Abstract
Purpose:
Genomic rearrangements can generate potent oncogenic drivers or disrupt tumor suppressor genes. This study examines the landscape of fusions and rearrangements detected by liquid biopsy (LBx) of circulating tumor DNA (ctDNA) across different cancer types.
Experimental Design:
LBx from 53,842 patients with 66 solid tumor types were profiled using FoundationOneLiquid CDx, a hybrid-capture sequencing platform that queries 324 cancer-related genes. Tissue biopsies (TBx) profiled using FoundationOneCDx were used as a comparator.
Results:
Among all LBx, 7,377 (14%) had ≥1 pathogenic rearrangement detected. A total of 3,648 (6.8%) LBx had ≥1 gain-of-function (GOF) oncogene rearrangement, and 4,428 (8.2%) LBx had ≥1 loss-of-function rearrangement detected. Cancer types with higher prevalence of GOF rearrangements included those with canonical fusion drivers: prostate cancer (19%), cholangiocarcinoma (6.4%), bladder (5.5%), and non–small cell lung cancer (4.4%). Although the prevalence of driver rearrangements was lower in LBx than TBx overall, the frequency of detection was comparable in LBx with a tumor fraction (TF) ≥1%. Rearrangements in FGFR2, BRAF, RET, and ALK, were detected across cancer types, but tended to be clonal variants in some cancer types and potential acquired resistance variants in others.
Conclusions:
In contrast to some prior literature, this study reports detection of a wide variety of rearrangements in ctDNA. The prevalence of driver rearrangements in tissue and LBx was comparable when TF ≥1%. LBx presents a viable alternative when TBx is not available, and there may be less value in confirmatory testing when TF is sufficient.
Translational Relevance.
The efficacy of liquid biopsies for fusion detection hinges on factors like ctDNA shedding (which is associated with tumor type and disease burden), as well as the testing platform employed. There is a widespread notion that ctDNA testing is suboptimal for detecting genetic fusions compared with tissue testing, but it is noteworthy that testing platforms have evolved over time, enhancing their performance. Our study highlights that when shedding reaches a specific threshold (a tumor fraction of ≥1%), liquid biopsies emerge as a viable alternative to tissue testing. Among the 53,842 examined liquid biopsies, 14% (7,377) contained at least one pathogenic rearrangement, a clinically significant discovery. These data suggest that liquid biopsies have the potential to bridge a crucial gap, expanding precision medicine opportunities, especially when acquiring tissue-based next-generation sequencing is impractical or delayed.
Introduction
Detection of genomic rearrangements in circulating tumor DNA (ctDNA) is heterogeneous across various commercially available and lab developed liquid biopsy (LBx) assays, with multiple reports of variable performance for the detection of these common genomic events (1–3). Some of these structural variants—deletions, duplications, inversions, and translocations—result in gene products that function as potent oncogenic drivers. Receptor tyrosine kinase (RTK) fusions and activating truncations are well-established targetable driver alterations in non–small cell lung cancer (NSCLC; ALK, RET, ROS1), cholangiocarcinoma (FGFR2), bladder cancer (FGFR3), and thyroid cancer (RET). Testing for activating fusions has become standard of care in these cancer types (4–8). Other kinase fusions such as BRAF have had limited actionability to date, but next-generation inhibitors are currently being tested in clinical trials (9). Some rearrangements activate nonenzymatic oncogenes such as transcription factors (TMPRSS2-ERG, EWSR1-FLI1) and have not been druggable, but proteolysis targeting chimeras (PROTACs) could represent a new therapeutic inroad to inhibit them (10). In addition, rearrangements can also disrupt tumor suppressor genes, including actionable genes (BRCA1/2) or clinically relevant biomarkers (RB1, STK11; refs. 11, 12). Detection of rearrangements and fusions is therefore critical for clinical decision making in cancer care.
Rearrangements are detected using multiple molecular pathology tests, including FISH, imbalance assays, and DNA- and RNA-based next-generation sequencing (NGS). NGS testing has seen recent increased uptake due to the ability to assess many relevant biomarkers simultaneously, but rearrangements present specific technical challenges for NGS platforms, and the NGS assay design dramatically affects performance. One obstacle for DNA-based assays is that rearrangement breakpoints can occur inside intronic regions that may be challenging to sequence, requiring intentional design and bioinformatics to ensure reliable detection. Some of the intronic regions are long and may have repetitive stretches. Although some genes have recurrent breakpoints in specific introns (e.g., ALK, FGFR2, RET), others have more flexible breakpoints and require sequencing across many introns (e.g., BRAF, ROS1, NTRK1). Sequencing RNA can allow breakpoint detection with a smaller sequencing footprint, but RNA has significant instability and high assay failure rates in real-world formalin-fixed, paraffin-embedded (FFPE) specimens. When tissue is unavailable, LBx-based NGS of ctDNA from peripheral blood is a pragmatic alternative sample type (13), but poses the additional challenge of detecting rearrangement events in samples with low tumor DNA relative to DNA from healthy cells (often <1%; ref. 14) compared with tissue biopsy where the specimen's tumor content is screened visually by a pathologist and required to be enriched to at least 20% prior to being advanced to DNA extraction. Sequencing must be performed to higher depth, which can increase the number of erroneous reads, and make it challenging to capture long intronic regions with sufficient depth in a targeted sequencing panel. ctRNA is swiftly digested by circulating ribonucleases and immune cells, and RNA from necrotic cells, which are a major contributor of tumor-derived circulating nucleic acids (15), can be degraded before it enters circulation. Clinical-grade interpretation of sequencing of this analyte is as yet unproven (16).
LBx have been widely considered to lack robust detection of fusions and gene rearrangements. In this study, we analyze LBx results from patients with solid tumors and report the detection of pathogenic rearrangements in a wide set of genes, including gain-of-function (GOF) fusions and rearrangements in kinase and transcription factor oncogenes, as well as truncating rearrangements in tumor suppressors. We compare detection of select rearrangements with tissue biopsies, including concordance analyses in specimens from the same patient, and report on rearrangements that appear in LBx as potential polyclonal resistance mechanisms.
Materials and Methods
Patient cohort
LBx from patients with solid tumors ordered within the United States between September 2020 and March 2023 during routine clinical care (N = 53,842) were retrospectively analyzed. For patients with multiple LBx results (2,594; 4.8%), one specimen was chosen on the basis of a heuristic that incorporates factors such as sample run date and quality metrics to choose a representative sample. The numbers of samples analyzed for each cancer type are provided in Supplementary Table S1. LBx submitted without a documented site of origin were designated as cancer of unknown primary (CUP) due to lack of a clear diagnosis and include true CUPs as well as samples for which tissue diagnostic workup may be ongoing or with inadequate information on the requisition form. Approval for this study, including a waiver of informed consent and Health Insurance Portability and Accountability Act (HIPAA) waiver of authorization, was obtained from the Western Institutional Review Board (protocol no. 20152817).
Comprehensive genomic profiling (CGP) of liquid biopsies
CGP was performed in a Clinical Laboratory Improvement Amendment–certified, College of American Pathologists–accredited, New York State–approved laboratory (Foundation Medicine). LBx samples were profiled using a validated, FDA-approved next-generation sequencing panel assay FoundationOneLiquid CDx (F1LCDx; ref. 17). Circulating cell-free DNA was extracted from peripheral whole blood and CGP was performed using hybridization-captured, adaptor ligation-based libraries. F1LCDx interrogates a total of 324 cancer-related genes for base substitutions, short insertions and deletions, copy-number amplifications and homozygous deletions, and large genomic rearrangements, as well as microsatellite instability, blood tumor mutational burden, and tumor fraction (TF) genomic signatures. Of the 324 genes in the panel, 309 are sequenced with complete exonic coverage, 20 of these with additional intronic coverage, and 15 with only select noncoding coverage. Targeted regions in 75 genes are sequenced with ultra-deep coverage for increased sensitivity. Importantly, F1CDx and F1LCDx have coverage of the same exons and introns of the 324 cancer-related genes (see Supplementary Table S2 for complete gene list).
De novo assembly is performed for detection of short variants, fusions, and other large-scale genomic rearrangements using proprietary algorithms which build de Bruijn graph models from k-mers spanning a variant candidate and consider the local coverage, number of supporting read clusters, read redundancy, and number of error-containing clusters for the mutant and reference alleles (17).
TF quantification and clonality assessment in liquid biopsy
Foundation Medicine's ctDNA TF on F1LCDx is a composite algorithm, rather than relying on only somatic variant allele frequencies (VAF). Because nearly all solid tumors have aneuploidy, the TF estimate prioritizes aneuploidy at higher levels. The algorithm prioritizes VAF of canonical alterations at lower levels when the fraction of cfDNA with aneuploidy cannot be reliably estimated. This composite approach avoids mistaking germline variants for somatic alleles and relying on VAF of variants in amplified genes. The purity assessment from a robust copy-number model, which accounts for both the observed coverage variation and allele frequencies of genome-wide SNP allele frequencies, is used to determine the TF estimate from aneuploidy. When aneuploidy is below the limit of reliable estimation, the allele frequencies of short variants and rearrangements known to be somatic through heuristics are used to estimate TF. In addition, a multiomic assessment of cfDNA is used both to exclude clonal hematopoiesis-derived aneuploidy from copy-number modeling and to positively identify the somatic status of short variants in this analysis. In this study, the clonality of a rearrangement variant was calculated as the ratio of percent reads of a rearrangement event to the TF of the sample (with a maximum set at 1).
CGP of tissue biopsies
Tissue biopsies from patients with the same cancer types as those analyzed for LBx that were ordered within the United States between January 2017 and January 2023 during routine clinical care (N = 295,592) were analyzed using FoundationOneCDx as described previously (18, 19). Briefly, the pathologic diagnosis of tissue biopsy was confirmed on routine hematoxylin and eosin–stained slides. Samples with a minimum of 20% tumor nuclei underwent DNA extraction and underwent hybrid capture-based sequencing of the same 324 cancer-related genes interrogated by F1LCDx. Some differences in sample preparation between the tissue and liquid platforms include fragmentation by sonication for DNA extracted from tissue, as well as more uniform and lower sequencing depth (18).
Concordance analysis between tissue and liquid biopsy
For patients with CCA or an unknown primary with CGP results available for both tissue biopsy and LBx (collected a median 34 days apart), agreement was assessed for detection of driver rearrangements. Sensitivity or percent positive agreement (PPA), and negative predictive value (NPV), were calculated with tissue as standard; 95% binomial confidence intervals were calculated using the Wilson score method with continuity correction. For FGFR2 rearrangement detection concordance, PPA was calculated among pairs submitted as CCA (N = 201) and CUP (N = 160), and NPV was calculated only among the CCA pairs.
Statistical tests
Comparisons for VAF/TF in Fig. 2B were made using the Kruskal–Wallis test in a pairwise fashion among the cancer types with ≥10 rearrangements in the analyzed gene. Comparisons between prevalence of rearrangements in KRAS-positive and -negative samples in Fig. 6E were done using the Fisher exact test. FDR was calculated using the Benjamini–Hochberg correction for multiple testing.
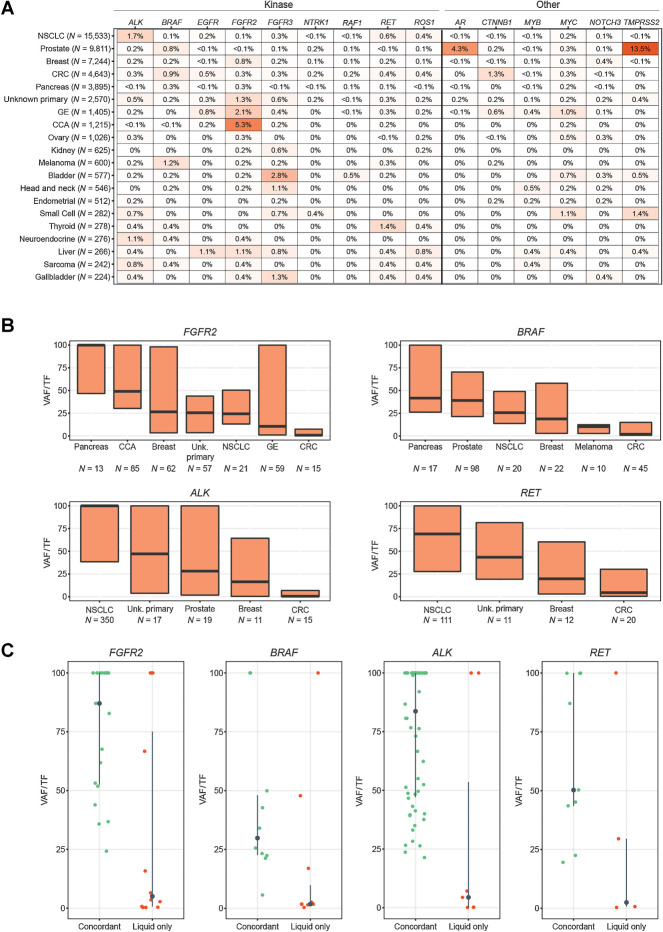
Figure 2.
GOF fusions and rearrangements. A, Heatmap of the prevalence of the most frequently rearranged oncogenes in the pan-tumor cohort (cancer types with ≥200 LBx shown). Kinase genes on the left, transcription factors on the right (only genes rearranged in ≥0.5% of any cancer type shown). B, The clonal fraction (VAF/TF) of gene rearrangements considered to be potential pan-tumor biomarkers and which appear in multiple cancer types. Median and interquartile range shown. Cancer types (n ≥ 10 with a rearrangement in the gene) are arranged in order of median VAF/TF. C, In tissue and liquid biopsies from the same patient, the VAF/TF of each rearrangement that was detected in both tissue and liquid (concordant) or in the liquid biopsy only. Tissue/liquid pairs where liquid was collected up to 90 days earlier and up to 2 years later than the tissue specimen were considered. Additional information about this analysis is available in Supplementary Table S6.
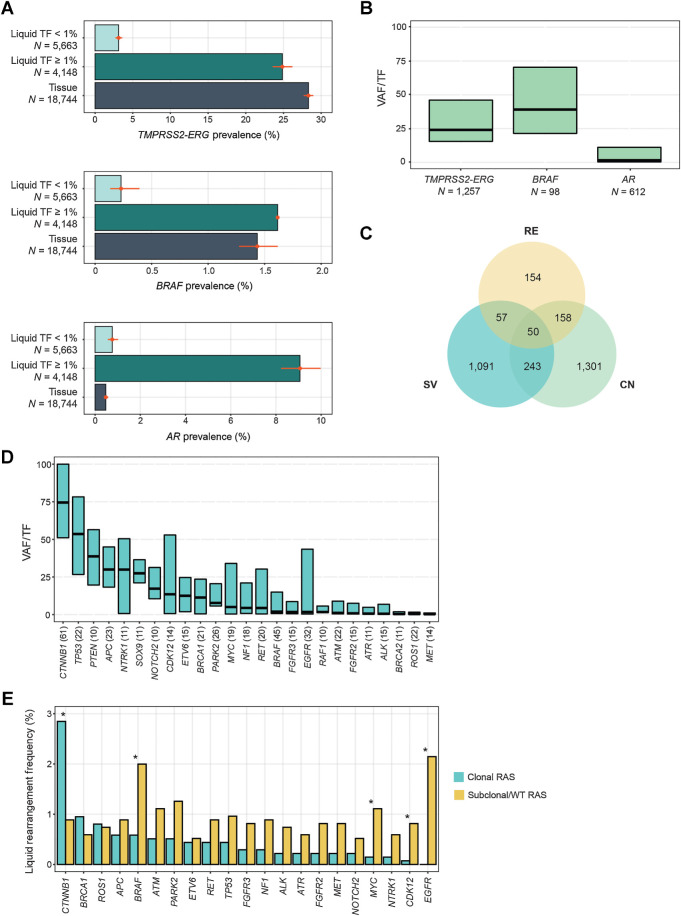
Figure 6.
Rearrangements in prostate and colorectal cancers. A, Comparison of the prevalence of activating rearrangements among prostate cancer tissue biopsies and liquid biopsies with TF ≥1% and <1%. B, The clonality (VAF/TF) of driver rearrangements like TMPRSS2-ERG and BRAF, and putative acquired resistance rearrangements in AR in prostate cancer LBx. C, The overlapping appearance of AR variants in the same prostate cancer LBx. CN, copy-number amplification; RE, rearrangement; SV, short variant (mutation or insertion/deletion). All of the variants are predicted to be activating and appear as acquired resistance after exposure to androgen deprivation therapy and AR inhibitors. D, The clonality (VAF/TF) of the most common pathogenic rearrangements detected in colorectal cancer LBx (N ≥ 10), in order of median VAF/TF. E, A comparison of the rearrangement frequencies among colorectal cancer LBx that have no clonal KRAS/NRAS/BRAF V600E mutations (N = 1,352) versus colorectal cancer LBx and a clonal KRAS or NRAS mutation (with VAF/TF of at least 25%; N = 1,370). Only LBx with TF≥1% were included, and LBx with clonal BRAF V600E mutations were excluded from this analysis. Asterisks denote significant differences in prevalence (FDR < 0.05). Only genes occurring in ≥0.5% of clonal RAS or subclonal/WT RAS samples are presented, but all genes were included in the Benjamini–Hochberg correction of FDR.
Data availability
All relevant data are provided within the article and its accompanying Supplementary Data. Because of HIPAA requirements, we are not consented to share individualized patient genomic data, which contains potentially identifying or sensitive patient information. Foundation Medicine is committed to collaborative data analysis, and have well-established, and widely utilized mechanisms by which investigators can query their core genomic database of >600,000 deidentified sequenced cancers to obtain aggregated datasets. More information and mechanisms for data access can be obtained by contacting the corresponding authors or the Foundation Medicine Data Governance Council at data.governance.council@foundationmedicine.com.
Results
Detection of pathogenic rearrangements in liquid biopsies
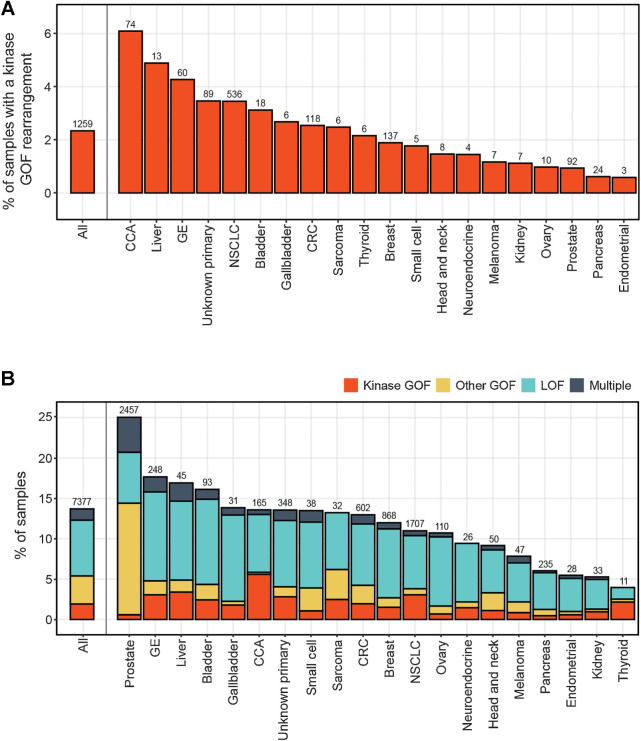
Among 53,842 LBx, 7,377 (14%) had at least one pathogenic gene rearrangement detected. GOF fusions and rearrangements were detected in 16 RTK and downstream kinases (ALK, FGFR2, BRAF, RET, FGFR3, ROS1, EGFR, NTRK1, RAF1, MET, NTRK3, ERBB2, FGFR1, PDGFRA, NTRK2, and NRG1; Fig. 1A). Gastrointestinal cancer types had the highest frequencies of kinase rearrangements detected: cholangiocarcinoma (CCA; 6.1%), liver (4.9%), and gastroesophageal cancers (4.3%). GOF rearrangements were also detected in genes encoding transcription factors (TMPRSS2-ERG and EWSR1-FLI1/ATF1/WT1 fusions, rearrangements of AR, MYC, CTNNB1, MYB, ESR1). Loss-of-function (LOF) rearrangements predicted to truncate tumor suppressor genes, including DNA repair components and cell-cycle regulators, were even more prevalent. A total of 3,648 (6.8%) LBx had at least one GOF rearrangement, 4,428 (8.2%) LBx had at least one LOF rearrangement detected, and 699 (1.3%) harbored both types (Fig. 1B). The frequency of rearrangement detection differed across cancer types. Cancer types with higher prevalence of GOF rearrangements included cancers with canonical fusion drivers: prostate cancer (19%), bladder (5.5%), and CCA (6.4%), as well as cancer types with abundant amplifications like liver (7.1%) and gastroesophageal (6.6%). Cancer types with lower detection rates of pathogenic rearrangements included pancreas (6.0%), endometrial (5.5%), kidney (5.3%), and thyroid (4.0%), some of which are also cancer types that shed less ctDNA (14). Rearrangement events were detected at a median VAF of 2.2% but ranged from 0.02% for an EGFR activating rearrangement to as high as 52% for an RB1 LOF rearrangement in breast cancer.
Figure 1.
Prevalence of pathogenic rearrangements across different cancer types. A, Frequency of detection of GOF rearrangements in kinase genes across different cancer types. B, As in A, but including rearrangements that were categorized as GOF in nonkinase oncogenes, and LOF in tumor suppressor genes. Cancer types with >200 LBx profiled are shown. Number above each bar corresponds to the number of cases with detected fusions. CUP, carcinoma of unknown primary; GE, gastroesophageal cancer.
GOF rearrangements in oncogenes
Many of the expected enrichments in oncogenic fusions/rearrangements were observed in this cohort: ALK, RET, and ROS1 fusions in NSCLC (20); TMPRSS2–ERG fusions and AR rearrangements in prostate cancer (21, 22); FGFR2 fusions/truncations in CCA (23–25); FGFR3 fusions/truncations in bladder (26) and head and neck cancer (27); BRAF fusions in melanoma (28, 29); and RET fusions in thyroid cancers (ref. 30; Fig. 2A).
Some oncogenic rearrangements that have been explored as pan-tumor biomarkers were detected across different cancer types. However, the clonality, that is, the ratio between the VAF and TF of the sample, of FGFR2, BRAF, RET, and ALK rearrangements was not uniform in these cancer types. FGFR2 had high clonality in pancreatic cancer and CCA, but tended to be a minor allele (VAF/TF ≥25%) in gastroesophageal and colorectal cancer (P < 0.05 for all pairwise comparisons). Pancreatic, prostate cancer, and NSCLC tended to have higher clonality BRAF rearrangements than melanoma and colorectal cancer (P < 0.001 for all comparisons). RET fusions tended to have higher clonality in NSCLC, whereas they tended to be subclonal variants in colorectal cancer (P < 0.0001) and breast cancer (P = 0.04). ALK fusions were found predominantly in NSCLC and were a major allele in 89% of cases, but tended to be a minor allele in breast cancer (P = 0.01) and colorectal cancer (P < 0.0001; Fig. 2B). These suggest a rearrangement of a particular oncogene may not always be a truncal oncogenic driver, especially in cancer types like colorectal cancer where fusions tended to be minor alleles. Examining tissue and LBx from the same patient, GOF rearrangements in these genes that were detected in both tissue and liquid had higher median VAF/TF than those detected only in the LBx: 87% versus 5.0% for FGFR2, 30% versus 1.8% for BRAF, 84% versus 4.4% for ALK, and 50% versus 2.5% for RET (Fig. 2C). Additional data on breakpoints, fusion partners, and comparison to tissue prevalence, and more details on Fig. 2C is provided in Supplementary Tables S3–S6.
Rare GOF fusions included NRG1 fusions (9): eight of these were detected in NSCLC, and seven were fusions to CD74. EWSR1 fusions (19) were detected fused to ATF1 (5) and FLI1 (5) and were detected among five soft tissue sarcoma, two Ewing sarcoma, three unknown primary, and nine other cancer types (Supplementary Fig. S1). Rearrangements in CD274 predicted to disrupt the 3′UTR, stabilize the transcript, and increase PD-L1 expression (31, 32) were detected among 42 LBx (9 NSCLC, 6 CUP, 5 head and neck, 5 liver, and 17 other types with N < 5 each).
Potential driver rearrangements that were found predominantly in one cancer type included TMPRSS2-ERG [1,440/1,432 (98%)] detected in prostate cancer, ALK [350/428 (82%)] detected in NSCLC, CTNNB1 [61/118 (52%)] detected in colorectal cancer, and FGFR2 [85/285 (30%)] detected in CCA (Supplementary Table S7).
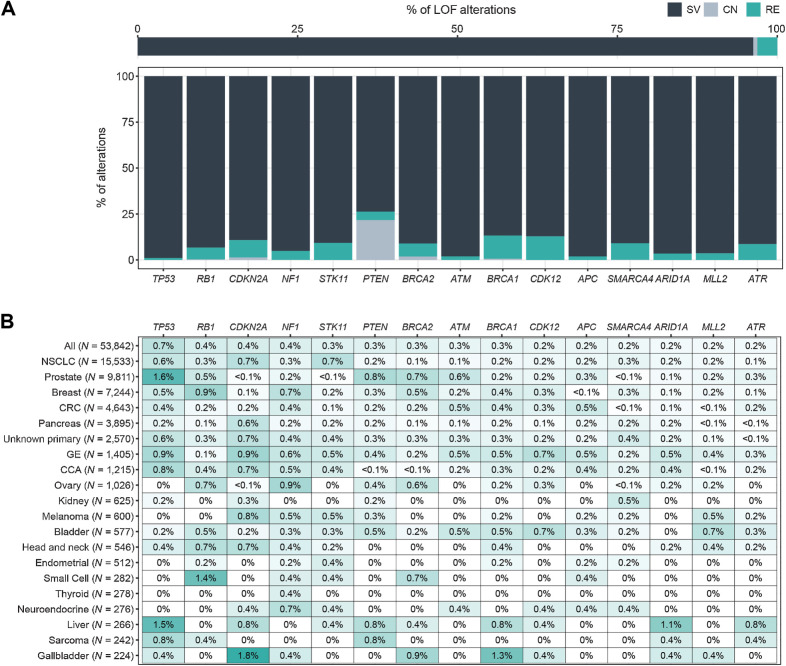
Pathogenic rearrangements in tumor suppressor genes
Although the majority of tumor suppressor gene disruptions detected by CGP are short variants (frameshift, nonsense mutations, splice site alterations), 3.1% of pathogenic variants in these genes were rearrangements (Fig. 3A). The tumor suppressor genes most frequently disrupted by large-scale rearrangements in this study were: TP53 (0.7% of all LBx), RB1 (0.4%), CDKN2A (0.4%), NF1 (0.4%), STK11 (0.3%), and PTEN (0.3%). These LOF rearrangements did not show patterns of cancer type enrichment as strong as GOF rearrangements. However, STK11 truncations were more common in NSCLC, RB1 in breast cancer and small cell lung carcinoma, NF1 in ovarian and breast cancers, PTEN in prostate cancer, and BRCA2 in prostate, breast, and ovarian cancers, consistent with the established roles of these tumor suppressors in the oncogenesis of the corresponding cancer types (Fig. 3B). Rare but potentially clinically actionable truncating rearrangements were also detected in MTAP (35 LBx, including 7 NSCLC and 5 CUP) and MEN1 (17 LBx, including 8 breast cancer and 3 NSCLC).
Figure 3.
LOF rearrangements. A, Relative prevalence of short variants (SV), copy-number deletions (CN), and rearrangements (RE) predicted to disrupt tumor suppressor genes: for all tumor suppressor genes (top), and for the top 15 rearranged tumor suppressor genes (bottom). CN is only reported for PTEN and BRCA1/2. B, Heatmap of the prevalence of the most frequently disrupted tumor suppressor genes in the pan-tumor cohort. Cancer types with ≥200 LBx and genes altered in >0.5% of at least 1 cancer type are shown.
In rare instances, rearrangements can restore function in tumor suppressor genes under therapeutic selective pressure, such as reversion events in BRCA1/2 (33–36). Rearrangements predicted to skip BRCA2 exons containing deleterious short variants detected in the same LBx were found in 26 LBx (12 in breast, 11 in prostate, 1 each in ovarian, pancreas, and CUP).
FGFR2 oncogenic rearrangements in cholangiocarcinoma
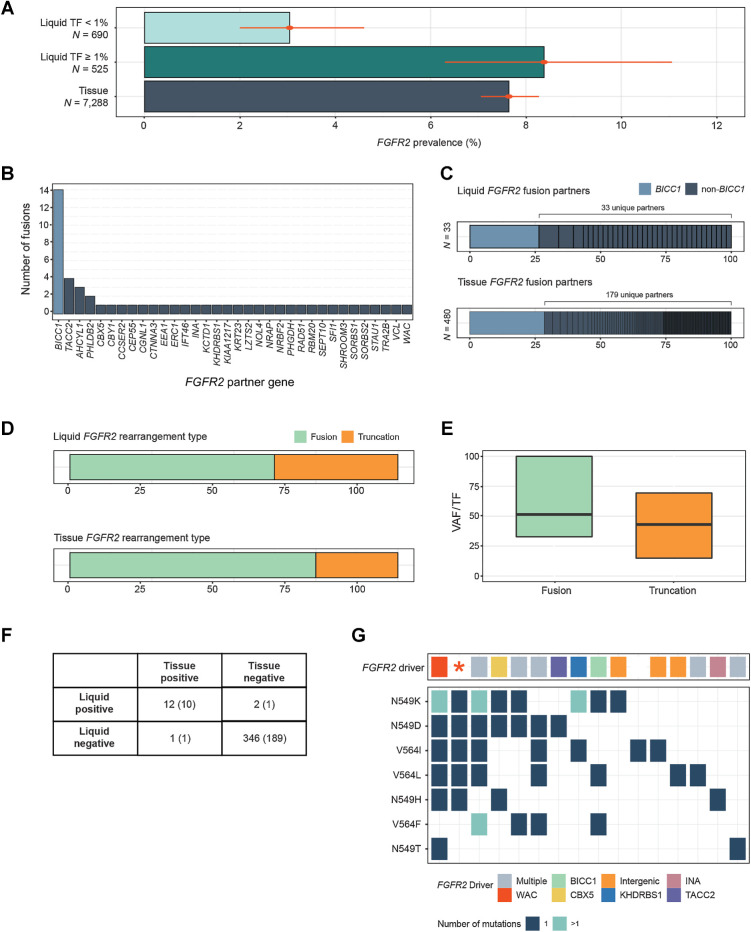
The overall prevalence of FGFR2 rearrangements in CCA LBx was 5.3% (Fig. 2A), lower than the 7.6% observed in tissue (P = 0.004). However, among LBx samples with TF ≥1% (525/1,215; 43%), the prevalence of 8.4% was comparable to tissue (Fig. 4A). FGFR2 fusions had breakpoints inside intron 17 or close to its junctions. Consistent with previous reports, BICC1, which resides near FGFR2 on chromosome 10, was the most common fusion partner gene (26% of FGFR2 fusions), but was only one of 34 partner genes found (Fig. 4B). This mirrors the distribution observed in tissue biopsies (Fig. 4C) and reported in the AACR GENIE database (37).
Figure 4.
Detection of FGFR2 rearrangements in cholangiocarcinoma. A, Comparison of the prevalence of FGFR2 activating rearrangements among CCA tissue biopsies and liquid biopsies with TF ≥1% and <1%. B, Gene partners in rearrangements predicted to encode FGFR2 fusion genes. C, A comparison between the diversity of FGFR2 fusion gene partners in CCA tissue and liquid biopsies. D, A comparison of the prevalence of FGFR2 fusions versus truncations in CCA tissue and liquid biopsies. E, The clonality (VAF/TF) of FGFR2 fusions and rearrangements in CCA liquid biopsies. F, Concordance of FGFR2 rearrangement detection in a set of samples from the same patient (201 CCA, 160 CUP pairs). Numbers in parentheses are the concordance results within CCA pairs alone. G, Results from 16 LBx where FGFR inhibitor resistance mutations were detected (one sample per vertical column). The top row shows the presumed FGFR2 driver, while the grid below shows the presence of particular FGFR inhibitor acquired resistance mutations. Colors indicate the gene fusion partners detected, “intergenic” indicates a truncation without a specific fusion partner, and the red asterisk denotes a sample with a FGFR2 C382R driver mutation. In 15 of 16 samples, a FGFR2 driver variant was detected alongside resistance mutations.
Of 85 FGFR2 pathogenic rearrangements, 53 (62%) were fusions and 32 (38%) were truncations or deletions of exon 18 (23, 38), all of which are predicted to encode a FGFR2 receptor that retains the kinase domain but lacks the regulatory C-terminal tail. This was somewhat higher than the relative prevalence of FGFR2 truncations in tissue (Fig. 4D). The median VAF/TF for these two types of rearrangements was similar and close to 50% (51% and 43%, respectively). Truncations were more likely to be found at lower allele frequency (P = 0.02). However, 25% of FGFR2+ LBx had multiple FGFR2 rearrangement events detected, versus 14% of tissue biopsies, and the samples with multiple events tended to have fusions and truncations present together.
In CCA and CUP paired samples from the same patient, the sensitivity of LBx to detect FGFR2 rearrangements detected in tissue was 92% (12/13; 95% CI, 67%–99%). The TF of the LBx that did not detect the FGFR2 variant from tissue was 0.5%. The two samples with detection in liquid but not tissue had subclonal rearrangements with VAF/TF 0.003% and 0.73%. The negative predictive value (NPV) of CCA LBx samples was 99% (189/190; 95% CI, 97%–99%; Fig. 4F).
Among 16 LBx where mutations associated with acquired resistance to FGFR inhibitors were detected, 14 also detected the driver FGFR2 fusion or truncation, 1 harbored a C382R mutation in FGFR2 functioning as a driver (its VAF was 24% whereas the resistance mutation VAFs ranged from 0.26% to 3.3%; ref. 39), and 1 LBx had no FGFR2 driver detected (the mutation's VAF was 0.13% and it was the sole variant detected in the sample). In total, an FGFR2 driver variant was detected in 94% (15/16) of samples where it was expected based on the presence of resistance mutations (Fig. 4G).
Driver fusion detection in NSCLC
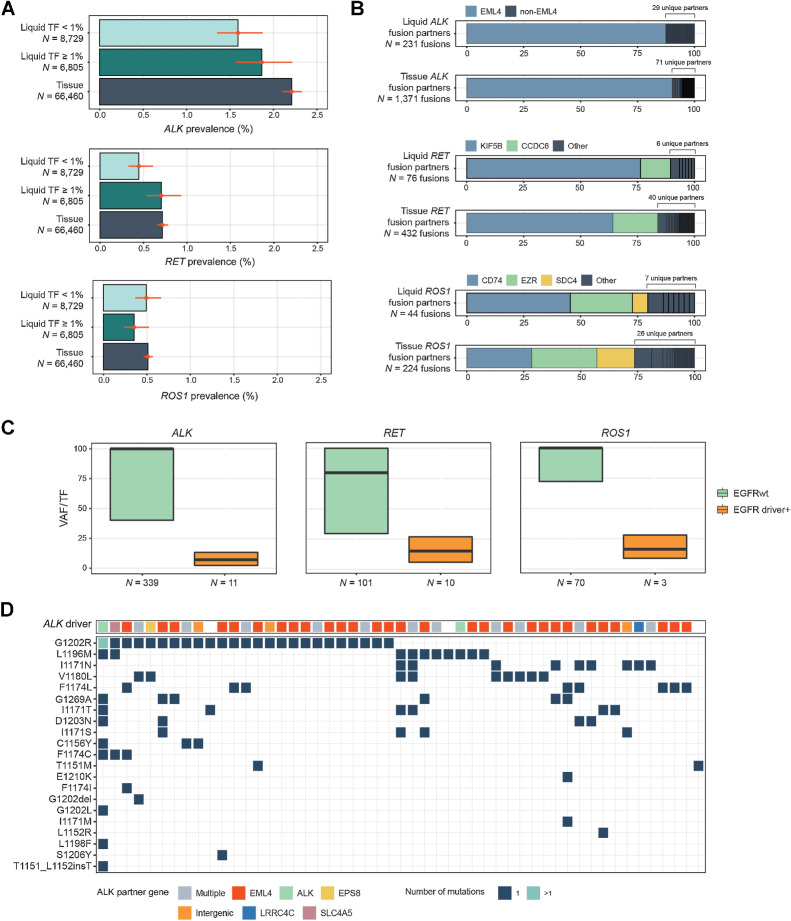
The prevalence of ALK and RET fusions in NSCLC LBx was 1.7% and 0.6%, respectively (Fig. 2A), which was lower than the prevalence detected in tissue: 2.2% and 0.7% (P = 0.0002; 0.05). However, the prevalence of these fusions in the 6,805/15,534 (44%) of LBx with TF ≥1% was more compatible with tissue: 1.9% and 0.7% (Fig. 5A). ROS1 fusion detection was comparable with tissue regardless of TF in this cohort (0.5% in tissue, 0.4% in LBx, P = 0.23). EML4 accounted for 87% of ALK fusion partners (30 unique genes), KIF5B (76%) and CCDC6 (13%) of RET fusion partners (8 unique genes), and CD74 (45%) and EZR (27%) of ROS1 fusion partners (10 unique genes). Tissue showed similar partner distributions (Fig. 5B).
Figure 5.
Detection of kinase rearrangements in NSCLC. A, Comparison of the prevalence of activating rearrangements in ALK, RET, and ROS1 among NSCLC tissue biopsies and liquid biopsies with TF ≥1% and <1%. B, A comparison of the diversity of fusion gene partners in ALK, RET, and ROS1 fusions in tissue and liquid biopsies. C, Comparison of the clonality of ALK, RET, and ROS1 rearrangements in samples with and without EGFR driver short variants (L858R, exon 19 deletion, or exon 20 insertion). D, Results from 51 LBx where ALK inhibitor resistance mutations were detected (one sample per vertical column). The top row shows the ALK fusion driver, while the grid below shows the presence of particular ALK inhibitor acquired resistance mutations. Colors indicate the gene fusion partners detected. In 47/51 samples; an ALK fusion was detected alongside resistance mutations.
ALK, RET, and ROS1 fusions tended to be a major allele in NSCLC. Notably, their VAF/TF ratio was lower when an EGFR driver mutation was also present in the sample (Fig. 5C). This finding could reflect fusions appearing as acquired resistance to EGFR inhibitors (40, 41), or the possible presence of multiple primaries (42).
Of 51 LBx with mutations associated with acquired resistance to ALK inhibitors, 47 (92%) also detected the ALK driver rearrangement (Fig. 5D). In 62 patients with an ALK fusion detected in tissue biopsy and a LBx collected, LBx detected the ALK fusion in 42 cases (68% sensitivity). However, in the LBx with TF ≥1%, sensitivity was 95% (20/21 concordantly detected). Similar patterns were observed for RET (23 RET+ tissue pairs) and ROS1 (19 ROS1+ tissue pairs) fusion detection (Supplementary Fig. S2). The sensitivity at several TF thresholds was examined in these NSCLC pairs. Sensitivity was 57% [95% confidence interval (95% CI), 47%–66%] if no threshold was applied, and rose sharply to 92% (95% CI, 79%–97%) even at the lower threshold of TF ≥0.5% (Supplementary Fig. S3). Frequency of TF ≥1% across different cancer types is provided in Supplementary Table S8.
Pathogenic rearrangements in prostate cancer
The prevalence of TMPRSS2–ERG fusions in prostate cancer LBx was 12% (Fig. 2A), which was lower than the 28% rate in tissue (P = 2E-220). Among the 4,148/9,811 (42%) of LBx with TF ≥1%, the prevalence was more comparable with tissue: 25% (Fig. 6A). BRAF rearrangements (43) were also identified at a similar rate (∼1.5%) in tissue and LBx with TF ≥1%. Rearrangements in the androgen receptor (AR) gene are a type of acquired resistance to androgen deprivation therapy or AR inhibitors (44). Consistent with this, we found the prevalence of AR rearrangements in LBx, which are often collected after androgen deprivation therapy exposure, was dramatically higher than in our tissue cohort (4% of all LBx, 9% of LBx with TF ≥1%, 0.5% in TBx, P = 3E−112; Fig. 6A). Although TMPRSS2–ERG fusions and BRAF rearrangements had a median VAF/TF of 24% and 39%, respectively, AR rearrangements were subclonal events (median VAF/TF 1.5%), consistent with their role as acquired resistance, which is often heterogeneous and polyclonal (Fig. 6B). Among 3,054 prostate cancer LBx with evidence of castrate-resistant AR variants, 419 (14%) had activating AR rearrangements. Of those, 154 (37%) were the sole detectable AR variant in the sample (Fig. 6C).
In 96 patients with TMPRSS2–ERG fusion detected in tissue biopsy and a LBx collected, LBx detected the ERG fusion in 52 cases (54% sensitivity). However, in the LBx with TF ≥1%, sensitivity was 87% (46/53 concordantly detected) versus 14% (6/43) in TF <1% (Supplementary Fig. S2).
Pathogenic rearrangements in colorectal cancer
Colorectal cancer had a high overall prevalence of pathogenic rearrangements (5.4% of LBx with GOF rearrangements, 8.6% with LOF, 1% with both; Fig. 1). However, some of the oncogenic rearrangements tended to be detected as minor alleles (Fig. 2B). Examining the clonality of rearrangements prevalent in colorectal cancer, CTNNB1, TP53, PTEN, APC, NTRK1, and SOX9 tended to be major alleles (median VAF/TF >25%), whereas GOF rearrangements in kinases (MET, ROS1, ALK, FGFR2/3, RAF1, EGFR, BRAF, RET) and LOF rearrangements in DNA repair genes (BRCA2, ATR, ATM) often represented subclones (Fig. 6D). MSI-H colorectal cancer tumors have been reported to be enriched for targetable kinase fusions (45), but MSI-H samples accounted for only 19/493 (3.9%) of the rearrangements in colorectal cancer liquid biopsies.
Because some of the kinase rearrangements potentially represent acquired resistance to anti-EGFR antibody therapy (46), we compared the prevalence of these rearrangements in LBx with a clonal KRAS/NRAS mutation (RAS-positive) versus LBx with no clonal KRAS/NRAS/BRAF mutations (RAS-negative; from patients who may have been treated with anti-EGFR antibody therapy prior to LBx collection) in LBx with TF ≥1% (Fig. 6E). Activating rearrangements in EGFR, BRAF, CDK12, and MYC were significantly more prevalent in RAS-negative LBx (FDR < 0.05). Other kinase genes more often rearranged in RAS-negative LBx were FGFR2/3, MET, RET, and NTRK1. The only rearrangement significantly enriched in RAS-positive LBx (FDR < 0.05) was CTNNB1. These CTNNB1 activating rearrangements, which are predicted to excise exon 3 and disrupt a degron motif and stabilize the β-catenin transcription factor (47), and we queried whether they co-occur with mutations in APC, another Wnt/β-catenin pathway component frequently mutated in colorectal cancer. CTNNB1 rearrangements tended to be found in APC-wild-type samples: 6% (3/49) of CTNNB1-rearranged LBx were APC-altered, versus 84% (2,399/2,865) APC-altered samples in colorectal cancer LBx with TF ≥1% overall.
Discussion
Detection of genomic rearrangements by next-generation sequencing of LBx has historically been considered difficult. Several reports interrogating commercially available and laboratory developed assays have found inferior detection of genomic rearrangements in LBx compared with DNA-based tissue biopsy assays, including lower rates of detection, low concordance in matched samples, and reduced diversity of fusion gene partners (2, 37, 48–53).
High confidence CGP results are critical to support evidence-based care of patients with cancer, but assay performance characteristics vary from manufacturer to manufacturer due to different design strategies and validation standards and the performance of one assay does not necessarily predict the accuracy of other tests using the same analyte (50). Well-designed DNA-based CGP platforms that can reliably detect fusion and rearrangement events must bait relevant intronic regions including in common partner genes, use hybrid-capture probes tiled at an appropriate density, sequence at a depth tailored to the application, and include an analytical pipeline that is capable of partner-gene-agnostic rearrangement calls. De novo assembly, rather than reliance on reference genomes, is especially important because breakpoints often feature novel sequences introduced by DNA repair. An additional consideration for LBx in particular is that the presence of ctDNA cannot be ascertained prior to sequencing. A post hoc estimate of the tumor content in the sample is valuable for the interpretation of a negative result. In this study, the sensitivity of the LBx platform to detect driver rearrangements was much higher in samples with TF ≥1% than those with TF <1%. In contrast to some prior literature, this study demonstrated detection of a wide variety of rearrangements in LBx, comparable prevalence of driver rearrangements in tissue and LBx when TF ≥1%, and distribution of fusion gene partners that resembles that of tissue biopsies.
In our study, cholangiocarcinoma was the cancer type with the highest prevalence of kinase fusions. This is a disease where there is often a lack of tissue to even make a diagnosis, and the utility of LBx to be able to reliably detect these actionable aberrations is of clinical value, given multiple approved FGFR inhibitors and available clinical trials (54–56). In parallel, the cancers of unknown primary that often present as a liver mass frequently turn out to be cholangiocarcinoma in the majority of cases. In these instances, the detection of FGFR2 fused with a common BICC1 partner not only has therapeutic potential but is diagnostic of the disease as well.
Kinase fusions remain the most actionable type of rearrangement (57) but transcription factor fusions represent attractive, largely still untapped therapeutic targets that could be exploited using novel modalities such as targeted protein degradation (58). Some GOF rearrangements are so typical of a particular cancer type (TMPRSS2-ERG, EML4-ALK) that they may aid in diagnosis when the site of origin of a tumor is uncertain, similar to the earlier discussion of FGFR2 and cholangiocarcinoma.
Although generally less actionable than GOF rearrangements, rearrangements that inactivate tumor suppressor genes may be overlooked biomarkers. RB1 rearrangements are enriched in neuroendocrine tumors in this study and could help detect neuroendocrine transformation. STK11 loss is a marker of immunotherapy resistance in NSCLC. Tumors with BRCA1/2 inactivating rearrangements may be more sensitive to PARP inhibitors due to a low likelihood of reversion, but BRCA1/2 rearrangements may also function as reversions themselves.
Intriguingly, this study found that certain rearrangements often considered to be pan-tumor biomarkers are more likely to be truncal drivers in some cancer types than in others. Rearrangements in FGFR2 in CCA and pancreatic cancers, BRAF in pancreatic and prostate cancers, RET in thyroid cancers and NSCLC, and ALK in NSCLC tended to have high clonality. In contrast, in colorectal cancer these potentially targetable rearrangements tended to be minor alleles and found in KRAS-wild-type samples, which may have been collected from patients treated with EGFR mAb therapy, consistent with prior reports that such fusions may be resistance mechanisms to anti-EGFR therapy (46, 59). Gastroesophageal cancers had frequent FGFR2 rearrangements but at variable clonality, suggesting they may sometimes be acquired or secondary events when the gene is amplified (60). This analysis underscores the utility of a TF estimate in LBx as a tool to determine clonality. Further investigation into the association of clinical outcomes with the clonality of putative driver variants is warranted because heterogeneous tumors where fusion genes are a minor variant may not be as sensitive to targeted monotherapy. Beyond distinguishing truncal and acquired variants, these findings intimate that serial LBx could be used to track changes in a tumor's predominant clones over time and inform treatment selection, since new clones can emerge and take over under therapeutic pressure. For detection of fusions that are acquired events, as in colorectal cancer post EGFR-blockade, LBx serve as a more practical tool since repeated tissue biopsies are not necessarily safe, feasible, or practical. These are clinically actionable events as shown by work from Clifton and colleagues (46).
It has been proposed that RNA-based methods of rearrangement detection have superior sensitivity, because they do not require sequencing intronic regions, and profile highly expressed transcripts, with less ambiguity about the resultant fusion genes than DNA-based methods. Although successful fusion detection using multiplex NGS of circulating tumor RNA (ctRNA) has been reported (61), it is a more unstable analyte than RNA extracted from FFPE specimens and the workflow has not been scaled for the analytical and repeatability rigor of an FDA-approved, globally available assay. In addition, although there is an advantage in preferentially detecting highly expressed GOF rearrangements, RNA profiling may not detect LOF rearrangements where the transcript becomes unstable.
There were certain limitations in this study. The cohort consisted of LBx submitted in the course of routine clinical care, thus there may be some inherent biases. For instance, patients who test positive for a fusion driver in a tissue biopsy or a single-gene LBx test may be less likely to have a LBx submitted for CGP. A limitation in our analysis was the lack of patient information beyond cancer type, age, and gender. The analysis of rearrangements as potential resistance mechanisms thus lacked data on intervening treatments in these cohorts. The findings of this analysis are intended to be hypothesis-generating for future studies. Detection rates were compared between tissue and liquid biopsies, but both assays were DNA-based. This study does not include data from RNA-based detection to compare the performance and sensitivity of the two methods. Future studies comparing DNA- and RNA-based hybrid capture targeted panels are warranted to address this question. When analyzing concordance, our paired analysis of sensitivity and negative predictive value for FGFR2 rearrangements was conducted using a convenience cohort of noncontemporaneously collected tissue and LBx, and reliance on tissue as standard presupposes that these rearrangements are truncal events.
These results show the ability of LBx to detect pathogenic rearrangements. Many of these alterations are targetable kinase fusions. LBx can report a rich, polyclonal landscape in patients with advanced, heavily treated disease. It can increase opportunities of precision medicine for patients for whom NGS of tissue may not be feasible or completed in a timely fashion. Because LBx have a faster turnaround and are non-invasive, a consideration for a liquid-first approach could also be a more practical and cost-effective consideration. Since some rearrangements are highly specific to certain cancer types, LBx could potentially be a powerful tool during diagnostic workup of advanced cancers where the site of origin is uncertain. Taken together, these results provide further evidence for LBx-based CGP by a well-designed assay to be a sensitive, pragmatic method for detection of rearrangements in solid tumors.
Supplementary Material
Supplementary Figures
Supplementary Tables
Acknowledgments
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
P.M. Kasi reports personal fees from Foundation Medicine, MSD Oncology/Merck, Tempus, Bayer, Lilly, Delcath Systems, QED Therapeutics, Servier, Taiho Oncology, Exact Sciences, Daiichi Sankyo/AstraZeneca, Eisai, Seattle Genetics, SAGA Diagnostics, Illumina, BostonGene, Guardant Health, and Taiho; personal fees and other support from Elicio Therapeutics; other support from Precision Biosensors Inc.; and grants from Merck, Novartis, and Agenus Bio outside the submitted work. J.K. Lee reports personal fees from Foundation Medicine and Roche during the conduct of the study. L.W. Pasquina reports other support from Foundation Medicine during the conduct of the study, as well as other support from Roche Holding outside the submitted work. B. Decker reports other support from Foundation Medicine and Roche during the conduct of the study, as well as other support from Barinthus Biotherapeutics outside the submitted work; B. Decker also has numerous patents related to detection of cancer in liquid biopsy pending. D.C. Pavlick reports personal fees from Foundation Medicine and F. Hoffmann-La Roche AG during the conduct of the study. J.M. Allen reports employment with Foundation Medicine and ownership of Foundation Medicine stock. C. Parachoniak reports personal fees from Foundation Medicine, as well as other support from Roche outside the submitted work. J.C.F. Quintanilha reports other support from Foundation Medicine, Inc. during the conduct of the study. R.P. Graf reports personal fees from Foundation Medicine, Inc. during the conduct of the study, as well as ownership of Roche stock. A.B. Schrock reports personal fees from Foundation Medicine and Roche during the conduct of the study. G.R. Oxnard reports personal fees and other support from Eli Lilly, as well as other support from Roche outside the submitted work. C.M. Lovly reports grants from GO2 Foundation/ALK Positive, LUNGevity/EGFR Resistors, and NCI during the conduct of the study, as well as personal fees from Amgen, AnHeart, Arrivent, AstraZeneca, Blueprints Medicine, BMS, Boehringer Ingelheim, Cepheid, D2G, Daiichi Sankyo, EMD Serono, Foundation Medicine, Genentech, Gilead, Indupro, Janssen, Novartis, Pfizer, Puma, Regeneron, Roche, Takeda, and Tempus outside the submitted work; C.M. Lovly also reports employment with Janssen. H. Tukachinsky reports personal fees from Foundation Medicine Inc. during the conduct of the study, as well as reports ownership of Roche stock. V. Subbiah reports travel fees and honorarium from Foundation Medicine for keynote speaker at the annual summit 2023. V. Subbiah also reports research funding/grant support (to institution: MD Anderson Cancer Center) for conducting clinical trials from AbbVie, Agensys, Inc., Alfasigma, Altum, Amgen, Bayer, BERG Health, Blueprint Medicines Corporation, Boston Biomedical, Inc., Boston Pharmaceuticals, Celgene Corporation, D3 Bio, Inc., Dragonfly Therapeutics, Inc., Exelixis, Fujifilm, GlaxoSmithKline, Idera Pharmaceuticals, Inc., Incyte Corporation, Inhibrx, Loxo Oncology, MedImmune, MultiVir, Inc., NanoCarrier, Co., National Comprehensive Cancer Network, NCI-CTEP, Northwest Biotherapeutics, Novartis, PharmaMar, Pfizer, Relay Therapeutics, Roche/Genentech, Takeda, Turning Point Therapeutics, UT MD Anderson Cancer Center, and Vegenics Pty Ltd; travel support from ASCO, ESMO, Helsinn Healthcare, Incyte Corporation, Novartis, and PharmaMar; consultancy/advisory board participation for Helsinn Healthcare, Jazz Pharmaceuticals, Incyte Corporation, Loxo Oncology/Eli Lilly, MedImmune, Novartis, QED Therapeutics, Relay Therapeutics, Daiichi-Sankyo, R-Pharm US, Illumina, Bayer, Medscape, and OncLive (Educational CME); a current relationship with Sarah Cannon Research Institute; scientific advisory board participation (to institution) for Relay Therapeutics, Pheon Therapeutics, Incyte, Novartis, Eli Lilly/Loxo Oncology, Roche, Pfizer, Bayer, AbbVie, Regeneron, and Clinical Care Communications (CME fees for education). No disclosures were reported by the other authors.
Authors' Contributions
P.M. Kasi: Conceptualization, writing–review and editing. J.K. Lee: Data curation, formal analysis, visualization, methodology, writing–original draft, writing–review and editing. L.W. Pasquina: Conceptualization, supervision, writing–review and editing. B. Decker: Data curation, writing–review and editing. P. Vanden Borre: Data curation, writing–review and editing. D.C. Pavlick: Writing–review and editing. J.M. Allen: Data curation, writing–review and editing. C. Parachoniak: Data curation, writing–review and editing. J.C.F. Quintanilha: Writing–review and editing. R.P. Graf: Writing–review and editing. A.B. Schrock: Writing–review and editing. G.R. Oxnard: Conceptualization, supervision, writing–review and editing. C.M. Lovly: Writing–review and editing. H. Tukachinsky: Conceptualization, visualization, methodology, writing–original draft, writing–review and editing. V. Subbiah: Conceptualization, writing–review and editing.
References
- 1. Guibert N, Pradines A, Favre G, Mazieres J. Current and future applications of liquid biopsy in non-small cell lung cancer from early to advanced stages. Eur Respir Rev 2020;29:190052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sugimoto A, Matsumoto S, Udagawa H, Itotani R, Usui Y, Umemura S, et al. A large-scale prospective concordance study of plasma- and tissue-based next-generation targeted sequencing for advanced non-small cell lung cancer (LC-SCRUM-Liquid). Clin Cancer Res 2023;29:1506–14. [DOI] [PubMed] [Google Scholar]
- 3. Tran MC, Strohbehn GW, Karrison TG, Rouhani SJ, Segal JP, Shergill A, et al. Brief report: discordance between liquid and tissue biopsy-based next-generation sequencing in lung adenocarcinoma at disease progression. Clin Lung Cancer 2023;24:e117–21. [DOI] [PubMed] [Google Scholar]
- 4. National Comprehensive Cancer Network. Non-small cell lung cancer. 2023. Available from:https://www.nccn.org/.
- 5. National Comprehensive Cancer Network. Biliary tract cancers. 2023. Available from:https://www.nccn.org/.
- 6. National Comprehensive Cancer Network. Thyroid carcinoma. 2023. Available from:https://www.nccn.org/.
- 7. National Comprehensive Cancer Network. Bladder cancer. 2023. Available from:https://www.nccn.org/.
- 8. National Comprehensive Cancer Network. Melanoma: cutaneous. 2023. Available from:https://www.nccn.org/. [Google Scholar]
- 9. Poulikakos PI, Sullivan RJ, Yaeger R. Molecular pathways and mechanisms of BRAF in cancer therapy. Clin Cancer Res 2022;28:4618–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shao J, Yan Y, Ding D, Wang D, He Y, Pan Y, et al. Destruction of DNA-binding proteins by programmable oligonucleotide PROTAC (O'PROTAC): effective targeting of LEF1 and ERG. Adv Sci (Weinh) 2021;8:e2102555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sluiter MD, van Rensburg EJ. Large genomic rearrangements of the BRCA1 and BRCA2 genes: review of the literature and report of a novel BRCA1 mutation. Breast Cancer Res Treat 2011;125:325–49. [DOI] [PubMed] [Google Scholar]
- 12. Davies HR, Broad KD, Onadim Z, Price EA, Zou X, Sheriff I, et al. Whole-genome sequencing of retinoblastoma reveals the diversity of rearrangements disrupting RB1 and uncovers a treatment-related mutational signature. Cancers (Basel) 2021;13:754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bekaii-Saab TS, Bridgewater J, Normanno N. Practical considerations in screening for genetic alterations in cholangiocarcinoma. Ann Oncol 2021;32:1111–26. [DOI] [PubMed] [Google Scholar]
- 14. Husain H, Pavlick DC, Fendler BJ, Madison RW, Decker B, Gjoerup O, et al. Tumor fraction correlates with detection of actionable variants across >23,000 circulating tumor DNA samples. JCO Precis Oncol 2022;6:e2200261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rostami A, Lambie M, Yu CW, Stambolic V, Waldron JN, Bratman SV. Senescence, necrosis, and apoptosis govern circulating cell-free DNA release kinetics. Cell Rep 2020;31:107830. [DOI] [PubMed] [Google Scholar]
- 16. Stejskal P, Goodarzi H, Srovnal J, Hajdúch M, van ’t Veer LJ, Magbanua MJM. Circulating tumor nucleic acids: biology, release mechanisms, and clinical relevance. Mol Cancer 2023;22:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Woodhouse R, Li M, Hughes J, Delfosse D, Skoletsky J, Ma P, et al. Clinical and analytical validation of FoundationOne Liquid CDx, a novel 324-Gene cfDNA-based comprehensive genomic profiling assay for cancers of solid tumor origin. PLoS One 2020;15:e0237802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Frampton GM, Fichtenholtz A, Otto GA, Wang K, Downing SR, He J, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol 2013;31:1023–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Milbury CA, Creeden J, Yip W-K, Smith DL, Pattani V, Maxwell K, et al. Clinical and analytical validation of FoundationOne(R)CDx, a comprehensive genomic profiling assay for solid tumors. PLoS One 2022;17:e0264138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Takeuchi K, Soda M, Togashi Y, Suzuki R, Sakata S, Hatano S, et al. RET, ROS1 and ALK fusions in lung cancer. Nat Med 2012;18:378–81. [DOI] [PubMed] [Google Scholar]
- 21. Scaravilli M, Koivukoski S, Latonen L. Androgen-driven fusion genes and chimeric transcripts in prostate cancer. Front Cell Dev Biol 2021;9:623809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ho Y, Dehm SM. Androgen receptor rearrangement and splicing variants in resistance to endocrine therapies in prostate cancer. Endocrinology 2017;158:1533–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zingg D, Bhin J, Yemelyanenko J, Kas SM, Rolfs F, Lutz C, et al. Truncated FGFR2 is a clinically actionable oncogene in multiple cancers. Nature 2022;608:609–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Arai Y, Totoki Y, Hosoda F, Shirota T, Hama N, Nakamura H, et al. Fibroblast growth factor receptor 2 tyrosine kinase fusions define a unique molecular subtype of cholangiocarcinoma. Hepatology 2014;59:1427–34. [DOI] [PubMed] [Google Scholar]
- 25. Wu Y-M, Su F, Kalyana-Sundaram S, Khazanov N, Ateeq B, Cao X, et al. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov 2013;3:636–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Williams SV, Hurst CD, Knowles MA. Oncogenic FGFR3 gene fusions in bladder cancer. Hum Mol Genet 2013;22:795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang Z, Anderson KS. Therapeutic targeting of FGFR signaling in head and neck cancer. Cancer J 2022;28:354–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hutchinson KE, Lipson D, Stephens PJ, Otto G, Lehmann BD, Lyle PL, et al. BRAF fusions define a distinct molecular subset of melanomas with potential sensitivity to MEK inhibition. Clin Cancer Res 2013;19:6696–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ross JS, Wang K, Chmielecki J, Gay L, Johnson A, Chudnovsky J, et al. The distribution of BRAF gene fusions in solid tumors and response to targeted therapy. Int J Cancer 2016;138:881–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vodopivec DM, Hu MI. RET kinase inhibitors for RET-altered thyroid cancers. Ther Adv Med Oncol 2022;14:17588359221101691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kelly AD, Murugesan K, Kuang Z, Montesion M, Ross JS, Albacker LA, et al. Pan-cancer landscape of CD274 (PD-L1) rearrangements in 283,050 patient samples, its correlation with PD-L1 protein expression, and immunotherapy response. J Immunother Cancer 2021;9:e003550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hoskins EL, Samorodnitsky E, Wing MR, Reeser JW, Hopkins JF, Murugesan K, et al. Pan-cancer landscape of programmed death ligand-1 and programmed death ligand-2 structural variations. JCO Precis Oncol 2023;7:e2200300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang Y, Bernhardy AJ, Nacson J, Krais JJ, Tan Y-F, Nicolas E, et al. BRCA1 intronic Alu elements drive gene rearrangements and PARP inhibitor resistance. Nat Commun 2019;10:5661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang Y, Krais JJ, Bernhardy AJ, Nicolas E, Cai KQ, Harrell MI, et al. RING domain-deficient BRCA1 promotes PARP inhibitor and platinum resistance. J Clin Invest 2016;126:3145–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Decker BJ, Domchek SM, Nathanson K, Bailey S, Danziger N, Thornton J, et al. Landscape of homologous recombination reversion mutations in pancreaticobiliary malignancies. J Clin Oncol 2022;40:4156.35998299 [Google Scholar]
- 36. Domchek SM, Reiss KA, Nathanson K, Bailey S, Danziger N, Thornton J, et al. Landscape of homologous recombination reversion mutations in gynecologic malignancies. J Clin Oncol 2022;40:5576. [Google Scholar]
- 37. Berchuck JE, Facchinetti F, DiToro DF, Baiev I, Majeed U, Reyes S, et al. The clinical landscape of cell-free DNA alterations in 1671 patients with advanced biliary tract cancer. Ann Oncol 2022;33:1269–83. [DOI] [PubMed] [Google Scholar]
- 38. Silverman IM, Hollebecque A, Friboulet L, Owens S, Newton RC, Zhen H, et al. Clinicogenomic analysis of FGFR2-rearranged cholangiocarcinoma identifies correlates of response and mechanisms of resistance to pemigatinib. Cancer Discov 2021;11:326–39. [DOI] [PubMed] [Google Scholar]
- 39. Hempel L, Lapa C, Dierks A, Gaumann A, Scheiber J, Veloso de Oliveira J, et al. A new promising oncogenic target (p.C382R) for treatment with pemigatinib in patients with cholangiocarcinoma. Ther Adv Med Oncol 2022;14:17588359221125096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schrock AB, Zhu VW, Hsieh W-S, Madison R, Creelan B, Silberberg J, et al. Receptor tyrosine kinase fusions and BRAF kinase fusions are rare but actionable resistance mechanisms to EGFR tyrosine kinase inhibitors. J Thorac Oncol 2018;13:1312–23. [DOI] [PubMed] [Google Scholar]
- 41. Piotrowska Z, Isozaki H, Lennerz JK, Gainor JF, Lennes IT, Zhu VW, et al. Landscape of acquired resistance to osimertinib in EGFR-mutant NSCLC and clinical validation of combined EGFR and RET inhibition with osimertinib and BLU-667 for acquired RET fusion. Cancer Discov 2018;8:1529–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fan J, Dai X, Wang Z, Huang B, Shi H, Luo D, et al. Concomitant EGFR mutation and EML4-ALK rearrangement in lung adenocarcinoma is more frequent in multifocal lesions. Clin Lung Cancer 2019;20:e517–30. [DOI] [PubMed] [Google Scholar]
- 43. Chehrazi-Raffle A, Tukachinsky H, Toye E, Sivakumar S, Schrock AB, Bergom HE, et al. Unique spectrum of activating BRAF alterations in prostate cancer. Clin Cancer Res 2023;29:3948–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jernberg E, Bergh A, Wikstrom P. Clinical relevance of androgen receptor alterations in prostate cancer. Endocr Connect 2017;6:R146–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cocco E, Benhamida J, Middha S, Zehir A, Mullaney K, Shia J, et al. Colorectal carcinomas containing hypermethylated mlh1 promoter and wild-type BRAF/KRAS are enriched for targetable kinase fusions. Cancer Res 2019;79:1047–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Clifton K, Rich TA, Parseghian C, Raymond VM, Dasari A, Pereira AAL, et al. Identification of actionable fusions as an anti-EGFR resistance mechanism using a circulating tumor DNA assay. JCO Precis Oncol 2019;3:PO.19.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yaeger R, Chatila WK, Lipsyc MD, Hechtman JF, Cercek A, Sanchez-Vega F, et al. Clinical sequencing defines the genomic landscape of metastatic colorectal cancer. Cancer Cell 2018;33:125–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mondaca S, Lebow ES, Namakydoust A, Razavi P, Reis-Filho JS, Shen R, et al. Clinical utility of next-generation sequencing-based ctDNA testing for common and novel ALK fusions. Lung Cancer 2021;159:66–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li BT, Janku F, Jung B, Hou C, Madwani K, Alden R, et al. Ultra-deep next-generation sequencing of plasma cell-free DNA in patients with advanced lung cancers: results from the actionable genome consortium. Ann Oncol 2019;30:597–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Supplee JG, Milan MSD, Lim LP, Potts KT, Sholl LM, Oxnard GR, et al. Sensitivity of next-generation sequencing assays detecting oncogenic fusions in plasma cell-free DNA. Lung Cancer 2019;134:96–9. [DOI] [PubMed] [Google Scholar]
- 51. Kwon M, Ku BM, Olsen S, Park S, Lefterova M, Odegaard J, et al. Longitudinal monitoring by next-generation sequencing of plasma cell-free DNA in ALK rearranged NSCLC patients treated with ALK tyrosine kinase inhibitors. Cancer Med 2022;11:2944–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bearz A, Martini J-F, Jassem J, Kim S-W, Chang G-C, Shaw AT, et al. Phase 3 trial of lorlatinib in treatment-naive patients (Pts) with ALK-positive advanced non–small cell lung cancer (NSCLC): comprehensive plasma and tumor genomic analyses. J Clin Oncol 2022;40:9070. [Google Scholar]
- 53. Cui S, Zhang W, Xiong L, Pan F, Niu Y, Chu T, et al. Use of capture-based next-generation sequencing to detect ALK fusion in plasma cell-free DNA of patients with non-small-cell lung cancer. Oncotarget 2017;8:2771–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Goyal L, Meric-Bernstam F, Hollebecque A, Valle JW, Morizane C, Karasic TB, et al. Futibatinib for FGFR2-rearranged intrahepatic cholangiocarcinoma. N Engl J Med 2023;388:228–39. [DOI] [PubMed] [Google Scholar]
- 55. Abou-Alfa GK, Sahai V, Hollebecque A, Vaccaro G, Melisi D, Al-Rajabi R, et al. Pemigatinib for previously treated, locally advanced or metastatic cholangiocarcinoma: a multicentre, open-label, phase 2 study. Lancet Oncol 2020;21:671–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Javle M, Roychowdhury S, Kelley RK, Sadeghi S, Macarulla T, Weiss KH, et al. Infigratinib (BGJ398) in previously treated patients with advanced or metastatic cholangiocarcinoma with FGFR2 fusions or rearrangements: mature results from a multicentre, open-label, single-arm, phase 2 study. Lancet Gastroenterol Hepatol 2021;6:803–15. [DOI] [PubMed] [Google Scholar]
- 57. Lee JK, Hazar-Rethinam M, Decker B, Gjoerup O, Madison RW, Lieber DS, et al. The pan-tumor landscape of targetable kinase fusions in circulating tumor DNA. Clin Cancer Res 2022;28:728–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bekes M, Langley DR, Crews CM. PROTAC targeted protein degraders: the past is prologue. Nat Rev Drug Discov 2022;21:181–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bray SM, Lee J, Kim ST, Hur JY, Ebert PJ, Calley JN, et al. Genomic characterization of intrinsic and acquired resistance to cetuximab in colorectal cancer patients. Sci Rep 2019;9:15365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Greer SU, Nadauld LD, Lau BT, Chen J, Wood-Bouwens C, Ford JM, et al. Linked read sequencing resolves complex genomic rearrangements in gastric cancer metastases. Genome Med 2017;9:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Choudhury Y, Cher CY, Ho JM, Chan C, Teh MW, Ngeow KC, et al. A cell-free RNA-based next-generation sequencing (NGS) assay for the detection of actionable gene fusions in patients with non–small cell lung cancer (NSCLC). J Clin Oncol 2022;40:3040. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures
Supplementary Tables
Data Availability Statement
All relevant data are provided within the article and its accompanying Supplementary Data. Because of HIPAA requirements, we are not consented to share individualized patient genomic data, which contains potentially identifying or sensitive patient information. Foundation Medicine is committed to collaborative data analysis, and have well-established, and widely utilized mechanisms by which investigators can query their core genomic database of >600,000 deidentified sequenced cancers to obtain aggregated datasets. More information and mechanisms for data access can be obtained by contacting the corresponding authors or the Foundation Medicine Data Governance Council at data.governance.council@foundationmedicine.com.