Abstract
The 39-kilodalton protein (P39) has previously been shown to be an immunodominant protein in Brucella infections. P39 gene deletion mutants of vaccine strains Brucella abortus S19 and Brucella melitensis Rev.1 were constructed by gene replacement. This deletion did not significantly modify the residual virulence of both vaccine strains in CD-1 mice. CD-1 mice vaccinated with the parent or mutant strains were protected against a virulent challenge. Mutant vaccine strains devoid of P39 could provide a means for differentiating vaccinated from infected animals.
Brucellae are facultative intracellular gram-negative bacteria that cause human disease and significant worldwide economic loss due to infection of livestock. Live attenuated Brucella abortus S19 and Brucella melitensis Rev.1 have served as efficacious vaccine strains for cattle and sheep, respectively (19). Current serologic tests are the major tools for brucellosis diagnosis and mainly detect antilipopolysaccharide antibodies. This dominant antigen is common to virulent and vaccine strains. Therefore, the distinction between infection and vaccination is difficult to make.
Over the past few years studies have been conducted on antiprotein antibody response elicited during brucellosis to identify potential diagnostic antigens (8, 9, 16–18, 21, 22). It appeared that the antibody response against most of the proteins identified was heterogeneous among infected animals and that only a combination of selected Brucella proteins could lead to a sensitive diagnostic test.
Another approach is based on the measure of the specific cellular immune response in infected animals. The delayed-type hypersensitivity (DTH) assay is extremely specific and is complementary to the serologic diagnosis of bovine brucellosis (2, 13). More recently, the gamma interferon (IFN-γ) assay was found to be a powerful diagnostic tool (23). The production of an allergen of defined composition could contribute to the improvement of the DTH test or the IFN-γ assay.
The P39 protein is one of the major components of the allergen manufactured by Rhône-Mérieux, Lyon, France (brucellergene). A brucellergene fraction containing the P39 induced a positive DTH reaction in infected guinea pigs and stimulated the production of IFN-γ by blood cells of infected cattle (12). In cows, DTH and lymphoblastogenesis tests with purified P39 seemed to be specific and sensitive (11). The gene encoding P39 has consequently been cloned and sequenced (11). Purified recombinant P39 also seemed to be a promising antigen for the serologic diagnosis of animal brucellosis (17). Thus, P39 appeared to be useful for the detection of both humoral and cellular immune responses of infected animals.
In the present report, we describe the deletion of the P39 gene from Brucella vaccine strains S19 and Rev.1 and the effect of this deletion on residual virulence and protection in a mouse model. Animals vaccinated with such an engineered vaccine strain would not develop an immune response to P39, and P39 could be further used as an antigen for the differentiation of vaccinated and infected animals.
Construction of P39 gene deletion mutants of B. abortus S19 and B. melitensis Rev.1.
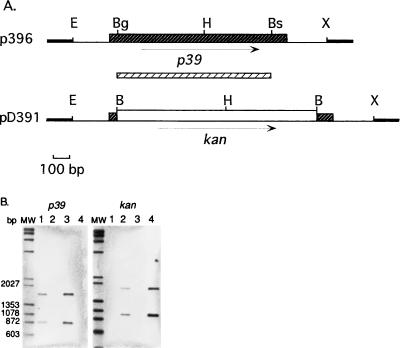
Construction of the deletion plasmid used for the P39 gene replacement in Brucella was done as follows (Fig. 1A). A 1.65-kb EcoRI-XbaI fragment encoding P39 was excised from pTZ1.2. (11) and cloned into the vector pBluescript SK(−) (Stratagene, La Jolla, Calif.) to create p396. The P39 gene open reading frame was deleted from a 1,008-nucleotide fragment by digestion of p396 at the BsmI and BglII unique sites. DNA ends were made blunt, ligated to BamHI linkers, digested with BamHI, and then ligated to the 1.3-kb BamHI kanamycin resistance cassette (kan) from vector pUC4K (Pharmacia P-L Biochemicals, Uppsala, Sweden). This generated the plasmid pD391. A 0.76-kb EcoRI fragment containing oriRK2 was excised from pTJS82 (kindly provided by G. Cornelis, Microbial Pathogenesis Unit, Institute of Cellular Pathology, Brussels, Belgium) and ligated into the EcoRI site of pD391, generating the deletion plasmid pD392. This plasmid was conjugated from Escherichia coli S17-1 into a variant of B. abortus S19 which is resistant to nalidixic acid (Nalr) and B. melitensis Rev.1 Nalr. Since pD392 is unable to replicate in Brucella, the vector-borne kan gene should be rescued by homologous recombination. A double crossover due to homologous recombination events in each of the P39 gene flanking arms resulted in replacement of the P39 gene coding sequence by the kan marker and loss of the vector-encoded bla gene. Brucella transconjugants were selected in the presence of nalidixic acid and kanamycin and further screened by replica plating for ampicillin-sensitive colonies. One Nalr, kanamycin-resistant, and ampicillin-sensitive colony of each vaccine strain was chosen for further study, and the strains were named S19ΔP39 and Rev.1ΔP39.
FIG. 1.
Construction of P39 deletion mutants by gene replacement. (A) Schematic restriction map of plasmid p396 and pD391 inserts. Black hatched box, P39 gene open reading frame; white hatched box, P39 gene probe. X, XbaI, B, BamHI; Bs, BsmI; H, HindIII; Bg, BglII; E, EcoRI. Arrows represent the direction of translation. (B) Southern hybridization of HindIII-digested parent and mutant genomic DNA with P39 gene or kan probes. Lanes 1, Rev.1; lanes 2, Rev.1ΔP39; lanes 3, S19; lanes 4, S19ΔP39; lanes MW, biotinylated lambda/HindIII and φX174/HaeIII digests.
To provide genetic evidence of P39 gene replacement by the kan cassette, DNA isolated from both mutant strains and parent vaccine strains was digested with HindIII and hybridized to P39 gene and kan probes (Fig. 1B). Chemiluminescent detection of biotinylated probes was performed according to the PolarPlex protocol (Millipore, Bedford, Mass.). The two bands (1,650 and 850 bp) characteristic of the presence of the P39 gene in the Rev.1 and S19 DNAs (15) were absent in their respective mutant DNAs. However, the two bands (1,800 and 950 bp) characteristic of the presence of the kan marker were visible only in mutant DNAs. These data indicated that the predicted recombination had occurred, resulting in the wild-type P39 gene being replaced by the kan cassette. Western blot analysis with the anti-P39 monoclonal antibody 5E1E8 (11) confirmed that P39 was not expressed in these gene replacement strains (data not shown). This result demonstrates that P39 is not essential for Brucella survival in vitro, which was also suggested by the absence of P39 protein in three B. abortus strains and in Brucella ovis and Brucella neotomae (11). In addition, deletion of the P39 gene had no detectable effect on conventional species and biovar phenotypic properties, differential characteristics of vaccine strains, or oxidative metabolic patterns (data not shown) (1). To evaluate the in vitro growth rate of the mutant and parent strains, impedance (capacitance) measurements (14) were carried out in Tryptic soy broth supplemented with 0.1% (wt/vol) yeast extract for 48 h and for two initial levels, i.e., 107 and 108 CFU. The generation time of strain Rev.1ΔP39 was significantly lower (4.34 h) than that of the parent Rev.1 strain (6.18 h). No significant difference was observed for the same parameter between S19ΔP39 (2.76 h) and S19 (2.95 h). The effect of P39 on the B. melitensis Rev.1 growth rate will be further analyzed by complementation and overexpression experiments. Absence of P39 expression could compensate for an uncharacterized mutation of the vaccine strain Rev.1.
Residual virulence of the P39 gene mutants in a mouse model.
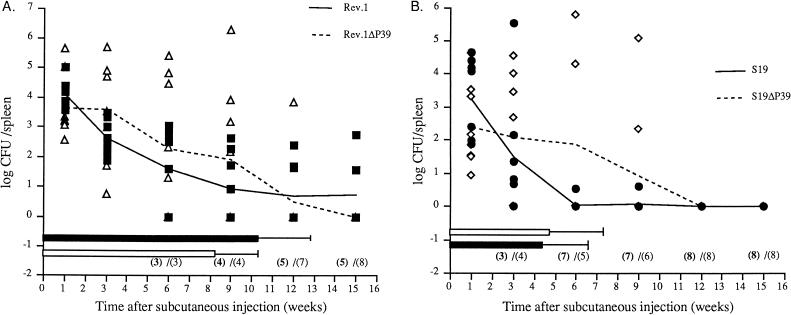
In order to determine the residual virulence of the P39 gene mutants compared to that of the parent strains, 6-week-old CD-1 female mice (eight per group) were injected subcutaneously with 0.2 ml of phosphate-buffered saline (PBS) containing 1.2 × 108 CFU of either B. abortus S19 or S19ΔP39 or B. melitensis Rev.1 or Rev.1ΔP39 (7). Mice were killed at 1, 3, 6, 9, 12, and 15 weeks after the challenge. Their spleens were homogenized in PBS, serially diluted, and plated on tryptic soy agar-yeast extract (TSA-YE). The numbers of CFU per organ were expressed as the log CFU to normalize the distribution of individual counts required for variance analysis (7). Means and standard deviations of transformed values per group were then computed. The 50% recovery times (RT50) and confidence limits (P = 0.95) were calculated at the end of the experiment from the accumulated numbers of Brucella-free spleens by the plotted probit method of Bonet-Maury et al. (3). The P39 gene replacement appeared to be stable because bacterial colonies recovered from mouse spleen at different times postinjection were found to retain kanamycin resistance. Brucella counts in spleens from mice injected with strain Rev.1ΔP39 decreased as regularly as counts of strain Rev.1 from week 3 to week 12 (Fig. 2A). Although the numbers of Brucella-infected spleens were similar at weeks 1, 3, 6, and 9 for both strains, strain Rev.1ΔP39 counts were higher than those of strain Rev.1. Three mice were still infected with Rev.1 at week 15, whereas all mice injected with strain Rev.1ΔP39 were Brucella free. RT50 calculated at the end of the experiment were 10.3 weeks for Rev.1 and 8.2 weeks for Rev.1ΔP39, with confidence limits of ±2.3 and ±2.7, respectively. The difference in RT50 between the two strains was not statistically significant.
FIG. 2.
Residual virulence of B. melitensis Rev.1 strains (A) and B. abortus S19 strains (B). Spleen infection of CD-1 mice after subcutaneous injection of B. melitensis Rev.1 (■) and Rev.1ΔP39 (▵) and B. abortus S19 (•) and S19ΔP39 (◊). Shown are individual results (symbols), mean time courses (lines), and RT50 (horizontal bars, with confidence limits, for mice receiving parent [black bars] and mutant [white bars] strains). Numbers in parentheses are numbers of negative mice out of eight receiving parent strains (bold numbers) and mutant strains (lightface numbers).
Although the number of S19ΔP39 Brucella-infected spleens decreased less rapidly than those infected with S19, all mice injected with either strain were Brucella free from week 12 on (Fig. 2B). As described for Rev.1, Brucella counts in spleens were higher in mice injected with strain S19ΔP39 than in mice injected with strain S19. The RT50, which were not significantly different, were 4.3 weeks for S19 and 4.8 weeks for S19ΔP39, with confidence limits of ±2.1 and ±2.4, respectively.
Although slight changes were observed in the spleen infection kinetics between P39 gene deletion mutants and their parent strains, no difference in residual virulence could be shown. Thus, the higher in vitro growth rate of strain Rev.1ΔP39 does not seem to affect its residual virulence. Even if a subtle effect of the mutation on virulence cannot be excluded by this experiment, our data strongly suggest that P39 is not a crucial virulence factor for the Brucella strains tested. The lack of phenotypes of the P39 gene mutants in vitro and in vivo does not give insights into the function of the P39 protein. However, evaluation of the effect of the P39 gene mutation in a wild-type background could be interesting.
Protection conferred by the B. abortus and B. melitensis vaccine strains with P39 gene deletions in CD-1 mice against the relevant virulent challenge.
P39 deletion mutants as well as the parent strains were tested in the CD-1 mouse model (5, 20) for their ability to protect against a virulent challenge. Mutant vaccine strains (105 CFU/0.2 ml), parent vaccine strains (105 CFU/0.2 ml) as positive controls, and PBS (0.2 ml) as a negative control were injected subcutaneously into 12 mice per group. Thirty days later, the virulent challenge strain (2 × 105 CFU of B. abortus 544 or 1 × 104 CFU of B. melitensis H38) was administered by the intraperitoneal route. Six mice from each group were randomly killed by cervical dislocation to isolate the spleens, 2 or 8 weeks postchallenge. Each spleen was weighed, homogenized, diluted, and spread on TSA-YE alone or TSA-YE plus 0.1% erythritol for differentiation of both B. abortus S19 strains from 544 or on TSA-YE containing 2.5 μg of streptomycin per ml for differentiation of both B. melitensis Rev.1 strains from H38 (S19 and 544 were also differentiated on the basis of CO2 requirement) (1). Colonies of Brucella were enumerated. The number of CFU per spleen was then transformed to y = log(x/log x). This transformation normalizes the distribution of individual counts as required for variance analysis (5, 6).
B. abortus S19ΔP39 induced significant protection against the B. abortus 544 challenge compared to the control PBS group 2 weeks postchallenge and 8 weeks postchallenge (Fig. 3). Mice immunized with the S19 vaccine strain were protected as expected. The same results were obtained in mice immunized with the B. melitensis Rev.1 (Fig. 3). Mice were significantly protected against the B. melitensis H38 virulent challenge.
FIG. 3.
Immune protection conferred in CD-1 mice by B. abortus S19 strains against a B. abortus 544 challenge and B. melitensis Rev.1 strains against a B. melitensis H38 challenge. Brucella counts in spleens of vaccinated mice are significantly different from those of the control mice: ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001. Data were obtained at week 2 (▩) and week 8 (□) postchallenge.
Other Brucella deletion mutants devoid of proteins with potential use in the diagnosis of brucellosis have recently been studied. Deletion of the gene encoding the bacterioferritin did not seem to modify the virulence of B. melitensis 16M in a mouse model (10). Also, the B. abortus S19 vaccine strain with a deletion of the gene encoding the periplasmic BP26 protein protected mice to the same extent as the parental S19 strain (4).
In this work, deletion of the P39 gene in the two current reference live Brucella vaccines for cattle and small ruminants, i.e., S19 and Rev.1, respectively, had no effect on the residual virulence or the protective capability of both vaccines as evaluated in the mouse model. Thus, deletion of genes encoding diagnostic proteins is a promising strategy for the relatively short-term development of live Brucella vaccines allowing differentiation between vaccinated and infected animals.
In addition, a multiprotein reagent for the serologic diagnosis of brucellosis could be evaluated by combining P39 and other antigens of interest, like P15, P17, and BP26 (8, 9, 17, 21). Indeed, in areas of high disease prevalence where vaccination is undertaken, an antiprotein enzyme-linked immunosorbent assay could be sensitive enough (17) and allow differentiation between natural infection and vaccination with an engineered strain. The availability of recombinant P39 protein (17) will also allow us to confirm the usefulness of P39 in the detection of cellular immunity to Brucella in cattle by stimulation of IFN-γ production or by DTH assay. Complementarity between P39 and BFR protein (described as an inducer of IFN-γ production [12]) for the cellular diagnosis of brucellosis will also be evaluated. The potential use of a multicomponent diagnostic antigen implies the construction and evaluation of a compatible vaccine strain mutated for all the corresponding genes.
Acknowledgments
We thank J. Limet, K. Kaniga, V. Weynants, J.-P. Matheise, and A. Cloeckaert for helpful discussions.
This work was supported by the Commission of the European Communities, contract Eclair AGRE-CT90-0049-C (EDB).
REFERENCES
- 1.Alton G, Jones L, Angus R, Verger J-M. Techniques for the brucellosis laboratory. Paris, France: INRA; 1988. [Google Scholar]
- 2.Berkovich Z, Ter Laak E A. An evaluation of the DTH test for diagnosing brucellosis in individual cattle: a field study. Vet Microbiol. 1990;22:241–248. doi: 10.1016/0378-1135(90)90111-8. [DOI] [PubMed] [Google Scholar]
- 3.Bonet-Maury P, Jude A, Servant P. La mesure statistique de la virulence et de l’immunité. Rev Immunol. 1954;18:21–49. [PubMed] [Google Scholar]
- 4.Boschiroli M L, Cravero S L, Arese A I, Campos E, Rossetti O L. Protection against infection in mice vaccinated with a Brucella abortusmutant. Infect Immun. 1997;65:798–800. doi: 10.1128/iai.65.2.798-800.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosseray N, Plommet A-M, Plommet M. Theoretical, practical and statistical basis for a general control method of activity for anti-Brucellavaccines. Dev Biol Stand. 1984;56:257–270. [PubMed] [Google Scholar]
- 6.Bosseray N, Plommet M. Transformation normalisant la distribution du nombre de Brucella dans la rate de souris inoculées par voie intrapéritonéale. J Biol Stand. 1976;4:341–351. doi: 10.1016/s0092-1157(76)80019-4. [DOI] [PubMed] [Google Scholar]
- 7.Bosseray N, Plommet M. Brucella suis S2, Brucella melitensis Rev.1 and Brucella abortus S19 living vaccines: residual virulence and immunity induced against three Brucellaspecies challenge strains in mice. Vaccine. 1990;8:462–468. doi: 10.1016/0264-410x(90)90247-j. [DOI] [PubMed] [Google Scholar]
- 8.Cloeckaert A, Debbarh H S-A, Vizcaino N, Saman E, Dubray G, Zygmunt M S. Cloning, nucleotide sequence, and expression of the Brucella melitensisbp26 gene coding for a protein immunogenic in infected sheep. FEMS Microbiol Lett. 1996;140:139–144. doi: 10.1016/0378-1097(96)00169-3. [DOI] [PubMed] [Google Scholar]
- 9.Debbarh H S A, Cloeckaert A, Zygmunt M S, Dubray G. Identification of seroreactive Brucella melitensiscytosoluble proteins which discriminate between antibodies elicited by infection and Rev.1 vaccination in sheep. Vet Microbiol. 1995;44:37–48. doi: 10.1016/0378-1135(94)00058-5. [DOI] [PubMed] [Google Scholar]
- 10.Denoel P A, Crawford R M, Zygmunt M S, Tibor A, Weynants V E, Godfroid F, Hoover D L, Letesson J-J. Survival of a bacterioferritin deletion mutant of Brucella melitensis16M in human monocyte-derived macrophages. Infect Immun. 1997;65:4337–4340. doi: 10.1128/iai.65.10.4337-4340.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denoel P A, Vo T K-O, Tibor A, Weynants V E, Trunde J-M, Dubray G, Limet J N, Letesson J-J. Characterization, occurrence, and molecular cloning of a 39-kilodalton Brucella abortuscytoplasmic protein immunodominant in cattle. Infect Immun. 1997;65:495–502. doi: 10.1128/iai.65.2.495-502.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denoel P A, Vo T K-O, Weynants V E, Tibor A, Gilson D, Zygmunt M S, Limet J N, Letesson J-J. Identification of the major T-cell antigens present in the Brucella melitensisB115 protein preparation, Brucellergene OCB. J Med Microbiol. 1997;46:801–806. doi: 10.1099/00222615-46-9-801. [DOI] [PubMed] [Google Scholar]
- 13.Fensterbank R. Le diagnostic allergique de la brucellose. Bull Acad Vet Fr. 1982;55:47–52. [Google Scholar]
- 14.Firstenberg-Eden R, Eden G. Impedance microbiology. Letchworth, England: Research Studies Press Ltd; 1985. [Google Scholar]
- 15.Grayon M, Verger J-M, Tibor A, Wansard V, Letesson J-J. 4ème Congrès National de la Société Française de Microbiologie. Tours, France: Société Française de Microbiologie; 1995. Polymorphisme de six gènes codant pour des protéines membranaires ou cytoplasmiques des Brucella, abstr. JE-74. [Google Scholar]
- 16.Hemmen F, Weynants V, Scarcez T, Letesson J-J, Saman E. Cloning and sequence analysis of a newly identified Brucella abortusgene and serological evaluation of the 17-kilodalton antigen that it encodes. Clin Diagn Lab Immunol. 1995;2:263–267. doi: 10.1128/cdli.2.3.263-267.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Letesson J J, Tibor A, van Eynde G, Wansard V, Weynants V, Denoel P, Saman E. Humoral immune responses of Brucella-infected cattle, sheep, and goats to eight purified recombinant Brucellaproteins in an indirect enzyme-linked immunosorbent assay. Clin Diagn Lab Immunol. 1997;4:556–564. doi: 10.1128/cdli.4.5.556-564.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Limet J N, Cloeckaert A, Bezard G, Van Broeck J, Dubray G. Antibody response to the 89-kDa outer membrane protein of Brucellain bovine brucellosis. J Med Microbiol. 1993;39:403–407. doi: 10.1099/00222615-39-6-403. [DOI] [PubMed] [Google Scholar]
- 19.Nicoletti P. Vaccination. In: Nielsen K, Duncan J R, editors. Animal brucellosis. Boca Raton, Fla: CRC Press, Inc.; 1990. pp. 283–299. [Google Scholar]
- 20.Plommet M, Bosseray N. Le contrôle des vaccins antibrucelliques par dénombrement des Brucella dans la rate de souris, vaccinées ou non, inoculées par voie intrapéritonéale. J Biol Stand. 1977;5:261–274. doi: 10.1016/s0092-1157(77)80011-5. [DOI] [PubMed] [Google Scholar]
- 21.Rossetti O L, Arese A I, Boschiroli M L, Cravero S L. Cloning of Brucella abortusgene and characterization of expressed 26-kilodalton periplasmic protein: potential use for diagnosis. J Clin Microbiol. 1996;34:165–169. doi: 10.1128/jcm.34.1.165-169.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tabatabai L B, Hennager S G. Cattle serologically positive for Brucella abortus have antibodies to B. abortusCu-Zn superoxide dismutase. Clin Diagn Lab Immunol. 1994;1:506–510. doi: 10.1128/cdli.1.5.506-510.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weynants V, Godfroid J, Limbourg B, Saegerman C, Letesson J-J. Specific bovine brucellosis diagnosis based on in vitro antigen-specific gamma interferon production. J Clin Microbiol. 1995;33:706–712. doi: 10.1128/jcm.33.3.706-712.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]