Abstract
Purpose:
Camonsertib is a highly selective and potent inhibitor of ataxia telangiectasia and Rad3-related (ATR) kinase. Dose-dependent anemia is a class-related on-target adverse event often requiring dose modifications. Individual patient risk factors for the development of significant anemia complicate the selection of a “one-size-fits-all” ATR inhibitor (ATRi) dose and schedule, possibly leading to suboptimal therapeutic doses in patients at low risk of anemia. We evaluated whether early predictors of anemia could be identified to ultimately inform a personalized dose-modification approach.
Patients and Methods:
On the basis of preclinical observations and a mechanistic understanding of ATRi-related anemia, we identified several potential factors to explore in a multivariable linear regression modeling tool for predicting hemoglobin level ahead of day 22 (cycle 2) of treatment.
Results:
In patients treated with camonsertib monotherapy (NCT04497116), we observed that hemoglobin decline is consistently preceded by reticulocytopenia, and dose- and exposure-dependent decreases in monocytes. We developed a nomogram incorporating baseline and day 8 hemoglobin and reticulocyte values that predicted the day 22 hemoglobin values of patients with clinically valuable concordance (within 7.5% of observations) 80% of the time in a cross-validation performance test of data from 60 patients.
Conclusions:
The prediction of future hemoglobin decrease, after a week of treatment, may enable a personalized, early dose modification to prevent development of clinically significant anemia and resulting unscheduled dose holds or transfusions.
Translational Relevance.
Camonsertib is a highly selective and potent inhibitor of ataxia telangiectasia and Rad3-related (ATR) kinase, the clinical and translational relevance of which was recently published. The safety profile of camonsertib is largely characterized by anemia, a class effect of ATR inhibitors. The development of a nomogram for camonsertib, described here, may instruct early dose adjustments prior to clinically significant anemia development, thereby preventing unscheduled dose holds and transfusions, and improving long-term tolerability. This nomogram may also provide a model for developing a similar decision tool for other ATR inhibitors and drugs with a similar mechanism of action.
Introduction
Ataxia telangiectasia and Rad3-related (ATR) kinase inhibitors are DNA damage response (DDR)-targeted agents and are currently in development for the treatment of various tumors (1–3). Camonsertib (RP-3500; RG-6526) is a highly potent and selective oral inhibitor of ATR kinase (4) that demonstrated efficacy in biomarker-selected patients with loss-of-function alterations in ataxia-telangiectasia mutated (ATM) and other DDR genes (5). The safety profile of camonsertib is characterized by mechanism-related anemia, which was reported in 68% of patients (32% grade 3) in the TRESR trial (NCT04497116). Anemia typically manifested after cycle 1 of treatment (21 days) and has been the primary determinant of long-term tolerability (5). While anemia is a key-related toxicity of ATR inhibitors (ATRi) in the clinic (2, 6), the mechanism leading to its development is poorly characterized.
Early monocytopenia has been previously described with the ATRi ceralasertib (7) and was hypothesized to be related to a deficiency in the base excision repair mechanism of monocytes (8). This observation was recently confirmed with a larger set of patients treated with various ATRis (9). A role for macrophages/monocytes in supporting erythropoiesis and iron hemostasis has been proposed previously (10). This link can be explained by a critical role of monocytes in the recycling of iron from senescent/damaged red blood cells into late-stage erythroid precursors for heme synthesis and hemoglobin production. Thus, the impaired viability of monocytes could contribute to the reticulocytopenia and hemoglobin decline (11).
Preclinical camonsertib dose/schedule-modeling studies have shown that intermittent dosing schedules (with a drug holiday of at least 4 consecutive days) enable prompt reticulocyte recovery and improve the therapeutic index (4); these studies instructed the evaluation of intermittent dosing schedules of camonsertib in the first-in-human studies (5). Preclinical, mechanistic studies are ongoing to evaluate the effects of camonsertib on red blood cell precursors in-depth.
Here, we describe how a mechanistic understanding of anemia based on preclinical and clinical studies of early hematologic changes informed the development of a nomogram to optimize personalized dosing. The nomogram was developed using day 8 changes in hematologic parameters to predict changes in hemoglobin levels at day 22. In this phase I population, the nomogram was able to predict the day 22 hemoglobin values within 7.5% of observed values 80% of the time. This nomogram could assist clinicians in the interpretation of early peripheral blood cytopenias, enabling preemptive dose modifications prior to the development of clinically significant anemia and alleviating the need for transfusions. This decision tool may facilitate early interventions for the management of anemia for this drug class, thereby minimizing dose holds or reductions, which may consequently improve drug efficacy.
Patients and Methods
Study design and treatment
The phase I/IIa TRESR study (NCT04497116) evaluated camonsertib monotherapy in 154 patients with molecularly selected advanced solid tumors. Patients received camonsertib doses ranging from 5 to 200 mg once daily or 40 to 80 mg twice daily on either 5 days on/2 days off (5/2) or 3 days on/4 days off (3/4) continuous weekly or as an intermittent weekly schedule of 2 weeks on/1 week off (Supplementary Table S1). Patients in an expansion module received camonsertib at the preliminary recommended phase II dose of 160 mg once daily, 3/4, every week. Evaluation of hematologic parameters was performed at a range of doses throughout the treatment period. The study was conducted in accordance with the Declaration of Helsinki and the Council for International Organizations of Medical Sciences International Ethical Guidelines, applicable International Conference on Harmonization Good Clinical Practice Guidelines, and applicable laws and regulations. All patients provided written informed consent to adhere to the clinical protocol, which was approved by the Institutional Review Board or ethics committee at each participating institution.
Study patients and methods
Inclusion and exclusion criteria were reported previously (5). In brief, eligibility criteria included adults with histologically confirmed solid tumors resistant or refractory to standard treatment and the presence of a gene alteration expected to sensitize tumors to ATR inhibition, including loss-of-function alterations in: ATM, ATRIP, BRCA1, BRCA2, CDK12, CHTF8, FZR1, MRE11, NBN, PALB2, RAD17, RAD50, RAD51B/C/D, REV3L, RNASEH2A, RNASEH2B, or SETD2. The requirement for baseline hemoglobin was 9.5 g/dL (10.0 g/dL in the expansion module). For the purpose of this analysis, we collected available relevant parameters guided by preclinical data and literature reports. Those included patient demographics and pharmacokinetics [previously reported (5) and reviewed in Supplementary Table S2] and baseline parameters such as Eastern Cooperative Oncology Group performance status (ECOG PS), body weight, and prior treatments (e.g., prior platinum or PARP inhibitor), as well as on-drug changes in several hematologic parameters including hemoglobin, monocytes, reticulocytes, neutrophils, and platelets on days 8, 15, and 22.
Statistical analysis
The primary objective of this study was to explore the predictors for hemoglobin reduction and occurrence of anemia, using baseline covariates and a nomogram approach. We first assessed whether any baseline patient features were associated with the emergence of treatment-related grade 3+ anemia. Those baseline covariates with a clinically plausible correlation with anemia were considered and included in the univariate logistic regression model with a response variable of grade 3+ treatment-related anemia. Covariates with marginal significance (P < 0.15) were included in the multivariate logistic regression analysis using a stepwise model selection method. The final model retained those covariates if the Wald test P value was <0.1, after adjusting for other covariates in the model, including initial dose groups assigned. Estimated ORs and 95% confidence intervals (CI) were obtained within the framework of these logistic regression models. P values reported were two-sided, without adjusting for multiplicity.
For changes between visits (baseline to post-baseline visit) for hematology lab tests, P value (two-sided) was based on a pairwise t test that included all evaluable patients within the group. Correlation between different hematology lab parameters and exposure were described with scatter plots and estimated Spearman rank correlation coefficients were provided.
Nomogram development
Each nomogram was composed by a single linear mathematical model for predicting the percent change from baseline of a patient's hemoglobin on day 22 using a set of patient input parameters (day 8 monocyte, reticulocyte, neutrophil, platelet, and hemoglobin levels). Nomogram weights were applied to each input parameter to minimize the error observed across the entire nomogram dataset.
All patient data were utilized to develop the nomogram, provided that their clinical observations met the following inclusion criteria: available baseline and day 22 ± 2 hemoglobin level; no dose modifications or withdrawals in the first 15 days of the study (ensuring that all patients had at least 2 weeks of dosing, regardless of schedule); and baseline and day 8 ± 2 hematology parameters (hemoglobin, monocytes, reticulocytes, and platelets and/or neutrophils). A summary of the population for each iteration of the nomogram is shown in Supplementary Fig. S1A and S1B.
The availability of laboratory values for each timepoint used in the nomogram (baseline, day 8, and day 22) resulted in differences in the number of patients available to develop the nomogram, depending on which input parameters were considered. For example, nomograms with input parameters of baseline and day 8 changes in hemoglobin values had the largest set of evaluable patient data (n = 109), whereas nomograms based on both day 8 changes in hemoglobin and reticulocytes had the smallest number of patients available (n = 60).
The sequential generic nomogram equations for predicting hemoglobin on day 22 are:
 |
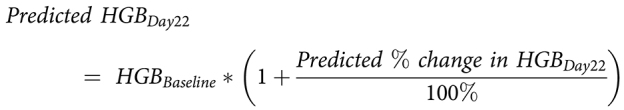 |
where input parameters were one or more parameters selected from a list, including the percent change from baseline in a patient's hematologic parameters (hemoglobin, monocytes, and/or reticulocytes) on day 8 ± 2. Int0 and InputParameterWt are weights derived from the fits to all qualifying individuals for a given nomogram. If a patient did not have a day 8 sample but did have one from within 2 days, the rate of change observed for that patient over the first 6–10 days for the parameter was scaled to day 8.
The performance of a nomogram was assessed to select the best performing structure. This was done by comparing the r2 correlation coefficient and root mean square error (RMSE) of observations versus predictions, as well as the fraction of patients’ predictions for day 22 hemoglobin that were within 7.5% of the observations, across nomograms using different inputs. These parameters were also compared with a non–nomogram-based approach (12), where predictions for the day 22 hemoglobin levels were generated by linear extrapolation of the percent change in hemoglobin observed at approximately day 7.
The nomogram performance was assessed using two approaches. First, a “validation-set approach” was used, where the entire set of patient data was randomly divided into a training set (n = two-thirds of the total available) and a testing set (the remaining one-third of the total available). Nomograms with the optimum structure were calibrated using a training set and performance was assessed on predictions made for individuals in the test set. This analysis was repeated for 100 iterations. Second, a “leave-one-out cross-validation approach” was used, where the complete set of patients was split into a training (n = total available minus 1) and a validation set (n = 1), and the goodness-of-fit scores were assessed. This process was repeated iteratively until all individuals had been the lone point in the validation set.
Data availability
To minimize the risk of patient reidentification, data will only be shared upon reasonable request. For eligible studies, qualified researchers may request access to individual patient-level clinical data through a data request platform. At the time of writing, this request platform is Vivli (https://vivli.org/ourmember/roche/). Datasets can be requested 18 months after a clinical study report has been completed and, as appropriate, once the regulatory review of the indication or drug has completed. Access to patient-level data from this trial can be requested and will be assessed by an independent review panel, which decides whether the data will be provided. Once approved, the data are available for up to 24 months. For up-to-date details on Roche's Global Policy on the Sharing of Clinical Information and how to request access to related clinical study documents, see https://go.roche.com/data_sharing. Anonymized records for individual patients across more than one data source external to Roche cannot, and should not, be linked owing to a potential increase in risk of patient reidentification.
Results
Correlates with grade 3+ anemia
We first assessed whether any patient baseline features were associated with the emergence of treatment-related grade 3+ anemia. Those baseline covariates with a clinically plausible correlation with anemia were considered and included in the univariate analysis shown in Table 1. To minimize the confounding with dose levels, this analysis included only those patients (n = 119) in the study that were treated with monotherapy at potential recommended phase II dose levels (120–160 mg 3/4 schedule). Without adjusting for other factors, univariate logistic regression models suggest that higher baseline hemoglobin values were associated with lower risk of the development of grade 3+ anemia (OR: 0.56; P = 0.001; Table 1). Patients with a lower performance status (ECOG PS score 1 vs. 0) were more likely to develop grade 3+ anemia (OR: 2.74; P = 0.01), as were females, likely due to the lower baseline hemoglobin level when compared with males (median hemoglobin 11.5 vs. 12.4 g/dL, respectively; P = 0.014). When adjusting for other baseline covariates, including dose levels, only baseline hemoglobin, ECOG PS, and number of prior regimens were independent baseline predictors of grade 3+ anemia.
Table 1.
Univariate and multivariate analysis of baseline covariates versus grade 3+ anemia.
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
| Individual predictor | OR (95% CI) | χ 2 P value | Adjusteda OR (95% CI) | AdjustedaP value |
| Baseline hemoglobin (g/dL) | 0.56 (0.40–0.79) | 0.001 | 0.51 (0.35–0.75) | <0.001 |
| Baseline monocytes (absolute) (109/L) | 0.83 (0.21–3.21) | 0.783 | ||
| Baseline neutrophils (absolute) (109/L) | 1.01 (0.84–1.21) | 0.908 | ||
| Baseline platelets (109/L) | 1.00 (1.00–1.01) | 0.203 | ||
| Age (years) | 1.01 (0.98–1.05) | 0.446 | ||
| Sex (male vs. female) | 0.32 (0.13–0.78) | 0.012 | ||
| Prior platinum use (Y vs. N) | 1.25 (0.56–2.77) | 0.589 | ||
| Prior PARP inhibitor (Y vs. N) | 0.61 (0.26–1.43) | 0.254 | ||
| Genotype category group | ||||
| ATM vs. other genesb | 0.70 (0.27–1.83) | 0.465 | ||
| BRCA1/BRCA2 vs. other genesb | 0.66 (0.26–1.69) | 0.391 | ||
| Primary tumor type | ||||
| Breast vs. other tumorsc | 1.68 (0.53–5.38) | 0.379 | ||
| Ovarian vs. other tumorsc | 1.44 (0.52–4.00) | 0.480 | ||
| Pancreatic vs. other tumorsc | 0.92 (0.26–3.24) | 0.895 | ||
| Number of regimens (>3 vs. ≤3) | 1.94 (0.87–4.31) | 0.105 | 2.52 (0.97–6.53) | 0.058 |
| Biallelic status (non-biallelic vs. biallelic) | 1.85 (0.70–4.86) | 0.214 | ||
| Baseline ECOG PS (1 vs. 0) | 2.74 (1.22–6.14) | 0.014 | 2.95 (1.15–7.56) | 0.025 |
| Baseline weight (kg) | 0.98 (0.96–1.01) | 0.174 | ||
| Germline status (somatic vs. germline) | 1.21 (0.49–3.00) | 0.676 | ||
Abbreviations: N, no; Y, yes.
aAdjusted ORs and P values are based on the final multivariate logistic model adjusted by dose levels and other covariates included in the final model.
bOther genes include all gene types except ATM, BRCA1, and BRCA2.
cOther tumors include all tumor types except breast, ovarian, and pancreatic (the most common tumor types).
In addition, exposure as measured by area under the curve (AUC)0–24 was also an independent predictor of grade 3+ anemia. In multivariable logistic regression analysis, higher exposure was associated with an increased chance of grade 3+ anemia (adjusted OR: 1.04; P = 0.005), after adjusting for baseline hemoglobin and ECOG PS.
Given the impact of various baseline features on the likelihood of development of clinically significant anemia upon ATRi treatment and the heterogeneity of patient characteristics, an individualized approach to dose selection may be warranted. We thus explored whether we could utilize both baseline features and early on-treatment patient laboratory data to reliably identify early predictors of hemoglobin decline.
Relationship between changes in hematologic parameters and anemia
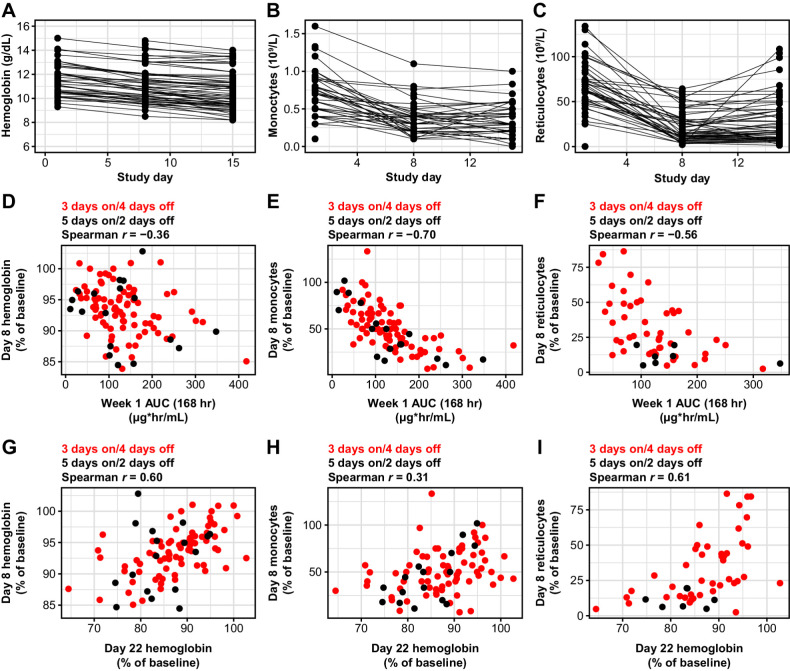
In the dose-escalation phase of the TRESR trial, we observed early decreases in monocytes and reticulocytes at day 8, with a more modest decline in hemoglobin at this timepoint (Fig. 1A–C). While the drop in monocytes and reticulocytes was close to maximal at day 8, hemoglobin levels further declined by day 15, though rarely approached grade 3+ levels. The absolute monocyte and reticulocyte counts consistently rebounded quickly (within 1 week) following drug holds (example patient shown in Supplementary Fig. S2) and both changes were exposure dependent. However, exposure in cycle 1 was not a predictor of hemoglobin changes on day 8 of treatment, likely due to the longer physiologic turnover time of erythrocytes (Fig. 1D–F). These findings are consistent with previously published work in mouse with camonsertib, where early changes in reticulocytes were observed with more intensive dosing of camonsertib with only modest changes in red blood cells, likely due to the turnover rate of red blood cells in the mouse (4). The dose/exposure dependence of the monocyte and reticulocyte changes, and quick rebound following drug hold, further support that these changes are drug specific and not an epiphenomenon.
Figure 1.
Hematology values in patients treated with camonsertib and the correlation between changes in monocytes and reticulocytes with exposure and hemoglobin levels. Longitudinal changes in hemoglobin (A), monocytes (B), and reticulocytes (C) in patients treated with either 120 or 160 mg of camonsertib during the first 15 days of treatment. The changes from baseline at day 8 and day 15 in hemoglobin, monocytes, and reticulocytes are statistically significant as a group (P < 0.001) based on pairwise t test. Day 8 hemoglobin (D), monocytes (E), and reticulocytes (F) versus camonsertib AUC. Day 8 changes in hemoglobin (G), monocytes (H), and reticulocytes (I) versus day 22 changes in hemoglobin.
Nomogram to predict hemoglobin changes
On the basis of the hypotheses of mechanism-based ATRi-induced anemia and these clinical observations, we developed a nomogram that leveraged several parameters at baseline and treatment day 8 for their predictive value to anticipate hemoglobin decrease on day 22. A total of 154 patients were treated in the monotherapy arms of the TRESR study. For the development of the nomogram, 45 patients were excluded because of early dose modifications, missed doses in cycle 1, and/or missing hemoglobin data throughout cycle 1. Of the remaining 109 patients, 2 were removed as they did not have assessments of both monocytes and hemoglobin in cycle 1 and an additional 47 were excluded as they did not have reticulocyte data.
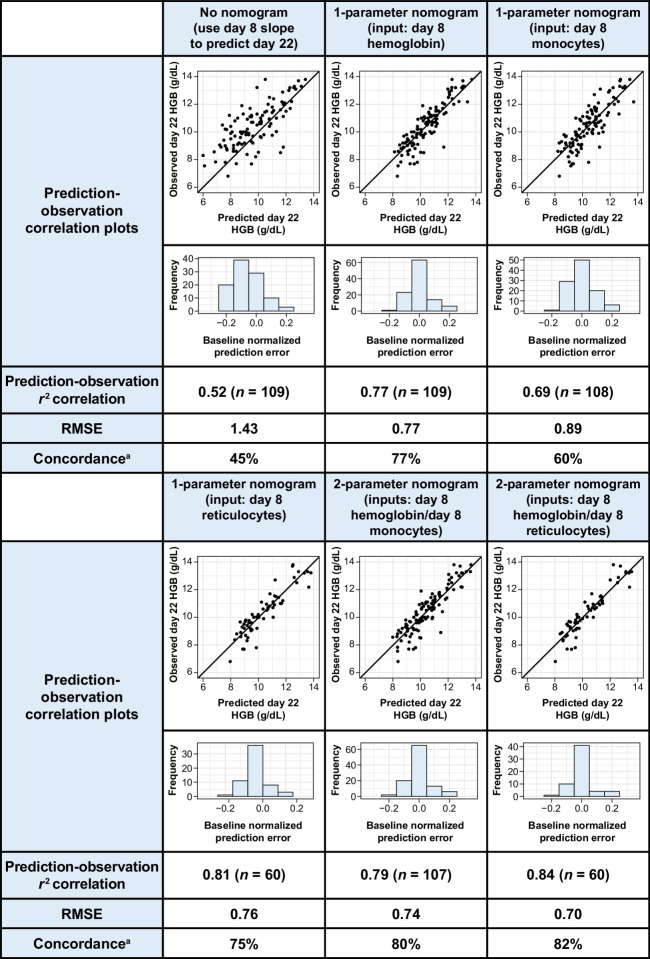
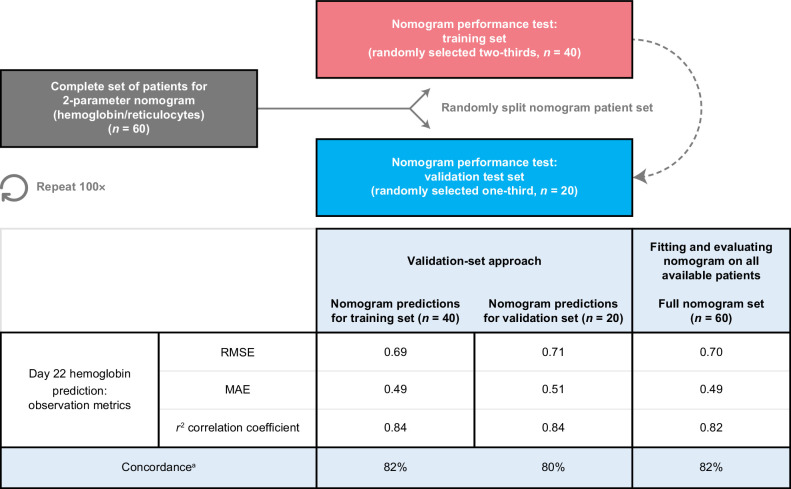
Changes in reticulocyte counts and hemoglobin on day 8 compared with baseline correlated well (Spearman rank correlations r = 0.61 and r = 0.60, respectively) with day 22 changes in hemoglobin, while monocyte changes were less strongly correlated (Spearman rank correlation r = 0.31; Fig. 1G–I). This observation led us to hypothesize that using baseline hematologic parameters, a mathematical model (a nomogram) could be developed to predict hemoglobin levels early in cycle 2. We iterated over combinations of baseline hematologic values and on-treatment hematologic changes for the nomogram, and identified the optimal input parameter set as baseline hemoglobin and reductions in hemoglobin and reticulocytes on day 8 (r2 = 0.82, RMSE = 0.70; Fig. 2). This nomogram provided a prediction accuracy with a concordance rate of 82%. When applied to the independent test data in the validation, a similar concordance rate of 80% was observed (Fig. 3). For the purposes of this analysis, we defined a prediction to be accurate when it was within 7.5% of the observed hemoglobin value, which has been used previously as a criterion of interchangeability of hemoglobin values (12).
Figure 2.
Nomogram-based predictions of cycle 2 day 1 hemoglobin. Summary of day 22 hemoglobin prediction-observation correlations and fit statistics for (left to right) predictions for day 22 using the daily rate of hemoglobin change through day 8, and five nomograms using different input parameters. HGB, hemoglobin. aPercent of predictions within 7.5% of day 22 observation.
Figure 3.
Assessment of nomogram. Summary of the validation-set approach, which randomly divided the total set of patients into a nomogram-training set (n = two-thirds of the total available) and a nomogram-testing set (n = one-third of the total available). The training set was used to calibrate the nomogram, and performance was assessed on predictions made for individuals in the test set (who were not used in calibration). Mean values across iterations are presented in the table. MAE, mean absolute error. aPercent of predictions within 7.5% of day 22 observation.
Furthermore, we investigated two predictive tests to assess the performance of the nomogram in patient populations not used in calibration. The nomogram performance in both the validation-set approach (Fig. 3) and the leave-one-out cross-validation approach (Supplementary Fig. S3) was very similar to what was observed when calibrating to the entire set, suggesting that the nomogram is robust and predictive of the population response.
Discussion
In summary, we developed a nomogram to accurately predict declines in hemoglobin in patients treated with the ATRi camonsertib. This nomogram utilizes standard clinical laboratory values (hemoglobin and reticulocyte count) and can easily be implemented in electronic medical record systems, similarly to other calculated parameters. As anemia is a class effect for ATRis and other agents, it would be of interest to test the applicability of such a nomogram to other ATRis in similar scenarios. Because of the different pharmacologic properties and dosing strategies used for the other ATRis, independent validation would be required. This decision tool may facilitate early interventions for anemia for this drug class, thereby reducing unplanned dose holds or reductions, and consequently, it may improve drug exposure and ultimately efficacy.
Patients treated with camonsertib monotherapy displayed rapid dose- and exposure-dependent decreases in reticulocytes and monocytes. Despite the hypothesis that monocyte levels would be critical for this nomogram, we found no significant correlation between monocyte decline on day 8 and later hemoglobin decline. This may be due to the different lifespans of the cell types (3 days for monocytes, <30 hours for reticulocytes), with reticulocyte changes providing an earlier readout.
Although this work has several limitations, including phase I patient selection criteria with a mandatory baseline hemoglobin cutoff of 9.5 g/dL and lack of validation in an independent dataset, we envision this work as proof of principle to optimize dosing of ATRi therapy to minimize clinically significant toxicity. This is additionally underscored by the low therapeutic index of ATRis, making the argument that nomograms are simple and practical for patients’ care, thus could be utilized for future dose selection and optimization of these therapeutic agents.
Supplementary Material
Supplementary Fig. S1. Summary of patient populations used in the analysis.
Supplementary Fig. S2. Absolute monocyte and reticulocyte counts rebound quickly following dose holds.
Supplementary Fig. S3. Summary statistics for the leave-one-out cross-validation of the nomogram.
Supplementary Table S1. Treatment regimens evaluated in TRESR.
Supplementary Table S2. Representativeness of study participants.
Acknowledgments
This study was funded by Repare Therapeutics, Inc. The authors would like to thank the patients, their families, and all investigators involved in this study. In addition, the authors would like to thank Hitesh Mistry of Systems Forecasting for his contribution to early iterations of the nomogram and subsequent discussions that shaped the final version, and Anne Roulston of Repare Therapeutics for several discussions on the mechanisms of anemia in preclinical studies. Editorial support, including figure preparation, formatting, proofreading, and submission, were provided by Allison Alwan TerBush, PhD, and Rosie Henderson, both of Onyx (a division of Prime, London, UK), supported by Repare Therapeutics according to Good Publication Practice guidelines. The sponsor was involved in the study design, collection, analysis, and interpretation of data, as well as data checking of information provided in the article. However, ultimate responsibility for opinions, conclusions, and data interpretation lies with the authors.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
T.A. Yap is an employee of the University of Texas MD Anderson Cancer Center. T.A. Yap also reports personal fees from AbbVie, Adagene, Almac, Aduro, Amphista, Astex, Athena, Atrin, Avenzo, Avoro, Axiom, Baptist Health Systems, BioCity Pharma, Boxer, Bristol Myers Squibb, C4 Therapeutics, Calithera, Cancer Research UK, Carrick Therapeutics, Circle Pharma, Cybrexa, Daiichi Sankyo, Dark Blue Therapeutics, Diffusion, Duke Street Bio, 858 Therapeutics, EcoR1 Capital, Ellipses Pharma, Entos, Genesis Therapeutics, Genmab, Glenmark, GLG, Globe Life Sciences, GSK, Guidepoint, Ideaya Biosciences, Idience, Ignyta, I-Mab, ImmuneSensor, Impact Therapeutics, Institut Gustave Roussy, Intellisphere, Janssen, Kyn, MEI Pharma, Mereo, Merit, Monte Rosa Therapeutics, Natera, Nested Therapeutics, Nexys, Nimbus, Novocure, Odyssey, OHSU, OncoSec, Ono Pharma, Onxeo, PanAngium Therapeutics, Pegascy, PER, Piper-Sandler, Pliant Therapeutics, Prolynx, Radiopharma Theranostics, resTORbio, Roche, Ryvu Therapeutics, SAKK, Sanofi, Schrodinger, Servier, Synnovation, Synthis Therapeutics, TCG Crossover, TD2, Terremoto Biosciences, Tessellate Bio, Theragnostics, Terns Pharmaceuticals, Tolremo, Tome, Thryv Therapeutics, Trevarx Biomedical, Varian, Veeva, Versant, Vibliome, Voronoi Inc., Xinthera, Zai Labs, and ZielBio; grants and personal fees from Acrivon, Artios, AstraZeneca, Bayer, BeiGene, Blueprint, Clovis, EMD Serono, F-star, Merck, Pfizer, Repare Therapeutics, and Tango; and grants from BioNTech, Bristol Meyers Squibb, Boundless Bio, Constellation, Cyteir, Eli Lilly, Forbius, GSK, Genentech, Haihe, Ideaya, ImmuneSensor, Insilico Medicine, Ionis, Ipsen, Jounce, Karyopharm, KSQ, Kyowa, Mirati, Novartis, Ribon Therapeutics, Regeneron, Rubius, Sanofi, Scholar Rock, Seagen, Seattle Genetics, Tesaro, Vivace, and Zenith during the conduct of the study. E.K. Lee reports other support from Merck, as well as personal fees from Aadi Biosciences outside the submitted work. M. Højgaard reports grants from Repare Therapeutics during the conduct of the study, as well as grants from Eli Lilly, AstraZeneca, Bristol Myers Squibb, Symphogen, Roche/Genentech, Genmab, Incyte Corporation, Pfizer, Bayer, Novartis, Alligator Bioscience, Bioinvent, Monta Bioscience, and Amgen outside the submitted work. N.B. Mettu reports grants from Repare Therapeutics during the conduct of the study, as well as grants from Sapience Therapeutics, Leap Therapeutics, Adanate, Actuate Therapeutics, Amgen, Amphivena Therapeutics, AstraZeneca, Bristol Myers Squibb, Erytech Pharma, Genentech-Roche, MedImmune, Incyte Corporation, Compass Therapeutics, BioMed Valley Discoveries, Syros Pharmaceuticals, Mereo BioPharma, Merck Sharpe & Dohme, Aravive, Inc, and Nucana outside the submitted work. S. Lheureux reports grants from Repare Therapeutics; grants and personal fees from AstraZeneca, GSK, and Roche; and personal fees from Eisai and Merck outside the submitted work. B.A. Carneiro reports research funding to institution from AstraZeneca, AbbVie, Actuate Therapeutics, Astellas, Agenus, Bayer, Dragonfly Therapeutics, Mink Therapeutics, and Pfizer, as well as payment or honoraria for advisory board participation from Foundation Medicine and Seagen. R. Plummer reports other support from Repare Therapeutics during the conduct of the study, as well as personal fees from Pierre Faber, Bayer, Novartis, Bristol Myers Squibb, Cybrexa, Ellipses, CV6 Therapeutics, Astex Pharmaceuticals, Medivir, Sanofi Aventis, AstraZeneca, MSD, Onexo, Genmab, Immunocore, Sotio Biotech AG, Alligator Biosciences, and GSK outside the submitted work. A.J. Fretland is a current employee and shareholder of Repare Therapeutics. D. Ulanet is an employee and shareholder of Repare Therapeutics. Y. Xu is a current employee and shareholder of Repare Therapeutics. R. McDougall is a current employee of Recursion Pharmaceuticals, former employee of Repare Therapeutics, and shareholder of Repare Therapeutics. M. Koehler is a shareholder and employee of Repare Therapeutics. E. Fontana reports personal fees and other support from Repair Therapeutics during the conduct of the study. E. Fontana also reports other support from Bicycle Therapeutics, Artios Pharma, Seagen, Amgen, Nurix Therapeutics, BioNTech SE, Relay Therapeutics, Taiho Pharmaceutical, Pfizer, Roche, Daiichi Sankyo, Gilead Sciences, Basilea Pharmaceutica, Jiangsu Hengrui Medicine, Mereo Biopharma, HUTCHMED, Merus, Crescendo Biologics, GSK plc, BeiGene, Turning Point Therapeutics, Sapience Pharma, and Nerviano outside the submitted work, as well as a leadership role for European Organisation for Research and Treatment of Cancer (EORTC) and GICTG secretary. No disclosures were reported by the other authors.
Authors' Contributions
E. Rosen: Conceptualization, investigation, writing–original draft, writing–review and editing. T.A. Yap: Investigation, writing–review and editing. E.K. Lee: Investigation, writing–review and editing. M. Højgaard: Investigation, writing–review and editing. N.B. Mettu: Investigation, writing–review and editing. S. Lheureux: Investigation, writing–review and editing. B.A. Carneiro: Investigation, writing–review and editing. R. Plummer: Investigation, writing–review and editing. A.J. Fretland: Conceptualization, formal analysis, visualization, writing–original draft, writing–review and editing. D. Ulanet: Conceptualization, data curation, visualization, writing–original draft, writing–review and editing. Y. Xu: Conceptualization, data curation, formal analysis, visualization, writing–original draft, writing–review and editing. R. McDougall: Conceptualization, data curation, formal analysis, visualization, methodology, writing–original draft, writing–review and editing. M. Koehler: Conceptualization, writing–original draft, writing–review and editing. E. Fontana: Conceptualization, investigation, writing–original draft, writing–review and editing.
References
- 1. Wengner AM, Siemeister G, Lücking U, Lefranc J, Wortmann L, Lienau P, et al. The novel ATR inhibitor BAY 1895344 is efficacious as monotherapy and combined with DNA damage-inducing or repair-compromising therapies in preclinical cancer models. Mol Cancer Ther 2020;19:26–38. [DOI] [PubMed] [Google Scholar]
- 2. Yap TA, Tan DSP, Terbuch A, Caldwell R, Guo C, Goh BC, et al. First-in-human trial of the oral ataxia telangiectasia and RAD3-related (ATR) inhibitor BAY 1895344 in patients with advanced solid tumors. Cancer Discov 2021;11:80–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yap TA, O'Carrigan B, Penney MS, Lim JS, Brown JS, de Miguel Luken MJ, et al. Phase I trial of first-in-class ATR inhibitor M6620 (VX-970) as monotherapy or in combination with carboplatin in patients with advanced solid tumors. J Clin Oncol 2020;38:3195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Roulston A, Zimmermann M, Papp R, Skeldon A, Pellerin C, Dumas-Berube E, et al. RP-3500: a novel, potent, and selective ATR inhibitor that is effective in preclinical models as a monotherapy and in combination with PARP inhibitors. Mol Cancer Ther 2022;21:245–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yap TA, Fontana E, Lee EK, Spigel DR, Hojgaard M, Lheureux S, et al. Camonsertib in DNA damage response-deficient advanced solid tumors: phase 1 trial results. Nat Med 2023;29:1400–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Martorana F, Da Silva LA, Sessa C, Colombo I. Everything comes with a price: the toxicity profile of DNA-damage response targeting agents. Cancers 2022;14:953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pierce A, Berges A, Cheung SA, Standifer N, Ross G, Smith S, et al. Dose-exposure-response relationship between AZD6738 and peripheral monocytes. J Clin Oncol 35:15s, 2017. (suppl; abstr e14063). [Google Scholar]
- 8. Bauer M, Goldstein M, Christmann M, Becker H, Heylmann D, Kaina B. Human monocytes are severely impaired in base and DNA double-strand break repair that renders them vulnerable to oxidative stress. Proc Natl Acad Sci U S A 2011;108:21105–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ngoi NY, Lin HY, Dumbrava E, Fu S, Karp DD, Naing A, et al. Dynamic changes in monocyte and reticulocyte counts predict mechanism-based anemia development and recovery during ATR inhibitor treatment in phase I/II trials. In:Proceedings of the American Association for Cancer Research Annual Meeting 2023; Part 1 (Regular and Invited Abstracts); 2023Apr 14–19; Orlando, FL. Philadelphia (PA): AACR; Cancer Res 2023;83(7_Suppl):Abstract nr 6181. [Google Scholar]
- 10. Jacobsen RN, Perkins AC, Levesque JP. Macrophages and regulation of erythropoiesis. Curr Opin Hematol 2015;22:212–9. [DOI] [PubMed] [Google Scholar]
- 11. Levy M, Ferraro G, Li L, Han Y, Varricchio L, Fournier S, et al. ATR inhibitor camonsertib (RP-3500) suppresses early stage erythroblasts by mediating ferroptosis. Blood 2022;140:8194–5. [Google Scholar]
- 12. Herman J, Park B, Awsare B, West F, Crittendon D, Evans L, et al. Point-of-care versus central testing of hemoglobin during large volume blood transfusion. BMC Anesthesiol 2019;19:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. S1. Summary of patient populations used in the analysis.
Supplementary Fig. S2. Absolute monocyte and reticulocyte counts rebound quickly following dose holds.
Supplementary Fig. S3. Summary statistics for the leave-one-out cross-validation of the nomogram.
Supplementary Table S1. Treatment regimens evaluated in TRESR.
Supplementary Table S2. Representativeness of study participants.
Data Availability Statement
To minimize the risk of patient reidentification, data will only be shared upon reasonable request. For eligible studies, qualified researchers may request access to individual patient-level clinical data through a data request platform. At the time of writing, this request platform is Vivli (https://vivli.org/ourmember/roche/). Datasets can be requested 18 months after a clinical study report has been completed and, as appropriate, once the regulatory review of the indication or drug has completed. Access to patient-level data from this trial can be requested and will be assessed by an independent review panel, which decides whether the data will be provided. Once approved, the data are available for up to 24 months. For up-to-date details on Roche's Global Policy on the Sharing of Clinical Information and how to request access to related clinical study documents, see https://go.roche.com/data_sharing. Anonymized records for individual patients across more than one data source external to Roche cannot, and should not, be linked owing to a potential increase in risk of patient reidentification.