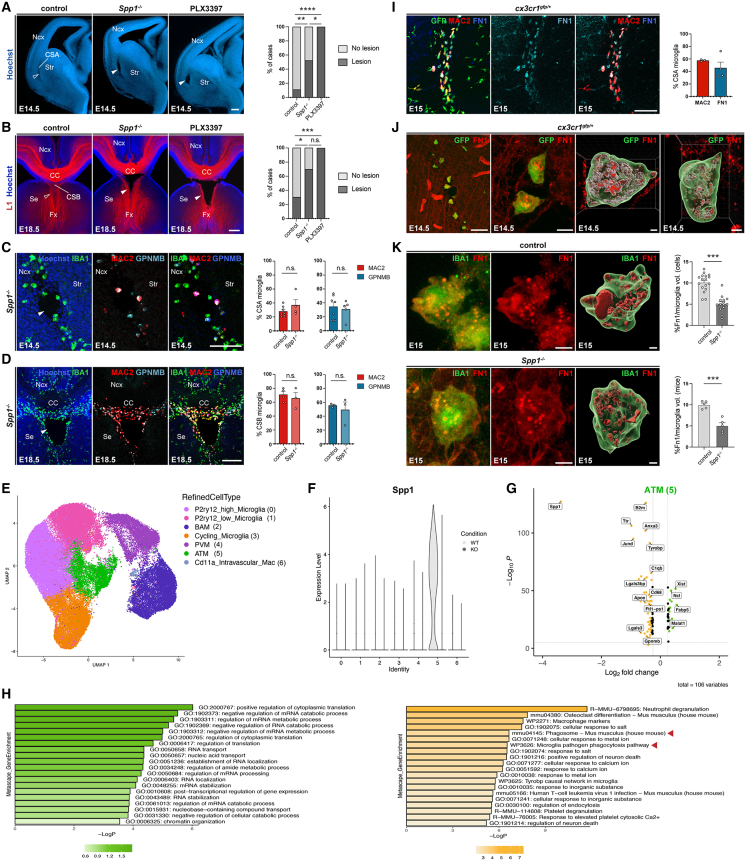
Figure 5.
ATM-core factor Spp1 contributes to tissue integrity at the CSA and CSB
(A) Coronal hemisections of E14.5 brains stained with Hoechst reveal CSA disruption in 50% of Spp1−/− mutants (solid arrowheads) compared to controls (open arrowheads) and in 100% of PLX3397-treated embryos (solid arrowheads) (ncontrols = 18, nSpp1KO = 21, nPLX3397 = 13). Quantification of CSA lesions across models.
(B) L1 immunolabeling enables axon visualization (open arrowheads) and midline lesions (solid arrowheads) in approximately 70% of Spp1−/− mutants compared with 100% in PLX3397-exposed embryos (ncontrols = 23, nSpp1KO = 20, nPLX3397 = 8). Quantification of CSB lesions across models.
(C and D) E14.5 (C) and E18.5 (D) coronal hemisections showing no differences in Mac2 and GPNMB co-expressing microglia at the CSA and CSB of controls (open arrowheads) and Spp1−/− embryos (ncontrols = 3, nSpp1KO = 3, at each stage from at least two distinct litters).
(E) UMAP visualization of single-cell RNA sequencing (scRNA-seq) data representing macrophage subsets extracted from wild-type (WT) and Spp1−/− E14.5 and E18.5 embryos colored by annotated clusters (RefinedCellType).
(F) Violin plot of normalized and scaled Spp1 expression across annotated clusters between wild-type (WT) (light gray) and Spp1−/− (KO)(dark gray) mice showing that Spp1 expression is largely restricted to WT ATM (cluster 5).
(G) Volcano plot of differentially expressed genes (DEGs) between WT and Spp1−/− conditions in ATM cells (false discovery rate[FDR]-adjusted p < 0.05 and avgerage_log2FC > 0.3). Genes downregulated in Spp1−/− embryos are displayed in orange, while the upregulated ones are shown in green, and some genes were manually annotated.
(H) Bar plots of top Metascape gene set enrichment of DEGs (G) in both WT or Spp1−/− conditions, highlighting upregulated (green) and downregulated pathways (orange) in Spp1−/− embryos versus controls, with pathways related to phagocytosis highlighted by a red arrowhead.
(I) Brain sections from E15 Cx3cr1gfp/+ mice showing specific fibronectin 1 (FN1) labeling within GFP- and Mac2-positive ATM microglia at the CSA (performed on brain sections of at least three mice from two distinct litters).
(J) High magnification confocal acquisition and 3D cell reconstructions (Imaris software) of immunolabeled sections from E14.5 embryonic brains showing Cx3cr1gfp-positive CSA microglia with FN1 signal inside cell bodies (performed on brain sections of at least three mice from two distinct litters).
(K) Comparison of the percentage of FN1 volume measured (Imaris software) inside individual CSA microglia shows a reduction in Spp1 mutants versus controls (ncontrols = 17, nSpp1KO = 14, from at least two distinct litters).
Graphs show percentages in (A) and (B) and means ± SEM for all others. Fisher’s exact test was performed to compare distributions of cases with lesions in controls, Spp1−/−, and PLX3397-exposed embryos (A and B), and Mann-Whitney U tests were performed for statistical comparison in all other graphs,∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ns, non significant (p > 0.5). Scale bars: 200 μm (A, B low magnification, and D); 100 μm (C and F); 5 μm (G, left); and 2 μm (3D reconstructed cells in F and G).
ATM, axon-tract-associated microglia; BAM, border associated macrophages; CC, corpus callosum; CSA, cortico-striato-amygdalar boundary; CSB, cortico-septal boundary; DEGs, differentially expressed genes; Fx, fornix; Intravasc mac, intravascular macrophages; Ncx, neocortex; PVM, perivascular macrophages; Se, Septum; Str, striatum; WT, wild type.
See also Figure S4 and Table S3. Genes defining the clusters identified by single-cell transcriptomic analyses on all the sorted cells from wild type and Spp1−/−, related to Figures 5 and S4, Table S4. Genes defining the clusters identified by single-cell transcriptomic analyses on the macrophages from wild type and Spp1−/−, related to Figure 5, Table S5. DEGs in ATM in wild type versus Spp1−/−, related to Figure 5, Table S6. Metascape analyses of ATM in wild type versus Spp1−/−, related to Figure 5.