Abstract
Serial crystallography (SX) is an emerging technique that can be used to determine the noncryogenic crystal structure of macromolecules while minimizing radiation damage. Applying SX using pump-probe or mix-and-inject techniques enables the observation of time-resolved molecular reactions and dynamics in macromolecules. After the successful demonstration of the SX experimental technique with structure determination in serial femtosecond crystallography using an X-ray free electron laser, this method was adapted to the synchrotron, leading to the development of serial synchrotron crystallography (SSX). SSX offers new opportunities for researchers to leverage SX techniques, contributing to the advancement of structural biology and offering a deeper understanding of the structure and function of macromolecules. This review covers the background and advantages of SSX and its experimental approach. It also discusses important considerations when conducting SSX experiments.
Keywords: X-ray crystallography, Serial crystallography, Serial synchrotron crystallography, Radiation damage, Room temperature, Time-resolved study
Graphical abstract
Highlights
-
•
The experimental limitation of macromolecular crystallography were introduced.
-
•
The background and advantages of serial synchrotron crystallography were reviewed.
-
•
The sample delivery techniques for serial synchrotron crystallography were reviewed.
-
•
The data processing procedures for serial crystallography were reviewed.
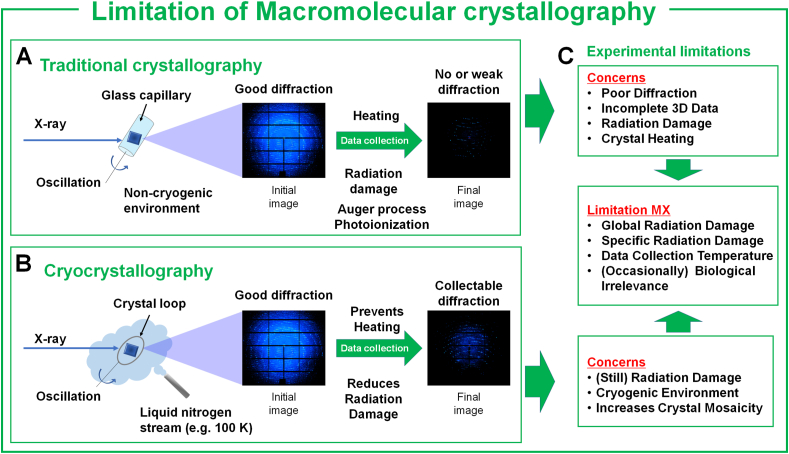
Conventional macromolecular crystallography (MX) using synchrotron X-rays with a single crystal is well established. It provides three-dimensional structural information, elucidating biological phenomena and enzymatic mechanisms at atomic resolution (Cachau et al., 2019; Bose et al., 2021; Bai et al., 2023). This fundamental scientific knowledge supports the development of new drugs (Batool et al., 2019; Thomas et al., 2023) and offers insights into protein engineering aimed at enhancing enzymes used in food, chemical production, and bioenergy industries (Tomadoni et al., 2020; Ao et al., 2023; Kim et al., 2023a; Watson et al., 2023). Although MX remains valuable for determining high-resolution structures, it is limited based on radiation damage and the need for cryogenic temperatures (Garman and Owen, 2005; Shelley and Garman, 2022; Garman and Weik, 2023). MX involves the collection of diffraction data while exposing single crystals to X-rays (Fig. 1A). However, exposed crystals undergo radiation-induced chemical reactions or physical damage, including K-shell photoionization and Auger decay (Garman and Owen, 2005; Shelley and Garman, 2022; Garman and Weik, 2023), thereby resulting in reduced diffraction or the potential for acquiring incomplete datasets. Specifically, when macromolecular crystals absorb X-rays, free radicals are generated within the crystals, leading to global or specific radiation damage (Murray and Garman, 2002). In global radiation damage, there is an overall increase in crystal nonisomorphism, resulting in reduced diffraction quality and an elevated Wilson B coefficient (Murray and Garman, 2002; Nam, 2023b). Specific radiation damage affects the electron density map, causing phenomena such as disulfide bond damage, decarboxylation of aspartic acid and glutamic acid side chains, and impacting the quality of high Z atoms, including metal centers or selenomethionine (Corbett et al., 2007; Nam, 2023a).
Fig. 1.
Limitations of conventional macromolecular crystallography. (A) Radiation damage during room temperature data collection: When X-rays are exposed to a single crystal at room temperature, the diffraction intensity is reduced due to X-ray radiation damage and heating effects. This causes the collection of incomplete data sets or poor diffraction quality. (B) Cryocrystallography experiment: Crystals were immersed in a cryoprotectant solution and exposed to X-rays in a cryogenic environment (e.g., 100 K under a liquid nitrogen stream). (C) Experimental limitations of macromolecular crystallography methods.
Cryocrystallography techniques have been developed to significantly reduce radiation damage (Watenpaugh, 1991; Garman and Weik, 2023) (Fig. 1B). Although this technique increases the success rate of collecting complete three-dimensional diffraction data, X-ray diffraction images still contain information on radiation damage resulting from extended X-ray exposure and the cryogenic temperature (Owen et al., 2006). Additionally, the cryogenic environment can reduce the flexibility of the macromolecule compared with room temperature (Weinert et al., 2017; Durdagi et al., 2021; Kim et al., 2023b), which may be biologically less relevant. Therefore, the crystal structures determined using MX may have accuracy issues stemming from radiation damage or temperature considerations (Fig. 1C). To overcome these challenges, the technique of small wedge synchrotron crystallography (SWSX) was developed, which is between SX and MX. This method involves the collection of multiple small-wedge (5–20° per crystal) dataset from tens or hundreds of crystals in different orientation. Subsequently, these individual data sets are merged to construct a complete dataset to determine the crystal structure (Baba et al., 2021).
The inherent problem of the traditional MX can be overcome with the application of serial crystallography (SX) using X-ray free electron laser (XFEL) or synchrotron X-rays. XFELs produce unprecedentedly bright, short pulse X-rays, avoiding radiation damage by following the “diffraction before destruction” principle (Chapman et al., 2011). In serial femtosecond crystallography (SFX), XFEL exposes the crystal only once during data collection, eliminating cryogenic conditions to prevent radiation damage and enabling structure determination at noncryogenic temperatures. The ultra-short pulse characteristics of XFEL (tens of femtoseconds) are invaluable for tracking fast reaction processes through time-resolved studies (Barends et al., 2022). Particularly, the photoactivation process of photoactive proteins can be tracked through pump-probe experiments using optical lasers (Pearson and Mehrabi, 2020; Poddar et al., 2021). In a pump-probe experiment, a photoactive protein or light-dependent caged molecules are exposed to targeted optical laser excitation with a specific wavelength. Subsequently, pulses of X-rays are directed at the reaction sample at desired time delay points, capturing various intermediate states. Additionally, for proteins interacting with substrates or other molecules, time-resolved studies using liquid applications (e.g., mixing and injection) aid the deeper understanding of macromolecular reaction mechanisms (Worrall and Hough, 2022). In a liquid mixing experiment, protein crystals are combined with a ligand (or inhibitor) in a mixing device. Subsequently, X-rays are directed at the mixing sample at desired time delay points, capturing various reaction states such as intermediate states or pre- and post-reaction states. Series datasets collected from different time-delay points in pump-probe experiments and liquid mixing experiments provide valuable insights into the dynamic behavior of molecules and the progression of the molecular reaction.
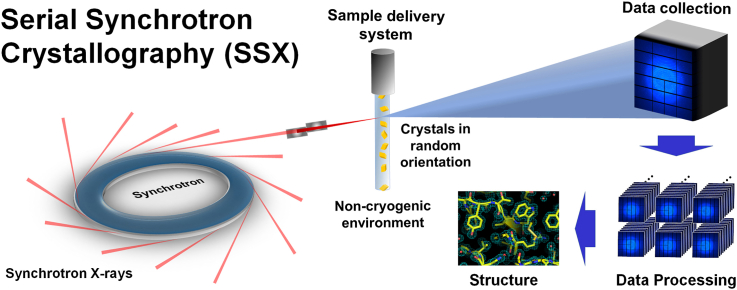
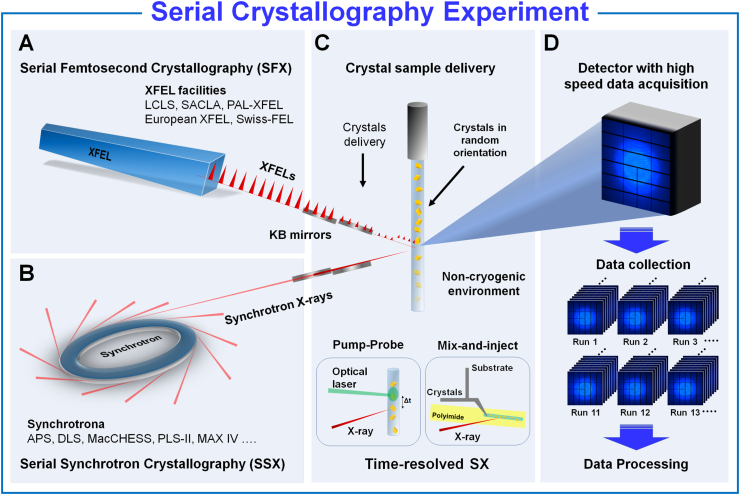
Although SFX offers scientific advantages over previous structural biology techniques, XFEL facilities are limited in number and inaccessible to many researchers. Only five XFELs are available worldwide (Fig. 2), resulting in limited beamtime for general SX studies. To address the beamtime limitation, it is widely established to use serial synchrotron crystallography (SSX) utilizing synchrotron X-rays (Gati et al., 2014; Weinert et al., 2017; Zhu et al., 2020) (Fig. 2). Globally, there are over 70 synchrotron facilities, each capable of serving multiple beamlines simultaneously. Synchrotrons can simultaneously deliver X-rays to different beamlines concurrently. Serial synchrotron crystallography (SSX) experiments have become possible in synchrotrons because of the development of technologies that can focus X-rays to several micrometers and detectors capable of rapid image acquisition. Technological advancements in synchrotron sources and the development of optical focusing mirror systems, such as K–B mirrors, have made the exposure of samples to high photon density beams technically feasible (Stellato et al., 2014). Recent improvements in commercial detector performance have also enabled data collection at several hundred Hz, facilitating SX experiments using synchrotron X-rays. In terms of beamtime accessibility, SSX is more readily available than SFX. XFEL beam time is crucial for time-resolved studies that observe fast molecular reactions in the seconds to femtosecond (10−14 s) timescale, while synchrotron-based, time-resolved Laue MX enables the observation of molecular reactions in the seconds to picosecond (10−10 s) timescale (Orville, 2020) However, synchrotron-based SX experiments are more practical for time-resolved studies involving slower molecular reactions (milliseconds to seconds 10−6 -101 s) or general noncryogenic structure determinations (Orville, 2020). The detailed timescale for the time-resolved study has been reviewed in previous studies (Orville, 2020).
Fig. 2.
Serial crystallography using an X-ray Free Electron Laser (XFEL) and synchrotron X-ray. (A) XFELs provide unprecedented intense X-ray pulses at the femtosecond level. Currently, there are five hard X-ray XFEL facilities worldwide capable of conducting serial femtosecond crystallography (SFX) experiments. One feature of XFELs is that X-rays can be delivered to only one beamline at a time. Although X-rays can be distributed to multiple beamlines using optical instruments, this reduces the photon flux. Consequently, the availability of XFEL beamtime for conducting SFX research is highly limited. (B) Synchrotrons are available in many countries, providing an advantage in terms of accessibility and beamtime allocation compared with XFEL facilities. (C) The experimental setup for the SFX and SSX is nearly identical. Crystals are continuously delivered to the X-ray interaction point in a noncryogenic environment. Time-resolved SX experiments using methods such as pump-probe or mix-and-inject allow the observation of reaction mechanisms. (D) Using high-speed detectors, numerous diffraction data were collected to determine the three-dimensional structure.
The progress of SSX experiments closely parallels that of SFX, and the structural results obtained from both techniques are highly similar (Mehrabi et al., 2021). However, SSX uses X-rays with lower brightness than XFEL, leading to longer X-ray exposure times than in SFX. The exposure time depends on the photon flux, the focused beam size (photon density), and the quality of the crystal. In a beamline with a high photon flux and a small beam, adequate diffraction data can be collected by exposing the crystal sample to X-rays for several picoseconds to milliseconds (Stellato et al., 2014; Meents et al., 2017; Weinert et al., 2017). If the photon flux or the photon density reaching the crystal is low, the exposure time required to achieve the necessary diffraction limit increases. The data collection time and diffraction intensity depend on the photon flux and density delivered to the crystal.
To increase photon flux, pink-beam SSX using a multilayer monochromator can be considered an alternative (Meents et al., 2017; Kim and Nam, 2022, 2023). Pink-beam presents a photon flux 10–102 times higher than that of a monochromatic beam and is advantageous in determining structures using a relatively small number of crystal samples. However, SSX using a pink-beam may require an additional experimental setup to lower the background scattering because the scattering increases in low-resolution range (Meents et al., 2017; Kim and Nam, 2022, 2023).
To conduct efficient SSX, it is crucial to deliver X-rays with a high photon density to the sample position. This not only enhances the intensity of crystal diffraction but also reduces the X-ray exposure time, enabling the collection of more data during the beam time. Additionally, an experimental environment (e.g., a vacuum or a helium environment) that minimizes air-scattering because of X-rays is advantageous for improving data quality by decreasing background scattering. Different synchrotron beamlines and sample environments exhibit unique features, indicating that the optimal specifications for SSX data collection vary for each beamline. Therefore, achieving efficient SSX data collection will necessitate the development of specialized devices and data collection strategies tailored to each beamline.
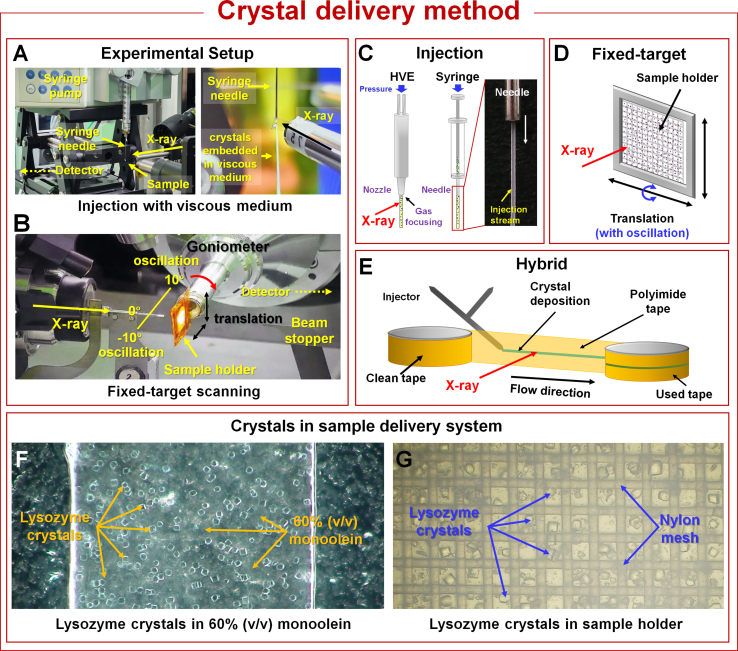
SX experiments involve exposing crystal samples to X-rays only once and require continuous delivery of multiple crystals to obtain complete three-dimensional structural data (Fig. 3). During SX data collection, numerous crystals are delivered to the X-ray interaction point in random orientations. The primary sample delivery techniques used in SX are injector-based and fixed-target (FT) scanning techniques (Zhao et al., 2019; Pearson and Mehrabi, 2020; Park and Nam, 2023) (Fig. 3). In the injector-based sample delivery technique, high viscous excluder (HVE) injectors or syringes with viscous media are widely used because they provide stable crystal sample delivery to X-ray interaction region, even at low flow rates, reducing sample consumption compared with liquid jet injectors (Weierstall et al., 2014; Nam, 2019; Park and Nam, 2019; Zhao et al., 2019; Pearson and Mehrabi, 2020). However, it is essential that the crystal samples remain stable in the viscous medium and do not react with it, as such reactions could damage the crystals and reduce the diffraction intensity. It is also important to ensure that the injection stream is stable to prevent extended or erratic exposure to X-rays, which could cause radiation damage at room temperature. Therefore, preliminary research is required to assess crystal stability in viscous materials and establish a stable injection process when using viscous media (Nam, 2019). Liquid jet injectors require high flow rates to maintain a stable injection stream, which is widely used in SFX using XFEL with high repetition rates. However, in SSX or SFX with low XFEL repetition rates, it is not preferred in terms of sample consumption because the number of samples not exposed to X-rays is overwhelmingly greater than those that are exposed to X-rays (Grünbein and Nass Kovacs, 2019; Zhao et al., 2019; Park and Nam, 2023). Conversely, the FT scanning method minimizes the physical effects on crystals during data collection and allows for precise crystal delivery to the desired positions by programing a translator. This method involves scanning in both vertical and horizontal directions and translation with oscillation using a goniometer or rotator (Wierman et al., 2019; Aumonier et al., 2020; Park et al., 2020). Oscillation during SSX data collection can theoretically provide more reflection information, thereby reducing data indexing ambiguity (Wierman et al., 2019; Aumonier et al., 2020; Park et al., 2020). To prevent radiation damage when scanning a fixed sample, the scanning interval should be configured to ensure that the crystal sample is exposed only once, after considering the beam size.
Fig. 3.
Overview of the experimental setup and sample delivery methods for serial synchrotron crystallography. (A) Photo of SSX experimental setup for injection-based sample delivery. The original figures were obtained from a previous study (Kim and Nam, 2022) and have been modified. This method uses a high-viscosity extruder (HVE) injector or a syringe to extrude an injection stream containing crystals embedded in a viscous medium. (B) Photo of SSX experimental setup for the fixed-target (FT) scanning experiment: Sample holders containing crystal samples are scanned in vertical and horizontal directions during data collection. The original figures were obtained from a previous study (Park et al., 2020) and have been modified. (C) Schematic drawing of the injection-based sample delivery devices. This method involves the use of a high-viscosity extruder (HVE) injector or a syringe to extrude an injection stream containing crystals embedded in a viscous medium. Typically, the HVE injector and syringe samples are extruded using an HPLC pump or mechanical pressure, respectively. The sample extruded from the HVE injector can be focused using the gas such as helium. Various factors, including crystal concentration, beam size, injection stream stability, and hit rate during data collection, determine the injection flow rate. (Inset: photo of an injection stream generated via a syringe) (D) Drawing of the fixed-target (FT) scanning device. This method uses sample holders with regular holes for crystals to settle into without relying on gravity. In general, these sample holders are enclosed in a film to prevent dehydration of the crystal solution. Sample holders containing crystals can be scanned vertically and horizontally, or translation with oscillation can be applied during data collection. (E) Tape-drive-based hybrid method: The crystal suspension was injected from the injector and placed on polyimide tape. The tape winds up, delivering the crystal to the X-ray interaction region. The sample delivery methods described above have been successfully applied to SSX and time-resolved SSX experiments with optical lasers. (F) Photo of lysozyme crystals in 60% (v/v) monoolein for the injection-based sample delivery method. (G) Photo of lysozyme crystals in nylon mesh-based sample holder for the FT scanning method.
Time-resolved studies can also be conducted using both injector and FT scanning methods. This involves collecting diffraction data with a time delay after exciting a photoactive crystal sample or a light-active caged compound that can interact with the crystal sample using an optical laser (Orville, 2020; Pearson and Mehrabi, 2020; Schulz et al., 2022). Additionally, ligand mixing-based time-resolved SSX can be performed using mix-and-diffuse or drop-on-tape approaches with a tape-drive system (Pearson and Mehrabi, 2020; Park and Nam, 2024, Schulz et al., 2022). A sample delivery device suitable for the beamline and an optical laser system capable of triggering the crystal sample must be developed and installed to conduct these time-resolved studies. In SX experiments, preparing many crystals for structure determination is a significant challenge (Beale et al., 2019; Nam, 2019; Mehrabi and Schulz, 2023). Therefore, it is preferable to select a sample delivery device that minimizes crystal consumption and provides a stable environment. Moreover, for mixing-based time-resolved SX, it is important to use thin crystals to account for the rapid diffusion time (Schmidt, 2013). Additionally, to reduce X-ray background scattering from the sample delivery device, it is advisable to expose the delivery material or sample holder to X-rays as thinly as possible.
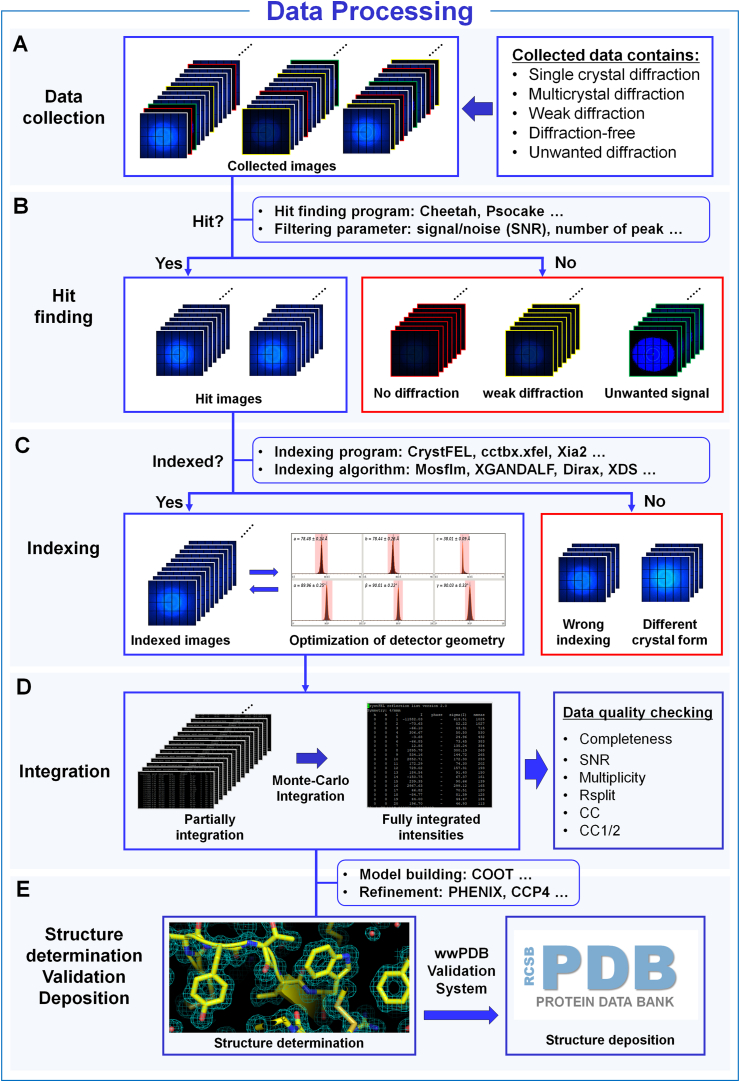
This results in the collection of crystal diffraction patterns and patterns with no interaction between X-rays and the crystal (Fig. 4). To enhance data processing efficiency and storage capacity, images with no or weak-intensity Bragg peaks from collected images should be filtered using hit-finding programs, such as Cheetah or Psocake (Barty et al., 2014, Yoon, 2020). Images with diffraction patterns were further indexed using different crystallographic indexing algorithms, such as MOSFL, XDS, Dirax, and XGANDALF (White, 2019). The precise optimization of detector geometry parameters, including crystal-to-detector distance and detector origin, is crucial for efficient data processing (Yefanov et al., 2015). MX processes images as a single dataset from one large crystal, SX collects data sequentially from multiple crystals, processes multiple datasets, and merges each data to obtain structural information. Specifically, the diffraction information from each image is then integrated, and Monte-Carlo integration is used to generate fully integrated intensities from partially integrated intensities (White, 2019). The resulting structure factor file determines the structure in the subsequent analysis. Collecting images with better diffraction intensity and maximizing the number of images is important because crystallographic statistics are directly proportional to the number of images. Commonly used programs and methods in MX, such as model building (e.g., COOT), structure refinement (e.g., PHENIX, CCP4), and structure visualization (e.g., PyMOL, Chimera), are equally applied for SSX structure determination.
Fig. 4.
General data processing procedure of the serial crystallography experiment. (A). Collected images from SSX experiment include crystal diffraction images and images without crystal diffraction or having weak diffraction intensity. (B). Hit-finding. Based on the hit-finding parameters (signal-to-noise ratio, number of peaks, etc), images with weak or no diffraction intensity and unwanted diffraction patterns were filtered through the hit-finding program such as Cheetah, or Psocake. Although this step is not absolutely essential, it can significantly reduce processing time for subsequent steps, such as indexing, and help maintain data storage efficiency by removing unnecessary data. (C) Indexing. Bragg peaks were indexed, and crystal information was obtained from the crystal diffraction pattern. No indexing or diffraction data with different crystal forms were excluded. (D) Integration. Fully integrated intensities were obtained using Monte-Calro integration with partially integrated intensities. The hkl file containing reflection information is utilized to determine the crystal structure. (E) Structure determination, validation, and deposition of structure factors and coordinates.
SSX continually evolves and promises to revolutionize and introduce new paradigms in structural biology. This experimental technique and SFX offer structural insights different from other structural biology methods, as it can reduce radiation damage and conduct experiments at noncryogenic temperatures.
CRediT authorship contribution statement
Ki Hyun Nam: Conceptualization, Writing – original draft, Visualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was funded by the National Research Foundation of Korea (NRF) (NRF-2017M3A9F6029736 and NRF-2021R1I1A1A01050838) and Korea Initiative for Fostering University of Research and Innovation (KIURI) Program of the NRF (NRF-2020M3H1A1075314). This study was supported by ProGen.
Handling editor: A Wlodawer
Data availability
Data will be made available on request.
References
- Ao Y.F., Pei S., Xiang C., Menke M.J., Shen L., Sun C., et al. Structure‐ and data‐driven protein engineering of transaminases for improving activity and stereoselectivity. Angew. Chem. Int. Ed. 2023;62(23) doi: 10.1002/anie.202301660. [DOI] [PubMed] [Google Scholar]
- Aumonier S., Santoni G., Gotthard G., von Stetten D., Leonard G.A., Royant A. Millisecond time-resolved serial oscillation crystallography of a blue-light photoreceptor at a synchrotron. IUCrJ. 2020;7(4):728–736. doi: 10.1107/s2052252520007411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba S., Matsuura H., Kawamura T., Sakai N., Nakamura Y., Kawano Y., et al. Guidelines for de novo phasing using multiple small-wedge data collection. J. Synchrotron Radiat. 2021;28(5):1284–1295. doi: 10.1107/s1600577521008067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X.-C., Gonen T., Gronenborn A.M., Perrakis A., Thorn A., Yang J. Challenges and opportunities in macromolecular structure determination. Nat. Rev. Mol. Cell Biol. 2023;25(1):7–12. doi: 10.1038/s41580-023-00659-y. [DOI] [PubMed] [Google Scholar]
- Barends T.R.M., Stauch B., Cherezov V., Schlichting I. Serial femtosecond crystallography. Nat. Rev. Methods Primers. 2022;2(1):59. doi: 10.1038/s43586-022-00141-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barty A., Kirian R.A., Maia F.R., Hantke M., Yoon C.H., White T.A., et al. Cheetah: software for high-throughput reduction and analysis of serial femtosecond X-ray diffraction data. J. Appl. Crystallogr. 2014;47(Pt 3):1118–1131. doi: 10.1107/S1600576714007626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batool M., Ahmad B., Choi S. A structure-based drug discovery paradigm. Int. J. Mol. Sci. 2019;20(11):2783. doi: 10.3390/ijms20112783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beale J.H., Bolton R., Marshall S.A., Beale E.V., Carr S.B., Ebrahim A., et al. Successful sample preparation for serial crystallography experiments. J. Appl. Crystallogr. 2019;52(6):1385–1396. doi: 10.1107/s1600576719013517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose K., Rathore I., Mishra V., Bhaumik P. Advancements in macromolecular crystallography: from past to present. Emerg. Top. Life Sci. 2021;5(1):127–149. doi: 10.1042/etls20200316. [DOI] [PubMed] [Google Scholar]
- Cachau R.E., Zhu J., Nicklaus M.C. The upcoming subatomic resolution revolution. Curr. Opin. Struct. Biol. 2019;58:53–58. doi: 10.1016/j.sbi.2019.05.013. [DOI] [PubMed] [Google Scholar]
- Chapman H.N., Fromme P., Barty A., White T.A., Kirian R.A., Aquila A., et al. Femtosecond X-ray protein nanocrystallography. Nature. 2011;470(7332):73–77. doi: 10.1038/nature09750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett M.C., Latimer M.J., Poulos T.L., Sevrioukova I.F., Hodgson K.O., Hedman B. Photoreduction of the active site of the metalloprotein putidaredoxin by synchrotron radiation. Acta Crystallogr. D Biol. Crystallogr. 2007;63(9):951–960. doi: 10.1107/s0907444907035160. [DOI] [PubMed] [Google Scholar]
- Durdagi S., Dağ Ç., Dogan B., Yigin M., Avsar T., Buyukdag C., et al. Near-physiological-temperature serial crystallography reveals conformations of SARS-CoV-2 main protease active site for improved drug repurposing. Structure. 2021;29(12):1382. doi: 10.1016/j.str.2021.07.007. 1396.e1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garman E.F., Owen R.L. Cryocooling and radiation damage in macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2005;62(1):32–47. doi: 10.1107/s0907444905034207. [DOI] [PubMed] [Google Scholar]
- Garman E.F., Weik M. Radiation damage to biological macromolecules. Curr. Opin. Struct. Biol. 2023;82 doi: 10.1016/j.sbi.2023.102662. [DOI] [PubMed] [Google Scholar]
- Gati C., Bourenkov G., Klinge M., Rehders D., Stellato F., Oberthur D., et al. Serial crystallography on in vivo grown microcrystals using synchrotron radiation. IUCrJ. 2014;1(Pt 2):87–94. doi: 10.1107/S2052252513033939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünbein M.L., Nass Kovacs G. Sample delivery for serial crystallography at free-electron lasers and synchrotrons. Acta Crystallogr. D Biol. Crystallogr. 2019;75(2):178–191. doi: 10.1107/s205979831801567x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I.J., Kim S.R., Bornscheuer U.T., Nam K.H. Engineering of GH11 xylanases for optimal pH shifting for industrial applications. Catalysts. 2023;13(11):1405. doi: 10.3390/catal13111405. [DOI] [Google Scholar]
- Kim I.J., Kim S.R., Kim K.H., Bornscheuer U.T., Nam K.H. Characterization and structural analysis of the endo-1,4-β-xylanase GH11 from the hemicellulose-degrading Thermoanaerobacterium saccharolyticum useful for lignocellulose saccharification. Sci. Rep. 2023;13(1) doi: 10.1038/s41598-023-44495-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Nam K.H. Pink-beam serial synchrotron crystallography at pohang light source II. Crystals. 2022;12(11):1637. doi: 10.3390/cryst12111637. [DOI] [Google Scholar]
- Kim Y., Nam K.H. Fixed-target pink-beam serial synchrotron crystallography at pohang light source II. Crystals. 2023;13(11):1544. doi: 10.3390/cryst13111544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meents A., Wiedorn M.O., Srajer V., Henning R., Sarrou I., Bergtholdt J., et al. Pink-beam serial crystallography. Nat. Commun. 2017;8(1):1281. doi: 10.1038/s41467-017-01417-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrabi P., Bücker R., Bourenkov G., Ginn H.M., von Stetten D., Müller-Werkmeister H.M., et al. Serial femtosecond and serial synchrotron crystallography can yield data of equivalent quality: a systematic comparison. Sci. Adv. 2021;7(12) doi: 10.1126/sciadv.abf1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrabi P., Schulz E.C. In: Sousa Â., Passarinha L., editors. Vol. 2652. 2023. Sample preparation for time-resolved serial crystallography: practical considerations; pp. 361–379. (Adv. Protein Chem. Struct. Biol. In Methods Mol. Biol.). Humana, New York, NY. [DOI] [PubMed] [Google Scholar]
- Murray J., Garman E. Investigation of possible free-radical scavengers and metrics for radiation damage in protein cryocrystallography. J. Synchrotron Radiat. 2002;9(6):347–354. doi: 10.1107/s0909049502014632. [DOI] [PubMed] [Google Scholar]
- Nam K.H. Sample delivery media for serial crystallography. Int. J. Mol. Sci. 2019;20(5):1094. doi: 10.3390/ijms20051094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam K.H. Radiation damage on selenomethionine-substituted single-domain substrate-binding protein. Crystals. 2023;13(12):1620. doi: 10.3390/cryst13121620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam K.H. Radiation damage on thaumatin: a case study of crystals that are larger than the microfocusing X-ray beam. Appl. Sci. 2023;13(3):1876. doi: 10.3390/app13031876. [DOI] [Google Scholar]
- Orville A.M. Recent results in time resolved serial femtosecond crystallography at XFELs. Curr. Opin. Struct. Biol. 2020;65:193–208. doi: 10.1016/j.sbi.2020.08.011. [DOI] [PubMed] [Google Scholar]
- Owen R.L., Rudino-Pinera E., Garman E.F. Experimental determination of the radiation dose limit for cryocooled protein crystals. Proc. Natl. Acad. Sci. U. S. A. 2006;103(13):4912–4917. doi: 10.1073/pnas.0600973103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Nam K.H. Sample delivery systems for serial femtosecond crystallography at the PAL-XFEL. Photonics. 2023;10(5) doi: 10.3390/photonics10050557. [DOI] [Google Scholar]
- Park S.Y., Choi H., Eo C., Cho Y., Nam K.H. Fixed-target serial synchrotron crystallography using nylon mesh and enclosed film-based sample holder. Crystals. 2020;10(9):803. doi: 10.3390/cryst10090803. [DOI] [Google Scholar]
- Park S.Y., Nam K.H. Sample delivery using viscous media, a syringe and a syringe pump for serial crystallography. J. Synchrotron Radiat. 2019;26(Pt 5):1815–1819. doi: 10.1107/S160057751900897X. [DOI] [PubMed] [Google Scholar]
- Park J., Nam K.H. Recent chemical mixing devices for time-resolved serial femtosecond crystallography. TrAC, Trends Anal. Chem. 2024;172:117554. doi: 10.1016/j.trac.2024.117554. [DOI] [Google Scholar]
- Pearson A.R., Mehrabi P. Serial synchrotron crystallography for time-resolved structural biology. Curr. Opin. Struct. Biol. 2020;65:168–174. doi: 10.1016/j.sbi.2020.06.019. [DOI] [PubMed] [Google Scholar]
- Poddar H., Heyes D.J., Schirò G., Weik M., Leys D., Scrutton N.S. A guide to time‐resolved structural analysis of light‐activated proteins. FEBS J. 2021;289(3):576–595. doi: 10.1111/febs.15880. [DOI] [PubMed] [Google Scholar]
- Schmidt M. Mix and inject: reaction initiation by diffusion for time-resolved macromolecular crystallography. Adv. Condens. Matter Phys. 2013;2013:1–10. doi: 10.1155/2013/167276. [DOI] [Google Scholar]
- Schulz E.C., Yorke B.A., Pearson A.R., Mehrabi P. Best practices for time-resolved serial synchrotron crystallography. Acta Crystallogr. D Struct. Biol. 2022;78(1):14–29. doi: 10.1107/s2059798321011621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelley K.L., Garman E.F. Quantifying and comparing radiation damage in the protein data bank. Nat. Commun. 2022;13(1):1314. doi: 10.1038/s41467-022-28934-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellato F., Oberthür D., Liang M., Bean R., Gati C., Yefanov O., et al. Room-temperature macromolecular serial crystallography using synchrotron radiation. IUCrJ. 2014;1(4):204–212. doi: 10.1107/s2052252514010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M., Bender A., de Graaf C. Integrating structure-based approaches in generative molecular design. Curr. Opin. Struct. Biol. 2023;79 doi: 10.1016/j.sbi.2023.102559. [DOI] [PubMed] [Google Scholar]
- Tomadoni B., Capello C., Valencia G.A., Gutiérrez T.J. Self-assembled proteins for food applications: a review. Trends Food Sci. Technol. 2020;101:1–16. doi: 10.1016/j.tifs.2020.04.015. [DOI] [Google Scholar]
- Watenpaugh K.D. Macromolecular crystallography at cryogenic temperatures. Curr. Opin. Struct. Biol. 1991;1(6):1012–1015. doi: 10.1016/0959-440x(91)90099-F. [DOI] [Google Scholar]
- Watson J.L., Juergens D., Bennett N.R., Trippe B.L., Yim J., Eisenach H.E., et al. De novo design of protein structure and function with RFdiffusion. Nature. 2023;620(7976):1089–1100. doi: 10.1038/s41586-023-06415-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weierstall U., James D., Wang C., White T.A., Wang D., Liu W., et al. Lipidic cubic phase injector facilitates membrane protein serial femtosecond crystallography. Nat. Commun. 2014;5:3309. doi: 10.1038/ncomms4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert T., Olieric N., Cheng R., Brunle S., James D., Ozerov D., et al. Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons. Nat. Commun. 2017;8:542. doi: 10.1038/s41467-017-00630-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T.A. Processing serial crystallography data with CrystFEL: a step-by-step guide. Acta Crystallogr. D Struct. Biol. 2019;75(Pt 2):219–233. doi: 10.1107/S205979831801238X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierman J.L., Pare-Labrosse O., Sarracini A., Besaw J.E., Cook M.J., Oghbaey S., et al. Fixed-target serial oscillation crystallography at room temperature. IUCrJ. 2019;6(Pt 2):305–316. doi: 10.1107/S2052252519001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worrall J.A.R., Hough M.A. Serial femtosecond crystallography approaches to understanding catalysis in iron enzymes. Curr. Opin. Struct. Biol. 2022;77 doi: 10.1016/j.sbi.2022.102486. [DOI] [PubMed] [Google Scholar]
- Yefanov O., Mariani V., Gati C., White T.A., Chapman H.N., Barty A. Accurate determination of segmented X-ray detector geometry. Opt Express. 2015;23(22):28459–28470. doi: 10.1364/OE.23.028459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon C.H. Psocake: GUI for Making Data Analysis a Piece of Cake. Handbook on Big Data and Machine Learning in the Physical Sciences. 2020. pp. 169–178. [DOI] [Google Scholar]
- Zhao F.Z., Zhang B., Yan E.K., Sun B., Wang Z.J., He J.H., et al. A guide to sample delivery systems for serial crystallography. FEBS J. 2019;286(22):4402–4417. doi: 10.1111/febs.15099. [DOI] [PubMed] [Google Scholar]
- Zhu L., Chen X., Abola E.E., Jing L., Liu W. Serial crystallography for structure-based drug discovery. Trends Pharmacol. Sci. 2020;41(11):830–839. doi: 10.1016/j.tips.2020.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.