Abstract
The tailorable properties of synthetic polyethylene glycol (PEG) hydrogels make them an attractive substrate for human organoid assembly. Here, we formed human neural organoids from iPSC-derived progenitor cells in two distinct formats: (i) cells seeded on a Matrigel surface; and (ii) cells seeded on a synthetic PEG hydrogel surface. Tissue assembly on synthetic PEG hydrogels resulted in three dimensional (3D) planar neural organoids with greater neuronal diversity, greater expression of neurovascular and neuroinflammatory genes, and reduced variability when compared with tissues assembled upon Matrigel. Further, our 3D human tissue assembly approach occurred in an open cell culture format and created a tissue that was sufficiently translucent to allow for continuous imaging. Planar neural organoids formed on PEG hydrogels also showed higher expression of neural, vascular, and neuroinflammatory genes when compared to traditional brain organoids grown in Matrigel suspensions. Further, planar neural organoids contained functional microglia that responded to pro-inflammatory stimuli, and were responsive to anti-inflammatory drugs. These results demonstrate that the PEG hydrogel neural organoids can be used as a physiologically relevant in vitro model of neuro-inflammation.
Keywords: PEG hydrogels, neural tissue engineering, organoids, neuroinflammation, disease modeling
Graphical abstract.
Introduction
Neural tissue-like constructs have received significant attention in recent years, mainly because of their potential use in predicting neural toxicity, modeling neural tissue disorders, and regenerating neural tissue.1 –3 In vivo neural applications typically rely on conventional two-dimensional (2D) cell culture techniques. However, 2D cell cultures provide only limited physiological relevance.4 –8 Thus, 3D neural cell culture models including organotypic cultures such as organoids, were developed to more closely mimic in vivo conditions.9 –14 Human organoid models are highly complex and feature multiple cell types to better mimic aspects of neural structure and physiology.15 –19 Organoids are typically derived from stem cells in the presence of defined growth factors specific to the organ.20 –22 Researchers have successfully developed human organoids to model the intestine,23 –26 kidney,27,28 pancreas,29 –33 colon,34 –37 liver,38 –44 brain,45 –54 and heart55 –59 from adult-derived stem cells, embryonic stem cells (ESC), or induced pluripotent stem cells (iPSCs).
The culture and expansion of organoids has mostly relied on animal-derived extracellular matrices such as Matrigel, which is a natural basement-membrane-like gel secreted by Engelbreth-Holm-Swarm mouse sarcoma cells. 60 Commercially available Matrigel compositions are often poorly defined 61 with significant batch-to-batch variability, leading to a lack of reproducibility in cell culture experiments.62,63 These limitations of Matrigel paved the way for developing synthetic 3D matrices which can be manipulated as required for the culture of human organoids.64 –67 In particular, synthetic poly (ethylene glycol; PEG) hydrogels have emerged as a Matrigel alternative, as they provide control over mechanical stiffness, biochemical composition, and degradability.68,69 As a result, PEG hydrogels have been widely used as a synthetic matrix for culture and expansion of organoids.54,70,71
In this study, we used either Matrigel or a synthetic PEG hydrogel as matrices for the generation and culture of neural organoids. The neural organoids assembled on the synthetic PEG hydrogel surface were planar and more enriched with neuro-vascular markers than the neural organoids assembled on the Matrigel surface. Planar neural organoids (PNOs) formed on PEG hydrogels showed greater expression of neural, vascular, and neuroinflammatory genes when compared to traditional brain organoids formed in Matrigel suspension. In addition, neural tissues formed on PEG hydrogels contained active microglia capable of producing anti-inflammatory cytokines (TNFα and IL-6) in response to lipopolysaccharide (LPS) stimulation. Treatment with anti-inflammatory drugs Celecoxib and Donepezil 72 reduced the production of both TNFα and IL-6 in the neural tissue culture, suggesting utility of these neural organoids in screening for neuroinflammatory diseases and associated therapeutics.
Aims
The development of physiologically relevant in vitro models of human neural tissue is of utmost importance to study poorly understood neural disorders and diseases. This paper aims to establish an open cell culture system to generate planar neural organoids from human induced pluripotent stem cells for possible applications in disease modeling. We explored both Matrigel and PEG hydrogel as matrices for the culture and expansion of the organoids and found that the neural organoids assembled on the PEG hydrogel surface were planar in morphology and more enriched with neuro-vascular markers than that assembled on the Matrigel surface. Our results demonstrate that the planar neural organoids can be used as a physiologically relevant in vitro model of neuro-inflammation.
Methodology
Cell culture
Culture of human induced pluripotent stem cells (iPSCs)
Essential 8 (E8) medium: The Essential 8 (E8) complete medium was prepared by adding 1 mL of E8 supplement (Fisher #A1517001) to 50 mL of base E8 media (Fisher #A1517001).
Human induced Pluripotent Stem Cells (iPSCs) were maintained in E8 medium in presence of 10 µM Y27632 (Rock inhibitor; R&D Systems #1254) on Matrigel (Corning #354277) coated culture plates. Cells were grown at 37°C, 5% CO2 atmosphere and cells were passaged when 70% confluent with Versene (Gibco #15040066).
Differentiation of endothelial cells (ECs)
E7VI medium: DF3S base media (Gibco #ME110262L1), SB341542 (5 µM), VEGFA165 (50 ng/mL), FGF2 (100 ng/mL), Transferrin (10.7 µg/mL), Insulin (20 µg/mL).
E8BAC medium: E8 base media (Gibco #A1517001), BMP4 (5 ng/mL), Activin A (25 ng/mL), CHIR 99021 (1 µM).
Differentiation of endothelial cells (ECs) was carried out following our previously published protocol. 73 Human induced pluripotent stem cells (iPSCs) were grown in E8 media on Matrigel coated plate. Next, cells were dissociated using Versene (Gibco #15040066) and incubate for 3–5 min at 37°C until colonies detach. Cells were cultured in Matrigel coated wells of 6 well plates in E8BAC media for 2 days (to 100% confluence) with the addition of 10 µM Y27632 (R&D Systems #1254/10). Cells were then grown in E7VI medium for an additional 3 days. Cells were then harvested with Accutase (Gibco #A1110501) to produce a single cell suspension and centrifuged at 300×g for 5 min and resuspend 1 × 108 cells in 300 µL of cold MACS buffer consisting of 1× PBS, 0.5% BSA (Fisher #BP1600), and 2 mM EDTA (Invitrogen #15575020). A 100 µL of FcR blocking reagent and 100 µL of CD34 microbeads (Miltenyi Biotec #130-046-702) were added and the mixture was incubated at 4°C for 30 min. Endothelial cells (ECs) were then isolated with CD34 microbeads by auto MACS (Miltenyi) to yield purified populations of CD34+CD31+ cells, which were cryopreserved.
Differentiation of pericyte cells (PC)
Differentiation of pericyte cells (PCs) was carried out following our previously published protocol. 73 Human induced pluripotent stem cells (iPSCs) were grown in E8 media (50%–60% confluent) on Matrigel coated plate. Next, cells were treated with Versene (Gibco #15040066) and incubated for 3–5 min at 37°C until colonies detach. Cells were transferred in Matrigel coated wells of 6 well plates and cultured in E8BAC media for 2 days (to 100% confluence) with the addition of 10 µM Y27632 (R&D Systems #1254/10). Cells were then grown in E7VI medium for an additional 3 days. Next, cells were harvested with Accutase (Gibco #A1110501) to produce a single cell suspension and centrifuged at 300×g for 5 min and resuspend 1 × 108 cells in 300 µL of cold MACS buffer consisting of 1× PBS, 0.5% BSA (Fisher #BP1600), and 2 mM EDTA (Invitrogen #15575020). A 100 µL of FcR blocking reagent and 100 µL of CD34 microbeads (Miltenyi Biotec #130-046-702) were added and the mixture was incubated at 4°C for 30 min. The unbound CD34- cells from the flow through were collected. CD34- pericyte precursor cells were cultured in 6 well plates pre-coated with 0.25 µg/cm2 of a 1:1 mixture of Fibronectin (Corning #356008) and Collagen IV (Corning #354245) using complete endothelial cell growth media 2 (EGM-2) media (Promocell #C-22111) with a minimum of 2 x 104 cells per cm2. These PCs cells were cells passaged 1–3 days later and then grown in PM media until day 9 when they were frozen and/or used for subsequent expansion and formation of neural organoids.
Differentiation of neural progenitor cells (NPCs)
SB Medium: DF3S base medium (Gibco ME110262L1), Insulin (5 µg/mL), Transferrin (10.7 µg/mL), SB431542 (10 µM)
SBF2 Medium: DF3S base medium (Gibco ME110262L1), Insulin (5 µg/mL), SB431542 (10 µM), Transferrin (100.7 ng/mL), FGF2 (100 ng/mL).
SBNog Medium: DF3S base medium (Gibco ME110262L1), Insulin (5 µg/mL), Transferrin (10.7 µg/mL), SB431542 (10 µM), Noggin (100 ng/mL).
N2B27(F) medium: DF3S base medium (Gibco ME110262L1), 1× N2 Supplement, 1× B27 Supplement, FGF2 (5 ng/mL).
Differentiation of neural progenitor cells (NPCs) was carried out following our previously published protocol. 54 Approximately, 10 × 105 human induced pluripotent stem cells (iPSCs) per well were seeded in Matrigel (Corning #354277) coated wells of a 6 well plate (Corning #3516) with 2 mL of SBF2 media per well with the addition of 10 µM Y27632. On day 1, cells were treated with 2 mL per well of SBF2 media without Y27632. On day 2, cells were treated with 1:1 SB and SBNog media. Cells were treated with 2 mL per well of either SB or SBNog medium with daily medium change for seven more days. On day 9, cells were washed with 1× PBS and incubated with 1 mL per well of Accutase (Gibco #A1110501) for 6–8 min at 37°C until the cells are singularized. Cells were transferred in 10 % FBS in 1× PBS and centrifuged at 300×g for 5 min and resuspend in N2B27(F) media and transferred to new Matrigel coated 6 well plates with approximately 1.6 × 106 cells per well in 2 mL N2B27(F). Cells were cultured for up to day 32 with 2 mL per well of N2B27(F) and passaged when confluent. Differentiation into neural progenitor cells (NPCs) was observed on day 35 or so. These NPCs cells were used for subsequent expansion and formation of brain organoids.
Differentiation of microglia (MG)
FVI medium: DF3S base medium (Gibco #ME110262L1), Insulin (5 µg/mL), VEGFA165 (50 ng/mL), FGF2 (100 ng/mL), SB431542 (10 µM).
HPC medium: DF3S base medium (Gibco #ME110262L1), Insulin (5 µg/mL), TPO (50 ng/mL), IL6 (50 ng/mL), SCF (10 ng/mL), IL-3 (10 ng/mL), FGF2 (50 ng/mL), and VEGFA165 (50 ng/mL).
MP medium: DF3S base medium (Gibco ME110262L1), Insulin (5 µg/mL), and GMCSF (200 ng/mL).
PMG medium: IMDM base medium (Gibco#12440053), HI FBS (10%), IL-1β (10 ng/mL), and MCSF (20 ng/mL).
Differentiation of microglia (MG) was carried out following our previously published protocol. 54 Human induced pluripotent stem cells (iPSCs) were seeded (1 × 105 cells per well) in Matrigel (Corning #354277) coated wells of a 6 well plate (Corning #3516) with 2 mL of E8BAC medium per well with the addition of 10 µM Y27632. On day 1, cells were treated with 2 mL per well of E8BAC medium without Y27632. On day 2, E8BAC medium was aspirated and replaced with FVI medium and treatment was continued for another day. On day 4, FVI medium was aspirated and replaced with HPC medium, which was used to grow the cells for up to day 12. Next, the blooms from the wells were removed without disturbing the underlying layers of cells and without detaching the large clumps of cells that constitute the upper layer and cells were placed into wells of low attachment plates in PM medium for 2 days. On day 14, PM medium was aspirated and replaced with PMG medium. When cells were about 70 % confluent, adherent cells were harvested for freezing and the non-adherent cells were harvested and transferred to new wells of a low attachment plate in PMG medium and they will attach to provide more microglia.
PEG hydrogel sample preparation
Polyethylene glycol (PEG) hydrogels were formed using thiol-ene photopolymerization chemistry. 54 PEG-8-arm poly (ethylene glycol) norbornene (tripentaerythritol; JenKem Technology #A10037-10) was resuspended in 1× PBS at a final concentration of 150 mg/mL and sterile-filtered through a 0.2 μm nylon syringe filter (Fisher Scientific). Matrix metalloproteinase (MMP) analog such as Tryp-MMP with amino acid sequence KCGGPQGIWGQGCK (Genscript) was used to prepare MMP degradable PEG hydrogels. Tryp-MMP was resuspended in 1× PBS at a stock of 200 mM and sterile-filtered through a 0.2 μm nylon syringe filter (Fisher Scientific). CRGDS (5′ C-terminal modification of amidation; Genscript) peptide was incorporated for cell adhesion. CRGDS was resuspended in 1× PBS at a concentration of 200 nM and sterile-filtered through a 0.2 μm nylon syringe filter (Fisher Scientific). Finally, a stock of 0.5 % of photoinitiator I2959 – Irgacure D2959 (Sigma #410896) was made in 1× PBS and sterile-filtered through a 0.2 μm nylon syringe filter (Fisher Scientific). For planar organoids formation, 40 mg/mL of 8-arm PEG- norbornene monomer formulation, 60 % Tryp-MMP-peptide crosslinker (60% molar ratio relative to norbornene arms), 2 mM CRGDS, and 0.05% (w/w) Irgacure 2959 photoinitiator stock solutions were used. PEG Hydrogels were prepared by adding the appropriate volumes of PEG in 1× PBS and then adding Tryp-MMP, followed by CRGDS and the resulting mixture was placed on ice. Next, appropriate volume of ice cold diluted I2959 was mixed with the gel solution. A 10 μL of the resulting mixture of PEG solution and I2959 was added to each well of the plate in such a way that the solution covers the bottom of the well evenly. The plate containing the gel solutions was placed on the top shelf of a UV light for 5 min. After polymerization, PEG hydrogels were incubated at 5 % CO2, 37°C in 50 μL DME/F12 (HyClone #SH3002301) medium overnight to allow swelling and equilibration. To measure the thickness of the PEG hydrogel on the angiogenesis 96 well plate, we added Fluorescein (FITC-488) dye to this gel mixture and prepared the PEG Hydrogels as described above. Next, we acquired Confocal microscopy images of Fluorescein (FITC-488) encapsulated PEG Hydrogel made on the angiogenesis 96 well plate and measured the thickness of the PEG Hydrogel layer. Average thickness of PEG Hydrogel layer was found to be 90 ± 10 µm (Supplemental Figure S1). We also measured the modulus of the PEG Hydrogel used for the culture and expansion of the organoids. The modulus of PEG Hydrogel was 1137.70 ± 38.15 Pa while that of Matrigel was 95.50 ± 6.80 Pa (Supplemental Figure S2).
Matrigel sample preparation
A 10 μL of Matrigel (Corning #354277) suspension was added to each well of the angiogenesis 96 well plate and incubated at incubator at 37°C for 30 min to ensure semi-solid gel formation. Next, 50 μL DME/F12 (HyClone #SH3002301) medium was added on top of the semi-solid Matrigel and further incubated at 5% CO2, 37°C in overnight to allow swelling of the gels.
Rheological methodology
Both the Matrigel and PEG samples were prepared as per the procedure mention in the Method section. 54 The stiffness of both Matrigel and PEG samples were measured by dynamic frequency sweep rheological studies. Elastic modulus G′ and viscous modulus G″ were plotted against strain at constant frequency 1 Hz at 25°C.
Culture of Matrigel suspension neural organoids
Preparation of cerebral organoid differentiation medium (CDM): For 250 mL CDM medium, 125 mL of DMEM-F12, 125 mL of Neurobasal medium, 1.25 mL of N2 supplement, 62.5 μL of insulin, 2.5 mL of GlutaMaX supplement, 1.25 mL of MEM-NEAA, and 2.5 mL of penicillin-streptomycin were added. Next, we prepared a 1:100 dilution of 2-mercaptoethanol in DMEM-F12 and 87.5 μL of it was added to the medium followed by addition of 2.5 mL of B27 with or without vitamin A (retinoic acid) supplement.
A 9000 human induced pluripotent stem cells (iPSCs) were seeded per well in 200 μL of TeSR1-E8 medium in low-attachment 96 well plate, which was then centrifuged at 270g for 5 min at room temperature and placed in the incubator at 37°C and 5% CO2. Cells were treated every other day by gently aspirating approximately half of the medium. When iPSCs are ~500–600 μm in diameter with smooth edges, the iPSCs were transferred with a cut 200-μL pipette tip to one well of a low-attachment 24-well plate containing 500 μL of neural induction medium. After 2 days, another 500 μL of neural induction medium was added to the 24-well plate. Next, Matrigel was thawed and dimpled Parafilm substrate was prepared for the generation of Matrigel droplets by layering a square of Parafilm over an empty tip tray for size 200-μL tips following a published protocol. 74 A grid of 4 × 4 dimples was made and the Parafilm was trimmed with sterile scissors to a small square containing this grid, which was then placed into a 60-mm tissue culture dish. The neuroepithelial tissues were transferred using a cut 200-μL tip 1 × 1 to each dimple in the Parafilm. Media was removed carefully and the droplets of Matrigel was added to each aggregate by dripping ~30 μL onto each tissue so that the droplet fills the Parafilm dimple. Next, the 60-mm dish containing droplets on Parafilm was put into the 37 °C incubator, and incubated for 20–30 min to allow the Matrigel to polymerize. A 5 mL of cerebral organoid differentiation medium was added without vitamin A to the 60-mm dish. Matrigel droplets were removed from Parafilm sheet and the tissue droplets were cultured in a CO2 incubator for 2 days before put on a shaker in the incubator. Medium was changed every 3 days with 5 mL cerebral organoid differentiation medium without vitamin A for two to three times, then changed to normal CDM.
Culture of Matrigel and PEG hydrogel surface neural organoids
On the day before generation of the organoids, Matrigel and/or PEG hydrogel matrices were formed in an angiogenesis plate (Ibidi #89646). About 2 × 104 wild type C11 (WTC11) EC cells and 4 × 103 WTC11 PC cells were added onto the PEG hydrogels with a final volume of 40 µL, in E7V medium. Cells were fed daily for 7 days by removing approximately 30 µL of medium and replacing with 40 µL fresh E7V medium. On day 7, 2.5 × 104 WTC11 NPC cells were added onto the existing WTC11 EC/PC networks with a final volume of 40 µL per well in N2B27(NG) medium. The organoids were refed daily with 40 µL of fresh N2B27(NG). On day 12, 625 WTC11 MG cells were added onto the existing EC/PC/NPC networks with a final volume of 40 µL per well in N2B27(NG) medium. The organoids were refed daily with fresh N2B27(NG) for up to 28 days to allow network formation.
Immunofluorescence imaging
Antibody staining
Matrigel and PEG hydrogel surface neural tissue were fixed with 4% PFA in 1× PBS for 4 h at room temperature (RT) and then stored in 1× PBS ready for staining. First, the wells containing the neural organoids were washed three times with 50 µL 1× PBS per well to ensure all traces of fixative are removed. About 50 μL of 0.25% Triton X-100 (Fisher #BP151) made in 1× PBS was added to each well and incubated for 15 min at RT to permeabilize the cells. Triton X-100 was removed and the cells were blocked with 50 µL per well of blocking buffer consisting of 1× PBS, 10% (Normal Donkey Serum) NDS, 1% Bovine Serum Albumin (BSA; Fisher #BP1600), 0.25% Triton X-100 for 4 h at RT. Blocking buffer was removed and 50 μL of primary antibody solution consisting of 1× PBS, 1% NDS, 1% BSA, and 0.25% Triton X-100 per well was added at 1:50 dilutions and incubated overnight at 4°C. After that, primary antibody solution was removed and each well was washed with 50 μL 1× PBS per well six times for 30 min. Next, 50 μL per well of secondary antibody solution consisting of 1× PBS, 1% NDS, 1% BSA, and 0.25% Triton X-100 was added at either 1:1000 or 1 in 100 dilutions and incubated for 4 h at RT in the dark. For the last 30 min of the incubation, cells were treated with DAPI (Fisher #574810) at 5 µg/mL to each well. The antibody solution was removed and 50 µL 1× PBS was added per well and incubated for 30 min at RT followed by 50 µL fresh 1× PBS for an overnight wash at 4°C. Finally, cells were washed with 1× PBS for six times for 30 min at RT and stored at 4°C in the dark for imaging. Sources of primary anti-bodies: Mouse anti-CD31 (Dako #M0823); Mouse anti-β3 tubulin (R&D #MAB1195); Mouse anti-SOX2 (Invitrogen #14-9811-82); Rat anti-GFAP (Thermo #13-0300); Chicken anti-GFAP (Sigma #AB5541), Rabbit anti-Iba1 (FUJIFILM #019-19741); Mouse anti-SNAP-25 (BioLegend #836303); Mouse anti-PSD-95 (Antibodies Incorporated #75-082-020). Sources of secondary anti-bodies: Donkey anti-mouse 488 (Life Technologies #A21202); Donkey anti-rat 488 (Life Technologies #A21208); Donkey anti-rabbit 488 (Life Technologies #A21206); Donkey anti-mouse 568 (Life Technologies #A10037); Donkey anti-chicken 647 (Jackson Immuno Research #703-605-155).
Image acquisition
Immunofluorescence images were collected using a Nikon A1RS HD confocal microscope with 4× Air (Nikon Instruments, NA = 0.2) and 20× Air (Nikon Instruments, NA = 0.75) objectives. Images were collected using GaAsP PMT detectors.
Additional images were acquired using a Nikon A1R MP+ HD upright multiphoton/confocal microscope and a 25× water immersion long working distance objective (Nikon Instruments, NA = 1.1) and GaAsP PMT detectors. Images were acquired at 1024 × 1024 pixels. Z-stacks were acquired with a step size of 0.375 µm for Nyquist sampling. Each slice was acquired using a combination of resonance scanning and built-in NIS Elements AI denoising software.
Image processing
Images were processed in NIS Elements and Fiji. 75 3D volumes of whole mount organoids were captured in NIS Elements volume viewer. Images acquired with the A1R MP+ microscope were further processed in Fiji (v. 1.53t). Images were filtered with a 1× median filter, then thresholded using a Triangle-based segmentation algorithm. The analyze particles plug-in was applied to filter particulates below 20 px in size for select channels. The final processed z-stacks were turned into maximum intensity projections. Further microglia segmentation and visualization was performed using Imaris software (Oxford Instruments, v. 9.9.1).
RNA isolation and next generation sequencing
First, 50 µL RLT + BME was added to each well of an angiogenesis 96 well plate containing the neural tissue. The plate was incubated for 2 min at RT to lyze the cells. Lyzed cells were harvested by gently pipetting up and down several times while minimizing the amount of PEG hydrogel in the sample. The RLT lysate was transferred to the labeled Eppendorf, vortexed for 10 s at top speed and placed on ice after pooling four identical wells to one Eppendorf. About 350 µL of 70% ethanol was added to each sample and mixed well by pipetting, then transferred to a labeled RNeasy MinElute spin column in a 2 mL collection tube before centrifugation for 15 s at 8000×g. The flow through the column was discarded and the column was washed with 350 µL of RW1 buffer and centrifuged for 15 s at 8000×g. The flow through the column was again discarded and 80 µL of DNase I incubation mix (10 µL DNase I + 70 µL RDD buffer mixed gently by pipetting) was added to the column membrane and incubated at RT for 15 min. About 350 µL of RW1 buffer was added to the column and centrifuged for 15 s at 8000×g. The column was then placed into a new 2 mL collection tube and 500 µL of RPE buffer was added to the column and centrifuged for 15 s at 8000×g. The flow through the column was discarded and 500 µL of 80% ethanol was added to the column and centrifuged for 2 min at 8000×g. The column was placed into a new 2 mL collection tube and centrifuged at full speed for 5 min with the lid of the column open to minimize ethanol transfer. The column was placed into a labeled 1.5 mL collection tube and 14 µL of RNase-free water was added to the membrane and incubated on the bench at RT for 10 min for maximum elution. The column was then centrifuged at full speed for 1 min. The elution RNA sample recovered was collected and stored at −80°C.
RNA was analyzed via capillary gel electrophoresis on a fragment analyzer (Agilent, Santa Clara, CA) and quantified by fluorometric quantification using a Quant-iT™ RNA assay kit (Thermo). About 10 ng of RNA was utilized to generate sequencing libraries using the TrueSeq RNA Exome workflow and reagents (Illumina, San Diego, CA). Libraries were analyzed via capillary gel electrophoresis for size distribution and quality and quantified by fluorometric detection using a Quant-iT™ DNA assay kit (Thermo). Sequencing libraries were pooled and sequenced on a NovaSeq 6000 (Illumina). Sequencing data was analyzed through Illumina Base Space applications. Samples were de-multiplexed and read quality was assessed by use of bcl2fastq conversion. Read mapping was performed using the STAR aligner with the human reference genome GRCh38 73 Differential expression analysis was performed using DESeq2. Differentially expressed genes were thresholded for Gene Ontology analysis with genes demonstrating fold changes of >1.2 fold or <−1.2-fold differences and with p-adj values of <0.05 carried into gene ontology analysis. 76 Gene ontology analysis was performed using the Correlation Engine platform (Illumina) which curates publicly available data sets.
ELISA assays
All the ELISA assays were performed as per the manufacturer’s protocol. Sources of all the ELISA Kits: Human BDNF ELISA Kit (Cat# RAB0026, Sigma Aldrich); Human VEGF Quantikine ELISA Kit (Cat# DVE00, R&D Sytem); Glutamate Assay Kit (Cat # ab83389, Abcam); Human IL-6 Quantikine ELISA Kit (Cat# D6050, R&D System); and Human TNF-alpha Quantikine ELISA Kit (Cat # DTA00D, R 7 D System).
Statistical analysis
All results are reported as means ± s.e.m. and all experiments were repeated at least three times.
Statistical analysis on the significance between data sets was calculated using a one-way or two-way ANOVA, followed by Tukey’s multiple comparisons test. Statistical significance is indicated as *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. Statistical analyzes were carried out using GraphPad Prism 8.0 (GraphPad Software, La Jolla, CA).
Results and discussion
Development of planar neural organoids (PNOs)
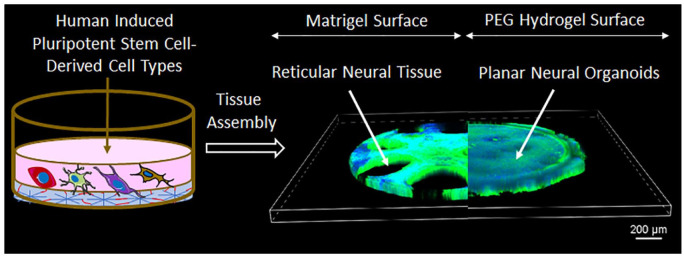
Human iPSC-derived progenitor cells cultured on the surface of synthetic PEG hydrogels formed planar neural organoids when seeded in a controlled time course. Human iPSC-derived endothelial cells (iPSC-ECs), pericytes (iPSC-PCs), neural progenitor cells (iPSC-NPCs), and microglia (iPSC-MG) were differentiated as described previously.54,76,77 Seeding iPSC-ECs and iPSC-PCs on day 0 led to formation of vascular assemblies on the surface of Matrigel and PEG hydrogels over a 7-day timeframe (Supplemental Figure S3). iPSC-NPCs and iPSC-MG were then seeded on day 7 and day 12, respectively. Culture of these neural tissues was continued for 28 days in serum-free and antibiotic-free growth medium on the surface of Matrigel or PEG hydrogels (Supplemental Figure 1A). Confocal microscopy images indicated formation of planar neural tissue on the surface of PEG hydrogels, which we refer to as “planar neural organoids” (PNOs; Figure 1(a)–(c); Supplemental Figure S4). Notably, the PNOs formed on PEG hydrogels were composed of more continuous, thinner cellular material compared to neural tissue grown on Matrigel. Therefore, PNOs were more amenable to imaging with a standard confocal approach versus the denser, reticular cellular organization formed on Matrigel. In a separate set of experiments, we added all the iPSC-derived cell types simultaneously on day 0 and continued the culture for 28 days. Simultaneous seeding of all cell types led to formation of clustered tissue morphology on both the Matrigel and PEG hydrogel surfaces (Supplemental Figure S5A and B) as well as formation of less interconnected CD31 vascular networks on the Matrigel surface than that of PEG hydrogel surface (Supplemental Figure S5C).
Figure 1.
Schematic representation showing the culture of neural tissues on Matrigel or PEG hydrogels surfaces: (a) schematic protocol for the culture of neural tissues on the Matrigel or PEG hydrogels surfaces. PEG hydrogels were formed through photopolymerization to crosslink PEG molecules with matrix metalloproteinase (MMP)-degradable peptides and include a pendant cell adhesion peptide. Human induced pluripotent stem cells (iPSCs)-derived precursor cells were co-cultured on Matrigel and/or PEG hydrogel surface in angiogenesis 96 well plates. Endothelial cells (ECs) and pericytes (PCs) were seeded on day 0, followed by neural progenitor cells (NPCs) on day 7 and microglia (MGs) on day 12. Brightfield images of organoids cultured in a angiogenesis 96 well plate display planar morphology for tissues formed on PEG, and reticular morphology from tissues grown on Matrigel (b and c). 3D confocal images of neural tissues grown on Matrigel (top) or PEG hydrogel (bottom) surfaces (scale bar 500 and 100 µm). Tissues on PEG hydrogel surfaces displayed planar morphology while those on the Matrigel surface showed reticular morphology.
Characterization of vascular assembly
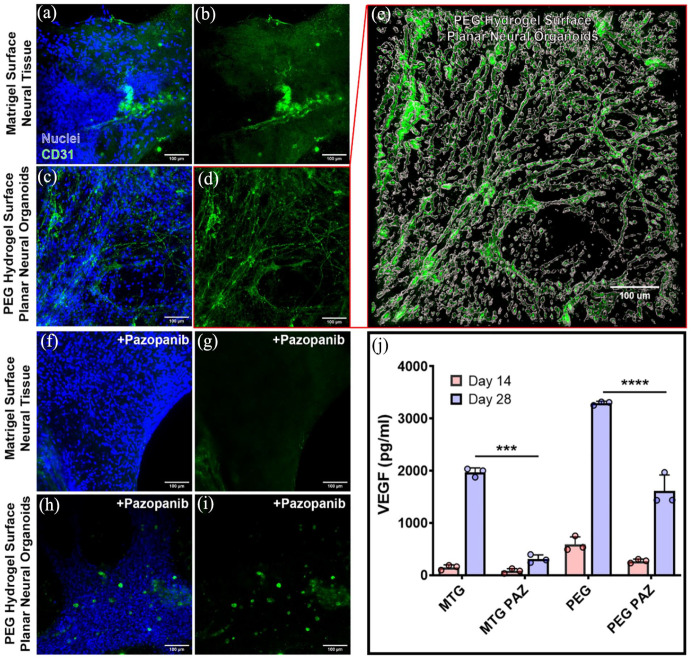
PNOs formed on PEG hydrogels showed more interconnected vascular assembly with higher expression of vascular marker protein than neural tissues formed on the Matrigel surface. Tissues formed on Matrigel did not contain a vascular network structure (Figure 2(a) and (b)), while PNOs formed on PEG hydrogels had an interconnected CD31+ vascular network (Figure 2(c)–(e)). Similarly, PNOs showed higher levels of vascular endothelial growth factor (VEGF) production than tissues formed on Matrigel at each time point (Figure 2(j)). Treatment of neural tissues with the VEGF-inhibitor pazopanib (PAZ) for 7 days reduced the growth of the CD31 vascular networks in the PNOs on PEG surfaces (Figure 2(f)–(i), respectively), and significantly decreased VEGF production on both PEG and Matrigel surfaces (Figure 2(j)).
Figure 2.
Characterization of vascular tissues in the neural organoids. Confocal images show CD31+ endothelial cells (green) in tissues formed on either Matrigel (a and b) or PEG hydrogels (c and d). (e) 3D reconstructions of CD31 vascular network within the planar neural organoid. Planar neural organoids on PEG hydrogels displayed denser and interconnected vascular network formation when compared to tissues formed on Matrigel at day 28. The vascular endothelial growth factor (VEGF) inhibitor pazopanib (PAZ) had a significant effect on CD31+ vascular networks (green) in neural tissues formed on Matrigel (e and f) or PEG hydrogels (g and h). (i) Measurement of VEGF protein from the culture samples of neural organoids formed on Matrigel (MTG) or PEG hydrogels (***p < 0.001; ****p < 0.0001). Scale bar 100 μm.
Characterization of neuronal and astrocyte phenotypes
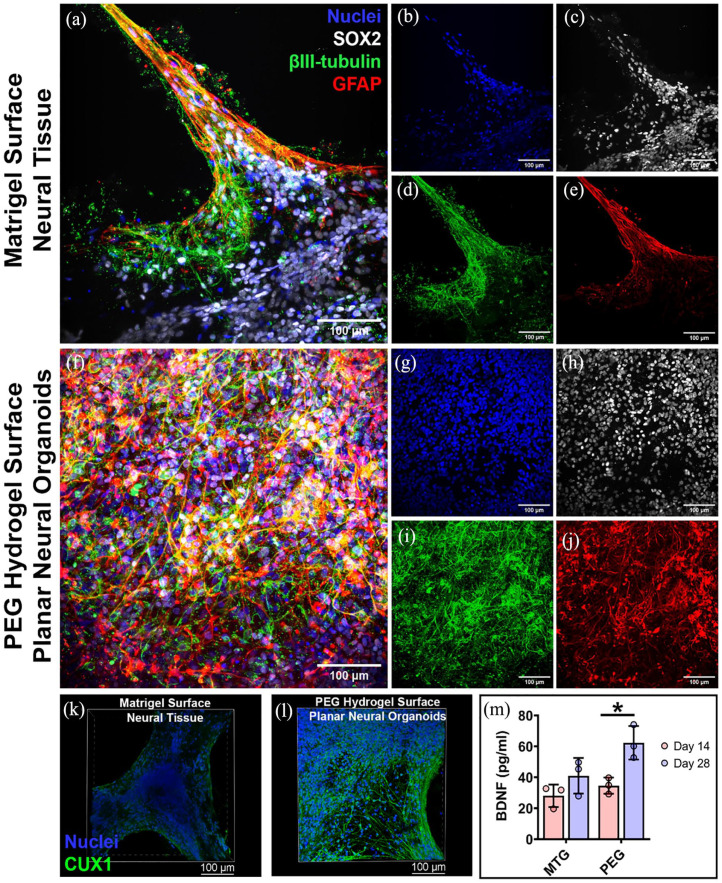
Immunofluorescence imaging showed the neural marker protein βIII-tubulin and neural precursor cell marker protein SOX2 were apparent throughout the neural tissues by 28 days. βIII-tubulin+ and SOX2+ cells appeared more abundant in the PNOs when compared to neural tissues formed on Matrigel (Figure 3(a)–(d) versus Figure 3(f)–(i), respectively; Supplemental Figure S6). Immunofluorescence imaging also revealed the presence of GFAP+ cells, suggesting radial glia accumulation within the neural tissues (Figure 3(e) and (j); Supplemental Figure S7). Presence of the human brain cortical neural marker protein CUX1 was also observed in the PNOs (Figure 3(k) and (l); Supplemental Figure S8). PNOs produced higher amounts of BDNF protein when compared to neural tissues formed on Matrigel, further indicating increased neural cell content on the PNOs relative to tissues formed on Matrigel. We further observed higher production of BDNF protein for the day 28 PNOs than that of day 14 PNOs (Figure 3(m)).
Figure 3.
Characterization of neural marker proteins in the organoids. Immunofluorescence staining of neural tissues formed on Matrigel surface (a) and PEG hydrogel surface (f). Distribution of nuclei (b and g) with antibodies against neural marker proteins SOX2 (c and h), βIII-tubulin (d and i), and GFAP (e and j). Scale bar 100 μm. Immunofluorescence staining of neural organoids with antibodies against human brain cortical neural marker protein CUX1 (green; k and l); Scale bar 100 μm. (m) Measurement of brain-derived neurotrophic factor (BDNF) protein secreted by the neural tissues (*p < 0.05).
Characterization of synaptic markers and microglia phenotypes
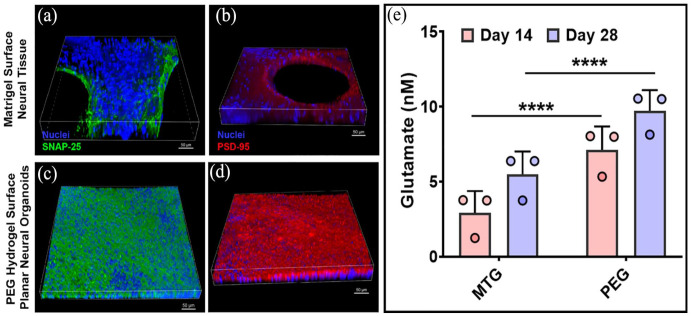
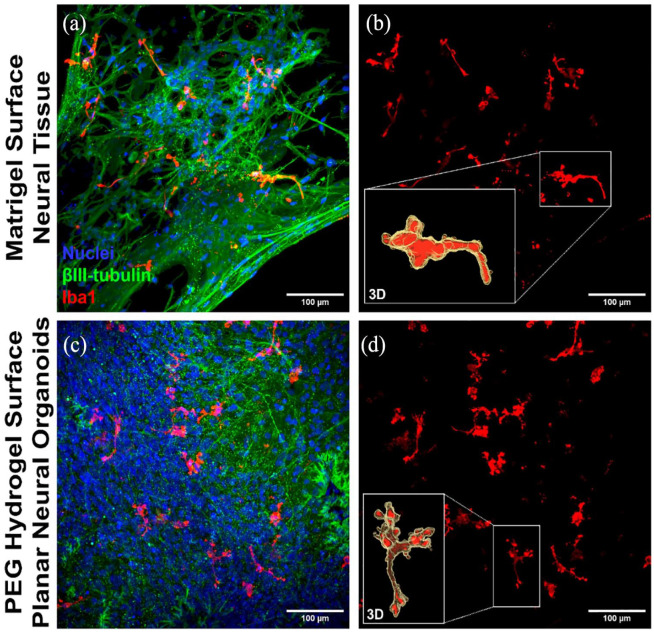
Immunofluorescence imaging showed increased appearance of both the pre-synaptic marker protein SNAP-25 and the post-synaptic marker protein PSD-95 in PNOs, and less appearance of these markers in neural tissues formed on Matrigel (Figure 4(a)–(d); Supplemental Figure S9). PNOs produced higher amounts of the neurotransmitter glutamate when compared to neural tissues formed on Matrigel (Figure 4(e)). PNOs also showed increased appearance of the microglia marker protein IBA1 when compared to neural tissues formed on Matrigel (Figure 5(a) and (c); Supplemental Figure S10). IBA1+ microglia-like cells were interspersed throughout the tissue and showed a ramified morphology consistent with homeostatic microglial morphology in the human brain (Figure 5(b) and (d)).
Figure 4.
Characterization of synaptic marker proteins in the neural organoids. Immunofluorescence staining for the pre-synaptic marker protein (a and c) SNAP-25 (green) and post-synaptic marker protein (b and d) PSD-95 (red). Scale bar 50 μm. (e) Measurement of glutamate from the culture samples of neural tissues (****p < 0.0001).
Figure 5.
Characterization of microglia phenotypes in the neural organoids. Confocal images of the 3D neural tissues showing presence of the microglia marker protein IBA1 (red) and neural marker βIII-tubulin (green; a and c). A 3D reconstructions of segmented microglia show ramified morphology within the tissues (b and d). Scale bar 100 μm.
Gene expression comparisons of planar neural organoids compared with traditional brain organoid aggregates
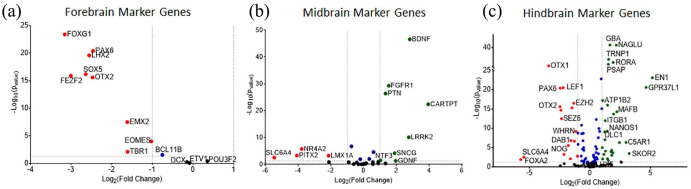
Planar neural organoids formed on PEG hydrogels ex-pressed several neural markers, including CD-31 (Figure 2(d)), βIII-tubulin (Figure 3(i); Supplemental Figure S6), GFAP (Figure 3(j); Supplemental Figure S7), and IBA1 (Figure 5; Supplemental Figure S10). Direct comparisons of PNOs formed on PEG hydrogels to traditional brain organoids formed in suspension culture using a commonly used, previously published protocol 78 revealed a number of important distinctions. First, traditional brain organoids were of variable and larger size and shape, and were not translucent (Figure 6(a); Supplemental Figure S11). This is in contrast to the PNOs, which had their shape and geometry defined by the underlying substrate, and were translucent and amenable to imaging. RNA-seq analysis revealed that 2456 genes were significantly upregulated in PNOs when compared to traditional brain organoids, while 1297 genes were upregulated in traditional brain organoids versus PNOs (Figure 6(b)). Importantly, genes associated with specific neuronal cell types, including GABAergic and glutamatergic neurons, and synaptic genes were found to be upregulated in PNOs versus traditional brain organoids (Figure 6(c)). Supplemental Figure S12 showed Heatmap for expression levels of various type of neurovascular genes in traditional brain organoids and planar neural organoids. Interestingly, forebrain (Figure 7(a)), midbrain (Figure 7(b)), and hindbrain (Figure 7(c); Supplemental Figure S13A) marker genes were also upregulated in the PNOs when compared to traditional brain organoids.
Figure 6.
Gene expression and gene ontology analysis for planar neural organoids versus traditional brain organoids: (a) microscopic images displaying morphological features of the neural organoids assembled in traditional Matrigel suspension (top) or on a PEG hydrogel surface (bottom), (b) global patterns of gene expression in traditional brain organoids formed in Matrigel suspension versus planar neural organoids formed on PEG hydrogels, and (c) Gene ontology analysis for biological processes related to the growth of neural tissues.
Figure 7.
Analysis of differentially expressed genes of the (a) forebrain, (b) midbrain, and (c) hindbrain in planar neural organoids formed on PEG hydrogels versus traditional brain organoids formed in Matrigel suspension. Fold change (Log2) plotted versus −Log10 of p-values of differential expression analysis. Green color points indicated positive fold change of those genes, red color points indicate negative fold change of the genes in the plots.
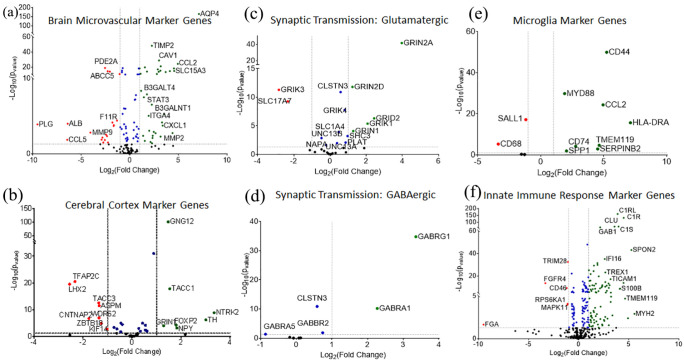
Planar neural organoids expressed higher levels of brain microvascular marker genes (Figure 8(a); Supplemental Figure S13B) and cerebral cortex marker genes (Figure 8(b)) when compared to traditional brain organoids. RNA-Seq results revealed upregulation of the genes associated with Glutamatergic and GABAergic synaptic transmissions in PNOs compared to traditional brain organoids (Figure 8(c) and (d)). Microglial cell marker genes were also upregulated in PNOs when compared to traditional suspension organoids (Figure 8(e)). Similarly, expression of microglial cell marker genes associated with the innate immune response were higher in PNOs (Figure 8(f); Supplemental Figure S13C). This result is not surprising, as the PNOs incorporate microglia progenitor cells during their assembly process, while the traditional brain organoids are not designed to include microglial phenotypes.
Figure 8.
Analysis of differentially expressed genes. Fold change (Log2) plotted versus −Log10 of p-values of marker genes between planar neural organoids compared to traditional brain organoids: (a) brain micro-vascular markers, (b) cerebral cortex markers, (c and d) Synaptic transmission associated genes, (e) microglial cell marker genes, and (f) innate immune response marker genes. Green color points indicated positive fold change of those genes, red color points indicate negative fold change of the genes in the plots.
Figure 9.
Lipopolysaccharide (LPS) induced inflammation in planar neural organoids: (a) LPS-induced TNF-α protein production in the media of planar neural organoid cultures, (b) LPS-induced IL-6 protein production in the media of the neural tissue culturesTreatment with the anti-inflammatory drugs Celecoxib (CEL) or Donepezil (DON) reduced the production of TNF-α and IL-6 in the culture medium. (C) Confocal microscopy images of the 3D neural organoids showing the ramified type morphology of microglia for the control treated group and ameboid type morphology of microglia for the LPS treated group. Scale bar 50 μm.
MTG: Matrigel; PEG: polyethyleneglycol; LPS: lipopolysaccharide.
**p < 0.01. ***p < 0.001. ****p < 0.0001).
Lipopolysaccharide induced inflammation
Neuroinflammation is a principal pathological event in neurodegenerative diseases. 79 We sought to test the sensitivity of PNOs to neuroinflammatory perturbations using lipopolysaccharide (LPS), as LPS-induced inflammation has been widely used to model neuroinflammation associated with neurodegenerative diseases. 80 We investigated LPS-induced inflammation using PNOs formed on PEG hydrogels. Neuroinflammation was induced at day 26 via addition of 200 ng/mL of LPS, followed by treatment with 1000 nM of the anti-inflammatory drugs Celecoxib or Donepezil on day 27. The experiment continued until day 28 when secreted anti-inflammatory cytokines were measured in the media (Figure 9(a) and (b)). Planar neural organoids produced elevated TNFα and IL-6 upon LPS stimulation compared to neural tissues formed on Matrigel. Treatment with the anti-inflammatory drugs Celecoxib or Donepezil reduced the production of both TNF-α and IL-6. We also measured LPS induced production of an anti-inflammatory cytokine such as IL-10 in the organoid samples. Our results showed anti-inflammatory drugs Celecoxib or Donepezil reduced the production of IL-10 as compared to that of the no drug treatment samples (Supplemental Figure S14). Confocal microscopy images revealed presence of ameboid microglia morphologies in the LPS-stimulated PNOs versus the ramified microglia morphologies observed without LPS stimulation (Figure 9(c)). This type of microglia morphological transition has also been observed in the human brain as a result of neuroinflammation. 81 Taken together, these results suggested that PNOs can be useful as an in vitro model to study therapeutic approaches to influence neuroinflammation.
Conclusions
We describe a physiologically-relevant in vitro model for the culture and expansion of 3D human planar neural organoids on synthetic PEG hydrogels. Immunofluorescence, RNA-Sequencing, and ELISA showed higher levels of neural, vascular, and neuroinflammatory markers in the PNOs when compared to either traditional brain organoids formed in Matrigel suspension or neural tissues formed on Matrigel surfaces. Planar neural organoids produced elevated TNF-α and IL-6 upon LPS stimulation when compared to neural tissues formed on Matrigel, and treatment with anti-inflammatory drugs Celecoxib or Donepezil significantly reduced TNF-α and IL-6 production. The PNOs formed in the current study have an open format that allows for addition of extraneous biologics as well as monitoring of secreted signals. In addition, their planar structure facilitates microscopic analysis of cell behaviors. Taken together, these observations suggest that the PNOs could be a uniquely valuable tool for in vitro disease modeling.
Supplemental Material
Supplemental material, sj-docx-1-tej-10.1177_20417314241230633 for Human induced pluripotent stem cell-derived planar neural organoids assembled on synthetic hydrogels by Joydeb Majumder, Elizabeth E Torr, Elizabeth A Aisenbrey, Connie S Lebakken, Peter F Favreau, William D Richards, Yanhong Yin, Qiang Chang and William L Murphy in Journal of Tissue Engineering
Footnotes
Author contributions: W.L.M. conceived the concept of the study. W.L.M. and Q.C. supervised the study. E.E.T., E.E.A., J.M., Y.Y., W.L.M., and Q.C. designed the experimental protocol. J.M., E.E.T., E.E.A., C.S.L., P.F.F, W.D.R., and Y.Y. carried out all the experiments. J.M. and W.L.M. prepared the initial draft of the manuscript. All the authors reviewed and edited the paper.
The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: W.L.M. and C.S.L. are co-founders and shareholders in Stem Pharm, Inc., which is focused on commercial applications of neural organoids. C.S.L., P.F.F., and W.D.R. are employees of Stem Pharm, Inc.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This research was funded by the US Environmental Protection Agency (STAR Grant no. 83573701), the US National Institutes of Health (award nos 1U01TR002383, R01HL093282, 1R01NS109427, 1R43ES029898, and 1R43ES029897) and the US National Science Foundation (award nos. EEC1648035 and DMR 170179).
ORCID iD: William L Murphy  https://orcid.org/0000-0001-5577-7739
https://orcid.org/0000-0001-5577-7739
Supplemental material: Supplemental material for this article is available online.
References
- 1. Phillips JB. ‘EngNT’-engineering live neural tissue for nerve replacement. Emerg Top Life Sci 2021; 5: 699–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Heusinkveld HJ, Staal YCM, Baker NC, et al. An ontology for developmental processes and toxicities of neural tube closure. Reprod Toxicol 2021; 99: 160–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yan Y, Shin S, Jha B, et al. Efficient and rapid derivation of primitive neural stem cells and generation of brain subtype neurons from human pluripotent stem cells. Stem Cells Transl Med 2013; 2: 862–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yamada KM, Cukierman E, Modeling tissue morphogenesis and cancer in 3D. Cell 2007; 130: 601–610. [DOI] [PubMed] [Google Scholar]
- 5. Pampaloni F, Reynaud EG, Stelzer EH. The third dimension bridges the gap between cell culture and live tissue. Nat Rev Mol Cell Biol 2007; 8: 839–845. [DOI] [PubMed] [Google Scholar]
- 6. Baker BM, Chen CS. Deconstructing the third dimension: how 3D culture microenvironments alter cellular cues. J Cell Sci 2012; 125: 3015–3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hickman JA, Graeser R, de Hoogt R, et al. Three-dimensional models of cancer for pharmacology and cancer cell biology: capturing tumor complexity in vitro/ex vivo. Biotechnol J 2014; 9: 1115–1128. [DOI] [PubMed] [Google Scholar]
- 8. Kapalczynska M, Kolenda T, Przybyla W, et al. 2D and 3D cell cultures – a comparison of different types of cancer cell cultures. Arch Med Sci 2018; 14: 910–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jensen C, Teng Y. Is it time to start transitioning from 2D to 3D Cell Culture? Front Mol Biosci 2020; 7, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hartung T. 3D - a new dimension of in vitro research. Adv. Drug Deliv Rev 2014, 69-70, vi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Breslin S, O’Driscoll L. Three-dimensional cell culture: the missing link in drug discovery. Drug Discov Today 2013; 18: 240–249. [DOI] [PubMed] [Google Scholar]
- 12. Huh D, Hamilton GA, Ingber DE. From 3D cell culture to organs-on-chips. Trends Cell Biol 2011; 21: 745–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Langhans SA. Three-dimensional in vitro cell culture models in drug discovery and drug repositioning. Front Pharmacol 2018; 9: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Matsusaki M, Case CP, Akashi M, et al. Three-dimensional cell culture technique and pathophysiology. Adv Drug Deliv Rev 2014; 74: 95–103. [DOI] [PubMed] [Google Scholar]
- 15. Tuveson D, Clevers H. Cancer modeling meets human organoid technology. Science 2019; 364: 952–955. [DOI] [PubMed] [Google Scholar]
- 16. Li Y, Tang P, Cai S, et al. Organoid based personalized medicine: from bench to bedside. Cell Regen 2020; 9: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xu H, Lyu X, Yi M, et al. Organoid technology and applications in cancer research. J Hematol Oncol 2018; 11: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yuki K, Cheng N, Nakano M, et al. Organoid models of tumor immunology. Trends Immunol 2020; 41: 652–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science 2014; 345: 1247125. [DOI] [PubMed] [Google Scholar]
- 20. Lewis A, Keshara R, Kim YH, et al. Self-organization of organoids from endoderm-derived cells. J Mol Med 2021; 99: 449–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brassard JA, Lutolf MP. Engineering stem cell self-organization to build better organoids. Cell Stem Cell 2019; 24: 860–876. [DOI] [PubMed] [Google Scholar]
- 22. Don FK, Huch M. Organoids, where we stand and where we go. Trends Mol Med 2021; 27: 416–418. [DOI] [PubMed] [Google Scholar]
- 23. Barker N, Huch M, Kujala P, et al. Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell 2010; 6: 25–36. [DOI] [PubMed] [Google Scholar]
- 24. Pompaiah M, Bartfeld S. Gastric organoids: an emerging model system to study helicobacter pylori pathogenesis. Curr Top Microbiol Immunol 2017; 400: 149–168. [DOI] [PubMed] [Google Scholar]
- 25. Zhang M, Liu Y, Chen YG. Generation of 3D human gastrointestinal organoids: principle and applications. Cell Regen 2020; 9: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Eicher AK, Kechele DO, Sundaram N, et al. Functional human gastrointestinal organoids can be engineered from three primary germ layers derived separately from pluripotent stem cells. Cell Stem Cell 2022; 29: 36–51 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu M, Hu T, Zhu P, et al. Kidney organoids as a promising tool in nephrology. Genes Dis 2022; 9: 585–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Maass C, Sorensen NB, Himmelfarb J, et al. Translational assessment of drug-induced proximal tubule injury using a kidney microphysiological system. CPT Pharmacometrics Syst Pharmacol 2019; 8: 316–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huch M, Bonfanti P, Boj SF, et al. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J 2013; 32: 2708–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Moreira L, Bakir B, Chatterji P, et al. Pancreas 3D organoids: current and future aspects as a research platform for personalized medicine in pancreatic cancer. Cell Mol Gastroenterol Hepatol 2018; 5: 289–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Georgakopoulos N, Prior N, Angres B, et al. Long-term expansion, genomic stability and in vivo safety of adult human pancreas organoids. BMC Dev Biol 2020; 20: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Driehuis E, Gracanin A, Vries RGJ, et al. Establishment of pancreatic organoids from normal tissue and tumors. STAR Protoc 2020; 1: 100192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Koike H, Iwasawa K, Ouchi R, et al. Engineering human hepato-biliary-pancreatic organoids from pluripotent stem cells. Nat Protoc 2021; 16: 919–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jung P, Sato T, Merlos-Suarez A, et al. Isolation and in vitro expansion of human colonic stem cells. Nat Med 2011; 17: 1225–1227. [DOI] [PubMed] [Google Scholar]
- 35. d’Aldebert E, Quaranta M, Srbert M, et al. Characterization of human colon organoids from inflammatory bowel disease patients. Front Cell Dev Biol 2020; 8: 363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Crespo M, Vilar E, Tsai SY, et al. Colonic organoids derived from human induced pluripotent stem cells for modeling colorectal cancer and drug testing. Nat Med 2017; 23: 878–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Michels BE, Mosa MH, Grebbin BM, et al. Human colon organoids reveal distinct physiologic and oncogenic wnt responses. J Exp Med 2019; 216: 704–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Huch M, Dorrell C, Boj SF, et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature 2013; 494: 247–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ye S, Boeter JWB, Mihajlovic M, et al. A chemically defined hydrogel for human liver organoid culture. Adv Funct Mater 2020; 30: 2000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Prior N, Inacio P, Huch M. Liver organoids: from basic research to therapeutic applications. Gut 2019; 68: 2228–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Harrison SP, Baumgarten SF, Verma R, et al. Liver organoids: recent developments, limitations and potential. Front Med 2021; 8: 574047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lam D, Dan YY, Chan YS, Emerging liver organoid platforms and technologies. Cell Regen 2021; 10: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Crignis ED, Hossain T, Romal S, et al. Application of human liver organoids as a patient-derived primary model for HBV infection and related hepatocellular carcinoma. eLife 2021; 10: e60747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Guan Y, Enejder A, Wang M, et al. A human multi-lineage hepatic organoid model for liver fibrosis. Nat Commun 2021; 12: 6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mansour AA, Goncalves JT, Bloyd CW, et al. An in vivo model of functional and vascularized human brain organoids. Nat Biotechnol 2018; 36: 432–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cui K, Chen W, Cao R, et al. Brain organoid-on-chip system to study the effects of breast cancer derived exosomes on the neurodevelopment of brain. Cell Regen 2022; 11: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mason JO, Price DJ. Building brains in a dish: prospects for growing cerebral organoids from stem cells. Neuroscience 2016; 334: 105–118. [DOI] [PubMed] [Google Scholar]
- 48. Agboola OS, Hu X, Shan Z, et al. Brain organoid: a 3D technology for investigating cellular composition and interactions in human neurological development and disease models in vitro. Stem Cell Res Ther 2021; 12: 430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chiaradia I, Lancaster MA. Brain organoids for the study of human neurobiology at the interface of in vitro and in vivo. Nat. Neurosci 2020; 23:1496–1508. [DOI] [PubMed] [Google Scholar]
- 50. Shou Y, Liang F, Xu S, et al. The application of brain organoids: from neuronal development to neurological diseases. Front Cell Dev Biol 2020; 8: 579659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Quadrato G, Brown J, Arlotta P. The promises and challenges of human brain organoids as models of neuropsychiatric disease. Nat Med 2016; 22: 1220–1228. [DOI] [PubMed] [Google Scholar]
- 52. Kelava I, Lancaster MA. Dishing out mini-brains: current progress and future prospects in brain organoid research. Dev Biol 2016; 420: 199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Passaro AP, Stice SL. Electrophysiological analysis of brain organoids: current approaches and advancements. Front Neurosci 2021; 14: 622137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schwartz MP, Hou Z, Propson NE, et al. Human pluripotent stem cell-derived neural constructs for predicting neural toxicity. Proc Natl Acad Sci USA 2015; 112: 12516–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lewis-Israeli YR, Wasserman AH, Gabalski MA, et al. Self-assembling human heart organoids for the modeling of cardiac development and congenital heart disease. Nat Commun 2021; 12: 5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lewis-Israeli YR, Wasserman AH, Aguirre A. Heart organoids and engineered heart tissues: Novel Tools for Modeling Human Cardiac Biology and Disease. Biomolecules 2021; 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pang JKS, Ho BX, Chan WK, et al. Insights to heart development and cardiac disease models using pluripotent stem cell derived 3D organoids. Front Cell Dev Biol 2021; 9: 788955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Miyamoto M, Nam L, Kannan S, et al. Heart organoids and tissue models for modeling development and disease. Semin Cell Dev Biol 2021; 118: 119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hoang P, Wang J, Conklin BR, et al. Generation of spatial-patterned early-developing cardiac organoids using human pluripotent stem cells. Nat Protoc 2018; 13: 723–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Benton G, Arnaoutova I, George J, et al. Matrigel: from discovery and ECM mimicry to assays and models for cancer research. Adv Drug Deliv Rev 2014; 79–80: 3–18. [DOI] [PubMed] [Google Scholar]
- 61. Hughes CS, Postovit LM, Lajoie GA. Matrigel: a complex protein mixture required for optimal growth of cell culture. Proteomics 2010; 10: 1886–1890. [DOI] [PubMed] [Google Scholar]
- 62. Kleinman HK, Martin GR. Matrigel: basement membrane matrix with biological activity. Semin Cancer Biol 2005; 15: 378–386. [DOI] [PubMed] [Google Scholar]
- 63. Vukicevic S, Kleinman HK, Luyten FP, et al. Identification of multiple active growth factors in basement membrane matrigel suggests caution in interpretation of cellular activity related to extracellular matrix components. Exp Cell Res 1992; 202: 1–8. [DOI] [PubMed] [Google Scholar]
- 64. Kozlowski MT, Crook CJ, Ku HT. Towards organoid culture without matrigel. Commun Biol 2021; 4: 1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Aisenbrey EA, Murphy WL. Synthetic alternatives to matrigel. Nat Rev Mater 2020; 5: 539–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gjorevski N, Sachs N, Manfrin A, et al. Designer matrices for intestinal stem cell and organoid culture. Nature 2016; 539: 560–564. [DOI] [PubMed] [Google Scholar]
- 67. Funfak A, Bouzhir L, Gontran E, et al. Biophysical control of bile duct epithelial morphogenesis in natural and synthetic scaffolds. Front Bioeng Biotechnol 2019; 7: 417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Magno V, Meinhardt A, Werner C. Polymer hydrogels to guide organotypic and organoid cultures. Adv Funct Mater 2020; 30: 2000097. [Google Scholar]
- 69. Cruz-Acuna R, Quiros M, Huang S, et al. PEG-4MAL hydrogels for human organoid generation, culture, and in vivo delivery. Nat Protoc 2018; 13: 2102–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kofman S, Mohan N, Sun X, et al. Human mini brains and spinal cords in a dish: Modeling strategies, current challenges, and prospective advances. J Tissue Eng 2022; 13: 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sun XY, Ju XC, Li Y, et al. Generation of vascularized brain organoids to study neurovascular interactions. eLife 2022; 11: e76707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yoshiyama Y, Kojima A, Ishikawa C, et al. Anti-inflammatory action of donepezil ameliorates tau pathology, synaptic loss, and neurodegeneration in a tauopathy mouse model. J Alzheimers Dis 2010; 22: 295–306. [DOI] [PubMed] [Google Scholar]
- 73. Aisenbrey EA, Torr E, Johnson H, et al. A protocol for rapid pericyte differentiation of human induced pluripotent stem cells. STAR Protoc 2021; 2: 100261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lancaster MA, Knoblich JA. Generation of cerebral organoids from human pluripotent stem cells. Nat Protoc 2014; 9: 2329–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Schindelin J, Arganda-Carreras I, Frise E, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 2012; 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kaushik G, Gupta K, Harms V, et al. Engineered perineural vascular plexus for modeling developmental toxicity. Adv Healthc. Mater 2020; 9: e2000825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Nosi D, Lana D, Giovannini MG, et al. Neuroinflammation: integrated nervous tissue response through intercellular interactions at the “whole system” scale. Cells 2021; 10: 1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Yücel G, Zhao Z, El-Battrawy I, et al. Lipopolysaccharides induced inflammatory responses and electrophysiological dysfunctions in human-induced pluripotent stem cell derived cardiomyocytes. Sci Rep 2017; 7: 2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Torres-Platas SG, Comeau S, Rachalski A, et al. Morphometric characterization of microglial phenotypes in human cerebral cortex. J Neuroinflammation 2014; 11: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013; 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 2014; 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-tej-10.1177_20417314241230633 for Human induced pluripotent stem cell-derived planar neural organoids assembled on synthetic hydrogels by Joydeb Majumder, Elizabeth E Torr, Elizabeth A Aisenbrey, Connie S Lebakken, Peter F Favreau, William D Richards, Yanhong Yin, Qiang Chang and William L Murphy in Journal of Tissue Engineering