ABSTRACT
Human activity affects the quality of potable water sources and their associated bacterial communities. Here, we discuss the heterotrophic Bacillus altitudinis 2R-9 draft isolated from the raw source of a drinking water distribution system in South Africa.
KEYWORDS: Bacillus, genome analysis, water treatment plants, South Africa
ANNOUNCEMENT
Bacillus altitudinis is a bacterial species isolated from various sources, including honey, bee bread, propolis, plant roots, and fish intestines (1). B. altitudinis has potential biotechnological applications, such as enhancing the accumulation of ginsenosides in Panax ginseng (2) and inhibiting the growth of Alternaria alternata in vitro (3). It has also been discovered to be beneficial for P. ginseng growth and morbidity reduction (4). Furthermore, it has been demonstrated that B. altitudinis can produce thermostable β−1,3–1,4-glucanase (5), and its genome has been sequenced (6). Lastly, research on antimicrobial resistance has illustrated B. altitudinis could be resistant to some antibiotics (7).
The B. altitudinis strain reported was isolated from the untreated water source for a drinking water distribution plant in the North-West province of South Africa in August 2016. The strain was inoculated on nutrient agar and incubated at 37℃ for 24 hours. The genomic DNA was extracted using the Chemagic DNA Bacteria Kit (PerkinElmer, Germany) following the manufacturer’s protocol. DNA concentration was quantified using a Nanodrop 1000 spectrophotometer (Thermo Fisher Scientific, USA), and the library was performed using the Nextera XT kit (Illumina Inc., San Diego, CA) according to the instructions provided by the manufacturer and sequenced using Illumina’s MiSeq300 in paired-end reads at the North-West University sequencing facility. The generated raw paired-end fastq reads (2 × 300 bp) were quality checked using FastQC v.0.11.7 (8) followed by trimming of low-quality bases using Trimmomatic v.0.39 (9), and the data quality was rechecked using FastQC v.0.11.7 (8). The cleaned reads were assembled using SPAdes v.3.15.5 (10). Quast (v.5.0.2) (11) was used to evaluate the genome assembly quality. The completeness and contamination were assessed with CheckM (v.1.1.6) (12). The assembled draft genome was annotated on the Rapid Annotation System Technology (RAST) Pipeline (13) and NCBI Prokaryotic Genome Annotation Pipeline (PGAP) v6.5(14). The annotated genomes were assessed against the Genome Taxonomy Database (GTDB) using GTDB-Tk software v1.7.0 (15). Default parameters were used for all software except where otherwise noted.
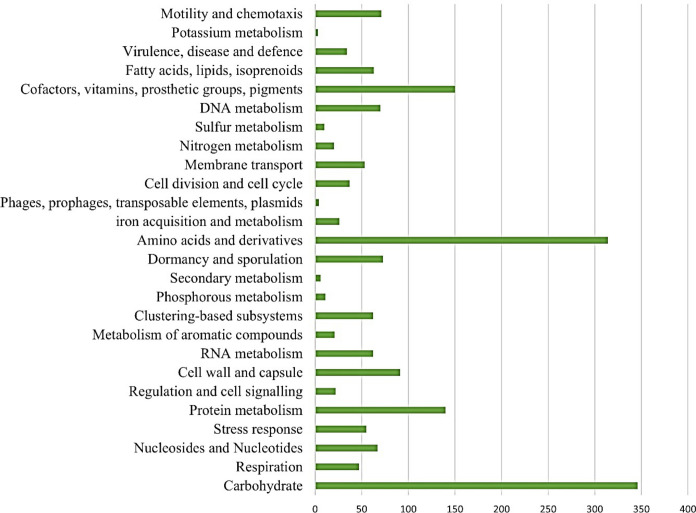
A total of 2,356,906 read counts and coverage of 130.7× were generated. The total length of the draft genome was 3,762,390 bp; it included 81 contigs, 41.1% GC content, N50 and L50 of 375,146 and 3, respectively. According to CheckM, the genome is 99.59% complete. The annotation revealed various functional genes showing important functions, including motility, virulence, and presence of mobile genetic elements such as phages, prophages, plasmids, and transposable elements (Fig. 1). The prediction of gene functions based on PGAP annotation revealed 3,897 genes, including 3,772 protein-coding genes and 3,795 coding DNA sequences (CDSs), 102 total rRNAs, and 74 tRNAs.
Fig 1.
Functional annotation categories in the genome based on RAST annotation.
In conclusion, the pathogenicity of this bacterial species on humans has not been established; therefore, this new genome will contribute further insight into the biology and diversity of this heterotrophic bacterial species.
ACKNOWLEDGMENTS
This work is based on research supported in part by the National Research Foundation of South Africa grant no. 109207 and 14226 (Bursary for RK), The Water Research Commission (WRC) of South Africa K5/2585//3, and NWU Postgraduate funding. O.S.O. would like to thank the NWU for the Postdoctoral fellowship.
The views expressed are those of the authors and not of the funding agencies.
Contributor Information
Lesego G. Molale-Tom, Email: Lesego.MolaleTom@nwu.ac.za.
Simon Roux, DOE Joint Genome Institute, Berkeley, California, USA.
DATA AVAILABILITY
The whole-genome shotgun project for Bacillus altitudinis 2R-9 has been deposited at DDBJ/ENA/GenBank under the accession JASCXE000000000, and the version described in this paper is version JASCXE010000000. The raw reads are available under the BioProject accession number PRJNA968034, and the BioSample accession number is SAMN34894825. The sequence data obtained in this work have been deposited in the NCBI Sequence Read Archive under the accession number SRR24490156.
REFERENCES
- 1. Ngalimat MS, Raja Abd Rahman RNZ, Yusof MT, Syahir A, Sabri S. 2019. Characterisation of bacteria isolated from the stingless bee, Heterotrigona itama, honey, bee bread and propolis. PeerJ 7:e7478. doi: 10.7717/peerj.7478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dong L, Cheng R, Xiao L, Wei F, Wei G, Xu J, Wang Y, Guo X, Chen Z, Chen S. 2018. Diversity and composition of bacterial endophytes among plant parts of Panax notoginseng. Chin Med 13:41. doi: 10.1186/s13020-018-0198-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sun M, Ye S, Xu Z, Wan L, Zhao Y. 2021. Endophytic Bacillus altitudinis Q7 from Ginkgo biloba inhibits the growth of Alternaria alternata in vitro and its inhibition mode of action. Biotechnology & Biotechnological Equipment 35:880–894. doi: 10.1080/13102818.2021.1936639 [DOI] [Google Scholar]
- 4. Wei G, Zhang G, Li M, Liu C, Wei F, Wang Y, Huang Z, Chen Z, Zheng Y, Chen S, Dong L. 2022. Core rhizosphere microbiome of Panax notoginseng and its associations with belowground biomass and saponin contents. Environ Microbiol 24:6238–6251. doi: 10.1111/1462-2920.16245 [DOI] [PubMed] [Google Scholar]
- 5. Mao S, Lu Z, Zhang C, Lu F, Bie X. 2013. Purification, characterization, and heterologous expression of a thermostable β-1,3-1,4-glucanase from Bacillus altitudinis YC-9. Appl Biochem Biotechnol 169:960–975. doi: 10.1007/s12010-012-0064-3 [DOI] [PubMed] [Google Scholar]
- 6. Cortese IJ, Castrillo ML, Onetto AL, Bich GÁ, Zapata PD, Laczeski ME. 2021. De novo genome assembly of Bacillus altitudinis 19Rs3 and Bacillus Altitudinis T5S-T4, two plant growth-promoting bacteria isolated from ilex paraguariensis St. Hil. (Yerba mate). PLoS One 16:e0248274. doi: 10.1371/journal.pone.0248274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lobova TI, Yemelyanova E, Andreeva IS, Puchkova LI, Repin VY. 2015. Antimicrobial resistance and plasmid profile of bacterial strains isolated from the urbanized Eltsovka-1 river (Russia). Microb Drug Resist 21:477–490. doi: 10.1089/mdr.2014.0203 [DOI] [PubMed] [Google Scholar]
- 8. Andrews S. 2010. Fastqc: a quality control tool for high throughput sequence data [Google Scholar]
- 9. Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. doi: 10.1093/bioinformatics/btt086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. doi: 10.1101/gr.186072.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. 2016. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44:6614–6624. doi: 10.1093/nar/gkw569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chaumeil P-A, Mussig AJ, Hugenholtz P, Parks DH, Hancock J. 2019. GTDB-Tk: a toolkit to classify genomes with the genome taxonomy database. Bioinformatics 36:1925–1927. doi: 10.1093/bioinformatics/btz848 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The whole-genome shotgun project for Bacillus altitudinis 2R-9 has been deposited at DDBJ/ENA/GenBank under the accession JASCXE000000000, and the version described in this paper is version JASCXE010000000. The raw reads are available under the BioProject accession number PRJNA968034, and the BioSample accession number is SAMN34894825. The sequence data obtained in this work have been deposited in the NCBI Sequence Read Archive under the accession number SRR24490156.