Abstract
Infection by hepatitis B virus (HBV) is responsible for approximately 296 million chronic cases of hepatitis B, and roughly 880,000 deaths annually. The global burden of HBV is distributed unevenly, largely owing to the heterogeneous geographic distribution of its subtypes, each of which demonstrates different severity and responsiveness to antiviral therapy. It is therefore crucial to the global public health response to HBV that the spatiotemporal spread of each genotype is well characterized. In this study, we describe a collection of 133 newly sequenced HBV strains from recent African immigrants upon their arrival in Belgium. We incorporate these sequences—all of which we determine to come from genotypes A, D, and E—into a large-scale phylogeographic study with genomes sampled across the globe. We focus on investigating the spatio-temporal processes shaping the evolutionary history of the three genotypes we observe. We incorporate several recently published ancient HBV genomes for genotypes A and D to aid our analysis. We show that different spatio-temporal processes underlie the A, D, and E genotypes with the former two having originated in southeastern Asia, after which they spread across the world. The HBV E genotype is estimated to have originated in Africa, after which it spread to Europe and the Americas. Our results highlight the use of phylogeographic reconstruction as a tool to understand the recent spatiotemporal dynamics of HBV, and highlight the importance of supporting vulnerable populations in accordance with the needs presented by specific HBV genotypes.
Keywords: HBV, evolution, genome diversity, dispersal, genotype, subgenotype, immigration, Africa, African, migration, antiviral therapy, phylogenetic, spatio-temporal, origin, ancient, sequences
Introduction
Hepatitis B virus (HBV) imposes a significant global burden to public health (Pourkarim et al. 2011; Malik et al. 2022), causing about 880,000 deaths per year (Revill et al. 2020) despite over two decades of efforts by the global community under the banner of the World Health Organization (Pourkarim, Lemey, and Van Ranst 2018). The efficient implementation of prevention and control measures to combat the spread of HBV requires detailed insight into the epidemiology of the separate lineages circulating in a given region (Pourkarim and Van Ranst 2011; Schweitzer et al. 2015). Such information at both country and continent levels is pivotal for implementing effective intervention strategies and estimating the burden of disease accurately (Pourkarim et al. 2011). Both effective antiviral therapies and a widely available vaccine have contributed to significant decreases in the risk of HBV infection globally (Lu et al. 2020); however, their effectiveness can vary significantly by HBV genotype (Bottecchia et al. 2011).
The global genetic diversity of HBV is classified into eight approved genotypes (A–H) and two tentative genotypes (I and J). These genotypes are further subdivided into more than forty-five subgenotypes and quasi-subgenotypes (Pourkarim et al. 2014; Thijssen et al. 2020). Increasing evidence suggests that its genotypes and subgenotypes are determinants for patients’ disease progression and may prompt heterogeneous responses to antiviral therapy (McMahon 2009; Pourkarim et al. 2014; Shen and Yan 2014; Croagh, Desmond, and Bell 2015; Mina et al. 2015a; Zhang et al. 2020). The genotypes and subgenotypes of HBV show distinct geographical distributions. For example, genotype A (HBV-A) subgenotype A2 is dominant in Europe, whereas subgenotype A1 is mostly prevalent in Africa and East Asia. Genotypes B and C circulate with high frequency in Southeast Asia and the Pacific Islands (Norder et al. 2004). While genotype D (HBV-D) is distributed worldwide, subgenotype D1 is the most prevalent HBV subgenotype in western Asia and the Mediterranean Basin (Zehender et al. 2012; Al-Qahtani et al. 2020; Trovão et al. 2022), and subgenotypes D2 and D3 are prevalent in eastern Europe (Pourkarim et al. 2014; Pineda-Peña et al. 2015). Genotype E (HBV-E) is mainly prevalent in West Africa (Ingasia et al. 2020), whereas genotype F is one of the most prevalent genotypes circulating in South and Central America (Pujol et al. 2020). Genotype G is a rarely isolated genotype in France, the USA, and Belgium (Kay and Zoulim 2007; Mina et al. 2015b). Genotype H has been occasionally reported in Japan but is commonly encountered in Central and South America. Recently, two new tentative genotypes (I and J) have been proposed, which are most likely recombinant strains of the previously identified genotypes (Pourkarim et al. 2014).
The global epidemiology of HBV infection is characterized by hepatitis B surface Antigen (HBsAg) seroprevalence, which displays strong geographical variation. Regions may be classified as low seroprevalence (seroprevalence <2 %), such as European countries, intermediate seroprevalence (2 % to 7 %; e.g. Alaska, the Mediterranean basin, and India), or high seroprevalence (seroprevalence >8 %), such as the South Pacific and countries of sub-Saharan Africa. In high seroprevalence regions, HBV is considered to be endemic. Viral transmission routes largely depend on regional prevalence (Pourkarim et al. 2011). A study by the WHO in 2015 showed that global migration trends are crucial to the disease burden and gradual increase of the chronic reservoir in industrialized, low-endemicity countries receiving new waves of migrants from high- and intermediate-endemicity countries without obligatory vaccination (Sharma et al. 2015). Consequently, large migration events have a considerable impact on the prevalence of communicable and contagious diseases like HBV, since there are significant differences of HBV prevalence in ‘source’ and ‘sink’ populations (Chu et al. 2013). It has been frequently reported that non-endemic HBV strains carried by streams of immigrants are changing the viral epidemiological profile in the destination countries (Khan et al. 2008; Pourkarim et al. 2011; Chu et al. 2013; Coppola et al. 2017; Mina et al. 2017; Thijssen et al. 2019). Besides their different geographical distribution, genotypes of HBV possess genotype-specific mutations in their open reading frames (ORFs). The introduction of these strains into countries where prophylaxis, diagnostic, and therapeutic protocols have been implemented based on native HBV strains could potentially raise serious concerns (Lampertico, Maini, and Papatheodoridis 2015; Limeres et al. 2019; Velkov, Protzer, and Michler 2020). Furthermore, mutations associated with antiviral resistance have been reported in HBV strains isolated from patients who had recently migrated into Europe, even before exposure to treatment (Selabe et al. 2007; Bottecchia et al. 2011). European countries are now faced with new public health challenges to control HBV incidence and prevalence, necessitating updated policies for screening, monitoring, and preventing transmission from large reservoirs (Schweitzer et al. 2015; Thijssen et al. 2019).
Historically, Belgium has witnessed high rates of immigration, particularly from African countries, with intermediate to high HBV seroprevalence (Schweitzer et al. 2015). However, until now there has been a dearth of comprehensive evolutionary analyses of non-domestic HBV strains in Belgium. In this study, we describe a collection of HBV strains isolated from patients who had recently immigrated to Belgium from a number of African countries. Many of the viruses may potentially carry medically important genomic mutations, complicating public health interventions. Due to the limited global epidemiological insights and the potential impact of continued spread of such strains, we here conduct a molecular epidemiological study investigating the spatio-temporal processes shaping the HBV genotypes imported to Belgium, as well as their clinical characterization in a sample collection spanning half a decade. Our analyses differ from other recent studies such as that performed by Kocher et al. 2021 in that we analyze each genotype independently in order to understand the spatiotemporal factors that have contributed to the global proliferation of each genotype.
Materials and methods
Study population & supplemental sequence selection
One hundred and thirty-three surface antigen (HBsAg) positive individuals, who immigrated to Belgium from Africa between 2005 and 2010, were enrolled in this study. Serum samples were collected from patients upon their first clinical visit during the study. Virological and serological markers were assayed, and demographic information like sex, age, origin, possible transmission routes, and antiviral therapy were collected. For our analyses, we supplemented these novel sequences with all HBV-A, HBV-D, and HBV-E sequences available from the NCBI genome database (Sayers et al. 2022) during our study period. Supplemental sequences were screened for quality and excluded if they showed signatures of recombination or gaps longer than fifty base pair (bp) in length. This study was approved by the Ethical Committee of University hospital of KU Leuven (S56121/ML10229).
Extraction, amplification, and complete genome sequencing
Viral DNA was extracted from sera using the QIAmp1 Viral DNA mini kit (Qiagen, Hilden, Germany). Complete HBV genomes were amplified and sequenced using a previously described method (Pourkarim et al. 2009, 2011). To eliminate any recombinant isolates that may influence the topology of phylogenetic trees of HBV, we screened for recombination events among our newly sequenced strains and strains retrieved from the database using the HBV NCBI genotyping tool (https://www.ncbi.nlm.nih.gov/projects/genotyping/formpage.cgi). This tool provides an estimation of recombination signatures along with the full-length sequences of HBV strains. If there was any indication of genome hybridization, further investigation was conducted using SimPlot software, version 3.5.1, and bootScan (Lole et al. 1999). Detected recombinant strains were then confirmed through the construction of phylogenetic trees using different targeted segments.
Genotyping and subgenotyping
We aligned genomic sequences using MAFFT v7.0 (Katoh and Standley 2013), and used the same genome alignments for subsequent analyses. In order to determine genotype and subgenotype of sequenced strains, maximum-likelihood phylogenetic trees were constructed using IQ-TREE v1.6.12 (Nguyen et al. 2015) under an HKY nucleotide substitution model (Hasegawa, Kishino, and Yano 1985), accommodating among-site rate heterogeneity through a discretized gamma distribution (Yang 1994). Clade support was verified using UFBoot2 with 1,000 replicates (Hoang et al. 2018). We assigned new sequences genotype and subgenotype designations based on their phylogenetic placement.
HBV mutational patterns
Newly sequenced HBV full-length genomes are genotyped through phylogenetic analysis before genome mapping analysis. Subsequently, the different ORFs of these strains are aligned with corresponding ORFs of reference genomes (the same genotype and subgenotype) at the nucleotide or protein level. Reference strains retrieved from GenBank are typically associated with papers that present correlations between mutations and conditions such as phenotypically or genotypically antiviral resistance, vaccine, or diagnostic escape strains in vivo and in vitro. These mutations are considered as ‘medically important mutations’ in the literature. Needless to say, in the assessment of HBV mutational patterns, variations at the nucleotide level are studied in the precore, core, and X genes, while mutations related to the Pol gene and S gene are identified at the protein level.
Bayesian phylogenetic inference
We started by investigating the temporal signal of each genotype using TempEst (Rambaut et al. 2016). We first estimated an unrooted phylogeny for each genotype using maximum-likelihood inference produced in the previous step. Sequences that showed incongruent temporal patterns were excluded from further analyses. Because of spatial heterogeneity and clinical differences exhibited by each of HBV’s genotypes, we chose to analyze each genotype independently. This allowed us to not only more specifically characterize the spatiotemporal dynamics of each genotype of interest independently, but also split the computational load between three separate analyses, affording greater scale in each individual analysis. This resulted in a dataset with 583 HBV-A sequences (dated 1979–2014), 764 HBV-D sequences (dated 1975–2012), and 234 HBV-E sequences (dated 1994–2010). Phylogenetic relationships were inferred for each of the datasets separately by performing Bayesian phylogenetic inference using Markov chain Monte Carlo (MCMC), as available via the BEAST v1.10 package (Suchard et al. 2018). We used an uncorrelated relaxed molecular clock with branch rates drawn from an underlying lognormal distribution to account for evolutionary rate variation among lineages, with a constant demographic model as the tree prior (Kingman 1982; Drummond et al. 2002). To accommodate the impact of the overlapping regions in the HBV genome on the substitution process of each codon position, we assumed an HKY substitution model (Hasegawa, Kishino, and Yano 1985) without codon partitioning, with among-site rate heterogeneity through a discretized gamma distribution (Yang 1994). We performed at least three independent replicates of our MCMC analysis to ensure proper convergence, with each replicate having a different starting seed to achieve proper statistical mixing and running for at least 500 million iterations to achieve convergence. Chains were sampled every 10,000 iterations, and we combined all replicates to form a single posterior distribution. We assumed default priors for all other parameters in BEAST for these analyses and used the BEAGLE v3 (Suchard and Rambaut 2009) high-performance computational library to speed up the likelihood calculations. All relevant parameters exhibited proper statistical mixing, reporting ESS values over 200 as assessed using Tracer v.1.7 (Rambaut et al. 2018), with statistical uncertainty reflected by the 95 % highest posterior density (HPD) interval.
Following the analysis of the contemporary genomic data sets, and to ensure sufficient temporal signal in the data, we repeated the analysis of HBV-A and HBV-D using available ancient genomes, collected from mummified tissue ranging in age from 450 years old to ∼4,200 years old and spanning from Italy, across Eastern Europe, and into Central Asia (Mühlemann et al. 2018; Ross et al. 2018). This resulted in the addition of four ancient genomes to the HBV-A dataset and five ancient genomes to the HBV-D dataset. No ancient sequences were available to add to the HBV-E dataset. Phylogenetic inference was performed under the same model parameters as analyses without ancient genomes, but used an updated lognormal evolutionary rate prior (μ = –11.3474, σ = 1.22) derived from the HBV evolutionary rate estimated by Muhlemann et al. (2018) of 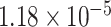 substitutions per site per year, based on the inclusion of ancient sequences. Default priors were used for all other parameters. For each subtype, MCMC chains were run for 500 million iterations and were sampled every 25,000th iteration. Three replicates of each subtype were run using different starting seeds to ensure proper convergence and statistical mixing. A single combined posterior distribution was formed by combining all replicates for each subtype, removing an appropriate proportion of each chain as burn-in. Maximum clade credibility (MCC) trees were summarized using TreeAnnotator v1.10 (Suchard et al. 2018) and the resulting trees were visualized in FigTree v1.4.3 and baltic v.0.1.5 (https://github.com/evogytis/baltic accessed on 10 June 2020).
substitutions per site per year, based on the inclusion of ancient sequences. Default priors were used for all other parameters. For each subtype, MCMC chains were run for 500 million iterations and were sampled every 25,000th iteration. Three replicates of each subtype were run using different starting seeds to ensure proper convergence and statistical mixing. A single combined posterior distribution was formed by combining all replicates for each subtype, removing an appropriate proportion of each chain as burn-in. Maximum clade credibility (MCC) trees were summarized using TreeAnnotator v1.10 (Suchard et al. 2018) and the resulting trees were visualized in FigTree v1.4.3 and baltic v.0.1.5 (https://github.com/evogytis/baltic accessed on 10 June 2020).
Phylogeographic inference
From the combined posterior distribution of each genotype’s phylogenetic reconstruction, 1,000 trees were sampled and used to perform subsequent Bayesian phylogeographic reconstruction in BEAST using empirical tree distributions (Pagel, Meade, and Barker 2004). This analysis modeled geographic locations as discrete traits, and allowed for different migration rates to and from each location (Lemey et al. 2009). This approach conditions on the geographic locations recorded at the tips of the trees and models the transition history among those locations as a time-reversible continuous-time Markov chain (CTMC) process to infer the unobserved locations at the internal nodes of each tree. BEAST default priors were used for each parameter. For these reconstructions, MCMC chains were run for 10 million iterations and sampled every 10,000th iteration. The links between locations that contribute significantly to explaining the migration history were identified using a Bayesian stochastic search variable selection (BSSVS) procedure (Lemey et al. 2009), and SpreaD3 (Bielejec et al. 2016) was used to calculate the Bayes factor (BF) support for these links. We quantify geographic transitions by logging Markov jumps and rewards through the phylogeographic analysis. We note that the data consist of a heterogeneous number of sequences for each location (Supplementary Tables 1, 2, 3) which show strong sampling bias between individual countries that can strongly impact the outcome of a discrete phylogeography study (Maio et al. 2015). To mitigate this issue, we opted to group sequences into continental regions, aiming to contain a number of sequences of the same order of magnitude where possible: Africa, the Americas, East & South Asia, Europe, and West & Central Asia. We note that for HBV-E, no samples from East & South Asia or West & Central Asia were available.
Statistical analysis
Statistical analyses were performed using Stata software version 16.1 for Windows. Categorical variables are expressed as frequencies; continuous variables are expressed as either means and SDs or medians and interquartile ranges, according to their distributions. Chi-squared test and Fisher’s exact test were applied to assess the association between categorical variables. T-test and Mann-Whitney-Wilcoxon test were used to assess continuous variables. The significance level was set to 0.05.
Results
Lineages circulating in Belgium and new isolates
We originally investigated 133 individuals coming from fifteen countries in sub-Saharan Africa. The majority are originally from the Republic of the Congo (n = 44; 33 %), Rwanda (n = 23; 17.25 %), and Guinea (n = 21; 15.75 %). The country of origin for one individual was unknown. Most of the individuals were male (n = 96; 72 %) and the median age was 30 years [interquartile range (IQR) = 25–37]. Nineteen individuals (14 %) were co-infected with human immunodeficiency virus (HIV), while three (2 %) were hepatitis C virus (HCV) carriers. Positive e-antigen (eAg) was present in 78 cases (58.65 %) (Supplementary Table 1). No significant differences were observed between HBV/HIV co-infected patients and HBV mono-infected patients in different genotype groups, in terms of medically important mutations at different strain ORFs.
Phylogenetic analysis and nucleotide divergence analysis of sequences with full-length genomes (n = 121) and large S genes (n = 12) revealed three distinct HBV genotypes circulating in African immigrants in Belgium. The most frequently identified genotypes were HBV-E (n = 67, 50.4 %) and HBV-A (n = 55, 41.4 %). Recombination was explored using SimPlot and BootScan (Supplementary Table 2 and Supplementary Figure S1). The genomes of three isolates (ID: MB-58, MB-56, and MB-92), from one patient of Guinea and two patients of Rwanda, showed recombination: between HBV-A and HBV-E in MB-58, and between HBV-D and HBV-E in both MB-56 and MB-92 (Supplementary Figure S1). MB-58 harbors 900 bp of HBV-E (1,100–2,000 bp) in the context of HBV-A. MB-92 carries 600 bp (1,800–2,400 bp) of HBV-E within HBV-D, with the wild type length of 3,182 bp. However, MB-56 harbors the same piece of HBV-E in the context of HBV-D with 3,212 bp of wild-type genome length of HBV-E.
HBV mutational patterns
For our new sequences, single nucleotide polymorphisms and amino acid polymorphisms were assessed throughout the genome. We further evaluated if the genotypes identified were associated with the presence of specific drug resistance mutations in the pol gene and medically important mutations at core and X genes. HBV-A was associated with the presence of core mutations at positions 1,762, 1,896, 1,899 and double mutations 1,653 + 1,762, 1,764 + 1,766 and 1,896 + 1,899, and with X mutations at position 130. HBV-D was positively associated with the presence of a mutation at position 180 of the pol gene. We found that HBV-E was associated with single mutations in the core gene at positions 1,653, 1,762, 1,858, 1,896, 1,899, and double mutations at positions 1,653 + 1,762, 1,653 + 1,764, and 1,764 + 1,766 (Supplementary Table 3).
We quantified strains with substitutions in different S ORFs for all three genotypes (Supplementary Table 4). Clinically important mutations which were mostly located at the major hydrophilic region (MHR) of HBsAg including sP120T, sT131K, sM133T, and sG145A were detected in all strains. Additionally, strains that harbored antiviral resistant mutations in the pol ORF showed corresponding mutations E164D/G/K in the MHR region. We found no strains with evidence of genomic insertions. In thirty-one strains, we found evidence of deletions, primarily in the HBV pre-S1 and Pre-S2 region.
Phylogenetic inference and subgenotyping
After filtering recombinants from our dataset, 118 complete genome sequences across HBV-A (n = 47), HBV-D (n = 7), and HBV-E (n = 64) were retained and combined with sequences from across the world to construct time-stamped phylogenies for each of the genotypes. For the genotypes analyzed, fifty-four belong to HBV-A and HBV-D, the only genotypes we consider for which subgenotype definitions exist. The remaining sixty-four new sequences belong to HBV-E.
Using maximum-likelihood phylogenetic inference, we inferred subgenotypes for novel HBV-A and HBV-D sequences. Among HBV-A, the identified subgenotypes were A1 (n = 29, 61.7 %), A2 (n = 1, 2.1 %), A3 (n = 9, 19.1 %), A5 (n = 2, 4.2 %), and A6 (n = 7, 12.72 %) (Fig. 1). Among HBV-D, we identified the subgenotypes of seven sequences; these belong to D1 (n = 4, 57.1 %), D2 (n = 2, 28.6 %), and D7 (n = 1, 14.3 %) (Fig. 2). While subgenotype definitions do not exist for HBV-E, we note that our sequences span the entire diversity of the genotype (Fig. 3).
Figure 1.
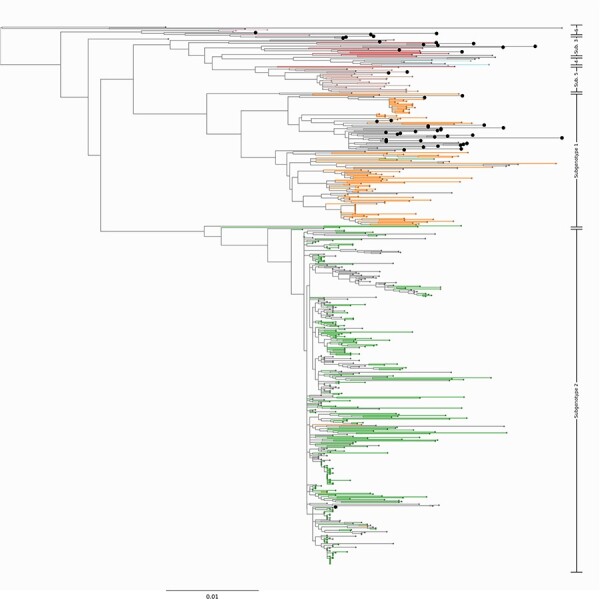
Newly sequenced HBV-A genomes in this study. Midpoint rooted phylogenetic tree representing the diversity of the novel HBV-A sequences from this study. The sequences from our study are shown as enlarged tips on the phylogeny. Branches leading to taxa with a known subgenotype are colored by their subgenotype. Out of a total of forty-seven new genomes, we introduce twenty-nine genomes that cluster most closely with subgenotype 1. Additionally, we introduce one genome that falls within subgenotype 2, nine new genomes that lie within the diversity of subgenotype 3, two genomes that cluster with subgenotype 5, and six genomes that cluster most closely with subgenotype 6. Branches leading to ancient genomes have been pruned to preserve scale.
Figure 2.
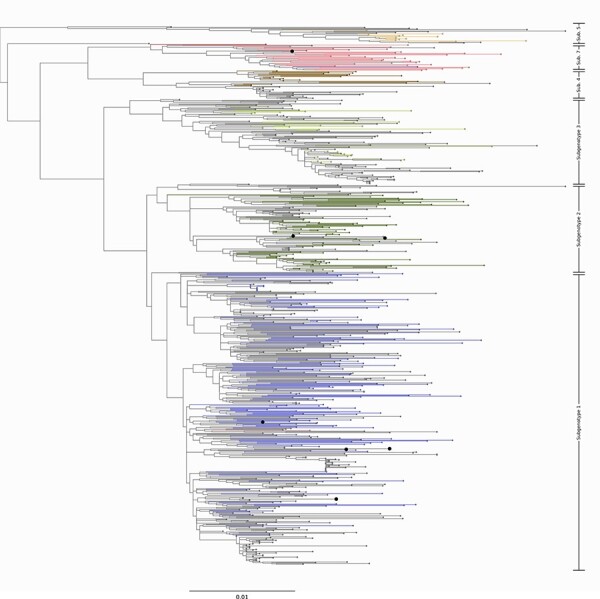
Newly sequenced HBV-D genomes in this study. Midpoint rooted phylogenetic tree representing the diversity of the novel HBV-D sequences from this study. The sequences from our study are shown as enlarged tips on the phylogeny. Branches leading to taxa with a known subgenotype are colored by their subgenotype. Here, we introduce four novel genomes that fall within the known diversity of subgenotype 1, two genomes that fall within subgenotype 2, and one genome in subgenotype 7. Branches leading to ancient genomes have been pruned to preserve scale.
Figure 3.
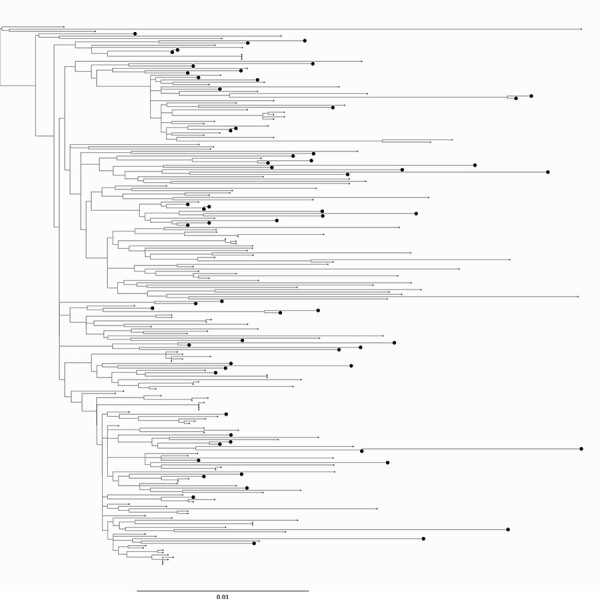
Newly sequenced HBV-E genomes in this study. Midpoint rooted phylogenetic tree representing the diversity of the novel HBV-E sequences from this study. The sequences from our study are shown as enlarged tips on the phylogeny. Novel genomes introduced here represent over a quarter (64/234) of the total number of available HBV-E sequences.
Following maximum-likelihood inference, we performed Bayesian phylogenetic inference on all three datasets. The mean evolutionary rate and time to the most recent common ancestor (tMRCA) were first estimated for all data sets without including any ancient sequences (and without any informative prior information). We estimated HBV-A to evolve at a mean rate of 5.75 × 10–4 [95 % HPD: 3.81 × 10−4–7.90 × 10–4] substitutions (subst)/site/year, HBV-D at a mean rate of 1.27 × 10–3 [95 % HPD: 1.12 × 10−3–1.43 × 10–3] subst/site/year, and HBV-E at a mean rate of 6.84 × 10–4 [95 % HPD: 2.08 × 10−4–1.16 × 10–3] subst/site/year. However, the lack of temporal signal in HBV data sets is often problematic, making it difficult to accurately estimate the time scale onto an HBV phylogeny and to hence reconstruct accurate dates for historical migrations of the virus. Mühlemann et al. (2018) have shown that including ancient genomic sequences can provide the required additional information to warrant the use of molecular clock models to reconstruct time-stamped phylogenetic trees for HBV. For HBV-A and HBV-D, we created new datasets that include all previous data supplemented by the relevant ancient sequences. For these new datasets containing ancient genomes, we set an informative prior distribution on the mean of the underlying lognormal distribution for the uncorrelated relaxed clock model, based on the reported estimate from Mühlemann et al. (2018). For HBV-A, this leads us to a somewhat increased mean rate of 7.32 × 10–4 [95 % HPD: 4.14 × 10−4–1.06 × 10–3] subst/site/year, while for HBV-D we obtained a much decreased mean rate of 9.65 × 10–5 [95 % HPD: 5.97 × 10−5–1.36 × 10–4] subst/site/year.
Discrete phylogeography
We performed discrete phylogeographic reconstruction using MCMC, conditional on the posterior tree distributions from our Bayesian phylogenetic inference with ancient genomes. Fig.4 shows the discrete phylogeographic reconstruction for HBV-A, including the relevant ancient genomic sequences from Mühlemann et al. (2018). We estimate HBV-A to have originated in East/South Asia around 2500 bce, after which it spread to Europe circa 2000 bce, and onwards to Africa between 500 and 1500 ce. Following the jump to Africa, the topology of trees generated both with and without ancient genomes is well conserved. Including the ancient sequences resulted in a more recent estimate of the first occurrence of HBV-A in Africa by about 50 years (from the 1740s to the 1790s). We observed at least five different introductions from Africa to the Americas taking place after the year 1700, consistent with the timing of the transatlantic slave trade which took place between 1520s and 1850s. Following 1960, we observed a marked increase in the number of viral transmission events between the Americas, Europe, and Western Asia. During this time period, we infer the majority of viral migration to be of European origin, which seeded introductions primarily to the Americas and East/South Asia (avg. 13 and 12 Markov jumps, respectively). We find that even with relatively few ancient genomes available, our phylogeographic analysis is more robust for their inclusion, as they contribute considerable temporal signal to our dataset (Supplementary Figure S2), which was otherwise lacking in the dataset in the absence of ancient genomes.
Figure 4.
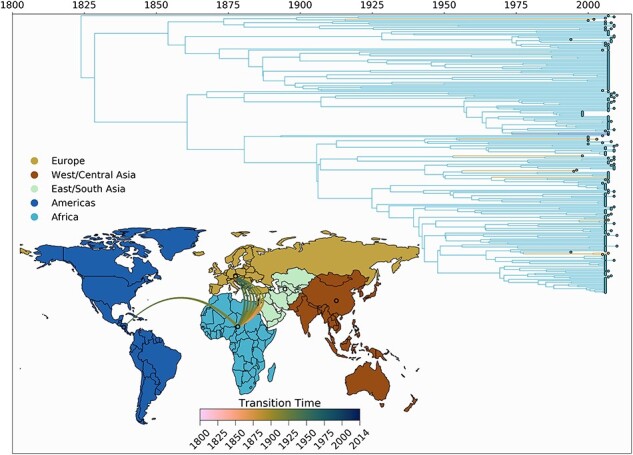
Spatio-temporal transmission dynamics of HBV-A. Time-scaled maximum clade credibility tree representing the spatio-temporal history of HBV genotype A. The time of origin is inferred to be at 2504 bce. Tips and branches are colored according to inferred location. Breaks in the time axis represent long periods of time covered by individual branches. (inset) World map colored according to the geographic regions used for the discrete phylogeographic analysis. Each line represents an inferred transmission event between two regions (55 in total). Line colors denote timing intervals of each introduction. Concave up lines represent west to east movement, concave down lines represent east to west movement, concave right lines represent north to south movement, and concave left lines represent south to north movement. Europe shows the greatest number of outgoing introductions. West/Central Asia is the inferred root location of the tree, with one ancient transmission to Europe inferred to have taken place between 2243 and 876 bce.
Using the same methods as for HBV-A, we performed discrete phylogeographic inference for HBV-D. We estimate HBV-D to have originated in East/South Asia around 1779 bce, after which it rapidly spread to West/Central Asia. We observe much more of HBV-D’s evolutionary history taking place in East/South Asia and West/Central Asia, with the former spawning introductions into Africa, the Americas, and Europe. Most of the introductions into the Americas occur from Europe however, with Europe in turn being mostly seeded from East/South Asia (Fig. 5). For HBV-D, we observe a large impact of including the ancient sequences into our data set. While we have shown that this leads to a large decrease in evolutionary rate, Fig. 5 shows much older divergence times being estimated compared to a data set with only modern-day sequences (data not shown). The tMRCA of the largest predominantly African lineage is estimated to be around 550 ce, with the oldest tMRCA of a mixed European/American clade estimated to be around 1100 ce. We infer the MRCA of this clade to have existed within the Americas; however, this timing does not correspond to any know human movement patterns, so we believe this inference to be an artifact of either heterogeneous sampling or the weak temporal signal of HBV-D. As with genotype A, individual clades show a strong local geographic structuring, with few individual location changes occurring within the most recent 50 years.
Figure 5.
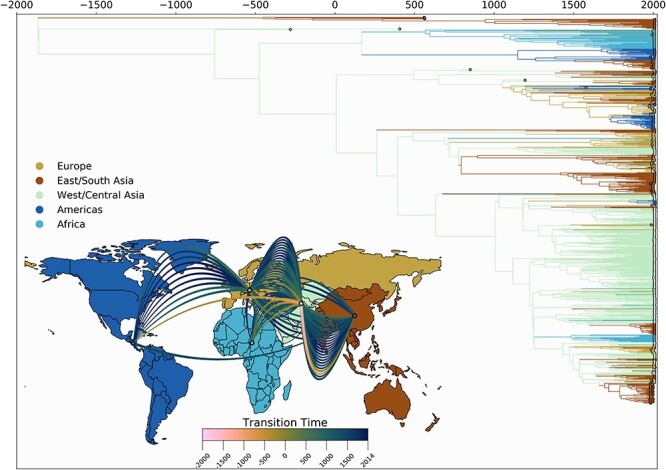
Spatio-temporal transmission dynamics of HBV-D. Time-scaled maximum clade credibility tree representing the evolutionary history of HBV genotype D. The time of origin is inferred to be at 1779 bce. Tips and branches are colored according to inferred locations. (inset) World map colored according to the geographic regions used for the discrete phylogeographic analysis. Each curved line represents an inferred transmission event between two regions (ninety-two in total). Line colors denote timing intervals of each introduction. Concave up lines represent west to east movement, concave down lines represent east to west movement, concave right lines represent north to south movement, and concave left lines represent south to north movement. West/Central Asia shows the greatest number of outgoing transmission events, as well as being the most likely location of the most recent common ancestor to sampled modern sequences.
We also performed similar phylogeographic reconstruction for HBV-E, however without any ancient genomes, as none were publicly available (Fig. 6). We estimated the tMRCA of HBV-E to 1825 ce, with the introductions into Europe and the Americas estimated to have come directly from the African continent. Given that nearly all European sequences in the tree appear as singletons, dating these introductions would be accompanied with much uncertainty.
Figure 6.
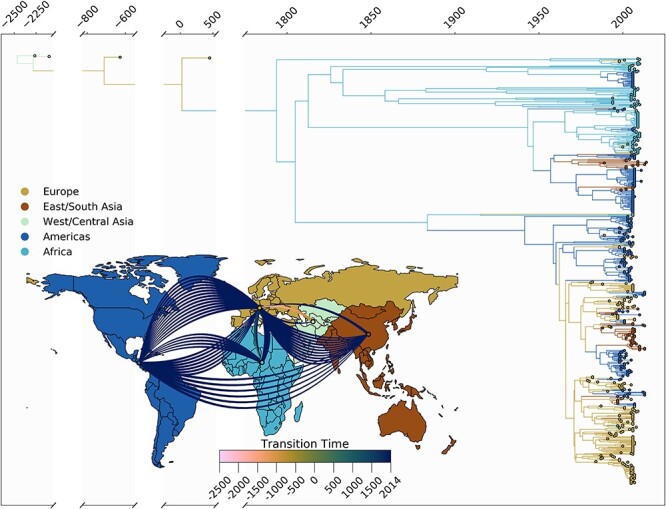
Spatio-temporal transmission dynamics of HBV-E. Time-scaled maximum clade credibility tree representing the evolutionary history of HBV genotype E. The time of origin is inferred to be at 1824 ce. Tip and branches are colored according to inferred location. (inset) World map colored according to the geographic regions used for the discrete phylogeographic analysis. Each counterclockwise arrow represents an inferred transmission event between two regions. Arrow colors denote timing intervals of each introduction. All migration events (eight in total) are inferred to have originated in Africa; Europe was the destination of seven migrations, and the Americas were the destination of one.
We summarized the strongest supported rates of discrete location transitions between pairs of regions for each dataset (Fig. 7). For HBV-A, we find very strong BF support (Kass and Raftery 1995) for movements from Europe to East/South Asia and the Americas, as well as strong support for movements from the Americas and Africa to Europe. For HBV-D, we observe very strong support for outgoing movements from West/Central Asia to Europe, Africa, and East/South Asia. We also find very strong support for outgoing movements from Europe to East/South Asia, West/Central Asia, and the Americas. Finally, for HBV-E, we only find very strong support for movements from Africa into Europe and strong support for movements from Africa to the Americas. For each genotype, we estimated Markov jump counts to quantify total geographic transitions. We estimated 62 [95 % HPD: 51–72] transitions in the evolutionary history of HBV-A, 92 [95 % HPD: 85–100] transitions in the evolutionary history of HBV-D, and 8 [95 % HPD: 8–10] transitions in the evolutionary history of HBV-E.
Figure 7.
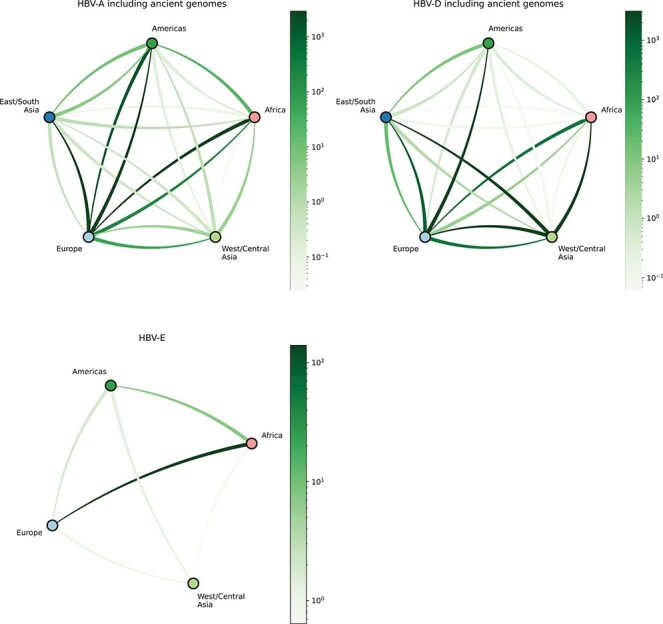
Bayes factor support for HBV’s geographic transitions. Networks representing posterior support for migrations between geographic regions as determined by BSSVS analysis. Inferred migrations are represented as curved lines. Bayes factors are represented by color intensity; darker lines depict higher Bayes factors, therefore higher posterior support. Direction of movement is represented by anticlockwise curvature of each line. In HBV-A, we estimate strongly supported migrations out of Europe to all other regions, as well as high support for inferred migrations into Europe from Africa and the Americas. In HBV-D, we observe well-supported migration history from Africa to Europe, and from Europe to the Americas and Asia. Finally, in HBV-E, we observe very strong support for migration from Africa to Europe, as well as relatively high support for migration from Africa to the Americas.
Discussion and conclusion
Human mobility exerts an immense pressure on the infectious disease profile and public health landscape of countries who receive significant numbers of immigrants (Thijssen et al. 2019). The prevalence and diversity of HBV are disproportionately affected by immigration when compared with other bloodborne viruses such as HIV and HCV. A recent large study showed that the frequency of HBV infection in immigrants in parts of Europe is three times greater than for HIV and four times greater than for HCV (González et al. 2020). Specific virological characteristics of HBV such as a long window of infectiousness, possibilities of occult infection promoted by mutations (Raimondo et al. 2019; Pronier et al. 2020; Olusola et al. 2021), and undetectable levels of genome amplification without antigen production have raised concerns about cryptic movement of specific HBV genotypes from countries of high endemicity to those of low endemicity (Zhu et al. 2016; Azarkar et al. 2019; Bedi et al. 2021). Systemic inequities in access to health services faced by immigrants in higher income countries (Smith 2018) compound with these virological characteristics to impair the implementation of effective public health responses to HBV.
Despite the significant public health burden of HBV worldwide, there is limited insight into the evolutionary history of its different genotypes. To the best of our knowledge, we present the most comprehensive investigation to date on the origins of HBV genotypes A, D, and E, owing to the large number of full-length African HBV genomes and broad spatiotemporal distribution of sequences we consider. In this study, we focused on HBV genotypes obtained from African immigrants in Belgium upon their arrival from 2005 to 2010, along with a broad selection of other publicly available sequences, both contemporary and ancient. Almost 16 % of HBV carriers from our study cohort showed co-infection with two other bloodborne viruses (HIV and HCV), which makes the design of antiviral therapies more complicated in destination countries (Torimiro et al. 2018; Arora et al. 2021). Three distinct genotypes were detected in immigrants from fifteen African countries: HBV-E (50.4 %), HBV-A (41.4 %), and HBV-D (8.6 %). This finding agrees with previous reports of HBV genotype diversity in sub-Saharan countries (Andernach et al., 2009; Forbi et al. 2013). The diversity of imported genotypes underscores the potential for changes in the epidemiological landscape of HBV in recipient countries (Mina et al. 2017; Aguilera et al. 2020). The HBV-A strains that we generated belong to the subgenotypes A1, A3, and quasi-subgenotypes A4, A5, and A7 that have already been isolated from African HBV carriers living in different parts of the world (Andernach et al., 2009; Kurbanov et al. 2005; Olinger et al. 2006; Pourkarim et al. 2010b). The recent discovery of novel subgenotype A8 from our study cohort (Thijssen et al. 2020) highlights the lack of comprehensive information about HBV in African immigrants in Belgium.
There is a well-documented association between HBV genotype and clinical outcome; this relation is further documented for individual HBV ORF mutations (Liu et al. 2009; Mello de et al. 2014; Mina et al. 2015a, 2017; Hou 2019). These associations between genotype-specific mutations in circulating African strains and clinical severity have been demonstrated, for instance with particular medically important mutations being mostly detected in the core gene of HBV strains isolated from patients with end stage liver diseases such as hepatocellular carcinoma (HCC) (Amougou et al. 2019; Mak and Kramvis 2020; Mbamalu et al. 2021). The association between A1762T, G1764A, and A1762T/G1764A, G1896A, and an increased risk of HCC has been previously shown (Wei et al. 2017). High frequency of pre-core and core mutations detected in HBV-E and HBV-A in our study highlights the importance of investigating individual origins for HBV cases, so that clinical interventions can be targeted appropriately. Importantly, we identified clinically and para-clinically related mutations at ORF S, as previously described (Mak and Kramvis 2020; Lin et al. 2021; Olusola et al. 2021).
Excepting their most ancient origins, our results agree with the work of others, with HBV-A demonstrating an African origin (Pourkarim et al. 2010a, 2010b, 2011; Toyé et al. 2021). Our results underscore the African continent as the evolutionary cradle of HBV-A diversification (Andernach et al., 2009; Toyé et al. 2021), and we date the proliferation and worldwide dispersal of this genotype to coincide with the transatlantic slave trade (1520s–1850s). Similar to HBV-A, we find the most ancient origins of HBV-D to be in Central Asia; however, we also infer the majority of its evolutionary history to remain in the region, with much later introductions into other global regions (Pourkarim et al. 2014). For both HBV-A and HBV-D, the location that we infer for the MRCA of the genotype corresponds to the location of the most ancient genomes that we included in the study. As more ancient HBV genomes from different locations and times become available, we propose incorporating them into a study such as ours to reduce the uncertainty of MRCA location and time inference.
Almost half of the HBV strains that we generated in this study were classified as HBV-E (Supplementary Table 2). This high proportion is supported by other studies from West African countries (Andernach et al., 2009; Assih et al. 2018). Because of low diversity along its entire length of genome, HBV-E does not classify into subgenotypes (Forbi et al. 2010) which may point to HBV-E having evolved more recently than other genotypes (Ingasia et al. 2020). In this study dataset, three strains with recombination signature were detected (Supplementary Figure S1) as previously described (Forbi et al. 2010; Pourkarim et al. 2010a; Mina et al. 2015a; Liu et al. 2020). Recombination can happen in over-populated locales with high prevalence of HBV, mostly occurring in East Asia and Africa (Pourkarim et al. 2014; Liu et al. 2020).
We observed that HBV-D was the fastest evolving among the genotypes we encountered. HBV-E is the most recently diverged genotype; however, both its divergence and evolutionary rate estimates have large 95 % HPD intervals, indicating that these estimates must be cautiously interpreted. A plausible explanation for why we observe such wide confidence interval is that our HBV-E dataset is considerably smaller than his (234 sequences), compared to the dataset sizes for the other genotypes (583 HBV-A and 764 HBV-D sequences). The combination of fewer sequences and no available ancient genomes may combine to provide more uncertainty in the evolutionary rate estimate. As future ancient HBV-E genomes are sequenced, their addition to studies like this may add certainty to future analyses that may help validate our inferred tMRCA estimate.
There is recent evidence that HBV has limited temporal signal warranting temporal and evolutionary estimation not reliable, particularly for very long timeframes, with the risk of underestimating when the MRCA of each genotype diverged from the ancestral HBV lineage (Ross et al. 2018). To avoid the inherent biases of estimating these measures for viruses that have an ancient time of divergence, we opted for analyzing each genotype separately. Additionally, we include ancient HBV genomes when possible to provide increased temporal signal in our dataset, thus making the estimation of evolutionary rate and tMRCA more reliable. A recent study by Kocher et al. (Kocher et al. 2021) performed similar temporal phylogenetic analysis of HBV and found MRCA estimates for each genotype different from ours by approximately a factor of two. This discrepancy may be explained by their use of an epoch-based time-dependent rate model, while we use a relaxed clock model for each genotype that we consider. Epoch-based time-dependent rate models require sufficient data to calibrate the time-dependent rate phenomenon. As we include only four HBV-A and five HBV-D ancient genomes in our genotype-split analysis, we find a relaxed clock model to be a more appropriate model for our datasets. Additionally, differences may arise as a result of our analysis of each HBV genotype separately, whereas Kocher et al. consider the entire unified HBV phylogeny. We find different mean evolutionary rates for each genotype, suggesting that a single estimate for all genotypes may not fully capture the different evolutionary dynamics affecting each, not even when using a time-dependent rate model.
Rather than employing each individual country as a different location for our discrete phylogeographic reconstruction, we have opted to perform these analyses on the regional level in order to deal with sampling bias among the different countries in our data sets. Our choice of continental groups for discrete phylogeographic reconstruction serves two purposes simultaneously. First, by reducing from fifty-four countries present between our three datasets to five continental regions we are able to significantly reduce the otherwise computationally infeasible reconstruction to one in which we are able to accumulate sufficient sampling from the posterior distribution to make significant inferences—an end that would not be possible with such a large and sparse sampling of countries. Secondly, because our phylogenetic reconstructions of the HBV genotypes range from hundreds to thousands of years, we find modern political designations to be unsatisfactory designations to describe historical patterns of viral evolution and spread; by keying our analysis to geographic regions, we find that our results have increased interpretability in a historical context. Unfortunately, to maintain consistent geographic subdivisions across datasets there remains significant heterogeneity in the number of sequences between regions for some analyses. Phylogeographic inference using structured coalescent models (Maio et al. 2015; Müller, Rasmussen, and Stadler 2018) offers a potential solution to deal with sampling bias, at the expense of much increased computational demands; however, we have instead opted to use continent-level analysis here to reduce computational complexity (Hong et al. 2020).
This study highlights the importance of genotype-aware treatment of HBV interventions, particularly in the case of individuals who have recently migrated from regions where HBV diversity is high. Because HBV genotype and specific mutational profile can have a significant impact on clinical outcomes, we find it imperative that these characteristics be taken into account so that individuals receive appropriate clinical and public health support for their infecting HBV genotype, and so that local diversity of HBV is well characterized so that policymakers and public health institutions can create the most effective intervention strategies possible. We believe this effort can be significantly assisted by both genetic analyses for individual-level interventions and by large-scale phylogenetic and phylogeographic analyses to inform population-level interventions that can lessen the overall burden of hepatitis B.
Supplementary Material
Acknowledgements
BIP and GB acknowledge support from the Internal Funds KU Leuven (Grant No. C14/18/094). GB acknowledges support from the Research Foundation—Flanders (‘Fonds voor Wetenschappelijk Onderzoek—Vlaanderen’, G0E1420N, G098321N). PL acknowledges support from the Research Foundation—Flanders (‘Fonds voor Wetenschappelijk Onderzoek—Vlaanderen’, G0D5117N, G0B9317N, G051322N). The opinions expressed in this article are those of the authors and do not reflect the view of the National Institutes of Health, the Department of Health and Human Services, or the United States government.
Contributor Information
Barney I Potter, Department of Microbiology, Immunology and Transplantation, KU Leuven, Rega Institute, Laboratory for Clinical and Epidemiological Virology, Herestraat 49, Leuven BE-3000, Belgium.
Marijn Thijssen, Department of Microbiology, Immunology and Transplantation, KU Leuven, Rega Institute, Laboratory for Clinical and Epidemiological Virology, Herestraat 49, Leuven BE-3000, Belgium.
Nídia Sequeira Trovão, Division of International Epidemiology and Population Studies, Fogarty International Center, National Institutes of Health, Bethesda, MD 20892, United States.
Andrea Pineda-Peña, Global Health and Tropical Medicine, GHTM, Instituto de Higiene e Medicina Tropical, IHMT; Universidade Nova de Lisboa, UNL, Portugal Rua da Junqueira No 100, Lisbon 1349-008, Portugal; Molecular Biology and Immunology Department, Fundacion Instituto de Inmunología de Colombia (FIDIC); Faculty of Animal Science, Universidad de Ciencias Aplicadas y Ambientales (U.D.C.A.), Avenida 50 No. 26-20, Bogota 0609, Colombia.
Marijke Reynders, Department of Laboratory Medicine, Medical Microbiology, AZ Sint-Jan Brugge-Oostende AV, Ruddershove 10, Bruges B-8000, Belgium.
Thomas Mina, Nonis Lab Microbiology—Virology Unit, Gregori Afxentiou 5, Limassol 4003, Cyprus.
Carolina Alvarez, Department of Microbiology, Immunology and Transplantation, KU Leuven, Rega Institute, Laboratory for Clinical and Epidemiological Virology, Herestraat 49, Leuven BE-3000, Belgium.
Samad Amini-Bavil-Olyaee, Cellular Sciences Department, Process Virology, Amgen Inc., One Amgen Center Drive, Thousand Oaks, CA 91320, USA.
Frederik Nevens, Department of Gastroenterology and Hepatology, University Hospital Leuven, KU Leuven, Herestraat 49, Leuven 3000, Belgium.
Piet Maes, Department of Microbiology, Immunology and Transplantation, KU Leuven, Rega Institute, Laboratory for Clinical and Epidemiological Virology, Herestraat 49, Leuven BE-3000, Belgium.
Philippe Lemey, Department of Microbiology, Immunology and Transplantation, KU Leuven, Rega Institute, Laboratory for Clinical and Epidemiological Virology, Herestraat 49, Leuven BE-3000, Belgium.
Marc Van Ranst, Department of Microbiology, Immunology and Transplantation, KU Leuven, Rega Institute, Laboratory for Clinical and Epidemiological Virology, Herestraat 49, Leuven BE-3000, Belgium.
Guy Baele, Department of Microbiology, Immunology and Transplantation, KU Leuven, Rega Institute, Laboratory for Clinical and Epidemiological Virology, Herestraat 49, Leuven BE-3000, Belgium.
Mahmoud Reza Pourkarim, Department of Microbiology, Immunology and Transplantation, KU Leuven, Rega Institute, Laboratory for Clinical and Epidemiological Virology, Herestraat 49, Leuven BE-3000, Belgium; Health Policy Research Centre, Institute of Health, Shiraz University of Medical Sciences, Shiraz 71348-14336, Iran; Blood Transfusion Research Centre, High Institute for Research and Education in Transfusion, Hemmat Exp.Way, Tehran 14665-1157, Iran.
Supplementary data
Supplementary data is available at VEVOLU Journal online.
Conflict of interest:
None declared.
References
- Aguilera A. et al. (2020) ‘GEHEP 010 Study: Prevalence and Distribution of Hepatitis B Virus Genotypes in Spain (2000–2016)’, Journal of Infection, 81: 600–6. [DOI] [PubMed] [Google Scholar]
- Al-Qahtani A. A. et al. (2020) ‘Molecular Epidemiology, Phylogenetic Analysis and Genotype Distribution of Hepatitis B Virus in Saudi Arabia: Predominance of Genotype D1’, Infection, Genetics and Evolution : Journal of Molecular Epidemiology and Evolutionary Genetics in Infectious Diseases, 77: 104051. [DOI] [PubMed] [Google Scholar]
- Amougou M. A. et al. (2019) ‘Enrichment in Selected Genotypes, Basal Core and Precore Mutations of Hepatitis B Virus in Patients with Hepatocellular Carcinoma in Cameroon’, Journal of Viral Hepatitis, 26: 1086–93. [DOI] [PubMed] [Google Scholar]
- Andernach I. E. et al. (2009) ‘Slave Trade and Hepatitis B Virus Genotypes and Subgenotypes in Haiti and Africa’, Emerging Infectious Diseases, 15: 1222–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora U. et al. (2021) ‘Complexities in the Treatment of Coinfection with HIV, Hepatitis B, Hepatitis C, and Tuberculosis’, The Lancet Infectious Diseases, 21: e399–406. [DOI] [PubMed] [Google Scholar]
- Assih M. et al. (2018) ‘Genetic Diversity of Hepatitis Viruses in West-African Countries from 1996 to 2018’, World Journal of Hepatology, 10: 807–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azarkar Z. et al. (2019) ‘Epidemiology, Risk Factors, and Molecular Characterization of Occult Hepatitis B Infection among Anti-hepatitis B Core Antigen Alone Subjects’, Journal of Medical Virology, 91: 615–22. [DOI] [PubMed] [Google Scholar]
- Bedi H. K. et al. (2021) ‘Occult Hepatitis B Reactivation after Liver Transplant: The Role of a Novel Mutation in the Surface Antigen’, Journal of Clinical and Translational Hepatology, 9: 136–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielejec F. et al. (2016) ‘SpreaD3: Interactive Visualization of Spatiotemporal History and Trait Evolutionary Processes’, Molecular Biology and Evolution, 33: 2167–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottecchia M. et al. (2011) ‘Detection of Hepatitis B Virus Genotype A3 and Primary Drug Resistance Mutations in African Immigrants with Chronic Hepatitis B in Spain’, Journal of Antimicrobial Chemotherapy, 66: 641–4. [DOI] [PubMed] [Google Scholar]
- Chu J. J. et al. (2013) ‘Changing Epidemiology of Hepatitis B and Migration—A Comparison of Six Northern and North-Western European Countries’, The European Journal of Public Health, 23: 642–7. [DOI] [PubMed] [Google Scholar]
- Coppola N. et al. (2017) ‘Hepatitis B Virus Infection in Undocumented Immigrants and Refugees in Southern Italy: Demographic, Virological, and Clinical Features’, Infectious Diseases of Poverty, 6: 58–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croagh C. M., Desmond P. V., Bell S. J. (2015) ‘Genotypes and Viral Variants in Chronic Hepatitis B: A Review of Epidemiology and Clinical Relevance’, World Journal of Hepatology, 7: 289–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A. J. et al. (2002) ‘Estimating Mutation Parameters, Population History and Genealogy Simultaneously from Temporally Spaced Sequence Data’, Genetics, 161: 1307–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbi J. C. et al. (2010) ‘Epidemic History and Evolutionary Dynamics of Hepatitis B Virus Infection in Two Remote Communities in Rural Nigeria’, PLOS ONE, 5: e11615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbi J. C. et al. (2013) ‘Disparate Distribution of Hepatitis B Virus Genotypes in Four sub-Saharan African Countries’, Journal of Clinical Virology, 58: 59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González R. et al. (2020) ‘Overt and Occult Hepatitis B among Immigrants and Native Blood Donors in Madrid, Spain’, Therapeutic Advances in Infectious Disease, 7: 2049936120982122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M., Kishino H., Yano T. (1985) ‘Dating of the Human-ape Splitting by a Molecular Clock of Mitochondrial DNA’, Journal of Molecular Evolution, 22: 160–74. [DOI] [PubMed] [Google Scholar]
- Hoang D. T. et al. (2018) ‘UFBoot2: Improving the Ultrafast Bootstrap Approximation’, Molecular Biology and Evolution, 35: 518–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S. L. et al. (2020) ‘In Search of Covariates of HIV-1 Subtype B Spread in the United States—A Cautionary Tale of Large-Scale Bayesian Phylogeography’, Viruses, 12: 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou W. (2019) ‘HCC-Associated Viral Mutations in Patients with HBV Genotype F1b Infection’, Clinical Laboratory, 65. [DOI] [PubMed] [Google Scholar]
- Ingasia L. A. O. et al. (2020) ‘Global and Regional Dispersal Patterns of Hepatitis B Virus Genotype E from and in Africa: A Full-genome Molecular Analysis’, PLOS ONE, 15: e0240375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass R. E., Raftery A. E. (1995) ‘Bayes Factors’, Journal of the American Statistical Association, 90: 773–95. [Google Scholar]
- Katoh K., Standley D. M. (2013) ‘MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability’, Molecular Biology and Evolution, 30: 772–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay A., Zoulim F. (2007) ‘Hepatitis B Virus Genetic Variability and Evolution’, Virus Research, 127: 164–76. [DOI] [PubMed] [Google Scholar]
- Khan A. et al. (2008) ‘Transmission of Hepatitis B Virus (HBV) Genotypes among Japanese Immigrants and Natives in Bolivia’, Virus Research, 132: 174–80. [DOI] [PubMed] [Google Scholar]
- Kingman J. F. C. (1982) ‘The Coalescent’, Stochastic Processes and Their Applications, 13: 235–48. [Google Scholar]
- Kocher A. et al. (2021) ‘Ten Millennia of Hepatitis B Virus Evolution’, Science. 374: 182–188. [DOI] [PubMed] [Google Scholar]
- Kurbanov F. et al. (2005) ‘A New Subtype (Subgenotype) Ac (A3) of Hepatitis B Virus and Recombination between Genotypes A and E in Cameroon’, Journal of General Virology, 86: 2047–56. [DOI] [PubMed] [Google Scholar]
- Lampertico P., Maini M., Papatheodoridis G. (2015) ‘Optimal Management of Hepatitis B Virus Infection – EASL Special Conference’, Journal of Hepatology, 63: 1238–53. [DOI] [PubMed] [Google Scholar]
- Lemey P. et al. (2009) ‘Bayesian Phylogeography Finds Its Roots’, PLoS Computational Biology, 5: e1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limeres M. J. et al. (2019) ‘Impact of Hepatitis B Virus Genotype F on in Vitro Diagnosis: Detection Efficiency of HBsAg from Amerindian Subgenotypes F1b and F4’, Archives of Virology, 164: 2297–307. [DOI] [PubMed] [Google Scholar]
- Lin Y.-T. et al. (2021) ‘Hepatitis B Virus Pre-S Gene Deletions and Pre-S Deleted Proteins: Clinical and Molecular Implications in Hepatocellular Carcinoma’, Viruses, 13(5): 862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. et al. (2009) ‘Associations between Hepatitis B Virus Mutations and the Risk of Hepatocellular Carcinoma: A Meta-Analysis’, JNCI Journal of the National Cancer Institute, 101: 1066–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. et al. (2020) ‘Complete Genome Analysis of Hepatitis B Virus in Qinghai-Tibet Plateau: The Geographical Distribution, Genetic Diversity, and Co-existence of HBsAg and anti-HBs Antibodies’, Virology Journal, 17: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lole K. S. et al. (1999) ‘Full-Length Human Immunodeficiency Virus Type 1 Genomes from Subtype C-Infected Seroconverters in India, with Evidence of Intersubtype Recombination’, Journal of Virology, 73: 152–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y. et al. (2020) ‘Virus-like Particle Vaccine with B-cell Epitope from Porcine Epidemic Diarrhea Virus (PEDV) Incorporated into Hepatitis B Virus Core Capsid Provides Clinical Alleviation against PEDV in Neonatal Piglets through Lactogenic Immunity’, Vaccine, 38: 5212–8. [DOI] [PubMed] [Google Scholar]
- Maio N. D. et al. (2015) ‘New Routes to Phylogeography: A Bayesian Structured Coalescent Approximation’, PLOS Genetics, 11: e1005421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak D., Kramvis A. (2020) ‘Molecular Characterization of Hepatitis B Virus Isolated from Black South African Cancer Patients, with and without Hepatocellular Carcinoma’, Archives of Virology, 165: 1815–25. [DOI] [PubMed] [Google Scholar]
- Malik G. F. et al. (2022) ‘Viral Hepatitis - the Road Traveled and the Journey Remaining’, Hepatic Medicine: Evidence and Research, 14: 13–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbamalu C. et al. (2021) ‘Hepatitis B Virus Precore/core Region Mutations and Genotypes among Hepatitis B Virus Chronic Carriers in South-Eastern, Nigeria’, International Journal of Health Science, 15: 26–38. [PMC free article] [PubMed] [Google Scholar]
- McMahon B. J. (2009) ‘The Influence of Hepatitis B Virus Genotype and Subgenotype on the Natural History of Chronic Hepatitis B’, Hepatology International, 3: 334–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello de F. M. M. A. et al. (2014) ‘Hepatitis B Virus Genotypes and Mutations in the Basal Core Promoter and Pre-core/core in Chronically Infected Patients in Southern Brazil: A Cross-sectional Study of HBV Genotypes and Mutations in Chronic Carriers’, Revista da Sociedade Brasileira de Medicina Tropical, 47: 701–8. [DOI] [PubMed] [Google Scholar]
- Mina T. et al. (2015a) ‘Genomic Diversity of Hepatitis B Virus Infection Associated with Fulminant Hepatitis B Development’, Hepatitis Monthly, 15: e29477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mina T. et al. (2015b) ‘A Rare Case of HBV Genotype Fluctuation (Shifting and Reversion) after Liver Transplantation’, Journal of Clinical Virology, 71: 93–7. [DOI] [PubMed] [Google Scholar]
- Mina T. et al. (2017) ‘15 Year Fulminant Hepatitis B Follow-up in Belgium: Viral Evolution and Signature of Demographic Change’, Infection Genetics & Evolution, 49: 221–5. [DOI] [PubMed] [Google Scholar]
- Mühlemann B. et al. (2018) ‘Ancient Hepatitis B Viruses from the Bronze Age to the Medieval Period’, Nature, 557: 418–23. [DOI] [PubMed] [Google Scholar]
- Müller N. F., Rasmussen D., Stadler T. (2018) ‘MASCOT: Parameter and State Inference under the Marginal Structured Coalescent Approximation’, Bioinformatics, 34: 3843–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L.-T. et al. (2015) ‘IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies’, Molecular Biology and Evolution, 32: 268–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norder H. et al. (2004) ‘Genetic Diversity of Hepatitis B Virus Strains Derived Worldwide: Genotypes, Subgenotypes, and HBsAg Subtypes’, Intervirology, 47: 289–309. [DOI] [PubMed] [Google Scholar]
- Olinger C. M. et al. (2006) ‘Phylogenetic Analysis of the Precore/core Gene of Hepatitis B Virus Genotypes E and A in West Africa: New Subtypes, Mixed Infections and Recombinations’, Journal of General Virology, 87: 1163–73. [DOI] [PubMed] [Google Scholar]
- Olusola B. A. et al. (2021) ‘Profiles of Mutations in Hepatitis B Virus Surface and Polymerase Genes Isolated from Treatment-naïve Nigerians Infected with Genotype E’, Journal of Medical Microbiology, 70: 001338. [DOI] [PubMed] [Google Scholar]
- Pagel M., Meade A., Barker D. (2004) ‘Bayesian Estimation of Ancestral Character States on Phylogenies’, Systematic Biology, 53: 673–84. [DOI] [PubMed] [Google Scholar]
- Pineda-Peña A.-C. et al. (2015) ‘Epidemiological History and Genomic Characterization of non-D1 HBV Strains Identified in Iran’, Journal of Clinical Virology, 63: 38–41. [DOI] [PubMed] [Google Scholar]
- Pourkarim M. R. et al. (2009) ‘Phylogenetic Analysis of Hepatitis B Virus Full-length Genomes Reveals Evidence for a Large Nosocomial Outbreak in Belgium’, Journal of Clinical Virology, 46: 61–8. [DOI] [PubMed] [Google Scholar]
- Pourkarim M. R. et al. (2010a) ‘Are Hepatitis B Virus “Subgenotypes” Defined Accurately?’, Journal of Clinical Virology, 47: 356–60. [DOI] [PubMed] [Google Scholar]
- Pourkarim M. R. et al. (2010b) ‘Novel Hepatitis B Virus Subgenotype A6 in African-Belgian Patients’, Journal of Clinical Virology, 47: 93–6. [DOI] [PubMed] [Google Scholar]
- Pourkarim M. R. et al. (2011) ‘Molecular Characterization of Hepatitis B Virus Strains Circulating in Belgian Patients Co-infected with HIV and HBV: Overt and Occult Infection’, Journal of Medical Virology, 83: 1876–84. [DOI] [PubMed] [Google Scholar]
- Pourkarim M. R. et al. (2014) ‘Molecular Characterization of Hepatitis B Virus (HBV) Strains Circulating in the Northern Coast of the Persian Gulf and Its Comparison with Worldwide Distribution of HBV Subgenotype D1’, Journal of Medical Virology, 86: 745–57. [DOI] [PubMed] [Google Scholar]
- Pourkarim M. R. et al. (2014) ‘Molecular Identification of Hepatitis B Virus Genotypes/subgenotypes: Revised Classification Hurdles and Updated Resolutions’, World Journal of Gastroenterology, 20: 7152–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourkarim M. R., Lemey P., Van Ranst M. (2018) ‘Iran’s Hepatitis Elimination Programme Is under Threat’, The Lancet, 392: 1009. [DOI] [PubMed] [Google Scholar]
- Pourkarim M. R., Van Ranst M. (2011) ‘Guidelines for the Detection of a Common Source of Hepatitis B Virus Infections’, Hepatitis Monthly, 11: 783–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pronier C. et al. (2020) ‘The Contribution of More Sensitive Hepatitis B Surface Antigen Assays to Detecting and Monitoring Hepatitis B Infection’, Journal of Clinical Virology, 129: 104507. [DOI] [PubMed] [Google Scholar]
- Pujol F. et al. (2020) ‘Hepatitis B Virus American Genotypes: Pathogenic Variants?’, Clinics and Research in Hepatology and Gastroenterology, 44: 825–35. [DOI] [PubMed] [Google Scholar]
- Raimondo G. et al. (2019) ‘Update of the Statements on Biology and Clinical Impact of Occult Hepatitis B Virus Infection’, Journal of Hepatology, 71: 397–408. [DOI] [PubMed] [Google Scholar]
- Rambaut A. et al. (2016) ‘Exploring the Temporal Structure of Heterochronous Sequences Using TempEst (Formerly Path-O-Gen)’, Virus Evolution, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A. et al. (2018) ‘Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7’, Systematic Biology, 67: 901–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revill P. A. et al. (2020) ‘The Evolution and Clinical Impact of Hepatitis B Virus Genome Diversity’, Nature Reviews Gastroenterology and Hepatology, 17: 618–34. [DOI] [PubMed] [Google Scholar]
- Ross Z. P. et al. (2018) ‘The Paradox of HBV Evolution as Revealed from a 16th Century Mummy’, PLOS Pathogens, 14: e1006750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers E. W. et al. (2022) ‘Database Resources of the National Center for Biotechnology Information’, Nucleic Acids Research, 50: D20–D26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer A. et al. (2015) ‘Estimations of Worldwide Prevalence of Chronic Hepatitis B Virus Infection: A Systematic Review of Data Published between 1965 and 2013’, The Lancet, 386: 1546–55. [DOI] [PubMed] [Google Scholar]
- Selabe S. G. et al. (2007) ‘Mutations Associated with Lamivudine-resistance in Therapy-naïve Hepatitis B Virus (HBV) Infected Patients with and without HIV Co-infection: Implications for Antiretroviral Therapy in HBV and HIV Co-infected South African Patients’, Journal of Medical Virology, 79: 1650–4. [DOI] [PubMed] [Google Scholar]
- Sharma S. et al. (2015) ‘Immigration and Viral Hepatitis’, Journal of Hepatology, 63: 515–22. [DOI] [PubMed] [Google Scholar]
- Shen T., Yan X.-M. (2014) ‘Hepatitis B Virus Genetic Mutations and Evolution in Liver Diseases’, World Journal of Gastroenterology, 20: 5435–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. (2018) ‘Migrant Health Is Public Health, and Public Health Needs to Be Political’, The Lancet Public Health, 3: e418. [DOI] [PubMed] [Google Scholar]
- Suchard M. A. et al. (2018) ‘Bayesian Phylogenetic and Phylodynamic Data Integration Using BEAST 1.10’, Virus Evolution, 4: vey016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchard M. A., Rambaut A. (2009) ‘Many-core Algorithms for Statistical Phylogenetics’, Bioinformatics, 25: 1370–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thijssen M. et al. (2019) ‘Mass Migration to Europe: An Opportunity for Elimination of Hepatitis B Virus?’, The Lancet Gastroenterology & Hepatology, 4: 315–23. [DOI] [PubMed] [Google Scholar]
- Thijssen M. et al. (2020) ‘Novel Hepatitis B Virus Subgenotype A8 and Quasi-subgenotype D12 in African–Belgian Chronic Carriers’, International Journal of Infectious Diseases, 93: 98–101. [DOI] [PubMed] [Google Scholar]
- Torimiro J. N. et al. (2018) ‘Rates of HBV, HCV, HDV and HIV Type 1 among Pregnant Women and HIV Type 1 Drug Resistance-associated Mutations in Breastfeeding Women on Antiretroviral Therapy’, BMC Pregnancy & Childbirth, 18: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyé R. M. et al. (2021) ‘Hepatitis B Virus Genotype Study in West Africa Reveals an Expanding Clade of Subgenotype A4’, Microorganisms, 9: 623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trovão N. S. et al. (2022) ‘Reconstruction of the Origin and Dispersal of the Worldwide Dominant Hepatitis B Virus Subgenotype D1’, Virus Evolution, 8: veac028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velkov S., Protzer U., Michler T. (2020) ‘Global Occurrence of Clinically Relevant Hepatitis B Virus Variants as Found by Analysis of Publicly Available Sequencing Data’, Viruses, 12: 1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei F. et al. (2017) ‘The Association between Hepatitis B Mutants and Hepatocellular Carcinoma: a meta-analysis’, Medicine (Baltimore), 96: e6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. (1994) ‘Maximum Likelihood Phylogenetic Estimation from DNA Sequences with Variable Rates over Sites: Approximate Methods’, Journal of Molecular Evolution, 39: 306–14. [DOI] [PubMed] [Google Scholar]
- Zehender G. et al. (2012) ‘Spatial and Temporal Dynamics of Hepatitis B Virus D Genotype in Europe and the Mediterranean Basin’, PLOS ONE, 7: e37198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M. et al. (2020) ‘Rapidly Decreased HBV RNA Predicts Responses of Pegylated Interferons in HBeAg-positive Patients: A Longitudinal Cohort Study’, Hepatology International, 14: 212–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H.-L. et al. (2016) ‘Genetic Variation of Occult Hepatitis B Virus Infection’, World Journal of Gastroenterology, 22: 3531–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


