Abstract
Although the structural rearrangement of the membrane-bound matrix (MA) protein trimers upon HIV-1 maturation has been reported, the consequences of MA maturation on the MA-lipid interactions are not well understood. Long-timescale molecular dynamics simulations of the MA multimeric assemblies of immature and mature virus particles with our realistic asymmetric membrane model have explored MA-lipid interactions and lateral organization of lipids around MA complexes. The number of stable MA-phosphatidylserine and MA-phosphatidylinositol 4,5-bisphosphate (PIP2) interactions at the trimeric interface of the mature MA complex is observed to be greater compared to that of the immature MA complex. Our simulations identified an alternative PIP2-binding site in the immature MA complex where the multivalent headgroup of a PIP2 lipid with a greater negative charge binds to multiple basic amino acid residues such as ARG3 residues of both the MA monomers at the trimeric interface and highly basic region (HBR) residues (LYS29, LYS31) of one of the MA monomers. Our enhanced sampling simulations have explored the conformational space of phospholipids at different binding sites of the trimer-trimer interface of MA complexes that are not accessible by conventional unbiased molecular dynamics. Unlike the immature MA complex, the 2′ acyl tail of two PIP2 lipids at the trimeric interface of the mature MA complex is observed to sample stable binding pockets of MA consisting of helix-4 residues. Together, our results provide molecular-level insights into the interactions of MA trimeric complexes with membrane and different lipid conformations at the specific binding sites of MA protein before and after viral maturation.
Significance
HIV-1 maturation facilitates the conversion of a newly formed immature virus particle to a mature infectious virion and initiates a new round of infection. The contributions of specific protein-lipid interactions in the HIV-1 assembly process are well recognized; however, the interactions of matrix protein lattice with the membrane before and after HIV-1 maturation are yet to be fully understood. Based on our simulated data, supported by prior experimental observations, the present study provides a molecular-level understanding of a possible altered binding mode of PIP2 lipids after viral maturation. Identification of protein-lipid-specific interactions and lipid-sorting data obtained from our long-time and large-scale atomistic molecular dynamics simulations advance the understanding of the HIV-1 matrix and membrane maturation.
Introduction
During the late phase of the human immunodeficiency virus type 1 (HIV-1) replication cycle, the newly synthesized major structural protein, Gag, binds to and multimerizes on the inner leaflet of the host cell plasma membrane (PM), leading to viral assembly, budding, and release of immature virus particles (1). Subsequent to the budding process, newly formed immature virions undergo the proteolytic maturation process, resulting in the cleavage of Gag into its structural proteins (2,3). Gag protein is composed of several structural domains having different roles in viral replication and infectivity. N-terminal myristoylated matrix (Myr-MA) domain mediates membrane binding of Gag and envelope (Env) glycoprotein incorporation into virus particles (4,5,6,7,8,9). Past studies have indicated that the capsid (CA) domain plays a critical role in Gag oligomerization, whereas the nucleocapsid (NC) domain promotes higher-order multimerization of Gag through the binding to viral genomic RNA during the viral assembly process (10,11,12,13,14). The virion maturation process leads to a major structural rearrangement of all three folded domains and a dramatic change in viral morphology that alters the protein-protein, protein-lipid, and protein-RNA interactions (1,11,15,16,17,18,19). In the mature infectious virion, the matrix (MA) domain remains associated with the virion membrane, whereas CA protein forms the outer shell of the core CA structure, and the NC domain condenses with the viral genomic RNA. Furthermore, maturation of the matrix domain is believed to be associated with the rearrangement of the Env glycoprotein and membrane maturation (19,20).
It has been recognized that the HIV-1 membrane differs notably from the producer cell PM. The viral membrane is enriched in anionic phospholipids, e.g., phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2 or PIP2), which enhances the membrane-binding affinity of the MA domain (21,22). Experimental data reported an exposure of phosphatidylserine (PS) and phosphatidylethanolamine (PE) lipids on the external surface of mature virions, and there may therefore be at least partial loss of the asymmetric lipid distribution of the host cell PM in the virus particles (23,24,25). MA-membrane interactions are facilitated by several key factors: 1) nonspecific electrostatic interactions between basic amino acid residues of MA and anionic membrane lipids (phosphatidylinositol (PI), PS), 2) specific interactions between MA protein and PIP2, and 3) hydrophobic interactions with lipid tails mediated by the N-terminal Myr group of MA, a post-translational modification covalently attached to the N-terminal glycine residue (26,27,28,29,30,31,32,33). This Myr moiety can adopt two different conformations: sequestered in a hydrophobic pocket of MA and a Myr-exposed state (34). Nuclear magnetic resonance (NMR) experiments (9,32) and molecular dynamics (MD) simulations (35) have revealed that MA-PIP2 interactions and MA trimerization facilitate Myr exposure. Although the inner leaflet of the membrane forms microdomains of anionic phospholipids induced by the MA-lipid interactions, the outer leaflet forms raft-like lipid domains, enriched in sphingomyelin (SM) and cholesterol (36). Further, these nanodomains in the two leaflets of the asymmetric bilayer are proposed to be correlated by trans-bilayer coupling (37,38). The impact of the altered MA-MA interactions after viral maturation on the membrane lateral organization is yet to be explored.
In a prior cryoelectron tomography (cryo-ET) study of MA lattice organization in the immature and mature HIV-1 particles, a rearrangement of the MA trimer-trimer interface upon maturation was reported, and altered MA-PIP2 interactions were observed (19). In the immature virion, MA trimer-trimer interactions are governed by N-terminal residues, helix-1, and 310 helix residues. On the other hand, in the mature virion, the MA trimer-trimer interface is stabilized by the interactions between the basic residues of the highly basic region (HBR) loop with the acidic residues of the N terminus of helix-4 and the 310 helices of adjacent MA monomers. Unlike the immature virion, the cryo-ET density map (Electron Microscopy Data Bank (EMDB): 13088) of the mature virus particle shows lipid density at the PIP2-binding pockets; however, cryo-ET data do not allow us to identify specific lipid molecules. In the past, experimental studies by Saad et al., Shkriabai et al., and Chukkapalli et al. demonstrated different MA-PIP2-binding modes (32,39,40). Among these, the extended PIP2 lipid configuration reported by the NMR study is consistent with the electron density map (32). In this lipid configuration, the 2′-acyl chain, being partially removed from the bilayer, binds to a hydrophobic cleft of the MA protein. In another recent experimental study, Saad and coworkers characterized a myristoylated MA lattice structure that has some features similar to those in the immature HIV-1 particles and verified protein-protein binding in the MA assembly structure using X-ray crystallography and NMR spectroscopy (41). They also interrogated an alternate PIP2-binding site in that MA lattice using NMR titration with inositol 1,4,5-triphosphate, the headgroup of the PIP2 lipid.
Unlike a system of integral membrane proteins, the determination of the specific protein-lipid binding properties of a peripheral membrane protein (PMP) such as HIV-1 MA remains an outstanding challenge both for experimental and computational studies because of their transient nature (32,42,43,44,45,46,47,48,49). Long-timescale MD simulations are needed to sample PMP binding with a membrane, which are computationally expensive even for a small monomeric PMP (35,50,51,52). In a previous all-atom MD (AAMD) study, using microsecond-long trajectories, the membrane-binding mechanism of MA monomers was characterized, including Myr insertion events and the initial membrane response to MA binding (35). An enrichment of PIP2 at the MA-binding site was reported, as has been suggested several times before (31,53,54). The analyses by Monje-Galvan et al. revealed that initial membrane targeting is mediated by the HBR domain, whereas Myr insertion plays an important role in sorting of membrane lipids around the protein binding site and maintaining the stable membrane binding of MA. Interestingly, the cryo-ET density maps suggest that all the Myr groups predominantly remain in the exposed state while being inserted into the membrane. Therefore, characterization of membrane-bound MA assembly structures employing AAMD simulations requires Myr groups of all the MA monomers stably inserted into the membrane, which makes the present study even more challenging.
To gain molecular-level insights into the altered MA-MA and MA-lipid interactions upon HIV-1 maturation, we applied long-timescale AAMD simulations using cryo-ET fitted atomic model structures of MA assemblies and a realistic asymmetric membrane model. We also performed biased free-energy sampling simulations using a combination of steered MD (SMD) and restrained MD (rMD) simulations to sample the conformational space of the MA-bound lipids. Some of the key findings of our study are as follows:
-
(1)
Our AAMD simulations verified the stability of MA-MA interactions reported by cryo-ET data and identified additional interactions in the trimeric interfaces.
-
(2)
Simulations explored PIP2/PS-binding sites in MA protein complexes. Here we report an alternate PIP2-binding site at the immature MA trimer-trimer interface.
-
(3)
Enhanced sampling simulations explore novel conformations of PIP2/PS lipids at the trimeric interface of both complexes and obtain a stable binding of two PIP2 lipid molecules at extended conformations where the 2′ acyl tail of PIP2 interacts with helix-4 residues of MA, unlike in the immature MA complex.
-
(4)
Time-averaged lipid density maps and quantitative analyses of lipid count/fraction data in the vicinity of the MA trimeric interfaces suggest an enrichment of inner-leaflet anionic phospholipids (PIP2) as well as outer-leaflet cholesterol lipids near the MA trimer-trimer interface in the mature MA complex compared to immature MA complex.
The following sections report detailed analyses of our simulated data as well as our simulation methodology.
Materials and methods
All-atom models of the MA trimeric assemblies
The initial all-atom protein configuration was generated by fitting an atomic model of MA trimer (PDB: 1HIW) to the cryo-ET density maps, EMDB-13087 and EMDB-13088, for the immature and mature MA lattice structures, respectively. The monomeric MA structure (PDB: 1UPH) (9) with all MA residues up to TYR132 was then superimposed to MA assemblies to obtain the final dimer-of-trimers MA structures to simulate. The Myr group, in an exposed conformation, was covalently attached to its N-terminal GLY1 residue using CHARMM-GUI input generator (55). In the simulated model, residue 1 is composed of the Myr group and GLY residue. Therefore, the residuen of the experimental PDB structure (PDB: 1UPH) corresponds to residuen-1 of the simulated models. A fully hydrated bilayer was built and equilibrated using CHARMM-GUI Membrane Builder and Quick-Solvator, respectively (55,56,57,58,59). Next, protein coordinates were positioned close to the inner leaflet of the membrane using VMD software (60) and those lipid molecules overlapping with Myr groups were removed. The protein-membrane merged structures for both the immature and mature MA were solvated in 150 mM aqueous KCl solution with initial box lengths of 15 × 15 × 25 nm to allow enough water layers to be present between the protein units and the periodic image of the outer leaflet. The total size of both systems was ∼580,000 atoms.
Membrane model and protein-membrane system setup
The membrane model designed for this study was based on lipidomic analysis of HIV-1 particles produced in HeLa cells reported by Lorizate et al. (61). The approximate composition of the extracellular leaflet was 35% cholesterol, 30% phosphatidylcholine (PC), and 35% SM and the cytoplasmic leaflet was 15% cholesterol, 40% PC, 15% PE, 15% PS, and 15% phosphatidylinositol (PIP2). This model mimics the asymmetric nature of the PM and allows us to examine the effect of trans-bilayer interactions that become operational upon protein binding. We have used the same membrane model to study MA-lipid interactions in immature and mature virions. As discussed earlier, the membrane of mature virions differs from the PM in terms of lipid asymmetry in the two leaflets. However, we do not have quantitative information about the leaflet asymmetry in the mature HIV-1 membrane. The asymmetric bilayer with a surface area of 15 × 15 nm was built using CHARMM-GUI Membrane Builder and Quick-Solvator (55,56,57,58) following a multistep minimization and equilibration protocol. This bilayer system was further equilibrated for 400 ns before merging the equilibrated protein coordinates using VMD software (60). All the data shown in the manuscript were computed for the membrane system with DOPC (Dioleoylphosphatidylcholine), POPE (1-Palmitoyl-2-oleoylphosphatidylethanolamine), DOPS (Dioleoylphosphatidylserine), SAPI25 (phosphatidylinositol), LSM (sphingolipid), and cholesterol. However, we have verified MA-MA and MA-lipid interactions with another membrane model with POPC, POPE, POPS, SAPI, LSM, and cholesterol. We have considered 15% PIP2 in the inner leaflet, which is 6% of the total number of lipid molecules in the bilayer (please note here that the outer leaflet contains more lipid molecules than the inner leaflet of our model membrane due to the difference in lipid packing). The PIP2 concentration in our model membrane has been chosen to be higher than the HIV-1 membrane (∼2% (22)) deliberately so that it allows us to study specific MA-PIP2 interactions within the simulation timescale. Despite that, 4-μs trajectories did not attain symmetric MA-PIP2 interactions for two MA trimers in the immature MA complex (Fig. 3). The lipid-sorting behavior reported in the manuscript was verified using multiple trajectories. We have also compared MA trimer-membrane binding with different concentrations of PIP2 and the binding sites remain unchanged at lower PIP2 concentration (Fig. S7).
Figure 3.
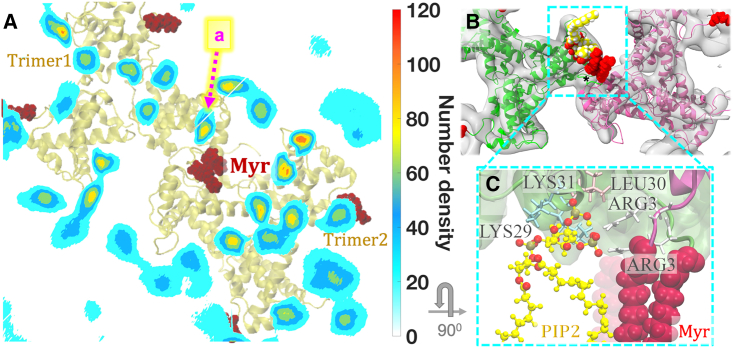
Alternate PIP2-binding site at the immature MA trimer-trimer interface. (A) Density map of the PIP2 lipid headgroups in the bilayer plane overlapped with immature MA complex, as viewed from the top toward the virus center. The alternate stable binding site at the trimer-trimer interface is denoted as “a.” (B) Acyl tails of PIP2 lipid molecule (yellow) at this binding site transiently interact with Myr groups (red). Cryo-ET density marked with an asterisk corresponds to N-terminal residues linked to Myr groups. (C) A close-up view of the residues involved in the alternate PIP2-binding site, which includes positively charged LYS29 and LYS31 residues of the HBR domain of MA and N-terminal ARG3 residues of both the MA monomers at trimer-trimer interface. To see this figure in color, go online.
MD simulations and analysis
We have used the CHARMM36m force field (62) for protein and lipids and CHARMM TIP3P for water (63,64). Simulations were performed in GROMACS 2019 MD software (65). Energy minimization was performed using the steepest descent algorithm until the maximum force was less than 1000 kJ mol−1nm−1. Then, equilibration was performed with harmonic restraints (using a 1000 kJ mol−1nm−2 spring constant) on each heavy atom throughout the protein for 1 ns in the constant NVT (canonical) ensemble with a time step of 1 fs, followed by a 15-ns constant NVT MD run with a time step of 2 fs and a 15-ns MD run in the constant NPT ensemble with a time step of 2 fs. Next, each system was allowed to run for a total of 2000 and 3000 ns, respectively, for immature and mature MA complexes in the constant NPT ensemble, following the parallel cascade selection MD procedure, to allow Myr insertion to occur. During this phase, MA assembly structures were kept intact by applying harmonic restraints (1000 kJ mol−1nm−2) on Cα-backbone atoms of protein monomers, except for the first 10 N-terminal residues including the Myr group. Next, 4-μs trajectories were generated for both the MA dimer-of-trimer complexes with all the six Myr groups inserted into the membrane. Previous restraints on the protein backbones were removed for production runs. For the immature MA complex, to mimic the effect of uncleaved Gag protein, we have applied small harmonic restraints (50 kJ mol−1nm−2) to the Cα atom of ALA119 residues (C-terminal residue of helix 5). To further confer stability to the immature MA dimer-of-trimers assembly structure, harmonic restraints (100 kJ mol−1nm−2) were applied to the N-terminal residues (residues 4–8) of the four peripheral MA monomers (not at the trimer-trimer interface) to emulate the binding of immature MA lattice. No such restraints were applied to the mature MA complex. Among the five replicas of the production runs, the attainment of specific protein-lipid interactions ensured the stability of the MA complexes.
Throughout this procedure, the temperature was kept constant at 310.15 K using the Nosé -Hoover thermostat with a 1.0-ps coupling constant (66,67), and the pressure was set at 1 bar and controlled using the Parinello-Rahman barostat semi-isotropically due to the presence of the membrane. The compressibility factor was set at 4.5 × 10−5 bar−1 with a coupling time constant of 5.0 ps (68,69). van der Waals interactions were computed using a force-switching function between 1.0 and 1.2 nm, whereas long-range electrostatics were evaluated using particle mesh Ewald (70) with a cutoff of 1.2 nm, and hydrogen bonds were constrained using the LINCS algorithm (71).
In addition, we performed biased MD simulations to explore the conformational space of PS and PIP2 lipids at the trimer-trimer interface of immature and mature MA complexes. Model fitting was done in ChimeraX. Protein-protein and protein-lipid interactions were visualized using VMD 1.9.3. (60). Analyses of the trajectories were performed using GROMACS 2019 (65) and VMD Tcl scripts (60). Lipid density maps were generated using the Python package Matplotlib (72).
Biased MD simulation details
To investigate the “extended lipid conformations” predicted by NMR experiments and cryo-ET density map, we have explored PIP2 and PS lipid conformational space employing SMD and rMD simulations. PIP2 and PS lipids at the trimer-trimer interface for immature and mature MA assemblies were pulled out of the membrane from multiple initial configurations selected from unbiased MD trajectory. SMD simulations were carried out for 10 ns using constant velocity pulling (0.5 nm per nanosecond) using a biasing force constant of 1000 kJ mol−1 nm−2. In these simulations, pulling the headgroup of lipids away from the membrane bilayer was not successful to sample extended lipid conformation. However, pulling the center of mass (COM) of each lipid molecule along the membrane normal, maintaining the MA-lipid (headgroup) interactions, sampled such extended lipid conformations for some selected PIP2 lipids. From each SMD trajectory, multiple configurations were selected depending on the distance between the COM of lipids and membrane bilayer, and rMD simulations were performed for 150 ns using a harmonic potential of 1000 kJ mol−1 nm−2 along the reaction coordinate. A total of ∼20 μs of SMD and rMD simulations were carried out and, finally, the stability of extended lipid conformations obtained for mature and immature MA assemblies was assessed by performing unbiased MD simulations (1 μs).
Results
MA trimer-trimer interactions in immature and mature virion states
As discussed earlier, MA trimers in the immature virion get rearranged upon HIV-1 maturation. Our simulations explored atomic-level stable and transient protein-protein interactions between the membrane-bound MA trimers in the immature and the mature MA complexes (Figs. 1 and 2). MA monomers consist of five α helices and one shorter 310 helix between helix-2 and helix-3 (Fig. S1 in supporting material). A flexible loop domain, connecting helix-1 and helix-2, is enriched with basic residues, denoted as HBR. As mentioned earlier, the HBR domain and Myr group play a crucial role in the membrane targeting and stable membrane binding of MA. Myr-MA protein primarily oligomerizes as trimers and further multimerizes in vitro and in virus to form a lattice of hexamer of trimers (19,31). To assess trimer-trimer interactions (TTIs) of MA assembly in the immature and the mature virions, we performed AAMD simulations with a dimer of MA trimers (derived from cryo-ET density map (19)) bound to an asymmetric membrane. The study of protein-protein interactions (PPIs) in such a PMP complex is challenging mainly for two reasons: 1) we simulate only a part of the continuous lattice structure, which renders instability or distortion to the simulated structures; 2) myristoyl insertion of the MA monomers requires long-timescale simulations. However, in both the assemblies, a comparison of the simulated structures and the cryo-ET density map verifies that Myr groups in both immature and mature MA lattice structures remained inserted into the membrane. Although the MA trimer-trimer interface structure is stable during simulations, simulated trajectories captured some deviations of the MA structures from cryo-ET density maps, especially for the membrane-bound N-terminal helix-1 in both immature and mature MA models.
Figure 1.
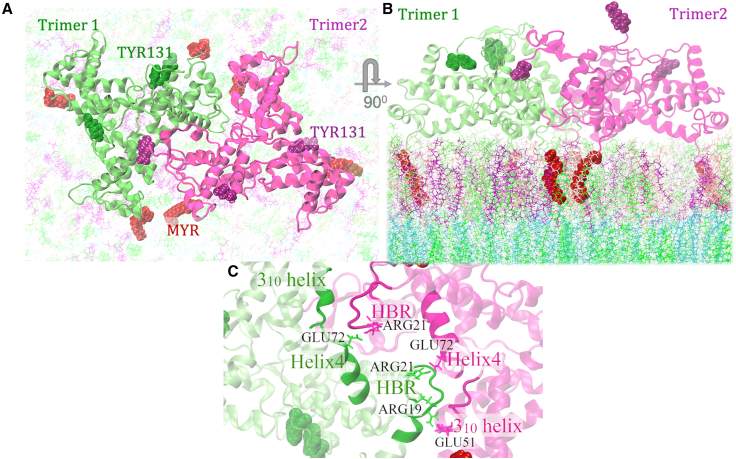
MA TTIs in the immature HIV-1 particles. (A and B) Bottom (as viewed from the virus center) and side views of MA protein assembly (dimer of trimers) with asymmetric membrane at the end of 4-μs simulation. Solvent molecules have been excluded for clarity. N-terminal Myr groups are inserted into the inner leaflet of the bilayer, which is consistent with the cryo-ET density map (EMDB: 13087). (C) MA domains participating in TTIs are mainly N-terminal domains, H1, H4, and 310 helices. To see this figure in color, go online.
Figure 2.
MA TTIs in the mature HIV-1 particles. (A and B) Bottom (as viewed from virus center) and side views of MA protein assembly(dimer of trimers) with asymmetric membrane at the end of 4-μs simulation. Solvent molecules have been excluded for clarity. N-terminal Myr groups are inserted into the cytoplasmic leaflet of the membrane. (C) TTIs are mainly mediated by highly basic region (HBR), H4, and 310 helices. Important residues stabilizing the trimer-trimer interface through electrostatic interactions are highlighted. To see this figure in color, go online.
Figs. 1 and 2 present the snapshots of MA complexes at the end of 4-μs simulations. The production runs of both systems started with MA dimer of trimers already attached to the membrane with the Myr groups being inserted into the inner leaflet of the bilayer in a stable configuration. Before the production runs, spontaneous Myr insertion of all MA monomers was achieved in long-timescale simulations when MA assembly structures were kept intact by applying harmonic restraints on Cα-backbone atoms of protein monomers (see “materials and methods” section for details). Unlike in the mature MA assembly, only two MA monomers, belonging to different trimers, form the trimer-trimer interface in the immature MA assembly. The cryo-ET-derived structural model reported that TTIs in the immature MA assembly are mainly mediated by the N-terminal residues and the residues from H1 and 310 helices (19). Throughout our simulations, these interactions remained stable. N-terminal residues of each MA monomer interact with N-terminal residues and H1 helix residues of the other MA monomer in the immature MA assembly. These interactions are mainly mediated by hydrophobic residues ALA2, ALA4, VAL6, and GLY9. Some of the stable contacts explored by our simulations are ALA2:ALA2, ALA2:ALA4, GLY9:VAL6, SER5:GLY9, and ALA4:GLY9. Myr residues of both the MA monomers remain inserted into the membrane in a stable configuration and interact with each other, rendering stabilization to the trimer-trimer interface (Figs. 1 and S2). The N-terminal residues, including ARG3, are observed to offer a stable binding site for PIP2 lipid in the immature assembly, which is discussed later in detail. Interestingly, in the immature MA assembly, N-terminal residues, which assist in the conformational switch during Myr insertion, mediate TTIs. As observed in our simulated trajectories, LEU30 of the flexible HBR loop transiently interacts with hydrophobic residues of the N-terminal domain of the same and the other MA monomer. Apart from these, our simulations explored a stable electrostatic interaction between ARG90 (the C-terminal residue of H4) of one MA monomer with the acidic residue of 310 helix, GLU51, of the other MA monomer across the trimeric interface. Although mutagenesis studies suggested that mutation of residues, important for MA trimerization, blocks Env incorporation and replication (30), GLU51 mutant was observed to have no effect on virus replication kinetics (73). Mutation of LEU30 does not affect viral particle production but blocks the Env incorporation and results in the production of noninfectious virions (74). As mentioned earlier, structures of MA monomers at the trimer-trimer interface throughout the simulated trajectories show some deviations from the cryo-ET density map. Interestingly, the membrane-bound helices of MA trimer 1 in the immature MA complex (with a bound PIP2 molecule at the alternate binding site) fit better to the cryo-ET density map than the other MA trimer. MA-PIP2 binding is discussed in the next section in detail.
By contrast, in the mature MA assembly, the reorganization of trimers enables positively charged residues from HBR domains to participate in the TTIs (19). Unlike the immature MA assembly, two monomers of each trimer mediate TTIs. HBR residues of one MA monomer of trimer 1 bind to residues of helix-4 and 310 helices of the other two monomers of trimer 2 (19). In our simulations, arginine residues from the HBR domain (ARG19 and ARG21) bind to acidic glutamate residues (GLU51 and GLU72) of 310 and helix-4 (Figs. 2 and S3). Unlike the ARG21:GLU72 interaction, our simulations did not sample ARG19:GLU51 interaction in both pairs of monomers at the trimer-trimer interface. Our simulations explored additional TTIs mediated by C-terminal cleaved end and helix 5 residues. In our simulated systems of mature MA, TYR131 of trimer 2 (shown as purple beads in Fig. 2) binds to ARG57 of trimer 1 through stable cation-pi interactions. Apart from ARG57, the binding pocket contains LYS112, GLY61, ILE103, TYR78, ALA114, LEU60, GLN58, GLN115, and GLN64. It should be noted that we report here stable MA-MA interactions as sampled by our simulated model systems. However, the protein crowding effect in a mature virion can affect this kind of protein-PPI mediated by TYR131 and, therefore, this mode of MA-MA interactions might not play an important role in the virion.
Overall, AAMD simulations with the membrane-bound dimer of MA trimers suggest Myr groups of two MA monomers at the dimeric interface, while inserted into the bilayer, interact with each other, thus stabilizing the MA trimer-trimer association in the immature virion. As reported by cryo-ET data, N-terminal residues and the residues of H1 and 310 helices mediate TTIs in the immature MA assembly. Besides these interactions, primarily hydrophobic in nature, our simulations suggest that electrostatic interactions between GLU51 (310 helix residue) and ARG90 (H4 helix residue) play an important role in stabilizing the trimer-trimer interface of immature MA assembly (Fig. 1). On the other hand, in the mature virion, two MA monomers from each trimer comprise the inter-trimer interface, and the interactions are mediated mainly by the basic residues of HBR domains and the acidic residues of 310 and H4 helices. The interactions between ARG21 (HBR residue) and GLU72 (H4 helix residue) in both pairs of MA monomers at the trimer-trimer interface were maintained throughout our simulated trajectories (Fig. S3). Apart from these, GLU51 was observed to interact with ARG19 of the HBR domain in our simulations. The reorganization of MA trimers is presumed to affect the MA-lipid interactions, which is discussed in the following sections.
Specific interactions of the protein complexes with PIP2 lipids
PIP2, a minor phospholipid component of the PM, plays a crucial role in the HIV-1 assembly process (27,75,76,77). A common feature of all eukaryotic PMs is the asymmetric lipid distribution in its two leaflets, PIP2 being primarily concentrated on the cytoplasmic (inner) leaflet of PM. This lipid molecule contains a negatively charged multivalent inositol head group that remains a few angstroms above the membrane surface. Furthermore, with one saturated tail (C18:0, 1′ acyl chain) and one polyunsaturated tail (C20:4, 2′ acyl chain), this lipid molecule can cause lipid packing defects in the membrane (78).
Long-timescale unbiased AAMD simulations of Myr-MA assemblies bound to the asymmetric model membrane revealed specific lipid-binding sites at the interface of two MA trimers, where MA-membrane binding is mediated by the electrostatic interactions of the lipid headgroups with basic amino acid residues. Time-averaged number density profiles of PIP2 headgroup atoms are computed with the last 200 ns of trajectories. The lipid density map reports a low PIP2 lipid density at the trimer-trimer interface of immature MA assembly. An alternate binding site of PIP2 was observed (denoted as “a” in Fig. 3) in which a multivalent inositol headgroup interacts with ARG3 of both the monomers at the interface and LYS29, LYS31 of T1:M3 (monomer 3 of trimer 1) (Fig. S4). Simulated data agree with an NMR titration experiment with inositol 1,4,5-triphosphate (the headgroup of PIP2 lipid), which reported an alternate group of signals corresponding to N-terminal residues and residues 27–32 for immature MA-like crystal structure (41). The acyl chains of this PIP2 lipid have been observed to interact transiently with Myr groups at the trimer-trimer interface. It should be noted here that our simulations did not attain symmetric MA-PIP2 interactions, especially for the binding site “a,” although we have considered higher PIP2 concentration in our model membrane (∼6%) compared to HIV-1 virion (∼2%) deliberately so that the MA-PIP2 contacts can be established in the stable binding sites of MA complexes within the simulation timescale.
For the mature MA assembly, our AAMD simulations revealed that the trimer-trimer interface is enriched with PIP2 lipids. Inositol headgroups of PIP2 lipids bind to the charged residues of the HBR domain at the trimer-trimer interface. As discussed earlier, using an NMR approach, Saad et al. demonstrated one possible mode of direct binding of MA with a truncated, soluble PIP2 derivative (tr-PI(4,5)P2), in which tr-PI(4,5)P2 adopts an extended conformation with 2′-acyl chain bound to a hydrophobic cleft of MA containing ARG76, SER77, THR81, and TRP36 (32). This NMR structure is consistent with the cryo-ET density map (EMDB-13088), which contains density components at those PIP2-binding pockets of mature MA (19). Taken together, NMR and the cryo-ET experimental data lead to an important question: besides the role of PIP2 in membrane targeting and stable membrane binding of MA, does it play a crucial role in HIV-1 membrane maturation?
To examine this hypothesis, we aimed to explore the conformational space of MA-bound anionic phospholipid molecules, mainly multivalent PIP2 and monovalent PS lipids at the trimer-trimer interface of both the immature and mature MA complexes. To investigate possible extended lipid conformations, we performed nonequilibrium SMD simulations with multiple initial structures selected from the last 500 ns of 4-μs trajectory for both the complexes (see “materials and methods” section for details). The COM of the selected lipid molecules was pulled along the z direction away from the bilayer center using SMD simulations, followed by rMD and unbiased MD simulations (1 μs) to examine the lipid conformational stability for these two MA complexes.
Among several selected PIP2- and PS-binding sites, PIP2 lipids in two of them (the combined density is denoted as “a” in Fig. 4 A) sampled a stable extended conformation in which 2′-fatty acid chain binds to the MA protein, whereas the 1′ lipid tail is still inserted into the membrane. We performed multiple umbrella sampling simulations with reaction coordinate (z distance between COM of the lipid and membrane center) ranging between 2.9 and 3.2 nm when the lipid explores binding pockets of the proteins. For the final structure shown in Fig. 4, the restraint potential of the umbrella sampling simulation was applied at 2.97 nm. As estimated from the umbrella sampling simulations, free-energy change for lipid pulling is 10–15 kcal/mol for these two PIP2 lipids. Therefore, in the absence of any mechanical stress of membrane and/or other stabilizing interactions present in the actual virion environment, it is not possible for the partial removal of lipids to occur spontaneously in unbiased MD simulations. The driving force for such lipid removal in an actual virion environment remains unknown.
Figure 4.
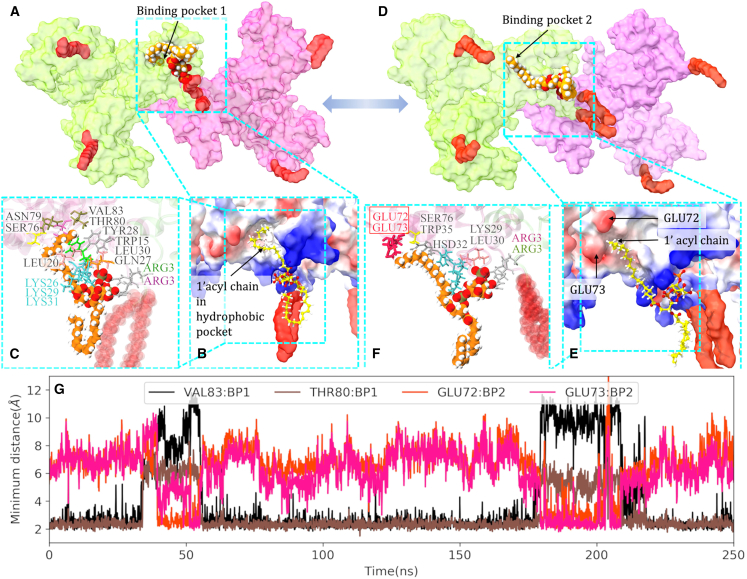
PIP2-binding sites at the mature MA trimer-trimer interface. (A) The density map of PIP2 lipids in the bilayer plane. Stable binding sites at the trimer-trimer interface are denoted as “a,” where two PIP2 lipid molecules bind MA proteins with their inositol headgroup as well as the 2′ acyl chain. (B) A close-up view of the PIP2-binding site in the mature MA complex. PIP2 lipid tails and headgroups are shown in orange and red, respectively. (C) Simulated MA-PIP2 structure is fitted to the cryo-EM density map of the mature MA complex (pink; EMDB-13088). Although the PIP2 lipid, especially its 2′ acyl tail, is close to the cryo-ET observed density, they do not superimpose with each other. (D) The residues at the mature MA-PIP2-binding site, conferring stability and specificity to the complex, are ARG21, LYS25, LYS26 (bind the headgroup of PIP2), GLU72, ARG75, SER76, and ASN79 (bind the 2′ acyl chain). To see this figure in color, go online.
In the extended conformation of MA-bound PIP2, the inositol headgroup of PIP2 interacts with positively charged residues such as LYS25, LYS26, and ARG21, and 2′ acyl chains of PIP2 molecules, being partially removed from the membrane, bind to helix-4 residues such as GLU72, ARG75, SER76, and ASN79 (Fig. 4 D). Note that ARG21 residues at the interface are predominantly occupied by interaction with the PIP2 lipid headgroup; however, PS lipids bind to LYS26, LYS29, and LYS31 of the HBR domain (Fig. 8). None of the PS lipids at the trimeric interface of the mature MA complex sampled such extended lipid conformation. The interactions between MA residues, SER76 and ASN79, interacting with the 2′ acyl tail of PIP2 lipid observed in our simulations agree with NMR predictions (Fig. S5) (32). Nevertheless, in the simulated trajectories, these PIP2 lipids occupy a different position within the MA trimer-trimer interface from that observed by NMR and cryo-ET. Here we report a pair of PIP2 lipids that bind the HBR domain with their multivalent headgroups, which can access the 2′ acyl tail binding pocket in a stable configuration. However, further simulations and higher-resolution cryo-ET data are required to confirm which PIP2 lipid (in terms of head group binding) accesses the 2′ acyl tail binding pocket in the actual virion environment. Similar to the mature MA complex, we also explored the possible extended conformations of the PIP2 lipids close to the trimeric interface in the immature MA complex, which is discussed in the next section.
Figure 8.
Frequency of PIP2 and PS interactions with each residue of the MA monomers (M1–4) at the trimer-trimer interface of the immature (A and C) and the mature (E and G) MA assembly. Snapshots show the MA-bound PIP2 and PS lipids and MA residues interacting with them in the immature (B and D) and mature (F and H) MA assembly. The Cα atoms of those MA residues are highlighted with red spheres. To see this figure in color, go online.
Simulated MA-PIP2 interactions are consistent with previous photocrosslinking data
In a previous set of experiments, a functionalized PI(4,5)P2 derivative (f-PI(4,5)P2) was incorporated in the immature and mature virus particles to study MA-PIP2 interactions (19). The f-PI(4,5)P2 contains a diazirine group covalently linked to its 1′ acyl chain that can be photo-crosslinked to nearby MA protein. Upon photoactivation, f-PI(4,5)P2 was observed to be efficiently crosslinked to MA within immature particles but not in mature particles.
Similar to the mature MA complex, unbiased AAMD simulations (1 μs) performed after SMD and rMD simulations explored extended conformations of PIP2 lipids in the immature MA complex and probable binding pockets of MA (see “materials and methods” section for details). Our findings reveal that, in immature MA assembly, unlike mature MA, the stable extended conformation of PIP2 has its 1′ acyl chain bind to MA while partially removed from the membrane. Simulated trajectories sampled two binding pockets, one of them (binding pocket 2 or BP2 as shown in Fig. 5) contains acidic glutamate residues (GLU72, GLU73), which are able to be crosslinked to f-PI(4,5)P2 upon photoactivation (79). The other binding pocket is enriched with hydrophobic residues, such as VAL83, TYR28, TRP15, LEU30, LEU20, and THR80. For both the binding modes, the inositol headgroup of the PIP2 lipid binds to ARG3 of both the MA monomers at the trimeric interface and LYS29. Fig. 5 G shows the switch of PIP2 lipid between two binding pockets as obtained from an unbiased MD trajectory. The 1′ acyl chain binding to the hydrophobic cleft (VAL83:BP1) is observed to be more probable than binding to pocket 2 (BP2). Further, in the mature MA structure, GLU72 and GLU73 are occluded by the 2′ acyl tail of bound PIP2 (Fig. 4). This provides a possible explanation for the experimental observation that f-PI(4,5)P2 was efficiently crosslinked to MA upon photoactivation within immature particles but not to MA within mature particles.
Figure 5.
Accessible binding pockets for 1′ acyl chain of PIP2 in immature MA complex. (A–C). Binding pocket 1 (BP1) contains hydrophobic residues, VAL83, TYR28, TRP15, LEU30, and LEU20. (D–F) Binding pocket 2 (BP2) contains acidic GLU residues (GLU72, GLU73), shown in red in (F), which preferentially react with alkyl diazirine moiety attached to 1′ acyl chain of f-PI(4,5)P2 (79). (B and E) Electrostatic surface potential map of immature MA protein. Red and blue colors represent negatively and positively charged electrostatic potential, respectively. (G) Minimum distance of 1′ acyl chain of the PIP2 lipid in an extended conformation, from VAL83, THR80 residues in BP1 and GLU72, GLU73 residues in BP2. To see this figure in color, go online.
In summary for this section, our enhanced sampling simulations explored stable binding pockets for PIP2 lipid tails close to the immature and mature MA trimer-trimer interface. In the mature MA assembly, the 2′ acyl tail of PIP2 lipid is observed to sample a stable binding pocket of MA consisting of helix-4 residues such as GLU72, ARG75, SER76, and ASN79. In contrast, in the immature MA assembly, the 1′ acyl tail sampled two binding pockets, whereas the headgroup of the PIP2 lipid binds to an alternate site containing N-terminal residues and HBR residues such as LYS29 and LYS31. These results provide a possible structural explanation of how PIP2 lipids bind immature and mature virions with different binding modes.
Lipid sorting around MA complexes
Next, we investigated the sorting of other membrane lipids around the HIV-1 MA trimer-trimer interface in the immature and mature virus particles. Although HIV-1 immature particles bud from the PM of the infected cell, the lipid composition of the viral particle membrane differs significantly from the host cell PM, as discussed earlier. The host cell PM is asymmetric in lipid distribution and lipid unsaturation. The inner leaflet is enriched in unsaturated phospholipids, such as PE, PS, PC, and PI lipids, and cholesterol, whereas the outer leaflet mainly contains PC, saturated SM lipids, and cholesterol. Experimental studies suggest that, during the HIV-1 assembly process, Gag multimerization occurs in the specialized microdomains of the inner leaflet of PM, enriched in PIP2 and cholesterol (21,22,28,61). SM and cholesterol in the outer bilayer form densely packed liquid-ordered phases, i.e., “lipid rafts,” whereas PIP2 in the inner bilayer prefers the liquid-disordered phase due to its unsaturated tail.
Fig. 6 presents the time-averaged lipid density maps for PS in the inner leaflet, cholesterol in both the leaflets, and SM lipids in the outer leaflet for both the MA complexes. Number density profiles of lipid headgroup atoms are computed with the last 200 ns of the trajectories. As already discussed, MA trimer reorganization is observed to alter specific interaction sites of PIP2 lipids (Figs. 3 and 4). Also, for PS lipids, the computed lipid density profiles show high-density domains at the trimer-trimer interface of the mature MA complex, especially close to the HBR loops. The headgroups of acidic PS lipid molecules bind to the basic lysine residues of the HBR domains, whereas specific binding sites of PIP2 lipid molecules include ARG21 of the two HBR domains (Fig. 8). On the other hand, the trimer-trimer interface of the immature MA complex contains a lower number of high-density domains of PS lipids compared to the mature MA assembly (Fig. 6 A and B). In the computed lipid density profile for the immature MA assembly, PS lipid density is observed near the HBR domain; however, those interactions are not maintained throughout the trajectories as captured by contact frequency calculation (Fig. 8). Also, PS lipid molecules are observed to interact with N-terminal residues in the immature MA assembly. On the other hand, PIP2 lipid molecules bind the HBR domains of trimer 1, trimer 2, and an alternate binding site consisting of N-terminal residues of both the trimers and the HBR domain of trimer 1 (Figs. 3 and 8). We further looked into the distribution of PS lipids around PIP2 lipids in the mature MA assembly and studied the effect of MA-membrane binding on such distribution. The radial distribution functions shown in Fig. S6 suggest an enrichment of PS lipids around PIP2 lipids at the specific binding sites of the MA trimer-trimer interface of mature MA complex, compared to the other PIP2 lipids in the simulated system.
Figure 6.
2D density maps of lipid bilayer species at the trimer-trimer interface of the immature and mature MA assembly. (A and B) Phosphatidylserine (PS) in the inner leaflet, (C–F) cholesterol in the inner and outer leaflets, and (G and H) sphingomyelin (SM) in the outer leaflet. MA complex (as viewed from the virus center) is shown in black with Myr groups (dark red). The density of SM lipids in the outer leaflet, marked with an asterisk in (H), is located near the density of PS and PIP2 lipids in the inner leaflet of the mature MA complex as shown in (B) and Fig. 4A. To see this figure in color, go online.
Cholesterol molecules constitute a major population (up to ∼30%) of the total species in the HIV-1 viral membrane (21,22), which helps counteract unfavorable packing of saturated and unsaturated lipids in membranes in general and maintain structural integrity and membrane fluidity. Time-averaged density maps for raft-forming cholesterol and SM in the outer leaflet, obtained from our simulated AAMD trajectory, exhibit high-density domains of those components near the MA complexes. In the past, multiple studies proposed that outer-leaflet rafts could be trapped by inner-leaflet PIP2/PS-Gag nanodomains through the trans-bilayer coupling (37,80,81,82). Our trajectories show transient interactions between inner-leaflet MA-bound anionic phospholipids and outer-leaflet SM; however, the contribution of inter-leaflet coupling and protein-mediated lipid raft formation in such systems is beyond the scope of the present study and needs to be carefully examined in the future by computational and experimental studies.
Quantitative characterization of MA-lipid interactions and lateral organization of the membrane
We examined the MA-lipid interactions and lipid arrangement in the vicinity of MA complexes, in further detail. In the last section, time-averaged lipid density maps are reported. The high-density domains in those maps may occur either by a greater number of lipid molecules in that particular domain or the dynamic localization of lipids. Here, we have computed the number/fraction of lipid molecules around MA monomers at the trimeric interface (TI) of both immature and mature MA complexes to compare their lipid environment quantitatively, as captured in our simulations. Here, we have used the last 200 ns of the trajectories, similar to the lipid density profile calculation (Fig. 6). For the PIP2 and PS lipid count calculation, we have considered a distance cutoff of 10 Å between the phosphorus (P) atoms of phospholipid headgroups and the Cα atoms of the MA monomers adjacent to the TI. Fig. 7 A and C show the probability of PIP2 and PS lipid count values for immature and mature MA trimer-trimer interfaces, respectively. According to the histogram data, the maximum probability is observed for two or three PIP2 and three PS lipids close to the two MA monomers in the immature MA TI and eight or nine PIP2 and five PS lipid molecules close to the four MA monomers in the mature MA TI. Therefore, the number of MA-PS interactions per MA monomer at TI is not different in these two MA complexes, as sampled in our simulations. However, these data suggest the binding of more PIP2 lipids to the MA monomers at the TI in the mature MA complex. The fluctuation in the lipid count data depends on the measurement criteria and the dynamic MA-lipid interactions. However, at a time instant, the number of PIP2/PS lipids in the vicinity of MA monomers at the TI should follow these statistics. We further investigated the binding frequency at different interaction sites to determine the stability of those interactions, which is discussed later in this section.
Figure 7.
Probability distribution of the number of lipid molecules/fraction of lipid molecules below the trimer-trimer interface of (A and B) the immature MA and (C and D) mature MA complexes. To see this figure in color, go online.
For the calculations of SM and cholesterol lipid count at the outer leaflet of the bilayer, we have considered different parameters and selection criteria. As the number of MA monomers at the TI is different, the surface area under those MA monomers is considerably different. Therefore, instead of lipid count, here we compare the fraction of lipid molecules near the MA monomers with respect to the total number of lipid molecules in the selected domains. Using the same trajectory segments, the number of SM/cholesterol lipids at the outer leaflet is computed using a distance cutoff of (1) 55 Å between the Cα atoms of MA monomers and the P atoms of SM lipids, and (2) 50 Å between the Cα atoms of MA monomers and oxygen atoms of the hydroxyl group of outer-leaflet cholesterol molecules. Figs. 7B and 7ure D present the probability distribution of the fraction of SM/cholesterol lipid near the MA monomers at the TI of immature and mature MA complexes, respectively. Although the fluctuation of the fraction of SM lipids is more in the immature MA complex, the probability distribution profiles of SM lipids in the immature and mature MA complexes overlap with each other. However, we observe significantly different distribution profiles for the fraction of outer-leaflet cholesterol. This result suggests an enrichment of cholesterol lipids in the outer leaflet near the MA monomers at the TI of mature MA complex, compared to that of immature MA complex. It should be noted here that a single MA monomer or MA trimer also contributes to the lipid enrichment; however, both of these contributions are present in immature and mature MA complexes and by comparing these two systems one can determine the effect of TTIs on the MA-membrane interactions for the MA monomers at the TI. The trend of lipid binding/sorting data presented in Fig. 7 has been reproduced using multiple simulation trajectories and different time domains of a particular trajectory. In our membrane model, long-chain C24:0 sphingolipids in the outer leaflet can interact with inner-leaflet PS/PIP2 lipids. Stable MA-PIP2/PS interactions can cause localization of such lipids compared to other PS/PIP2 lipids in the system with higher lateral mobility. This can affect the dynamic organization of the outer-leaflet SM lipids and create a high-density domain of SM lipids beneath the mature MA trimer-trimer interface as observed in Fig. 6 H. However, quantification of such interdigitation is beyond the scope of this current study.
Next, we examined the strength of PIP2/PS binding at different sites of MA monomers at the TI of immature and mature MA complexes by computing binding frequency. For this purpose, we have used a distance cutoff of 2.5 Å for the minimum distance between MA residues and PIP2/PS lipids. The data presented in Fig. 8 A and E suggest that N-terminal basic residues (ARG3), HBR residues (ARG19, ARG21, LYS25, LYS26, LYS29, LYS31), and helix-2 residues (ARG38, ARG42) exhibit stable interactions with PIP2 in both systems. Most of these residues show a contact frequency of >100% for PIP2 binding, which signifies more than one PIP2 lipid binds/remains in contact (within the cutoff distance) with a single amino acid residue. Unlike this, MA-PS binding frequencies suggest stable binding of only one PS with basic residues of MA. As we discussed earlier, our simulations revealed an alternative binding site for PIP2 at the TI of immature MA complex, which contains HBR (LYS29, LYS31) residues and ARG3 of monomer 1 (M1) and ARG3 of monomer 2 (M2) (as shown in Fig. 3). The contact frequency data verify the stability of such binding (Fig. 8 A). In the immature MA complex, most of the PS lipids are observed to interact transiently with MA residues (Fig. 8 C) and the contact frequency data for two MA monomers change at different segments of our trajectories. On the other hand, our simulations sampled stable binding between PS lipids and HBR residues in the mature MA complex (Fig. 8 G). Fig. 8 B, D, F, and H present snapshots of the MA-bound PIP2/PS lipids and the corresponding Cα atoms of MA protein involved in those protein-lipid interactions. Please note that our simulations did not attain symmetric MA-lipid interactions for all the MA monomers at the trimer-trimer interface in both systems. For such a PMP complex, the attainment of perfect lipid-sorting behavior by MA protein complexes demands a much longer simulation timescale. However, the computation of binding frequency allows us to distinguish between stable and transient interactions, explored in our simulations.
Altogether, the long-timescale MD simulations distinguished the lipid sorting around the MA trimer-trimer interface in the immature and the mature MA complexes. As captured in the time-averaged lipid density map, the mature MA trimer-trimer interface is observed to be enriched with anionic phospholipids such as PIP2, compared to the immature MA trimer-trimer interface. The enrichment has been quantified by computing the probability of the number of bound-PIP2/PS lipids at the TI. The contact frequency data analyzed the stable and transient MA-PIP2 and MA-PS interactions. The stability of MA-PIP2 binding at the alternative binding site is verified using the contact frequency data (Figs. 3 and 8 A). Further, the lipid fraction data revealed enrichment of cholesterol at the outer leaflet below the MA monomers at the TI of the mature MA complex compared to the immature MA complex.
Discussion
In this work, we have presented a detailed structural analysis of the membrane-bound HIV-1 MA protein complexes as found in the immature and the mature virions. The role of PM PIP2 lipids in the HIV-1 assembly process is well recognized (22,40,76); however, its function in the maturation process is still unknown. Using atomistic MD simulations here we interrogated altered MA-lipid-specific interactions and lateral organization of lipids in the vicinity of MA complexes. Different modes of direct binding of PIP2 lipids with HIV-1 MA have been reported by several experimental techniques, including NMR (32,41) and mass spectrometric protein footprinting analysis (39). In this study, we have identified the favorable MA-PIP2-binding modes and specific PIP2-binding sites in the immature and mature MA complexes. The binding affinity and arrangement of PIP2, PS, and raft-forming SM, cholesterol at the two different trimer-trimer interfaces were quantified using different computational analyses.
The cryo-ET density map captured different organizations of MA trimers in the immature and mature MA lattice structures (19). The electrostatic surface potential of MA proteins facing the central hole of hexamer of trimers in the lattice was observed to be altered from positive (in immature MA lattice) to neutral or negative in the mature MA assembly. In addition, the hole becomes smaller in the mature MA lattice. Previously, these holes have been hypothesized to mediate interactions of MA with the C-terminal tail of the HIV-1 Env protein and promote Env incorporation into the virions (30,83); however, using high Env VLPs (hVLPs) with truncated gp41 C-terminal domain, a recent cryo-ET experiment suggested that the trimeric Env C-terminal domain does not lie in the central hole formed by the immature Gag lattice but instead binds to its hexameric rim, at the MA trimer-trimer interface (20). On the other hand, in the mature virions, the MA layer was observed to be fragmented and Env trimers were observed to gain significant mobility after structural rearrangement.
In the present study, MD simulations with the MA assembly structures derived from cryo-ET density maps explored molecular-level MA-MA interactions at the TI in immature and mature virions. The characterization of protein-PPI remains a challenge for computational studies (84,85,86). Here, we start our simulations from cryo-ET structures of MA multimers and by performing strategic simulations we obtain stable membrane-bound dimer-of-trimer structures of mature and immature MA complexes. We have studied the stability of MA-MA interactions at the TIs predicted by cryo-ET structures and explored other possible interactions that might play a role in rendering stability to the protein complexes. As cryo-ET structures reported (19), in the immature MA complex, TTIs are mainly governed by the N-terminal residues, helix-1, and 310 helix residues. On the other hand, in the mature MA complex, MA TTIs are mediated by the basic residues of HBR domains and the acidic residues of 310 and helix 4. All the crucial MA TTIs reported by cryo-ET data remained stable in our long-timescale simulations. Further, our simulated data revealed that Myr groups at the trimer-trimer interface of immature MA complex, being inserted into the membrane, remain in contact with each other and the hydrophobic interactions between them render stability to the TI (Fig. 3). Apart from the hydrophobic interactions mediated by N-terminal residues and H1 residues, our simulated data suggest electrostatic interactions between ARG90 of helix-4 and GLU51 of 310 helix stabilize MA TTIs in the immature MA complex (Fig. 1 and S2). A comparison of the structures of MA monomers in our simulated trajectories with the cryo-ET-derived atomic structure shows deviations, especially for the membrane-bound helices of MA monomers at the TI. Interestingly, the structural deviation for the membrane-bound helices in the MA monomer at the TI of the immature MA complex is lower when a PIP2 lipid headgroup binds to the alternate site shown in Fig. 3. This indirectly verifies this mode of MA-PIP2 interaction in the immature virion. In the mature MA complex, our simulations explored stable electrostatic interactions between a pair of residues, ARG19:GLU51. Apart from that, cleaved end residues and more flexible H5 helix residues (LYS97 and LYS113) are observed to participate in the TTIs in our simulated MD trajectories of mature MA complexes. In summary, the MA trimer-trimer interface changes its nature significantly, as it involves more charged residues upon maturation. This inevitably modifies lipid binding sites across the MA TI upon MA maturation.
Our long-timescale AAMD simulations characterized the lipid binding to the MA monomers at the trimer-trimer interface of both the MA complexes. Due to a large free-energy barrier (as estimated by umbrella sampling simulations), in the absence of any mechanical stress of membrane and/or other stabilizing interactions present in the actual virion environment, the partial removal of lipids does not occur spontaneously in unbiased MD simulations. Biased MD simulations have explored the conformational space of MA-bound anionic phospholipids and obtained stable extended conformations for PIP2 lipid in the mature MA assembly, in which the 2′ acyl chain of the PIP2 molecules is outside the hydrophobic membrane core and is bound to the protein. Although some of the MA residues (SER76, ASN79) interacting with the 2′ acyl tail of PIP2 lipid in the extended conformation in our simulations match NMR data (Figs. 4 and S5), the overall position of the PIP2 lipid is distinct from the positions as previously suggested by NMR or electron microscopy experiments. However, the binding sites of two PIP2 lipids in the extended conformations, as obtained in our simulations, are also at the MA trimer-trimer interface close to the position of the cryo-ET density as well as the NMR structure. Thus, our simulations suggest that the MA trimer-trimer interface, as in the mature virion, can stabilize extended conformations of PIP2 lipid where the 2′ acyl chain, being partially removed from the bilayer, can interact with the MA protein. Cryo-ET density map of mature MA lattice predicted the partial removal of up to ∼2500 PIP2 lipids in the mature virion (19). However, the driving force for such PIP2 removal could not be recapitulated in our simulations and its impact on the membrane structural and mechanical properties in the mature virion is still an open question. Further, the difference between the positions of PIP2 lipid as obtained in our simulations and cryo-ET data suggests that there may be other stabilizing interactions that are not well represented in the simulations.
Interestingly, this mechanism of MA-PIP2 binding is not operational for the immature MA assembly, but simulations revealed two specific binding pockets of MA for the 1′-acyl chain of PIP2 in the immature MA trimer-trimer interface. One of them is the hydrophobic cleft containing VAL83 and THR80, and another contains the acidic residues GLU72 and GLU73 (Fig. 5). These results provide a possible explanation for the experimental observations using the photocrosslinking approach (19). A functionalized PIP2 derivative (f-PI(4,5)P2) with a diazirine group covalently linked to the 1′ acyl chain was crosslinked to the immature MA assembly but not to the mature MA lattice. A recent study has shown the reactivity of such a diazirine moiety with acidic amino acids such as GLU (79), which are present in one of the binding pockets of MA sampled by 1′ acyl chain of PIP2 in the simulated trajectories of immature MA assembly. On the other hand, these GLU72 and GLU73 residues are preoccupied with the 2′ acyl chain of PIP2 in the mature MA assembly (Fig. 4). Thus, they cannot bind the 1′ acyl tail of f-PI(4,5)P2 in the photocrosslinking experiment. Therefore, our AAMD simulations add to the observations of photocrosslinking experiments and suggest an altered binding mode of PIP2 in the mature MA complex.
Further, we quantified the binding affinity of MA-PIP2/PS interactions and verified the stable binding sites for the PIP2 and PS lipids including the alternative binding site of PIP2 at the TI of immature MA complex (Figs. 3 and 8 A) revealed by our simulations. Snapshots of all the MA residues at the trimer-trimer interfaces interacting with PIP2 and PS lipids are shown in Fig. 8 B, D, F and H. Figs. 3 C and 4 D present a close-up view of important MA-PIP2 interactions in the immature and mature MA complexes, respectively. Additional MA-PIP2 binding, as explored by our simulations, are shown in Fig. S8. For most of our simulations, we observe three PIP2 lipids binding to the central ARG ring of the MA trimer, which consists of ARG38, and ARG42 of all three MA monomers (shown in Fig. S8 C). There are other stable MA-binding sites for multivalent PIP2 binding, consisting of 1) LYS29, LYS31, ARG2, and ARG3, and 2) LYS31, TRP35, ARG38, ARG42 (shown in Fig. S8 A and B). Although our simulations identified MA residues important for PIP2 binding, such as ARG19, ARG21, LYS25, LYS26, LYS29, LYS31, ARG38, ARG42, and ASN79, whether these MA-lipid binding sites play roles in the maturation and subsequent viral processes remains to be elucidated. For example, mutagenesis studies have shown that substitution of ARG19, ARG42, and ASN79 does not affect viral infectivity (73). This result may imply that other residues rescue the PIP2 binding in these mutants.
Our lipid count/fraction data suggest that the simulated system of mature MA trimer-trimer interface is enriched in the anionic phospholipids, PIP2 on the inner leaflet, and cholesterol on the outer leaflet of the bilayer, compared to the MA TI of immature MA complex (Fig. 7). We have also shown that, in the mature MA complex, the density of PS lipids around PIP2 lipids at the specific binding sites of the MA trimer-trimer interface is enhanced compared to the other PIP2 lipids in the simulated system (Fig. S7). This enrichment of inner-leaflet anionic phospholipids in the mature MA complex increases the negative surface charge density of the membrane near the MA trimer-trimer interface. The negative surface charge density of the membrane generates a negative zeta potential, which may facilitate the accumulation of positive ionic species in the mature HIV-1 particles on the anionic membrane surface. Furthermore, an increase in the outer-leaflet cholesterol concentration, as observed for the mature MA simulations, may affect the mechanical properties of the membrane, such as stiffness. The stiffening effect of cholesterol on different membrane compositions continues to be a matter of extensive debate (87,88,89,90,91). Although the bending rigidity of bilayers with partially or fully saturated lipids (such as POPC) increases with the enrichment of cholesterol, little or no effect was observed with (poly)unsaturated lipids such as DOPC (92,93). Further investigation is required to elucidate the impact of cholesterol on the mechanical properties of asymmetric viral membranes carrying a mixture of saturated and unsaturated lipids. Experimental studies reported the modulation of mechanical properties of the HIV-1 membrane, such as stiffening during viral budding and softening during entry. Other factors in mature HIV-1 virions have been suggested to regulate the rigidity of the membrane, such as the Env cytoplasmic tails (94). In our simulations, we have not included the Env tail, therefore the role of the Env tail in modulating MA-membrane interactions and lipid-sorting behavior around MA protein remains unknown from our work.
Conclusion
In summary, we have performed long-time and large-scale AAMD simulations of HIV-1 Myr-MA dimer of trimers bound to the inner leaflet of an asymmetric membrane model. We have considered two different Myr-MA structures as reported for immature and mature virions by cryo-ET experiment. A series of carefully performed simulations explored the MA-MA and specific MA-lipid interactions at the interface of MA trimers when Myr groups of all MA proteins remain inserted into the membrane. Our simulations explored electrostatic interactions between ARG90 of helix-4 and GLU51 of 310 helix and hydrophobic interactions between Myr groups providing additional stabilization to the immature MA TI, besides the hydrophobic interactions mediated by N-terminal residues. Here we report an alternative PIP2-binding site in the immature MA complex where the headgroup of multivalent PIP2 lipid binds to ARG3 residues of both MA monomers at the trimer-trimer interface and the LYS29 and LYS31 residues of the HBR domain of one of the monomers. We further quantified the stability of such interactions and other MA-PIP2/PS interactions by contact frequency data. Our time-averaged lipid density maps and quantitative calculations of the lipid environment below the MA monomers at the TIs of both the complexes suggest an enrichment of PIP2 at the inner leaflet and cholesterol at the outer leaflet of the membrane in the mature MA system, compared to the immature MA system. We explored PIP2 conformational change at the specific binding sites of MA in immature and mature MA complexes using biased MD simulations. Simulated trajectories explored stable binding pockets for the 2′ acyl chain of PIP2 at the trimer-trimer interface of mature MA complex, unlike immature MA complex. In the immature MA complex, our enhanced sampling simulations sampled two binding pockets for 1′ acyl chain; one of them contains hydrophobic residues VAL83 and THR80, and another binding pocket contains the acidic residues GLU72 and GLU73. This finding provides a possible structural explanation for the observation of the photocrosslinking experiment. Altogether, the present simulation results, combined with prior experimental observations, provide molecular-level insights into the altered binding site and binding mode for PIP2 lipids upon MA maturation. Therefore, specific MA-PIP2 interactions are not merely important for the stable MA-membrane binding during the viral assembly process; they show interesting characteristics in the virions before and after the maturation process. However, whether the maturation process of MA protein is aided by such PIP2-specific interactions is beyond the scope of our current study. As such, this study reports important insights into altered MA-MA and MA-lipid interactions and fills a significant gap in our understanding of the HIV-1 matrix and membrane maturation.
Data and code availability
The analysis code, plotting script, data files, and coordinate files of membrane-bound immature and mature MA systems are available in the GitHub repository (https://github.com/pujabanerjee91/HIV1_MA_Maturation). Additional details are provided within the article or supporting material.
Author contributions
P.B., J.A.G.B., and G.A.V. designed the research. P.B. prepared simulation models, designed and performed simulations, analyzed the data, prepared the figures, and wrote the manuscript. K.Q. contributed to the discussions and manuscript preparation. J.A.G.B. and G.A.V. supervised the research and the writing process and contributed to the discussions to finalize the manuscript.
Acknowledgments
This research was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (NIH grant R01AI178550) to G.A.V. and J.A.G.B. and the Max Planck Society to J.A.G.B. The authors acknowledge computational resources provided by the University of Chicago Research Computing Center, the Frontera supercomputer at the Texas Advanced Computer Center funded by the National Science Foundation (OAC-1818253), the Stampede2 supercomputer at the Texas Advanced Computing Center, the Bridges2 supercomputer at the Pittsburgh Supercomputing Center through allocation MCA94P017 with resources provided by the Extreme Science and Engineering Discovery Environment (XSEDE) supported by NSF grant ACI-1548562, and the NIH-funded Beagle-3 computer (NIH award 1S10OD028655-01).
Declaration of interests
The authors declare no competing interests.
Editor: Chris Neale.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2024.01.006.
Supporting material
References
- 1.Sundquist W.I., Kräusslich H.G. HIV-1 assembly, budding, and maturation. Cold Spring Harb. Perspect. Med. 2012;2:a006924. doi: 10.1101/cshperspect.a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pettit S.C., Lindquist J.N., et al. Swanstrom R. Processing sites in the human immunodeficiency virus type 1 (HIV-1) Gag-Pro-Pol precursor are cleaved by the viral protease at different rates. Retrovirology. 2005;2:1–6. doi: 10.1186/1742-4690-2-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Könnyű B., Sadiq S.K., et al. Müller V. Gag-Pol processing during HIV-1 virion maturation: a systems biology approach. PLoS Comput. Biol. 2013;9 doi: 10.1371/journal.pcbi.1003103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chukkapalli V., Ono A. Molecular determinants that regulate plasma membrane association of HIV-1 Gag. J. Mol. Biol. 2011;410:512–524. doi: 10.1016/j.jmb.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lalonde M.S., Sundquist W.I. How HIV finds the door. Proc. Natl. Acad. Sci. USA. 2012;109:18631–18632. doi: 10.1073/pnas.1215940109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hill C.P., Worthylake D., et al. Sundquist W.I. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc. Natl. Acad. Sci. USA. 1996;93:3099–3104. doi: 10.1073/pnas.93.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang C., Ndassa Y., Summers M.F. Structure of the N-terminal 283-residue fragment of the immature HIV-1 Gag polyprotein. Nat. Struct. Biol. 2002;9:537–543. doi: 10.1038/nsb806. [DOI] [PubMed] [Google Scholar]
- 8.Dick R.A., Vogt V.M. Membrane interaction of retroviral Gag proteins. Front. Microbiol. 2014;5:187. doi: 10.3389/fmicb.2014.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang C., Loeliger E., et al. Summers M.F. Entropic switch regulates myristate exposure in the HIV-1 matrix protein. Proc. Natl. Acad. Sci. USA. 2004;101:517–522. doi: 10.1073/pnas.0305665101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan A., Pak A.J., et al. Briggs J.A.G. Immature HIV-1 assembles from Gag dimers leaving partial hexamers at lattice edges as potential substrates for proteolytic maturation. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2020054118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schur F.K.M., Obr M., et al. Briggs J.A.G. An atomic model of HIV-1 capsid-SP1 reveals structures regulating assembly and maturation. Science. 2016;353:506–508. doi: 10.1126/science.aaf9620. [DOI] [PubMed] [Google Scholar]
- 12.Chen J., Rahman S.A., et al. Hu W.-S. HIV-1 RNA genome dimerizes on the plasma membrane in the presence of Gag protein. Proc. Natl. Acad. Sci. USA. 2016;113:E201–E208. doi: 10.1073/pnas.1518572113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carlson L.-A., Bai Y., et al. Hurley J.H. Reconstitution of selective HIV-1 RNA packaging in vitro by membrane-bound Gag assemblies. Elife. 2016;5 doi: 10.7554/eLife.14663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pak A.J., Grime J.M.A., et al. Voth G.A. Immature HIV-1 lattice assembly dynamics are regulated by scaffolding from nucleic acid and the plasma membrane. Proc. Natl. Acad. Sci. USA. 2017;114:E10056–E10065. doi: 10.1073/pnas.1706600114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mattei S., Tan A., et al. Briggs J.A.G. High-resolution structures of HIV-1 Gag cleavage mutants determine structural switch for virus maturation. Proc. Natl. Acad. Sci. USA. 2018;115:E9401–E9410. doi: 10.1073/pnas.1811237115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Konvalinka J., Kräusslich H.G., Müller B. Retroviral proteases and their roles in virion maturation. Virology. 2015;479–480:403–417. doi: 10.1016/j.virol.2015.03.021. [DOI] [PubMed] [Google Scholar]
- 17.Deshmukh L., Tugarinov V., et al. Clore G.M. Binding kinetics and substrate selectivity in HIV-1 protease− Gag interactions probed at atomic resolution by chemical exchange NMR. Proc. Natl. Acad. Sci. USA. 2017;114:E9855–E9862. doi: 10.1073/pnas.1716098114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mattei S., Schur F.K., Briggs J.A. Retrovirus maturation—an extraordinary structural transformation. Curr. Opin. Virol. 2016;18:27–35. doi: 10.1016/j.coviro.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 19.Qu K., Ke Z., et al. Briggs J.A.G. Maturation of the matrix and viral membrane of HIV-1. Science. 2021;373:700–704. doi: 10.1126/science.abe6821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prasad V.M., Leaman D.P., et al. Lee K.K. Cryo-ET of Env on intact HIV virions reveals structural variation and positioning on the Gag lattice. Cell. 2022;185:641–653.e617. doi: 10.1016/j.cell.2022.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan R., Uchil P.D., et al. Wenk M.R. Retroviruses human immunodeficiency virus and murine leukemia virus are enriched in phosphoinositides. J. Virol. 2008;82:11228–11238. doi: 10.1128/JVI.00981-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mücksch F., Citir M., Kräusslich H.G., et al. Quantification of phosphoinositides reveals strong enrichment of PIP2 in HIV-1 compared to producer cell membranes. Sci. Rep. 2019;9:17661–17673. doi: 10.1038/s41598-019-53939-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Callahan M.K., Popernack P.M., et al. Henderson A.J. Phosphatidylserine on HIV envelope is a cofactor for infection of monocytic cells. J. Immunol. 2003;170:4840–4845. doi: 10.4049/jimmunol.170.9.4840. [DOI] [PubMed] [Google Scholar]
- 24.Chua B.A., Ngo J.A., et al. Morizono K. Roles of phosphatidylserine exposed on the viral envelope and cell membrane in HIV-1 replication. Cell Commun. Signal. 2019;17:132–139. doi: 10.1186/s12964-019-0452-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huarte N., Carravilla P., et al. Nieva J.L. Functional organization of the HIV lipid envelope. Sci. Rep. 2016;6 doi: 10.1038/srep34190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Waheed A.A., Freed E.O. The role of lipids in retrovirus replication. Viruses. 2010;2:1146–1180. doi: 10.3390/v2051146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ono A., Ablan S.D., et al. Freed E.O. Phosphatidylinositol (4, 5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc. Natl. Acad. Sci. USA. 2004;101:14889–14894. doi: 10.1073/pnas.0405596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brügger B., Glass B., et al. Kräusslich H.G. The HIV lipidome: a raft with an unusual composition. Proc. Natl. Acad. Sci. USA. 2006;103:2641–2646. doi: 10.1073/pnas.0511136103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mercredi P.Y., Bucca N., et al. Summers M.F. Structural and molecular determinants of membrane binding by the HIV-1 matrix protein. J. Mol. Biol. 2016;428:1637–1655. doi: 10.1016/j.jmb.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tedbury P.R., Novikova M., et al. Freed E.O. Biochemical evidence of a role for matrix trimerization in HIV-1 envelope glycoprotein incorporation. Proc. Natl. Acad. Sci. USA. 2016;113:E182–E190. doi: 10.1073/pnas.1516618113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alfadhli A., Barklis R.L., Barklis E. HIV-1 matrix organizes as a hexamer of trimers on membranes containing phosphatidylinositol-(4, 5)-bisphosphate. Virology. 2009;387:466–472. doi: 10.1016/j.virol.2009.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saad J.S., Miller J., et al. Summers M.F. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc. Natl. Acad. Sci. USA. 2006;103:11364–11369. doi: 10.1073/pnas.0602818103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yandrapalli N., Muriaux D., Favard C. Lipid domains in HIV-1 assembly. Front. Microbiol. 2014;5:220. doi: 10.3389/fmicb.2014.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saad J.S., Ablan S.D., et al. Summers M.F. Structure of the myristylated human immunodeficiency virus type 2 matrix protein and the role of phosphatidylinositol-(4, 5)-bisphosphate in membrane targeting. J. Mol. Biol. 2008;382:434–447. doi: 10.1016/j.jmb.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Monje-Galvan V., Voth G.A. Binding mechanism of the matrix domain of HIV-1 gag on lipid membranes. Elife. 2020;9 doi: 10.7554/eLife.58621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ono A., Freed E.O. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc. Natl. Acad. Sci. USA. 2001;98:13925–13930. doi: 10.1073/pnas.241320298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sengupta P., Lippincott-Schwartz J. Revisiting membrane microdomains and phase separation: a viral perspective. Viruses. 2020;12:745. doi: 10.3390/v12070745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sengupta P., Seo A.Y., et al. Lippincott-Schwartz J. A lipid-based partitioning mechanism for selective incorporation of proteins into membranes of HIV particles. Nat. Cell Biol. 2019;21:452–461. doi: 10.1038/s41556-019-0300-y. [DOI] [PubMed] [Google Scholar]
- 39.Shkriabai N., Datta S.A.K., et al. Kvaratskhelia M. Interactions of HIV-1 Gag with assembly cofactors. Biochemistry. 2006;45:4077–4083. doi: 10.1021/bi052308e. [DOI] [PubMed] [Google Scholar]
- 40.Chukkapalli V., Hogue I.B., et al. Ono A. Interaction between the human immunodeficiency virus type 1 Gag matrix domain and phosphatidylinositol-(4, 5)-bisphosphate is essential for efficient gag membrane binding. J. Virol. 2008;82:2405–2417. doi: 10.1128/JVI.01614-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Samal A.B., Green T.J., Saad J.S. Atomic view of the HIV-1 matrix lattice; implications on virus assembly and envelope incorporation. Proc. Natl. Acad. Sci. USA. 2022;119 doi: 10.1073/pnas.2200794119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lai C.-L., Landgraf K.E., et al. Falke J.J. Membrane docking geometry and target lipid stoichiometry of membrane-bound PKCα C2 domain: a combined molecular dynamics and experimental study. J. Mol. Biol. 2010;402:301–310. doi: 10.1016/j.jmb.2010.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lai C.-L., Srivastava A., et al. Voth G.A. Molecular mechanism of membrane binding of the GRP1 PH domain. J. Mol. Biol. 2013;425:3073–3090. doi: 10.1016/j.jmb.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stahelin R.V. Methods in Cell Biology. Elsevier; 2013. Monitoring peripheral protein oligomerization on biological membranes; pp. 359–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Larsen A.H., John L.H., et al. Corey R.A. Specific interactions of peripheral membrane proteins with lipids: what can molecular simulations show us? Biosci. Rep. 2022;42 doi: 10.1042/BSR20211406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Monje-Galvan V., Klauda J.B. Peripheral membrane proteins: Tying the knot between experiment and computation. Biochim. Biophys. Acta. 2016;1858:1584–1593. doi: 10.1016/j.bbamem.2016.02.018. [DOI] [PubMed] [Google Scholar]
- 47.Pu M., Orr A., et al. Roberts M.F. Defining specific lipid binding sites for a peripheral membrane protein in situ using subtesla field-cycling NMR. J. Biol. Chem. 2010;285:26916–26922. doi: 10.1074/jbc.M110.123083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eells R., Barros M., et al. Lösche M. Structural characterization of membrane-bound human immunodeficiency virus-1 Gag matrix with neutron reflectometry. Biointerphases. 2017;12 doi: 10.1116/1.4983155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Banerjee P., Voth G.A. Conformational transitions of the HIV-1 Gag polyprotein upon multimerization and gRNA binding. Biophys. J. 2023;122:1–15. doi: 10.1016/j.bpj.2023.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rogaski B., Klauda J.B. Membrane-binding mechanism of a peripheral membrane protein through microsecond molecular dynamics simulations. J. Mol. Biol. 2012;423:847–861. doi: 10.1016/j.jmb.2012.08.015. [DOI] [PubMed] [Google Scholar]
- 51.Wang Q., Pechersky Y., et al. Shaw D.E. Structural mechanism for Bruton’s tyrosine kinase activation at the cell membrane. Proc. Natl. Acad. Sci. USA. 2019;116:9390–9399. doi: 10.1073/pnas.1819301116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ryckbosch S.M., Wender P.A., Pande V.S. Molecular dynamics simulations reveal ligand-controlled positioning of a peripheral protein complex in membranes. Nat. Commun. 2017;8:6–10. doi: 10.1038/s41467-016-0015-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barros M., Heinrich F., et al. Lösche M. Membrane binding of HIV-1 matrix protein: dependence on bilayer composition and protein lipidation. J. Virol. 2016;90:4544–4555. doi: 10.1128/JVI.02820-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Charlier L., Louet M., et al. Floquet N. Coarse-grained simulations of the HIV-1 matrix protein anchoring: revisiting its assembly on membrane domains. Biophys. J. 2014;106:577–585. doi: 10.1016/j.bpj.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee J., Cheng X., et al. Im W. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theor. Comput. 2016;12:405–413. doi: 10.1021/acs.jctc.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jo S., Kim T., Im W. Automated builder and database of protein/membrane complexes for molecular dynamics simulations. PLoS One. 2007;2 doi: 10.1371/journal.pone.0000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jo S., Kim T., et al. Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 58.Brooks B.R., Brooks C.L., III, et al. Karplus M. CHARMM: the biomolecular simulation program. J. Comput. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu E.L., Cheng X., et al. Im W. 2014. CHARMM-GUI Membrane Builder toward Realistic Biological Membrane Simulations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 61.Lorizate M., Sachsenheimer T., et al. Brügger B. Comparative lipidomics analysis of HIV-1 particles and their producer cell membrane in different cell lines. Cell Microbiol. 2013;15:292–304. doi: 10.1111/cmi.12101. [DOI] [PubMed] [Google Scholar]
- 62.Huang J., Rauscher S., et al. MacKerell A.D. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods. 2017;14:71–73. doi: 10.1038/nmeth.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jorgensen W.L., Chandrasekhar J., et al. Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 64.MacKerell A.D., Jr., Bashford D., et al. Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 65.Abraham M.J., Murtola T., et al. Lindahl E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. [Google Scholar]
- 66.Hoover W.G. Canonical dynamics: Equilibrium phase-space distributions. Phys. Rev. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 67.Nosé S. A molecular dynamics method for simulations in the canonical ensemble. Mol. Phys. 1984;52:255–268. [Google Scholar]
- 68.Nosé S., Klein M. Constant pressure molecular dynamics for molecular systems. Mol. Phys. 1983;50:1055–1076. [Google Scholar]
- 69.Parrinello M., Rahman A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981;52:7182–7190. [Google Scholar]
- 70.Darden T., York D., Pedersen L. Particle mesh Ewald: An N· log (N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 71.Hess B., Bekker H., et al. Fraaije J.G.E.M. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- 72.Hunter J.D. Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 2007;9:90–95. [Google Scholar]
- 73.Freed E.O., Orenstein J.M., et al. Martin M.A. Single amino acid changes in the human immunodeficiency virus type 1 matrix protein block virus particle production. J. Virol. 1994;68:5311–5320. doi: 10.1128/jvi.68.8.5311-5320.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Freed E.O., Martin M.A. Virion incorporation of envelope glycoproteins with long but not short cytoplasmic tails is blocked by specific, single amino acid substitutions in the human immunodeficiency virus type 1 matrix. J. Virol. 1995;69:1984–1989. doi: 10.1128/jvi.69.3.1984-1989.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mücksch F., Laketa V., et al. Kräusslich H.G. Synchronized HIV assembly by tunable PIP2 changes reveals PIP2 requirement for stable Gag anchoring. Elife. 2017;6 doi: 10.7554/eLife.25287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Favard C., Chojnacki J., et al. Muriaux D. HIV-1 Gag specifically restricts PI (4, 5) P2 and cholesterol mobility in living cells creating a nanodomain platform for virus assembly. Sci. Adv. 2019;5 doi: 10.1126/sciadv.aaw8651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wen Y., Feigenson G.W., et al. Dick R.A. Mechanisms of PI (4, 5) P2 enrichment in HIV-1 viral membranes. J. Mol. Biol. 2020;432:5343–5364. doi: 10.1016/j.jmb.2020.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bigay J., Antonny B. Curvature, lipid packing, and electrostatics of membrane organelles: defining cellular territories in determining specificity. Dev. Cell. 2012;23:886–895. doi: 10.1016/j.devcel.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 79.West A.V., Muncipinto G., et al. Woo C.M. Labeling preferences of diazirines with protein biomolecules. J. Am. Chem. Soc. 2021;143:6691–6700. doi: 10.1021/jacs.1c02509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kerviel A., Thomas A., et al. Muriaux D. Virus assembly and plasma membrane domains: which came first? Virus Res. 2013;171:332–340. doi: 10.1016/j.virusres.2012.08.014. [DOI] [PubMed] [Google Scholar]
- 81.Blosser M.C., Honerkamp-Smith A.R., et al. Keller S.L. Transbilayer colocalization of lipid domains explained via measurement of strong coupling parameters. Biophys. J. 2015;109:2317–2327. doi: 10.1016/j.bpj.2015.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Raghupathy R., Anilkumar A.A., et al. Mayor S. Transbilayer lipid interactions mediate nanoclustering of lipid-anchored proteins. Cell. 2015;161:581–594. doi: 10.1016/j.cell.2015.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tedbury P.R., Freed E.O. The role of matrix in HIV-1 envelope glycoprotein incorporation. Trends Microbiol. 2014;22:372–378. doi: 10.1016/j.tim.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Banerjee P., Mondal S., Bagchi B. Insulin dimer dissociation in aqueous solution: A computational study of free energy landscape and evolving microscopic structure along the reaction pathway. J. Chem. Phys. 2018;149:114902. doi: 10.1063/1.5042290. [DOI] [PubMed] [Google Scholar]
- 85.Banerjee P., Mondal S., Bagchi B. Effect of ethanol on insulin dimer dissociation. J. Chem. Phys. 2019;150 doi: 10.1063/1.5079501. [DOI] [PubMed] [Google Scholar]
- 86.Banerjee P., Bagchi B. Dynamical control by water at a molecular level in protein dimer association and dissociation. Proc. Natl. Acad. Sci. USA. 2020;117:2302–2308. doi: 10.1073/pnas.1908379117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Henriksen J., Rowat A.C., et al. Ipsen J.H. Universal behavior of membranes with sterols. Biophys. J. 2006;90:1639–1649. doi: 10.1529/biophysj.105.067652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pan J., Mills T.T., et al. Nagle J.F. Cholesterol perturbs lipid bilayers nonuniversally. Phys. Rev. Lett. 2008;100 doi: 10.1103/PhysRevLett.100.198103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chakraborty S., Doktorova M., et al. Ashkar R. How cholesterol stiffens unsaturated lipid membranes. Proc. Natl. Acad. Sci. USA. 2020;117:21896–21905. doi: 10.1073/pnas.2004807117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nagle J.F., Evans E.A., et al. Dimova R. A needless but interesting controversy. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2025011118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fiorin G., Forrest L.R., Faraldo-Gómez J.D. Membrane free-energy landscapes derived from atomistic dynamics explain nonuniversal cholesterol-induced stiffening. bioRxiv. 2023 doi: 10.1101/2023.02.02.525347. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pan J., Tristram-Nagle S., Nagle J.F. Effect of cholesterol on structural and mechanical properties of membranes depends on lipid chain saturation. Phys. Rev. 2009;80 doi: 10.1103/PhysRevE.80.021931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gracià R.S., Bezlyepkina N., et al. Dimova R. Effect of cholesterol on the rigidity of saturated and unsaturated membranes: fluctuation and electrodeformation analysis of giant vesicles. Soft Matter. 2010;6:1472–1482. [Google Scholar]
- 94.Kol N., Shi Y., et al. Rousso I. A stiffness switch in human immunodeficiency virus. Biophys. J. 2007;92:1777–1783. doi: 10.1529/biophysj.106.093914. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The analysis code, plotting script, data files, and coordinate files of membrane-bound immature and mature MA systems are available in the GitHub repository (https://github.com/pujabanerjee91/HIV1_MA_Maturation). Additional details are provided within the article or supporting material.