Abstract
Ribosome biogenesis is among the most resource-intensive cellular processes, with ribosomal proteins accounting for up to half of all newly synthesized proteins in eukaryotic cells. During stress, cells shut down ribosome biogenesis in part by halting rRNA synthesis, potentially leading to massive accumulation of aggregation-prone ‘orphan’ ribosomal proteins (oRPs). Here we show that, during heat shock in yeast and human cells, oRPs accumulate as reversible peri-nucleolar condensates recognized by the Hsp70 co-chaperone Sis1/DnaJB6. oRP condensates are liquid-like in cell-free lysate but solidify upon depletion of Sis1 or inhibition of Hsp70. When cells recover from heat shock, oRP condensates disperse in a Sis1- and Hsp70-dependent manner, and the oRP constituents are incorporated into functional ribosomes in the cytosol, enabling cells to efficiently resume growth. Preserving biomolecules in reversible condensates–like mRNAs in cytosolic stress granules and oRPs at the nucleolar periphery–may be a primary function of the Hsp70 chaperone system.
Cells must double their ribosome content each division cycle, and ribosome biogenesis (RiBi) may be rate-limiting for proliferation1–4. In yeast, where eukaryotic RiBi has been most thoroughly studied, ribosomal proteins (RPs) account for up to half of all newly synthesized proteins under nutrient-rich conditions5,6. This huge investment of resources results in the synthesis of more than 105 RPs each minute1,6.
To assemble a yeast ribosome, 79 RPs must stoichiometrically associate with four ribosomal RNA (rRNA) molecules to form the large 60S and small 40S subunits7. Many RPs cannot adopt their native structures and are highly aggregation prone in the absence of rRNA8. This poses a challenge in the compartmentalized eukaryotic cell: rRNA is transcribed in the nucleolus, while RPs are synthesized on ribosomes in the cytosol. Most RPs translocate from the cytosol into the nucleolus, and specific karyopherins and dedicated chaperones known as escortins mediate their transport9. Transcription of rRNA and the messenger RNAs encoding RPs is coordinately regulated and rapidly repressed upon nutrient limitation or other environmental stressors such as heat shock10–13. However, while this transcriptional attenuation occurs even under modest stress conditions in yeast such as 37 °C, protein synthesis remains active and is only repressed at temperatures above 40 °C (refs. 14,15). Given the rate of RP synthesis, imperfect coordination between transcription and translation could result in a massive buildup of excess RPs relative to rRNA.
Several cellular mechanisms have been described that respond to ‘orphan’ RPs (oRPs), that is, RPs that are not bound to rRNA and/or otherwise mislocalized in the cell16. Two distinct pathways, mediated by the Tom1/HUWE1 and UBE2O ubiquitin ligases, recognize oRPs and target them for proteasomal degradation17,18. Loss-of-function mutations in RP genes—which result in stoichiometric imbalance among RPs and accumulation of oRPs—lead to a class of human diseases known as ‘ribosomopathies’19. In yeast, genetic disruptions that result in the accumulation of oRPs activate a ribosome assembly stress response that results in downregulation of RP genes and induction of the heat shock response (HSR), a transcriptional regulon encoding protein homeostasis (proteostasis) factors20–22. The ability of oRPs to activate the HSR suggests that ribosome assembly is under surveillance by the proteostasis network.
In this Article, we demonstrate that oRPs accumulate and drive key early events in the cellular response to heat shock. We find that oRPs interact with the J-domain protein (JDP) Sis1, an essential regulator of the chaperone Hsp70. oRPs trigger the localization of Sis1 in yeast, and its homologue DnaJB6 in human cells, to the periphery of the nucleolus. Rather than targeting oRPs for degradation, Sis1 and Hsp70 maintain oRPs in dynamic condensates that remain liquid-like in cell-free lysate. Following recovery from heat shock, oRP condensates disperse, the RPs are incorporated into functional ribosomes in the cytosol, and cells efficiently resume proliferation. By actively maintaining condensate reversibility, Sis1 and Hsp70 preserve oRP functionality and conserve cellular resources during stress.
Results
Sis1 localizes to the nucleolar periphery during heat shock
To define the molecular species that drive the spatial re-organization of the proteostasis network during heat shock, we focused on the JDP Sis1. Subcellular re-localization of Sis1 represents the earliest known cell biological event following heat shock in yeast23. We constructed a four-colour yeast strain to monitor Sis1–mVenus localization in live cells with respect to the nucleolus (Nsr1–mScarlet-I), the nuclear boundary/cortical endoplasmic reticulum (Sec61–HaloTagJF646) and cytosolic protein aggregates (Hsp104–mTFP1) (Supplementary Table 1). We imaged this strain in three dimensions over a heat shock time course using lattice light sheet microscopy (Supplementary Video 1). Before heat shock, Sis1 was concentrated in the nucleoplasm and diffuse throughout the cytosol (Fig. 1a). Acute heat shock at 39 °C resulted in re-localization of Sis1 from the nucleoplasm to a region surrounding the nucleolus within 2.5 min (Fig. 1a). Image analysis revealed increased Sis1 peri-nucleolar localization in nearly all cells following 2.5 and 10 min of heat shock (Fig. 1b, Extended Data Fig. 1a and Methods). We additionally observed overlapping cytosolic foci containing Sis1 and Hsp104 that formed with delayed kinetics compared with Sis1 peri-nucleolar localization (Fig. 1a and Extended Data Fig. 1b). Ongoing protein synthesis contributes to activation of the heat shock transcriptional response and to re-localization of Sis1 and Hsp104 (refs. 23–27). Indeed, Sis1 re-localization was abolished following heat shock upon pre-treatment with the translation elongation inhibitor cycloheximide (CHX) (Fig. 1a,b and Extended Data Fig. 1b). By contrast, the nucleolus expands during heat shock even in the presence of CHX (Fig. 1a), consistent with mature proteins localizing to the nucleolus upon heat shock in human cells and fission yeast28,29. Thus, ongoing translation drives Sis1 spatial dynamics during heat shock–in the cytosol and within the nucleus.
Fig. 1 |. Sis1 localizes to the nucleolar periphery and interacts with RPs during heat shock.
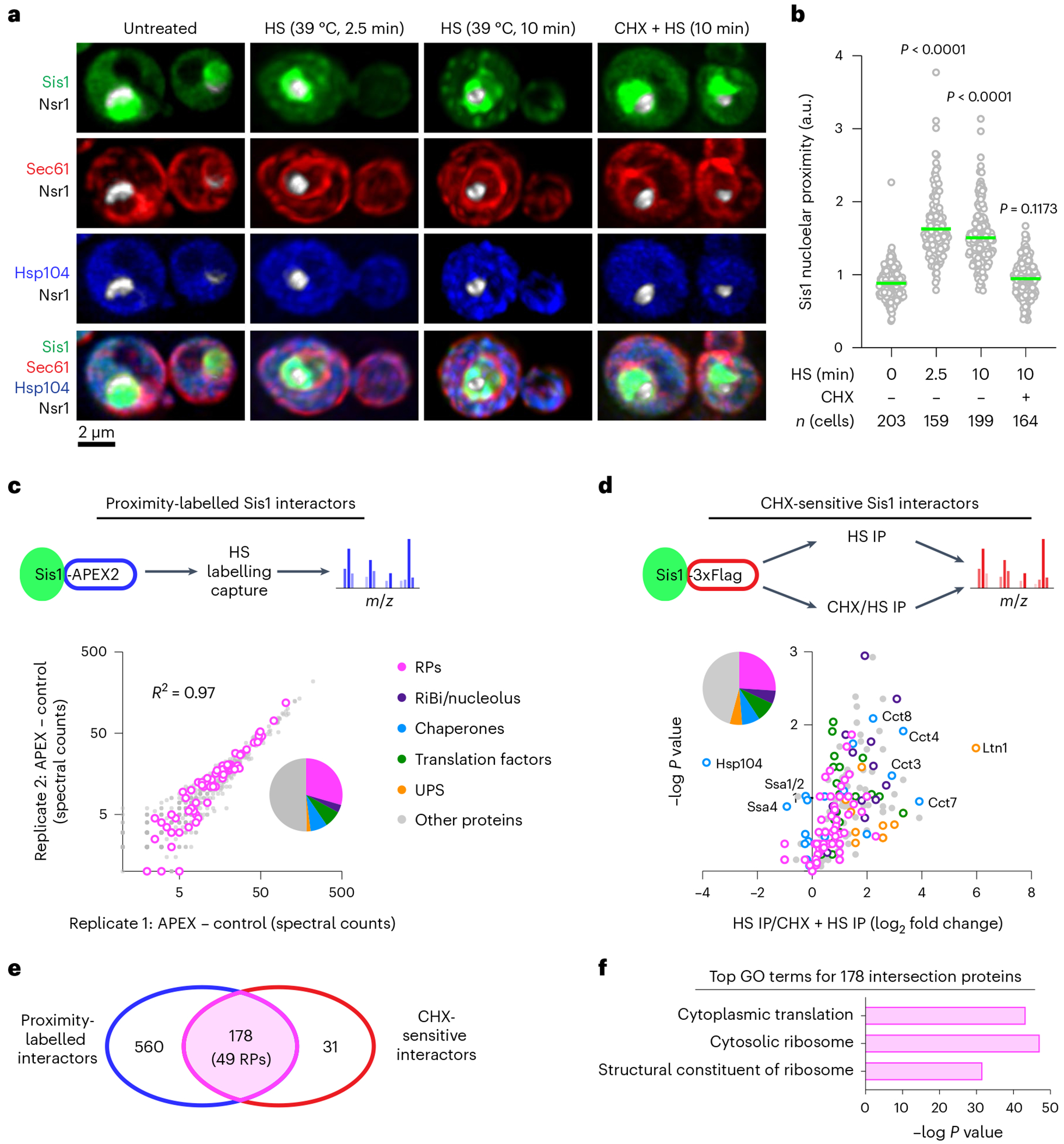
a, Lattice light sheet live imaging of yeast cells with endogenously tagged Sis1–mVenus (green), Hsp104–TFP (blue), Sec61–Halo (red) and Nsr1–mScarlet-I (white) under non-stress (30 °C), heat shock (39°C, 2.5 and 10 min) and pre-treatment with CHX (50 μg ml−1, 5 min) followed by heat shock (39 °C, 10 min). b, Single-cell quantification of Sis1 nucleolar proximity defined as the ratio of mean Sis1 intensity in the half of the nucleus containing the nucleolus to the mean Sis1 intensity in the other half of the nucleus. Statistical significance was determined by Brown–Forsythe and Welch’s one-way ANOVA analysis, coupled with Games–Howell post hoc multiple comparisons. n denotes number of cells, pooled from four biologically independent replicates for 0, 2.5 and 10 min heat shock (HS) and from five biologically independent replicates for CHX pre-treatment followed by the 10 min heat shock (CHX + 10 min HS) conditions. c, Top: schematic of in vivo proximity labelling of Sis1–APEX2 followed by MS analysis. Bottom: enrichment of proteins labelled by Sis1–APEX2 following HS in two biological replicates. RPs highlighted in magenta. UPS, ubiquitin-proteasome system. d, Top: workflow to IP Sis1–3xFlag following heat shock (39°C, 10 min) pre-treated with either vehicle or CHX (50 μg ml−1, 5 min). Bottom: volcano plot demonstrating magnitude and statistical significance of CHX sensitivity of Sis1 interactors. e, Venn diagram of proteins identified in c and d. f, Gene Ontology (GO) terms enriched among the 178 intersection proteins from e. Yeastmine (http://yeastmine.yeastgenome.org/yeastmine) was employed to carry out GO analysis and Bonferroni test corrections were used to account for multiple testing and P value determination.
RPs are a major class of Sis1 interactors during heat shock
To identify proteins that Sis1 interacts with during heat shock, we utilized proximity-dependent labelling and affinity capture coupled to mass spectrometry (MS). We endogenously fused Sis1 to the engineered peroxidase APEX2 (ref. 30), heat-shocked cells expressing Sis1–APEX2, and performed proximity labelling in the presence and absence of a pulse of H2O2 to distinguish specifically and non-specifically labelled proteins (Methods). Biological replicates showed strong concordance in the abundance of the 698 proximity-labelled proteins reproduced across replicates and quantifiable above background (Fig. 1c and Supplementary Table 2: APEX data). The data revealed that, during heat shock, Sis1 is proximal to other chaperones, metabolic enzymes, translation factors, RiBi factors and–most prominently–RPs (Fig. 1c and Extended Data Fig. 1c). Comparing Sis1 interactors amino acids with yeast proteome revealed enrichment in alanine, glycine, valine, aspartate, arginine and lysine (Extended Data Fig. 1d). Sis1 interactors were substantialy depleted for intrinsically disordered regions (IDRs) relative to the whole proteome (Extended Data Fig. 1e).
To complement the proximity ligation approach, we performed immunoprecipitation (IP)–MS of Sis1–3xFlag following heat shock in the absence and presence of CHX. Of the 247 proteins we reproducibly pulled down during heat shock with Sis1, 209 were depleted in the CHX-treated cells relative to cells that were heat shocked in the absence of CHX (Fig. 1d, Extended Data Fig. 1f and Supplementary Table 3: IP–MS data). Notably, interactions between Sis1 and cytosolic chaperones such as Hsp70 and Hsp104 increased in cells treated with CHX, while interactions with factors involved in co-translational protein folding and degradation–including members of the TRiC/CCT complex and the ubiquitin ligase Ltn1–were sensitive to CHX. However, the most abundant class of CHX-sensitive Sis1 interactors was RPs. Comparison of the set of APEX2-labelled Sis1 interactors with the set of CHX-sensitive Sis1 interactors revealed an intersection of 178 proteins, 49 of which are RPs (Fig. 1e,f). These proteomic results indicate that RPs are a major class of CHX-sensitive Sis1 interactors.
Sis1 interacts with oRPs at the nucleolar periphery upon heat shock
The CHX sensitivity of the interaction between Sis1 and RPs could result either from interactions of Sis1 with mature ribosomes or from interactions of Sis1 with newly synthesized RPs. To distinguish these possibilities, we performed in vivo pulse-chase experiments to selectively label either mature or new RPs. We endogenously tagged RPs of the large (Rpl26a and Rpl25) and small ribosomal subunits (Rps4b and Rps9a) with the HaloTag to enable labelling with cell-permeable haloalkanes, either a non-fluorescent ‘blocker’ (7-bromoheptanol) or fluorescent dye (JF646) (Fig. 2a). To label mature RPs, we first pulsed with JF646 and subsequently chased with blocker to mask any new RPs. Conversely, we first added blocker to mask all the mature RPs and then added JF646 to label new RPs. We pulled down Sis1–3xFlag under these two labelling schemes in the presence or absence of heat shock. Sis1 co-precipitated only with the new RPs and only during heat shock (Fig. 2b,c and Extended Data Fig. 2a).
Fig. 2 |. oRPs interact with Sis1/DnaJB6 at the nucleolar periphery.
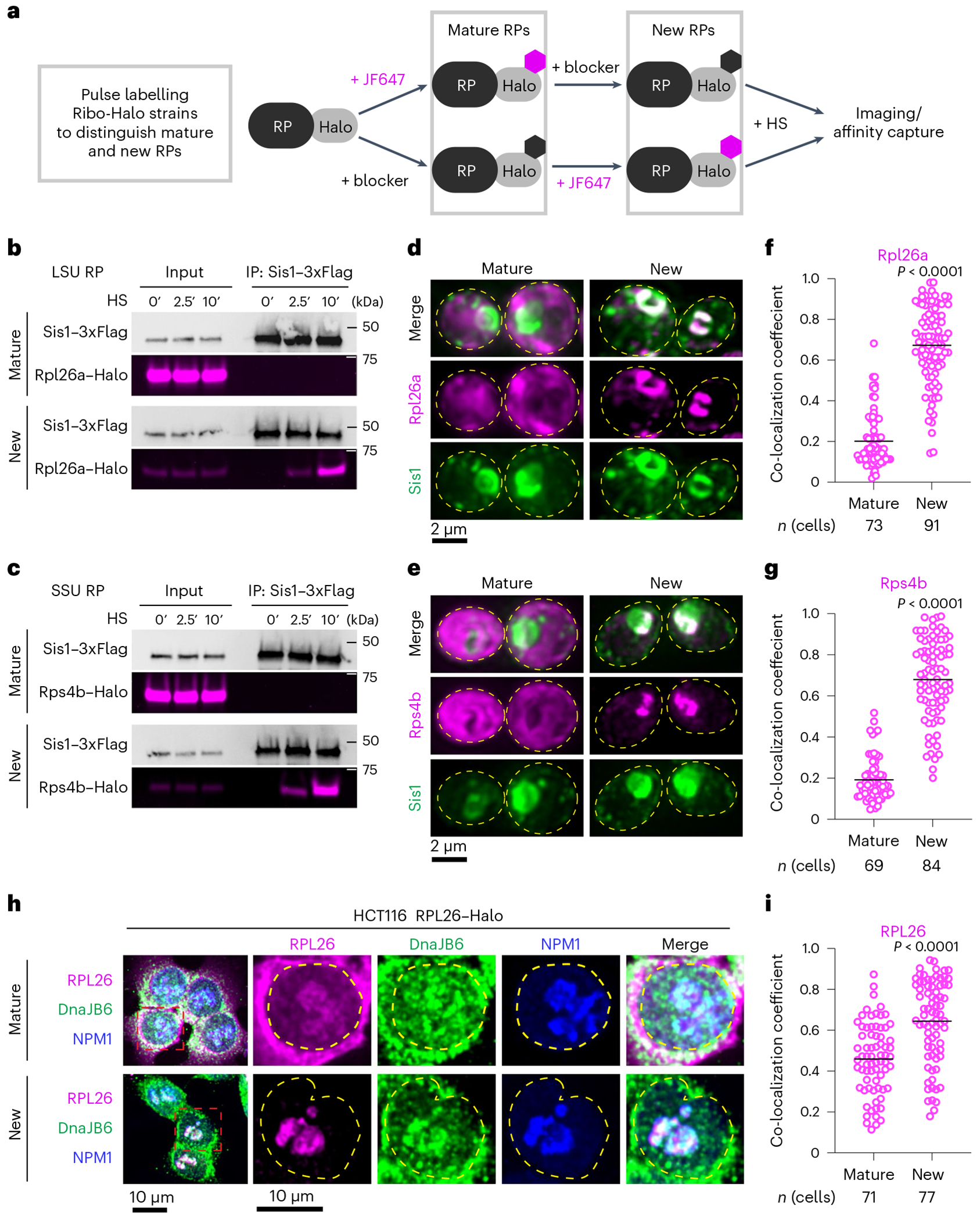
a, Workflow for in vivo pulse-labelling of mature and new RPs. b, IP of Sis1–3xFlag and either mature or new Rpl26a–Halo from cells left unstressed or heat shocked at 39 °C for the indicated times. n = 2 biologically independent experiment. LSU, 60S large subunit. c, As in b, but for Rps4b–Halo. n = 2 biologically independent experiment. SSU, 40S small subunit. d, Lattice light sheet live imaging of yeast under heat shock (39 °C, 10 min) expressing Sis1–mVenus and labelled for either new or mature Rpl26a–Halo. The dashed line indicates the cellular boundary. e, As in c, but for Rps4b–Halo. f, Co-localization (Mander’s overlap coefficient) of Sis1–mVenus with either mature or new Rpl26a–Halo in heat-shocked cells (39 °C, 10 min). P values were calculated with unpaired two-tailed Welch’s t-test. n indicates number of cells, pooled from four independent biological replicates. g, As in f, but for Rps4b–Halo. P values were calculated with unpaired two-tailed Welch’s t-test. n indicates number of cells, pooled from four independent biological replicates. h, Human HCT116 cells stably expressing RPL26–Halo labelled for mature or new RPL26 and heat shocked (43 °C for 30 min). Cells were fixed and immunostained for DnaJB6 and NPM1. Dashed line indicates the nuclear boundary. i, Co-localization (Mander’s overlap coefficient) of DnaJB6 with either mature or new RPL26–Halo in heat-shocked cells (43 °C, 30 min). P values were calculated with unpaired two-tailed Welch’s t-test. n indicates number of cells, pooled from three independent biological replicates.
Next, we imaged RPs with respect to Sis1. We endogenously tagged three large subunit RPs (Rpl26a, Rpl25 and Rpl29) and three small subunit RPs (Rps4b, Rps9a and Rps3) with the HaloTag in the Sis1–mVenus background (Supplementary Table 4). Using the pulse-chase labelling scheme described above, we imaged either mature or new RPs in live cells. Under non-stress conditions, all newly synthesized RPs were immediately localized to the cytosol, demonstrating the rapidity of RiBi (Extended Data Fig. 2b). We heat shocked the cells for 10 min to cause both Sis1 cytosolic foci formation and peri-nucleolar Sis1 accumulation. Mature RPs formed neither cytosolic foci nor peri-nucleolar structures, and the Sis1 cytosolic foci localized away from mature ribosomes (Fig. 2d–g and Extended Data Fig. 2c–f). By contrast, new Rpl26a, Rpl25, Rps4b and Rps9a–which are all incorporated into ribosomal subunits in the nucleolus7–accumulated at the nucleolar periphery as mislocalized oRPs that co-localized with Sis1 (Fig. 2d–g and Extended Data Fig. 2c,d). However, new Rpl29 and Rps3–which are incorporated into their respective ribosomal subunits at a late stage of biogenesis which occurs in the cytosol7–neither localized to the nucleolar periphery nor co-localized with Sis1 (Extended Data Fig. 2e,f). These observations suggest that oRPs accumulate at the nucleolar periphery with Sis1 during heat shock, provided that the RP is incorporated into ribosome assembly in the nucleolus.
Select RiBi factors co-localize with Rpl26a at the nucleolar periphery
To determine whether RiBi factors co-localize with oRPs, we tagged 15 different 60S RiBi factors with mScarlet-I in a strain background expressing Rpl26a–HaloTag. Assembly of the 60S particle involves a series of interconnected steps taking place sequentially in the nucleolus, nucleoplasm and cytosol (Extended Data Fig. 3a)7,31. RPs imported into the nucleolus associate with rRNA co-transcriptionally, but we found no evidence that rRNA biogenesis factors are associated with oRPs. We did not observe co-localization of Rpl26a–HaloTagJF646 during heat shock with either Pol I subunit Rpa190 or Nsr1, which bridges the interaction of RPs with rRNA (Extended Data Fig. 3b,c). By contrast, we did observe co-localization of Rpl26a with early nucleolar assembly factors Rpf1, Ytm1, Nop16, Tif6 and Nog1, but not Nop15 (Extended Data Fig. 3d–i). However, late nucleolar factors Noc3 and Nop2 did not co-localize with Rpl26a (Extended Data Fig. 3j,k). Like-wise, the nucleoplasmic assembly factor Arx1 did not co-localize with Rpl26a, though Nog2 did (Extended Data Fig. 3l,m). Finally, the export complex factor Nmd3, the cytoplasmic assembly factor Drg1 and the karyopherin Kap123 all remained cytosolic (Extended Data Fig. 3n–p). The role, organization and constellation of RiBi factors that localize to the nucleolar periphery remains unclear.
DnaJB6 co-localizes with newly synthesized RPL26 in human cells during heat shock
Do oRPs also accumulate during heat shock in human cells? To test this, we obtained HCT116 human cells with a Halo-tagged copy of RPL26 integrated into the genome32,33. The human JDP DnaJB6 is likely to be the functional homologue of Sis1 in human cells34. To determine whether DnaJB6 co-localizes with RPs during heat shock, we followed a similar pulse-labelling protocol in the HCT116 cells as we used in yeast (Fig. 2a), fixed the cells following heat shock, and stained with antibodies against DnaJB6 and NPM1 to mark the periphery of the nucleolus. As in yeast, mature RPL26 was primarily localized to the cytosol, though some was detectible co-localizing with NPM1 (Fig. 2h). Mature RPL26 showed partial co-localization with DnaJB6 during heat shock (Fig. 2h,i). However, newly synthesized RPL26 was highly concentrated at the periphery of the nucleolus and co-localized with DnaJB6 (Fig. 2h,i). Thus, in yeast and human cells, heat shock triggers the peri-nucleolar accumulation of oRPs that co-localize with homologous JDPs.
RP production is required for peri-nucleolar recruitment of Sis1/DnaJB6
To established whether oRPs are necessary for Sis1 re-localization, we utilized auxin-inducible degradation to acutely, rapidly and quantitatively deplete Ifh1 (Extended Data Fig. 4a), a transcription factor required for the expression of RPs in yeast35. Depletion of Ifh1 impaired cell growth, and mRNA deep sequencing under non-stress conditions revealed that acute loss of Ifh1 resulted in reduced expression of RP mRNAs with few other changes to the transcriptome; importantly, RiBi factor mRNA levels remained unchanged (Fig. 3a,b). Upon heat shock, cells depleted for Ifh1 showed reduced transcriptional induction of Hsf1-regulated genes without alterations in other stress response genes (Fig. 3c and Extended Data Fig. 4b,c). On average, Hsf1 targets were reduced by 25% in Ifh1-depleted cells, suggesting that newly synthesized RPs contribute to activation of the HSR. While the effect of Ifh1 depletion on the HSR was relatively modest, loss of Ifh1 nearly abolished Sis1 localization to the nucleolar periphery during heat shock in most cells (Fig. 3d,e). These findings demonstrate that newly synthesized RPs play a crucial role in recruiting Sis1 to the nucleolar periphery relative to RiBi factors. However, Ifh1 depletion did not prevent formation of Sis1 cytosolic foci during heat shock, demonstrating a specific effect on peri-nucleolar Sis1 (Extended Data Fig. 4d,e). In HCT116 cells, we used the mTOR inhibitor torin-1 to reduce expression of RPs36, and this treatment prevented heat shock-induced localization of DnaJB6 to the nucleolar periphery in most cells (Fig. 3f,g). These data suggest that oRPs recruit Sis1/DnaJB6 to the nucleolar periphery during heat shock.
Fig. 3 |. RPs drive Sis1/DnaJB6 localization to the nucleolar periphery.
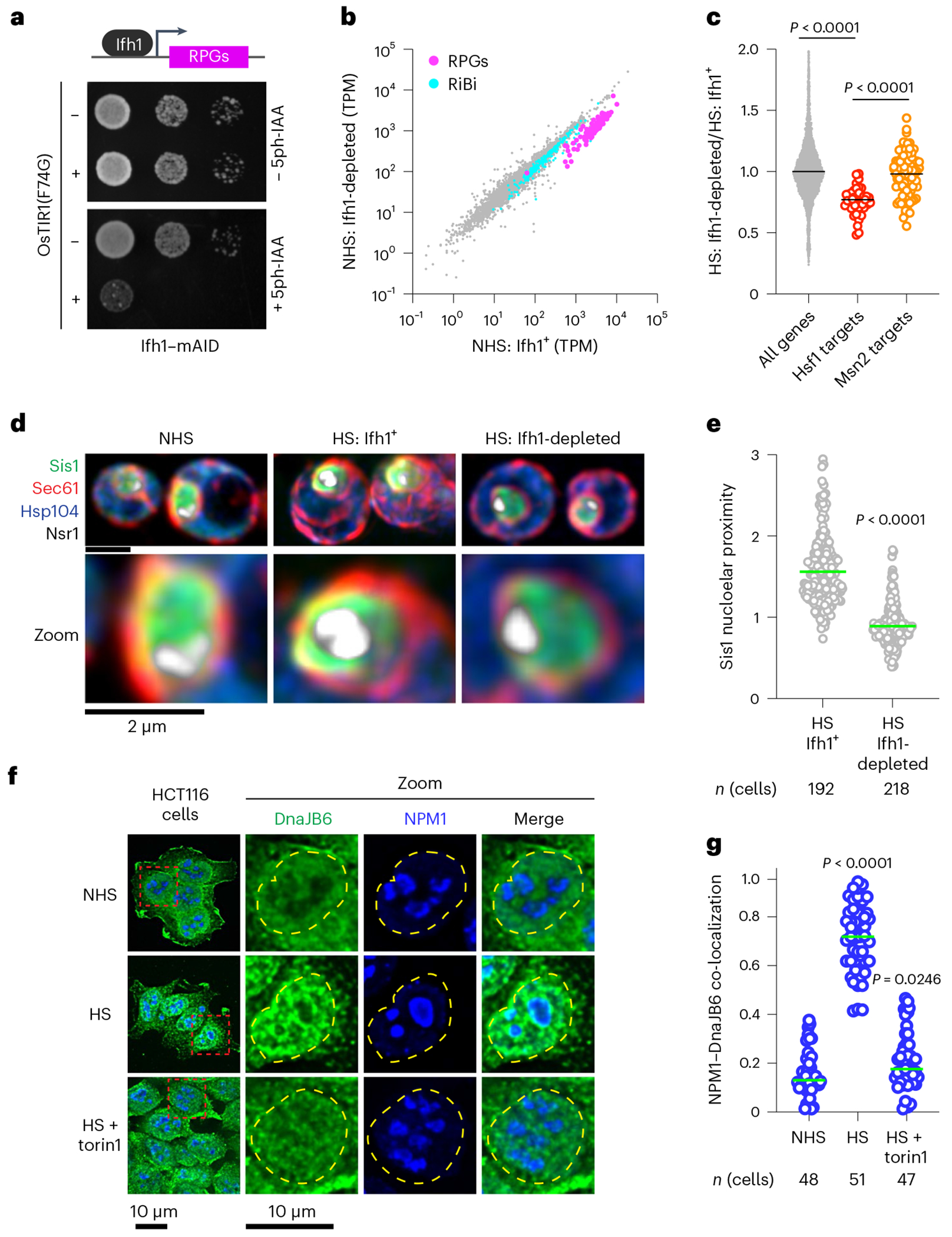
a, Dilution series spot assay of yeast cells expressing Ifh1–mAID with or without β-oestradiol-inducible OsTIR1(F74G) spotted on to rich medium supplemented with either β-oestradiol (1 μM) alone or with 5ph-IAA (5 μM) grown for 48 h. b, Scatter plot of RNA-seq data from Ifh1–mAID/OsTIR1(F74G) treated with β-oestradiol alone or along with 5phIAA for 30 min. RiBi factors (cyan) and ribosomal protein genes (RPGs, magenta) are highlighted. NHS, non heat shock c, Ifh1-dependent fold change in expression in heat-shocked cells showing a reduction in Hsf1 target gene induction but not Msn2 target induction. Statistical significance was assessed using the Brown–Forsythe and Welch one-way ANOVA, followed by Dunnett T3 multiple comparison tests. Each dot symbolizes individual genes, where n = 4,983, 42 and 90 for all genes, Hsf1 target genes and Msn2 target genes, respectively. Gene expression analysis was carried out across two biologically independent experiments. d, Lattice light sheet imaging of the distribution of Sis1–mVenus in non-stressed cells, heat-shocked cells and heat-shocked cells depleted for Ifh1. e, Quantification of Sis1 nucleolar proximity during heat shock (HS) in cells with Ifh1 and following Ifh1 depletion. Statistical significance was determined by Brown–Forsythe and Welch’s one-way ANOVA analysis combined with Games–Howell post hoc multiple comparisons. n indicates number of cells, pooled from four and five biologically independent replicates for Ifh1+ and Ifh1-depleted conditions, respectively. f, Immunostaining of HCT116 cell lines for DnaJB6 (green) and NPM1 (blue) under non-stressed conditions, heat shock (43 °C, 30 min) and heat shock of cells pre-treated with Torin1 (300 nM, 30 min). g, Quantification of co-localization (Mander’s overlap coefficient) of NPM1 and DnaJB6 in single cells in the conditions in f. Statistical significance was assessed using the Brown–Forsythe and Welch one-way ANOVA, followed by Dunnett T3 multiple comparison tests. n indicates number of cells, pooled from three, four and three biologically independent replicates for NHS, HS, and HS + torin1 conditions, respectively.
oRPs form dynamic condensates with Sis1 during heat shock
We next performed time-lapse 3D lattice light-sheet imaging of newly synthesized Rpl26a to observe spatiotemporal dynamics of oRPs during heat shock with respect to the nucleolus and Sis1. Over a sustained 10-min heat shock, new Rpl26a and Sis1 remained localized at the nucleolar periphery in dynamic clusters that rapidly re-organized via fission and fusion (Fig. 4a and Supplementary Video 2). The fluorescence signal remained nearly constant over time, suggesting that these clusters are stable and are not sites of degradation (Extended Data Fig. 5a). We will refer to these clusters as ‘oRP condensates’. To probe the sensitivity of oRP condensates to alcohols known to disrupt certain other biomolecular condensates, we treated cells with 2,5-hexanediol (HD) and 1,6-HD at carefully optimized concentrations37. 2,5-HD had no effect on Sis1 or new Rpl26a localization (Fig. 4b). However, while treatment with 1,6-HD did not disrupt Rpl26a clustering, it resulted in loss of Sis1 peri-nucleolar localization (Fig. 4b). This differential sensitivity to 1,6-HD suggests that the interactions that target Sis1 to oRPs are biophysically distinct from the interactions among the oRPs themselves. Remarkably, Sis1 localization to oRPs was restored upon washout of 1,6-HD (Fig. 4b).
Fig. 4 |. oRPs form dynamic condensates that are stable in cell-free extract.
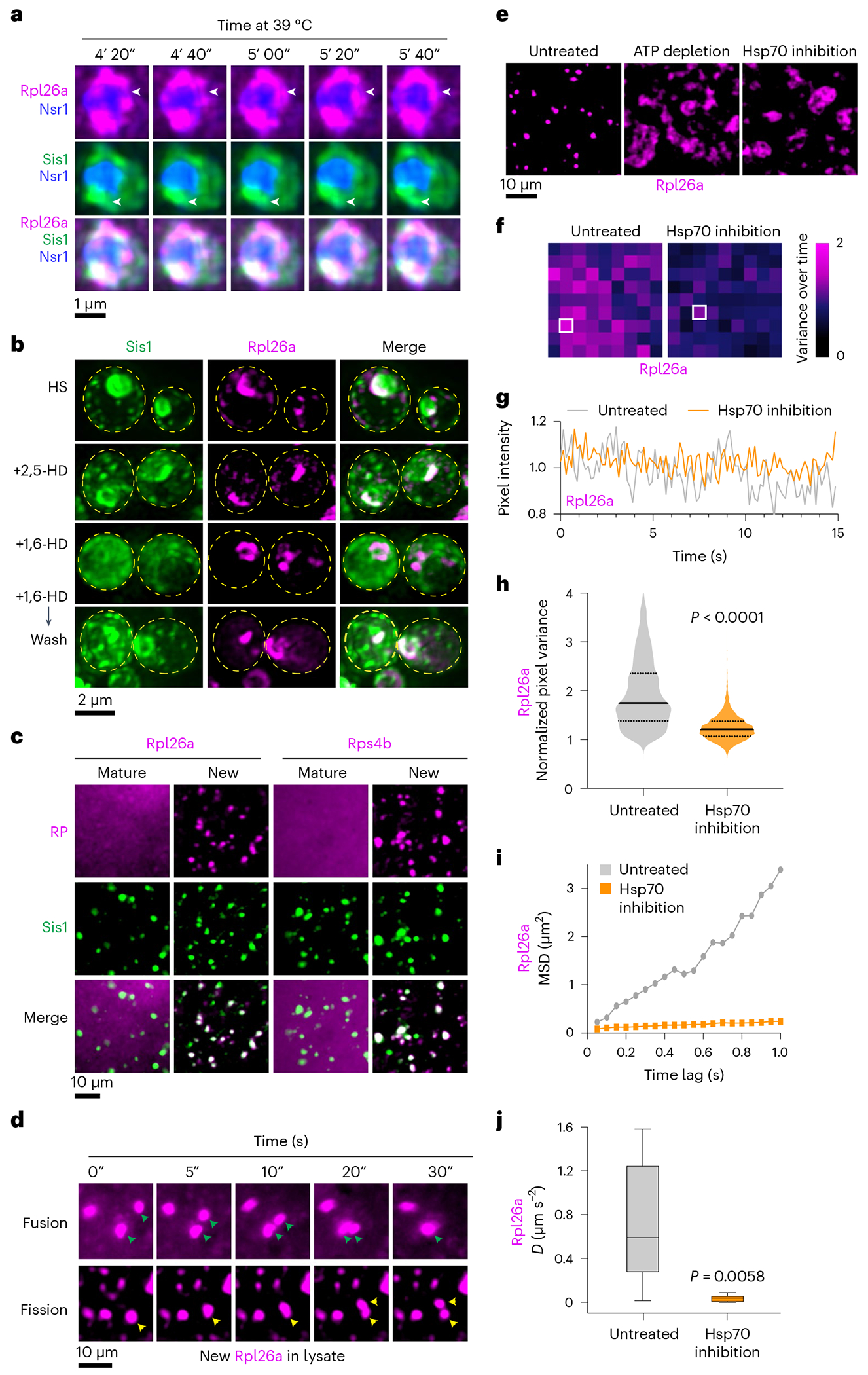
a, 4D LLS imaging of Sis1–mVenus (green) and oRpl26a (magenta). b, Effect of 1,6-HD and 2,5-HD on Sis1 localization and oRP condensates. Cells were heat shocked for 10 min and incubated with either 5% 1,6 HD or 2,5 HD for 2 min. 1,6-HD was washed out for an additional 2 min at heat shock temperature, and cells were imaged again. Micrograph represents data of three biologically independent experiment. c, Imaging of mature and new Rpl26a–Halo and Rps4b–Halo along with Sis1–mVenus in cell-free lysate from heat-shocked cells. Micrograph represents data of four independent experiments. d, Time lapse imaging of heat-shocked cell-free lysate depicting fission and fusion of oRpl26a droplets. e, Depletion ofATP (apyrase, 0.05 units μl−1) or inhibition of Hsp70 (VER-155008, 50 μM) activity in the lysate resulted in the formation of irregular clumps of oRpl26a. The micrograph illustrates data from three independent experiments. f, Heat map representing the normalized variance of pixel photon count over time within the heat shock-induced oRpl26a condensate and upon Hsp70 inhibition. Data are representative of seven and five condensates of untreated and Hsp70 inhibited conditions pooled from three biologically independent experiments. g, Shot noise-normalized intensity over time of the most variable pixel from an oRpl26a condensate in lysate with versus without Hsp70 inhibition. Data are representative of seven and five condensates of untreated and Hsp70 inhibited conditions pooled from three biologically independent experiments. h, Violin plots showing the distribution of normalized pixel variance of oRpl26a in condensates over time in lysate with and without Hsp70 inhibition. P values were calculated with unpaired two-tailed Welch’s t-test. n = 7 and 5 condensates in untreated and Hsp70 inhibited conditions pooled from three biologically independent experiments. i, MSD traces have decreased slope for beads in Hsp70-inhibited condensates. n = 7 condensates for each condition pooled from three biologically independent experiments. j, Inhibition of Hsp70 activity in lysate leads to 20× decrease in the effective diffusion coefficient (D) of beads inside the condensates (mean D untreated 0.73 μm2 s−1, s.d. 0.55 μm2 s−1; mean D inhibited 0.038 μm2 s−1, s.d. 0.031 μm2 s−1). Box plots display medians at their centres and are enclosed by the first and third quartiles. Whiskers extend to 1.5 times the interquartile range (IQR) on both ends. Each condition was analysed using seven condensates, sourced from three biologically independent experiments. P values were calculated with unpaired two-tailed Student’s t-test.
Hsp70 activity maintains the liquid-like dynamics of oRP condensates in cell-free extract
To interrogate the biochemical and biophysical properties of oRP condensates, we developed an assay to observe them in crude cell lysate. We labelled mature and new Rpl26a and Rps4b in cells co-expressing Sis1–mVenus, heat shocked the cells, cryo-milled and resuspended in buffer with a chemical composition like the yeast cytoplasm (Methods). In the lysate, mature RPs showed diffuse signal, but new Rpl26a and Rps4b were concentrated in round condensates that co-localized with Sis1–mVenus (Fig. 4c). Condensates were nearly absent in lysate from non-heat-shocked cells (Extended Data Fig. 5b). Even when the lysate from non-stressed samples was heated to extreme heat shock temperatures of 46 °C and 50 °C, condensates did not form (Extended Data Fig. 6a,b). However, heating the condensates derived from heat-shocked lysate at temperatures of 46 °C or higher resulted in a change in their morphology to an amorphous appearance (Extended Data Fig. 6b). Incubation with RNA-specific dye revealed that the condensates appear to exclude RNA, and addition of RNAse to the lysate did not disrupt condensate morphology or number, suggesting that the RPs are orphaned from rRNA (Extended Data Fig. 6c–e).
We observed both fusion and fission of oRP condensates in the lysate (Fig. 4d and Supplementary Video 3). To determine whether maintenance of oRP condensate dynamics required ATP, we added apyrase to hydrolyse ATP in the lysate. ATP depletion resulted in loss of round condensates and the formation of larger amorphous aggregates (Fig. 4e). Since Sis1, like all JDPs, activates ATP hydrolysis by Hsp70 (ref. 38), we wondered whether Hsp70 activity might contribute to the requirement for ATP. Indeed, addition of the Hsp70 inhibitor VER-155008 phenocopied ATP depletion, resulting in the formation of large irregular clumps of Rpl26a (Fig. 4e). Yet, in conditions where Hsp70 activity was inhibited, Sis1 continued to associate with the amorphous aggregates (Supplementary Fig. 7a), consistent with Sis1 acting upstream of Hsp70. Furthermore, when the oRP condensates were incubated with 1,6-HD, the condensates were effectively dissolved. It is unclear why 1,6-HD works in the lysate to disperse oRP condensates but not in cells, but it could simply be due to the effective concentration in cells. Moreover, prior research has elucidated the pleiotropic impact of 1,6-HD on kinase activities and various other biological molecules39,40. Regardless, 1,6-HD failed to dissolve the condensates when Hsp70 was inhibited, indicating that the ability of 1,6-HD to dissolve oRP condensates in lysate depends on Hsp70 activity (Extended Data Fig. 7b).
We collected videos of the oRP condensates in lysate in the presence and absence of the Hsp70 inhibitor to evaluate their homogeneity and measure the dynamics of oRPs moving within the condensates. A solid sample or a homogeneous liquid is expected to exhibit a variance in photon count equal to its average photon count due to the underlying Poisson statistics of arriving photons41. Samples that do vary, such as an inhomogeneous liquid, will exhibit higher-than-expected variance at each pixel. We quantified the Poisson-normalized variance in Rpl26a fluorescence over time (see ‘Condensate pixel variance analysis’ in Methods)42. High normalized variance (>1) indicates that a condensate is dynamically rearranging and therefore more liquid-like, while normalized variance close to 1 indicates that a condensate is less dynamic and more solid-like. Pixels in individual condensates showed large variance in untreated lysate and much lower variance in the Hsp70-inhibited lysate (Fig. 4f,g). Collective analysis across all pixels revealed that Hsp70 inhibition significantly reduced the pixel variance (Fig. 4h). Thus, Hsp70 inhibition in lysate makes oRP condensates less dynamic and more solid-like.
To directly measure diffusion within the oRP condensates, we employed a bead tracking protocol (Methods). We incubated fluorescent beads with the lysate, allowing them to interact with the condensates. High-frame-rate video analysis revealed that the mean squared displacement (MSD) of beads within the condensates was reduced upon Hsp70 inhibition (Fig. 4i). This reduction in MSD indicates a decrease in the effective diffusion coefficient of beads within the condensates, and quantification of the diffusion coefficient is consistent with a transition from a liquid-like state to a solid-like state when Hsp70 activity is inhibited (Fig. 4j). Supporting the functional relevance of these Hsp70 inhibitor experiments, we found that Hsp70 (marked by HaloTagJF646-–Ssa1) co-localizes with Sis1 at the nucleolar periphery during heat shock, suggesting that Hsp70 is involved in regulating oRP condensates in cells (Supplementary Video 4). Together, these data demonstrate that the Sis1/Hsp70 chaperone system maintains liquid-like dynamics of oRP condensates.
oRP condensates are reversible upon recovery from heat shock
The liquid-like character of the oRP condensates in lysate prompted us to ask whether they might be reversible upon recovery from stress in cells. We pulse labelled new Rpl26a and Rps4b with JF646, heat shocked the cells for 5 min, and chased with excess blocker in medium either at 39 °C for a sustained heat shock or at 30 °C to monitor recovery (Fig. 5a). Imaging revealed that, while the oRP condensates persisted at 39 °C, cells allowed to recover at 30 °C showed Rpl26a and Rps4b signal disperse to the cytosol within 7.5 min (Fig. 5b–d and Extended Data Fig. 8a,b). The fluorescence signal of Rpl26a and Rps4b localized at the nucleolar periphery during heat shock can be quantitatively accounted for in the cytosol after recovery, suggesting there is little to no degradation of oRPs (Extended Data Fig. 8c). We observed the same re-localization of the signal of pulse-labelled RPL26 from the nucleolar periphery to the cytosol in human HCT116 cells following recovery (Fig. 5g,h).
Fig. 5 |. oRP condensates are reversible upon recovery from heat shock.
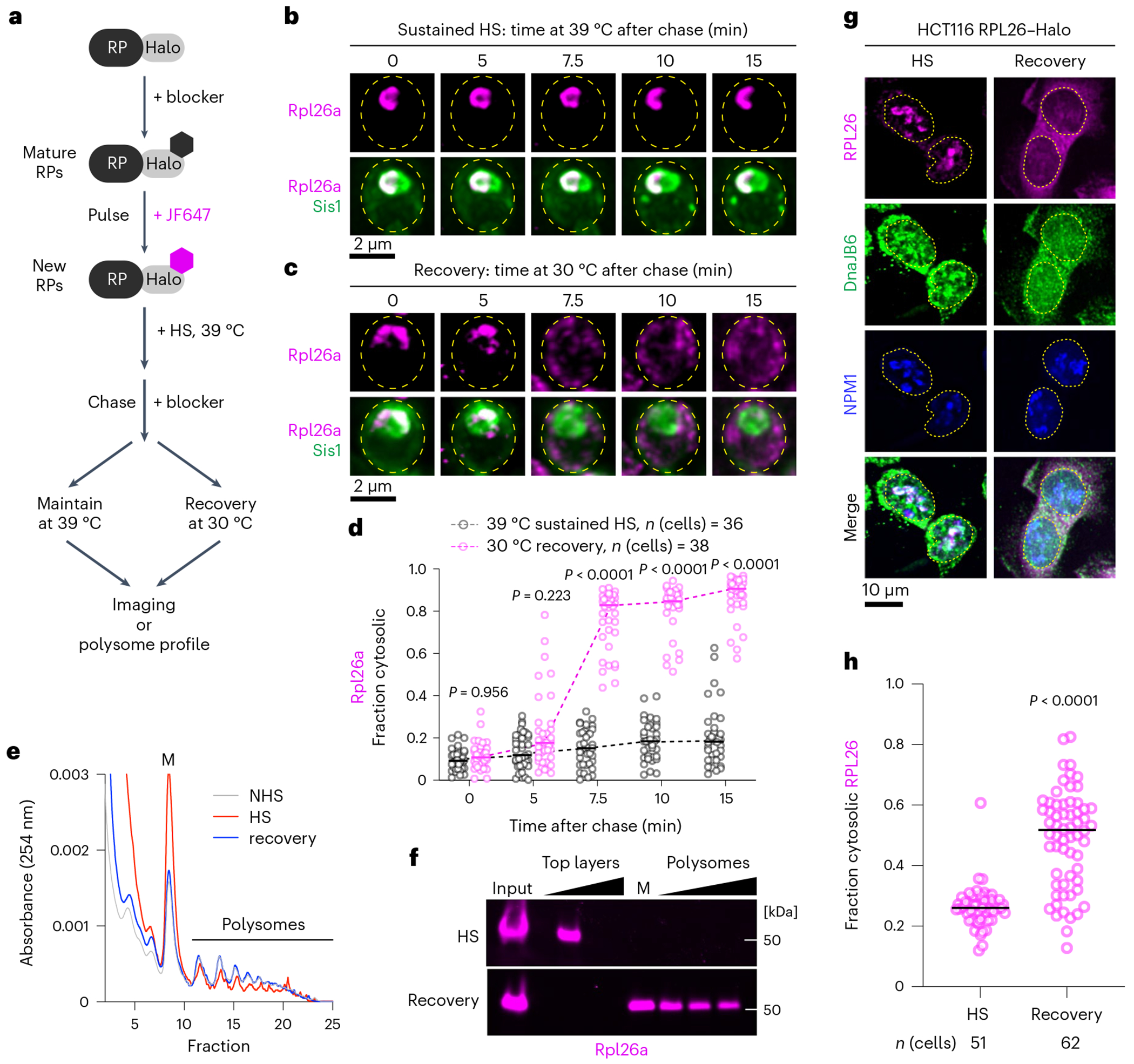
a, Workflow to evaluate the fate of oRPs during sustained heat shock (HS) or recovery. b, Representative live-cell time lapse images of the spatial distribution of oRpl26a (magenta) and Sis1–mVenus (green) during sustained heat shock. c, As in b but following recovery from heat shock. d, Quantification of the fraction of cytosolic Rpl26a signal under sustained heat shock or recovery. Statistical significance was assessed using the Brown–Forsythe and Welch one-way ANOVA test, followed by Dunnett T3 multiple comparison tests. n indicates number of cells pooled from three biologically independent replicates. e, Polysome profiles of yeast expressing Rpl26a–Halo during non-stress, heat shock and recovery conditions. f, In-gel fluorescence of oRpl26a across the polysome profile in heat shock and recovery. g, Localization of oRPL26 (magenta) in HCT116 cells following heat shock and recovery. Cells were fixed and immunostained for DnaJB6 (green) and NPM1 (blue). h, Quantification of the fraction of cytosolic RPL26 in HCT116 cells under sustained heat shock or recovery. P values were calculated with unpaired two-tailed Welch’s t-test. n indicates number of cells pooled from three biologically independent replicates.
Given the cytosolic localization of pulse-labelled RPs following recovery, we hypothesized that the RPs were being incorporated into mature ribosomes. To test this, we performed polysome profiling of lysate from unstressed yeast cells, heat-shocked cells and cells that been heat shocked and allowed to recover. Consistent with previous reports43,44, we observed an increase in monosomes and a decrease in polysomes in heat shocked cells compared with unstressed cells that was completely reversed upon recovery (Fig. 5e). In the heat-shocked sample, we observed pulse-labelled Rpl26a in the top layer fraction of cellular components not incorporated in or associated with ribosomes, consistent with it being an oRP (Fig. 5f). By contrast, Rpl26a sedimented in the heavy polysome fractions in cells allowed to recover after heat shock (Fig. 5f). Together, the cytosolic localization and co-sedimentation with polysomes suggest that RPs orphaned at the nucleolar periphery during heat shock are incorporated into functional ribosomes upon recovery.
oRP condensate reversibility requires Sis1/Hsp70 and promotes fitness
Since Sis1 localizes to the oRP condensates, and Hsp70 activity was required to maintain the liquid-like character of the oRP condensates in lysate, we next asked whether the Sis1/Hsp70 chaperone system was also required for the dispersal of the condensates when cells recover from heat shock. In yeast, we utilized the ‘anchor away’ approach we had previously established to conditionally deplete Sis1 from the nucleus23. We generated RP–Halo strains in the Sis1 anchor away background and either left the cells untreated (Sis1+) or depleted Sis1 from the nucleus (Sis1-depleted) (Extended Data Fig. 9a,b). Then we pulse labelled new Rpl26a and Rps4b, heat shocked the cells at 39 °C, returned the cells to the recovery temperature of 30 °C, and imaged the RPs with respect to the nucleolus over time. In the Sis1+ condition, Rpl26a and Rps4b rapidly dispersed from the nucleolar periphery into the cytosol (Fig. 6a,b and Extended Data Fig. 9e,f). However, in the Sis1-depleted cells, the RPs failed to disperse to the cytosol and remained adjacent to the nucleolus upon recovery (Fig. 6a,b and Extended Data Fig. 9c,d). Supporting a connection between the reversibility of oRP condensates and their liquid-like internal dynamics, analysis of pixel-by-pixel normalized variance showed that new Rpl26a formed amorphous solid-like aggregates in lysate from Sis1-depleted cells (Fig. 6c,d). Consistent with these results, when we inhibited Hsp70 activity in human cells, export of RPL26 to the cytoplasm upon recovery from heat shock was significantly impaired (Fig. 6e,f). These data suggest that Sis1/Hsp70 act at the nucleolar periphery during heat shock to maintain the liquid-like properties of oRP condensates, enabling them to be rapidly dispersed and utilized to restart RiBi upon recovery from stress.
Fig. 6 |. Sis1 and Hsp70 promote oRP condensate reversibility.
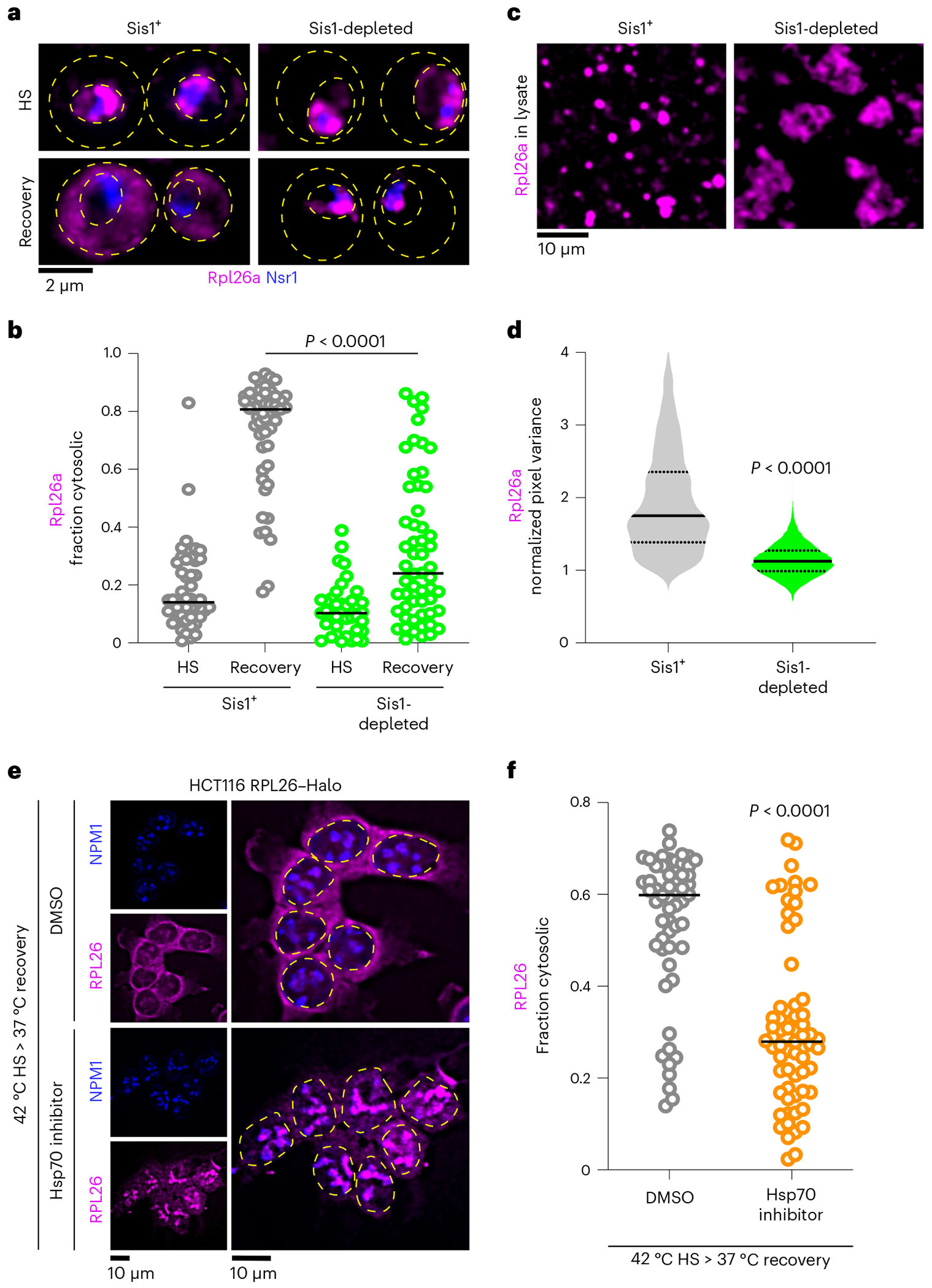
a, LLS imaging of oRpl26a (magenta) and the nucleolar marker Nsr1 (blue) during heat shock and recovery in the absence or presence of Sis1 depletion. b, Quantification of the fraction of cytosolic Rpl26a under sustained HS or recovery in the absence or presence of Sis1 depletion. P values were calculated with unpaired two-tailed Welch’s t-test. n indicates number of cells pooled from three biologically independent replicates. c, Depletion of Sis1 resulted in formation of irregular clumps of Rpl26a in lysate. d, Distribution of normalized pixel variance of Rpl26a in condensates over time in control lysate and lysate with Sis1 depletion. Control lysate distribution replotted from Fig. 4a. P values were calculated with unpaired two-tailed Welch’s t-test. n = 7 and 5 condensates of SIS1+ and Sis1-depleted conditions pooled from three biologically independent experiments. e, Micrograph representing the localization of oRPL26 (magenta) in NPM1-immunostained HCT116 cells following recovery from heat shock pre-treated for DMSO or Hsp70 inhibitor (VER-155008, 50 μM). f, Quantification of the fraction of cytosolic RPL26 from e. P values were calculated with unpaired twotailed Welch’s t-test. n indicates number of cells pooled from three biologically independent replicates.
To determine whether Sis1 would be able to promote dispersal of the solid-like oRP condensates, we performed transient Sis1 depletion during heat shock, releasing Sis1 to return to the nucleus upon recovery (Fig. 7a). Following release and recovery, Sis1 was able to re-enter the nucleus as marked by the nuclear pore protein Nup49 within 30 min, and it immediately homed to co-localize with Rpl26a (Fig. 7b). Notably, as we previously reported, cytosolic Hsp104 foci failed to form during heat shock under Sis1-depleted conditions23, and even upon the release of Sis1, these cytosolic foci of Hsp104 do not appear (Extended Data Fig. 9e,f). Back in the nucleus, despite its co-localization with Sis1, Rpl26a remained condensed for more than 105 min, suggesting that Sis1 was unable to immediately initiate dispersal. However, after 120 min, the Rpl26a signal became cytosolic while showing no appreciable degradation (Fig. 7c). This result indicates that eventually the Sis1/Hsp70 chaperone system promotes dissolution of the solid-like condensates so the constituent RPs can be utilized.
Fig. 7 |. oRP condensate reversibility promotes growth recovery following stress.
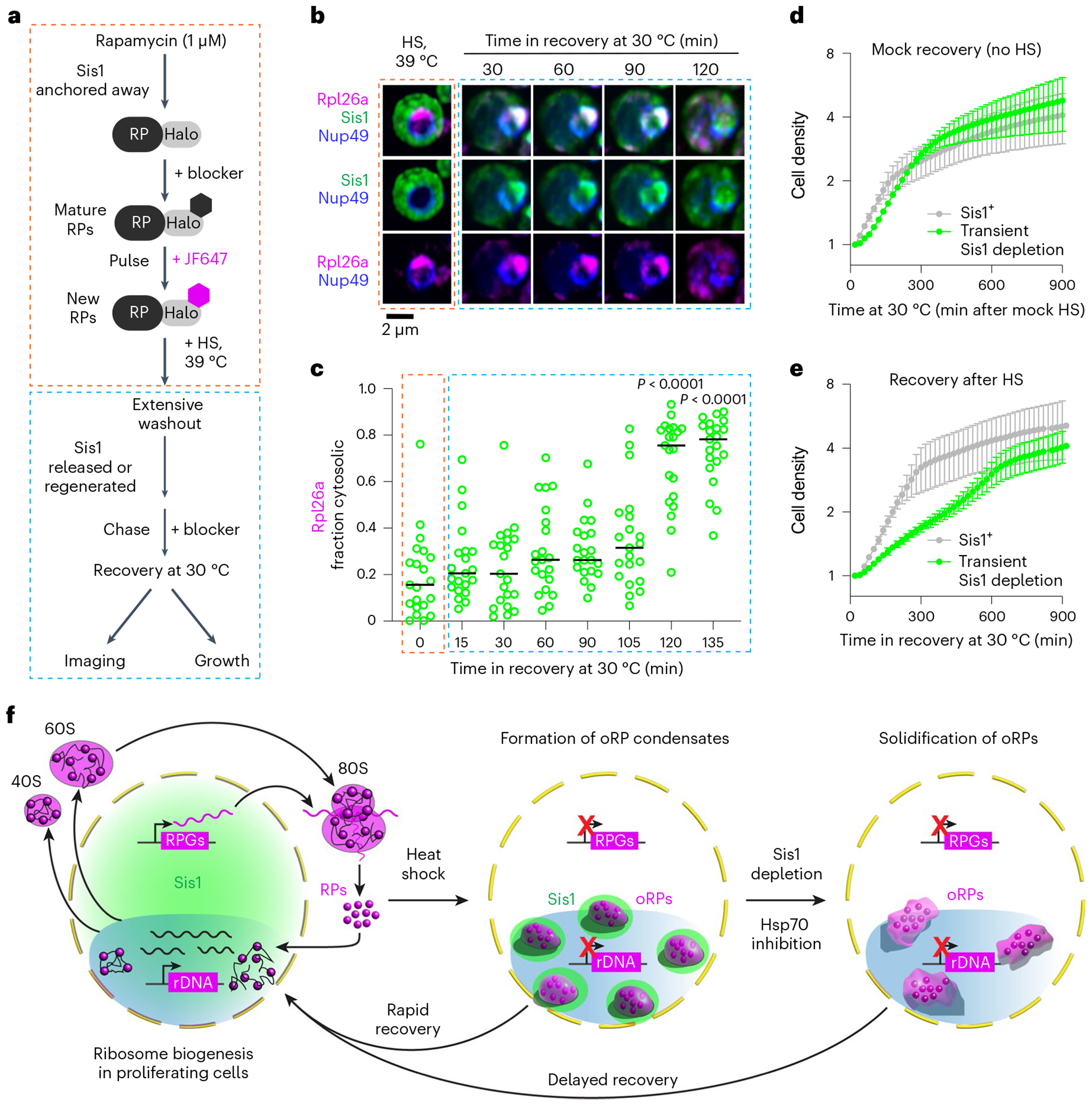
a, Workflow of transient Sis1 depletion in oRpl26a-labelled yeast. b, LLS imaging of oRPl26a (magenta) and the nuclear membrane marker Nup49 (blue), in Sis1 (green) transient depleted condition during the period of heat shock (HS) and recovery. c, Quantification of the fraction of cytosolic oRpl26a from b. n denotes number of cells pooled from three biologically independent replicates. P values were computed using a Brown–Forsythe and Welch one-way ANOVA test, followed by Dunnett T3 multiple comparison tests. d, Growth curve of cells following ‘recovery’ from mock heat shock in the absence or presence of transient Sis1 depletion. Mean and standard deviation of three biological replicates are plotted. e, Growth curve of cells following recovery from heat shock in the absence or presence of transient Sis1 depletion. Mean and standard deviation of three biological replicates are plotted. f, Model of oRP preservation during stress in chaperone-stirred condensates. RPG, ribosomal protein gene.
Finally, we performed growth measurements of cells during recovery from heat shock. We either left Sis1 in the nucleus or transiently depleted it and heat shocked the cells as above. Cells that were mock heat shocked showed no difference in growth in the Sis1+ and Sis1-depleted conditions (Fig. 7d). In the Sis1+ cells following heat shock and recovery, we observed immediate resumption of growth at a constant doubling rate that persisted until the cells plateaued after 300 min (Fig. 7e). By contrast, the cells in which Sis1 was transiently depleted during heat shock but released upon recovery showed slower growth and did not plateau until after 600 min (Fig. 7e). This suggests that the immediate reversibility of oRP condensates contributes to resumption of rapid growth upon recovery from stress (Fig. 7f).
Discussion
Upon exposure to a broad range of environmental stressors, cells rapidly inactivate RiBi by repressing transcription of rRNA and RP mRNAs10–13. Limiting production of ribosomes limits proliferation1, providing a general mechanism for cells to slow growth during stress and liberate resources. Here we show that heat shock triggers the accumulation of oRPs at the periphery of the nucleolus in yeast and human cells. Rather than being degraded or forming toxic aggregates, oRPs are preserved for use upon recovery from stress.
Like stress-induced condensates formed by the stress granule factors Pab1 and Ded1 (refs. 14,45), oRP condensates play an adaptive role in the stress response. For Pab1 and Ded1, the ability to undergo temperature-dependent phase separation has been linked to fitness. In the case of oRPs, potential benefits of condensate formation include protection from degradation and prevention of toxic gain of function effects. Moreover, their reversibility may confer fitness by enabling cells to rapidly resume proliferation. We envision that a sudden buildup of oRPs drives initial condensation, but such assemblies would quickly become amorphous aggregates without Sis1 and Hsp70. The Hsp70 system has been shown to efficiently disperse Pab1 condensates46, stress-induced amyloids of pyruvate kinase47, and transcriptional condensates formed by the HSR regulator Hsf1 (refs. 37,48). The additional role in maintenance of oRP condensates suggests that Sis1 and Hsp70 may broadly surveil and remodel biomolecular condensates during stress.
Basic questions about the composition, properties, and regulation of oRP condensates remain open. For example, the relationship between oRP condensates and the nucleolus–which has itself been interpreted as functioning as a proteostasis compartment28–will be interesting to resolve. Biochemically. it will be particularly interesting to understand whether Hsp70 is acting as a multiple-turnover enzyme that repeatedly disrupts interactions among RPs within the condensates or more as a holdase that blocks exposure of aggregation-prone regions on RPs.
Our data suggest a model in which heat shock inhibits production of rRNA, resulting in the buildup of oRPs on the surface of the nucleolus. Supporting this model, cells are known to repress rRNA production during stress12,13, and inhibition of rRNA production has been shown to lead to accumulation of oRPs and activation of the HSR21,22. While the mechanism by which heat shock inhibits transcription is unclear, it is known to be independent of the HSR49. Given that many nucleolar factors form reversible aggregates during heat shock50, condensation of rRNA biogenesis factors may inactivate rRNA production.
Our data support a direct connection between the biophysical properties of oRP condensates and fitness. We established this connection by depleting Sis1 from the nucleus, which solidified oRP condensates, delayed their reversibility, and slowed proliferation upon recovery from heat shock. Remarkably, when we allow Sis1 to return to the nucleus after the condensates have become solid-like, we find that they remain reversible. By extension, perhaps counteracting the known age-associated loss of chaperone expression in the brain51 may help to sustain condensate liquidity in neurons where loss of liquid-like properties has been associated with neurodegenerative disease52,53. In healthy cells, oRP condensates define proteostasis hubs that play an adaptive role in the stress response. The mechanisms that sustain the liquid-like properties of oRP condensates could function more broadly to prevent–and perhaps be therapeutically harnessed to reverse–the transformation of adaptive condensates into maladaptive aggregates.
Methods
Yeast strain construction and cell growth
Yeast strains used in this study are catalogued in Supplementary Table 1. All strains are derivatives of W303, and fluorescent protein and epitope tags are integrated into the genome using scarless CRISPR–Cas9 gene editing unless otherwise stated. Single guide RNA (sgRNA) was designed using CRISPOR tool. All sgRNA and repair template oligo used are listed in Supplementary Table 4. Yeast was cultured in rich glucose medium (YPD) or appropriate synthetic complete drop-out medium at 30 °C. For lattice light-sheet imaging, cells were cultured in synthetic dextrose complete (SDC) media without riboflavin and folic acid to minimize autofluorescence.
Pulse labelling of Halo-tagged RPs (yeast)
To visualize newly synthesized RPs, corresponding strains with RPs fused with HaloTag7 (codon optimized for Saccharomyces cerevisiae) were grown to (optical density) OD600 0.3–0.6. Cells were incubated with non-fluorescent Halo tag ligand 7-bromo-1-heptanol (7BRO, 100 μM, VWR #AAH54762) for 10 min to irreversibly mask pre-existing RPs. After three washes with fresh medium (5× volume, 1 min shaking), cells were pelleted, redissolved to original volume and incubated with Janelia Fluor 646 Halo tag ligand (JF646, 1 μM, Promega #GA1120) at 30 °C or 39 °C for 7 min. Subsequently, cells were washed (5× volume) with pre-warmed medium (30 °C/39 °C) for additional 3 min before imaging. For mature RP labelling, yeast was incubated with 10 μM JF646 Halo dye for 10 min. Excess dyes were washed away, and further labelling with dye is prevented by addition of 100 μM 7BRO.
Acute depletion of Ifh1
Genomic IFH1 was fused in frame with mini auxin-inducible degron (miniAID) at its C termini using CRISPR–Cas9 gene editing. Oestradiol-inducible OsTIR1(F74G) was introduced at the leu2 locus. Acute and non-leaky depletion of Ifh1 is achieved by co-treatment with β-oestradiol (1 μM, Sigma #E8875) and 5ph-IAA (5 μM, MedChem Express #HY-134653) for 30 min.
RNA sequencing
Sequencing of polyA+ mRNA from yeast was performed at the North-western University Genomics Core and analysed as described23,54.
Transient Sis1 depletion and quantitative growth assay
For anchoring away Sis1–FRB in a rapamycin-resistant background (TOR1-1 fpr1Δ) with Rpl13a–FKBP, 1 μM rapamycin for 10 min (Sigma #R8781) was applied for imaging experiments23. For growth assay biological triplicates of Sis1–FRB/Rpl13a–FKBP strain were grown overnight at 30 °C in 2× SDC media, then diluted to OD600 0.05. After 5 h at 30 °C to reach OD600 0.2, each replicate was split into two groups: (1) DMSO-pretreated or (2) rapamycin-pretreated (1 μM, 10 min) followed by heat shock (39 °C, 20 min). Samples were quickly washed, resuspended in 2× SDC (final OD600 0.1), and OD600 measured every 20 min for 24 h at 30 °C using an automated plate reader.
Human cell culture
HCT116 and HCT116–RPL26–Halo (kind gift from H. An and W. Harper) were grown in Dulbecco’s modified Eagle medium (high glucose and pyruvate) supplemented with 10% (vol/vol) foetal bovine serum, 1× GlutaMAX and 100 units of penicillin and streptomycin, and maintained in a 5% CO2 incubator at 37 °C. All cell lines were found to be free of mycoplasma using the MycoAlert PLUS Mycoplasma Detection Kit (Lonza #LT07-703). Heat shock with conditioned media was performed essentially as described55. Two identically grown cell plates at around 70–80% confluence, were used. Conditioned media from the first plate was collected, heated to 43 °C and then used to replace the medium on the second plate, which was placed in a 43 °C incubator with 5% CO2.
Pulse labelling of Halo-tagged RPs (human cell lines)
To visualize newly synthesized RPL26 cells, pre-existing RPL26 were masked by incubation with non-fluorescent irreversible Halo ligand (7BRO, 100 μM) for 15 min. Cells were washed with complete Dulbecco’s modified Eagle medium three times under shaking for 2 min. Cells were then treated with JF646 Halo dye (1 μM) mixed with pre-warmed conditioned medium and heat shocked as indicated. Conversely, to label pre-existing RPL26, cells were incubated with JF646 Halo dye (10 μM) for 15 min. After extensive washing, further labelling was prevented by addition of 100 μM 7BRO.
Immunofluorescence of HCT116 cells
HCT116 RPL26–Halo cells were seeded at 0.8 × 105 cells ml−1 on polylysine-coated coverslips 30 h before experiment. Cells were fixed (4% paraformaldehyde and 4% sucrose), quenched (125 mM glycine) and then permeabilized with 0.1% Triton X-100. DnaJB6 was stained by incubating overnight with 1:100 dilution polyclonal rabbit DnaJB6 antibody (ThermoFisher #PA5-27577). Nucleolus was stained by incubating with 1:1,000 dilution of mouse monoclonal NPM1 antibody (ThermoFisher #FC-61991). Cells were then washed with 1× phosphate-buffered saline and fluorescently labelled with goat–anti-rabbit 488 (Invitrogen #A32731) and goat–anti-mouse 594 secondary antibodies (Invitrogen #A-11032,). Cells were then washed with 1× phosphate-buffered saline, mounted on microscope slides using ProLong Gold Antifade Mountant with 4’,6-diamidino-2-phenylindole (DAPI; Invitrogen #P36930) and imaged the following day. Images were taken using a 3i Marianas Spinning Disk Confocal with 100× oil objective.
Lattice light-sheet imaging and analysis
Lattice light-sheet imaging of live yeast was executed using a phase 2 system manufactured by Intelligent Imaging Innovations (3i) and run with SlideBook 6.0 software. The design is a commercially produced clone of the original56. The imaging camera used was a Hamamatsu Fusion chilled sCMOS with annulus mask set for a 20-μm beam length (outer numerical aperture (NA), 0.55; inner NA, 0.493) with 400-nm thickness, with dither set at 9 μm. Temperature was controlled by a built-in Peltier device (empirically set using a probe thermometer to indicated temperatures). Optics were aligned daily before the experiment, and the bead PSFs were collected before imaging to ensure that the setup has consistent resolution in XYZ. Graphics processing unit-based Richardson–Lucy deconvolution was with measured PSFs via Brian Northan’s ‘Ops’ implementation57. Three-dimensional reconstructions and videos were assembled using MPI-CBG developed ClearVolume plugin of Fiji58,59.
APEX2 proximity labelling
Sis1–Apex2 strains grew in 300 ml SDC medium to OD600 0.3–0.6. Heat shock at 39 °C for 15 min was followed by fixation with 1% paraformaldehyde, and quenching with 125 mM glycine. Cell walls were lysed in KpiS buffer with 100T zymolyse, and membrane was permeabilized with 0.1% Triton X-100. Spheroplasts were incubated with 0.5 mM biotin phenol and 1 mM hydrogen peroxide for 20 min, which activated APEX2. For background correction, one set without peroxide treatment was identically processed. Reactions were quenched with a cocktail of Trolox (2.5 mM), sodium ascorbate (10 mM) and sodium azide (10 mM). Cells were cryo-lysed, and proteins were precipitated with methanol. Precipitates dissolved in 2% sodium dodecyl sulfate (SDS) RIPA Buffer, and biotinylated proteins captured with streptavidin beads. Beads were washed and eluted with hexafluoro isopropanol.
Sis1 co-IP from cells labelled for mature and new RPs
Corresponding Rp–Halo strains with Sis1–3xFlag were grown to mid-log phase in 300 ml SDC medium. Cells were labelled for mature or new RPs (as in pulse labelling section). and kept unstressed or heat shocked. After treatment, cells were vacuum filtered, flash frozen (liquid nitrogen) and cryo-lysed. Samples were dissolved 1:2 w/v in physiological buffer (as mentioned above). Sis1–3xFlag was immunoprecipitated using anti-Flag M2 magnetic beads (Sigma #M8823) from the cell lysates. Bead-bound complexes were released using 3xFlag peptide (Sigma #F4799), resloved on SDS–polyacrylamide gel electrophoresis gel and visualized using a fluorescence gel imager (Invitrogen iBright CL1500). Gels were then blotted and probed with anti-Flag antibody (Sigma #F3165).
Proteomics sample preparation
Samples underwent SDS–polyacrylamide gel electrophoresis and gel bands were in-gel digested. Gel bands were washed with 100 mM ammonium bicarbonate/acetonitrile, reduced with 10 mM dithiothreitol (50 °C, 45 min) and alkylated with 100 mM iodoacetamide (50 °C, 45 min). Trypsin (Promega #V5111) digestion occurred at 37 °C, followed by peptide extraction using varying acetonitrile concentrations. Peptides were dried and then reconstituted for liquid chromatography–MS/MS.
Liquid chromatography–MS/MS analysis
Performed at Northwestern University Proteomics Core using Dionex nanoLC-QE mass spectrometer. Peptides were analysed with a gradient elution, and MS/MS was recorded in positive-ion mode. Mascot was used for protein ID with modifications considered. A 1% false discovery rate cut-off was applied, and Scaffold software was used for visualization.
Proteins were identified from the MS raw files using the Mascot search engine (Matrix Science, version 2.5.1). MS/MS spectra were searched against the SwissProt Saccharomyces cerevisiae database. All searches included carbamidomethyl cysteine as a fixed modification and oxidized methionine, deamidated asparagine and aspartic acid, and acetylated N-term as variable modifications. Three missed tryptic cleavages were allowed. A 1% false discovery rate cut-off was applied at the peptide level. Only proteins with a minimum of two peptides above the cut-off were considered for further study. Identified peptides/protein were visualized by Scaffold software (version 5.0, Proteome Software).
Polysome profiling
Polysome analyses by sucrose gradient fractionation were performed as described60. Rpl26a–Halo strains were cultured in SDC medium to mid-log phase. Pre-existing ribosomes were masked by incubating with 7BRO (100 μM, 10 min). After washing cells three times with fresh medium (2 min shaking), newly synthesized Rpl26a were labelled using JF646 Halo ligand (2 μM). Heat shock at 39 °C for 10 min occurred by mixing cells with an equal volume of 50 °C medium. Half of the cells were vacuum filtered to denote the heat shock sample. The other half was allowed to recover at 30 °C with 7BRO blocker (100 μM) for 30 min before vacuum filtering to create the recovery sample. Cell pellets were flash frozen, cryo-lysed and resuspended in lysis buffer (20 mM HEPES-KOH (pH 7.4), 100 mM KCl, 5 mM MgCl2, 200 μg ml−1 heparin (Sigma #H3149), 1% Triton X-100, 0.5 mM tris(2-carboxyethyl)phosphine, 100 μg ml−1 CHX, 20 U ml−1 superase-IN (Invitrogen #AM2696) and 1:200 Millipore protease inhibitor IV #539136). The lysate was clarified, and the supernatant was frozen in aliquots. A sucrose gradient (10–50%) in polysome gradient buffer (5 mM HEPES–KOH (pH 7.4), 140 mM KCl, 5 mM MgCl2, 100 μg ml−1 CHX, 10 U ml−1 superase-in and 0.5 mM tris(2-carboxyethyl) phosphine) was prepared in SW 28.1 tubes. Lysate was layered onto the gradient and spun. Fractions were collected, frozen and normalized to the 40S peak. For in-gel fluorescence, samples were treated with 0.02% sodium deoxy cholate, TCA precipitated, washed with ice-cold acetone, and resuspended in 2× Laemmli sample buffer.
Cell-free lysate droplet assay
Corresponding newly synthesized or matured RP-stained yeast cultures were heat shocked, filter collected, cryo-milled and dissolved in 1:2 (w/v) in physiological buffer (20 mM NaCl, 50 mM KCl, 150 mM K+ glutamate, 50 mM HK2PO4, 0.5 mM MgCl2, 25 mM HEPES–KOH, 25 mM MES pH 7.4, 5 mM phenylmethylsulfonyl fluoride and 1:200 Millipore Protease Inhibitor Cocktail). Cellular debris and un-lysed cells were clarified by centrifugation at 3,500g for 5 min. Cleared lysates were immediately loaded onto a homemade microfluidic chamber comprising a glass slide or coverslip with a top coverslip attached by two parallel strips of double-sided tape to form a single central channel61. Slides were then imaged with a custom wide-field microscope with high-NA objective (Olympus PlanApo N 60×/1.42), excited with a 637 nm continuous laser at ~10 W cm−2 (Coherent Obis LX) and sCMOS camera detection (Photometrics Prime95B) at 66 fps.
Sis1 nucleolar proximity analysis
From the maximum intensity projections, nuclei were segmented by drawing oval masks around peri-nuclear ring of Sec61–Halo (Fiji). This was used as an input to custom-written Python code, which carried out the rest of the analysis. For each nucleus, the geometric centre was computed from the mask, and a family of lines drawn at varying angles to bisect the nuclear area into two halves. For each nucleus, the fold change in Nsr1 and Sis1 intensity between the two halves was computed, and the angle with a maximal fold change in Nsr1 intensity was identified as splitting the nucleus into Nsr1-proximal and Nsr1-distal. The fold-change in Sis1 signal along the bisecting angle was recorded for each nucleus.
Yeast cytosol image segmentation
Deconvolved z-stack images were subjected to maximum intensity projection. For whole cell segmentation, the minimum error thresholding (MET) with watershed algorithm was applied using the Sis1 signal. Yeast nucleoli were segmented using the Nsr1 signal. The nucleolar mask underwent binary dilation to cover the proximal region. By subtracting nucleolar and peri-nucleolar signals from whole cell signal, cytosolic signals were isolated. Cytosolic signal fraction was determined by dividing whole cell signal’s integrated density by cytosol signal.
Human cell cytosol image segmentation
The nuclei were masked and segmented using DAPI signal with Li-threshold, watershed and fill hole algorithm in Fiji. DAPI masks were binary dilated for whole cell segmentation. Cytosolic RPL26 signal fraction was determined by dividing whole cell integrated density by cytosol integrated density.
Co-localization analysis
For quantifying co-localization of Sis1 and RP signals, the Coloc2 plugin in Fiji software was used. A mask was generated for each cell using the MET with watershed algorithm, utilizing the Sis1 signal. Automatic Otsu thresholding segmented high-intensity peri-nucleolar Sis1 and cytosolic foci from low-intensity diffuse signal. The Mander’s overlap coefficient was then calculated to assess the extent of overlap between the mature or new form of RP and Sis1 signals. For human cells, Huang thresholding with fill hole and watershed algorithms created a mask from DAPI signal for nuclei, followed by binary dilation to segment the cytosol. ROIs were used to calculate Mander’s overlap coefficient for RPL26 (mature or new form) and DnaJB6 signals.
Bead tracking
We utilized FluoSpheres Carboxylate-Modified Microspheres, 0.2 μm in size, with a yellow-green, fluorescent marker (505/515) (Thermo Fisher Scientific #F8811). The microspheres were mixed with cell lysate at a 500× dilution. During the process of droplet formation, fusion and settling onto the cover slide, beads become randomly distributed within the droplets. To visualize the beads within nsRP condensates, we employed the same wide-field microscope as described above for the cell-free lysate droplet assay. The condensates were excited using continuous wave lasers at wavelengths of 488 nm and 637 nm, and the resulting images were captured in the red and green colour channels. To adjust for the brightness of the beads, an OD filter was employed, setting the intensity of the 488 nm laser to approximately 1 W cm−2. Videos were of 15-s duration (exposure time of 50 ms).
To align the red and green colour channels, beads with two different colours were imaged, and their centroid positions were determined by fitting two-dimensional Gaussian functions to their intensity profiles in each channel. These localizations were used to generate an affine matrix transformation, which was then applied to the bead tracking data to visualize beads (green channel) co-localized with condensates (red channel).
Only beads that remained within the boundaries of a condensate for all 300 frames were included in the analysis. Due to diffusion in the Z axis, during some frames beads drifted out of focus before re-appearing. Bead locations were identified through thresholding and centroid localization for groups of three or more adjacent pixels, applied in ImageJ. Centroids were calculated from the binary mask for each frame containing a bead using the arithmetic mean of the zeroth order moment of the array.
To assess bead movement, the MSD of the bead centroid was calculated using the following equation62:
In the equation, time lags τ range from 50 ms up to 1 s, and n(τ) defines the number of frames corresponding to time lag τ. Cx(i) and Cy(i) represent the coordinates of the bead centroid in frame i, τ is the time lag corresponding to a lag of n frames, and N is the total number of frames. The MSD(τ) vector was computed for each bead, and the average MSD value for each time lag was determined on the basis of data from seven beads per treatment condition. These values were converted to μm2 and plotted against τ. A linear model was fitted to the data using the Statsmodels module in Python. The slope of the line, measured in μm2 s−1, corresponds to four times the diffusion coefficient (4 × D) according to the Einstein–Smoluchowski equation MSD = 4Dτ, where D is the diffusion coefficient of the particle. For a 200-nm bead in water at 20 °C, the expected diffusion coefficient is 2.145 × 10−12 m2 s−1 based on the Stokes–Einstein equation D = kBT/(6πηr). Any decrease in D, provided that the temperature T and bead radius r remain constant, indicates an increase in the effective viscosity of the material (ηeff).
Condensate pixel variance analysis
To assess condensate dynamics, high-frame-rate video was captured and fluctuation in brightness variance over time were measured. A custom Python script analysed condensate videos with consistent illumination conditions. Pixel count values were converted to photon counts according to a previously calibrated gain matrix unique to the sCMOS camera used (Photometrics Prime95B). Converted videos were cropped to include only areas within the condensate. For each pixel at location (i,j) in the condensate, normalized pixel variance over time was calculated from the photon counts observed in each of N frames, xij(t) and the expected variance of the signal according to a Poisson distribution of photon counts, .
The resulting two-dimensional array of normalized variances for each condensate, , was generated and displayed as a heat map. Normalized variance values from multiple condensate videos of each condensate type were aggregated to enable robust comparison among treatment conditions, displayed as a violin plot. The expected normalized variance for solid objects is 1, since any variability in the signal would be due to shot noise generated from photon counting by the detector. Similarly, fluctuations above the expected shot noise variance would have values greater than 1, indicating that the object is exhibiting fluctuations in addition to those predicted from shot noise.
Statistics and reproducibility
Experiments were conducted using two to five biological replicates and were independently reproduced as indicated in the figure legends. While statistical methods were not employed to pre-determine sample sizes, our sample sizes closely resembled those reported in prior publications33,47,63. Although we assumed a normal distribution of the data, formal testing for this assumption was not carried out and individual data points are reported. While the investigators were aware of the experimental design, the execution of data analysis and acquisition was performed by separate individuals throughout the procedure and data analysis.
For graphs with two conditions, the significance of pairwise comparisons was assessed through an unpaired two-tailed Student’s t-test with Welch’s correction. In cases where graphs displayed three or more conditions, the significance of pairwise comparisons with their corresponding controls was calculated using a Brown–Forsythe and Welch’s one-way analysis of variance (ANOVA). Subsequently, post-hoc Dunnett’s T3 test (when n < 50 per group) or Games–Howell multiple comparisons test (when n > 50 per group) was applied, considering individual variances.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Extended Data
Extended Data Fig. 1 |. Sis1 localization and interactions during heat shock.
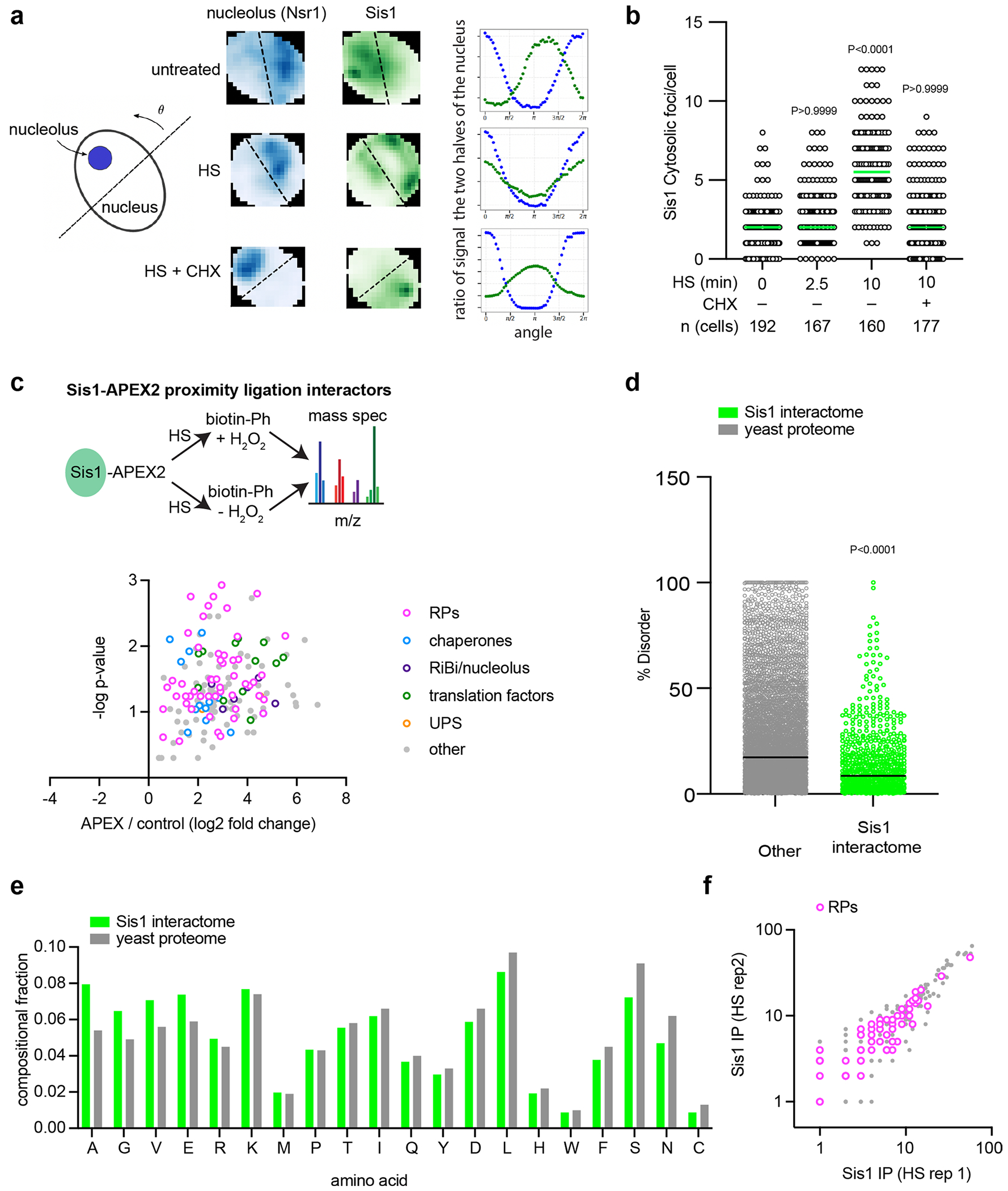
(a) Left: Schematic of how to bisect the nucleus with the nucleolus on one side by finding the line angle with the maximum difference in signal of the nucleolar marker in the two halves. Middle: Representative 2D projections of cells showing Nsr1 (blue) to mark the nucleolus and Sis1 (green). Line is set to maximize the difference in nucleolar signal and the ratio of Sis1 in the two halves is calculated. Right: Sis1 ratio as a function of the line angle rotated as depicted in the schematic to the left. (b) Quantification of Sis1 cytosolic foci per cell in the conditions listed. Foci were identified using the FindFoci plugin in ImageJ. Statistical significance was determined by Brown-Forsythe and Welch one-way ANOVA test followed by Games-Howell multiple comparison tests. n obtained from 3 independent experiment. (c) Volcano plot of Sis1-APEX2 interactors during heat shock. (d) Scatter plot showing the percentage of disorder in Sis1 interactors relative the whole proteome. P values were calculated with unpaired two-tailed Welch’s t-test. n = 2 biological replicates. Each dot symbolizes individual proteins, with ‘n’ representing 5151 proteins for the entire yeast proteome and 731 proteins for the Sis1 interactors induced by heat shock. Data is representative of 2 biologically independent experiments. (e) Bar plots representing the amino acid sequences enrichment of the Sis1 interactors compared to the yeast proteome. Data is representative of 2 biologically independent experiments. (f) Biological replicates of Sis1-3xFlag IP interactors.
Extended Data Fig. 2 |. Interaction and localization of pulse-labeled ribosomal proteins with Sis1.
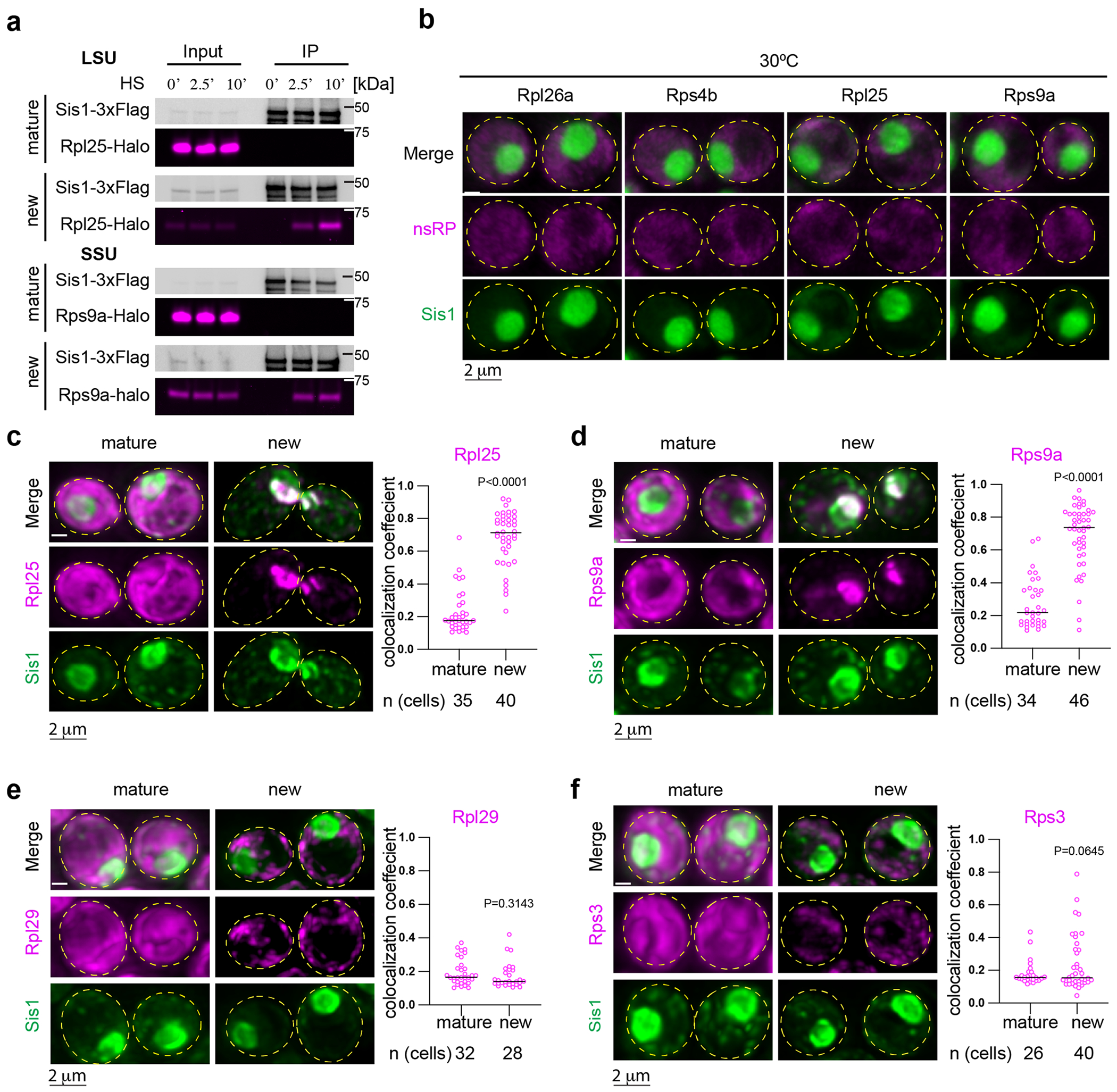
(a) IP of Sis1-3xFlag and either mature or new Rpl25-Halo and Rps9a-Halo from cells left unstressed or heat shocked at 39 °C for the indicated times. n = 2 biologically independent experiment. (b) In the absence of heat shock, pulse-labeled RPs localize immediately to the cytosol. Micrograph represents data obtained from 3 biologically independent experiments. (c) Left Panel: Lattice light sheet live imaging of yeast under heat shock (39 °C, 10 min) expressing Sisl-mVenus and labeled for either new or mature Rpl25-Halo. Right Panel: Dot plot representing the colocalization coefficient (Mander’s overlap coefficient) of Sis1-mVenus with either mature or new Rpl25-Halo in heat shocked cells (39 °C, 10 min). n = number of cells pooled from 3 biologically independent replicates. (d) As in (c) but for Rps9a-Halo. n = number of cells pooled from 3 biologically independent replicates. (e) As in (c) but for the latejoining subunit Rpl29-Halo. n = number of cells pooled from 3 biologically independent replicates. (f) As in (c) but for the late joining subunit Rps3-Halo. n = number of cells pooled from 3 biologically independent replicates. P values were calculated with unpaired two-tailed Welch’s t-test.
Extended Data Fig. 3 |. Localization of pre-60S ribosome biogenesis factors during heat shock.
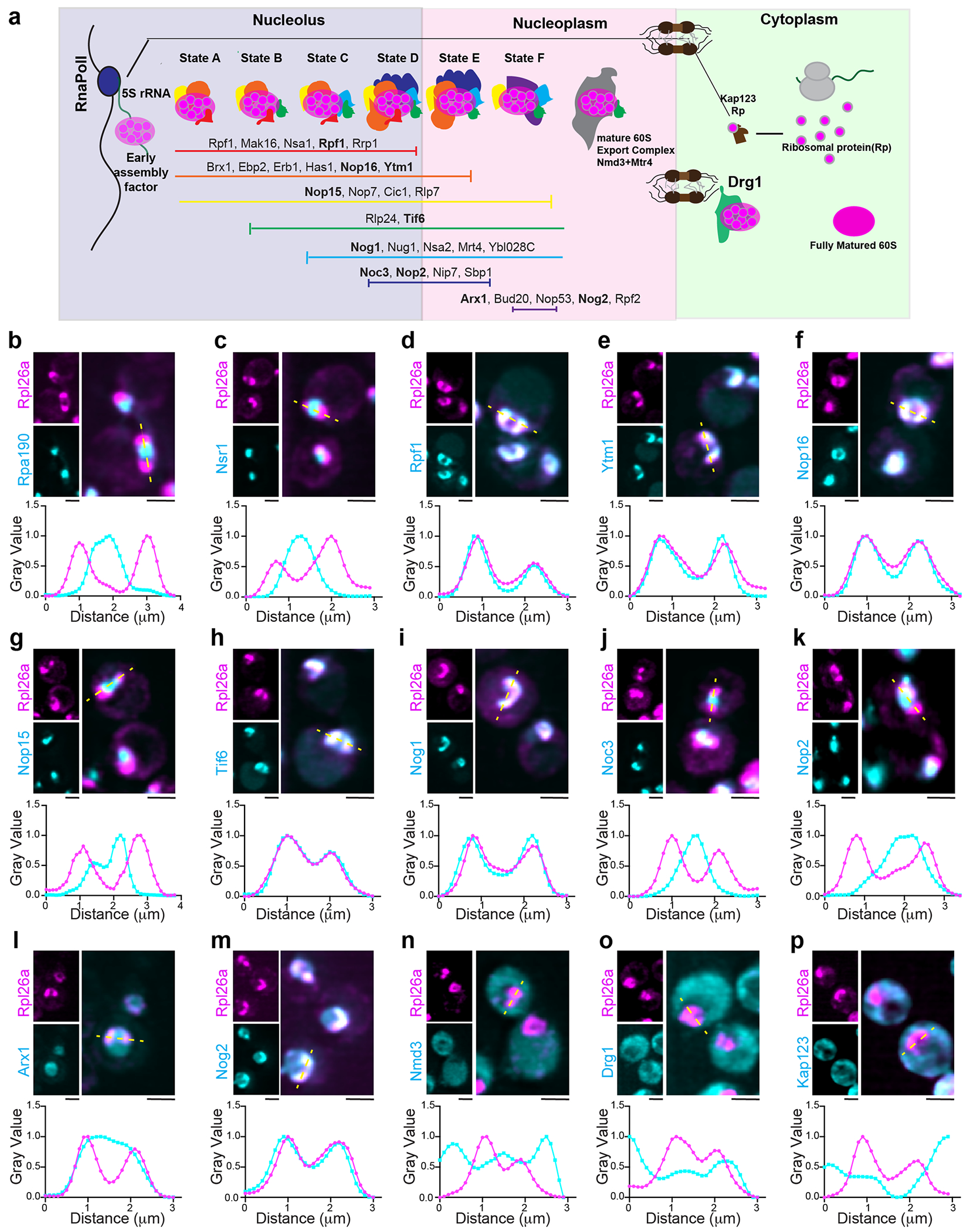
(a) Illustrate showing the association of assembly factors with various states of pre60S maturation. Clustering and coloration in the diagram indicate the time points of stable association and dissociation from the maturing particle, as denoted by the horizontal lines. (b-p) LLS imaging of Live cells representing the localization of oRpl26a during heat shock in context of pre-60S ribosome assembly factors as depicted in (a). Scale bar = 2 μm. Inset shows the normalized line scan graph of representing assembly factors across the oRpl26a signal.
Extended Data Fig. 4 |. Cell biological and transcriptional effects of Ifh1 depletion during heat shock.
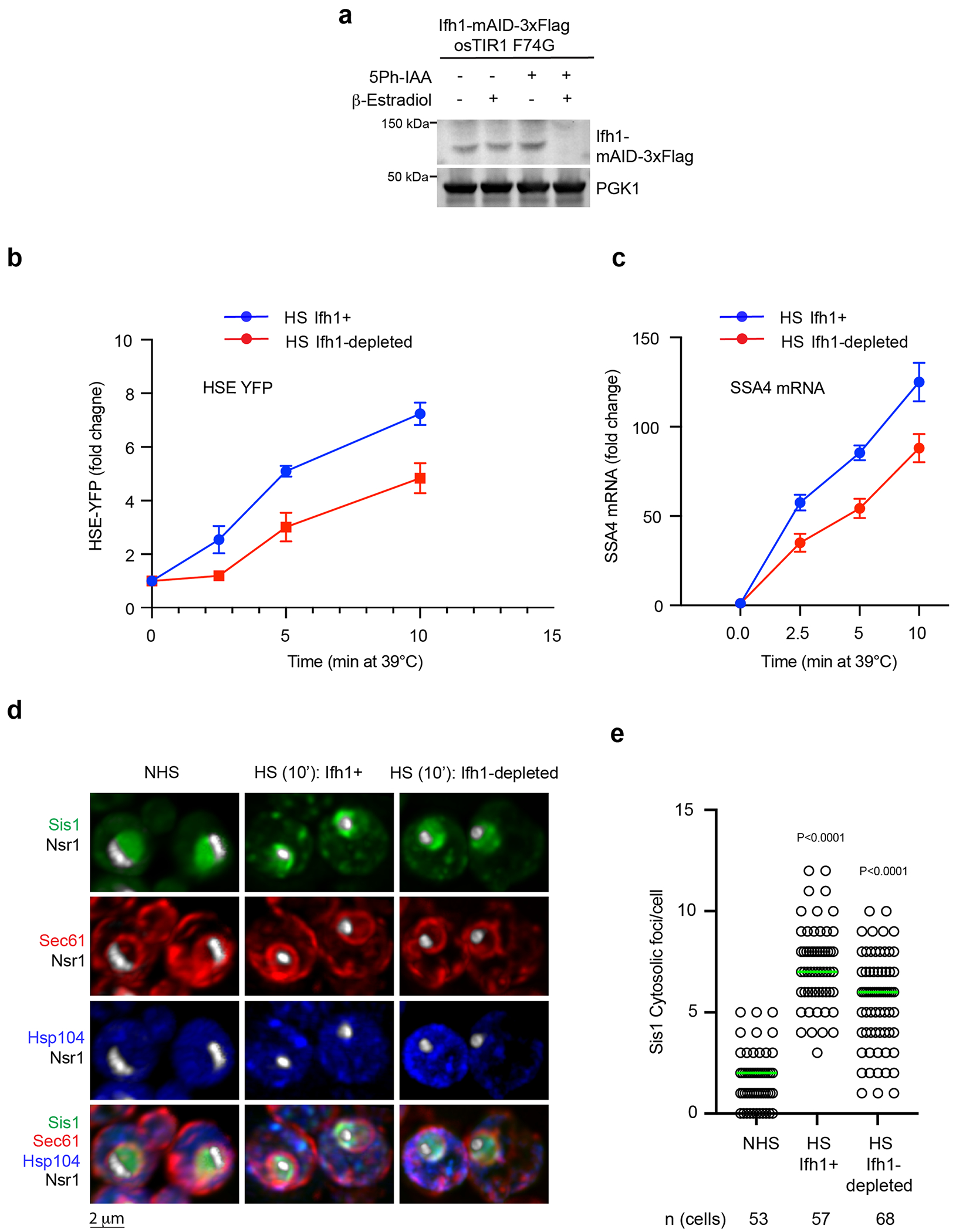
(a) Immunoblot showing of the level of Ifh1-mAID-3xFlag upon incubation with 5ph-IAA and β-estradiol. PGK1 level is used as loading control between the samples. (b) HSE-YFP reporter heat shock time course showing reduced HSR induction when Ifh1 is depleted. Data are presented as mean ± S.D. n = 3 biologically independent sample. (c) RT-qPCR of the HSR target gene transcript SSA4 over a heat shock time course in the absence and presence of Ifh1 depletion. Data are presented as mean ± S.D n = 3 biologically independent sample. (d) LLS live-imaging of yeast cells with endogenously tagged Sis1-mVenus (green), Hsp104-TFP (blue), Sec61-Halo (red) and Nsr1-mScarlet-I (white) under non-stress (30 °C) and heat shock (39 °C, 10 min) in the absence and presence of Ifh1 depletion. (e) Quantification of Sis1 cytosolic foci per cell in the conditions shown in (d). Statistical significance was assessed using the Brown-Forsythe and Welch ANOVA test, along with Games-Howell multiple post hoc comparisons. n denotes number of cells from 3 independent experiment.
Extended Data Fig. 5 |. oRP condensates are stable and heat shock-dependent.
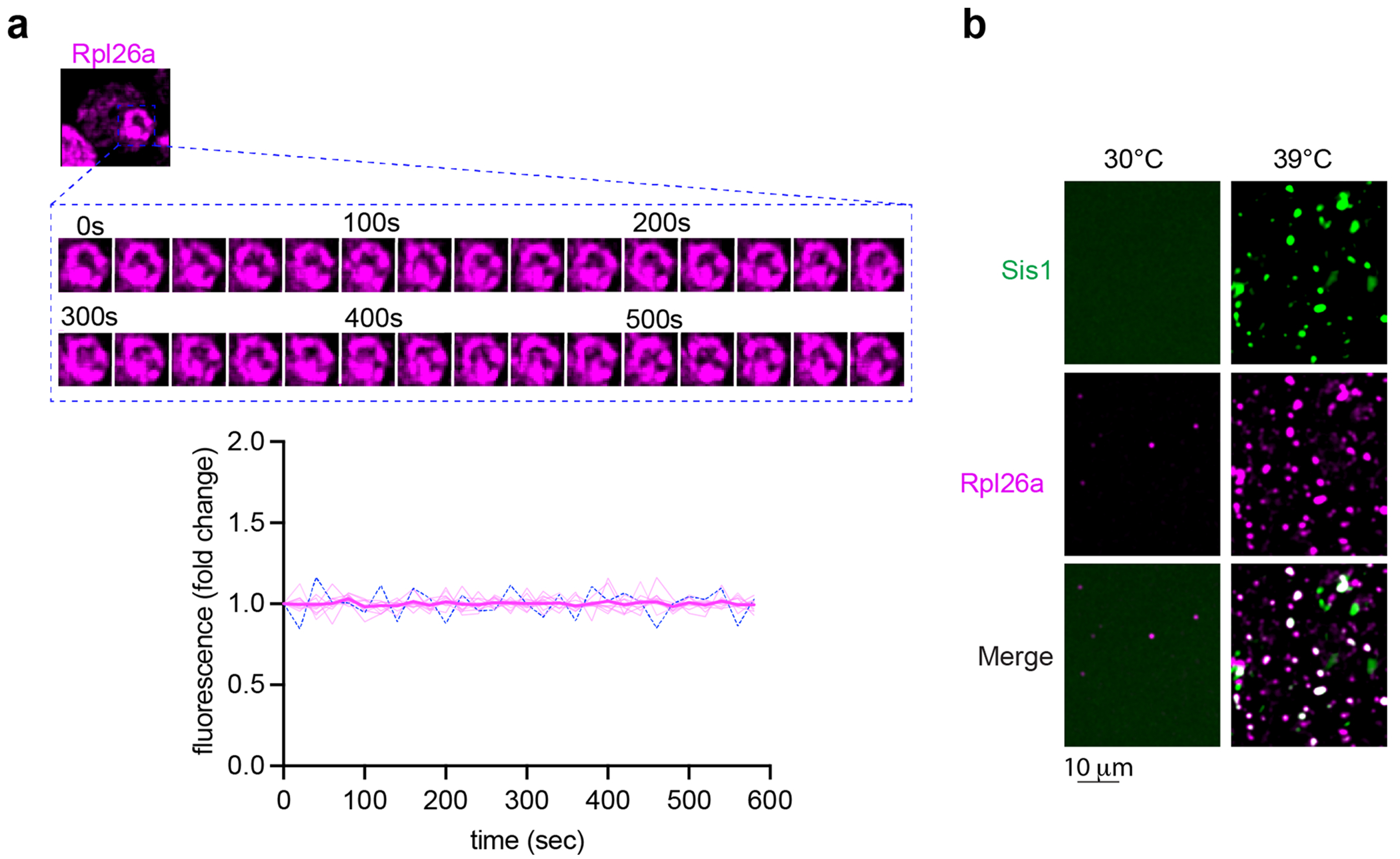
(a) oRP proteins are stable (not degraded) in condensates in cells. (b) oRP condensates are more abundant in lysate from heat-shocked cells. Micrograph represents data of 3 biologically independent experiments.
Extended Data Fig. 6 |. Temperature scan and RNA assessment of oRP condensates.
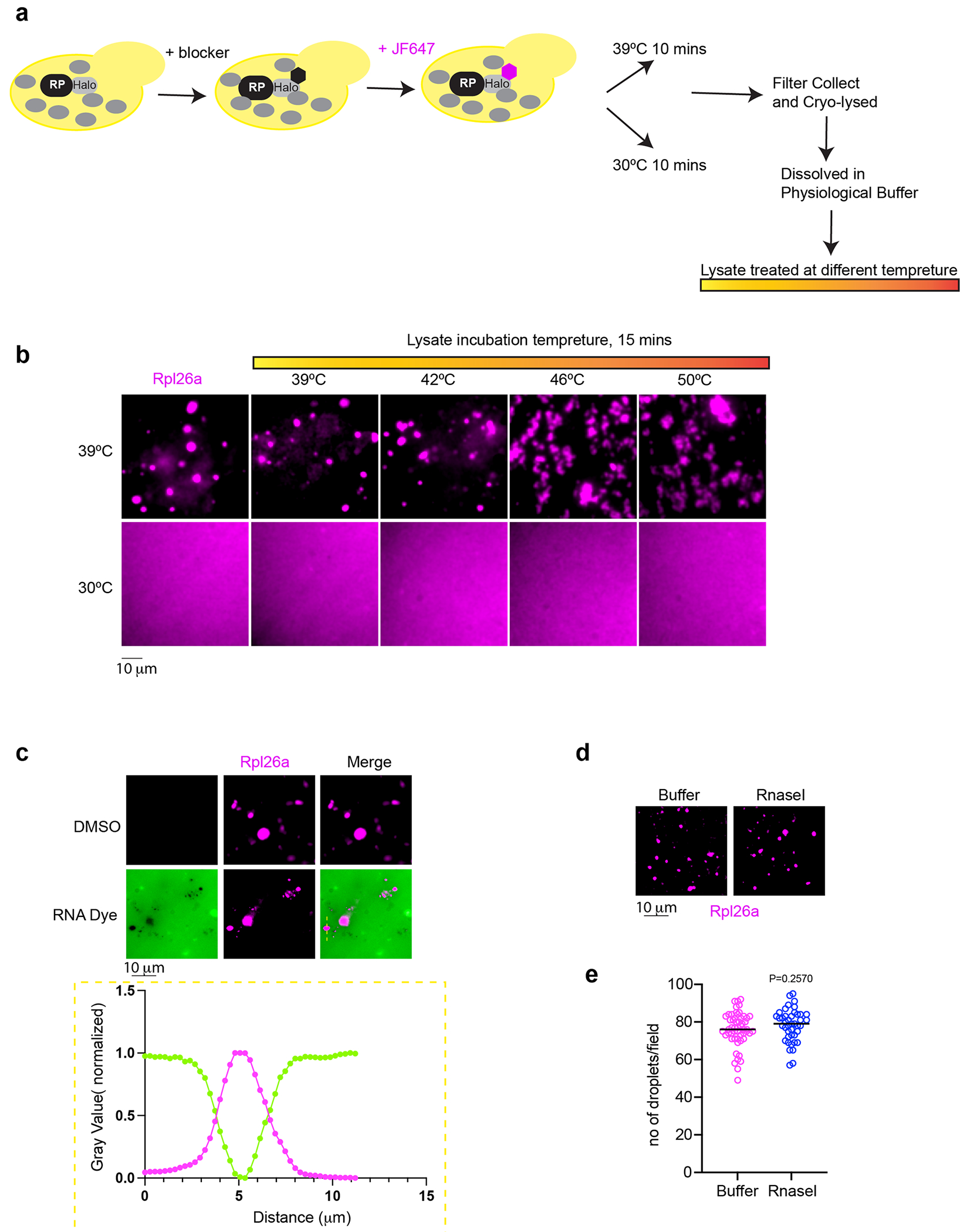
(a) Illustrate depicting the workflow to label newly synthesized RP in yeast and lysate preparation to conduct temperature scan. (b) Micrograph of oRP condensate prepared from non-stressed or heat shocked yeast and upon incubation at indicated temperature. n = 3 biologically independent experiments. (c) RNA dye (SYTO RNASelect, 0.5 mM, 10 min) is excluded from the oRP condensate. n = 3 biologically independent experiments. (d) oRP condensates are resistant to RNaself (5units/μl, 15 min, 25 °C). (e) Quantification of number of droplets per field in buffer or RNaself treatment to the lysate. P values were calculated with unpaired two-tailed Welch’s t-test. n is representing number of droplets quantified in microscopic field of 53 and 42 for Buffer and RNasel conditions respectively.
Extended Data Fig. 7 |. Effect of Hsp70 inhibition and hexanediol on oRP condensates in lysate.
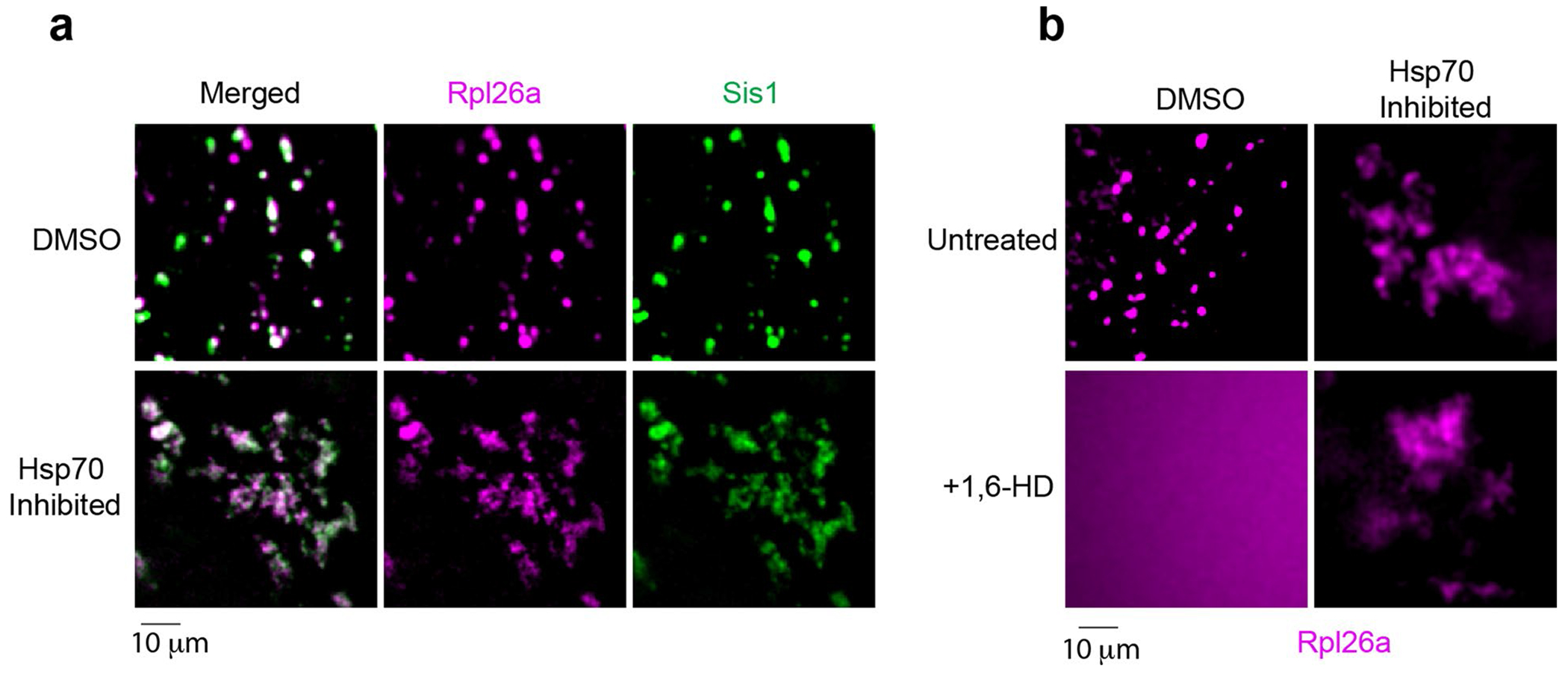
(a) Effect of Hsp70 inhibition in the morphology of Rpl26a and Sis1 condensates. (b) Effect of 5% 1,6-HD upon the condensates with or without Hsp70 inhibition. The micrograph represents data derived from 3 independent experiments.
Extended Data Fig. 8 |. oRPs in condensates are not degraded and are transported to the cytosol upon recovery.
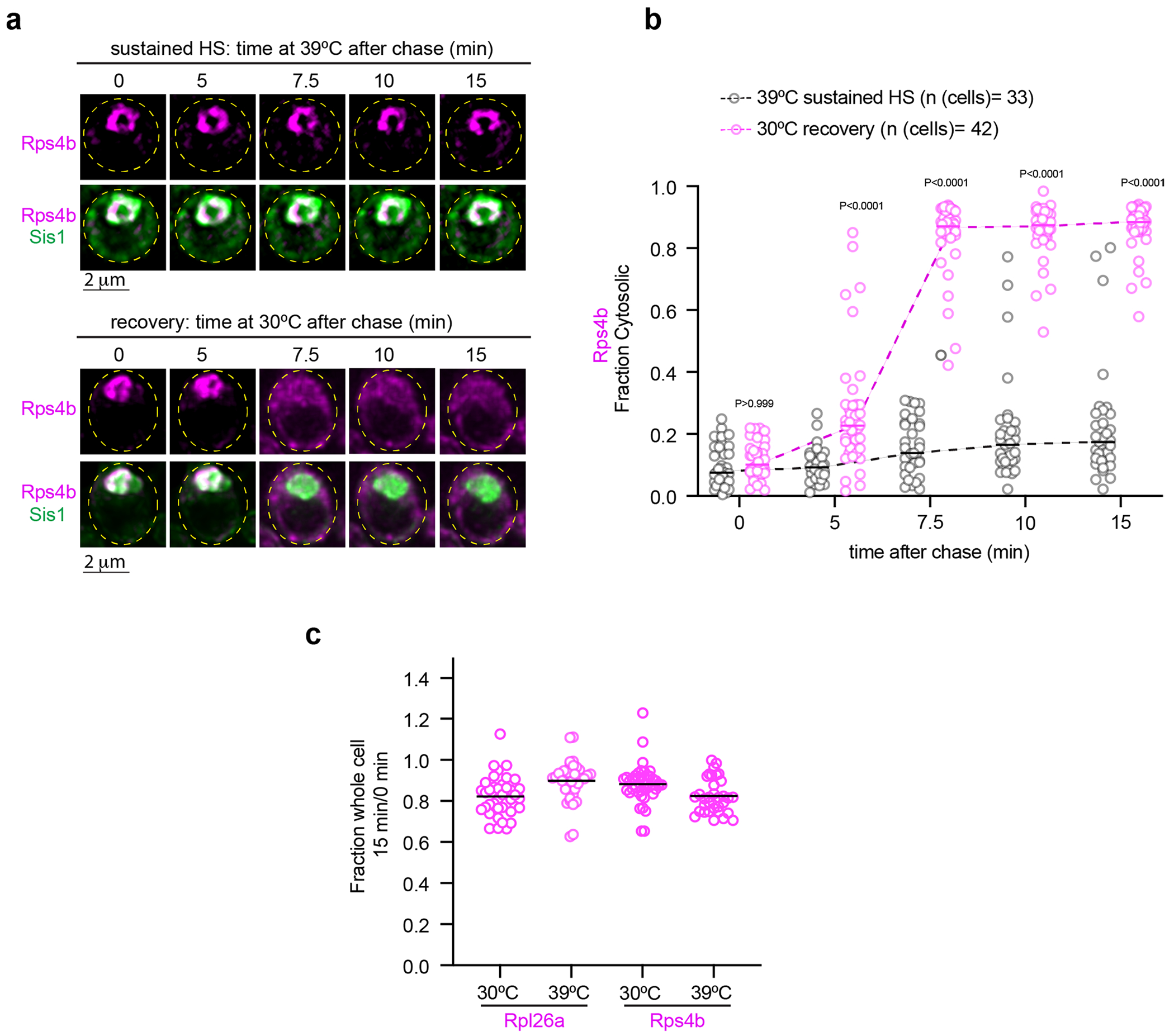
(a) Live cell time lapse imaging of the spatial distribution of Rps4b (magenta) and Sis1-mVenus (green) during sustained heat shock and recovery. (b) Quantification of the fraction of cytosolic Rps4b signal under sustained HS or recovery. Statistical significance was established using the Brown-Forsythe and Welch one-way ANOVA test, followed by Dunnett T3 multiple comparison analyses. n = number of cells pooled from 3 biologically independent replicates. (c) Fraction of total pulse labeled Rpl26a or Rps4b remaining after chase for 15 minutes at indicated temperature. n=number of cells pooled from 3 biologically independent replicates.
Extended Data Fig. 9 |. oRP condensate reversibility depends upon Sis1 availability.
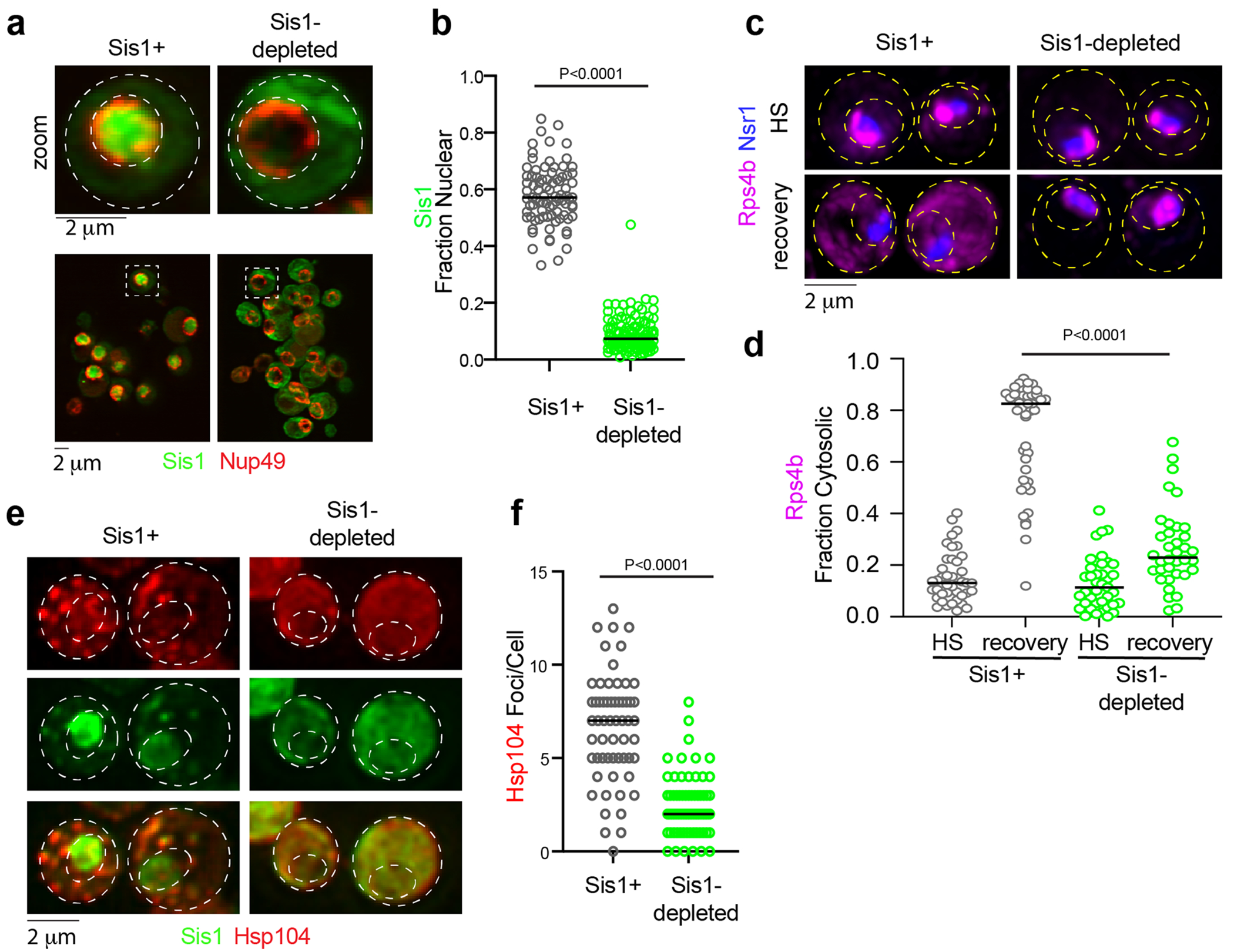
(a) imaging of Sis1-mVenus (green) and Nup49-mScarlet-I (red) following Sis1 depletion or not. (b) Quantification of fraction of nuclear Sis1 upon Sis1 depletion or not. P values were calculated with unpaired two-tailed Welch’s t-test. n denotes number of cells as obtained from 4 independent experiment. n = number of cells pooled from 4 biologically independent replicates. (c) LLS imaging of Hsp104-mKate2 during heat shock (39 °C, 15 mins) pre-depleted for Sis1 (green) or not. (d) Quantification of Hsp104-mKate2 foci per cell for (c). P values were calculated with unpaired two-tailed Welch’s t-test. n denotes number of cells from 3 independent experiments. n = number of cells pooled from 3 biologically independent replicates. (e) LLS live cell imaging of Rps4b (magenta) and the nucleolar marker Nsr1 (blue) during heat shock and recovery in the absence or presence of Sis1 depletion. (f) Quantification of the fraction of cytosolic Rps4b under sustained HS or recovery in the absence or presence of Sis1 depletion. P values were calculated with unpaired two-tailed Welch’s t-test. n denotes number of cells obtained from 3 independent experiment. n = number of cells pooled from 3 biologically independent replicates.
Supplementary Material
Acknowledgements
We are grateful to H. An and W. Harper for sharing the HCT116 RPL26–HaloTag cell line. We thank V. Bindokas and C. Labno at the University of Chicago Integrated Light Microscopy Core (RRID: SCR_019197) for imaging assistance. We are especially grateful to B. Glick and lab members for the yeast HaloTag construct and advice on using JF dyes in yeast. We thank S. Kron for use of gel imaging instruments, and K. Lin for assistance aligning the Squires Lab custom wide-field setup and camera calibration. We also thank members of the Pincus, Squires and Drummond labs for helpful discussions. J.A.M.B. acknowledges fellowship support from the Helen Hay Whitney Foundation. This work was supported by NIH grants R01 GM138689 to D.P., R35 GM144278 to D.A.D., support from the Neubauer Family Foundation to A.H.S., and NSF QLCI QuBBE grant OMA-2121044 to D.P. and A.H.S.
Footnotes
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41556-023-01253-2.
Competing interests
We declare that none of the authors has competing financial or non-financial interests.
Extended data is available for this paper at https://doi.org/10.1038/s41556-023-01253-2.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41556-023-01253-2.
Peer review information Nature Cell Biology thanks the anonymous reviewers for their contribution to the peer review of this work.
Code availability
Custom code used for the image analysis are deposited at Zenodo (https://doi.org/10.5281/zenodo.8076227).
Data availability
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD039068 and PXD039134. RNA-seq raw sequence files and processed data were deposited in the Gene Expression Omnibus (accession no. GSE237174). Source data are provided with this paper. Any other potential type of data used to interpret the finding can be provided upon request to corresponding author.
References
- 1.Warner JR The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci 24, 437–440 (1999). [DOI] [PubMed] [Google Scholar]
- 2.Maaløe O & Kjeldgaard NO Control of macromolecular synthesis; a study of DNA, RNA, and protein synthesis in bacteria. (W. A. Benjamin, 1966). [Google Scholar]
- 3.Scott M, Klumpp S, Mateescu EM & Hwa T Emergence of robust growth laws from optimal regulation of ribosome synthesis. Mol. Syst. Biol 10, 747 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lempiainen H & Shore D Growth control and ribosome biogenesis. Curr. Opin. Cell Biol 21, 855–863 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Ingolia NT, Ghaemmaghami S, Newman JR & Weissman JS Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shore D & Albert B Ribosome biogenesis and the cellular energy economy. Curr. Biol 32, R611–R617 (2022). [DOI] [PubMed] [Google Scholar]
- 7.Woolford JL Jr. & Baserga SJ Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics 195, 643–681 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jakel S, Mingot JM, Schwarzmaier P, Hartmann E & Gorlich D Importins fulfil a dual function as nuclear import receptors and cytoplasmic chaperones for exposed basic domains. EMBO J. 21, 377–386 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pillet B, Mitterer V, Kressler D & Pertschy B Hold on to your friends: dedicated chaperones of ribosomal proteins: dedicated chaperones mediate the safe transfer of ribosomal proteins to their site of pre-ribosome incorporation. Bioessays 39, 1–12 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Gasch AP & Werner-Washburne M The genomics of yeast responses to environmental stress and starvation. Funct. Integr. Genomics 2, 181–192 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Gasch AP et al. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11, 4241–4257 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sawarkar R. Transcriptional lockdown during acute proteotoxic stress. Trends Biochem. Sci 47, 660–672 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shore D, Zencir S & Albert B Transcriptional control of ribosome biogenesis in yeast: links to growth and stress signals. Biochem. Soc. Trans 49, 1589–1599 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iserman C. et al. Condensation of Ded1p promotes a translational switch from housekeeping to stress protein production. Cell 181, 818–831 e819 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muhlhofer M. et al. The heat shock response in yeast maintains protein homeostasis by chaperoning and replenishing proteins. Cell Rep. 29, 4593–4607 e4598 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Juszkiewicz S & Hegde RS Quality control of orphaned proteins. Mol. Cell 71, 443–457 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yanagitani K, Juszkiewicz S & Hegde RS UBE2O is a quality control factor for orphans of multiprotein complexes. Science 357, 472–475 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sung MK et al. A conserved quality-control pathway that mediates degradation of unassembled ribosomal proteins. eLife 5, e19105 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Narla A & Ebert BL Ribosomopathies: human disorders of ribosome dysfunction. Blood 115, 3196–3205 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pincus D. Regulation of Hsf1 and the heat shock response. Adv. Exp. Med Biol 1243, 41–50 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Albert B. et al. A ribosome assembly stress response regulates transcription to maintain proteome homeostasis. eLife 8, e45002 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tye BW et al. Proteotoxicity from aberrant ribosome biogenesis compromises cell fitness. eLife 8, e43002 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feder ZA et al. Subcellular localization of the J-protein Sis1 regulates the heat shock response. J. Cell Biol 220, e202005165 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garde R, Singh A, Ali A & Pincus D Transcriptional regulation of Sis1 promotes fitness but not feedback in the heat shock response. eLife 12, e79444 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masser AE et al. Cytoplasmic protein misfolding titrates Hsp70 to activate nuclear Hsf1. eLife 8, e47791 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Triandafillou CG, Katanski CD, Dinner AR & Drummond DA Transient intracellular acidification regulates the core transcriptional heat shock response. eLife 9, e54880 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tye BW & Churchman LS Hsf1 activation by proteotoxic stress requires concurrent protein synthesis. Mol. Biol. Cell 32, 1800–1806 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frottin F. et al. The nucleolus functions as a phase-separated protein quality control compartment. Science 365, 342–347 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Gallardo P, Real-Calderon P, Flor-Parra I, Salas-Pino S & Daga RR Acute heat stress leads to reversible aggregation of nuclear proteins into nucleolar rings in fission yeast. Cell Rep. 33, 108377 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Hung V. et al. Spatially resolved proteomic mapping in living cells with the engineered peroxidase APEX2. Nat. Protoc 11, 456–475 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sailer C. et al. A comprehensive landscape of 60S ribosome biogenesis factors. Cell Rep. 38, 110353 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.An H, Ordureau A, Korner M, Paulo JA & Harper JW Systematic quantitative analysis of ribosome inventory during nutrient stress. Nature 583, 303–309 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garshott DM et al. iRQC, a surveillance pathway for 40S ribosomal quality control during mRNA translation initiation. Cell Rep. 36, 109642 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klaips CL, Gropp MHM, Hipp MS & Hartl FU Sis1 potentiates the stress response to protein aggregation and elevated temperature. Nat. Commun 11, 6271 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schawalder SB et al. Growth-regulated recruitment of the essential yeast ribosomal protein gene activator Ifh1. Nature 432, 1058–1061 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Thoreen CC et al. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485, 109–113 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chowdhary S, Kainth AS, Paracha S, Gross DS & Pincus D Inducible transcriptional condensates drive 3D genome reorganization in the heat shock response. Mol. Cell 82, 4386–4399.e7 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Craig EA & Marszalek J How do J-proteins get Hsp70 to do so many different things? Trends Biochem. Sci 42, 355–368 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duster R, Kaltheuner IH, Schmitz M & Geyer M 1,6-Hexanediol, commonly used to dissolve liquid–liquid phase separated condensates, directly impairs kinase and phosphatase activities. J. Biol. Chem 296, 100260 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muzzopappa F. et al. Detecting and quantifying liquid–liquid phase separation in living cells by model-free calibrated half-bleaching. Nat. Commun 13, 7787 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lakowicz JR Principles of Fluorescence Spectroscopy 3rd Edn (Springer, 2006). [Google Scholar]
- 42.Linsenmeier M. et al. Dynamic arrest and aging of biomolecular condensates are modulated by low-complexity domains, RNA and biochemical activity. Nat. Commun 13, 3030 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cherkasov V. et al. Coordination of translational control and protein homeostasis during severe heat stress. Curr. Biol 23, 2452–2462 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Grousl T. et al. Heat shock-induced accumulation of translation elongation and termination factors precedes assembly of stress granules in S. cerevisiae. PLoS ONE 8, e57083 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Riback JA et al. Stress-triggered phase separation is an adaptive, evolutionarily tuned response. Cell 168, 1028–1040 e1019 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoo H, Bard JAM, Pilipenko EV & Drummond DA Chaperones directly and efficiently disperse stress-triggered biomolecular condensates. Mol. Cell 82, 741–755 e711 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cereghetti G. et al. Reversible amyloids of pyruvate kinase couple cell metabolism and stress granule disassembly. Nat. Cell Biol 23, 1085–1094 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kainth AS, Chowdhary S, Pincus D & Gross DS Primordial super-enhancers: heat shock-induced chromatin organization in yeast. Trends Cell Biol. 31, 801–813 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rawat P. et al. Stress-induced nuclear condensation of NELF drives transcriptional downregulation. Mol. Cell 81, 1013–1026 e1011 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wallace EW et al. Reversible, specific, active aggregates of endogenous proteins assemble upon heat stress. Cell 162, 1286–1298 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brehme M. et al. A chaperome subnetwork safeguards proteostasis in aging and neurodegenerative disease. Cell Rep. 9, 1135–1150 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alberti S & Dormann D Liquid–liquid phase separation in disease. Annu. Rev. Genet 53, 171–194 (2019). [DOI] [PubMed] [Google Scholar]
- 53.Alberti S & Hyman AA Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nat. Rev. Mol. Cell Biol 22, 196–213 (2021). [DOI] [PubMed] [Google Scholar]
- 54.Zheng X. et al. Dynamic control of Hsf1 during heat shock by a chaperone switch and phosphorylation. eLife 5, e18638 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mahat DB & Lis JT Use of conditioned media is critical for studies of regulation in response to rapid heat shock. Cell Stress Chaperones 22, 155–162 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen BC et al. Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science 346, 1257998 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Northan B Ops-experiments. GitHub; https://github.com/imagej/ops-experiments (2022). [Google Scholar]
- 58.Royer LA et al. ClearVolume: open-source live 3D visualization for light-sheet microscopy. Nat. Methods 12, 480–481 (2015). [DOI] [PubMed] [Google Scholar]
- 59.Schindelin J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aboulhouda S, Di Santo R, Therizols G & Weinberg D Accurate, streamlined analysis of mRNA translation by sucrose gradient fractionation. Bio Protoc. 7, e2573 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roy R, Hohng S & Ha T A practical guide to single-molecule FRET. Nat. Methods 5, 507–516 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Michalet X. Mean square displacement analysis of single-particle trajectories with localization error: Brownian motion in an isotropic medium. Phys. Rev. E 82, 041914 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Enkler L. et al. Arf1 coordinates fatty acid metabolism and mitochondrial homeostasis. Nat. Cell Biol 25, 1157–1172 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD039068 and PXD039134. RNA-seq raw sequence files and processed data were deposited in the Gene Expression Omnibus (accession no. GSE237174). Source data are provided with this paper. Any other potential type of data used to interpret the finding can be provided upon request to corresponding author.


