Abstract
Objectives
The use of multiparametric magnetic resonance imaging (mpMRI) has been widely adopted in the diagnostic work‐up for suspicious prostate cancer (PCa) and is recommended in most current guidelines. However, mpMRI lesions are often indeterminate and/or turn out to be false‐positive on prostate biopsy. The aim of this work was to evaluate Proclarix, a biomarker test for the detection of relevant PCa, regarding its diagnostic value in all men before biopsy and in men with indeterminate lesions on mpMRI (PI‐RADS 3) during work‐up for PCa.
Materials and Methods
Men undergoing mpMRI‐targeted and systematic biopsy of the prostate were prospectively enrolled. The Proclarix test was evaluated for the detection accuracy of clinically significant PCa (csPCa) defined as Grade Group ≥ 2 and its association to mpMRI results. Further, Proclarix's performance was also tested when adapted to prostate volume (Proclarix density) and performance compared to PSA density (PSAD).
Results
A total of 150 men with a median age of 65 years and median PSA of 5.8 ng/mL were included in this study. CsPCa was diagnosed in 65 (43%) men. Proclarix was significantly associated with csPCa and higher PI‐RADS score (p < 0.001). At the pre‐defined cut‐off of 10%, Proclarix's sensitivity for csPCa was 94%, specificity 19%, negative predictive value 80% and positive predictive value 47%. Proclarix density showed the highest AUC for the detection of csPCa of 0.77 (95%CI: 0.69–0.85) compared to PSA, PSAD and Proclarix alone. Proclarix was able to identify all six csPCa in men with PI‐RADS 3 lesions (n = 28), whereas PSAD missed two out of six. At optimized cut‐offs, Proclarix density outperformed PSAD by potentially avoiding 41% of unnecessary biopsies.
Conclusion
Proclarix demonstrates high sensitivity in detecting csPCa but may still result in unnecessary biopsies. However, Proclarix density was able to outperform PSAD and Proclarix and was found to be useful in men with PI‐RADS 3 findings by safely avoiding unnecessary biopsies without missing csPCa.
Keywords: biomarkers; biopsy; cathepsin D, CTSD; Proclarix; prostate cancer; thrombospondin 1, THBS1
1. INTRODUCTION
In recent years, the diagnostic work‐up for prostate cancer (PCa) has emerged to incorporate the routine use of multiparametric magnetic resonance imaging (mpMRI) in men with an elevated prostate specific antigen (PSA) level. 1 Although it is clear that men with a PI‐RADS 4 or 5 lesion require a diagnostic prostate biopsy, it remains controversial as to whether patients with a grade 3 lesion require the same. 2 , 3 This is because only 4%–27% of men with a PI‐RADS 3 lesion will ultimately be found to have clinically significant PCa (csPCa). 3 , 4 , 5 In contrast, 22%–57% and 72%–79% of men with a PI‐RADS 4 or 5 lesion will be found to have csPCa, respectively. 6 , 7
Considering the high infectious complication rate from prostate biopsy 8 and the potentially negative downstream consequences of needlessly diagnosing low grade PCa, 9 it is of critical importance to more accurately risk stratify patients with an equivocal finding on mpMRI prior to moving forward with a biopsy. Although prior work has shown that the incorporation of PSA density (PSAD) into the decision to perform a prostate biopsy in men with a PI‐RADS 3 lesion on mpMRI improves the diagnostic yield for csPCa, 10 , 11 the discriminatory ability of this parameter alone still leads to a significant number of unnecessary prostate biopsies. 12
Thus, additional markers are needed to further estimate the risk of csPCa in men with a PI‐RADS 3 lesion on mpMRI.
Proclarix (Proteomedix AG, Switzerland) is a novel serum‐based assay that was developed to aid in the risk stratification of men with an elevated PSA level. 13 This test, which incorporates serum total PSA (tPSA), free PSA (fPSA), cathepsin D (CTSD), thrombospondin 1 (THBS1) and patient age, 14 has been shown to detect csPCa with more favourable diagnostic performance characteristics as compared to percent fPSA (%fPSA) and the ERSPC risk calculator. 15 , 16 , 17 , 18 , 19 , 20 , 21 , 22 One limitation of the available literature on the Proclarix test is that all papers relied upon the results of a 10–14 core systematic biopsy of the prostate with or without targeted biopsy core. 15 , 18 , 19 , 20 Although this practice is typical in the prostate cancer biomarker literature, concerns remain regarding the accuracy of this somewhat limited method of sampling the prostate. 6 , 23 For example, at the time of radical prostatectomy, up to 30% of patients will be upgraded or downgraded following a 12–14 core biopsy. 24 In light of this, it is possible that prior analyses of the Proclarix test were impacted by sampling bias.
In this study, we aimed to compare the performance of the Proclarix test to standard clinical parameters in a contemporary biopsy cohort of men who underwent an mpMRI followed by a transperineal saturation biopsy of the prostate. Importantly, we perform a subset analysis of the diagnostic yield of this test in patients with an indeterminate mpMRI result.
2. PATIENTS AND METHODS
2.1. Study design
This is a single‐centre, prospective study designed to evaluate the diagnostic performance of the Proclarix test for diagnosing csPCa (grade group ≥ 2 PCa) among men undergoing a combined systematic and targeted prostate biopsy. The study is in line with the STARD 2015 reporting guidelines 25 and was approved by the local ethics committee (KEK Nr. 2016‐00075). All participants of the study provided written informed consent.
2.2. Participants and setting
The study involved 150 men who underwent a prostate biopsy at the University Hospital of Zurich between January 2019 and March 2022. Patients were referred for a prostate biopsy on the basis of an elevated or rising PSA level. Findings on digital rectal exam and family history of prostate cancer also informed the decision as to whether to perform a prostate biopsy. Additionally, this decision took into account patient life expectancy and concomitant illnesses.
Before undergoing a biopsy, each patient underwent mpMRI and provided a blood sample for Proclarix testing. Importantly, the decision to do a biopsy was not based on Proclarix or MRI results. Exclusion criteria included patients having a history of PCa, serious illness (such as dementia or severe cardiovascular disease), treatment with 5‐reductase inhibitors, or contraindication to undergoing an mpMRI.
2.3. Proclarix testing
Serum samples for Proclarix testing were prospectively collected prior to plan biopsies. Samples were stored at −80°C, and the Proclarix assay was performed in batch for the entire patient cohort at a centralized lab (Proteomedix AG, Switzerland). Aside from patient age, the laboratory performing the assay was blinded to all clinical parameters and biopsy results. Performance of the Proclarix assay and subsequent risk score calculation was performed according to the manufacturer's instruction for use. 14 The cut‐off used to evaluate the clinical performance of Proclarix was set at 10%.
2.4. Prostate MRI and biopsy procedures
mpMRIs were conducted on a 3T MAGNETOM Skyra MRI system and used T2‐weighted, diffusion‐weighted and dynamic contrast‐enhanced sequences. All mpMRI exams were read by board‐certified radiologists using the Prostate Imaging‐Reporting and Data System (PI‐RADS) v.2.0 (2). Prostate volume was calculated in millilitres (mL) and was determined using the formula height × length × width × π/6.
Prostate biopsies were performed using the transperineal approach under general anaesthesia with the BiopSee platform (MedCom GmbH, Darmstadt, Germany). A saturation biopsy was performed on all patients with 1–3 biopsies cores obtained from each of the 20 modified Barzell zones. 26 Additional targeted biopsy cores were taken of any suspicious lesion found on mpMRI (2–3 biopsy cores per lesion). Each collected biopsy core was individually reviewed by a board‐certified uropathologist, and in the case of PCa, a second pathologist verified the diagnosis and grade. csPCa was defined as grade group ≥ 2.
2.5. Statistical analysis
Sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) were calculated for the Proclarix test as well as PSA, PSA density and Proclarix density. For calculations of PSA and Proclarix density, prostate volume was determined by mpMRI. To compare the patient characteristics including results of the above mentioned parameters in the groups of patients with and without PCa, the Mann–Whitney U test was used for continuous variables and the Fisher's exact test for categorical variables. To assess differences between sensitivity and specificity, the McNemar test was used. p‐values for NPV and PPV were calculated according to Moskowitz and Pepe. 27 , 28 Receiver operating characteristic (ROC) curves were constructed, and areas under the curve (AUCs) were calculated by using the trapezoidal rule. The statistical analysis was carried out using SPSS version 28.0. (IBM Corp). p‐values < 0.05 were considered as statistically significant.
3. RESULTS
The baseline characteristics of the study cohort are summarized in Table 1. A total of 65 (43%) was diagnosed with csPCa. Grade group 2 PCa was found in 27 (18%), grade group 3 in 16 (10%), grade group 4 in 18 (12%) and grade group 5 in 4 (3%) of the patients. Age, volume, PSAD, Proclarix and Proclarix density did significantly differ between patients with and without csPCa (p < 0.001 for all comparisons), whereas tPSA was not significantly different (p = 0.227).
TABLE 1.
Patient characteristics.
| Characteristics | Total | No cancer or ciPCa | csPCa | p‐value |
|---|---|---|---|---|
| Patients, n (%) | 150 (100) | 85 (57) a | 65 (43) | ‐ |
| Age (year) |
65 (60–71) |
62 (59–68) |
68 (63–73) |
<0.001 |
| tPSA (ng/mL) |
5.8 (3.7–9.8) |
4.9 (3.1–7.8) |
7.5 (4.3–11.5) |
0.227 |
| Volume (mL) |
45.9 (34.8–64.9) |
50.1 (38.5–69.8) |
41.8 (29.0–56.6) |
0.003 |
| PSA density (ng/mL2) |
0.12 (0.08–0.20) |
0.10 (0.07–0.14) |
0.17 (0.12–0.29) |
<0.001 |
| Proclarix score |
30.8 (16.9–46.0) |
23.9 (14.7–36.7) |
40.4 (21.3–58.4) |
<0.001 |
| Proclarix density (%/mL) |
0.44 (0.25–0.71) |
0.90 (0.57–1.45) |
0.59 (0.31–1.03) |
<0.001 |
Note: Data presented in median and interquartile ranges (IQR).
Abbreviations: ciPCa, grade group < 2; csPCa, clinically significant prostate cancer (defined as grade group ≥ 2); tPSA, total prostate specific antigen.
63 (42%) pts no cancer, 22 (15%) pts ciPCa.
The Proclarix risk score (score range from 0% to 100%) was significantly associated with higher PI‐RADS score and cancer grade (for both p < 0.001) (Figure 1). When applying a risk score cut‐off of 10%, the Proclarix test was found to have a sensitivity of 94% (95%CI: 88%–100%) and a specificity of 19% (95%CI: 11%–27%) for the diagnosis of csPCa. Values of NPV and PPV were found to be of 80% (95%CI: 62%–98%) and 47% (95%CI: 38%–56%), respectively (Table 2). Using this cut‐off to determine the need for a prostate biopsy, the Proclarix test would avoided the need for a prostate biopsy in 16 patients (11%), with four (6%) of cases of csPCa being missed. This included three patients with grade group 2 PCa and one with grade group 4 disease.
FIGURE 1.
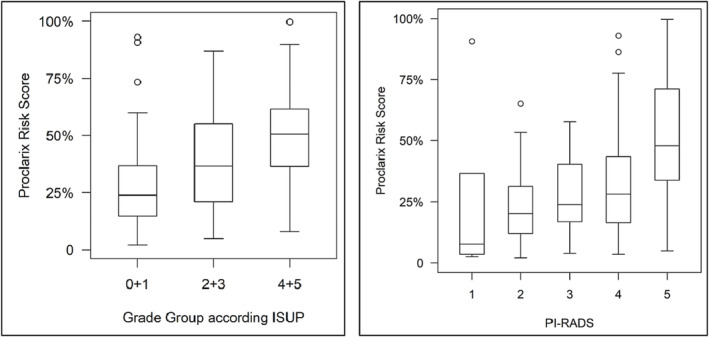
Proclarix risk score (0%–100%) in association with grade group according to ISUP (left) and with PI‐RADS (right).
TABLE 2.
Performance of Proclarix and PSA density across the whole cohort.
| Parameter | Proclarix | PSA density | p‐value |
|---|---|---|---|
| Cut‐off | 10 | 0.15 | ‐ |
| Sensitivity, % | 94 (88–100) | 58 (46–70) | <0.001 |
| Specificity, % | 19 (11–27) | 82 (74–90) | <0.001 |
| NPV, % | 80 (62–98) | 72 (63–81) | 0.343 |
| PPV, % | 47 (38–56) | 72 (60–84) | <0.001 |
Compared to Proclarix, PSAD with a cut‐off of 0.15 ng/mL2 demonstrated a sensitivity of 58% (95%CI: 46%–70%) and a specificity of 82% (95%CI: 74%–90%). The PPV was 72% (95%CI: 60%–84%), and NPV was 72% (95%CI: 63%–81%). Based on these data, PSAD would have avoided 70 (47%) patients of unnecessary biopsies, with 27 out of 65 (42%) patients with csPCa being missed, including seven patients with grade group 5 PCa and seven patients with a grade group 4 cancer.
To test discrimination for csPCa, ROC curves for Proclarix, PSAD and Proclarix density for the whole cohort were applied (Figure 2). Proclarix density showed the highest level of discrimination, with an area under the curve (AUC) of 0.77 (95%CI: 0.69–0.85), compared to Proclarix (AUC of 0.71; 95%CI: 0.62–0.79) and PSA density (AUC of 0.73; 95%CI: 0.65–0.82).
FIGURE 2.
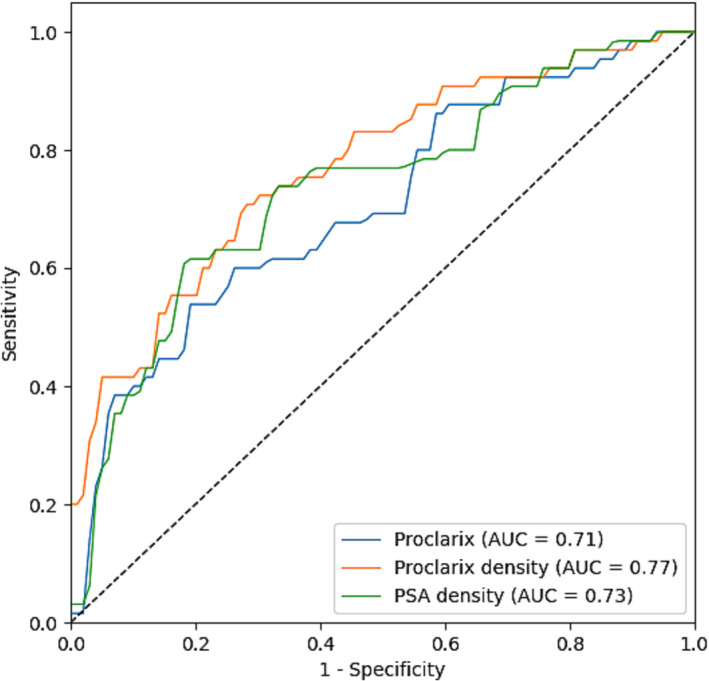
ROC curves for Proclarix, PSA density and Proclarix density with AUC of 0.71, 0.73 and 0.77, respectively.
The performance of Proclarix, PSAD and Proclarix density was evaluated in a subgroup of men with indeterminate PI‐RADS 3 lesion seen on mpMRI (n = 28). Six out of 28 (21%) men in this subgroup were diagnosed with csPCa, whereas the other 22 (79%) had either a negative biopsy or grade group 1 PCa. In this subgroup of men, the Proclarix test would have avoided three (11%), without missing any of the six cases of csPCa. In contrast, PSAD would have avoided 18 biopsies (64%), while missing four cases of csPCa. When Proclarix density was compared with PSAD using optimized cut‐offs to yield similar sensitivity of 100% each, Proclarix density showed a specificity of 41% (95%CI: 20–61), while PSAD demonstrated a specificity of 14% (95%CI: 0–28, p = 0.03).
Furthermore, Proclarix density could significantly discriminate ciPCa/noPCa from csPCa in the PI‐RADS 3 subpopulation (p‐values of 0.021 for Proclarix density and 0.256 for PSAD, respectively) (Figure 3). Results of this analysis are summarized in Table 3.
FIGURE 3.
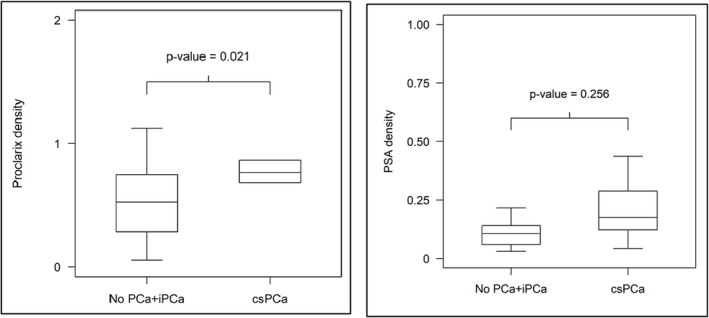
Boxplots show discrimination of no PCa and iPCa versus csPCa in patients with PI‐RADS 3 findings by Proclarix density (left) and PSA density (right).
TABLE 3.
Comparison of the clinical performance for the detection of csPCa of Proclarix, PSAD and Proclarix density in the patient population with PI‐RADS 3 findings (n = 28).
| Parameter | Proclarix | PSA density | p‐value a | Proclarix density | PSA density | p‐value b |
|---|---|---|---|---|---|---|
| Cut‐off | 10 | 0.15 | ‐ | 0.395 c | 0.043 c | ‐ |
| Sensitivity, % |
100 (100–100) |
67 (29–100) |
0.157 |
100 (100–100) |
100 (100–100) |
1.000 |
| Specificity, % |
14 (0–24) |
82 (66–98) |
<0.001 |
41 (20–61) |
14 (0–28) |
0.034 |
| NPV, % |
100 (100–100) |
90 (77–100) |
0.157 |
100 (100–100) |
100 (100–100) |
1.000 |
| PPV, % |
24 (7–41) |
50 (15–85) |
0.012 |
32 (11–52) |
24 (7–41) |
0.034 |
Comparison between Proclarix and PSA density.
Comparison between Proclarix density and PSA density at optimal cut‐offs.
Optimized cut‐offs for this patient population with PI‐RADS 3 findings.
4. DISCUSSION
The purpose of this study was to assess the performance of Proclarix, a novel blood‐based biomarker for the detection of csPCa, in a contemporary cohort of men who underwent an mpMRI followed by saturation biopsy of the prostate. In all cases, the decision to perform a biopsy was made independent of the results of the mpMRI or Proclarix test results, allowing for an unbiased assessment of the diagnostic accuracy of this novel biomarker against a highly rigorous truth standard. In our study, Proclarix demonstrated a sensitivity of 94% and a NPV of 80% for identifying csPCa using a 10% risk score cut‐off, missing four out of 65 csPCa cases. The specificity of 19% was rather low, translating in only 16 out of 85 unnecessary biopsies that could have been avoided. Compared to PSA density and Proclarix alone, Proclarix density showed the highest level of discrimination with an AUC of 0.77. In the subgroup of men with indeterminate PI‐RADS 3 lesions, Proclarix achieved a sensitivity and NPV of 100%, outperforming PSAD, while its specificity at 14% was again relatively low.
Our study confirms Proclarix's high sensitivity for csPCa and shows the correlation with the results of mpMRI. The results were similar to those previously published in other studies. 15 , 17 , 18 , 29 When the manufacturer‐recommended cut‐off of 10% was applied, Proclarix would have missed only 6% of csPCa, while PSAD would have missed 41% csPCa. These results are consistent with recently published data by Campistol et al., 19 where Proclarix sensitivity outperformed PSAD by missing only 6, respectively, 23 out of 232 csPCa patients. However, Proclarix showed only moderate performance for the avoidance of unnecessary biopsies of men harbouring either only ciPCa or no cancer (specificity of 19%), meaning only 16 out of 85 unnecessary biopsies could have been avoided. However, by including prostate volume in the Proclarix test as Proclarix density, we could show the highest discriminatory power for csPCa compared to PSAD or Proclarix alone, with an AUC of 0.77. These results are in line with a recent study by Steuber et al. 15 on 121 mpMRI‐fusion biopsies. The study found that the specificity increased from 22% when using Proclarix alone to 33% with Proclarix density, while PSAD showed a significantly lower specificity of only 8% (p < 0.001). By utilizing Proclarix density, unnecessary biopsies could be avoided in up to one‐third of cases.
Men with indeterminate PI‐RADS 3 findings might often be candidates for continued PSA surveillance rather than biopsy, but appropriate selection for biopsy avoidance in these subpopulation is challenging. Hence, we performed a sensitivity analysis for Proclarix performance in this specific group and found Proclarix density with a specific cut‐off of 0.395 to maintain optimal sensitivity but gaining relevant specificity of 41% compared to PSAD (only 14% specificity). Therefore, Proclarix density could potentially reduce the need for unnecessary prostate biopsies in this subgroup, while still allowing for sufficient detection of csPCa.
Our suggested diagnostic pathway would be to use Proclarix density when the volume of the prostate can be established either by abdominal or transrectal ultrasound. A prostate volume estimation from a DRE is not recommended. When dealing with a PI‐RADS 3 lesion, the volume should ideally be calculated from MRI data.
Despite these promising results, our study is not without limitations. Most relevant is the relative small population size and the restriction of testing to one Western European clinical site with mostly Caucasian patients. Furthermore, our study did not include a validation cohort, limiting the generalizability of our findings.
5. CONCLUSION
Proclarix shows good performance in detecting csPCa but is hampered by a relatively low specificity, which would still result in many unnecessary biopsies. Proclarix density outperformed Proclarix alone for csPCa detection. In the subgroup of men with PI‐RADS 3 lesions on mpMRI, Proclarix density showed very good accuracy in detecting all csPCa cases while saving up to 40% of unnecessary biopsies.
AUTHOR CONTRIBUTIONS
Conceptualization: Basil Kaufmann and Cédric Poyet. Data collection: Sharon Fischer. Data analysis: Basil Kaufmann. Writing—original draft: Basil Kaufmann and Cédric Poyet. Writing—review and editing: all authors. All authors have read and agreed to the published version of the manuscript.
CONFLICT OF INTEREST STATEMENT
A. A. and R. S. received/held stock options and salaries and founder shares (R. S.) of Proteomedix AG. A. A. and R. S. are inventors of the following patent application (WO2018011212) as well as R. S. on patent application (WO2009138392). All other authors declare no conflicts of interest.
ACKNOWLEDGEMENTS
We thank Alexandra Veloudios for her role in study coordination, sample collection, and storage.
Kaufmann B, Fischer S, Athanasiou A, Lautenbach N, Wittig A, Bieri U, et al. Evaluation of Proclarix in the diagnostic work‐up of prostate cancer. BJUI Compass. 2024;5(2):297–303. 10.1002/bco2.293
Funding information This research was funded by Proteomedix AG, which provided the Proclarix test and lab analysis for free. The study was conducted independently by the authors, and Proteomedix AG had no influence over the study design, data collection, interpretation of the results or manuscript preparation.
DATA AVAILABILITY STATEMENT
The data presented in this study are available on request from the corresponding author.
REFERENCES
- 1. Mottet N, van den Bergh RC, Briers E, van den Broeck T, Cumberbatch MG, de Santis M, et al. EAU‐EANM‐ESTRO‐ESUR‐SIOG guidelines on prostate cancer—2020 update. Part 1: screening, diagnosis, and local treatment with curative intent. Eur Urol. 2021;79(2):243–262. 10.1016/j.eururo.2020.09.042 [DOI] [PubMed] [Google Scholar]
- 2. Van der Kwast TH, Roobol MJ. Defining the threshold for significant versus insignificant prostate cancer. Nat Rev Urol. 2013;10(8):473–482. 10.1038/nrurol.2013.112 [DOI] [PubMed] [Google Scholar]
- 3. Schoots IG. MRI in early prostate cancer detection: how to manage indeterminate or equivocal PI‐RADS 3 lesions? Transl Androl Urol. 2018;7(1):70–82. 10.21037/tau.2017.12.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Venderink W, van Luijtelaar A, Bomers JG, van der Leest M, Hulsbergen‐van de Kaa C, Barentsz JO, et al. Results of targeted biopsy in men with magnetic resonance imaging lesions classified equivocal, likely or highly likely to be clinically significant prostate cancer. Eur Urol. 2018;73(3):353–360. 10.1016/j.eururo.2017.02.021 [DOI] [PubMed] [Google Scholar]
- 5. Schoots IG, Osses DF, Drost F‐JH, Verbeek JFM, Remmers S, van Leenders GJLH, et al. Reduction of MRI‐targeted biopsies in men with low‐risk prostate cancer on active surveillance by stratifying to PI‐RADS and PSA‐density, with different thresholds for significant disease. Transl Androl Urol. 2018;7(1):132–144. 10.21037/tau.2017.12.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kaufmann B, Saba K, Schmidli TS, Stutz S, Bissig L, Britschgi AJ, et al. Prostate cancer detection rate in men undergoing transperineal template‐guided saturation and targeted prostate biopsy. Prostate. 2022;82(3):388–396. 10.1002/pros.24286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mehralivand S, Bednarova S, Shih JH, et al. Prospective evaluation of PI‐RADS™ version 2 using the international society of urological pathology prostate cancer grade group system. J Urol. 2017;198(3):583–590. 10.1016/j.juro.2017.03.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bennett HY, Roberts MJ, Doi SAR, Gardiner RA. The global burden of major infectious complications following prostate biopsy. Epidemiol Infect. 2015;144(8):1784–1791. 10.1017/S0950268815002885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Aizer AA, Gu X, Chen M‐H, Choueiri TK, Martin NE, Efstathiou JA, et al. Cost implications and complications of overtreatment of low‐risk prostate cancer in the United States. J Natl Compr Canc Netw. 2015;13(1):61–68. 10.6004/jnccn.2015.0009 [DOI] [PubMed] [Google Scholar]
- 10. Frisbie JW, van Besien AJ, Lee A, Xu L, Wang S, Choksi A, et al. PSA density is complementary to prostate MP‐MRI PI‐RADS scoring system for risk stratification of clinically significant prostate cancer. Prostate Cancer Prostatic Dis. 2022;26(2):1–6. 10.1038/s41391-022-00549-y [DOI] [PubMed] [Google Scholar]
- 11. Görtz M, Radtke JP, Hatiboglu G, Schütz V, Tosev G, Güttlein M, et al. The value of prostate‐specific antigen density for prostate imaging‐reporting and data system 3 lesions on multiparametric magnetic resonance imaging: a strategy to avoid unnecessary prostate biopsies. Eur Urol Focus. 2021;7(2):325–331. 10.1016/j.euf.2019.11.012 [DOI] [PubMed] [Google Scholar]
- 12. Washino S, Okochi T, Saito K, Konishi T, Hirai M, Kobayashi Y, et al. Combination of prostate imaging reporting and data system (PI‐RADS) score and prostate‐specific antigen (PSA) density predicts biopsy outcome in prostate biopsy naïve patients. BJU Int. 2017;119(2):225–233. 10.1111/bju.13465 [DOI] [PubMed] [Google Scholar]
- 13. Endt K, Goepfert J, Omlin A, Athanasiou A, Tennstedt P, Guenther A, et al. Development and clinical testing of individual immunoassays for the quantification of serum glycoproteins to diagnose prostate cancer. PLoS ONE. 2017;12(8):e0181557. 10.1371/journal.pone.0181557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Macagno A, Athanasiou A, Wittig A, Huber R, Weber S, Keller T, et al. Analytical performance of thrombospondin‐1 and cathepsin D immunoassays part of a novel CE‐IVD marked test as an aid in the diagnosis of prostate cancer. PLoS ONE. 2020;15(5):e0233442. 10.1371/journal.pone.0233442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Steuber T, Heidegger I, Kafka M, Roeder MA, Chun F, Preisser F, et al. PROPOSe: a real‐life prospective study of proclarix, a novel blood‐based test to support challenging biopsy decision‐making in prostate cancer. Eur Urol Oncol. 2022;5(3):321–327. 10.1016/j.euo.2020.12.003 [DOI] [PubMed] [Google Scholar]
- 16. Morote J, Pye H, Campistol M, Celma A, Regis L, Semidey M, et al. Accurate diagnosis of prostate cancer by combining Proclarix with magnetic resonance imaging. BJU Int. 2023;132(2):188–195. 10.1111/bju.15998 [DOI] [PubMed] [Google Scholar]
- 17. Terracciano D, la Civita E, Athanasiou A, Liotti A, Fiorenza M, Cennamo M, et al. New strategy for the identification of prostate cancer: the combination of Proclarix and the prostate health index. Prostate. 2022;82(15):1469–1476. 10.1002/pros.24422 [DOI] [PubMed] [Google Scholar]
- 18. Morote J, Campistol M, Celma A, Regis L, de Torres I, Semidey ME, et al. The efficacy of Proclarix to select appropriate candidates for magnetic resonance imaging and derived prostate biopsies in men with suspected prostate cancer. World J Men's Health. 2022;40(2):270–279. 10.5534/wjmh.210117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Campistol M, Morote J, Triquell M, Regis L, Celma A, de Torres I, et al. Comparison of Proclarix, PSA density and MRI‐ERSPC risk calculator to select patients for prostate biopsy after mpMRI. Cancer. 2022;14(11):2702. 10.3390/cancers14112702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Campistol M, Morote J, Regis L, Celma A, Planas J, Trilla E. Proclarix, a new biomarker for the diagnosis of clinically significant prostate cancer: a systematic review. Mol Diagn Ther. 2022;26(3):1–9. 10.1007/s40291-022-00584-4 [DOI] [PubMed] [Google Scholar]
- 21. Morote RJ, Campistol TM, Celma A, Regis L, Planas J, Santamaria A, et al. Retrospective evaluation of Proclarix as a companion to mpMRI for the detection of clinically significant prostate cancer. Paper presented at: EUROPEAN UROLOGY 2021.
- 22. Pye H, Ahmed H, Heavey S, Stopka‐Farooqui U, Johnston E, Schiess R, et al. Evaluation of Proclarix, a prostate cancer risk score, used together with magnetic resonance imaging for the diagnosis of clinically significant prostate cancer. Am Soc Clin Oncol. 2020;38(6_suppl):278. 10.1200/JCO.2020.38.6_suppl.278 [DOI] [Google Scholar]
- 23. Moon K, Theodorescu D. Does saturation biopsy improve prostate cancer detection? Nat Clin Pract Urol. 2006;3(9):468–469. 10.1038/ncpuro0552 [DOI] [PubMed] [Google Scholar]
- 24. Epstein JI, Feng Z, Trock BJ, Pierorazio PM. Upgrading and downgrading of prostate cancer from biopsy to radical prostatectomy: incidence and predictive factors using the modified Gleason grading system and factoring in tertiary grades. Eur Urol. 2012;61(5):1019–1024. 10.1016/j.eururo.2012.01.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bossuyt PM, Reitsma JB, Bruns DE, Gatsonis CA, Glasziou PP, Irwig L, et al. STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. Clin Chem. 2015;61(12):1446–1452. 10.1373/clinchem.2015.246280 [DOI] [PubMed] [Google Scholar]
- 26. Ahmed HU, Hu Y, Carter T, Arumainayagam N, Lecornet E, Freeman A, et al. Characterizing clinically significant prostate cancer using template prostate mapping biopsy. J Urol. 2011;186(2):458–464. 10.1016/j.juro.2011.03.147 [DOI] [PubMed] [Google Scholar]
- 27. Moskowitz CS, Pepe MS. Comparing the predictive values of diagnostic tests: sample size and analysis for paired study designs. Clin Trials. 2006;3(3):272–279. 10.1191/1740774506cn147oa [DOI] [PubMed] [Google Scholar]
- 28. Tango T. Equivalence test and confidence interval for the difference in proportions for the paired‐sample design. Stat Med. 1998;17(8):891–908. [DOI] [PubMed] [Google Scholar]
- 29. Klocker H, Golding B, Weber S, Steiner E, Tennstedt P, Keller T, et al. Development and validation of a novel multivariate risk score to guide biopsy decision for the diagnosis of clinically significant prostate cancer. BJUI Compass. 2020;1(1):15–20. 10.1002/bco2.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.


