Abstract
Motivation
Transcriptomic long-read (LR) sequencing is an increasingly cost-effective technology for probing various RNA features. Numerous tools have been developed to tackle various transcriptomic sequencing tasks (e.g. isoform and gene fusion detection). However, the lack of abundant gold-standard datasets hinders the benchmarking of such tools. Therefore, the simulation of LR sequencing is an important and practical alternative. While the existing LR simulators aim to imitate the sequencing machine noise and to target specific library protocols, they lack some important library preparation steps (e.g. PCR) and are difficult to modify to new and changing library preparation techniques (e.g. single-cell LRs).
Results
We present TKSM, a modular and scalable LR simulator, designed so that each RNA modification step is targeted explicitly by a specific module. This allows the user to assemble a simulation pipeline as a combination of TKSM modules to emulate a specific sequencing design. Additionally, the input/output of all the core modules of TKSM follows the same simple format (Molecule Description Format) allowing the user to easily extend TKSM with new modules targeting new library preparation steps.
Availability and implementation
TKSM is available as an open source software at https://github.com/vpc-ccg/tksm.
1 Introduction
Long-read (LR) sequencing technologies have become a cost-effective alternative to short-read (SR) sequencing for many genomic and transcriptomic sequencing tasks (Amarasinghe et al. 2020). LRs are shown to be useful for many transcriptomic tasks such as alternative isoform detection (Kovaka et al. 2019, Tang et al. 2020, Orabi et al. 2023), gene fusion detection (Liu et al. 2020, Karaoglanoglu et al. 2022), transcript-level expression analysis (Hu et al. 2021), or single-cell transcriptomic analysis (Tian et al. 2021, Ebrahimi et al. 2022, You et al. 2023).
However, due to the nature of LR sequencing as an emerging technology, there are very few well established benchmark datasets or gold-standard datasets to assess transcriptomic LR bioinformatics tools. Such bioinformatics tools targeting these tasks require realistic simulations in order to assess their accuracy and performance. This includes the ability to simulate explicitly target specific library or cellular processes such as single-cell barcoding and UMI tagging, PCR, or molecule truncation.
Existing LR simulators such as Badread (Wick 2019), DeepSimulator (Li et al. 2020), Icarust (Munro et al. 2023), PBSIM3 (Ono et al. 2022), and Nanosim (Yang et al. 2017), typically focus on simulating the sequencing process, i.e., the point of contact of sequencing platform with the RNA/DNA molecule. Some have extensions focusing on specific sequencing libraries such as Trans-Nanosim transcriptomic and plasmid simulation (Hafezqorani et al. 2020), Meta-Nanosim metagenomic simulation (Yang et al. 2023), SLSim single-cell simulation (You et al. 2023a), and SQANTI-SIM alternative splicing simulation (Mestre-Tomás et al. 2023). However, these tools are not designed with modularity in mind and cannot be easily modified to address changes in the library preparation protocols such as adding a barcode tag or simulating the PCR process. A comprehensive survey of long-read tools, including simulation tools, is available at Long-read Tools catalogue (Amarasinghe et al. 2021).
We describe TKSM, a software that simulates realistic transcriptomic long-read datasets. TKSM modular design allows to target a wide range of library/cell processes. The power of TKSM lies in two key aspects: (i) the ease with which its simulation pipeline can be modified to cater to specific sequencing designs and (ii) high performance in terms of time and memory use. TKSM is open source, accessible via GitHub.
2 Methods
TKSM is flexible, both in that it can simulate a wide variety of datasets, and it is extendable. It is composed of several independent modules, each representing a cellular (e.g. polyadenylation) or a library preparation (e.g. PCR) process that modifies a nucleic acid molecule. This design allows the user to simulate different sequencing protocols by using TKSM’s modules in various arrangements, imitating the different steps in the desired sequencing protocol. Additionally, this modular design allows TKSM to be easily extendable with future modules targeting additional library and cellular processes. To enable this modularity, we designed TKSM’s modules to take and generate files in the same format that we call Molecule Description Format (MDF). An MDF file is a tabular file that describes molecules by listing for each molecule its genomic intervals alongside any sequence-level modifications to these intervals (e.g. substitutions). The rationale for using a tabular format is that write their own scripts that can generate or modify intermediate MDF files. We expand on the details of MDF files in Supplementary Section S1. The only exceptions to this design pattern are the entry module which generates the initial set of molecules from a transcript abundance profile and the exit module which generates the reads obtained by simulating the sequencing of the given molecules.
Each of TKSM’s modules can be run as a separate process (tksm < module_name >). We also provide as part of TKSM a Snakemake (Mölder et al. 2021) script which can be configured by the user to specify a wide range of simulation experiments and run them all as a single command. Additionally, to optimize the computation time, we take advantage of Snakemake’s piped input/output feature to allow modules to start running the moment they receive any input from a previous module, rather than having to wait for the preceding module to terminate.
TKSM can use real sequencing datasets to parameterize the behaviour of its modules, or alternatively, these parameters can be specified manually by the user. For example, TKSM contains preprocessing modules to compute the expression profile of transcripts from a given real sample which is then used to generate the molecules in the initial MDF file, whose sequencing according to a chosen protocol will be simulated by the next modules.
2.1 TKSM modules
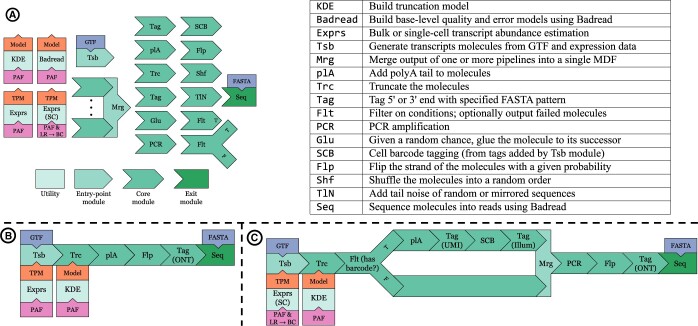
TKSM contains three classes of modules, defined by features of their input and output: (i) entry-point modules start a TKSM pipeline and output an MDF file, (ii) core modules take an MDF file as input and output another MDF file, and (iii) exit (sequencing) modules take an MDF file as input and generate FASTA/FASTQ file(s) as output. Additionally, some preprocessing utilities in TKSM can take a real sequencing dataset and output model parameters for some of TKSM modules. A list of the implemented TKSM modules is presented in Fig. 1A and detailed in Supplementary Section S1. Additional modules and utilities can be implemented and easily integrated into TKSM in order to target specific steps in alternate sequencing protocols.
Figure 1.
(A) Existing TKSM modules and utilities alongside their high-level descriptions. TKSM is designed with modularity in mind; the user can specify a simulation pipeline of their choosing by chaining any number of TKSM modules including the possibility of using the same module multiple times. (B) Typical RNA-seq simulation pipeline that imitates Trans-Nanosim’s workflow. (C) Single-cell long-read simulation pipeline. The pipeline makes use of the Filtering and Merging modules to add the short-read Illumina adapter and 10× Genomics cellular barcodes only to molecules that have a tag indicating that they should have a cellular barcode.
2.2 Customizable TKSM pipelines using Snakemake
An important design choice for TKSM is to make it easily customizable by the user, i.e. to make it easy to build a, possibly complex, simulation pipeline using the TKSM modules. To achieve that, we packaged TKSM with Snakemake and configuration scripts that can be edited by the user to add new modules or to define simulation experiments using any arrangement of TKSM modules. To define a simulation pipeline, the user lists the names of required TKSM modules and specifies, for the modules that require model construction, the real samples to build such models on. Additionally, using the Merging module, the user may build complex pipelines that are composed of different linear pipelines. An example of the configuration script is presented in Supplementary Listing S1.
3 Results
To illustrate TKSM and assess its performances, we designed three simulation pipelines to emulate examples of standard transcriptomic sequencing protocols. Specifically, we present simulations of a standard bulk RNA sequencing experiment, a hybrid long-short read single-cell RNA sequencing (scRNA-seq) experiment, and an RNA sequencing experiment similar to the bulk RNA sequencing experiment but with 100 random gene fusion events added. The Snakemake configuration files that specify these simulation pipelines are presented in Supplementary Listings S2–S4.
In the standard bulk RNA-seq experiment, we primarily compare against Trans-Nanosim (Hafezqorani et al. 2020) and try to conform to its pipeline design using TKSM modules. For both the bulk and gene fusion experiments, we use an RNA-seq sample generated from the MCF7 cell line by Chen et al. (2021) (direct RNA, replicate 1, run 2). We first accessed the SG-NEx data on 2020–06-17 via https://registry.opendata.aws/sgnex/. For the scRNA-seq experiment, we used an in-house dataset, named N1, first described by Ebrahimi et al. (2022). N1 follows the short-long single-cell hybrid protocol described previously in the literature (Gupta et al. 2018, Singh et al. 2019, Tian et al. 2021). In this manuscript, we use a random subsample of N1 with 1M long-reads. The three TKSM pipelines are illustrated in Fig. 1B and C and Supplementary Figure S11.
Using these experiments, our goal is to assess TKSM on multiple metrics: (i) the similarity of the simulated data compared to the input real data on measures such as transcript expression, molecule sequence truncation, single cell barcode detection rates, and gene fusion generation, (ii) the time and memory footprint of various steps, and (iii) the ability to generate gene fusion events that can be detected by standard gene fusion tools. The results of all these experiments are presented in Supplementary Section S2. Note that all these results are reproducible using Snakemake scripts provided on the TKSM GitHub repository.
4 Conclusion
TKSM is a modular, accurate, and efficient transcriptomic LR sequencing simulator. Its modular design enables the user to construct a large verity of sequencing experiments with minimal effort. TKSM’s standardized input and output for its modules allow the users of TKSM to add new modules that target existing and future library preparation techniques that TKSM currently does not target. For example, it is easy to envision an alternative entry-point module to the Transcribing module that generates nucleic acid molecules from DNA fragmentation while still making use of the rest of TKSM modules. TKSM also performs well in terms of generating realistic datasets with characteristics matching the real datasets it is simulating. Additionally, TKSM is engineered with efficient CPU and memory use in mind and its performance on those metrics is excellent.
Supplementary Material
Contributor Information
Fatih Karaoğlanoğlu, Computing Science Department, Simon Fraser University, Burnaby, BC V5A 1S6, Canada.
Baraa Orabi, Department of Computer Science, the University of British Columbia, Vancouver, BC V6T 1Z4, Canada.
Ryan Flannigan, Department of Urologic Sciences, the University of British Columbia, Vancouver, BC V5Z 1M9, Canada; Vancouver Prostate Centre, Vancouver, BC V6H 3Z6, Canada.
Cedric Chauve, Department of Mathematics, Simon Fraser University, Burnaby, BC V5A 1S6, Canada.
Faraz Hach, Department of Computer Science, the University of British Columbia, Vancouver, BC V6T 1Z4, Canada; Department of Urologic Sciences, the University of British Columbia, Vancouver, BC V5Z 1M9, Canada; Vancouver Prostate Centre, Vancouver, BC V6H 3Z6, Canada.
Author contributions
Fatih Karaoglanoglu (Conceptualization [equal], Data curation [Supporting], Formal analysis [equal], Investigation [equal], Methodology [equal], Software [equal], Visualization [equal], Writing—original draft [equal], writing—review & editing [equal]), Baraa Orabi (Conceptualization [equal], Data curation [equal], Formal analysis [equal], Funding acquisition [supporting], Investigation [equal], Methodology [equal], Software [equal], Visualization [equal], Writing—original draft [equal], writing—review & editing [equal]), Ryan Flannigan (Data curation [lead], Resources [supporting]), Cedric Chauve (Funding acquisition [lead], Project administration [equal], Supervision [equal], Writing—review & editing [equal]), and Faraz Hach (Data curation [lead], Funding acquisition [lead], Project administration [equal], Resources [lead], Supervision [equal], Writing—review & editing [equal]).
Supplementary data
Supplementary data are available at Bioinformatics online.
Conflict of interest
None declared.
Funding
This work was supported by the National Science and Engineering Council of Canada (NSERC) Discovery Grants [grant numbers RGPIN-05952 to F.H. and RGPIN-03986 to C.C.]; the Michael Smith Foundation for Health Research (MSFHR) Scholar Award [grant number SCH-2020–0370 to F.H.]; and the NSERC Alexander Graham Bell Canada Graduate Scholarship-Doctoral (CGS D) to B.O.
Data availability
TKSM is available as an open source software at github.com/vpc-ccg/tksm. Datasets used in this manuscript are publicly available via registry.opendata.aws/sgnex/for the MCF7 dataset and at doi.org/10.6084/m9.figshare.23155145 for the N1 sample. An archive of the TKSM version used in this manuscript is available at doi.org/10.6084/m9.figshare.24970317.v1.
References
- Amarasinghe SL, Su S, Dong X. et al. Opportunities and challenges in long-read sequencing data analysis. Genome Biol 2020;21:30–16. 10.1186/s13059-020-1935-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarasinghe SL, Ritchie ME, Gouil Q.. long-read-tools.org: an interactive catalogue of analysis methods for long-read sequencing data. GigaScience 2021;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Davidson NM, Wan YK. et al. A systematic benchmark of nanopore long read RNA sequencing for transcript level analysis in human cell lines. bioRxiv, 10.1101/2021.04.21.440736, 2021, preprint: not peer reviewed. [DOI] [Google Scholar]
- Ebrahimi G, Orabi B, Robinson M. et al. Fast and accurate matching of cellular barcodes across short-reads and long-reads of single-cell RNA-seq experiments. iScience 2022;25:104530. 10.1016/j.isci.2022.104530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta I, Collier PG, Haase B. et al. Single-cell isoform RNA sequencing characterizes isoforms in thousands of cerebellar cells. Nat Biotechnol 2018;36:1197–202. 10.1038/nbt.4259. [DOI] [PubMed] [Google Scholar]
- Hafezqorani S, Yang C, Lo T. et al. Trans-NanoSim characterizes and simulates nanopore RNA-sequencing data. GigaScience 2020;9. 10.1093/gigascience/giaa061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Fang L, Chen X. et al. LIQA: long-read isoform quantification and analysis. Genome Biol 2021;22:182. 10.1186/s13059-021-02399-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaoglanoglu F, Chauve C, Hach F.. Genion, an accurate tool to detect gene fusion from long transcriptomics reads. BMC Genomics 2022;23:129. 10.1186/s12864-022-08339-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovaka S, Zimin AV, Pertea GM. et al. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol 2019;20:278–13. 10.1186/s13059-019-1910-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wang S, Bi C. et al. DeepSimulator1.5: a more powerful, quicker and lighter simulator for nanopore sequencing. Bioinformatics 2020;36:2578–80. 10.1093/bioinformatics/btz963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Hu Y, Stucky A. et al. LongGF: computational algorithm and software tool for fast and accurate detection of gene fusions by long-read transcriptome sequencing. BMC Genomics 2020;21:793–12. 10.1186/s12864-020-07207-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestre-Tomás J, Liu T, Pardo-Palacios F. et al. SQANTI-SIM: a simulator of controlled transcript novelty for lrRNA-seq benchmark. bioRxiv preprint: not peer reviewed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mölder F, Jablonski KP, Letcher B. et al. Sustainable data analysis with Snakemake. F1000Res 2021;10:33. 10.12688/f1000research.29032.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro RJ, Payne A, Loose MW.. Icarust, a real-time simulator for Oxford Nanopore adaptive sampling. bioRxiv 2023.05.16.540986, preprint: not peer reviewed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono Y, Hamada M, Asai K.. PBSIM3: a simulator for all types of PacBio and ONT long reads. NAR Genom Bioinform 2022;4:. 10.1093/nargab/lqac092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orabi B, Xie N, McConeghy B. et al. Freddie: annotation-independent detection and discovery of transcriptomic alternative splicing isoforms using long-read sequencing. Nucleic Acids Res 2023;51:e11. 10.1093/nar/gkac1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M, Al-Eryani G, Carswell S. et al. High-throughput targeted long-read single cell sequencing reveals the clonal and transcriptional landscape of lymphocytes. Nat Commun 2019;10:3120–13. 10.1038/s41467-019-11049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang AD, Soulette CM, van Baren MJ. et al. Full-length transcript characterization of SF3B1 mutation in chronic lymphocytic leukemia reveals downregulation of retained introns. Nat Commun 2020;11:1438. 10.1038/s41467-020-15171-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Jabbari JS, Thijssen R. et al. Comprehensive characterization of single-cell full-length isoforms in human and mouse with long-read sequencing. Genome Biol 2021;22:310–24. 10.1186/s13059-021-02525-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick RR. Badread: simulation of error-prone long reads. JOSS 2019;4:1316. 10.21105/joss.01316. [DOI] [Google Scholar]
- Yang C, Chu J, Warren RL. et al. NanoSim: nanopore sequence read simulator based on statistical characterization. GigaScience 2017;6:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Lo T, Nip KM. et al. Characterization and simulation of metagenomic nanopore sequencing data with Meta-Nanosim. GigaScience 2023;12:giad013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Y, Prawer YD, De Paoli-Iseppi R. et al. Identification of cell barcodes from long-read single-cell RNA-seq with BLAZE. Genome Biol 2023;24:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
TKSM is available as an open source software at github.com/vpc-ccg/tksm. Datasets used in this manuscript are publicly available via registry.opendata.aws/sgnex/for the MCF7 dataset and at doi.org/10.6084/m9.figshare.23155145 for the N1 sample. An archive of the TKSM version used in this manuscript is available at doi.org/10.6084/m9.figshare.24970317.v1.