Abstract
Objectives
In this study, we aimed to develop a multiparameter prediction model to improve the diagnostic accuracy of invasive adenocarcinoma in pulmonary pure glass nodules.
Method
We included patients with pulmonary pure glass nodules who underwent lung resection and had a clear pathology between January 2020 and January 2022 at the Qilu Hospital of Shandong University. We collected data on the clinical characteristics of the patients as well as their preoperative biomarker results and computed tomography features. Thereafter, we performed univariate and multivariate logistic regression analyses to identify independent risk factors, which were then used to develop a prediction model and nomogram. We then evaluated the recognition ability of the model via receiver operating characteristic (ROC) curve analysis and assessed its calibration ability using the Hosmer-Lemeshow test and calibration curves. Further, to assess the clinical utility of the nomogram, we performed decision curve analysis.
Result
We included 563 patients, comprising 174 and 389 cases of invasive and non-invasive adenocarcinoma, respectively, and identified seven independent risk factors, namely, maximum tumor diameter, age, serum amyloid level, pleural effusion sign, bronchial sign, tumor location, and lobulation. The area under the ROC curve was 0.839 (95% CI: 0.798–0.879) for the training cohort and 0.782 (95% CI: 0.706–0.858) for the validation cohort, indicating a relatively high predictive accuracy for the nomogram. Calibration curves for the prediction model also showed good calibration for both cohorts, and decision curve analysis showed that the clinical prediction model has clinical utility.
Conclusion
The novel nomogram thus constructed for identifying invasive adenocarcinoma in patients with isolated pulmonary pure glass nodules exhibited excellent discriminatory power, calibration capacity, and clinical utility.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12893-024-02341-2.
Keywords: Pulmonary pure glass nodule, Invasive adenocarcinoma, Prediction, Logical model, Nomogram
Introduction
Owing to advances in diagnostic imaging and the widespread use of low-dose computerized tomography (LDCT) screening, an increasing number of pulmonary pure gross glass nodules (pGGNs) are being detected, causing alarm to patients [1–4]. A ground-glass nodule (GGN) is defined as a nodule with a slightly increased density that does not obscure underlying bronchial structures or vascular margins in high-resolution CT images [5]. Depending on the presence or absence of solid components, GGN can be classified as pure GGN (pGGN) and partially solid GGN. Notably, pGGNs are defined as GGNs without solid components [6].
Reportedly, the development of pGGNs progresses slowly, and at different pathological stages, they exhibit different growth patterns and show varying degrees of invasiveness. Some pGGN stages include: atypical adenomatous hyperplasia (AAH), adenocarcinoma in situ (AIS), microinvasive adenocarcinoma (MIA), and invasive adenocarcinoma (IAC) [7–10]. Currently, there are no uniform guidelines for GGN surgery. Some studies have shown that sublobar resection is acceptable for AAH, AIS, and MIA, but is unsuitable for IAC [11, 12]. With adequate surgical resection, patients with AIS and MIA exhibit approximately 100% disease-free survival [13–15], whereas the 5-year survival rate of patients with localized IAC have varies in the range70–90% [16, 17]. Therefore, the preoperative identification of the IAC is essential to help clinicians choose the correct procedure for their patients.
In clinical practice, identifying IAC manifesting as a pGGN is challenging. Attempts have been made in previous studies to distinguish between preinvasive adenocarcinomas and IACs presenting as pGGN [18–26]. Further, the correlation between pathological manifestations and nodules presenting as solid, partially solid, and pGGN have been investigated in other studies [27, 28]. However, in these previous studies, a wide variety of assessment factors were not identified. Additionally, the validity of some of these studies is limited by the small number of patients included.
In this study, we aimed to retrospectively analyze a relatively large number of patients with pGGN < 2 cm and develop a multiparametric predictive model and nomogram using patients’ clinical information, hematological findings, and imaging features to improve the ability of clinicians to diagnose IAC in pGGN and provide a basis for rational clinical decision-making.
Materials and methods
This study was approved by the Ethics Committee of Qilu Hospital, Shandong University (registration number: KYLL-202008-023-1), and all the patients read and signed the informed consent form prior to this study, approving the use of their clinical information.
Patient selection
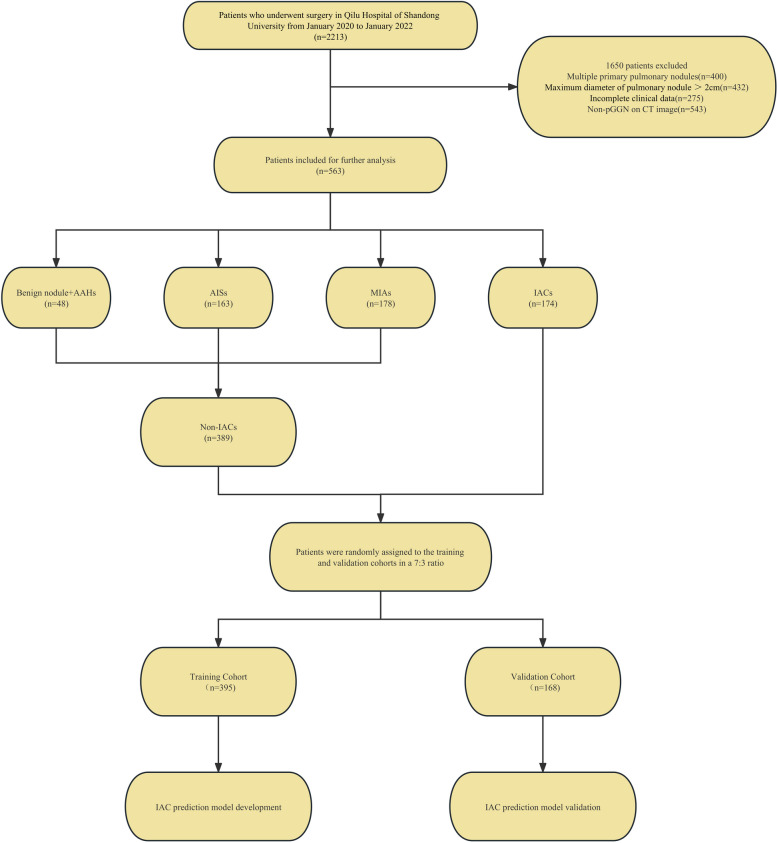
In this study, we retrospectively evaluated patients who underwent minimally invasive pneumonectomy with a clear pathology of pulmonary nodules at the Qilu Hospital of Shandong University between January 2020 and January 2022. The inclusion criteria were as follows: (1) patients with a single intrapulmonary nodule based on a chest CT scan performed within 1 month before surgery; (2) patients with lung nodules with maximum diameter ≤ 2 cm; (3) patients with pGGN without any solid component as indicated via CT imaging; (4) absence of pulmonary atelectasis and active inflammation based on lung images; (5) clear pathological findings obtained after surgical resection; (6) asymptomatic at diagnosis; and (7) no preoperative treatment. The exclusion criteria were as follows: (1) aged < 18 years, (2) history of thoracic surgery, (3) incomplete perioperative data, (4) history of malignant disease within the past 5 years, and (5) metastatic tumors. The patients included in our study were screened according to the inclusion and exclusion criteria (Fig. 1). Further, we used a random split-sample approach to randomly assign all the enrolled patients to either the training cohort or validation cohort at a ratio of 7:3. The training cohort was used to develop the prediction nomogram, while the validation cohort was used to verify the performance of the nomogram.
Fig. 1.
Flow diagram of patient selection through the study. AAH, atypical adenomatous hyperplasia; AIS, adenocarcinoma in situ; MIA, microinvasive adenocarcinoma; IAC, invasive adenocarcinoma
Data collection and variable definitions
The following data were collected from the hospital database for all the eligible patients: (1) demographic data: sex, age, smoking history, body mass index (BMI), and preoperative comorbidities [hypertension, diabetes, and chronic obstructive pulmonary disease (COPD)]; (2) preoperative assessment data: American Society of Anesthesiologists (ASA) score, percentage of the predicted forced expiratory volume in one second (FEV1% predicted), and percentage of the predicted value of maximal voluntary ventilation (MVV% predicted); (3) laboratory blood test indicators: blood type, serum complement C1q, lactate dehydrogenase (LDH), serum amyloid (SA), serum 5’-nucleotidase (5’-NT), blood sugar, albumin, neutrophil, eosinophil, basophil, monocyte, lymphocyte, erythrocyte, hemoglobin, and platelet levels, derivative prognostic nutritional index (PNI), neutrophil-lymphocyte ratio (NLR), derived neutrophil-to-lymphocyte ratio (dNLR), platelet-lymphocyte ratio (PLR), monocyte-lymphocyte ratio (MLR), neutrophil-to-lymphocyte and platelet ratio (NLPR), systemic inflammatory response syndrome (SIRS), the aggregate index of systemic inflammation (AISI), systemic inflammation index (SII), and pan-immune-inflammation value (PIV); (4) lung cancer tumor markers: pro-gastrin-releasing peptide (pro-GRP), carcinoembryonic antigen (CEA), squamous cell carcinoma antigen (SCC), cytokeratin 19-fragment (cyfra21-1), carcinoma antigen 125 (CA125), and neuron-specific enolase (NSE) levels; (5) CT image characteristics: nodule location (central or peripheral), nodule shape (regular or irregular), spiculation (sunburst appearance), calcification, lobulation, cavitation signs, pleural adhesions, vascular penetration signs, bronchus signs, lymph node enlargement signs, pleural effusion signs, and maximum tumor diameter; and (6) postoperative pathological results. PNI, NLR, dNLR, MLR, NLPR, SIRI, AISI, SII, and PIV were calculated using the following expressions:
All chest CT examinations included the complete thorax and were performed in the supine position. Scans were obtained during deep inspiration and the patients holding their breath. The CT images were examined and interpreted by two radiologists with more than 5 years of experience in thoracic radiology. The two radiologists independently determined the characteristics of each image, while a third radiologist, with over 20 years of experience in thoracic radiology reassessed the measurements to check for discrepancies. Any discrepancies noted were resolved through discussion among the three radiologists. Central nodules were defined as those located in the bronchi, lobular bronchi, or segmental bronchi of the lungs, while peripheral nodules were defined as those located below the tertiary bronchi. Spiculation was defined as diffusion from the nodule edge into the lungs without contact with the pleural surface. Further, cavitation was defined as the presence of spaces filled with gas and considered regions of transparency or low attenuation. Pleural adhesion was defined as the linear attenuation of pleura or fissures from the nodule. Patterns in CT images, including delamination, central nodule, diffusion, or a popcorn pattern, were considered signs of calcification. A pulmonary artery passing through the nodule, as observed in the CT images was indicative of vascular penetration. Further, bronchial signs on the CT image showed direct bronchial engagement of the nodule. Lobulation was defined as a wavy or scalloped portion on the surface of a lesion, with strands stretching from the nodal edge to the lung parenchyma. Pleural effusion was defined based on a blunted angle of the rib diaphragm in the CT image. Lymph node enlargement was defined as a > 1-cm long lymph node axis in the CT image.
All pathological samples were fixed in formalin, stained with hematoxylin and eosin (H&E), and assessed by two experienced lung pathologists. The histopathological evaluation was performed by examining H&E-stained slides under a light microscope. All registered GGNs had clear pathological diagnoses. Pathological findings were divided into four groups: benign, AAH, AIS, MIA, and IAC. AAH, AIS, MIA, and IAC were based on the International Association for the Study of Lung Cancer (IASLC)/American Thoracic Society (ATS)/European Respiratory Society (ERS) classifications of lung in resected specimens [29]. Additionally, owing to the poor prognosis of patients with IAC, patients with benign lesions, AAH, AIS, and MIA were designated as the non-IAC group.
Establishment of the predictive model
First, data for the training cohort were analyzed using univariate analysis to assess all the factors affecting IAC in pGGNs. Thereafter, all factors with P < 0.2 in the univariate analysis were included for further multivariate logistic regression analyses. A predictive model and nomogram were constructed using R statistical software (Windows version 4.2.1, http://www.r-project.org. The scores for each variable were calculated using a regression model, and the predicted probability of IAC in pGGNs was derived by summing the scores for each variable.
Predictive model and nomogram performance
The performance of the predictive nomogram was evaluated based on its discriminatory power, calibration ability, and clinical utility. Discrimination describes the ability of a model to properly distinguish between incidents and non-incidents. Receiver operating characteristic (ROC) curves were used to evaluate the efficiency of the predictive nomograms [30]. Calibration gauges the extent to which predicted probabilities correspond to actual results. We used the Hosmer-Lemeshow test to assess the calibration capability of the model, and P > 0.05 was indicative of a satisfactory calibration ability [31]. Further evaluation of the calibration ability of the model was performed by constructing nomogram calibration curves. Internal verification was performed by repeating the bootstrap method 1,000 times [32]. Decision curve analysis (DCA) was used to assess the clinical utility of the predictive nomogram based on the net benefit of different threshold probabilities [33]. We determined the optimal cutoff value based on ROC curve analysis results for the training cohort when the Youden index (sensitivity + specificity − 1) reached its maximum value.
Results
Patient characteristics
The procedure for identifying and selecting eligible patients is illustrated in Fig. 1.
A total of 563 eligible patients were included in this study. Among these patients, there were 48 cases of benign nodules and AAH, 163 were AIS, 178 cases of MIA, and 174 cases of IAC. Further, we classified all the cases under the non-IACs (n = 389) or IACs (n = 174) groups, according to the criterion, “whether the nodule had IAC characteristics.” We then randomly assigned the enrolled patients to either the training cohort (n = 395) or validation cohort (n = 168) in a 7:3 ratio; no significant differences existed between the two cohorts with respect to any of the variable (Table 1). Thus, there were 273 non-IAC and 122 IAC cases in the training cohort and 116 non-IAC and 52 IAC cases in the validation cohort. The characteristics of the training and validation cohorts are presented in Table 2.
Table 1.
Patients’ characteristics of the training cohort and validation cohort
| Characteristics | All cohort (N = 563) |
Validation cohort (N = 168) |
Training cohort (N = 395) |
p |
|---|---|---|---|---|
| IAC, n (%) | 0.988 | |||
| No | 389 (69.1) | 116 (69.0) | 273 (69.1) | |
| Yes | 174 (30.9) | 52 (31.0) | 122 (30.9) | |
| Gender, n (%) | 0.785 | |||
| Female | 360 (63.9) | 106 (63.1) | 254 (64.3) | |
| Male | 203 (36.1) | 62 (36.9) | 141 (35.7) | |
| Hypertension, n (%) | 0.368 | |||
| No | 423 (75.1) | 122 (72.6) | 301 (76.2) | |
| Yes | 140 (24.9) | 46 (27.4) | 94 (23.8) | |
| Diabetes, n (%) | 0.221 | |||
| No | 503 (89.3) | 146 (86.9) | 357 (90.4) | |
| Yes | 60 (10.7) | 22 (13.1) | 38 (9.6) | |
| COPD, n (%) | 0.629 | |||
| No | 558 (99.1) | 167 (99.4) | 391 (99.0) | |
| Yes | 5 (0.9) | 1 (0.6) | 4 (1.0) | |
| Smoking history, n (%) | 0.78 | |||
| Non-smoker | 463 (82.2) | 137 (81.5) | 326 (82.5) | |
| Smoker | 100 (17.8) | 31 (18.5) | 69 (17.5) | |
| Blood type, n (%) | 0.435 | |||
| A | 166 (29.5) | 47 (28.0) | 119 (30.1) | |
| B | 179 (31.8) | 60 (35.7) | 119 (30.1) | |
| AB | 76 (13.5) | 18 (10.7) | 58 (14.7) | |
| O | 142 (25.2) | 43 (25.6) | 99 (25.1) | |
| ASA, n (%) | 0.239 | |||
| 1 | 77 (13.7) | 17 (10.1) | 60 (15.2) | |
| 2 | 481 (85.4) | 150 (89.3) | 331 (83.8) | |
| 3 | 5 (0.9) | 1 (0.6) | 4 (1.0) | |
| Location, n (%) | 0.5 | |||
| Centrality | 47 (8.3) | 12 (7.1) | 35 (8.9) | |
| Peripherality | 516 (91.7) | 156 (92.9) | 360 (91.1) | |
| Shape, n (%) | 0.995 | |||
| Regularity | 335 (59.5) | 100 (59.5) | 235 (59.5) | |
| Irregularity | 228 (40.5) | 68 (40.5) | 160 (40.5) | |
| Spiculation, n (%) | 0.882 | |||
| No | 311 (55.2) | 92 (54.8) | 219 (55.4) | |
| Yes | 252 (44.8) | 76 (45.2) | 176 (44.6) | |
| Cavitation sign, n (%) | 0.599 | |||
| No | 500 (88.8) | 151 (89.9) | 349 (88.4) | |
| Yes | 63 (11.2) | 17 (10.1) | 46 (11.6) | |
| Calcification, n (%) | 0.356 | |||
| No | 561 (99.6) | 168 (100.0) | 393 (99.5) | |
| Yes | 2 (0.4) | 0 (0.0) | 2 (0.5) | |
| Vascular penetration sign, n (%) | 0.234 | |||
| No | 207 (36.8) | 68 (40.5) | 139 (35.2) | |
| Yes | 356 (63.2) | 100 (59.5) | 256 (64.8) | |
| Pleural adhesions, n (%) | 0.688 | |||
| No | 365 (64.8) | 111 (66.1) | 254 (64.3) | |
| Yes | 198 (35.2) | 57 (33.9) | 141 (35.7) | |
| Bronchus sign, n (%) | 0.27 | |||
| No | 458 (81.3) | 132 (78.6) | 326 (82.5) | |
| Yes | 105 (18.7) | 36 (21.4) | 69 (17.5) | |
| Lobulation, n (%) | 0.7 | |||
| No | 448 (79.6) | 132 (78.6) | 316 (80.0) | |
| Yes | 115 (20.4) | 36 (21.4) | 79 (20.0) | |
| Lymph node enlargement sign, n (%) | 0.422 | |||
| No | 508 (90.2) | 149 (88.7) | 359 (90.9) | |
| Yes | 55 (9.8) | 19 (11.3) | 36 (9.1) | |
| Pleural effusion sign, n (%) | 0.356 | |||
| No | 561 (99.6) | 168 (100.0) | 393 (99.5) | |
| Yes | 2 (0.4) | 0 (0.0) | 2 (0.5) | |
| Albumin (g/L), median (IQR) | 60.10 (58.00, 62.05) | 60.10 (57.98, 62.30) | 60.10 (58.00, 61.90) | 0.825 |
| Lymphocyte (×109/L), median (IQR) | 1.84 (1.49, 2.23) | 1.81 (1.48, 2.19) | 1.86 (1.50, 2.26) | 0.517 |
| PNI (%), median (IQR) | 69.35 (66.93, 71.90) | 69.45 (66.99, 71.48) | 69.30 (66.85, 72.03) | 0.958 |
| Neutrophil (×109/L), median (IQR) | 2.97 (2.43, 3.66) | 2.95 (2.45, 3.90) | 2.97 (2.43, 3.50) | 0.375 |
| Eosinophil (×109/L), median (IQR) | 0.10 (0.06, 0.16) | 0.10 (0.07, 0.16) | 0.09 (0.06, 0.16) | 0.573 |
| Basophil (×109/L), median (IQR) | 0.03 (0.02, 0.04) | 0.03 (0.02, 0.04) | 0.03 (0.02, 0.04) | 0.508 |
| Monocyte (×109/L), median (IQR) | 0.40 (0.33, 0.49) | 0.42 (0.35, 0.52) | 0.40 (0.33, 0.49) | 0.089 |
| Erythrocyte (×1012/L), median (IQR) | 4.48 (4.20, 4.83) | 4.48 (4.16, 4.81) | 4.48 (4.23, 4.84) | 0.722 |
| Hemoglobin (g/L), median (IQR) | 136.00 (128.00, 146.50) | 135.50 (127.00, 144.25) | 136.00 (129.00, 147.00) | 0.351 |
| Platelet (×109/L), median (IQR) | 236.00 (206.00, 272.50) | 237.00 (202.75, 272.50) | 235.00 (207.50, 272.50) | 0.991 |
| NLR (%), median (IQR) | 1.64 (1.27, 2.09) | 1.70 (1.25, 2.15) | 1.62 (1.28, 2.06) | 0.423 |
| PLR (%), median (IQR) | 130.00 (105.50, 158.60) | 133.50 (104.57, 160.84) | 129.21 (106.15, 156.53) | 0.531 |
| MLR (%), median (IQR) | 0.22 (0.18, 0.27) | 0.22 (0.18, 0.28) | 0.21 (0.17, 0.27) | 0.149 |
| dNLR (%), median (IQR) | 1.25 (1.00, 1.54) | 1.26 (0.96, 1.57) | 1.25 (1.00, 1.53) | 0.753 |
| NLPR (%), median (IQR) | 0.01 (0.01, 0.01) | 0.01 (0.01, 0.01) | 0.01 (0.01, 0.01) | 0.6 |
| SIRI (%), median (IQR) | 0.63 (0.47, 0.92) | 0.65 (0.47, 1.03) | 0.62 (0.47, 0.90) | 0.153 |
| AISI (%), median (IQR) | 147.97 (103.82, 226.98) | 156.63 (104.56, 253.41) | 146.32 (103.60, 220.58) | 0.155 |
| SII (%), median (IQR) | 379.78 (288.68, 505.98) | 387.88 (281.58, 537.30) | 375.44 (290.75, 488.74) | 0.399 |
| PIV (%), median (IQR) | 147.97 (103.82, 226.98) | 156.63 (104.56, 253.41) | 146.32 (103.60, 220.58) | 0.155 |
| Blood sugar(mmol/L), median (IQR) | 5.09 (4.70, 5.58) | 5.11 (4.71, 5.62) | 5.07 (4.70, 5.58) | 0.591 |
| Complement C1q(mg/L), median (IQR) | 171.00 (153.05, 190.85) | 167.00 (151.10, 190.50) | 171.60 (153.85, 190.85) | 0.341 |
| LDH (U/L), median (IQR) | 191.00 (169.50, 215.50) | 195.00 (166.00, 217.00) | 191.00 (171.00, 215.00) | 0.991 |
| SA (mg/dL), median (IQR) | 53.10 (48.90, 57.30) | 52.70 (48.80, 57.90) | 53.20 (48.95, 57.20) | 0.928 |
| 5’-NT (U/L), median (IQR) | 4.00 (3.00, 5.00) | 4.00 (3.00, 5.00) | 4.00 (3.00, 5.00) | 0.801 |
| Pro-GRP (pg/mL), median (IQR) | 41.96 (33.02, 45.84) | 41.96 (32.49, 45.51) | 41.96 (33.05, 46.17) | 0.483 |
| SCC (ng/mL), median (IQR) | 1.05 (0.80, 1.70) | 1.07 (0.73, 1.83) | 1.03 (0.80, 1.69) | 0.735 |
| Cyfra21-1 (ng/mL), median (IQR) | 2.28 (1.62, 2.54) | 2.20 (1.60, 2.43) | 2.31 (1.64, 2.56) | 0.57 |
| CEA (ng/mL), median (IQR) | 2.30 (1.41, 2.60) | 2.26 (1.39, 2.54) | 2.32 (1.42, 2.61) | 0.613 |
| CA125 (U/mL), median (IQR) | 10.72 (7.50, 11.55) | 10.70 (7.40, 11.60) | 10.72 (7.53, 11.35) | 0.663 |
| NSE (ng/mL), median (IQR) | 19.45 (15.85, 21.00) | 19.30 (15.57, 20.33) | 19.45 (16.30, 21.05) | 0.359 |
| Age (years), median (IQR) | 56.00 (48.00, 63.00) | 57.00 (49.00, 63.00) | 56.00 (48.00, 63.00) | 0.464 |
| BMI (kg/m2), median (IQR) | 24.57 (22.48, 26.43) | 24.76 (22.58, 26.46) | 24.45 (22.42, 26.34) | 0.391 |
| FEV1% predicted (%), median (IQR) | 105.97 (95.56, 114.79) | 104.89 (94.26, 113.56) | 106.05 (96.50, 115.71) | 0.129 |
| MVV% predicted (%), median (IQR) | 104.79 (91.40, 116.59) | 103.80 (90.24, 113.51) | 105.00 (91.90, 118.06) | 0.288 |
| Maximum diameter (cm), median (IQR) | 1.00 (0.70, 1.40) | 1.00 (0.80, 1.30) | 1.00 (0.70, 1.40) | 0.383 |
IAC Invasive adenocarcinomam, COPD Chronic obstructive pulmonary diseases, ASA American Society of Anesthesiologists, PNI Prognostic nutritional index, NLR Neutrophil-lymphocyte ratio, PLR Platelet-lymphocyte ratio, MLR Monocyte-lymphocyte ratio, dNLR Derived neutrophil-to-lymphocyte ratio, NLPR Neutrophil to lymphocyte and platelet ratio, SIRI Systemic inflammatory response syndrome, AISI Aggregate index of systemic inflammation, SII Systemic inflammation index, PIV Pan-immune-inflammation value, LDH Lactate dehydrogenase, SA Serum amyloid, 5'-NT 5'-nucleotidase, Pro-GRP Pro-gastrin-releasing peptide, SCC Squamous cell carcinoma, Cyfra21-1 Cytokeratin 19-fragments, CEA Carcinoembryonic antigen, CA125 Carcinoma antigen 125, NSE Neuron-specific enolase, BMI Body mass index, FEV1 Forced expiratory volume in one second, MVV Maximal voluntary ventilation
P-value for the comparison between training cohort and validation cohort
Table 2.
Clinical characteristics of patients with IACs and Non-IACs in the training and validation cohorts
| Characteristics | Training Cohort(n = 395) | Validation cohort(n = 168) | ||||
|---|---|---|---|---|---|---|
| Non-IACs(n = 273) | IACs(n = 122) | P | Non-IACs(n = 116) | IACs(n = 52) | P | |
| Gender, n (%) | 0.009 | 0.779 | ||||
| Female | 187 (68.5) | 67 (54.9) | 74 (63.8) | 32 (61.5) | ||
| Male | 86 (31.5) | 55 (45.1) | 42 (36.2) | 20 (38.5) | ||
| Hypertension, n (%) | 0.075 | 0.643 | ||||
| No | 215 (78.8) | 86 (70.5) | 83 (71.6) | 39 (75.0) | ||
| Yes | 58 (21.2) | 36 (29.5) | 33 (28.4) | 13 (25.0) | ||
| Diabetes, n (%) | 0.786 | 0.114 | ||||
| No | 246 (90.1) | 111 (91.0) | 104 (89.7) | 42 (80.8) | ||
| Yes | 27 (9.9) | 11 (9.0) | 12 (10.3) | 10 (19.2) | ||
| COPD, n (%) | 0.406 | 0.134 | ||||
| No | 271 (99.3) | 120 (98.4) | 116 (100.0) | 51 (98.1) | ||
| Yes | 2 (0.7) | 2 (1.6) | 0 (0.0) | 1 (1.9) | ||
| Smoking history, n (%) | 0.013 | 0.798 | ||||
| Non-smoker | 234 (85.7) | 92 (75.4) | 94 (81.0) | 43 (82.7) | ||
| Smoker | 39 (14.3) | 30 (24.6) | 22 (19.0) | 9 (17.3) | ||
| Blood type, n (%) | 0.032 | 0.92 | ||||
| A | 88 (32.2) | 31 (25.4) | 31 (26.7) | 16 (30.8) | ||
| B | 74 (27.1) | 45 (36.9) | 41 (35.3) | 19 (36.5) | ||
| AB | 35 (12.8) | 23 (18.9) | 13 (11.2) | 5 (9.6) | ||
| O | 76 (27.8) | 23 (18.9) | 31 (26.7) | 12 (23.1) | ||
| ASA, n (%) | 0.541 | 0.262 | ||||
| 1 | 44 (16.1) | 16 (13.1) | 13 (11.2) | 4 (7.7) | ||
| 2 | 227 (83.2) | 104 (85.2) | 103 (88.8) | 47 (90.4) | ||
| 3 | 2 (0.7) | 2 (1.6) | 0 (0.0) | 1 (1.9) | ||
| Location, n (%) | < 0.001 | 0.853 | ||||
| Centrality | 15 (5.5) | 20 (16.4) | 8 (6.9) | 4 (7.7) | ||
| Peripherality | 258 (94.5) | 102 (83.6) | 108 (93.1) | 48 (92.3) | ||
| Shape, n (%) | < 0.001 | 0.007 | ||||
| Regularity | 189 (69.2) | 46 (37.7) | 77 (66.4) | 23 (44.2) | ||
| Irregularity | 84 (30.8) | 76 (62.3) | 39 (33.6) | 29 (55.8) | ||
| Spiculation, n (%) | < 0.001 | 0.03 | ||||
| No | 171 (62.6) | 48 (39.3) | 70 (60.3) | 22 (42.3) | ||
| Yes | 102 (37.4) | 74 (60.7) | 46 (39.7) | 30 (57.7) | ||
| Cavitation sign, n (%) | 0.543 | 0.336 | ||||
| No | 243 (89.0) | 106 (86.9) | 106 (91.4) | 45 (86.5) | ||
| Yes | 30 (11.0) | 16 (13.1) | 10 (8.6) | 7 (13.5) | ||
| Calcification, n (%) | 0.034 | NA | ||||
| No | 273 (100.0) | 120 (98.4) | 116 (100.0) | 52 (100.0) | ||
| Yes | 0 (0.0) | 2 (1.6) | 0(0) | 0(0) | ||
| Vascular penetration sign, n (%) | < 0.001 | 0.04 | ||||
| No | 112 (41.0) | 27 (22.1) | 53 (45.7) | 15 (28.8) | ||
| Yes | 161 (59.0) | 95 (77.9) | 63 (54.3) | 37 (71.2) | ||
| Pleural adhesions, n (%) | < 0.001 | 0.059 | ||||
| No | 195 (71.4) | 59 (48.4) | 82 (70.7) | 29 (55.8) | ||
| Yes | 78 (28.6) | 63 (51.6) | 34 (29.3) | 23 (44.2) | ||
| Bronchus sign, n (%) | < 0.001 | 0.005 | ||||
| No | 245 (89.7) | 81 (66.4) | 98 (84.5) | 34 (65.4) | ||
| Yes | 28 (10.3) | 41 (33.6) | 18 (15.5) | 18 (34.6) | ||
| Lobulation, n (%) | < 0.001 | 0.001 | ||||
| No | 236 (86.4) | 80 (65.6) | 99 (85.3) | 33 (63.5) | ||
| Yes | 37 (13.6) | 42 (34.4) | 17 (14.7) | 19 (36.5) | ||
| Lymph node enlargement sign, n (%) | 0.142 | 0.555 | ||||
| No | 252 (92.3) | 107 (87.7) | 104 (89.7) | 45 (86.5) | ||
| Yes | 21 (7.7) | 15 (12.3) | 12 (10.3) | 7 (13.5) | ||
| Pleural effusion sign, n (%) | 0.558 | NA | ||||
| No | 272 (99.6) | 121 (99.2) | 116 (100.0) | 52 (100.0) | ||
| Yes | 1 (0.4) | 1 (0.8) | 0(0) | 0(0) | ||
| Albumin (g/L), median (IQR) | 60.00 (57.90, 61.90) | 60.45 (58.42, 61.98) | 0.188 | 59.85 (57.58, 61.92) | 60.60 (58.58, 63.02) | 0.032 |
| Lymphocyte (×109/L), median (IQR) | 1.86 (1.53, 2.19) | 1.86 (1.45, 2.29) | 0.932 | 1.81 (1.47, 2.19) | 1.83 (1.49, 2.19) | 0.986 |
| PNI (%), median (IQR) | 69.20 (66.90, 71.90) | 69.68 (66.46, 72.35) | 0.278 | 69.47 (66.58, 71.18) | 69.40 (67.79, 72.03) | 0.21 |
| Neutrophil (×109/L), median (IQR) | 2.96 (2.42, 3.46) | 3.00 (2.48, 3.80) | 0.323 | 3.03 (2.55, 3.91) | 2.87 (2.38, 3.86) | 0.595 |
| Eosinophil (×109/L), median (IQR) | 0.09 (0.06, 0.15) | 0.12 (0.06, 0.21) | 0.033 | 0.10 (0.07, 0.16) | 0.09 (0.06, 0.16) | 0.603 |
| Basophil (×109/L), median (IQR) | 0.03 (0.02, 0.04) | 0.03 (0.02, 0.04) | 0.284 | 0.03 (0.02, 0.04) | 0.03 (0.02, 0.03) | 0.184 |
| Monocyte (×109/L), median (IQR) | 0.39 (0.32, 0.48) | 0.42 (0.33, 0.49) | 0.117 | 0.42 (0.35, 0.51) | 0.42 (0.33, 0.54) | 0.739 |
| Erythrocyte (×1012/L), median (IQR) | 4.46 (4.18, 4.75) | 4.54 (4.29, 4.93) | 0.059 | 4.44 (4.16, 4.78) | 4.55 (4.17, 4.86) | 0.291 |
| Hemoglobin (g/L), median (IQR) | 135.00 (128.00, 144.00) | 139.50 (130.00, 149.00) | 0.041 | 134.00 (127.00, 143.25) | 139.00 (126.75, 148.50) | 0.33 |
| Platelet (×109/L), median (IQR) | 232.00 (207.00, 272.00) | 239.00 (209.50, 276.00) | 0.622 | 237.50 (201.25, 281.50) | 234.50 (205.25, 257.25) | 0.616 |
| NLR (%), median (IQR) | 1.60 (1.28, 2.03) | 1.67 (1.27, 2.23) | 0.363 | 1.71 (1.35, 2.11) | 1.67 (1.22, 2.19) | 0.826 |
| PLR (%), median (IQR) | 128.66 (107.41, 153.85) | 131.50 (104.25, 166.56) | 0.587 | 137.07 (106.19, 162.07) | 130.97 (99.35, 159.77) | 0.746 |
| MLR (%), median (IQR) | 0.21 (0.17, 0.27) | 0.22 (0.18, 0.28) | 0.161 | 0.22 (0.18, 0.27) | 0.22 (0.19, 0.28) | 0.918 |
| dNLR (%), median (IQR) | 1.25 (1.00, 1.50) | 1.26 (1.01, 1.63) | 0.536 | 1.26 (1.00, 1.54) | 1.26 (0.91, 1.60) | 0.802 |
| NLPR (%), median (IQR) | 0.01 (0.01, 0.01) | 0.01 (0.00, 0.01) | 0.496 | 0.01 (0.01, 0.01) | 0.01 (0.01, 0.01) | 0.862 |
| SIRI (%), median (IQR) | 0.59 (0.46, 0.89) | 0.66 (0.48, 0.92) | 0.132 | 0.65 (0.49, 0.95) | 0.63 (0.45, 1.14) | 0.809 |
| AISI (%), median (IQR) | 143.50 (96.94, 213.11) | 161.29 (112.10, 227.89) | 0.119 | 161.00 (109.14, 259.27) | 152.14 (95.38, 228.30) | 0.869 |
| SII (%), median (IQR) | 362.80 (284.64, 487.08) | 392.56 (307.21, 513.13) | 0.208 | 393.73 (282.70, 541.46) | 369.95 (277.57, 516.84) | 0.588 |
| PIV (%), median (IQR) | 143.50 (96.94, 213.11) | 161.29 (112.10, 227.89) | 0.119 | 161.00 (109.14, 259.27) | 152.14 (95.38, 228.30) | 0.869 |
| Blood sugar(mmol/L), median (IQR) | 5.04 (4.69, 5.52) | 5.10 (4.71, 5.67) | 0.455 | 5.04 (4.69, 5.61) | 5.22 (4.89, 5.71) | 0.092 |
| Complement C1q(mg/L), median (IQR) | 171.60 (153.90, 190.10) | 171.80 (153.98, 192.02) | 0.811 | 169.50 (149.85, 190.50) | 166.70 (154.23, 188.52) | 0.76 |
| LDH (U/L), median (IQR) | 191.00 (172.00, 212.00) | 188.50 (167.25, 218.75) | 0.845 | 195.00 (167.50, 214.50) | 193.50 (161.50, 219.25) | 0.825 |
| SA (mg/dL), median (IQR) | 52.80 (48.80, 56.50) | 53.40 (49.02, 58.27) | 0.207 | 52.50 (48.77, 57.28) | 53.45 (49.53, 59.20) | 0.434 |
| 5’-NT (U/L), median (IQR) | 4.00 (3.00, 5.00) | 4.00 (3.00, 5.00) | 0.653 | 4.00 (3.00, 5.00) | 4.00 (3.00, 5.00) | 0.251 |
| Pro-GRP (pg/mL), median (IQR) | 41.96 (33.49, 46.73) | 41.83 (32.18, 45.64) | 0.479 | 41.96 (32.49, 45.72) | 41.64 (32.70, 42.81) | 0.724 |
| SCC (ng/mL), median (IQR) | 1.03 (0.82, 1.73) | 1.04 (0.72, 1.59) | 0.258 | 1.03 (0.76, 1.76) | 1.09 (0.70, 1.83) | 0.684 |
| Cyfra21-1 (ng/mL), median (IQR) | 2.28 (1.55, 2.37) | 2.32 (1.82, 2.73) | 0.035 | 2.19 (1.50, 2.44) | 2.27 (1.67, 2.43) | 0.563 |
| CEA (ng/mL), median (IQR) | 2.28 (1.36, 2.43) | 2.32 (1.65, 3.14) | 0.013 | 2.04 (1.26, 2.45) | 2.32 (1.91, 2.64) | 0.052 |
| CA125 (U/mL), median (IQR) | 10.72 (7.52, 11.20) | 10.72 (7.64, 12.28) | 0.366 | 10.72 (7.40, 11.72) | 10.22 (7.65, 10.72) | 0.613 |
| NSE (ng/mL), median (IQR) | 19.45 (16.70, 20.50) | 19.35 (14.90, 22.05) | 0.505 | 19.42 (15.75, 21.00) | 17.60 (15.40, 19.49) | 0.3 |
| Age (years), median (IQR) | 55.00 (46.00, 60.00) | 61.00 (52.00, 67.00) | < 0.001 | 56.00 (47.75, 62.00) | 60.00 (53.50, 65.25) | 0.041 |
| BMI (kg/m2), median (IQR) | 24.39 (22.27, 26.35) | 24.57 (22.62, 26.20) | 0.712 | 24.65 (22.48, 26.32) | 24.90 (22.65, 27.14) | 0.448 |
| FEV1% predicted (%), median (IQR) | 105.92 (96.93, 113.73) | 109.20 (95.46, 118.07) | 0.204 | 105.32 (94.40, 113.67) | 102.44 (93.88, 112.58) | 0.747 |
| MVV% predicted (%), median (IQR) | 103.87 (91.91, 116.64) | 106.58 (91.88, 119.90) | 0.183 | 103.80 (91.20, 115.39) | 103.85 (88.55, 112.29) | 0.717 |
| Maximum diameter (cm), median (IQR) | 0.80 (0.70, 1.10) | 1.40 (1.20, 1.70) | < 0.001 | 0.80 (0.70, 1.00) | 1.20 (1.00, 1.50) | < 0.001 |
IAC Invasive adenocarcinoma, COPD Chronic obstructive pulmonary diseases, ASA American Society of Anesthesiologists, PNI Prognostic nutritional index, NLR Neutrophil-lymphocyte ratio, PLR Platelet-lymphocyte ratio, MLR Monocyte-lymphocyte ratio, dNLR derived neutrophil-to-lymphocyte ratio, NLPR Neutrophil to lymphocyte and platelet ratio, SIRI Systemic inflammatory response syndrome, AISI Aggregate index of systemic inflammation, SII Systemic inflammation index, PIV Pan-immune-inflammation value, LDH Lactate dehydrogenase, SA Serum amyloid, 5'-NT 5'-nucleotidase, Pro-GRP Pro-gastrin-releasing peptide, SCC Squamous cell carcinoma, Cyfra21-1 Cytokeratin 19-fragments, CEA Carcinoembryonic antigen, CA125 Carcinoma antigen 125, NSE Neuron-specific enolase, BMI Body mass index, FEV1 Forced expiratory volume in one second, MVV Maximal voluntary ventilation
Identification of risk factors for pGGNs measuring ≤ 2 cm
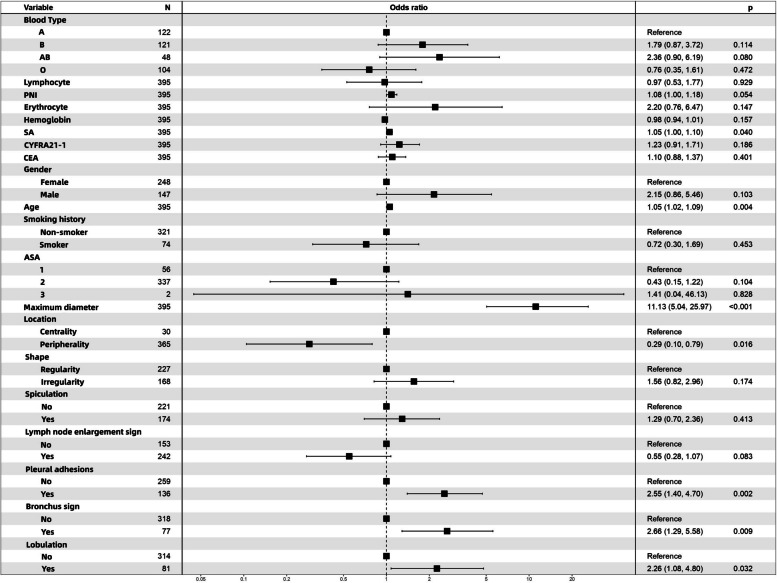
Univariate and multivariate logistic regression analyses were performed on the training cohort to explore the independent risk factors for IACs in pGGNs. Table 3 shows the results of the logistic regression analysis. Via univariate analysis, up to 20 factors were identified as potential risk factors for IACs in pGGNs measuring ≤ 2 cm (P < 0.2). Further multivariate logistic regression analysis based on these 20 univariate variables with P < 0.2 led to the identification of seven indicators, namely, maximum tumor diameter [odds ratio (OR) = 11.130; 95% confidence interval (CI): 5.044–25.966; P < 0.001]; age (OR = 1.054; 95% CI: 1.018–1.094; P = 0.004); SA (OR = 1.050; 95% CI:1.002–1.100; P = 0.04); pleural effusion sign (yes vs. no; OR = 2.548; 95% CI: 1.398–4.700; P = 0.002); bronchus sign (yes vs. no; OR = 2.662; 95% CI: 1.286–5.576; P = 0.009); tumor location (centrality vs. peripherality; OR = 0.288; 95% CI: 0.104–0.794; P = 0.016); and lobulation (yes vs. no; OR = 2.260; 95% CI: 1.078–4.799; P = 0.032). The forest plot for the multivariate logistic regression analysis is shown in Fig. 2.
Table 3.
Univariate and multivariate logistic regression analysis of IAC factors of pGGNs within 2 cm in a training cohort
| Variables | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| OR (95%CI) | P value | OR (95%CI) | P value | |
| Gender | ||||
| Female | Ref. | Ref. | Ref. | Ref. |
| Male | 1.615 (1.043, 2.499) | 0.031 | 2.154 (0.859, 5.458) | 0.103 |
| Smoking history | ||||
| Non-smoker | Ref. | Ref. | Ref. | Ref. |
| Smoker | 1.697 (0.999, 2.856) | 0.048 | 0.721 (0.304, 1.685) | 0.453 |
| Pleural effusion sign | ||||
| No | Ref. | Ref. | Ref. | Ref. |
| Yes | 2.779 (1.785, 4.344) | < 0.001 | 2.548 (1.398, 4.700) | 0.002 |
| Bronchus sign | ||||
| No | Ref. | Ref. | Ref. | Ref. |
| Yes | 4.103 (2.452, 6.935) | < 0.001 | 2.662 (1.286, 5.576) | 0.009 |
| Location | ||||
| Centrality | Ref. | Ref. | Ref. | Ref. |
| Peripherality | 0.415 (0.194, 0.884) | 0.022 | 0.288 (0.104, 0.794) | 0.016 |
| Lobulation | ||||
| No | Ref. | Ref. | Ref. | Ref. |
| Yes | 2.760 (1.669, 4.575) | < 0.001 | 2.260 (1.078, 4.799) | 0.032 |
| Vascular penetration sign | ||||
| No | Ref. | Ref. | Ref. | Ref. |
| Yes | 2.372 (1.492, 3.846) | < 0.001 | 0.549 (0.275, 1.074) | 0.083 |
| Shape | ||||
| Regularity | Ref. | Ref. | Ref. | Ref. |
| Irregularity | 3.798 (2.436, 5.993) | < 0.001 | 1.559 (0.819, 2.962) | 0.174 |
| Spiculation | ||||
| No | Ref. | Ref. | Ref. | Ref. |
| Yes | 3.105 (2.001, 4.865) | < 0.001 | 1.288 (0.699, 2.360) | 0.413 |
| Blood type | ||||
| A | Ref. | Ref. | Ref. | Ref. |
| B | 1.550 (0.898, 2.698) | 0.117 | 1.790 (0.875, 3.722) | 0.114 |
| AB | 2.009 (0.991, 4.055) | 0.051 | 2.356 (0.898, 6.187) | 0.08 |
| O | 0.986 (0.541, 1.788) | 0.964 | 0.757 (0.352, 1.609) | 0.472 |
| ASA | ||||
| 1 | Ref. | Ref. | Ref. | Ref. |
| 2 | 1.982 (1.020, 4.169) | 0.054 | 0.425 (0.153, 1.222) | 0.104 |
| 3 | 4.091 (0.154, 109.087) | 0.332 | 1.412 (0.045, 46.131) | 0.828 |
| Maximum diameter | 16.916 (8.943, 33.624) | < 0.001 | 11.130 (5.044, 25.966) | < 0.001 |
| Age | 1.056 (1.033, 1.081) | < 0.001 | 1.054 (1.018, 1.094) | 0.004 |
| SA | 1.046 (1.013, 1.081) | 0.006 | 1.050 (1.002, 1.100) | 0.04 |
| PNI | 1.063 (1.013, 1.120) | 0.016 | 1.084 (1.002, 1.183) | 0.054 |
| Erythrocyte | 1.801 (1.130, 2.895) | 0.014 | 2.196 (0.759, 6.471) | 0.147 |
| Hemoglobin | 1.015 (1.000, 1.030) | 0.048 | 0.975 (0.941, 1.010) | 0.157 |
| Cyfra21_1 | 1.438 (1.137, 1.846) | 0.003 | 1.232 (0.911, 1.705) | 0.186 |
| CEA | 1.198 (1.017, 1.421) | 0.032 | 1.099 (0.877, 1.366) | 0.401 |
| Lymphocyte | 1.445 (1.012, 2.068) | 0.043 | 0.973 (0.527, 1.769) | 0.929 |
| Hypertension | ||||
| No | Ref. | Ref. | ||
| Yes | 1.372 (0.836, 2.227) | 0.205 | ||
| Diabetes | ||||
| No | Ref. | Ref. | ||
| Yes | 0.966 (0.471, 1.887) | 0.922 | ||
| COPD | ||||
| NO | Ref. | Ref. | ||
| Yes | 2.258 (0.268, 19.002) | 0.418 | ||
| Calcification | ||||
| No | Ref. | Ref. | ||
| Yes | 4818859.448 (0.000, NA) | 0.98 | ||
| Cavitation sign | ||||
| No | Ref. | Ref. | ||
| Yes | 1.181 (0.581, 2.307) | 0.633 | ||
| Pleural effusion sign | ||||
| No | Ref. | Ref. | ||
| Yes | 0 | 0.981 | ||
| Lymph node enlargement sign | ||||
| No | Ref. | Ref. | ||
| Yes | 1.479 (0.715, 2.972) | 0.278 | ||
| AISI | 1.000 (0.999, 1.001) | 0.811 | ||
| Albumin | 1.037 (0.983, 1.099) | 0.204 | ||
| Basophil | 4.812 (0.191, 336.295) | 0.339 | ||
| Blood sugar | 1.064 (0.905, 1.248) | 0.432 | ||
| BMI | 1.014 (0.947, 1.086) | 0.681 | ||
| CA125 | 1.010 (0.978, 1.042) | 0.521 | ||
| Complement C1q | 1.000 (0.993, 1.007) | 0.996 | ||
| dNLR | 0.845 (0.536, 1.309) | 0.46 | ||
| Eosinophil | 1.588 (0.639, 4.419) | 0.312 | ||
| FEV1% predicted (%) | 1.005 (0.992, 1.019) | 0.438 | ||
| IDH | 1.000 (0.993, 1.006) | 0.917 | ||
| MLR | 1.054 (0.558, 1.769) | 0.83 | ||
| Monocyte | 1.205 (0.879, 1.952) | 0.279 | ||
| MVV% predicted (%) | 1.001 (0.998, 1.005) | 0.396 | ||
| Neutrophil | 1.077 (0.877, 1.317) | 0.473 | ||
| NLPR | 0.003 | 0.813 | ||
| NLR | 0.923 (0.685, 1.226) | 0.589 | ||
| NSE | 1.010 (0.975, 1.045) | 0.572 | ||
| PIV | 1.000 (0.999, 1.001) | 0.811 | ||
| Platelet | 1.000 (0.997, 1.004) | 0.81 | ||
| PLR | 0.998 (0.994, 1.002) | 0.41 | ||
| Pro-GRP | 0.997 (0.980, 1.011) | 0.663 | ||
| SCC | 0.849 (0.583, 1.167) | 0.361 | ||
| SII | 1.000 (0.999, 1.001) | 0.465 | ||
| SIRI | 1.028 (0.836, 1.236) | 0.748 | ||
| 5’-NT | 1.004 (0.886, 1.121) | 0.944 | ||
IAC Invasive adenocarcinoma, COPD Chronic obstructive pulmonary diseases, ASA American Society of Anesthesiologists, PNI Prognostic nutritional index, NLR Neutrophil-lymphocyte ratio, PLR Platelet-lymphocyte ratio, MLR Monocyte-lymphocyte ratio, dNLR derived neutrophil-to-lymphocyte ratio, NLPR Neutrophil to lymphocyte and platelet ratio, SIRI Systemic inflammatory response syndrome, AISI Aggregate index of systemic inflammation, SII Systemic inflammation index, PIV Pan-immune-inflammation value, LDH, Lactate dehydrogenase, SA Serum amyloid, 5'-NT 5'-nucleotidase, Pro-GRP Pro-gastrin-releasing peptide, SCC Squamous cell carcinoma, Cyfra21-1 Cytokeratin 19-fragments, CEA Carcinoembryonic antigen, CA125 Carcinoma antigen 125, NSE Neuron-specific enolase, BMI Body mass index, FEV1 Forced expiratory volume in one second, MVV Maximal voluntary ventilation
Fig. 2.
Multi-factor logistic regression analysis of forest plots. PNI, prognostic nutritional index; SA, serum amyloid; Cyfra21-1, cytokeratin 19-fragments; CEA, carcinoembryonic antigen; ASA, American Society of Anesthesiologists
Nomogram establishment
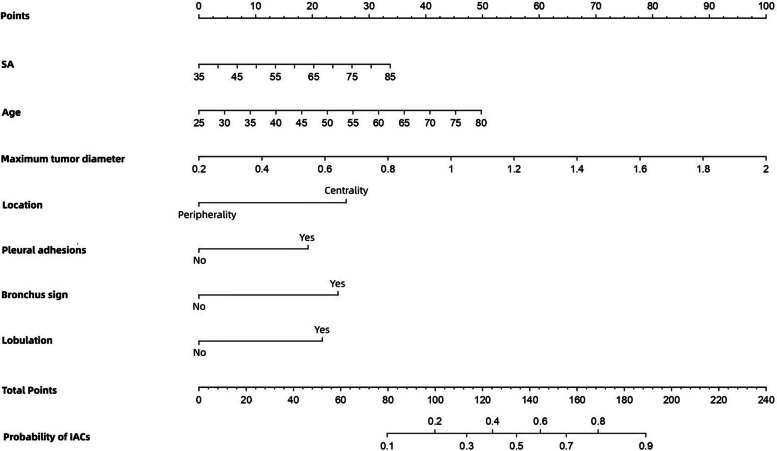
All seven independent risk factors for pGGNs measuring ≤ 2 cm were included in the logistic regression models. Details regarding the prediction model are presented in Table 4. The probability of the occurrence of IACs in small pGGNs was then calculated according to the following equation: ln (p/1-p) = 2.41 × maximum tumor diameter + 0.053 × age + 0.049 × SA – 1.245 × tumor location (centrality = 0; peripherality = 1) + 0.935 × pleural adhesions (no = 0; yes = 1) + 0.979 × bronchus sign (no = 0; yes = 1) + 0.815 × lobulation (no = 0; yes = 1) – 12.759. Further, we plotted the predicted nomogram for the probability of IACs in pGGNs of size ≤ 2 cm using R statistical software, based on the above equation (Fig. 3). The nomogram comprised 10 axes, with axes 2–8 representing the seven variables in the prediction model. By drawing a line vertically to the highest-point axis, the estimated score of each risk factor could be computed and added to obtain the total risk score, which was then used to predict the probability of pGGNs developing IACs before surgery. Thus, appropriate treatment and surgery modalities can be selected.
Table 4.
Details of the predictive model used to calculate the probability of IAC in pGGNs measurements ≤ 2 cm
| Risk factors | Estimate | Std. Error | Statistic | OR (95% CI) | p |
|---|---|---|---|---|---|
| Intercept | -12.759 | 4.13 | 0 | 0.002 | |
| Maximum diameter | 2.41 | 0.417 | 5.784 | 11.130 (5.044, 25.966) | < 0.001 |
| Age | 0.053 | 0.018 | 2.871 | 1.054 (1.018, 1.094) | 0.004 |
| Lymphocyte | -0.027 | 0.308 | -0.089 | 0.973 (0.527, 1.769) | 0.929 |
| PNI | 0.081 | 0.042 | 1.931 | 1.084 (1.002, 1.183) | 0.054 |
| Erythrocyte | 0.787 | 0.543 | 1.449 | 2.196 (0.759, 6.471) | 0.147 |
| Hemoglobin | -0.025 | 0.018 | -1.414 | 0.975 (0.941, 1.010) | 0.157 |
| SA | 0.049 | 0.024 | 2.049 | 1.050 (1.002, 1.100) | 0.04 |
| CYFRA21-1 | 0.209 | 0.158 | 1.321 | 1.232 (0.911, 1.705) | 0.186 |
| CEA | 0.094 | 0.112 | 0.84 | 1.099 (0.877, 1.366) | 0.401 |
| Gender | |||||
| Female | Ref. | ||||
| Male | 0.768 | 0.47 | 1.633 | 2.154 (0.859, 5.458) | 0.103 |
| Smoking history | |||||
| Non-smoker | Ref. | ||||
| Smoker | -0.327 | 0.435 | -0.751 | 0.721 (0.304, 1.685) | 0.453 |
| Blood type | |||||
| A | Ref. | ||||
| B | 0.582 | 0.368 | 1.582 | 1.790 (0.875, 3.722) | 0.114 |
| AB | 0.857 | 0.49 | 1.749 | 2.356 (0.898, 6.187) | 0.08 |
| O | -0.278 | 0.386 | -0.72 | 0.757 (0.352, 1.609) | 0.472 |
| ASA | |||||
| 1 | Ref. | ||||
| 2 | -0.855 | 0.526 | -1.625 | 0.425 (0.153, 1.222) | 0.104 |
| 3 | 0.345 | 1.586 | 0.218 | 1.412 (0.045, 46.131) | 0.828 |
| Location | |||||
| Centrality | Ref. | ||||
| Peripherality | -1.245 | 0.515 | -2.418 | 0.288 (0.104, 0.794) | 0.016 |
| Shape | |||||
| Regularity | Ref. | ||||
| Irregularity | 0.444 | 0.327 | 1.358 | 1.559 (0.819, 2.962) | 0.174 |
| Spiculation | |||||
| No | Ref. | ||||
| Yes | 0.253 | 0.309 | 0.819 | 1.288 (0.699, 2.360) | 0.413 |
| Vascular penetration sign | |||||
| No | Ref. | ||||
| Yes | -0.601 | 0.346 | -1.736 | 0.549 (0.275, 1.074) | 0.083 |
| Pleural adhesions | |||||
| No | Ref. | ||||
| Yes | 0.935 | 0.308 | 3.034 | 2.548 (1.398, 4.700) | 0.002 |
| Bronchus sign | |||||
| No | Ref. | ||||
| Yes | 0.979 | 0.373 | 2.625 | 2.662 (1.286, 5.576) | 0.009 |
| Lobulation | |||||
| No | Ref. | ||||
| Yes | 0.815 | 0.38 | 2.147 | 2.260 (1.078, 4.799) | 0.032 |
IAC Invasive adenocarcinoma, COPD Chronic obstructive pulmonary diseases, ASA American Society of Anesthesiologists, PNI Prognostic nutritional index, NLR Neutrophil-lymphocyte ratio, PLR Platelet-lymphocyte ratio, MLR Monocyte-lymphocyte ratio, dNLR derived neutrophil-to-lymphocyte ratio, NLPR Neutrophil to lymphocyte and platelet ratio, SIRI Systemic inflammatory response syndrome, AISI Aggregate index of systemic inflammation, SII Systemic inflammation index, PIV Pan-immune-inflammation value, LDH Lactate dehydrogenase, SA Serum amyloid, 5'-NT 5'-nucleotidase, Pro-GRP Pro-gastrin-releasing peptide, SCC Squamous cell carcinoma, Cyfra21-1 Cytokeratin 19-fragments, CEA Carcinoembryonic antigen, CA125 Carcinoma antigen 125, NSE Neuron-specific enolase, BMI Body mass index, FEV1 Forced expiratory volume in one second, MVV Maximal voluntary ventilation
Fig. 3.
Nomogram for predicting the probability of IAC for pGGN ≤ 2 cm. SA, serum amyloid
As shown in this nomogram, there are a total of 10 axes, and axes 2–8 represent the seven variables in the prediction model. By plotting a line perpendicular to the highest point axis, the estimated score for each risk factor can be calculated and can be further summed to obtain a total score. The total point axis was then used to predict the probability of IAC for pGGNs measuring ≤ 2 cm before surgery.
Predictive performance and nomogram validation
The discriminatory power of the prediction model and nomogram was assessed via ROC curve analyses. As shown in Fig. 4, the area under the ROC curve (AUC) for the training cohort was 0.839 (95% CI: 0.798–0.879) and for the validation cohort, it was 0.782 (95% CI: 0.706–0.858), indicating that nomogram showed good predictive accuracy. Further, the ROC curve for the training cohort had cutoff, sensitivity, and specificity values of 0.274, 0.811, and 0.733, respectively, indicating excellent performance (Table 5).
Fig. 4.
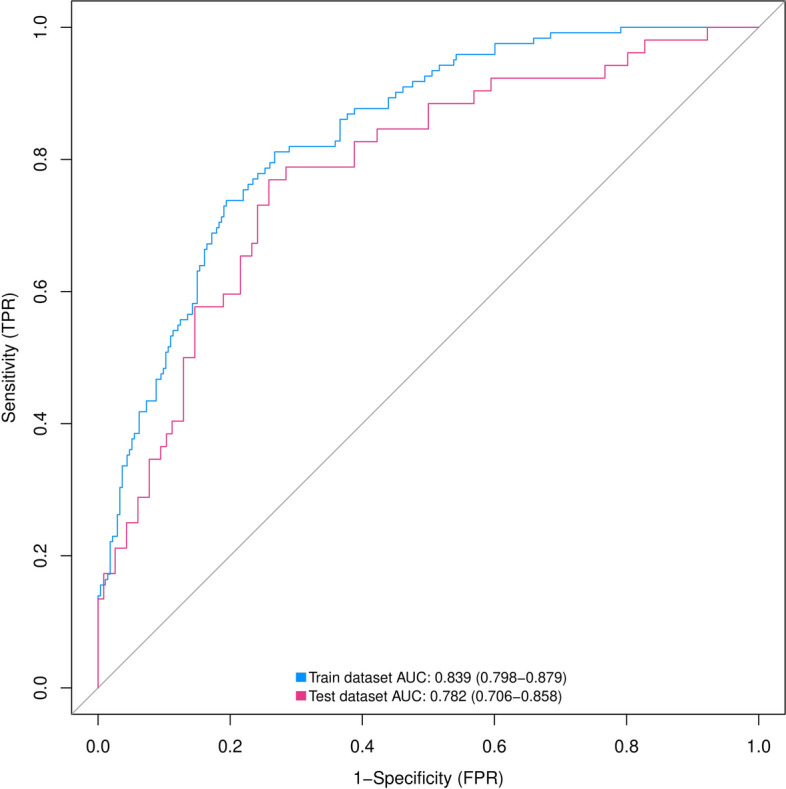
ROC curves of nomograms predicting IAC for pGGN ≤ 2 cm in the training and validation groups. ROC, receiver operating characteristic; AUC, area under the ROC curve; IAC, invasive adenocarcinoma; pGGN, pure ground glass nodule
Table 5.
Results of ROC curve for training cohort
| Characteristics | Value |
|---|---|
| Threshold | 0.274 |
| Specificity | 0.733 |
| Sensitivity | 0.811 |
| Accuracy | 0.757 |
| TN | 200 |
| TP | 99 |
| FN | 23 |
| FP | 73 |
| NPR | 0.897 |
| PPV | 0.576 |
| FDR | 0.424 |
| FPR | 0.267 |
| TPR | 0.811 |
| TNR | 0.733 |
| FNR | 0.189 |
| 1-specificity | 0.267 |
| 1-sensitivity | 0.189 |
| 1-accuracy | 0.243 |
| 1-NPV | 0.103 |
| 1-PPV | 0.424 |
| Precision | 0.576 |
| Recall | 0.811 |
| Youden index | 1.544 |
| Closest.topleft | 0.107 |
TP True positive, FP False positive, TN True negative, FN False negative, TPR True positive rate, FPR False positive rate, TNR True negative rate, FNR False negative rate, PPV Positive predict value, NPR Negative predict value, FDR False discovery rate
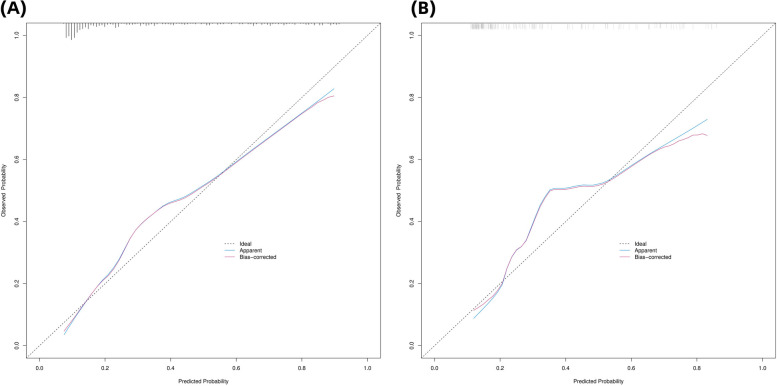
We used the Hosmer-Lemeshow test and calibration charts to assess the calibration capability of our model. Thus, the observed Hosmer-Lemeshow test P-values for the training and validation cohorts were 0.1071 and 0.2595, respectively, suggesting that the difference between the predicted and actual observed probabilities was not significant. Therefore, the nomogram showed good calibration as indicated by the calibration plots corresponding to the training (Fig. 5a) and validation cohorts (Fig. 5b). Further, the bias-corrected C-indices for the training and validation cohorts were 0.840 and 0.785, respectively.
Fig. 5.
(A, B): Calibration curves of the prediction nomogram in the training cohort (A) and validation cohort (B). IAC, invasive adenocarcinoma; pGGN, pure ground glass nodule
The X-axis represents the probability predicted by the nomogram and the Y-axis represents the actual probability of pGGN being an ICA within 2 cm. The black dashed line represents the ideal curve, the blue solid line represents the apparent curve (uncorrected), and the red solid line represents the deviation curve corrected by bootstrap method (B = 1000 times).
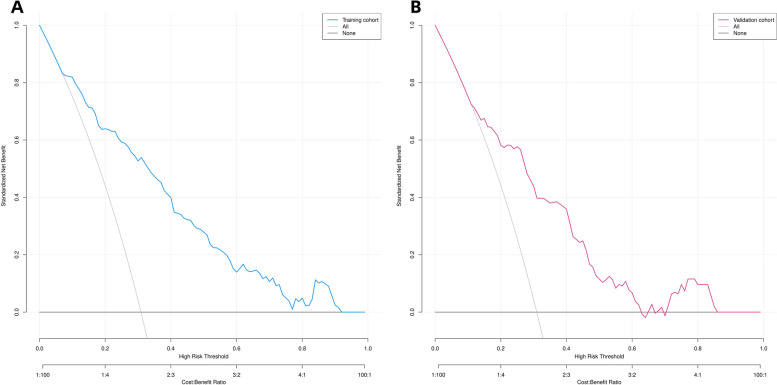
Clinical utility of the predictive nomogram
We performed DCA to assess the clinical utility of the predictive nomograms. As shown in Fig. 6a and b, the nomogram provided a greater net benefit and wider threshold probabilities for predicting the risk of IACs in pGGNs measuring ≤ 2 cm in both the training and validation cohorts, demonstrating its clinical utility. Additionally, our clinical impact curve (Fig. 7) showed that a high benefit ratio could be obtained within a probability threshold of 0.5–1.0. This observation suggested that the present model can be used in clinical practice to predict the probability of IACs developing in small pGGNs, helping surgeons make better clinical decisions.
Fig. 6.
(A, B): Decision curve analysis of predicted nomogram in the training cohort (A) and validation cohort (B)
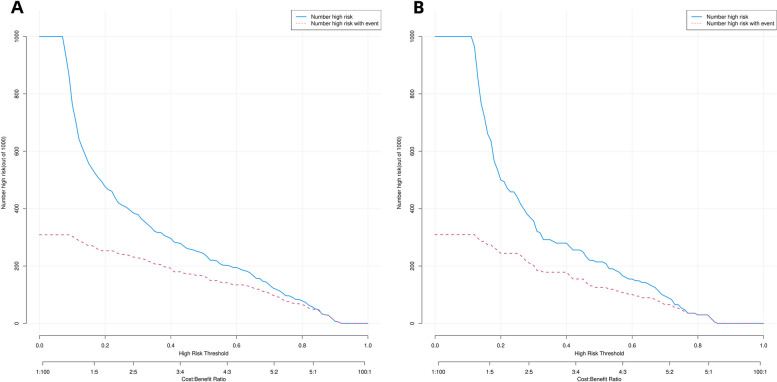
Fig. 7.
(A, B): Clinical impact curves of predicted nomogram in the training cohort (A) and validation cohort (B)
The y-axis measures the net gain, and the black line represents the hypothesis that pGGNs within 2 cm are non-IACs in nature, and the gray line represents the hypothesis that pGGNs measuring ≤ 2 cm are IACs. The blue line in Fig. 7A represents the training cohort, and the red line in Fig. 7B represents the validation cohort.
The horizontal coordinate is the probability threshold and the vertical coordinate is the number of people. The blue line indicates the number of people whose pGGNs were judged by the model to be IAC at different probability thresholds; the red line indicates the number of people whose pGGNs were judged by the model to be IAC and nodal true IAC at different probability thresholds. At the bottom, the cost: benefit ratio is also added, indicating the ratio of loss to benefit at different probability thresholds.
Discussion
The definitive pathological diagnosis of pGGN with diameter ≤ 2 cm is relatively challenging owing technical limitations and the potential risk of complications when performing coarse needle aspiration biopsy for pGGN [34, 35]. With the increasing number of cases of early stage lung cancer presenting as pulmonary pGGNs, it is crucial to determine whether a pulmonary pGGN is an IAC; such information is vital for selecting relevant treatment options for patients. In this study, we showed that the percentages of IAC in pGGN were 30.9%, 30.9%, and 31.0% for the total, training, and validation cohorts, respectively. Attempts have been made in previous studies to distinguish IAC from pre-infiltrative lesions; however, these studies did not include benign GGN [18–26]. Further, studies have also been conducted to analyze the differentiation of solid, partial, and GGN nodules [27, 28]. However, clearly distinguishing IAC from non-IAC in pGGN remains challenging. Therefore, in this study, we aimed to explore the potential predictors that can be used to distinguish IAC and non-IAC in pGGN. Thus, we identified seven correlated factors, namely, maximum tumor diameter, age, SA, pleural effusion sign, bronchial signs, tumor location, and lobulation.
Nodule size is an important parameter for assessing GGN invasiveness. Several investigations have shown that increasing adenocarcinoma aggressiveness is associated increasing pGGN lesion size increases [21, 36]. It has also been reported that a critical nodule size of 1 cm is optimal for predicting aggressive pGGN with 100% specificity [20]. However, Wu et al. demonstrated that nodule size cannot be used to distinguish between infiltrative and pre-infiltrative lesions; the mean nodule size in their study was < 1 cm [21]. In our predictive nomogram, the maximum tumor diameter was identified as the most significant risk factor, consistent with the results of previous studies [37, 38]. Notably, in China, the prevalence of IAC in pGGN is higher than that in pGGN of a similar size in Western countries (24%) [4]. These observation suggests that, owing to the high risk associated with IAC, biopsy or surgery should be considered as soon as possible when pGGN size is > 1 cm.
Hu et al. indicated that age ≥ 60 years is a risk factor for IAC [39]. Consistent with their findings, the results of this study suggested that the probability of IAC in pGGN increases with age. However, this risk factor is not well recognized. Therefore, clinicians should be cautious when pGGN is observed in older patients. Further, such patients should undergo more frequent follow-up CT scans and be considered for biopsy or surgery.
Previous studies have also demonstrated that the CT features of pulmonary nodules can be used to assess their aggressiveness. These imaging features included lobulation, spiculation, bronchus signs, cavitation signs, pleural adhesion signs, and nodule shape [24, 25, 27, 40–52]. Furuya et al. reported that 82% of lobulated nodules and 97% of acinar nodules are malignant [53]. Lobulation and spiculation of pGGN are also more common in invasive lesions than in pre-infiltrative lesions [18]. Further, Lee et al. found that lobulation is more common in IAC than in pre-infiltrative lesions [42]. However, lobulation was not included as a risk factor for malignant lung nodules in the Herder model [51]. In this study, we identified lobulation as a risk factor for the occurrence of IAC in pGGN; however, no significant differences were observed in this regard with respect to spiculation. A possible reason for this observation is the limited number of nodules with spiculations included in the study. The bronchus signs observed in this study constituted another CT feature that showed association with malignancy. Bronchial signs were more frequently observed in patients with malignant GGN than in those with benign GGN. Reportedly, patients with IAC present with bronchial signs more frequently than those with AIS [45, 54]. Thus, based on our results, bronchial signs were identified as significant predictors of IAC (P = 0.009).
Pleural effusions have rarely been associated with the aggressiveness of pGGN in the literature. Our study demonstrated that pleural effusion signs on CT images can be used as a predictor of IAC in pGGN. At initial diagnosis, approximately 15% of patients with lung cancer present with pleural effusion, while 50% of patients at the advanced stages of the disease present with pleural effusion [55, 56]. If the pleural effusion is malignant, the patient’s prognosis is poor. Therefore, if signs of pleural effusion are observed in CT images for patients with pGGN, treatment should be considered as soon as possible.
The distinction between the centrality and peripherality of nodules is also a significant indicator of the risk of IAC in pGGN. Our results suggested that central pGGNs are more likely to be aggressive than peripheral pGGNs. However, previous studies with a focus on the relationship between centrality and nodal aggressiveness did not show any significant association in this regard [57]. Therefore, a multicenter clinical study with a large sample size is required to validate our findings.
Amylase production in lung cancer has been identified via pathological or immunohistochemical analyses, and it has also been confirmed that its serum level decreases after resection. Previous pathological and biochemical studies also support the existence of a mechanism by which lung cancer tissues produce amylase [58, 59]. Further, several case reports have described high serum amylase levels in patients with lung cancer [60–70]. However, this study is the first identify SA as a risk factor for predicting the probability of IAC occurrence in pGGN. In an immunohistochemical study, amylase levels in lung cancer tissues were found to be higher than those in normal lung tissues [63]. In contrast, other studies have shown that inflammatory as well as normal lung tissue can also produce amylase [58, 71] and that increased positive staining for amylase in lung cancer tissue is not associated with hyperamylasemia [72]. Adenocarcinoma is the predominant histological type of amylase-producing lung cancer [59, 73]. Our results revealed a positive correlation between SA concentration in vivo and nodule invasiveness, i.e., a higher SA concentration resulted in a higher probability of IAC occurrence in pGGN. Therefore, SA may be a new indicator for monitoring and evaluating patient prognosis.
Additionally, this study showed that multiple serum tumor biomarkers (CEA, CYFRA21-1, SCC, NSE, and CA125) had no significant value in distinguishing IAC from pGGN. The reason for their ineffectiveness may be that in early-stage lung adenocarcinomas, tumor proteins are rarely secreted into the bloodstream.
The performance of our constructed predictive model was found to be comparable with those of previously published predictive models by a considerable margin. First, we introduced benign tumors and combined them in a non-IAC cohort. This grouping method showed great value for guiding clinical decision making given that pGGNs observed in clinical practice cannot be completely excluded as benign. Second, we randomly divided the collected cases into the training and internal validation cohorts, thereby strengthening our conclusions. Third, we collected comprehensive clinical and imaging data and provided a clear pathological diagnosis for each patient. Fourth, the corrected C-index value for the training cohort of the model was 0.837, and the Hosmer-Lemeshow test P-value was 0.1071, indicating good performance. Further, the ROC, calibration, and DCA curves performed well, and the accuracy and reliability of the model were satisfactory. Fifth, all the important risk factors in our nomogram are readily available and prevalent in clinical practice.
This study had some limitations. First, this was a single-center retrospective study that included only surgically resected pGGNs; therefore, validation bias was inherent in our study design. Second, this was a retrospective cross-sectional study. Until present, no longitudinal study assessing nodal growth has been reported. Thus, further studies are required in this regard. Third, the definition of pGGN and the criteria for pathological diagnosis may vary among physicians. Fourth, the subjectivity of radiologists may have led to different judgments regarding the characteristics of the examined pulmonary nodules. Fifth, only internal validation was conducted. Therefore, further studies with the recruitment of more patients for external validation are necessary. Additionally, the applicability of our predictive nomogram remains to be confirmed in multicenter prospective clinical trials with large sample sizes.
Conclusion
In this study, we showed that the maximum tumor diameter, age, SA, pleural effusion sign, bronchial sign, tumor location, and lobulation were predictors of IAC in patients with pGGN in the lungs. We also developed and validated a novel, easy-to-use nomogram for predicting the risk of IAC occurrence in patients with pGGN measuring ≤ 2 cm in diameter, based on certain influencing factors. This tool may be used to guide clinicians in developing specific and individualized treatment strategies for patients.
Supplementary Information
Acknowledgements
We would like to thank Editage for English language editing.
We thank the imaging physicians and pathologists at Qilu Hospital for their contributions to this study.
Authors’ contributions
Conceptualization, HT and MX. Methodology, RL. Software, MX. Validation, ML and RL. Formal analysis, MX. Investigation, ZL and JL. Resources, ZL and JL. Data curation, WL and HZ. Writing—original draft preparation, MX. Writing—review and editing, MX and RL. Visualization, MX and HT. Supervision, HT. Project administration, HT. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by National Key Research and Development Program (2021YFC2500904, and 2021YFC2500905) and Natural Science Foundation of Shandong Province (ZR2021LSW006).
Availability of data and materials
The data that support the findings of this study are available on request from the corresponding author.
Declarations
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Qilu Hospital, Shandong University (registration number: KYLL-202008-023-1), and all patients signed an informed consent form for the use of their clinical information prior to the procedure. All methods were performed in accordance with the Declaration of Helsinki.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer statistics 2020: GLOBOCAN estimates of incidence and Mortality Worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Wu FZ, Huang YL, Wu CC, Tang EK, Chen CS, Mar GY, et al. Assessment of Selection Criteria for low-dose lung screening CT among Asian ethnic groups in Taiwan: from Mass Screening to specific risk-based screening for Non-smoker Lung Cancer. Clin Lung Cancer. 2016;17(5):e45–e56. doi: 10.1016/j.cllc.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 3.Lin KF, Wu HF, Huang WC, Tang PL, Wu MT, Wu FZ. Propensity score analysis of lung cancer risk in a population with high prevalence of non-smoking related lung cancer. BMC Pulm Med. 2017;17(1):120. doi: 10.1186/s12890-017-0465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu FZ, Huang YL, Wu YJ, Tang EK, Wu MT, Chen CS, et al. Prognostic effect of implementation of the mass low-dose computed tomography lung cancer screening program: a hospital-based cohort study. Eur J Cancer Prev. 2020;29(5):445–51. doi: 10.1097/CEJ.0000000000000569. [DOI] [PubMed] [Google Scholar]
- 5.Austin JH, Müller NL, Friedman PJ, Hansell DM, Naidich DP, Remy-Jardin M, et al. Glossary of terms for CT of the lungs: recommendations of the Nomenclature Committee of the Fleischner Society. Radiology. 1996;200(2):327–31. doi: 10.1148/radiology.200.2.8685321. [DOI] [PubMed] [Google Scholar]
- 6.Kim HY, Shim YM, Lee KS, Han J, Yi CA, Kim YK. Persistent pulmonary nodular ground-glass opacity at thin-section CT: histopathologic comparisons. Radiology. 2007;245(1):267–75. doi: 10.1148/radiol.2451061682. [DOI] [PubMed] [Google Scholar]
- 7.Takahashi S, Tanaka N, Okimoto T, Tanaka T, Ueda K, Matsumoto T, et al. Long term follow-up for small pure ground-glass nodules: implications of determining an optimum follow-up period and high-resolution CT findings to predict the growth of nodules. Jpn J Radiol. 2012;30(3):206–17. doi: 10.1007/s11604-011-0033-8. [DOI] [PubMed] [Google Scholar]
- 8.Lee HW, Jin KN, Lee JK, Kim DK, Chung HS, Heo EY, et al. Long-term Follow-Up of Ground-Glass nodules after 5 years of Stability. J Thorac Oncol. 2019;14(8):1370–7. doi: 10.1016/j.jtho.2019.05.005. [DOI] [PubMed] [Google Scholar]
- 9.Cho J, Kim ES, Kim SJ, Lee YJ, Park JS, Cho YJ, et al. Long-term follow-up of small Pulmonary Ground-Glass nodules stable for 3 years: implications of the proper follow-up period and risk factors for subsequent growth. J Thorac Oncol. 2016;11(9):1453–9. doi: 10.1016/j.jtho.2016.05.026. [DOI] [PubMed] [Google Scholar]
- 10.Tang EK, Chen CS, Wu CC, Wu MT, Yang TL, Liang HL, et al. Natural history of Persistent Pulmonary Subsolid nodules: Long-Term Observation of different interval growth. Heart Lung Circ. 2019;28(11):1747–54. doi: 10.1016/j.hlc.2018.08.015. [DOI] [PubMed] [Google Scholar]
- 11.Sugi K, Kobayashi S, Sudou M, Sakano H, Matsuda E, Okabe K. Long-term prognosis of video-assisted limited surgery for early lung cancer. Eur J Cardiothorac Surg. 2010;37(2):456–60. doi: 10.1016/j.ejcts.2009.07.017. [DOI] [PubMed] [Google Scholar]
- 12.Koike T, Togashi K, Shirato T, Sato S, Hirahara H, Sugawara M, et al. Limited resection for noninvasive bronchioloalveolar carcinoma diagnosed by intraoperative pathologic examination. Ann Thorac Surg. 2009;88(4):1106–11. doi: 10.1016/j.athoracsur.2009.06.051. [DOI] [PubMed] [Google Scholar]
- 13.Ito M, Miyata Y, Kushitani K, Yoshiya T, Mimae T, Ibuki Y, et al. Prediction for prognosis of resected pT1a-1bN0M0 adenocarcinoma based on tumor size and histological status: relationship of TNM and IASLC/ATS/ERS classifications. Lung Cancer. 2014;85(2):270–5. doi: 10.1016/j.lungcan.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 14.Boland JM, Froemming AT, Wampfler JA, Maldonado F, Peikert T, Hyland C, et al. Adenocarcinoma in situ, minimally invasive adenocarcinoma, and invasive pulmonary adenocarcinoma–analysis of interobserver agreement, survival, radiographic characteristics, and gross pathology in 296 nodules. Hum Pathol. 2016;51:41–50. doi: 10.1016/j.humpath.2015.12.010. [DOI] [PubMed] [Google Scholar]
- 15.Van Schil PE, Asamura H, Rusch VW, Mitsudomi T, Tsuboi M, Brambilla E, et al. Surgical implications of the new IASLC/ATS/ERS adenocarcinoma classification. Eur Respir J. 2012;39(2):478–86. doi: 10.1183/09031936.00027511. [DOI] [PubMed] [Google Scholar]
- 16.Shah R, Sabanathan S, Richardson J, Mearns AJ, Goulden C. Results of surgical treatment of stage I and II lung cancer. J Cardiovasc Surg (Torino) 1996;37(2):169–72. [PubMed] [Google Scholar]
- 17.Goldstraw P, Chansky K, Crowley J, Rami-Porta R, Asamura H, Eberhardt WE, et al. The IASLC Lung Cancer Staging Project: proposals for revision of the TNM Stage groupings in the Forthcoming (Eighth) Edition of the TNM classification for Lung Cancer. J Thorac Oncol. 2016;11(1):39–51. doi: 10.1016/j.jtho.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 18.Chu ZG, Li WJ, Fu BJ, Lv FJ. CT characteristics for Predicting Invasiveness in pulmonary pure ground-glass nodules. AJR Am J Roentgenol. 2020;215(2):351–8. doi: 10.2214/AJR.19.22381. [DOI] [PubMed] [Google Scholar]
- 19.Xiang W, Xing Y, Jiang S, Chen G, Mao H, Labh K, et al. Morphological factors differentiating between early lung adenocarcinomas appearing as pure ground-glass nodules measuring ≤ 10 mm on thin-section computed tomography. Cancer Imaging. 2014;14(1):33. doi: 10.1186/s40644-014-0033-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee SM, Park CM, Goo JM, Lee HJ, Wi JY, Kang CH. Invasive pulmonary adenocarcinomas versus preinvasive lesions appearing as ground-glass nodules: differentiation by using CT features. Radiology. 2013;268(1):265–73. doi: 10.1148/radiol.13120949. [DOI] [PubMed] [Google Scholar]
- 21.Wu F, Tian SP, Jin X, Jing R, Yang YQ, Jin M, et al. CT and histopathologic characteristics of lung adenocarcinoma with pure ground-glass nodules 10 mm or less in diameter. Eur Radiol. 2017;27(10):4037–43. doi: 10.1007/s00330-017-4829-5. [DOI] [PubMed] [Google Scholar]
- 22.Lee GD, Park CH, Park HS, Byun MK, Lee IJ, Kim TH, et al. Lung adenocarcinoma invasiveness risk in pure ground-glass opacity lung nodules smaller than 2 cm. Thorac Cardiovasc Surg. 2019;67(4):321–8. doi: 10.1055/s-0037-1612615. [DOI] [PubMed] [Google Scholar]
- 23.Lim HJ, Ahn S, Lee KS, Han J, Shim YM, Woo S, et al. Persistent pure ground-glass opacity lung nodules ≥ 10 mm in diameter at CT scan: histopathologic comparisons and prognostic implications. Chest. 2013;144(4):1291–9. doi: 10.1378/chest.12-2987. [DOI] [PubMed] [Google Scholar]
- 24.Liang J, Xu XQ, Xu H, Yuan M, Zhang W, Shi ZF, et al. Using the CT features to differentiate invasive pulmonary adenocarcinoma from pre-invasive lesion appearing as pure or mixed ground-glass nodules. Br J Radiol. 2015;88(1053):20140811. doi: 10.1259/bjr.20140811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin X, Zhao SH, Gao J, Wang DJ, Wu J, Wu CC, et al. CT characteristics and pathological implications of early stage (T1N0M0) lung adenocarcinoma with pure ground-glass opacity. Eur Radiol. 2015;25(9):2532–40. doi: 10.1007/s00330-015-3637-z. [DOI] [PubMed] [Google Scholar]
- 26.Han L, Zhang P, Wang Y, Gao Z, Wang H, Li X, et al. CT quantitative parameters to predict the invasiveness of lung pure ground-glass nodules (pGGNs) Clin Radiol. 2018;73(5):504.e1-.e7. doi: 10.1016/j.crad.2017.12.021. [DOI] [PubMed] [Google Scholar]
- 27.Gao F, Sun Y, Zhang G, Zheng X, Li M, Hua Y. CT characterization of different pathological types of subcentimeter pulmonary ground-glass nodular lesions. Br J Radiol. 2019;92(1094):20180204. doi: 10.1259/bjr.20180204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aoki T, Tomoda Y, Watanabe H, Nakata H, Kasai T, Hashimoto H, et al. Peripheral lung adenocarcinoma: correlation of thin-section CT findings with histologic prognostic factors and survival. Radiology. 2001;220(3):803–9. doi: 10.1148/radiol.2203001701. [DOI] [PubMed] [Google Scholar]
- 29.Travis WD, Brambilla E, Noguchi M, Nicholson AG, Geisinger KR, Yatabe Y, et al. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol. 2011;6(2):244–85. doi: 10.1097/JTO.0b013e318206a221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Obuchowski NA, Bullen JA. Receiver operating characteristic (ROC) curves: review of methods with applications in diagnostic medicine. Phys Med Biol. 2018;63(7):07tr1. doi: 10.1088/1361-6560/aab4b1. [DOI] [PubMed] [Google Scholar]
- 31.Nattino G, Pennell ML, Lemeshow S. Assessing the goodness of fit of logistic regression models in large samples: a modification of the Hosmer-Lemeshow test. Biometrics. 2020;76(2):549–60. doi: 10.1111/biom.13249. [DOI] [PubMed] [Google Scholar]
- 32.Rosenfeld JP, Donchin E. Resampling (bootstrapping) the mean: a definite do. Psychophysiology. 2015;52(7):969–72. doi: 10.1111/psyp.12421. [DOI] [PubMed] [Google Scholar]
- 33.Vickers AJ, Holland F. Decision curve analysis to evaluate the clinical benefit of prediction models. Spine J. 2021;21(10):1643–8. doi: 10.1016/j.spinee.2021.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andrade JR, Rocha RD, Falsarella PM, Rahal Junior A, Santos RSD, Franceschini JP, et al. CT-guided percutaneous core needle biopsy of pulmonary nodules smaller than 2 cm: technical aspects and factors influencing accuracy. J Bras Pneumol. 2018;44(4):307–14. doi: 10.1590/s1806-37562017000000259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohno Y, Hatabu H, Takenaka D, Higashino T, Watanabe H, Ohbayashi C, et al. CT-guided transthoracic needle aspiration biopsy of small (< or = 20 mm) solitary pulmonary nodules. AJR Am J Roentgenol. 2003;180(6):1665–9. doi: 10.2214/ajr.180.6.1801665. [DOI] [PubMed] [Google Scholar]
- 36.Jones KD. Whence lepidic? The history of a Canadian neologism. Arch Pathol Lab Med. 2013;137(12):1822–4. doi: 10.5858/arpa.2013-0144-HP. [DOI] [PubMed] [Google Scholar]
- 37.Lee JH, Park CM, Lee SM, Kim H, McAdams HP, Goo JM. Persistent pulmonary subsolid nodules with solid portions of 5 mm or smaller: their natural course and predictors of interval growth. Eur Radiol. 2016;26(6):1529–37. doi: 10.1007/s00330-015-4017-4. [DOI] [PubMed] [Google Scholar]
- 38.Lee SW, Leem CS, Kim TJ, Lee KW, Chung JH, Jheon S, et al. The long-term course of ground-glass opacities detected on thin-section computed tomography. Respir Med. 2013;107(6):904–10. doi: 10.1016/j.rmed.2013.02.014. [DOI] [PubMed] [Google Scholar]
- 39.Hu F, Huang H, Jiang Y, Feng M, Wang H, Tang M, et al. Discriminating invasive adenocarcinoma among lung pure ground-glass nodules: a multi-parameter prediction model. J Thorac Dis. 2021;13(9):5383–94. doi: 10.21037/jtd-21-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gurney JW. Determining the likelihood of malignancy in solitary pulmonary nodules with bayesian analysis. Part I. Theory. Radiology. 1993;186(2):405–13. doi: 10.1148/radiology.186.2.8421743. [DOI] [PubMed] [Google Scholar]
- 41.Winer-Muram HT. The solitary pulmonary nodule. Radiology. 2006;239(1):34–49. doi: 10.1148/radiol.2391050343. [DOI] [PubMed] [Google Scholar]
- 42.Lee HJ, Goo JM, Lee CH, Park CM, Kim KG, Park EA, et al. Predictive CT findings of malignancy in ground-glass nodules on thin-section chest CT: the effects on radiologist performance. Eur Radiol. 2009;19(3):552–60. doi: 10.1007/s00330-008-1188-2. [DOI] [PubMed] [Google Scholar]
- 43.Liu LH, Liu M, Wei R, Jin EH, Liu YH, Xu L, et al. CT findings of persistent pure ground glass opacity: can we predict the invasiveness? Asian Pac J Cancer Prev. 2015;16(5):1925–8. doi: 10.7314/APJCP.2015.16.5.1925. [DOI] [PubMed] [Google Scholar]
- 44.Nambu A, Araki T, Taguchi Y, Ozawa K, Miyata K, Miyazawa M, et al. Focal area of ground-glass opacity and ground-glass opacity predominance on thin-section CT: discrimination between neoplastic and non-neoplastic lesions. Clin Radiol. 2005;60(9):1006–17. doi: 10.1016/j.crad.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 45.Yanagawa M, Johkoh T, Noguchi M, Morii E, Shintani Y, Okumura M, et al. Radiological prediction of tumor invasiveness of lung adenocarcinoma on thin-section CT. Med (Baltim) 2017;96(11):e6331. doi: 10.1097/MD.0000000000006331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takashima S, Maruyama Y, Hasegawa M, Yamanda T, Honda T, Kadoya M, et al. CT findings and progression of small peripheral lung neoplasms having a replacement growth pattern. AJR Am J Roentgenol. 2003;180(3):817–26. doi: 10.2214/ajr.180.3.1800817. [DOI] [PubMed] [Google Scholar]
- 47.Mei X, Wang R, Yang W, Qian F, Ye X, Zhu L, et al. Predicting malignancy of pulmonary ground-glass nodules and their invasiveness by random forest. J Thorac Dis. 2018;10(1):458–63. doi: 10.21037/jtd.2018.01.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weng Q, Zhou L, Wang H, Hui J, Chen M, Pang P, et al. A radiomics model for determining the invasiveness of solitary pulmonary nodules that manifest as part-solid nodules. Clin Radiol. 2019;74(12):933–43. doi: 10.1016/j.crad.2019.07.026. [DOI] [PubMed] [Google Scholar]
- 49.Xing Y, Li Z, Jiang S, Xiang W, Sun X. Analysis of pre-invasive lung adenocarcinoma lesions on thin-section computerized tomography. Clin Respir J. 2015;9(3):289–96. doi: 10.1111/crj.12142. [DOI] [PubMed] [Google Scholar]
- 50.Swensen SJ, Silverstein MD, Ilstrup DM, Schleck CD, Edell ES. The probability of malignancy in solitary pulmonary nodules. Application to small radiologically indeterminate nodules. Arch Intern Med. 1997;157(8):849–55. doi: 10.1001/archinte.1997.00440290031002. [DOI] [PubMed] [Google Scholar]
- 51.McWilliams A, Tammemagi MC, Mayo JR, Roberts H, Liu G, Soghrati K, et al. Probability of cancer in pulmonary nodules detected on first screening CT. N Engl J Med. 2013;369(10):910–9. doi: 10.1056/NEJMoa1214726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chung K, Ciompi F, Scholten ET, Goo JM, Prokop M, Jacobs C, et al. Visual discrimination of screen-detected persistent from transient subsolid nodules: an observer study. PLoS ONE. 2018;13(2):e0191874. doi: 10.1371/journal.pone.0191874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Furuya K, Murayama S, Soeda H, Murakami J, Ichinose Y, Yabuuchi H, et al. New classification of small pulmonary nodules by margin characteristics on high-resolution CT. Acta Radiol. 1999;40(5):496–504. doi: 10.3109/02841859909175574. [DOI] [PubMed] [Google Scholar]
- 54.Hu H, Wang Q, Tang H, Xiong L, Lin Q. Multi-slice computed tomography characteristics of solitary pulmonary ground-glass nodules: differences between malignant and benign. Thorac Cancer. 2016;7(1):80–7. doi: 10.1111/1759-7714.12280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anderson CB, Philpott GW, Ferguson TB. The treatment of malignant pleural effusions. Cancer. 1974;33(4):916–22. doi: 10.1002/1097-0142(197404)33:4<916::AID-CNCR2820330405>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 56.Memon A, Zawadzki ZA. Malignant effusions: diagnostic evaluation and therapeutic strategy. Curr Probl Cancer. 1981;5(8):1–30. doi: 10.1016/S0147-0272(81)80012-8. [DOI] [PubMed] [Google Scholar]
- 57.Yanagawa M, Tsubamoto M, Satoh Y, Hata A, Miyata T, Yoshida Y, et al. Lung adenocarcinoma at CT with 0.25-mm section thickness and a 2048 Matrix: high-spatial-resolution imaging for Predicting Invasiveness. Radiology. 2020;297(2):462–71. doi: 10.1148/radiol.2020201911. [DOI] [PubMed] [Google Scholar]
- 58.Seyama K, Nukiwa T, Takahashi K, Takahashi H, Kira S. Amylase mRNA transcripts in normal tissues and neoplasms: the implication of different expressions of amylase isogenes. J Cancer Res Clin Oncol. 1994;120(4):213–20. doi: 10.1007/BF01372559. [DOI] [PubMed] [Google Scholar]
- 59.Tomita N, Matsuura N, Horii A, Emi M, Nishide T, Ogawa M, et al. Expression of alpha-amylase in human lung cancers. Cancer Res. 1988;48(11):3292–6. [PubMed] [Google Scholar]
- 60.Casadei Gardini A, Mariotti M, Lucchesi A, Pini S, Valgiusti M, Bravaccini S, et al. Paraneoplastic lipase and amylase production in a patient with small-cell lung cancer: case report. BMC Cancer. 2016;16:118. doi: 10.1186/s12885-016-2167-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Katayama S, Ikeuchi M, Kanazawa Y, Akanuma Y, Kosaka K, Takeuchi T, et al. Amylase-producing lung cancer: case report and review of the literature. Cancer. 1981;48(11):2499–502. doi: 10.1002/1097-0142(19811201)48:11<2499::AID-CNCR2820481125>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 62.Morohoshi T, Nakamura N, Hayashi K, Kanda M. Amylase producing lung cancer. Electronmicroscopical and biochemical studies. Virchows Arch Pathol Anat Histol. 1980;387(2):125–32. doi: 10.1007/BF00430694. [DOI] [PubMed] [Google Scholar]
- 63.Tsukawaki M, Izawa M, Yoshida M, Araki N, Hashiba Y, Nakagawa H, et al. A case of amylase-producing lung cancer. Intern Med. 1992;31(1):60–3. doi: 10.2169/internalmedicine.31.60. [DOI] [PubMed] [Google Scholar]
- 64.Lenler-Petersen P, Vejby-Christensen H, Brock A, Hvidman LE, Axelgaard G. [Amylase-producing lung tumors] Ugeskr Laeger. 1989;151(2):83–5. [PubMed] [Google Scholar]
- 65.Yamazaki S, Ebisawa S, Yasuo M, Urushihata K, Koizumi T, Fujimoto K, et al. Small-cell lung carcinoma produces salivary-type amylase: a case report with review. Intern Med. 2007;46(12):883–7. doi: 10.2169/internalmedicine.46.6205. [DOI] [PubMed] [Google Scholar]
- 66.Minami S, Komuta K, Asai M. [A case of amylase-producing lung cancer] Nihon Kokyuki Gakkai Zasshi. 2003;41(10):717–21. [PubMed] [Google Scholar]
- 67.Kamio T, Sameshima K, Irie J, Shigematsu K, Kawai K, Tsuchiyama H, et al. [Two cases of amylase-producing lung cancer] Gan No Rinsho. 1989;35(6):735–40. [PubMed] [Google Scholar]
- 68.Kitazawa M, Nakagawa M, Baba O, Sumiyoshi K, Saito Y, Nishimura T, et al. [A case of amylase producing lung cancer] Kokyu Junkan. 1993;41(4):393–6. [PubMed] [Google Scholar]
- 69.Nakao M, Kodama T, Sasaki M, Kuroda K. [A case of ectopic amylase-producing lung cancer] Kyobu Geka. 1996;49(13):1069–73. [PubMed] [Google Scholar]
- 70.Nakata Y, Kubota A, Mori Y, Matsumoto A. [An autopsied case of an amylase-producing lung cancer] Gan No Rinsho. 1988;34(7):881–5. [PubMed] [Google Scholar]
- 71.Hayashi Y, Fukayama M, Koike M, Nakayama T. Amylase in human lungs and the female genital tract. Histochemical and immunohistochemical localization. Histochemistry. 1986;85(6):491–6. doi: 10.1007/BF00508431. [DOI] [PubMed] [Google Scholar]
- 72.Lenler-Petersen P, Grove A, Brock A, Jelnes R. Alpha-amylase in resectable lung cancer. Eur Respir J. 1994;7(5):941–5. doi: 10.1183/09031936.94.07050941. [DOI] [PubMed] [Google Scholar]
- 73.Martin PC, Sarma DP. Amylase-producing lung cancer. J Surg Oncol. 1982;21(1):30–2. doi: 10.1002/jso.2930210108. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author.