Abstract
Purpose:
The gut microbiota plays important roles in health and disease. We questioned whether the gut microbiota and related metabolites are altered in monoclonal gammopathies and evaluated their potential role in multiple myeloma and its response to treatment.
Experimental Design:
We used 16S rRNA sequencing to characterize and compare the gut microbiota of patients with monoclonal gammopathy of undetermined significance (n = 11), smoldering multiple myeloma (n = 9), newly diagnosed multiple myeloma (n = 11), relapsed/refractory multiple myeloma (n = 6), or with complete remission (n = 9). Short-chain fatty acids (SCFA) were quantified in serum and tested in cell lines. Relevant metabolites were validated in a second cohort of 62 patients.
Results:
Significant differences in alpha- and beta diversity were present across the groups and both were lower in patients with relapse/refractory disease and higher in patients with complete remission after treatment. Differences were found in the abundance of several microbiota taxa across disease progression and in response to treatment. Bacteria involved in SCFA production, including Prevotella, Blautia, Weissella, and Agathobacter, were more represented in the premalignant or complete remission samples, and patients with higher levels of Agathobacter showed better overall survival. Serum levels of butyrate and propionate decreased across disease progression and butyrate was positively associated with a better response. Both metabolites had antiproliferative effects in multiple myeloma cell lines.
Conclusions:
We demonstrate that SCFAs metabolites and the gut microbiota associated with their production might have beneficial effects in disease evolution and response to treatment, underscoring its therapeutic potential and value as a predictor.
Translational Relevance.
We have identified changes in gut microbiota composition in the progression of multiple myeloma and its response to treatment. Specifically, we observed a reduction in Short-chain fatty acid (SCFA) producers in patients with active disease and in those with poor prognosis after treatment, which was reflected in SCFA-related metabolic pathways. Importantly, some of these changes were associated with a greater overall survival. Moreover, serum levels of the SCFAs, specifically butyrate, inversely correlated with disease progression and were directly associated with a better response. These changes were validated in a larger cohort and might serve as predictors of disease progression and drug response.
Introduction
Multiple myeloma (MM) is a malignancy of terminally differentiated plasma cells and is the second most common hematologic malignancy after non-Hodgkin lymphoma (1). Most patients present an infiltration of clonal plasma cells in the bone marrow (BM) and monoclonal protein in the serum and/or urine and, accordingly, the disease is part of a class of disorders referred to as the monoclonal gammopathies. MM has an asymptomatic premalignant stage termed monoclonal gammopathy of undetermined significance (MGUS), which consistently precedes the development of the disease, with or without an identified asymptomatic stage referred to as smoldering multiple myeloma (SMM). Nearly 15% of patients with MGUS will progress to MM (2). The introduction of immunomodulatory drugs, proteasome inhibitors such as bortezomib, and CD38-targeting antibodies has extended survival, but ultimately most patients will die from their disease. Response to therapy is an important prognostic factor, with complete response significantly improving long-term survival. Disease progression and subsequent relapses are characterized by increasingly resistant disease (3). There is thus an urgent need for a better understanding of disease evolution to improve risk stratification, to define treatment strategies, and to guide the development of new therapies, including supportive care measures, which are of key importance in disease management.
New knowledge of the pathogenesis of monoclonal gammopathy has highlighted the possibility of disease initiation by microorganisms (4, 5). For example, recent findings in patients with hepatitis C virus infection have revealed that antiviral treatment leads to MGUS or MM regression only in those patients whose monoclonal immunoglobulin targets the virus (6). The gut microbiota is a relevant factor in this context, as it is the community of microorganisms that inhabit the intestinal tract and the most dominant population among other microbial niches in the body (7). There is compelling evidence that an imbalance (dysbiosis) in microbiota composition between individuals in a population can have a strong impact on cancer (8). This imbalance can modify the relationship between the colonizing microorganisms and epithelial/immune cells, leading to disturbances in inflammation, immunoregulation, epithelial cell cycle, proliferation, or mucus production (8). In turn, these disturbances can promote cell transformation or DNA damage, which are risk factors for developing precancerous lesions and cancer (8). Indeed, the microbiome has been recently included as one of “Hallmarks of Cancer” through its involvement in the development, progression, and response to the disease (9).
MM plasma cells can potentially survive in the gastrointestinal tract for long periods of time (10). The nature of the gut microbiota affects the degree of antigen stimulation of these cells and might influence mutation development and clonal evolution (10). Accordingly, strategies to modulate the microbiota with the aim of improving therapeutic response are being investigated in several clinical trials in MM (11). The gut microbiota is also involved in the regulation of hematopoiesis and the correct development of the immune system (12), but the mechanisms of this interaction remain unclear. Of note, microbiota-modulated immunity has been proposed as a mechanism of progression from SMM to MM (11), and an imbalance in the composition and diversity of the gut microbiota has been recently reported in patients with MM at diagnosis (13). In this regard, Calcinotto and colleagues reported an association between a commensal microorganism and disease progression and demonstrated that Prevotella heparinolytica promotes the differentiation and migration of gut Th17 cells toward the BM in a mouse model of myelomagenesis, favoring tumor development (14). Also, Jian and colleagues reported an increase in the relative abundance of nitrogen-recycling gut bacteria and urea nitrogen accumulation during MM disease progression (15). These studies support the influence of the gut microbiota on the myeloma microenvironment and raise questions about how the gut microbiota and its metabolites can affect myeloma development. Along this line, it has been recently proposed that the microbiota can positively influence the BM microenvironment through the production of bioactive metabolites, including short-chain fatty acids (SCFA) such as acetate, propionate, and butyrate, which induce local and systemic responses (16). SCFAs are produced in the colon through bacterial fermentation of dietary fibers (17) and are typically associated with beneficial properties (18–21). Indeed, butyrate producers were found in a higher relative abundance in patients with MM presenting with a negative minimal residual disease treatment response and reduced gastrointestinal toxicity, suggesting an association between microbial signatures and patient outcome (22, 23). In the context of CAR-T cell therapy, an enrichment of SCFA producers has also been observed in patients with hematologic disease achieving complete remission (24).
Despite the growing evidence of the role of the microbiota and its metabolites in MM, it is still unknown what effect the relation of these microorganisms has and their possible role in the progression, response to therapy, and outcome of the disease. In this work, we have identified differences in gut microbiota composition and their metabolites specifically related to the SCFAs production during the MM progression and response to the treatment.
Materials and Methods
Patients and samples
The study included 43 patients with MGUS (n = 11), SMM (n = 9), MM at diagnosis (NDMM; n = 11), with relapse or refractory disease (RRMM; n = 6), or with complete remission (CRMM; n = 9; three of them paired at diagnosis). The control group consisted of individuals (n = 8, median age 66 years; range 49–94 years) with no signs of the disease in BM. We collected fecal and serum samples from this cohort. We used a second independent cohort of patients (n = 62) from a randomized clinical trial (clinicaltrials.gov NCT02575144) for validation and analyzed serum samples at diagnosis. In this cohort, 34 patients received lenalidomide and dexamethasone (Rd) and 28 patients received lenalidomide and dexamethasone with clarithromycin (C-Rd), as previously explained (25). All clinical characteristics at the time of diagnosis are shown in Tables 1 and 2, respectively. The study was approved by the local ethics committee (21/580), and all patients and donors provided written informed consent in accordance with the Declaration of Helsinki.
Table 1.
Main clinical characteristics from cohort 1 patients at disease stage of each group.
| MGUS | SMM | NDMM | RRMM | CRMM | |
|---|---|---|---|---|---|
| Patients, n | 11 | 9 | 11 | 6 | 9 |
| Sex | |||||
| n | 11 | 9 | 10 | 7 | 9 |
| M/F (male %) | 45 | 67 | 70 | 71 | 78 |
| Age at diagnosis | |||||
| n | 11 | 9 | 10 | 7 | 9 |
| Median | 72 | 58 | 68 | 72 | 58 |
| Range | 51–85 | 28–86 | 49–85 | 42–80 | 49–75 |
| Amount of Mc Ig (g/L) | |||||
| n | 11 | 9 | 10 | 6 | 7 |
| Median | 0.48 | 1.2 | 2.61 | 0.875 | 0.07 |
| Range | 0–1.27 | 0.8–4.3 | 0.08–6.8 | 0.16–2.25 | 0–4.3 |
| BM plasma cells (%) | |||||
| n | 3 | 8 | 10 | 5 | 6 |
| Median | 7 | 21 | 29.5 | 26 | 1 |
| Range | 6–8 | 7–60 | 2–77 | 1–50 | 0–44 |
| β2-microglobulin (mg/L) | |||||
| n | 5 | 4 | 6 | 2 | 4 |
| Median | 2 | 2.45 | 4.45 | 3.05 | 2.7 |
| Range | 0.48–2.3 | 1.7–2.5 | 2.4–18.2 | 2.7–3.4 | 2.2–9.4 |
| n > 3.5 mg/L | 0 | 0 | 5 | 0 | 1 |
| Leukocytes (*1,000/μL) | |||||
| n | 11 | 8 | 10 | 7 | 9 |
| Median | 5.5 | 6.3 | 6.05 | 5 | 6.2 |
| Range | 4.1–8.6 | 4.9–7.3 | 2–12.7 | 1.8–13.8 | 0.78–13.4 |
| Hemoglobin (mg/dL) | |||||
| n | 11 | 9 | 10 | 7 | 9 |
| Median | 13.8 | 13.6 | 11.4 | 11.4 | 12.1 |
| Range | 11.1–16.1 | 10.2–16.2 | 7.4–16.7 | 7.6–12.7 | 10.7–14.9 |
| Platelets (*1,000/μL) | |||||
| n | 11 | 8 | 10 | 7 | 9 |
| Median | 210 | 263.5 | 217.5 | 148 | 197 |
| Range | 99–364 | 237–327 | 101–344 | 80–423 | 109–350 |
| Serum calcium (mg/dL) | |||||
| n | 9 | 8 | 9 | 7 | 8 |
| Median | 9.7 | 9.4 | 9.6 | 9.4 | 9.65 |
| Range | 9.4–10.1 | 6.4–9.9 | 9–11.2 | 8.3–9.8 | 8.3–14.08 |
| Creatinine (mg/dL) | |||||
| n | 11 | 9 | 10 | 7 | 8 |
| Median | 0.91 | 0.8 | 1.035 | 0.92 | 0.885 |
| Range | 0.59–1.74 | 0.54–5.36 | 0.69–2.01 | 0.77–4 | 0.73–1.87 |
| Bone lesions | |||||
| n | 9 | 5 | 10 | 6 | 4 |
| n with lesions (%) | 11.11 | 0 | 60 | 83.33 | 25 |
| ISS Stage | |||||
| n | 0 | 2 | 8 | 3 | 5 |
| Stage I | 0 | 1 | 3 | 0 | 3 |
| Stage II | 0 | 0 | 4 | 1 | 1 |
| Stage III | 0 | 0 | 1 | 2 | 1 |
Abbreviations: MGUS, monoclonal gammopathy of undetermined significance; SMM, smoldering myeloma; MM, multiple myeloma; NDMM, MM at diagnosis; RRMM, MM at relapse/refractory; CRMM, MM in complete remission; n, number of patients; M, male; F, female; Mc Ig, monoclonal immunoglobulin; BM, Bone marrow; ISS, Multiple Myeloma International Staging System.
Table 2.
Main patient characteristics of cohort 2 at diagnosis of MM, classified as achieving at least very good partial response or not.
| ≥VGPR | <VGPR | |
|---|---|---|
| Patients, n | 35 | 27 |
| Sex | ||
| n | 35 | 27 |
| M/F (male %) | 42.857143 | 37.037037 |
| Age at diagnosis | ||
| n | 35 | 27 |
| Median | 76 | 80 |
| Range | 66–88 | 68–88 |
| Amount of Mc Ig (g/L) | ||
| n | 35 | 27 |
| Median | 2.58 | 3 |
| Range | 0–8.84 | 0.1–6.01 |
| BM plasma cells (%) | ||
| n | 35 | 35 |
| Median | 22 | 31 |
| Range | 0–93 | 0.4–88 |
| β2-microglobulin (mg/L) | ||
| n | 35 | 27 |
| Median | 3.86 | 4.05 |
| Range | 0–23 | 0–13.4 |
| Nbr > 3.5 mg/L | 21 | 18 |
| Leukocytes (*1,000/μL) | ||
| n | 35 | 27 |
| Median | 6.52 | 5.9 |
| Range | 3–13.6 | 3–8.3 |
| Hemoglobin (mg/dL) | ||
| n | 35 | 27 |
| Median | 11.4 | 10.5 |
| Range | 7.6–14.7 | 8.2–13.8 |
| Platelets (*1,000/μL) | ||
| n | 35 | 27 |
| Median | 235 | 195 |
| Range | 144–395 | 95–378 |
| Calcemia (mg/dL) | ||
| n | 35 | 27 |
| Median | 9.36 | 9.4 |
| Range | 0–110 | 0–11.8 |
| Creatinine (mg/dL) | ||
| n | 35 | 35 |
| Median | 0.89 | 0.93 |
| Range | 0–5.11 | 0.38–3.48 |
| Bone lesions | ||
| n | 18 | 16 |
| n with lesions (%) | 55.6 | 56.3 |
| ISS stage | ||
| n | 35 | 27 |
| Stage I | 11 | 4 |
| Stage II | 12 | 13 |
| Stage III | 12 | 10 |
Note: Response was assessed following the guidelines of the International Myeloma Working Group.
Abbreviations: VGPR, very good partial response; n, number of patients; M, male; F, female; Mc Ig, monoclonal immunoglobulin; BM, bone marrow; ISS, Multiple Myeloma International Staging System.
Fecal total DNA extraction
Fecal and serum samples were collected and immediately stored at −80°C until further processing. Microbial DNA from fecal samples was extracted using the AllPrep PowerFecal DNA/RNA Kit (Qiagen). DNA concentration was determined with a Qubit 4.0 Fluorometer using the dsDNA HS Assay (Invitrogen, Thermo Fisher Scientific).
Targeted library preparation and sequencing
Bacterial 16S ribosomal RNA gene-targeted sequencing was performed using the Quick-16S NGS Library Prep Kit (Zymo Research). The bacterial 16S primers amplified the V1–V2 or V3–V4 regions of the 16S rRNA gene. Primers were custom-designed by Zymo Research to provide the best coverage of the 16S gene while maintaining high sensitivity.
The sequencing library was prepared using a process in which PCR reactions were performed in real-time PCR format to control cycles and prevent/limit PCR chimera formation. The final PCR products were quantified by qPCR fluorescence readings and pooled together based on equal molarity. The pooled library was cleaned up with the Select-a-Size DNA Clean and Concentrator (Zymo Research) and then quantified with TapeStation (Agilent Technologies) on the Qubit platform. The final library was sequenced on an Illumina MiSeq instrument (RRID:SCR_016379) using the v3 reagent kit (600 cycles). The sequencing was performed with >10% PhiX spike-in.
Bioinformatics analysis
Primer regions were removed, and sequences were trimmed to 150 bp using a self-developed Python pipeline. FASTQ files were analyzed using QIIME 2 software (v2019.7; RRID: SCR_018074) to dereplicate reads and remove singletons. Reference-based clustering of operational taxonomic units (OTUs) at 99% similarity was performed using the SILVA 16S database (v138; RRID:SCR_006423). Relative OTU abundances were computed as percent proportions based on the total number of reads per sample. Alpha- and beta-diversity analysis was performed using a Python pipeline. To assess alpha diversity, three metrics were calculated: Pielou's, Shannon's, and Chao's indices. To study beta diversity, the Bray-Curtis dissimilarity measure was computed. Principal coordinate analysis was then applied to the resulting dissimilarity matrices, and two-dimensional principal coordinate analysis plots were used for the visualization of the compositional dissimilarity between groups and samples.
Taxonomic assignment of the identified OTUs was performed using QIIME2 and SILVA 16S taxonomy. The PICRUST2 software package (v2.2.0-b; RRID:SCR_022647) was used to predict the functional content of microbial communities as Kyoto Encyclopedia of Genes and Genomes (KEGG) ortholog profiles and Metacyc pathways. PICRUSt2 inference relies on 16S ribosomal RNA gene sequences and associates OTUs with gene contents.
Biomarker discovery
The linear discriminant analysis effect size (LEfSe) analysis was used to identify differentially abundant taxa and functional pathways acting as biomarkers among groups according to the disease stage (classes). The α-value was fixed <0.05. The bacterial taxa or functions showing significant variations among groups were used to construct the linear discriminant analysis (LDA) model and to estimate its impact as a distinctive factor between them. A threshold of 2.5 was applied to identify discriminative features based on the logarithmic LDA score. Biomarker discovery was conducted at the genus level and across all hierarchical functional levels.
Analysis of metabolites and their effect on tumoral cell lines
Quantification of SCFAs in serum samples was performed by multiple reaction monitoring with liquid chromatography triple quadruple tandem mass spectrometry (LC-QQQ-MS). Proteins were precipitated with an equal volume of acetonitrile and H2O (1:1). After 10 minutes of centrifugation (4°C), derivatization was carried out by mixing 40 μL of samples with 20 μL of 3–3-nitrophenylhydrazine hydrochloride (3-NPH) and 20 μL of 120 mmol/L N-(3-dimethylaminopropy l)-N’-ethylcarbodiimide hydrochloride in 6% pyridine, followed by incubation for 30 minutes at 40°C. Samples were then diluted with 920 μL of 10% acetonitrile. Finally, samples were filtered using 0.22-μm PTFE filters and analyzed on an LC-ESI-QQQ 8030 Shimadzu mass spectrometer (Shimadzu). This analysis was carried out externally at the Mass Spectrometry Unit of the Research Assistance Center of the Universidad Complutense de Madrid (certified according to the ISO 9001:201 standard).
The effect of SCFAs on the proliferation of human MM cell lines was evaluated using a standard tetrazolium salt reduction assay. JJN3 (cat. # ACC541, RRID:CVCL_2078, from the BM of a 57-year-old woman with plasma cell leukemia) and U266B1 [U266] (cat. # TIB-196; RRID:CVCL_0566, from the peripheral blood of a 53-year-old, male patient with myeloma) were obtained in 2018 from DSMZ or ATCC repositories, respectively. After being obtained, both cell lines were lentivirally transduced with a third-generation pRLLluc/GFP virus in our laboratory. GFP+ cells were detected by flow cytometry and quantitative real-time PCR (qPCR), followed by sorting on a FACS BD INFLUX cell sorter (BD Biosciences). To minimize the number of passages after unfreezing (<6), a large stock of original cell lines was generated. Cell authentication and integrity of cell lines were tested by NGS in our laboratory and the Genomic Unit-CNIO. Mycoplasma was tested periodically by PCR in our laboratory. All cells were maintained at 37°C in a humidified incubator, in an atmosphere of 5% CO2, and passaged every 2–3 days.
Cell viability was determined after 48 hours of exposure to compounds using the Cell Counting Kit-8 reagent (Sigma-Aldrich). The dose range was 0.16–250 mmol/L for acetate, 0.23–20 mmol/L for butyrate, and 0–92–80 mmol/L for propionate. The IC50 values were determined according to a nonlinear regression program (GraphPad Prism 8.0.1; RRID: SCR_002798, GraphPad Software Inc.). For drug synergy calculation, a range of serial dilutions of butyrate (12–0.09 mmol/L) and propionate (50–0.39 mmol/L) were made across the IC50 value. The first-line drug bortezomib (5–1.25 nmol/L) was selected in a range of serial dilutions for the combinatory assays. The agents were then added simultaneously for 48 hours, and cell viability assays were performed. Potential synergistic or additive effects were calculated using CompuSyn software (ComboSyn Inc., Biosoft). Drug synergism, addition, and antagonism effects were defined by combination index values of <1.0, 1.0, and >1.0, respectively. The Chou–Talalay combination index theorem was used to calculate the combination index (CI) of bortezomib and butyrate or propionate, respectively (26).
Statistical analysis
We used the normality tests (Shapiro–Wilk, Kolmogorov–Smirnov, and D'Agostino–Pearson) to verify the normal distribution of the data. Normally distributed data are represented as mean ± SEM. For comparisons between two groups, we used the Student t test, and the one-way analysis of variances (ANOVA) with Bonferroni correction for comparisons between more than two groups. The effect of treatment in paired samples was analyzed with a paired t test. Nonnormally distributed data were analyzed by the Kruskal–Wallis one-way ANOVA with Bonferroni post hoc correction or the nonparametric Mann–Whitney U test. Differences among groups in community structure (beta diversity) were tested using a permutational multivariate ANOVA (PERMANOVA). Correlations in controls, MGUS, NDMM, and SMM were calculated with Spearman or Pearson tests, as appropriate. Statistical comparisons were performed using GraphPad Prism 8.0.1 software and Python. Differences were considered statistically significant at P < 0.05.
Regarding the analysis of the cohort of patients (n = 62) from the randomized NCT02575144 clinical trial, the strength of association of each factor with “very good partial response” was estimated using logistic regression models. Univariate and multivariate analyses including age, sex, International Staging System (ISS) stage, treatment, butyrate level, and propionate level were used to determine adjusted odds ratios and 95% confidence intervals (95% CI) for reaching a “very good partial response” relative to a reference category for each group of factors.
Data availability statement
All data generated or analyzed during this study are included in this published article and its supplementary information files. The 16S rRNA sequencing data performed in this study can be accessed under the Sequence Read Archive database (https://www.ncbi.nlm.nih.gov/sra, accession number: SRP473620, bioproject: PRJNA1043906).
Results
SCFA-producing gut microbiota associated with MM progression
First, we investigated the alpha and beta diversity of gut microbiota, which refer to the intrinsic biodiversity (richness) of each sample and the dissimilarity between different groups, respectively, in patients with MM at different stages (MGUS, SMM, and NDMM). No significant differences were found neither in alpha diversity (Pielou's and Shannon's indexes, P = 0.85 and P = 0.92, respectively; Supplementary Fig. S1A) nor beta diversity (Supplementary Fig. S1B) between groups.
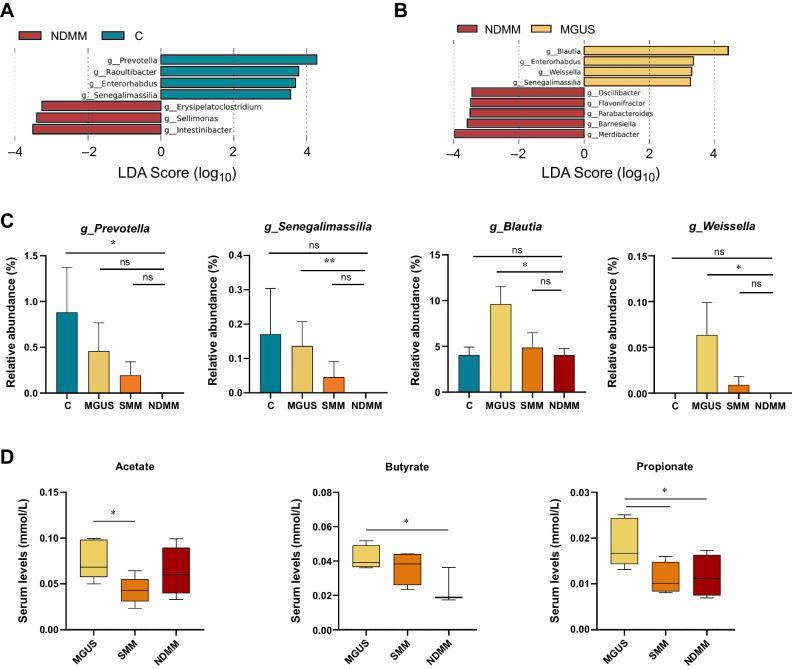
Nevertheless, the taxonomic composition of gut microbiota of patients with MM at different stages was found to be distinct, as presented in Supplementary Fig. S2. Thus, we investigated whether MM progression was associated with more specific changes in the gut microbiota by using the LEfSe biomarker discovery tool that elucidates which genera were driving divergence between the groups. LEfSe analysis allowed to find important SCFA-producing biomarkers such as Prevotella and Senegalimassilia in controls compared with NDMM (Fig. 1A) and Blautia and Weissella in MGUS compared with NDMM (Fig. 1B). Interestingly, their decrease was associated with MM progression. Prevotella and Senegalimassilia abundance decreased from controls to NDMM (Fig. 1C). Weissella and Blautia abundance decreased from MGUS to NDMM groups (P = 0.01 and P = 0.007, respectively; Fig. 1C).
Figure 1.
Gut microbiota associated with the progression of the disease. Taxonomic biomarkers identified by LEfSe analysis between control subjects (C) and multiple myeloma at diagnosis (NDMM; A) and between in patients with monoclonal gammopathy of undetermined significance (MGUS) and NDMM (B). C, Relative abundance (%) of SCFA-producing genera in C and in MGUS, smoldering multiple myeloma (SMM), and NDMM. The results are represented by mean ± SEM. *, P < 0.05; **, P < 0.01; ns, nonsignificant. D, Acetate, propionate and butyrate levels (mmol/L) in serum in MGUS, SMM, and NDMM groups. The results are represented by boxes and whiskers (minimum to maximum). *, P < 0.05.
To explore the functional potential of the microbial communities, we predicted metabolic pathways based on OTU profiles using LEfSe analysis to compare the KEGG metabolic pathways across the different groups. As expected, we also found changes in gut microbiota metabolic pathways associated with disease progression. Specifically, we found a decrease in the abundance of butyrate metabolic pathways in the NDMM group as compared with the control group (P = 0.0049 in the pathway from succinate and P = 0.028 in the pathway from acetyl CoA; Supplementary Fig. S3A), and also in the methanogenesis pathways involving the SCFA acetate when compared with the MGUS group (P = 0.015; Supplementary Fig. S3B).
To validate the association of the SCFAs related to these pathways with disease progression, we studied their levels in serum. We found that acetate was significantly lower in the SMM group than in the MGUS group (P = 0.01), and both butyrate and propionate were significantly lower in the NDMM group than in the MGUS group (P = 0.02 and P = 0.03, respectively; Fig. 1C).
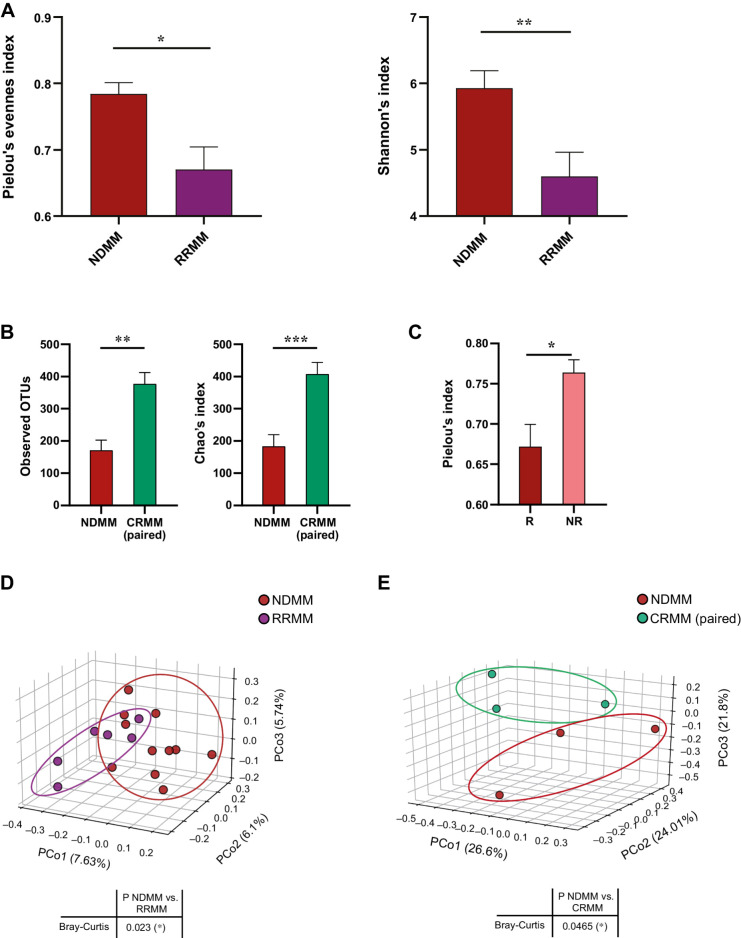
Gut microbiota diversity is altered in the response to treatment
The diversity of gut microbiota from NDMM patients and after response (complete response, CRMM) or not (relapse/refractory, RRMM) to treatment was estimated by alpha diversity. We observed that richness was significantly lower in the RRMM group than in the NDMM group (Pielou index P = 0.003; Shannon index P = 0.009; Fig. 2A). We also studied the alpha diversity in paired patient samples (n = 3) before and after treatment, finding a significant increase in richness in those patients who achieved complete remission (observed OTUs P = 0.003 and Chao's index P = 0.001 between CRMM and NDMM groups; Fig. 2B). Moreover, we found a decrease in the alpha diversity (Pielou's index P = 0.01) in those patients who relapsed compared with those who did not (Fig. 2C).
Figure 2.
The diversity of the gut microbiota in the response to the disease. A and B, Alpha diversity represented by (A) Pielou's and Shannon's indices in newly diagnosed multiple myeloma (NDMM) and multiple myeloma at relapse or refractory (RRMM); B, observed OTUs and Chao's index in paired samples at diagnosis (NDMM) and at complete remission (CRMM). C, Comparison of alpha diversity between patients who relapse (R) or not (NR). The results are represented by mean ± SEM. *, P < 0.05. D and E, Beta diversity represented by principal coordinate analysis plot of Bray-Curtis dissimilarity of (D) NDMM and RRMM and (E) NDMM and CRMM in paired samples. Principal coordinate 1 (PCo1), 2 (PCo2), and 3 (PCoa3) values for each sample are plotted with the percentage of explained variance shown in parentheses.
We next examined beta diversity using principal coordinates analysis to compare the groups according to their community structure. Patients with relapse/refractory disease clustered independently and the PERMANOVA test indicated a significant difference between the RRMM and NDMM groups (P = 0.023; Fig. 2D). Moreover, we observed significant differences in beta diversity between patients with complete remission and patients before treatment (P = 0.046), with CRMM paired samples clustering distinctly from NDMM samples (Fig. 2E).
We found differences in the abundance of some gut microbiota taxa in patients after treatment. At the phylum level, patients with relapse/refractory disease had a much lower relative abundance (%) of some bacterial phyla than the other groups (Supplementary Fig. S2).
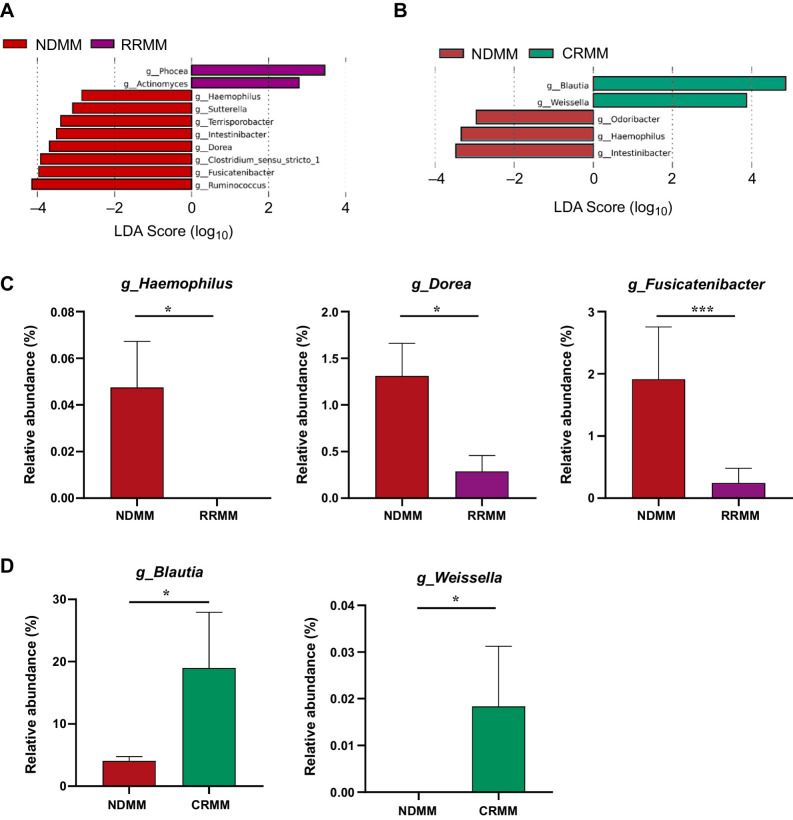
SCFA-producing gut microbiota involved in the response to treatment
In response to treatment, LEfSe analysis again detected differences in several gut microbiota genera involved in the production of SCFAs, both in RRMM (Fig. 3A) and CRMM compared with NDMM (Fig. 3B). Specifically, three bacteria that correlated positively with SCFA production, Haemophilus, Dorea, and Fusicatenibacter (27–29), were significantly lower in abundance in the RRMM group than in NDMM group (P = 0.03, P = 0.02, P = 0.007, respectively; Fig. 3A). At complete response, the levels of the SCFA producers Blautia and Weissella, which decreased in the NDMM group (P = 0.02 and P = 0.04), were restored (Fig. 3B).
Figure 3.
Gut microbiota associated with relapse and response to the disease. A and B, Taxonomic biomarkers identified by LEfSe analysis between multiple myeloma at diagnosis (NDMM) and (A) at relapse/refractory (RRMM) and (B) at complete remission (CRMM). C, Relative abundance (%) of the genus Haemophilus, Fusicatenibacter, and Dorea in NDMM compared with RRMM. D, Relative abundance (%) of the genus Blautia and Weisella in NDMM compared with CRMM. The results are represented by mean ± SEM. *, P < 0.05.
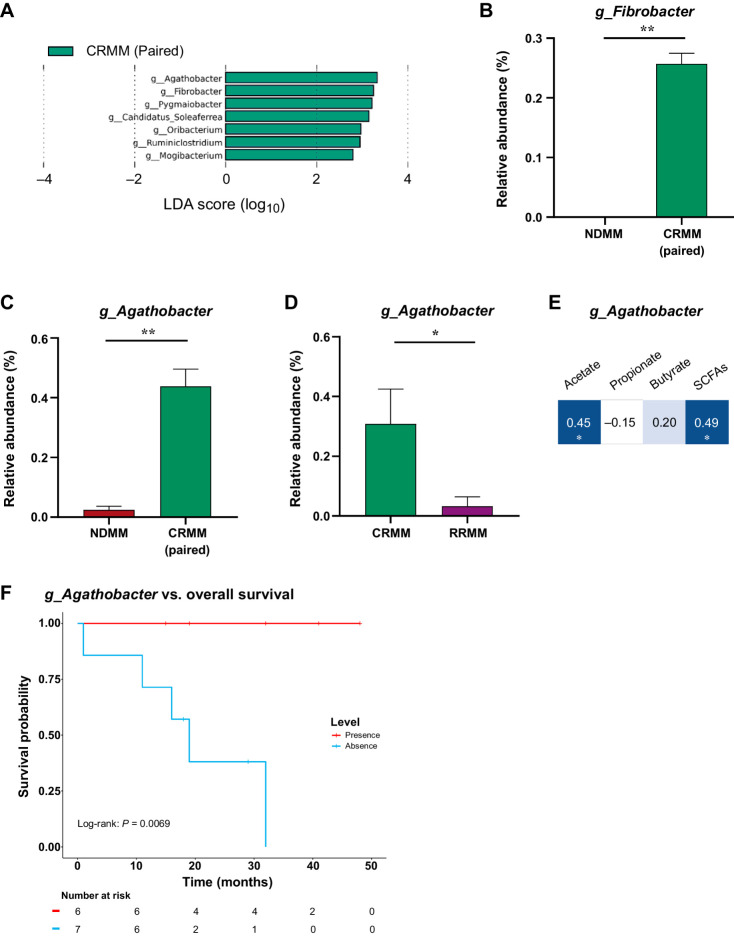
Using paired samples (n = 3), we also identified SCFA-producing genera associated with the CRMM (Fig. 4A). Indeed, we found an increase in the fiber producer Fibrobacter at complete response (P = 0.002; Fig. 4B). We identified the SCFA-producing genus Agathobacter as being potentially involved in the response to treatment. The relative abundance of Agathobacter was higher in the CRMM group than in the NDMM group (P = 0.009; Fig. 4C) and the RRMM group (P = 0.02; Fig. 4D). As Agathobacter has been previously related to SCFA production (30, 31), we analyzed its correlation with serum SCFAs, finding an overall significant positive association, particularly with acetate (Fig. 4E). As Agathobacter was strongly related to complete remission of the disease, we analyzed overall survival in patients after receiving treatment (CRMM and RRMM) with presence/absence of Agathobacter. Notably, the survival in patients with the presence of Agathobacter was significantly longer than those without that genus (P = 0.0069; Fig. 4F).
Figure 4.
Gut microbiota associated with the survival of the disease. A, Taxonomic biomarkers identified by LEfSe analysis between multiple myeloma at diagnosis (NDMM) and at complete remission (CRMM) in paired samples. B–D, Relative abundance (%) of the genus (B) Fibrobacter and (C) Agathobacter in paired samples and (D) Agathobacter in CRMM and RRMM. The results are represented by mean ± SEM. *, P < 0.05. E, Correlation between Agathobacter and SCFAs. Blue color represents a positive correlation (Pearson correlation) and the intensity is proportional to the correlation coefficients. Significant correlation is indicated by * (P < 0.05). F, Kaplan–Meier curve representation of survival probability in patients after receiving treatment (CRMM and RRMM) with presence (red) or absence (blue) levels of Agathobacter.
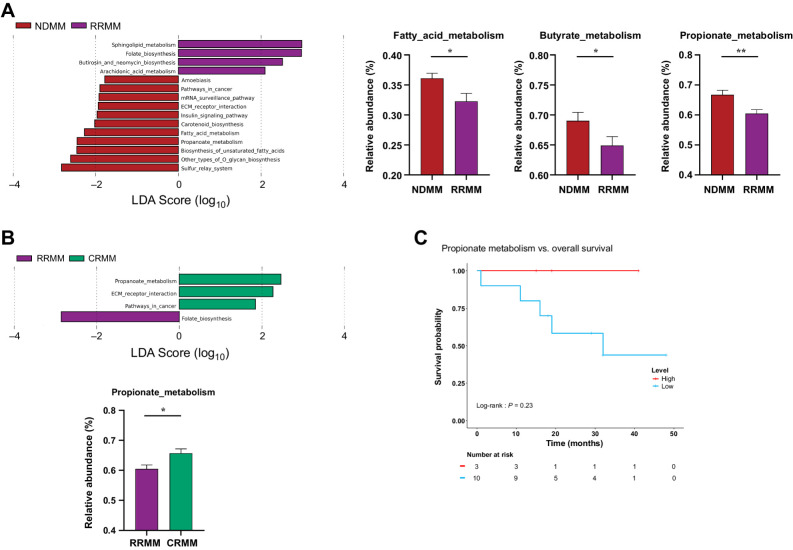
As expected, pathways involved in SCFA production, particularly the butyrate and propionate pathways, were lower in abundance (P = 0.03 and P = 0.008, respectively) in the RRMM group than in the NDMM group (Fig. 5A). Of note, the propionate metabolic pathway was also lower in the RRMM group than in the CRMM group (P = 0.017; Fig. 5B). Finally, we observed a trend that patients with a higher level of the propionate pathway had a higher probability of survival (P = 0.23; Fig. 5C).
Figure 5.
Gut metagenome metabolic pathways. A and B, Linear discriminant analysis (LDA) effect size (LEfSE) analysis showing discriminative Kyoto Encyclopedia of Genes and Genomes (KEGG) metabolic pathways among (A) NDMM and multiple myeloma at relapse or refractory (RRMM); B, RRMM compared with multiple myeloma at complete remission (CRMM). C, Kaplan–Meier curve representation of survival probability in patients after receiving treatment (CRMM and RRMM) with high (red) or low (blue) levels of the propionate metabolism pathway. All KEGG pathways showed statistically significant changes (P < 0.05), with an LDA score threshold set to 2.5. Relative abundance of metabolic pathways related to SCFAs is represented by mean ± SEM. *, P < 0.05; **, P < 0.01.
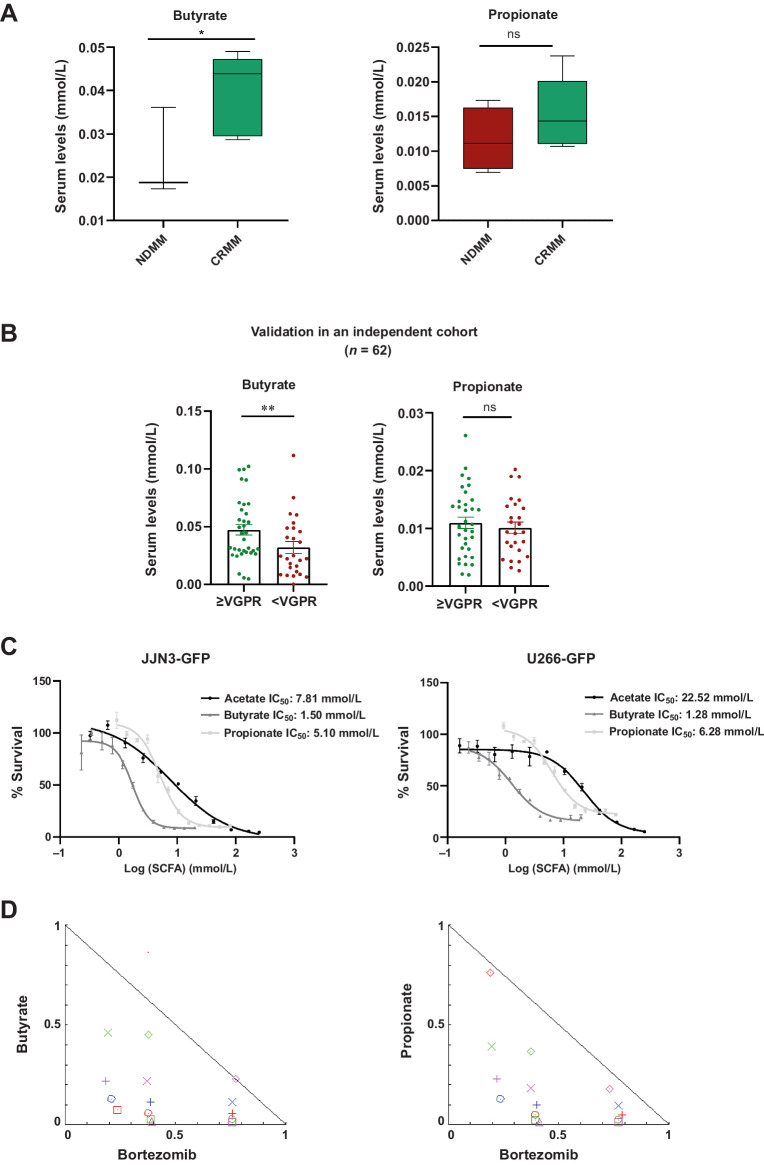
To validate these results, we examined the levels of SCFAs in response to treatment, finding that butyrate levels were significantly higher in the CRMM group than in the NDMM group (P = 0.03). The increase in propionate levels did not show significant differences (P = 0.15; Fig. 6A). Using serum from a larger cohort of patients with MM at diagnosis, we found that butyrate levels were significantly higher in those patients who had a response equal to or better than very good partial response (P = 0.005; Fig. 6B). However, we did not find these differences in propionate levels (P = 0.27; Fig. 6B). As it is shown in Supplementary Table S1, the multivariate analysis did not determine a significant influence of other variables (age, sex, ISS, and treatment) on “very good partial response.”
Figure 6.
The SCFA levels in the response to treatment and their antiproliferative effects in multiple myeloma cell lines. A, Butyrate levels and propionate (mmol/L) in serum in patients with NDMM compared with patients at complete remission (CRMM). B, Propionate and butyrate levels (mmol/L) in serum at the time of diagnosis in a cohort from the randomized clinical trial NCT02575144 according to the achievement of a response equal or better than very good partial response (≥VGPR) or not (<VGPR). The results are represented by box and whiskers (minimum to maximum) and by mean ± SEM. *, P < 0.05. C, Dose–response curves of SCFAs in JJN3-GFP and U266-GFP MM cell lines. The IC50 value is shown for acetate, butyrate, and propionate. D, Normalized isobolograms for butyrate and propionate in combination with bortezomib, respectively in the JJN3-GFP cell line.
Effect of SCFAs on tumoral cell lines
To explore the beneficial role of SCFAs, we performed in vitro assays in MM cell lines. Acetate, butyrate, and propionate efficiently inhibited cell growth in the millimolar range both in JJN3-GFP (IC50 acetate: 7.81 mmol/L, IC50 butyrate: 1.50 mmol/L, IC50 propionate: 5.10 mmol/L) and in U266-GFP (IC50 acetate: 22.52 mmol/L, IC50 butyrate: 1.28 mmol/L, IC50 propionate: 6.28 mmol/L) cell lines (Fig. 6C). These results indicate that some gut microbiota metabolites have antiproliferative effects on myeloma cell lines. Additionally, we tested the combination of butyrate or propionate with the first-line treatment bortezomib, revealing synergic effects in both cases (Fig. 6D).
Discussion
In the present study, we provide the first evidence for SCFAs (and associated bacteria and pathways) as predictors of MM evolution, including response to treatment. We also demonstrate a role for SCFAs, particularly butyrate and propionate, as factors associated with a better response and higher overall survival, respectively. In line with emerging data in other tumor types (32), we demonstrate that the proliferation of tumoral cells can be modulated by microbiota-related metabolites. In addition, our results confirm the recent findings of Sha and colleagues (23), who reported an association between butyrate in stool samples and a sustained negative minimal residual disease in MM (23).
We found that the community richness of the gut microbiota is reduced in patients with refractory or relapsed disease. The changes in the alpha diversity were not due to the treatment, which has been previously reported in acute myeloid leukemia (33), as patients in complete remission had a higher level of alpha diversity. Furthermore, although the sample size is very small, these differences were also observed when comparing paired samples of patients at diagnosis and in complete remission (n = 3). Likewise, we found differences in beta diversity in patients with relapsed or refractory disease, which clustered distinctly, and again we observed an enrichment of microbiota in patients with complete remission. Overall, our findings are consistent with previous studies in which a lower diversity of gut microbiota was associated with the disease (15) and with a higher risk of needing a transplant and poor survival (34).
To our knowledge, this is the first study to examine the gut microbiota in MM progression and response to treatment through the evaluation of different stages of premalignant or active disease. Future studies could establish typical taxa of each stage of MGUS and SMM to predict the evolution of these gammopathies prior to MM. Although the small size of the cohort limited the analysis of the differences in the diversity due to different types of treatment, our data suggest that studying gut microbiota diversity could be exploited to anticipate the evolution of disease and response to treatment. Indeed, our findings might open new horizons to ameliorate disease progression and poor response to treatment through the administration of prebiotics, probiotics, and postbiotics in patients with poor microbiota diversity. Along this line, dietary strategies to improve the response in hematologic diseases are being investigated in ongoing clinical trials (11). Although other studies focused on “negative” microorganisms enriched in MM, such as Bacteroides, Clostridium, Rothia (13), Klebsiella, and Streptococcus (15), here we have identified some potentially protective genera of the microbiota that are related to SCFA production.
We found that the progression of the disease was related to a decrease in the abundance of some SCFA producers such as Prevotella and Senegalimassilia, (27, 35). Prevotella is a well-known dietary fiber fermenter associated with a high production of total SCFAs, including propionate as the major product (36). As mentioned, Prevotella heparinolytica has been reported to accelerate the progression of MM (14). Nevertheless, the same authors recognized that Prevotella might exert pro- or antitumor effects depending on the dominant strain. Indeed, in preclinical models and humans, the presence of Prevotella melaninogenica in the gut is associated with a decreased number of proinflammatory Th17 cells in the intestine, joints, central nervous system, and BM, leading to suppression of arthritis (37), amelioration of autoimmune encephalomyelitis (38), absence of multiple sclerosis (39), and delayed MM progression (14). With respect to Senegalimassilia, both have been previously reported as decreased in abundance in different diseases (40, 41). We also observed the diminution of two genera, Blautia and Weissella, previously associated with SCFA production, especially propionate (28, 42), associated with MM progression. Although both genera were significantly more abundant in MGUS than in the control group, we could hypothesize a compensatory mechanism, because they have been linked to protection against different diseases (42–44).
Among the taxa that were underrepresented in patients with refractory or relapsed disease, we found the genera Fusicatenibacter, Haemophilus, and Dorea, which are all positively correlated with SCFAs (27–29). Contrastingly, we found a significant enrichment of microorganisms related to SCFA production in patients with complete remission, including the aforementioned Blautia and Weissella. Both bacteria have been previously related to cancer control (45, 46) and a superior response to allo-HSTC (47–49). Likewise, we found an increase in the abundance of Fibrobacter, which provides the substrates for SCFA production from cellulose (50), and the SCFA producer Agathobacter. Indeed, we found a positive correlation between Agathobacter and SCFAs, which has been previously reported (30, 31) and, importantly, survival was significantly longer survival in patients with a higher relative abundance of Agathobacter than in those with lower abundance, pointing to this microorganism as a potential biomarker of response. In fact, enrichment of Agathobacter has been previously observed in patients with favorable overall response rates and progression-free survival in advanced non–small cell lung cancer (51).
In summary, the present study strongly supports that SCFA production is associated with delayed MGUS progression to MM and a better response to therapy. Importantly, the study of the SCFA pathways and levels revealed that butyrate and propionate were associated with the progression and a better response. Due to the low number of patients in one of the branches, we have not been able to observe significant differences. However, the metabolism of propionate could potentially have an influence on overall survival. Further studies with a larger cohort are needed to validate this finding. The in vitro analysis in MM lines after treatment with these metabolites revealed an antiproliferative potential of these metabolites in cell lines and the combination of butyrate or propionate with bortezomib had a synergic effect which could be explored as a therapeutic option. These observations, corroborated by two independent cohorts, are in line with previous suggestions of a protective role of these compounds, particularly butyrate (19), based on three independent studies by Jian and colleagues (15), Pianko and colleagues (22), and Shah and colleagues (23). In the first study, Clostridium butyricum, a butyrate producer, was decreased in abundance in patients with MM. In the second and third studies, the butyrate producer Eubacterium hallii and butyrate levels were increased in MRD-negative patients with MM. Although further validation will be necessary, our data also point to the value of SCFA producers such as Agathobacter, as possible biomarkers of disease progression and response to treatment. In addition to the low diversity associated with poor responders, our results underscore the importance of the gut microbiota for MM evolution and its response to treatment.
Supplementary Material
Supplementary Material
Acknowledgments
This work was supported by grant PID2021-123056OA-I00 funded by MCIN/AEI/10.13039/501100011033 and ERDF A way of making Europe, the Instituto de Investigación Hospital 12 de Octubre (i+12), CIBERONC, AECC (Accelerator Award and Ideas Semilla, IDEAS20014LINA), and the CRIS foundation. A. Rodríguez-García is the recipient of a grant from the Sociedad Española de Hematología y Hemoterapia, and R. Garcia-Vicente holds a predoctoral fellowship (FPU) grant from the Ministry of Science, Innovation and Universities of Spain (FPU19/04933). We are particularly indebted to all the patients who participated in the study and to Dr. María Hernández-Sánchez for the careful revision of the manuscript.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
R. Garcia-Vicente reports grants from the Spanish Ministry of Science, Innovation and Universities (FPU19/04933) during the conduct of the study. N. Puig reports grants, personal fees, and other support from Amgen, BMS, Janssen, and Takeda; personal fees from The Binding Site and Sanofi; and grants and personal fees from Pfizer outside the submitted work. M. Mateos reports personal fees from Janssen, BMS-Celgene, Amgen, Takeda, GSK, AbbVie, Oncopeptides, Sanofi, Pfizer, Regeneron, Roche, Sea-Gen, Bluebird bio, and Adaptive outside the submitted work. J. Martinez-Lopez reports personal fees and nonfinancial support from Janssen; grants and nonfinancial support from Roche and BMS; and personal fees and nonfinancial support from Sanofi outside the submitted work. No disclosures were reported by the other authors.
Authors' Contributions
A. Rodríguez-García: Formal analysis, investigation, methodology, writing–original draft, writing–review and editing. A. Arroyo: Software, formal analysis, writing–review and editing. R. Garcia-Vicente: Methodology, writing–review and editing. M. Morales: Methodology, writing–review and editing. R. Gómez-Gordo: Software, formal analysis. P. Justo: Patients samples. C. Cuéllar: Patients samples. J. Sánchez-Pina: Patients samples. N. López: Patients samples. R. Alonso: Resources. N. Puig: Resources. M.-V. Mateos: Resources. R. Ayala: Funding acquisition. D. Gómez-Garre: Supervision, writing–review and editing. J. Martínez-López: Conceptualization, resources, supervision, funding acquisition, project administration, writing–review and editing. M. Linares: Conceptualization, resources, supervision, funding acquisition, project administration, writing–review and editing.
References
- 1. Kumar S, Paiva B, Anderson KC, Durie B, Landgren O, Moreau P, et al. International myeloma working group consensus criteria for response and minimal residual disease assessment in multiple myeloma. Lancet Oncol 2016;17:e328–46. [DOI] [PubMed] [Google Scholar]
- 2. Landgren O, Kyle RA, Pfeiffer RM, Katzmann JA, Caporaso NE, Hayes RB, et al. Monoclonal gammopathy of undetermined significance (MGUS) consistently precedes multiple myeloma: a prospective study. Blood 2009;113:5412–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. van de Donk NWCJ, Pawlyn C, Yong KL. Multiple myeloma. Lancet 2021;397:410–27. [DOI] [PubMed] [Google Scholar]
- 4. Bosseboeuf A, Feron D, Tallet A, Rossi C, Charlier C, Garderet L, et al. Monoclonal IgG in MGUS and multiple myeloma targets infectious pathogens. JCI Insight 2017;2:e95367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bosseboeuf A, Mennesson N, Allain-Maillet S, Tallet A, Piver E, Decaux O, et al. Characteristics of MGUS and multiple myeloma according to the target of monoclonal immunoglobulins, glucosylsphingosine, or Epstein-Barr virus EBNA-1. Cancers 2020;12:1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rodríguez-García A, Linares M, Morales ML, Allain-Maillet S, Mennesson N, Sanchez R, et al. Efficacy of antiviral treatment in hepatitis C virus (HCV)-driven monoclonal gammopathies including myeloma. Front Immunol 2022;12:797209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bull MJ, Plummer NT. Part 1: the human gut microbiome in health and disease. Integr Med (Encinitas) 2014;13:17–22. [PMC free article] [PubMed] [Google Scholar]
- 8. Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell 2014;157:121–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov 2022;12:31–46. [DOI] [PubMed] [Google Scholar]
- 10. Alkharabsheh O, Sidiqi MH, Aljama MA, Gertz MA, Frankel AE. The human microbiota in multiple myeloma and proteasome inhibitors. Acta Haematol 2020;143:118–23. [DOI] [PubMed] [Google Scholar]
- 11. Brevi A, Cogrossi LL, Lorenzoni M, Mattorre B, Bellone M. The insider: impact of the gut microbiota on cancer immunity and response to therapies in multiple myeloma. Front Immunol 2022;13:845422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yan H, Baldridge MT, King KY. Hematopoiesis and the bacterial microbiome. Blood 2018;132:559–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang B, Gu J, Liu J, Huang B, Li J. Fecal microbiota taxonomic shifts in Chinese multiple myeloma patients analyzed by Quantitative Polimerase Chain Reaction (QPCR) and 16S rRNA high-throughput sequencing. Med Sci Monit 2019;25:8269–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Calcinotto A, Brevi A, Chesi M, Ferrarese R, Garcia Perez L, Grioni M, et al. Microbiota-driven interleukin-17-producing cells and eosinophils synergize to accelerate multiple myeloma progression. Nat Commun 2018;9:4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jian X, Zhu Y, Ouyang J, Wang Y, Lei Q, Xia J, et al. Alterations of gut microbiome accelerate multiple myeloma progression by increasing the relative abundances of nitrogen-recycling bacteria. Microbiome 2020;8:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ahmed N, Ghannoum M, Gallogly M, de Lima M, Malek E. Influence of gut microbiome on multiple myeloma: friend or foe? J Immunother Cancer 2020;8:e000576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Koh A, De Vadder F, Kovatcheva-Datchary P, Bäckhed F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell 2016;165:1332–45. [DOI] [PubMed] [Google Scholar]
- 18. Zhang M, Zhou Q, Dorfman RG, Huang X, Fan T, Zhang H, et al. Butyrate inhibits interleukin-17 and generates Tregs to ameliorate colorectal colitis in rats. BMC Gastroenterol 2016;16:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Brevi A, Bellone M. Fatty is not that bad: feeding short-chain fatty acids to restrain autoimmunity. Cell Mol Immunol 2017;14:878–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Luu M, Pautz S, Kohl V, Singh R, Romero R, Lucas S, et al. The short-chain fatty acid pentanoate suppresses autoimmunity by modulating the metabolic-epigenetic crosstalk in lymphocytes. Nat Commun 2019;10:760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Maslowski KM, Mackay CR. Diet, gut microbiota and immune responses. Nat Immunol 2011;12:5–9. [DOI] [PubMed] [Google Scholar]
- 22. Pianko MJ, Devlin SM, Littmann ER, Chansakul A, Mastey D, Salcedo M, et al. Minimal residual disease negativity in multiple myeloma is associated with intestinal microbiota composition. Blood Advances 2019;3:2040–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shah UA, Maclachlan KH, Derkach A, Salcedo M, Barnett K, Caple J, et al. Sustained minimal residual disease negativity in multiple myeloma is associated with stool butyrate and healthier plant-based diets. Clin Cancer Res 2022;28:5149–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Smith M, Zakrzewski J, James S, Sadelain M. Posttransplant chimeric antigen receptor therapy. Blood 2018;131:1045–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Puig N, Hernández MT, Rosiñol L, González E, de Arriba F, Oriol A, et al. Lenalidomide and dexamethasone with or without clarithromycin in patients with multiple myeloma ineligible for autologous transplant: a randomized trial. Blood Cancer J 2021;11:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chou T-C. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res 2010;70:440–6. [DOI] [PubMed] [Google Scholar]
- 27. Zhao Y, Bi J, Yi J, Wu X, Ma Y, Li R. Pectin and homogalacturonan with small molecular mass modulate microbial community and generate high SCFAs via in vitro gut fermentation. Carbohydr Polym 2021;269:118326. [DOI] [PubMed] [Google Scholar]
- 28. Pang J, Zhou X, Ye H, Wu Y, Wang Z, Lu D, et al. The high level of xylooligosaccharides improves growth performance in weaned piglets by increasing antioxidant activity, enhancing immune function, and modulating gut microbiota. Front Nutr 2021;8:764556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. de la Cuesta-Zuluaga J, Mueller NT, Álvarez-Quintero R, Velásquez-Mejía EP, Sierra JA, Corrales-Agudelo V, et al. Higher fecal short-chain fatty acid levels are associated with gut microbiome dysbiosis, obesity, hypertension and cardiometabolic disease risk factors. Nutrients 2018;11:E51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Che L, Hu Q, Wang R, Zhang D, Liu C, Zhang Y, et al. Inter-correlated gut microbiota and SCFAs changes upon antibiotics exposure links with rapid body-mass gain in weaned piglet model. J Nutr Biochem 2019;74:108246. [DOI] [PubMed] [Google Scholar]
- 31. Oh JK, Vasquez R, Kim SH, Hwang I-C, Song JH, Park JH, et al. Multispecies probiotics alter fecal short-chain fatty acids and lactate levels in weaned pigs by modulating gut microbiota. J Anim Sci Technol 2021;63:1142–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Coutzac C, Jouniaux J-M, Paci A, Schmidt J, Mallardo D, Seck A, et al. Systemic short chain fatty acids limit antitumor effect of CTLA-4 blockade in hosts with cancer. Nat Commun 2020;11:2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Galloway-Peña JR, Smith DP, Sahasrabhojane P, Ajami NJ, Wadsworth WD, Daver NG, et al. The role of the gastrointestinal microbiome in infectious complications during induction chemotherapy for acute myeloid leukemia. Cancer 2016;122:2186–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Peled JU, Gomes ALC, Devlin SM, Littmann ER, Taur Y, Sung AD, et al. Microbiota as predictor of mortality in allogeneic hematopoietic-cell transplantation. N Engl J Med 2020;382:822–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nogal A, Louca P, Zhang X, Wells PM, Steves CJ, Spector TD, et al. Circulating levels of the short-chain fatty acid acetate mediate the effect of the gut microbiome on visceral fat. Front Microbiol. 2021;12:711359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen T, Long W, Zhang C, Liu S, Zhao L, Hamaker BR. Fiber-utilizing capacity varies in Prevotella- versus Bacteroides-dominated gut microbiota. Sci Rep 2017;7:2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Marietta EV, Murray JA, Luckey DH, Jeraldo PR, Lamba A, Patel R, et al. Suppression of inflammatory arthritis by human gut-derived prevotella histicola in humanized mice. Arthritis Rheumatol 2016;68:2878–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mangalam A, Shahi SK, Luckey D, Karau M, Marietta E, Luo N, et al. Human gut-derived commensal bacteria suppress CNS inflammatory and demyelinating disease. Cell Rep 2017;20:1269–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cosorich I, Dalla-Costa G, Sorini C, Ferrarese R, Messina MJ, Dolpady J, et al. High frequency of intestinal TH17 cells correlates with microbiota alterations and disease activity in multiple sclerosis. Sci Adv 2017;3:e1700492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tremlett H, Fadrosh D, Lynch S, Hart J, Graves J, Lulu S, et al. Gut microbiome in early pediatric multiple sclerosis: a case-control study (P4.027). Neurology 2015;84. Available from: https://n.neurology.org/content/84/14_Supplement/P4.027 [Google Scholar]
- 41. Jin M, Li D, Ji R, Liu W, Xu X, Li Y. Changes in intestinal microflora in digestive tract diseases during pregnancy. Arch Gynecol Obstet 2020;301:243–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Antoine Pepeljugoski C, Morgan G, Braunstein M. Analysis of intestinal microbiome in multiple myeloma reveals progressive dysbiosis compared to MGUS and healthy individuals. Blood 2019;134:3076. [Google Scholar]
- 43. Teixeira CG, Fusieger A, Milião GL, Martins E, Drider D, Nero LA, et al. Weissella: an emerging bacterium with promising health benefits. Probiotics & Antimicro Prot 2021;13:915–25. [DOI] [PubMed] [Google Scholar]
- 44. Takahashi K, Nishida A, Fujimoto T, Fujii M, Shioya M, Imaeda H, et al. Reduced abundance of butyrate-producing bacteria species in the fecal microbial community in Crohn's disease. Digestion 2016;93:59–65. [DOI] [PubMed] [Google Scholar]
- 45. Liu X, Mao B, Gu J, Wu J, Cui S, Wang G, et al. Blautia: a new functional genus with potential probiotic properties? Gut Microbes 2021;13:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kwak S-H, Cho Y-M, Noh G-M, Om A-S, Cancer preventive potential of kimchi lactic acid bacteria (Weissella cibaria, Lactobacillus plantarum). J Cancer Prev 2014;19:253–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Devaux CA, Million M, Raoult D, The butyrogenic and lactic bacteria of the gut microbiota determine the outcome of allogenic hematopoietic cell transplant. Front Microbiol 2020;11:1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dutta D, Lim SH, Bidirectional interaction between intestinal microbiome and cancer: opportunities for therapeutic interventions. Biomark Res 2020;8:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jenq RR, Taur Y, Devlin SM, Ponce DM, Goldberg JD, Ahr KF, et al. Intestinal blautia is associated with reduced death from graft-versus-host disease. Biol Blood Marrow Transplant 2015;21:1373–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chassard C, Delmas E, Robert C, Bernalier-Donadille A. The cellulose-degrading microbial community of the human gut varies according to the presence or absence of methanogens. FEMS Microbiol Ecol 2010;74:205–13. [DOI] [PubMed] [Google Scholar]
- 51. Hakozaki T, Richard C, Elkrief A, Hosomi Y, Benlaïfaoui M, Mimpen I, et al. The gut microbiome associates with immune checkpoint inhibition outcomes in patients with advanced non-small cell lung cancer. Cancer Immunol Res 2020;8:1243–50. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files. The 16S rRNA sequencing data performed in this study can be accessed under the Sequence Read Archive database (https://www.ncbi.nlm.nih.gov/sra, accession number: SRP473620, bioproject: PRJNA1043906).