotides were always superior (for example, see Table 2, experiments 2 to 4, and Table 3). In support of these results with bovine B cells, others have shown that both murine and human B-cell proliferative responses to phosphorothioate-modified oligonucleotides did not always require CpG motifs (35, 42). Furthermore, recent studies have shown that methylating the central cytosine of oligodeoxynucleotide 1916 (TCCTGACGTTGAAGT) reduced by one-half its ability to prevent apoptosis of murine splenic B cells, but importantly this did not abrogate activity (65). Together, these studies suggest that the low level of mitogenic activity that we have observed for bovine B cells with methylated DNA or oligonucleotides as well as non-CpG sequences is not unique to the bovine host.
Bovine B cells recognize at least two oligonucleotide sequences known to stimulate murine B cells, which argues that these two species can recognize the same or similar motifs. Oligonucleotides containing the sequence GGTCAACGTTGA used in our experiments stimulated murine B-cell proliferation, although the corresponding non-CpG oligonucleotides had no mitogenic activity (33). Bovine B cells were also stimulated by the phosphorothioate-modified oligonucleotide TTCCATGACGTTCCTGATGCT (data not presented) that induces murine B-cell proliferation (33). The precise mechanism by which CpG-containing oligonucleotides preferentially activate bovine B cells is not clear. Cellular uptake of the DNA fragment appears to be required for stimulation of murine B cells (33), whereas human B cells appear to be activated via surface receptor binding (35).
Leukocyte recognition of CpG motifs of microbial origin could represent an innate immune defense mechanism which would enable discrimination of pathogen from host DNA and trigger a selective immune response at the site of infection (42). Thus, an overwhelming parasite infection has a means of nonspecifically activating innate defense mechanisms that could enable host survival and result in persistent parasitic infection. In fact, humans who recover from malaria and cattle that recover from natural or experimental infection with T. parva or B. bovis remain persistently infected, thereby ensuring parasite survival by providing a reservoir for subsequent arthropod-vectored transmission (19, 36, 39, 66).
The immunostimulatory properties of protozoal DNA could also contribute to the pathology of acute protozoal infection, such as splenomegaly and hypergammaglobulinemia (6, 28, 37), conditions which could be provoked by hyperactivation of B cells and macrophages in response to DNA released from dying parasites. In fact, phosphorothioate-modified CpG-containing oligonucleotides induced splenomegaly and polyclonal B-cell activation in mice (42). Nonmethylated plasmid DNA is taken into macrophages by endocytosis or phagocytosis, where it can activate the transcription factor NF-κB, leading to downstream activation of inflammatory cytokine genes, including IL-1β and TNF-α (52). Among the array of cytokines induced by bacterial DNA, many are regulated by NF-κB (for a review, see reference 38). Thus, it is possible that DNA released by intracellular or extracellular parasites could activate B cells and phagocytic cells, leading to enhanced immunoglobulin secretion and production of inflammatory mediators. Malarial and babesial parasites induce TNF-α production by macrophages (2, 3, 51, 59), which together with IFN-γ, is believed to play a protective role in these infections. However, if overproduced or expressed late in infection, TNF-α and IFN-γ can enhance host pathology, leading to cerebral malaria and, by analogy, cerebral babesiosis (25, 27, 64). Nitric oxide, a key player in both scenarios, is induced by P. falciparum (47) and B. bovis (54) extracts and bacterial DNA (52), but it remains to be determined if protozoal DNA can similarly stimulate macrophages to produce nitric oxide and inflammatory cytokines involved in either protection or pathology.
The exceptional immunogenicity of DNA vaccines is attributed to specific CpG ISS in the backbone of the plasmid DNA vector which prime macrophages and other antigen-presenting cells towards a type-1 immune response (reviewed in references 46 and 60). In mice, this response is generally characterized by enhanced IFN-γ and IgG2a responses, which are mimicked by the use of CpG-containing oligonucleotides as adjuvants for protein vaccines (14, 16, 34, 48). Our results with two CpG ISS demonstrate the feasibility of similarly enhancing immune responses in cattle when used as adjuvants to deliver protein vaccines. In addition, our results support the use of plasmid vectors and genes containing ISS for nucleic acid-based immunization protocols (49). Experiments are planned to characterize the effect of CpG-containing oligonucleotides on macrophage activation, including cytokine and iNOS gene expression, and on priming for IFN-γ and IgG2-biased immune responses in cattle.
ACKNOWLEDGMENTS
We would like to thank Ruguang Oh and Debby Alperin for excellent technical assistance.
This research was supported in part by U.S. Department of Agriculture National Research Initiative Competitive Research Grants 95-37204-2347 (W.C.B.), 96-35204-3584 (D.M.E.), 97-35204-4513 (W.C.B.), and 98-35204-6462 (W.C.B.) and by National Institutes of Health grant R01-AI30136 (W.C.B.).
REFERENCES
- 1.Ballas Z K, Rasmussen W L, Krieg A M. Induction of NK activity in murine and human cells by CpG motifs in oligodeoxynucleotides and bacterial DNA. J Immunol. 1996;157:1840–1845. [PubMed] [Google Scholar]
- 2.Bate C A W, Taverne J, Playfair J H L. Malarial parasites induce TNF production by macrophages. Immunology. 1988;64:227–231. [PMC free article] [PubMed] [Google Scholar]
- 3.Bate C A W, Taverne J, Playfair J H L. Soluble malarial antigens are toxic and induce the production of tumor necrosis factor in vivo. Immunology. 1989;66:600–605. [PMC free article] [PubMed] [Google Scholar]
- 4.Bird A P. CpG islands as gene markers in the vertebrate nucleus. Trends Genet. 1987;3:342–347. [Google Scholar]
- 5.Branda R F, Moore A L, Mathews L, McCormack J J, Zon G. Immune stimulation by an antisense oligomer complementary to the rev gene of HIV-1. Biochem Pharmacol. 1993;45:2037–2043. doi: 10.1016/0006-2952(93)90014-n. [DOI] [PubMed] [Google Scholar]
- 6.Brener Z. Immunity to Trypanosoma cruzi. Adv Parasitol. 1980;18:247–292. doi: 10.1016/s0065-308x(08)60401-7. [DOI] [PubMed] [Google Scholar]
- 7.Brown W C, Davis W C, Choi S H, Dobbelaere D A E, Splitter G A. Functional and phenotypic characterization of WC1+ γ/δ T cells isolated from Babesia bovis-stimulated T cell lines. Cell Immunol. 1994;153:9–27. doi: 10.1006/cimm.1994.1002. [DOI] [PubMed] [Google Scholar]
- 8.Brown W C, Davis W C, Tuo W. Human IL-12 upregulates proliferation and IFN-γ production by parasite antigen-stimulated Th cell clones and γ/δ T cells of cattle. Ann N Y Acad Sci. 1996;795:321–324. doi: 10.1111/j.1749-6632.1996.tb52682.x. [DOI] [PubMed] [Google Scholar]
- 9.Brown W C, Logan K S. Bovine T-cell clones infected with Theileria parva produce a factor with IL-2-like activity. Parasite Immunol. 1986;8:189–192. doi: 10.1111/j.1365-3024.1986.tb00844.x. [DOI] [PubMed] [Google Scholar]
- 10.Brown W C, Logan K S. Babesia bovis: bovine helper T cell lines reactive with soluble and membrane antigens of merozoites. Exp Parasitol. 1992;74:188–199. doi: 10.1016/0014-4894(92)90046-d. [DOI] [PubMed] [Google Scholar]
- 11.Brown W C, Logan K S, Wagner G G, Tetzlaff C L. Cell-mediated immune responses to Babesia bovis merozoite antigens in cattle following infection with tick-derived or cultured parasites. Infect Immun. 1991;59:2418–2426. doi: 10.1128/iai.59.7.2418-2426.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown W C, Shkap V, Zhu D, McGuire T C, Tuo W, McElwain T F, Palmer G H. CD4+ T-lymphocyte and immunoglobulin G2 responses in calves immunized with Anaplasma marginale outer membranes and protected against homologous challenge. Infect Immun. 1998;66:5406–5413. doi: 10.1128/iai.66.11.5406-5413.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burge C, Campbell A M, Karlin S. Over- and under-representation of short oligonucleotides in DNA sequences. Proc Natl Acad Sci USA. 1992;89:1358–1362. doi: 10.1073/pnas.89.4.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chu R S, Targoni O S, Krieg A M, Lehmann P V, Harding C V. CpG oligodeoxynucleotides act as adjuvants that switch on T helper 1 (Th1) immunity. J Exp Med. 1997;186:1623–1631. doi: 10.1084/jem.186.10.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Currier J, Sattabongkot J, Good M F. “Natural” T cells responsive to malaria: evidence implicating immunological cross-reactivity in the maintenance of TCR αβ+ malaria-specific responses from non-exposed donors. Int Immunol. 1992;4:985–994. doi: 10.1093/intimm/4.9.985. [DOI] [PubMed] [Google Scholar]
- 16.Davis H L, Weeranta R, Waldschmidt T J, Tygrett L, Schorr J, Krieg A M. CpG DNA is a potent enhancer of specific immunity in mice immunized with recombinant hepatitis B surface antigen. J Immunol. 1988;160:870–876. [PubMed] [Google Scholar]
- 17.Devereaux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dobbelaere D A E, Coquerelle T M, Roditi I J, Eichorn M, Williams R O. Theileria parva infection induces autocrine growth of bovine lymphocytes. Proc Natl Acad Sci USA. 1988;85:4730–4734. doi: 10.1073/pnas.85.13.4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dolan T T. Chemotherapy of East Coast fever: the long-term weight changes, carrier state and disease manifestations of parvaquone-treated cattle. J Comp Pathol. 1986;96:137–146. doi: 10.1016/0021-9975(86)90004-6. [DOI] [PubMed] [Google Scholar]
- 20.Ellis J A, Davis W C, MacHugh N D, Emery D L, Kaushal A, Morrison W I. Differentiation antigens on bovine mononuclear phagocytes identified by monoclonal antibodies. Vet Immunol Immunopathol. 1988;19:325–340. doi: 10.1016/0165-2427(88)90118-3. [DOI] [PubMed] [Google Scholar]
- 21.Estes D M, Clossier N M, Allen G K. IFN-γ stimulates IgG2 production from bovine B cells costimulated with anti-μ and mitogen. Cell Immunol. 1994;154:287–295. doi: 10.1006/cimm.1994.1078. [DOI] [PubMed] [Google Scholar]
- 22.Estes D M, Hirano A, Heussler V T, Dobbelaere D A E, Brown W C. Expression and biological activities of bovine interleukin 4: effects of recombinant bovine interleukin 4 on T cell proliferation and B cell differentiation and proliferation in vitro. Cell Immunol. 1995;163:268–279. doi: 10.1006/cimm.1995.1126. [DOI] [PubMed] [Google Scholar]
- 23.Fell A H, Currier J, Good M F. Inhibition of Plasmodium falciparum growth in vitro by CD4+ and CD8+ T cells from nonexposed donors. Parasite Immunol. 1994;16:579–586. doi: 10.1111/j.1365-3024.1994.tb00313.x. [DOI] [PubMed] [Google Scholar]
- 24.Goddeeris B M, Morrison W I. The bovine autologous Theileria mixed lymphocyte reaction: influence of monocytes and phenotype of the parasitized stimulator cell on proliferation and parasite specificity. Immunology. 1987;60:63–69. [PMC free article] [PubMed] [Google Scholar]
- 25.Grau G E, Piguet P-F, Vassalli P, Lambert P-H. Tumor necrosis factor and other cytokines in malaria: experimental and clinical data. Immunol Rev. 1989;112:49–70. doi: 10.1111/j.1600-065x.1989.tb00552.x. [DOI] [PubMed] [Google Scholar]
- 26.Halpern M D, Kurlander R J, Pisetsky D S. Bacterial DNA induces murine IFN-γ production by stimulation of interleukin-12 and tumor necrosis factor-α. Cell Immunol. 1996;167:72–78. doi: 10.1006/cimm.1996.0009. [DOI] [PubMed] [Google Scholar]
- 27.Jacobs P, Radzioch D, Stevenson M M. A Th1-associated increase in tumor necrosis factor alpha expression in the spleen correlates with resistance to blood-stage malaria in mice. Infect Immun. 1996;64:535–541. doi: 10.1128/iai.64.2.535-541.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jayawardena A N. Immune responses in malaria. In: Mansfield J M, editor. Parasitic diseases. Vol. 1. New York, N.Y: Marcel Dekker; 1981. pp. 85–136. [Google Scholar]
- 29.Jones K R, Hickling J K, Targett G A T, Playfair J H L. Polyclonal in vitro proliferative responses from non-immune donors to Plasmodium falciparum malaria antigens require UCHL-1+ (memory) T cells. Eur J Immunol. 1990;20:307–315. doi: 10.1002/eji.1830200212. [DOI] [PubMed] [Google Scholar]
- 30.Karlin S, Ladunga I, Blaisdell B E. Heterogeneity of genomes: measures and values. Proc Natl Acad Sci USA. 1994;91:12837–12841. doi: 10.1073/pnas.91.26.12837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kemp M, Hansen M B, Theander T G. Recognition of Leishmania antigens by T lymphocytes from nonexposed individuals. Infect Immun. 1992;60:2246–2251. doi: 10.1128/iai.60.6.2246-2251.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klinman D M, Yi A-K, Beaucage S L, Conover J, Krieg A M. CpG motifs present in bacterial DNA rapidly induce lymphocytes to secrete interleukin 6, interleukin 12, and interferon γ. Proc Natl Acad Sci USA. 1996;93:2879–2883. doi: 10.1073/pnas.93.7.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krieg A M, Yi A-K, Matson S, Waldschmidt T J, Bishop G A, Teasdale R, Koretsky G A, Klinman D M. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature. 1995;374:546–549. doi: 10.1038/374546a0. [DOI] [PubMed] [Google Scholar]
- 34.Leclerc C, Deriaud E, Rojas M, Whalen R G. The preferential induction of a Th1 immune response by DNA-based immunization is mediated by the immunostimulatory effect of plasmid DNA. Cell Immunol. 1997;179:97–106. doi: 10.1006/cimm.1997.1161. [DOI] [PubMed] [Google Scholar]
- 35.Liang H, Nishioka Y, Reich C F, Pisetsky D S, Lipsky P E. Activation of human B cells by phosphorothioate oligodeoxynucleotides. J Clin Investig. 1996;98:1119–1129. doi: 10.1172/JCI118894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mahoney D F, Wright I G, Mirre G B. Bovine babesiosis: the persistence of immunity to Babesia argentina and B. bigemina in calves (Bos taurus) after naturally acquired infection. Ann Trop Med Parasitol. 1973;67:197–203. doi: 10.1080/00034983.1973.11686877. [DOI] [PubMed] [Google Scholar]
- 37.Mansfield J M. Immunology and immunopathology of African trypanosomiasis. In: Mansfield J M, editor. Parasitic diseases. Vol. 1. New York, N.Y: Marcel Dekker; 1981. pp. 167–226. [Google Scholar]
- 38.May M J, Ghosh S. Signal transduction through NFκB. Immunol Today. 1998;19:80–88. doi: 10.1016/s0167-5699(97)01197-3. [DOI] [PubMed] [Google Scholar]
- 39.Miller L H, Good M F, Milon G. Malaria pathogenesis. Science. 1994;264:1878–1883. doi: 10.1126/science.8009217. [DOI] [PubMed] [Google Scholar]
- 40.Pearson T W, Lundin L B, Dolan T T, Stagg D A. Cell-mediated immunity to Theileria-transformed cell lines. Nature. 1979;281:678–680. doi: 10.1038/281678a0. [DOI] [PubMed] [Google Scholar]
- 41.Pinder M, Kar S, Withey K S, Lundin L B, Roelants G E. Proliferation and lymphocyte stimulatory capacity of Theileria-infected lymphoblastoid cells before and after the elimination of intracellular parasites. Immunology. 1981;44:51–60. [PMC free article] [PubMed] [Google Scholar]
- 42.Pisetsky D. Immune activation by bacterial DNA: a new genetic code. Immunity. 1996;5:303–310. doi: 10.1016/s1074-7613(00)80256-3. [DOI] [PubMed] [Google Scholar]
- 43.Pisetsky D S, Reich C F. Stimulation of in vitro proliferation of murine lymphocytes by synthetic oligodeoxynucleotides. Mol Biol Rep. 1993;18:217–221. doi: 10.1007/BF01674433. [DOI] [PubMed] [Google Scholar]
- 44.Piuvezam M R, Russo D M, Burns J M, Jr, Skeiky Y A W, Grabstein K H, Reed S G. Characterization of responses of normal human T cells to Trypanosoma cruzi antigens. J Immunol. 1993;150:916–924. [PubMed] [Google Scholar]
- 45.Pollack Y, Kogan N, Golenser J. Plasmodium falciparum: evidence for a DNA methylation pattern. Exp Parasitol. 1991;72:339–344. doi: 10.1016/0014-4894(91)90079-c. [DOI] [PubMed] [Google Scholar]
- 46.Raz E. Introduction: gene vaccination, current concepts, and future directions. Springer Semin Immunopathol. 1997;19:131–137. doi: 10.1007/BF00870263. [DOI] [PubMed] [Google Scholar]
- 47.Rockett K A, Kwiatkowski D, Bate C A W, Awburn M M, Rockett E J, Clark I A. In vitro induction of nitric oxide by an extract of Plasmodium falciparum. J Infect. 1996;32:187–196. doi: 10.1016/s0163-4453(96)80018-1. [DOI] [PubMed] [Google Scholar]
- 48.Roman M, Martin-Orozco M, Goodman J S, Nguyen M-D, Sato Y, Ronaght A, Kornbluth R S, Richman D D, Carlson D A, Raz E. Immunostimulatory DNA sequences function as T helper-1-promoting adjuvants. Nat Med. 1997;3:849–854. doi: 10.1038/nm0897-849. [DOI] [PubMed] [Google Scholar]
- 49.Sato Y, Roman M, Tighe H, Lee D, Corr M, Nguyen M-D, Silverman G J, Lotz M, Carson D A, Raz E. Immunostimulatory DNA sequences necessary for effective intradermal gene immunization. Science. 1996;273:352–354. doi: 10.1126/science.273.5273.352. [DOI] [PubMed] [Google Scholar]
- 50.Severson C D, Burg D L, Lafrenz D E, Feldbush T L. An alternative method of panning for rat B lymphocytes. Immunol Lett. 1987;15:291–295. doi: 10.1016/0165-2478(87)90130-1. [DOI] [PubMed] [Google Scholar]
- 51.Shoda L K M, Stich R W, Dreewes M, Brown W C. Second Woods Hole Immunoparasitology Meeting, Woods Hole, Mass. 1998. Induction of IL-12 p35, IL-12 p40, TNF-α, and iNOS in bovine macrophages by Babesia bovis or fractionated antigen, abstr. 61. [Google Scholar]
- 52.Stacey K J, Sweet M J, Hume D A. Macrophages ingest and are activated by bacterial DNA. J Immunol. 1996;157:2216–2122. [PubMed] [Google Scholar]
- 53.Stich, R. W., and W. C. Brown. Unpublished observations.
- 54.Stich R W, Shoda L K M, Dreewes M, Adler B, Jungi T W, Brown W C. Stimulation of nitric oxide production in macrophages by Babesia bovis. Infect Immun. 1998;66:4130–4136. doi: 10.1128/iai.66.9.4130-4136.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suarez, C. E., G. H. Palmer, I. Hötzel, and T. F. McElwain. Structure, sequence, and transcriptional analysis of the Babesia bovis rap-1 multigene locus. Mol. Biochem. Parasitol., in press. [DOI] [PubMed]
- 56.Suarez C E, Palmer G H, Jasmer D P, Hines S A, Perryman L E, McElwain T F. Characterization of the gene encoding a 60-kilodalton Babesia bovis merozoite protein with conserved and surface-exposed epitopes. Mol Biochem Parasitol. 1991;46:45–52. doi: 10.1016/0166-6851(91)90197-e. [DOI] [PubMed] [Google Scholar]
- 57.Sun S, Beard C, Jaenisch R, Jones P, Sprent J. Mitogenicity of DNA from different organisms for murine B cells. J Immunol. 1997;159:3119–3125. [PubMed] [Google Scholar]
- 58.Sun S, Cai Z, Langlade-Demoyen P, Kosaka H, Brunmark A, Jackson M R, Peterson P A, Sprent J. Dual function of Drosophila cells as APCs for naive CD8+ T cells: implications for tumor immunity. Immunity. 1996;4:555–564. doi: 10.1016/s1074-7613(00)80482-3. [DOI] [PubMed] [Google Scholar]
- 59.Taverne J, Bate C A W, Kwiatkowski D, Jakobsen P H, Playfair J H L. Two soluble antigens of Plasmodium falciparum induce tumor necrosis factor release from macrophages. Infect Immun. 1990;58:2923–2928. doi: 10.1128/iai.58.9.2923-2928.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tighe H, Corr M, Roman M, Raz E. Gene vaccination: plasmid DNA is more than just a blueprint. Immunol Today. 1998;19:89–97. doi: 10.1016/s0167-5699(97)01201-2. [DOI] [PubMed] [Google Scholar]
- 61.Trinchieri G. Interleukin-12: a proinflammatory cytokine with immunoregulatory functions that bridge innate resistance and antigen-specific adaptive immunity. Annu Rev Immunol. 1995;13:251–276. doi: 10.1146/annurev.iy.13.040195.001343. [DOI] [PubMed] [Google Scholar]
- 62.Trueblood E S, Brown W C, Palmer G H, Davis W C, Stone D M, McElwain T F. B-lymphocyte proliferation during bovine leukemia virus-induced persistent lymphocytosis is enhanced by T-lymphocyte-derived IL-2. J Virol. 1998;72:3169–3177. doi: 10.1128/jvi.72.4.3169-3177.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Turrini F, Giribaldi G, Valente E, Arese P. Mycoplasma contamination of Plasmodium cultures: a case of parasite parasitism. Parasitol Today. 1997;13:367–368. doi: 10.1016/s0169-4758(97)01088-0. [DOI] [PubMed] [Google Scholar]
- 64.Wright I G, Goodger B V, Clark I A. Immunopathophysiology of Babesia bovis and Plasmodium falciparum infections. Parasitol Today. 1988;4:214–218. doi: 10.1016/0169-4758(88)90161-5. [DOI] [PubMed] [Google Scholar]
- 65.Yi A-K, Ming C, Peckham D W, Krieg A M, Ashman R F. CpG oligodeoxyribonucleotides rescue mature spleen B cells from spontaneous apoptosis and promote cell cycle entry. J Immunol. 1998;160:5898–5906. [PubMed] [Google Scholar]
- 66.Young A S, Leitch B L, Newson R M. The occurrence of a Theileria parva carrier state in cattle from an East Coast fever endemic area of Kenya. In: Irvin A D, Cunningham M P, Young A S, editors. Advances in the control of theileriosis: current topics in veterinary medicine and animal science. Vol. 14. The Hague, The Netherlands: Martinus Nijhoff; 1981. pp. 60–62. [Google Scholar]