Abstract
Brucella organisms are facultative intracellular bacteria that may infect many species of animals as well as humans. The smooth lipopolysaccharide (S-LPS) has been reported to be an important virulence factor of these organisms, but the genetic basis of expression of the S-LPS O antigen has not yet been described. Likewise, the role of the O side chain of S-LPS in the survival of Brucella has not been clearly defined. A mini-Tn5 transposon mutant library of Brucella melitensis 16M was screened by enzyme-linked immunosorbent assay (ELISA) with monoclonal antibodies (MAbs) directed against the O side chain of Brucella. One mutant, designated B3B2, failed to express any O side chain as confirmed by ELISA, Western blot analysis, and colony coloration with crystal violet. Nucleotide sequence analysis demonstrated that the transposon disrupted an open reading frame with significant homology to the putative perosamine synthetase genes of Vibrio cholerae O1 and Escherichia coli O157:H7. The low G+C content of this DNA region suggests that this gene may have originated from a species other than a Brucella sp. The survival of B. melitensis mutant strain B3B2 in the mouse model and in bovine macrophages was examined. The results suggested that S-LPS or, more precisely, its O side chain is essential for survival in mice but not in macrophages.
Brucella spp. are gram-negative, facultative intracellular bacteria that cause a zoonotic disease worldwide. Like other intracellular pathogens, brucellae are virulent mainly due to their ability to avoid the bactericidal phagocyte functions and to proliferate within macrophages, leading to the establishment of a chronic infection in the host.
As in other gram-negative bacteria, the lipopolysaccharide (LPS) in brucellae is one of the most biologically active and important components of the outer membrane. The smooth LPS (S-LPS) is composed of three domains: the lipid A, the core oligosaccharide, and the immunodominant portion of the molecule—the O side chain, also called the O antigen. The lipid A moiety forms the outer leaflet of the outer-membrane bilayer and is responsible for most of the biological activity of the S-LPS (40). The core of Brucella LPS contains mannose, glucose, quinovosamine, and 2-keto-3-deoxyoctulosonic acid (KDO) and corresponds to a region that links the other two parts of the molecule (12, 41). The O side chain of the Brucella S-LPS is made of a homopolymer of 4,6-dideoxy-4-formamido-α-d-mannopyranosyl units linked to α-1,2 in A-dominant smooth Brucella strains but linked with every fifth α-1,3 residue in M-dominant strains (7–9, 13). Because of its external position, the S-LPS plays an important role in many of the host-pathogen interactions and is the immunodominant antigen of Brucella. The presence of perosamine (4-amino, 4,6 dideoxymannose) in the LPS is responsible for the antigenic cross-reactivity with Escherichia hermanni, Escherichia coli O:157, Salmonella O:30, Stenotrophomonas maltophilia, Vibrio cholerae O1, and Yersinia enterocolitica O:9 LPS (43). Rough mutants, which lack the O antigen, are viable and not much reduced in growth rate in culture, although they are described as less virulent. Surprisingly, the two rough Brucella species, B. ovis and B. canis, remain fully virulent in their primary host despite their phenotype (53).
To date, little is known about the mechanism of intracellular survival of brucellae. In other gram-negative bacteria, the O side chain has been shown to function as a protective barrier to hydrophobic agents (42) and complement-mediated lysis (32, 33) and is implicated in resistance to killing by the microbicidal intracellular granules of polymorphonuclear leukocytes (55).
The genes encoding the enzymes involved in O antigen biosynthesis have been identified in many bacteria (47, 51). Most of these genes, usually 10 to 20, are clustered within a locus named rfb. In spite of the importance of LPS in the Brucella life cycle, very little is known about the metabolic pathways and enzymes required to synthesize it. In the present study, we started the molecular analysis of the genes required for the synthesis of the O antigen of Brucella melitensis 16M. After a rough transposon insertion mutant was identified and characterized, the disrupted open reading frame (ORF) was cloned and sequenced. Because this rough transposon insertion mutant had a well-defined nonreverting LPS-related phenotype, it was used to investigate the role of S-LPS in Brucella infections. This mutant was first tested for survival in the mouse model, which has been shown to correlate with virulence in the primary host (25). Because macrophages might play a central role in the pathogenesis of chronic brucellosis (10, 44), we also evaluated the survival of the rough mutant in bovine macrophages.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth media.
The bacterial strains and plasmids used in this study are listed in Table 1. All Brucella strains were grown on tryptic soy agar with yeast extract (0.1%) or in 2YT medium (10% yeast extract, 10 g of tryptone, 5 g of NaCl [per liter]). E. coli strains were grown on Luria-Bertani broth (50). Cell extracts were prepared by sonication as previously described (14). The antibiotic concentrations were as follows: for ampicillin, 50 μg/ml; for kanamycin, 50 μg/ml; for nalidixic acid, 25 μg/ml; and for tetracycline, 12.5 μg/ml.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype, serotype, or description | Reference or source |
|---|---|---|
| E. coli strains | ||
| XL1-Blue | recA endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac (F′ proAB lacIq ZΔM15Tn10 [Tetr]) | Stratagene |
| S17-1 | thi pro hsdR hsdM+ recA::RP4-2− Tc::Mu-Km::Tn7 | 52 |
| Brucella strains | ||
| B. melitensis 16Ma | Wild type | ATCC 23456 |
| B. ovis Reo 198a | Rough, CO2 independent | BCCN R22 1 |
| B. melitensis 16M Nalra | Spontaneous nalidixic acid-resistant mutant | 60 |
| B. melitensis B115a | Rough mutant, O side chain production in the cytoplasm | BCCN R19 1, 16 |
| B. melitensis H38Ra | Rough mutant, no O side chain production | BCCN V3r 6, 14 |
| B. melitensis B3B2 | Rough mini-Tn5 insertion mutant | This study |
| B. melitensis DR 1 to 4 | Rough double recombinants | This study |
| Plasmids | ||
| pUTmini-Tn5Kmcat | Suicide plasmid in Brucella spp. containing mini-Tn5 Kmcat | 20 |
| pBluescript KS(−) | Phagemid cloning vector, Ampr | Stratagene |
| pBluescript SK(−) oriT | Mobilizable phagemid cloning vector, Ampr, oriT | 59 |
| pKSTn5R | pBluescript KS derivative containing a 6.5-kb SalI chromosomal DNA fragment from the B3B2 insertion mutant; this fragment contains mini-Tn5 | This study |
| pSKoritTn5R | pBluescript SK(−) oriT derivative containing a 6.5-kb SalI chromosomal DNA fragment from the B3B2 insertion mutant; this fragment contains mini-Tn5 | This study |
| pCAT19 | pUC19 containing a chloramphenicol acetyltransferase-encoding cassette | 28 |
This strain was received from J.-M. Verger (Laboratoire de Pathologie Infectieuse et d’Immunologie, Institut National de Recherche Agronomique, Nouzilly, France).
Selection of a mini-Tn5Kmcat mutant of B. melitensis 16M defective in the expression of the O side chain.
The mini-Tn5Kmcat, a derivative of transposon mini-Tn5 (21) bearing the kanamycin resistance gene and the chloramphenicol acetyltransferase reporter gene, was used to mutagenize B. melitensis 16M (20). Briefly, the transposon carried on the suicide vector pUTmini-Tn5Kmcat was introduced by mating into a nalidixic acid-resistant (Nalr) strain of B. melitensis 16M, with E. coli S17-1 as the donor strain. Nalr Kmr Amps transconjugants were selected (20). These clones were individually stored in 2YT containing 30% glycerol at −80°C in microtiter plates. Mini-Tn5 mutants (3,040) of B. melitensis 16M were individually tested by enzyme-linked immunosorbent assay (ELISA) for loss of the O antigen.
MAbs.
The monoclonal antibodies (MAbs) against S-LPS, rough LPS (R-LPS), and peptidoglycan (PG) were produced as previously described (14). Supernatants of hybridoma cultures of the anti-R-LPS MAb A68/03F03/D05 (immunoglobulin G2b [IgG2b]) (17), anti-S-LPS MAb 12G12 (IgG1) and 2E11 (IgG3) (16, 37), and anti-PG MAb 3D6 (IgG3) (15) were used. The optimal dilution of these MAbs was tested by ELISA on whole cells and on whole-cell lysates of B. melitensis 16M. Dilutions used in this study were the highest dilutions reaching an optical density (OD) of 1.
ELISA.
For the selection of rough insertion mutants, mini-Tn5Kmcat mutants of B. melitensis 16M were grown individually at 37°C in microtiter plates (model 001-67008A; Nunc, Roskilde, Denmark) containing 2YT. After heat inactivation of the brucellae by incubation for 2 h at 80°C, the plates were washed six times with 0.15 M NaCl–0.01% Tween 20 (NaCl-Tween). MAbs 12G12 and 2E11 directed against the Brucella S-LPS O side chain were diluted in phosphate-buffered saline (PBS) containing 50 mM EDTA, 0.1% Tween, and 4% casein hydrolysate (PBS-EDTA-Tween-ch). Fifty microliters of diluted MAbs was added to the plates that were then incubated for 1 h at room temperature. The plates were washed six times with NaCl-Tween. The binding of the MAbs was revealed by incubation for 1 h at room temperature with a goat anti-mouse peroxidase conjugate (Amersham, Ghent, Belgium) diluted 1/1,000 in PBS-EDTA-Tween-ch. The excess reagents were removed with NaCl-Tween in six wash cycles. Citrate-phosphate buffer (0.05 Na2HPO4, 0.025 M citric acid [pH 5]) containing 0.4% o-phenylenediamine and 2 mM H2O2 was used to visualize the peroxidase activity. The difference in OD at 490 and at 630 nm was read with a Bio-Kinetics reader model EL-340 (Biotek Instruments, Winooski, Vt.).
For phenotypic characterization of the selected mutant (B3B2), the same protocol except the antigen coating and the MAb, was used. Freshly grown Brucella cells were heat inactivated. The plates were coated overnight at 4°C with bacterial suspensions, sonicated or not, and diluted in PBS (100 μl of a bacterial suspension at an OD600 of 1 per well). MAbs directed against S-LPS, R-LPS, and PG were all diluted in PBS-EDTA-Tween-ch.
SDS-polyacrylamide gel electrophoresis and Western blotting.
Whole-cell extracts were run by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (12% polyacrylamide) (36) and transferred to nitrocellulose membranes (Millipore). The blots were saturated for 1 h with TBS (10 mM Tris-HCl [pH 7.5], 150 mM NaCl) containing 3% bovine serum albumin (BSA) and then incubated overnight with MAbs diluted in TBS containing 1% BSA, 0.01% Tween 20 (TBS-BSA-Tween). After three washes with TBS–0.05% Tween (TTBS), the blots were incubated for 1 h with biotinylated goat anti-mouse IgG (Amersham) diluted 1/1,000 in TBS-BSA-Tween. The blots were then washed three times with TTBS and incubated with streptavidin-horseradish peroxidase (Amersham) diluted 1,000 times in TBS-BSA-Tween. After two washes in TTBS and one in TBS, the signals were revealed following incubation at room temperature in TBS containing 0.06% (wt/vol) 4-chloro-1-naphthol (Bio-Rad Laboratories, Richmond, Calif.) and 5 mM H2O2.
Morphologic characterization of the rough-mutant colonies.
The crystal violet method (63) was used to stain distinct colonies of B. melitensis 16M or the rough insertion mutant. In this test, the smooth colonies take up the dye, whereas the rough colonies do not.
Routine DNA procedures.
Restriction enzymes were used according to the manufacturer’s instructions (Boehringer, Mannheim, Germany). Procedures, including agarose gel electrophoresis, were performed as described previously (50).
Hybridization probe preparation.
To generate a DNA probe specific for the chloramphenicol acetyltransferase gene (cat), an 819-bp fragment containing the cat gene obtained by restriction of the pCAT19 plasmid (28) with TaqI was purified from agarose with JET sorb (Genomed).
For screening the λGem-12 library, a DNA probe specific for the genomic DNA region downstream from the transposon in the B3B2 rough insertion mutant was generated. A fragment of 1,650 bp was PCR amplified on pKSTn5R (primers GFP1 and PRS2). This fragment was purified by Wizard PCR preps (Promega Corp., Madison, Wis.).
DNA fragments were labelled with the random primer fluorescein DNA labeling kit (Tropix, Bedford, Mass.).
Southern blot hybridization.
B. melitensis 16M strain genomic DNA was obtained from J.-M. Verger and M. Grayon (Institut National de la Recherche Agronomique, Centre de Recherche de Tours, Nouzilly, France). Southern blot hybridization on positively charged nylon membranes (Amersham) was performed with a Hybaid vacuum blotter (Biozym). Membrane prehybridization, hybridization, and washes were performed under highly stringent conditions according to the Southern-Star protocol (Tropix). Briefly, the blots were hybridized overnight with the probe at 65°C and washed at 65°C with 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–1% SDS. The hybridization results were visualized by autoradiography.
Oligonucleotides.
All primers, including reverse and forward primers, were purchased from Eurogentec (Liège, Belgium). Primers specific for the transposon were kanex (5′-GGGGCGATTCAGGCCTGGTAT-3′) and catex (5′-GCCGGCATCAGGCGGGCAAGAATGTGAAT-3′). Two primers specific for the DNA region downstream from the transposon in the insertion mutant B3B2 (PRS2, 5′-CAGAGCGCACTAAGG-3′ and ABC1, 5′-CCGCGCTGCGCCGCATA-3′) were defined on the basis of the preliminary sequences of this region (Fig. 2a).
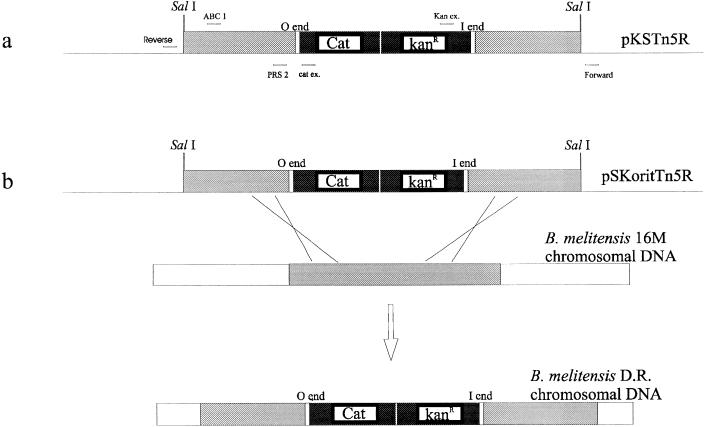
FIG. 2.
(a) pKSTn5R and primers used in this study. (b) Gene replacement strategy used to create B. melitensis 16M DR identical to the B3B2 insertion mutant. The construction of pKSTn5R and pSKoritTn5R is described in the text. Open regions, B. melitensis chromosomal DNA; light-grey regions, SalI chromosomal DNA containing the mini-Tn5 in the B3B2 mutant; solid regions, kanamycin resistance gene and cat reporter gene of the mini-Tn5.
Screening of the λGem-12 library and subcloning into pGEM plasmids.
A probe specific for the DNA region downstream from the transposon in the B3B2 insertion mutant was used to screen a B. melitensis 16M genomic DNA library constructed in the λ-Gem12 vector (Promega) (61). To screen the library, approximately 2,000 PFU was plated per 150-mm-diameter plate on E. coli KW251 (Promega). The phages were allowed to grow overnight at 37°C. After 1 h at 4°C, the plates were overlaid with a positively charged nylon membrane (Amersham) and incubated for 20 min at 37°C. The blots were air dried, denatured with 0.5 N NaOH and 1.5 M NaCl (denaturation solution) for 5 min, and equilibrated for 5 min in a neutralization solution (0.5 M Tris [pH 7.5], 1.5 M NaCl). The membranes were then baked for 1 h at 80°C. Membrane prehybridization, hybridization, and washes were performed under highly stringent conditions according to the Southern-Star protocol (Tropix) as described before.
The DNA of the recombinant positive phages was obtained as described by Sambrook et al. (50) for the rapid small-scale isolation of lambda DNA. The phage DNA was then cut with NotI, BamHI, EcoRI, or SacI. The restriction fragments of one clone containing an insert of approximately 12 kb which had been cleaved with NotI were ligated into pGEM-5Zf− (Promega). The DNA from a recombinant clone was extracted, restricted by HindIII, and analyzed by agarose gel electrophoresis.
DNA sequencing and sequence analysis.
The double-stranded DNA was prepared by the alkaline lysis method with a commercial kit (Qiagen Inc., Chatsworth, Calif.). DNA sequencing was performed by gene walking with the ABI PRISM dye terminator cycle sequencing ready reaction kit (Perkin-Elmer, Applied Biosystems Division). Reactions were analyzed with an ABI 373A DNA sequencer (Perkin-Elmer). Searches for nucleic acid and protein sequence similarities were performed at the National Center for Biotechnology Information with the BLAST network service (2, 29). Amino acid sequences were aligned with Match-box software (23, 24).
Gene replacement.
The strategy used was originally described by Halling et al. (30). In this study, the gene was replaced by its transposon-disrupted copy. The goal of this strategy was to recreate the disruption that occurred in the B3B2 insertion mutant. A fragment containing the mini-Tn5-disrupted gene was excised from pKS-Tn5R by SalI digestion and ligated into the corresponding site of the mobilizable vector pSKori (59). The construct, designated pSKoritTn5R, was transferred by conjugation into B. melitensis 16M Nalr as previously described (59). The recombinant clones were selected in the presence of kanamycin and nalidixic acid.
Survival of the rough mutant in the mouse model.
Groups of 7-week-old BALB/c mice were inoculated intraperitoneally (i.p.) with 104 CFU of B. melitensis 16M and the rough B3B2 mutant. At 1- and 4-week intervals postinoculation, six mice from each group were sacrificed for spleen collection. Bacterial survival was determined following homogenization of the mouse spleens in 5 ml of PBS with a Stomacher 80 homogenizer. Serial dilutions of the homogenized spleens were plated on tryptic soy agar with yeast extract to determine bacterial counts.
Survival of the rough mutant in macrophages.
Survival of the rough mutant was evaluated in an immortalized cell line of bovine peritoneal macrophages (54) by using a previously described procedure (22). Briefly, brucellae grown in liquid for 24 h were washed in saline and resuspended in a complete cell culture medium at 5 × 108 CFU/ml and then were added to bovine macrophages at a multiplicity of infection of 10:1. After 1 h, the monolayers were washed three times and the complete cell culture medium was added. At 1, 24, and 48 h, the monolayers were washed with the complete culture medium and lysed with 0.1% Triton X-100. The Triton lysates were then diluted serially and plated on brucella agar to determine the number of bacterial CFU per culture.
Nucleotide sequence accession number.
The nucleotide sequence presented in this study has been deposited in GenBank under accession no. AF036614.
RESULTS
Identification and characterization of a mini-Tn5Kmcat insertion mutant of B. melitensis 16M with defective O antigen expression.
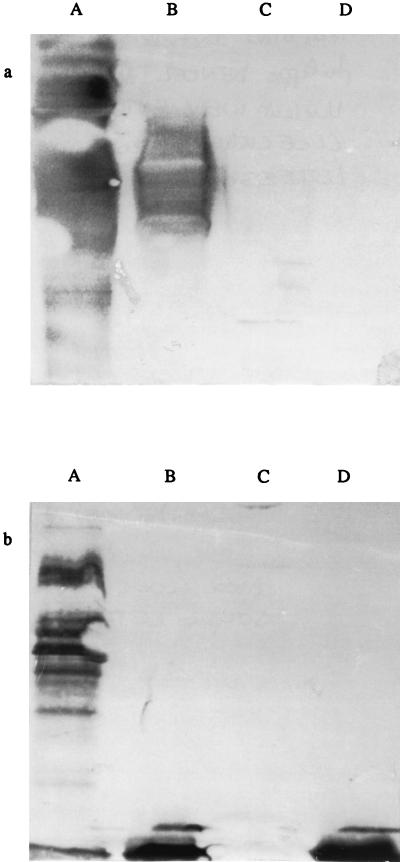
To identify regions of the B. melitensis genome containing rfb genes, we screened a library of B. melitensis 16M transposon mutants (20). A total of 3,040 insertion mutants were tested by ELISA for loss of the O antigen. For one mutant (B3B2), whole cells and whole-cell lysates failed to react, by ELISA, with MAbs directed against the S-LPS O side chain of Brucella species (S-LPS-specific MAbs) (Table 2). These results indicated that no O antigen was produced on the cell surface or in the cytoplasm of this insertion mutant. In this test, the cell wall integrity of whole cells was assessed with a MAb directed against the PG. The absence of an O side chain in the selected clone was also confirmed by Western blot analysis of whole-cell extracts with the same MAbs (Fig. 1) and by a differential colony coloration test with crystal violet. ELISA and Western blotting with MAb A68/036F03/D05 specific for Brucella R-LPS showed that this part of the molecule was still expressed in the B3B2 insertion mutant (Table 2 and Fig. 1b) and indicated that the insertion of the mini-Tn5 into the B3B2 genome had not occurred in genes involved in the lipid A or core biosynthesis.
TABLE 2.
ELISA binding of antibodies to whole cells and whole-cell lysates of three Brucella strains
| MAb | Specificity | OD490 after subtraction of blank value
|
|||||
|---|---|---|---|---|---|---|---|
| Whole cells
|
Whole-cell lysates
|
||||||
| B. melitensis 16Ma | B. melitensis B3B2b | B. ovis Reo 198c | B. melitensis 16Ma | B. melitensis B3B2b | B. ovis Reo 198c | ||
| 12G12 | S-LPS | 1.328 | 0.006 | 0.003 | 1.198 | 0.005 | 0.039 |
| 2E11 | S-LPS | 0.953 | 0.002 | 0.000 | 1.387 | 0.001 | 0.042 |
| A68/3F03/D05 | R-LPS | 0.124 | 0.212 | 0.281 | 0.443 | 1.197 | 1.117 |
| 3D6 | PG | 0.011 | 0.004 | 0.000 | 0.350 | 0.194 | 0.376 |
Parental strain.
Insertion mutant.
Rough strain.
FIG. 1.
Immunoblot analysis of Brucella whole-cell lysates probed with MAbs 12G12 and 2E11 (directed against Brucella S-LPS) (a) and MAb A68/3F03/D05 (directed against R-LPS of Brucella) (b). Lanes: A, B. melitensis 16M (parental strain); B, B. melitensis B115; C, B. melitensis H38 rough mutant; D, B. melitensis B3B2 mutant.
XbaI-digested chromosomal DNA from the B3B2 mutant was analyzed by Southern blotting with a probe specific for the cat reporter gene of the mini-Tn5. The hybridization of this probe to a single band demonstrated that a single insertion of mini-Tn5Kmcat had occurred in the genome (data not shown).
Cloning and sequencing of the DNA region adjacent to mini-Tn5Kmcat in the B3B2 mutant strain.
Genomic DNA isolated from the selected mutant was digested by SalI. The restriction products were ligated into the corresponding site of the pBluescript KS(−) plasmid. A 6.5-kb insert containing the transposon and the adjacent genomic regions was cloned. The resulting construction was designated pKSTn5R. With reverse and forward primers and two other primers specific for the transposon, sequences adjacent to mini-Tn5Kmcat were obtained (Fig. 2a). On the basis of these sequences, a fragment of the B. melitensis genomic DNA located downstream from the transposon insertion site in the B3B2 mutant was PCR amplified, fluorescein labelled, and used as a probe to screen a genomic library of B. melitensis 16M constructed in λ-GEM12. Five positive plaques were selected. These plaques were purified further, and the phage DNA was isolated. One of these clones, containing a 14-kb NotI Brucella genomic DNA fragment, was used for further analysis. This insert was subcloned into NotI-digested pGEM5, yielding pGfRI. The nucleotide sequence of the disrupted ORF in the B3B2 rough mutant was completed by gene walking on this plasmid.
Sequence analysis.
The nucleotide sequence and the deduced amino acid sequence of the transposon-disrupted gene are presented in Fig. 3. This ORF begins with an ATG codon 11 bp downstream from a potential Shine-Dalgarno ribosome binding site (AAGGA), extending to a stop codon (TAG). This ORF of 1,101 nucleotides encodes a putative polypeptide of 367 amino acids (Fig. 3). The amino acid sequence has 48 and 50% identity with the putative perosamine synthetase encoded by rfbE of V. cholerae O1 (57) and E. coli O157:H7 (5), respectively. To investigate the extent of the regions conserved between these three proteins, multiple simultaneous alignment was performed with Match-box software (23, 24). The boxes indicated very similar regions between the three sequences (Fig. 4). Because the structural sugar of the O side chain of Brucella LPS is perosamine, as in V. cholerae O1, and because perosamine is one of the components of the E. coli O157:H7 LPS O side chain, we assumed that the disrupted gene corresponds to the perosamine synthetase gene of B. melitensis 16M. The perosamine synthetase should catalyze the conversion of GDP-4-keto-6-deoxymannose to 4-NH2-4,6-dideoxymannose (perosamine) (56). We named this B. melitensis 16M gene rfbEBm16M (in accordance with the nomenclature proposed by Reeves et al. [47], this gene might also be named per). The GC content of rfbEBm16M is about 48%, which is rather low compared with the global GC content of the Brucella DNA (56 to 58%) (19).
FIG. 3.
The nucleotide sequence and the deduced amino acid sequence of the B. melitensis 16M perosamine synthetase gene. The putative ribosome binding site (RBS) is underlined. The asterisk denotes the termination codon. The arrow indicates the site of the mini-Tn5 insertion in strain B3B2.
FIG. 4.
Simultaneous multiple alignment of perosamine synthetase amino acid sequences from B. melitensis 16M (1), V. cholerae O1 (2), and E. coli O157:H7 (3). The matching regions for the three sequences are outlined by boxes. Amino acids are numbered above the sequence. Shaded boxes indicate identities. Lowercase letters indicate unaligned residues.
Gene replacement.
Because rough mutants can occur spontaneously under laboratory conditions, we used gene replacement (30) to demonstrate that the rough phenotype of B3B2 is due to the insertion of the transposon. The SalI fragment of plasmid pKS-Tn5R containing the mini-Tn5Kmcat-disrupted gene was cloned into the corresponding site of the mobilizable vector pSKori. The resulting vector (pSKoritTn5R) was transferred by conjugation into B. melitensis 16M. Because pSKoritTn5R does not replicate in Brucella species, kanamycin-resistant transformants result from the integration of the antibiotic resistance gene carried by the transposon into the Brucella chromosome via a homologous recombination. Single crossovers were distinguished from double crossovers by screening for vector-conferred ampicillin resistance (Fig. 2b). Kanr Amps transformants resulted from the replacement of the wild-type perosamine synthetase gene by the transposon-disrupted copy of the rough B3B2 mutant. Four of the 10 Kanr Amps transformants were selected for characterization and were designated double recombinants (DR) 1 to 4. As observed with the original B3B2 mutant, these recombinants failed to react with MAbs specific for S-LPS by ELISA on whole cells and by Western blot analysis of cell extracts. Colony staining with crystal violet also confirmed the rough phenotype of the four DR (data not shown). To confirm gene replacement, genomic DNA of DR 1 to 4 was isolated, digested by XbaI, and analyzed by Southern blotting. As expected, the probe specific for the cat reporter gene of mini-Tn5Kmcat hybridized with a single fragment which was the same size as the B3B2 mutant (data not shown).
Survival of the B. melitensis rough B3B2 mutant in the mouse model.
Groups of mice were inoculated i.p. with the rough B3B2 mutant and the parental smooth strain B. melitensis 16M. Six mice per strain were sacrificed at weeks 1 and 4 postinoculation, at which time their spleens were examined for Brucella proliferation (Table 3). As early as the first week, no bacteria (detection limit, 20 CFU) were detected in the spleens of five of the six mice infected with the rough B3B2 mutant. In contrast, all spleens from mice infected with B. melitensis 16M had >102 CFU per spleen during the entire 4-week period.
TABLE 3.
Bacterial counts in mouse spleens examined 1 and 4 weeks after i.p. infection
| Strain | Median no. of CFU/spleen at indicated weeka
|
|
|---|---|---|
| 1 | 4 | |
| Wild-type B. melitensis 16M | 6.2 × 105 (3.9 × 105– 9.5 × 105) | 1.8 × 103 (4.6 × 102– 1.8 × 103) |
| Rough B3B2 mutant | 5.0 × 102 (0–3 × 103) | 0 (0) |
Lowest and highest values are presented in parentheses.
Survival of the B. melitensis rough B3B2 mutant in bovine macrophages.
To evaluate the importance of the S-LPS O side chain in the intracellular survival of B. melitensis, we compared the survival of the rough insertion mutant and that of its parental strain in bovine macrophages. The number of viable brucellae in a monolayer of bovine macrophages was estimated 1, 24, and 48 h after infection (Fig. 5). One hour after infection, the numbers of intracellular B. melitensis 16M and B3B2 were 3.28 and 3.31 log10 units, respectively, indicating that the B3B2 rough mutant strain was internalized at the same rate as the parental strain. Within 48 h, the numbers of recoverable bacteria were 5.09 and 5.12 log10 units, indicating that the intracellular growth of B. melitensis 16M and that of the mutant strain were similar.
FIG. 5.
Growth of B. melitensis 16M (parental strain) and the rough insertion mutant B3B2 in bovine macrophages. The data presented are means ± standard deviations of quintuplicate plate counts and are representative of two experiments. p.i., postinfection.
DISCUSSION
In this study, we identify a rough insertion mutant of B. melitensis 16M and characterize the disrupted ORF. We also evaluate the survival of this rough mutant in the mouse model and in bovine macrophages.
The B3B2 mutant was selected by ELISA for the loss of O antigen production. The rough phenotype of the B3B2 insertion mutant was further characterized by different methods, including ELISA, Western blotting, and differential colony staining. The presence of entire lipid A core molecules seen in Western blot analysis demonstrated that the mutation did not take place in the lipid A or in the core biosynthesis pathway. The absence of a banding pattern by Western blotting with MAbs directed against the core indicated that no O antigen is ligated to the core in the rough B3B2 mutant and demonstrated that the inability of anti-S-LPS MAbs to recognize the B3B2 mutant is due to a total absence of its expression and not to an altered structure of the O antigen (loss of epitopes). Furthermore, our results clearly showed that no O antigen was produced in the B3B2 insertion mutant on the cell surface or in the cytoplasm, indicating that the mutation does not affect the transport of the O side chain to the outer membrane but does affect an earlier stage of biosynthesis. The mutation was recreated by gene replacement, indicating that the mutant phenotype was due to the transposon insertion rather than to spontaneous mutation. Deletion and homologous complementation experiments could be done to determine if the rough phenotype is due to the loss of the product of the disrupted ORF or to a polar effect on downstream gene expression.
The perosamine synthetase gene was cloned and sequenced. In V. cholerae O1, perosamine is synthesized from fructose 6-phosphate via four intermediates: mannose 6-phosphate, mannose 1-phosphate, GDP-mannose, and 4-keto-6-dideoxymannose. Ultimately, this final product is converted to GDP-perosamine by the perosamine synthetase (56). Because the last step of the perosamine synthesis pathway is identical for V. cholerae and B. melitensis, we assumed that the earlier steps might be similar or identical for these two organisms. In Brucella, the GDP-perosamine would then serve as a substrate for the addition of a formyl group and could then be polymerized into the O antigen, translocated to the periplasm, transferred to the lipid A-core oligosaccharide, and exported to the cell surface. The mapping of the transposon insertion site in the perosamine synthetase gene in the B3B2 mutant agreed with the phenotypic characteristics of this mutant. Indeed, such a disruption prevented any O side chain production, not only at the surface but also in the cytoplasm of the bacteria.
The coding sequence of rfbEBm16M has a lower G+C content (48%) than that typical of Brucella spp. (55 to 58%) (19). The low G+C content suggests that this gene resulted from the relatively recent acquisition of another microorganism of lower G+C content. The hypothesis of lateral transfer of rfb genes has been reported for many other gram-negative bacteria (4, 34, 38, 46, 58, 62). The presence of the rfbE gene and other rfb genes will be tested for in all species of Brucella, including the rough species B. ovis and B. canis, to determine if rfb gene transfer occurred in a common ancestor or after Brucella speciation.
The preliminary sequencing results indicated that the rfbE gene of B. melitensis 16M (rfbEBm16M) is surrounded by other rfb genes. Because the single-locus structure of a number of genes involved in the biosynthesis of polysaccharide is commonly encountered in many bacteria (45), we assumed that the rfbEBm16M gene is located in the rfb cluster of B. melitensis 16M.
The attenuated nature of Brucella rough mutants has been observed for many years (53). The isolation of a defined mutation in rfbEBm16M gave us an opportunity to analyze the effect of a specific defect in LPS biosynthesis on Brucella virulence.
The drastically decreased survival of the rough mutant in the mouse model confirmed the involvement of the O side chain in the ability of Brucella to resist the host’s killing mechanisms. These mechanisms may act on the extra- and intracellular steps of Brucella infection.
Corbeil et al. demonstrated that Brucella strains are sensitive to complement-mediated lysis via the classical pathway and that the lack of an O side chain increased bacterial sensitivity to the killing activity of the complement (18).
Survival and replication in host phagocytes, particularly macrophages, are critical to the pathogenesis of Brucella infections. The oxidative killing pathways are thought to be the primary mechanism by which host phagocytes eliminate intracellular brucellae (3, 10, 26). The O antigen of Brucella strains has been described as an important factor of resistance to phagocytosis. Investigators have shown that the B. abortus smooth strain 45/0 is more resistant to the intraleukocytic killing system and to oxygen-dependent killing by granule extracts from human and bovine polymorphonuclear leukocytes than is the B. abortus rough strain 45/20 (35, 48, 49). Likewise, strain 2308, a fully virulent strain of B. abortus, replicates within some bovine mammary gland macrophages, whereas the number of B. abortus 45/20 is reduced (31). Nevertheless, all these studies involved uncharacterized rough mutants. In these uncharacterized rough mutants, the absence of the O antigen might combine with other deficiencies to decrease the intracellular survival of Brucella.
To evaluate the involvement of the O antigen in the resistance to the killing mechanisms of macrophages, we analyzed the survival and growth of the rough insertion mutant B3B2 in bovine macrophages. Only minor differences in uptake and cellular growth were observed between the rough insertion mutant and its parental strain, suggesting that the O side chain of LPS is not essential for protecting B. melitensis against the cellular defenses of the host. These results agree with the comparative survival of B. abortus smooth strains 2308 and 19 (vaccine strain) and the corresponding rough transposon mutant strains (strain 2308::Tn5 LacZ and strain 19::Tn5 LacZ) in restrictive bovine mammary macrophages (44). A significant reduction in the survival of strain 19::Tn5 LacZ but not in that of strain 2308::Tn5 LacZ indicated that at least one factor other than S-LPS contributes to the intracellular survival of B. abortus in bovine macrophages.
The results presented here demonstrated that S-LPS or, more precisely, its O side chain was essential for survival in mice but not in the bovine macrophage. We do not rule out the possible involvement of the LPS O side chain in a protective mechanism of Brucella against the bactericidal actions of professional phagocytes. Indeed, Martinez de Tejada et al. have demonstrated that the LPS O side chain plays a role in the resistance of Brucella to polycationic compounds involved in the oxygen-independent systems of phagocytes. These researchers also demonstrated that the core lipid A plays a major role in this resistance (39).
Moreover, the B3B2 mutant is only deficient for the O side chain production and retains other mechanisms for the intracellular survival of Brucella. The mechanisms and virulence factors responsible for the ability of brucellae to escape the bactericidal effects of host phagocytes are not well understood. However, B. abortus in neutrophils has been shown to inhibit degranulation (11) and the oxidative burst (35), whereas it survives in macrophages principally by preventing phagolysosomal fusion (27). Under these conditions, the protective activity of the O side chain might not be required.
ACKNOWLEDGMENTS
We thank Vizcaino Nieves, Institut National de la Recherche Agronomique, Centre de Recherche de Tours, Nouzilly, France, for providing the B. melitensis genomic library. We gratefully acknowledge P. Michel, Centre d’Étude et de Recherche Vétérinaire et Agrochimique, Brussels, Belgium, for his help with the mouse model. We are grateful to D. Prozzi and Peter Flanagan for correcting and improving our English.
Fabrice Godfroid received a specialization grant from the Fonds pour la Formation à la Recherche dans l’Industrie et dans l’Agriculture.
REFERENCES
- 1.Alton G, Jones L, Angus R, Verger J-M, editors. Techniques for the brucellosis laboratory. Paris, France: INRA; 1988. [Google Scholar]
- 2.Altschul S, Gish W, Miller W, Myers E, Lipman D. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Baldwin C L, Winter A J. Macrophages and Brucella. In: Zwilling B S, Eisenstein T K, editors. Macrophage-pathogen interactions. 1st ed. New York, N.Y: Marcel Dekker, Inc.; 1993. pp. 363–380. [Google Scholar]
- 4.Bastin D A, Reeves P R. Sequence and analysis of the O antigen gene (rfb) cluster of Escherichia coli O111. Gene. 1995;164:17–23. doi: 10.1016/0378-1119(95)00459-j. [DOI] [PubMed] [Google Scholar]
- 5.Bilge S S, Vary J C, Dowell S F, Tarr P I. Role of the Escherichia coli O157:H7 O side chain in adherence and analysis of an rfb locus. Infect Immun. 1996;64:4795–4801. doi: 10.1128/iai.64.11.4795-4801.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowden R A, Cloeckaert A, Zygmunt M S, Bernard S, Dubray G. Surface exposure of outer membrane protein lipopolysaccharide epitopes in Brucella species studied by enzyme-linked immunosorbent assay and flow cytometry. Infect Immun. 1995;63:3945–3952. doi: 10.1128/iai.63.10.3945-3952.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bundel D R, Cherwonogrodzky J W, Carrof M, Perry M B. The lipopolysaccharides of Brucella abortus and B. melitensis. Ann Inst Pasteur Microbiol. 1987;138:92–98. doi: 10.1016/0769-2609(87)90083-4. [DOI] [PubMed] [Google Scholar]
- 8.Bundel D R, Cherwonogrodzky J W, Perry M B. Structural elucidation of the Brucella melitensis M antigen by high-resolution NMR at 500 MHz. Biochemistry. 1987;26:8717–8726. doi: 10.1021/bi00400a034. [DOI] [PubMed] [Google Scholar]
- 9.Bundel D R, Cherwonogrodzky J W, Perry M B. The structure of the lipopolysaccharide O-chain (M antigen) and polysaccharide B produced by Brucella melitensis 16M. FEBS Lett. 1987;216:261–264. doi: 10.1016/0014-5793(87)80702-0. [DOI] [PubMed] [Google Scholar]
- 10.Canning P C. Phagocyte function in resistance to brucellosis. In: Adams L G, editor. Advances in brucellosis research. 1st ed. Austin: Texas A&M University Press; 1990. [Google Scholar]
- 11.Canning P C, Roth J A, Deyoe B L. Release of 5′-guanosine monophosphate and adenine by Brucella abortus and their role in the intracellular survival of the bacteria. J Infect Dis. 1986;154:464–470. doi: 10.1093/infdis/154.3.464. [DOI] [PubMed] [Google Scholar]
- 12.Caroff M, Bundel D R, Perry M B, Cherwonogrodzky J, Duncan J R. Antigenic S-type lipopolysaccharide of Brucella abortus 1119-3. Infect Immun. 1984;46:384–388. doi: 10.1128/iai.46.2.384-388.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caroff M, Bundle D R, Perry M B. Structure of the O-chain of the phenol-phase soluble cellular lipopolysaccharide of Yersinia enterocolitica serotype O:9. Eur J Biochem. 1984;139:195–200. doi: 10.1111/j.1432-1033.1984.tb07994.x. [DOI] [PubMed] [Google Scholar]
- 14.Cloeckaert A, de Wergifosse P, Dubray G, Limet J N. Identification of seven surface exposed Brucella outer membrane proteins by use of monoclonal antibodies: immunogold labelling for electron microscopy and enzyme-linked immunosorbent assay. Infect Immun. 1990;58:3980–3987. doi: 10.1128/iai.58.12.3980-3987.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cloeckaert A, Zygmunt M S, de Wergifosse P, Dubray G, Limet J N. Demonstration of peptidoglycan-associated Brucella outer-membrane proteins by use of monoclonal antibodies. J Gen Microbiol. 1992;138:1543–1550. doi: 10.1099/00221287-138-7-1543. [DOI] [PubMed] [Google Scholar]
- 16.Cloeckaert A, Zygmunt M S, Dubray G, Limet J N. Characterization of O-polysaccharide specific monoclonal antibodies derived from mice infected with the rough Brucella melitensis strain B115. J Gen Microbiol. 1993;139:1551–1556. doi: 10.1099/00221287-139-7-1551. [DOI] [PubMed] [Google Scholar]
- 17.Cloeckaert A, Zygmunt M S, Nicolle J-C, Dubray G, Limet J N. O-chain expression in the rough Brucella melitensis strain B115: induction of O-polysaccharide-specific monoclonal antibodies and intracellular localization demonstrated by immunoelectron microscopy. J Gen Microbiol. 1992;138:1211–1219. doi: 10.1099/00221287-138-6-1211. [DOI] [PubMed] [Google Scholar]
- 18.Corbeil L B, Blau K, Inzana T I, Neilsen K H, Jacobson R H, R. C R, Winter A J. Killing of Brucella abortus by bovine serum. Infect Immun. 1988;56:3251–3261. doi: 10.1128/iai.56.12.3251-3261.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corbel M J. Recent advances in the study of Brucella antigens and their serological cross-reactions. Vet Bull. 1985;55:927–942. [Google Scholar]
- 20.Danese I, Tibor A, Denoel P A, Weynants V, Godfroid F, Letesson J-J. Transposition mutagenesis of Brucella melitensis 16M with a mini-Tn5Kmcat and evaluation of the reporter gene expression. Arch Physiol Biochem. 1996;104:45. [Google Scholar]
- 21.de Lorenzo V, Herrero M, Jakubzik U, Timmis K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Denoel P A, Crawford R M, Zygmunt M S, Tibor A, Weynants V E, Godfroid F, Hoover D L, Letesson J-J. Survival of a bacterioferritin deletion mutant of Brucella melitensis 16M in human monocyte-derived macrophages. Infect Immun. 1997;65:4337–4340. doi: 10.1128/iai.65.10.4337-4340.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Depiereux E, Feytmans E. MATCH-BOX: a fundamentally new algorithm for the simultaneous alignment of several protein sequences. CABIOS. 1992;8:501–509. doi: 10.1093/bioinformatics/8.5.501. [DOI] [PubMed] [Google Scholar]
- 24.Depiereux E, Feytmans E. Simultaneous and multivariate alignment of protein sequences. Correspondence between physicochemical profiles and structurally conserved regions (SCR’s) Protein Eng. 1991;4:603–613. doi: 10.1093/protein/4.6.603. [DOI] [PubMed] [Google Scholar]
- 25.Elzer P H, Jacobson R H, Nielsen K H, Douglas J T, Winter A J. BALB/c mice infected with Brucella abortus express protracted polyclonal responses of both IgG2a and IgG3 isotypes. Immunol Lett. 1994;42:145–150. doi: 10.1016/0165-2478(94)90078-7. [DOI] [PubMed] [Google Scholar]
- 26.Enright F M. The pathogenesis and pathobiology of Brucella infection in domestic animals. In: Nielsen K, Duncan J R, editors. Animal brucellosis. Boca Raton, Fla: CRC Press; 1990. pp. 301–320. [Google Scholar]
- 27.Frenchick P J, Markham R J F, Cochrane A H. Inhibition of phagosome-lysosome fusion in macrophages by soluble extracts of virulent Brucella abortus. Am J Vet Res. 1985;46:332–335. [PubMed] [Google Scholar]
- 28.Fuqua, C. W. 1992. An improved chloramphenicol resistance gene cassette for site-directed marker replacement mutagenesis. BioTechniques 12. [PubMed]
- 29.Gish W, States D J. Identification of protein coding regions by database similarity search. Nat Genet. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- 30.Halling S M, Detilleux P G, Tatum F M, Judge B A, Mayfield J E. Deletion of the BCSP31 gene of Brucella abortus by replacement. Infect Immun. 1991;59:3863–3868. doi: 10.1128/iai.59.11.3863-3868.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harmon B G, Adams L G, Frey M. Survival of rough and smooth strains of Brucella abortus in bovine mammary gland macrophages. Am J Vet Res. 1988;49:1092–1097. [PubMed] [Google Scholar]
- 32.Joiner K A. Studies of the mechanism of bacterial resistance to complement killing and on the mechanism of action of bacterial antibody. Curr Top Microbiol Immunol. 1985;121:99–133. doi: 10.1007/978-3-642-45604-6_6. [DOI] [PubMed] [Google Scholar]
- 33.Joiner K A, Grossman N, Schmetz M, Leive L. C3 binds preferentially to long-chain lipopolysaccharide during alternative pathway activation by Salmonella montevideo. J Immunol. 1986;136:710–715. [PubMed] [Google Scholar]
- 34.Klena J D, Schnaitman C A. Function of the rfb gene cluster and the rfe gene in the synthesis of O antigen by Shigella dysenteriae 1. Mol Microbiol. 1993;9:393–402. doi: 10.1111/j.1365-2958.1993.tb01700.x. [DOI] [PubMed] [Google Scholar]
- 35.Kreutzer D L, Dreyfus L A, Robertson D C. Interaction of polymorphonuclear leukocytes with smooth and rough strains of Brucella abortus. Infect Immun. 1979;23:737–742. doi: 10.1128/iai.23.3.737-742.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 37.Limet J N, Bosseray N, Garin-Bastuji B, Dubray G, Plommet M. Humoral immunity in mice mediated by monoclonal antibodies against the A and M antigens of Brucella. J Med Microbiol. 1989;30:37–43. doi: 10.1099/00222615-30-1-37. [DOI] [PubMed] [Google Scholar]
- 38.Marolda C L, Valvano M A. Identification, expression, and DNA sequence of the GDP-mannose biosynthesis genes encoded by the O7 rfb gene cluster of strain VW187 (Escherichia coli O7:K1) J Bacteriol. 1993;175:148–158. doi: 10.1128/jb.175.1.148-158.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martinez de Tejada G, Pizzaro-Cerda J, Moreno E, Moriyon I. The outer membranes of Brucella spp. are resistant to bactericidal cationic peptides. Infect Immun. 1995;63:3054–3061. doi: 10.1128/iai.63.8.3054-3061.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moreno E, Berman D T, Boettcher L A. Biological activities of Brucella abortus lipopolysaccharide. Infect Immun. 1981;31:362–368. doi: 10.1128/iai.31.1.362-370.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moreno E, Jones L M, Berman D T. Immunochemical characterization of rough Brucella lipopolysaccharides. Infect Immun. 1984;43:779–782. doi: 10.1128/iai.43.3.779-782.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nikaido H. Outer membrane of Salmonella typhimurium. Transmembrane diffusion of some hydrophobic substances. Biochim Biophys Acta. 1976;433:118–132. doi: 10.1016/0005-2736(76)90182-6. [DOI] [PubMed] [Google Scholar]
- 43.Perry M B, Bundle D R. Lipopolysaccharide antigens and carbohydrates of Brucella. In: Adams L G, editor. Advances in Brucellosis research. Austin: Texas A&M University; 1990. pp. 76–88. [Google Scholar]
- 44.Price R E, Templeton J W, Adams L G. Survival of smooth, rough and transposon mutant strains of Brucella abortus in bovine mammary macrophages. Vet Immunol Immunopathol. 1990;26:353–365. doi: 10.1016/0165-2427(90)90119-d. [DOI] [PubMed] [Google Scholar]
- 45.Reeves P. Biosynthesis and assembly of lipopolysaccharide. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier Science; 1994. pp. 281–317. [Google Scholar]
- 46.Reeves P. Evolution of Salmonella O antigen variation by interspecific gene transfer on a large scale. Trends Genet. 1993;9:17–22. doi: 10.1016/0168-9525(93)90067-R. [DOI] [PubMed] [Google Scholar]
- 47.Reeves P R, Hobbs M, Valvano M A, Skurnik M, Whitfield C, Coplin D, Kido N, Klena J, Maskel D, Raetz C R H, Rick P D. Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol. 1996;4:495–503. doi: 10.1016/s0966-842x(97)82912-5. [DOI] [PubMed] [Google Scholar]
- 48.Riley L K, Robertson D C. Brucellacidal activity of human and bovine polymorphonuclear leukocyte granule extracts against smooth and rough strains of Brucella abortus. Infect Immun. 1984;46:231–236. doi: 10.1128/iai.46.1.231-236.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Riley L K, Robertson D C. Ingestion and intracellular survival of Brucella abortus in human and bovine polymorphonuclear leukocytes. Infect Immun. 1984;46:224–230. doi: 10.1128/iai.46.1.224-230.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 51.Schnaitman C A, Klena J D. Genetics of lipopolysaccharide biosynthesis in enteric bacteria. Microbiol Rev. 1993;57:655–682. doi: 10.1128/mr.57.3.655-682.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;10:783–791. [Google Scholar]
- 53.Smith L D, Ficht T A. Pathogenesis of Brucella. Crit Rev Microbiol. 1990;17:209–230. doi: 10.3109/10408419009105726. [DOI] [PubMed] [Google Scholar]
- 54.Stabel J R, Stabel T J. Immortalization and characterization of bovine peritoneal macrophages transfected with SV40 plasmid DNA. Vet Immunol Immunopathol. 1995;45:211–220. doi: 10.1016/0165-2427(94)05348-v. [DOI] [PubMed] [Google Scholar]
- 55.Stinavage P, Martin L E, Spitznagel J K. O antigen and lipid A phosphoryl groups in resistance of Salmonella typhimurium LT-2 to nonoxidative killing in human polymorphonuclear neutrophils. Infect Immun. 1989;57:3894–3900. doi: 10.1128/iai.57.12.3894-3900.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stroeher U H, Karageorgos L E, Brown M H, Morona R, Manning P A. A putative pathway for perosamine biosynthesis is the first function encoded within the rfb region of Vibrio cholerae O1. Gene. 1995;166:33–42. doi: 10.1016/0378-1119(95)00589-0. [DOI] [PubMed] [Google Scholar]
- 57.Stroeher U H, Karageorgos L E, Morona R, Manning P A. Serotype conversion in Vibrio cholerae O1. Proc Natl Acad Sci USA. 1992;89:2566–2570. doi: 10.1073/pnas.89.7.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thorson J S, Lo S F, Ploux O, He X, Liu H-W. Studies of the biosynthesis of 3,6-dideoxyhexoses: molecular cloning and characterization of the asc (ascarylose) region from Yersinia pseudotuberculosis serogroup VA. J Bacteriol. 1994;176:5483–5493. doi: 10.1128/jb.176.17.5483-5493.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tibor A, Wansard V, Grayon M, Verger J-M, Letesson J-J. Abstracts of the 95th General Meeting of the American Society for Microbiology 1995. Washington, D.C: American Society for Microbiology; 1995. Positive selection of double recombinants in Brucella abortus using the Bacillus subtilis sacB gene, abstr. B-337; p. 224. [Google Scholar]
- 60.Verger J M, Grayon M, Chaslus-Dancla E, Meurisse M, Lafont J P. Conjugative transfer and in vitro/in vivo stability of the broad-host-range incP R751 plasmid in Brucella spp. Plasmid. 1993;29:142–146. doi: 10.1006/plas.1993.1016. [DOI] [PubMed] [Google Scholar]
- 61.Vizcaino N, Cloeckaert A, Zygmunt M S, Dubray G. Cloning, nucleotide sequence, and expression of the Brucella melitensis omp31 gene coding for an immunogenic major outer membrane protein. Infect Immun. 1996;64:3744–3751. doi: 10.1128/iai.64.9.3744-3751.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang L, Romana L K, Reeves P. Molecular analysis of Salmonella enterica group E1 rfb gene cluster: O antigen and the genetic basis of the major polymorphism. Genetics. 1992;130:429–443. doi: 10.1093/genetics/130.3.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.White P G, Wilson J B. Differentiation of smooth and non-smooth colonies of Brucellae. J Bacteriol. 1951;61:239–240. doi: 10.1128/jb.61.2.239-240.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]