Abstract
Emerging studies have shown that lipid metabolism plays an important role in aging. High resolution in situ imaging of lipid metabolic dynamics inside cells and tissues affords a novel and potent approach for understanding many biological processes such as aging. Here we established a new optical imaging platform that combines D2O-probed stimulated Raman scattering (DO-SRS) imaging microscopy and a Drosophila model to directly visualize metabolic activities in situ during aging. The sub-cellular spatial distribution of de novo lipogenesis in the fat body was quantitatively imaged and examined. We discovered a dramatic decrease in lipid turnover in 35-day-old flies. Decreases in protein turnover occurred earlier than lipids (25-day vs. 35-day), and there are many proteins localized on the cell and lipid droplet membrane. This suggests that protein metabolism may act as a prerequisite for lipid metabolism during aging. This alteration of maintenance of protein turnover indicates disrupted lipid metabolism. We further found a significantly higher lipid turnover rate in large LDs, indicating more active metabolism in large LDs, suggesting that large and small LDs play different roles in metabolism to maintain cellular homeostasis. This is the first study that directly visualizes spatiotemporal alterations of lipid (and protein) metabolism in Drosophila during the aging process. Our study not only demonstrates a new imaging platform for studying lipid metabolism, but also unravels the important interconnections between lipid metabolism and aging.
Introduction
Lipids and their metabolites play crucial roles in regulating and controlling cellular function and diseases.1,2 They not only serve as energy stores and fundamental constituents of cell membranes, but also function as the primary and secondary messengers in signal transduction and molecular recognition processes.3 Additionally, lipid metabolism regulates gene expressions that influence many aspects of development. Emerging data further suggested that lipid metabolism directly regulates the aging process,4 rather than only being associated with aging or age-related diseases. However, the lipid metabolic dynamics during the aging process is still unclear. Unraveling these will promote the design of therapeutic approaches for diet-regulated aging and age-related diseases such as obesity and Alzheimer’s disease (AD).4,5
Conventional methods for lipidomic analyses are usually mass spectrometry (MS)-based, such as electrospray ionization (ESI)-MS used for the analysis of fatty acids (FAs) and lipids with large molecular weights (e.g. phospholipids)6 and gas chromatography (GC)- or liquid chromatography (LC)-MS for the analysis of methanol-forming FA methyl esters.7 However, these methods cannot determine the spatial location of lipids in tissues. Matrix-assisted laser desorption/ionization (MALDI)-MS has been used for the analysis of blood lipids and highly hydrophobic lipids and provides the spatial distribution of lipids in tissues, but it is invasive and has relatively low spatial resolution (cellular)8 and imaging depth (<15 μm).9 Other imaging technologies have inherent limitations such as low spatial resolution (magnetic resonance imaging), loss of spatial information (nuclear magnetic resonance spectroscopy), radioactive labels (microautoradiography), or bulky fluorescent dyes (fluorescence microscopy) that will disturb the activities of native molecules.
As a quickly evolving optical imaging technology, Raman spectroscopy/microscopy surpasses these techniques in molecule-specific imaging with advantages including non-invasiveness, high chemical specificity, high resolution, and multiplex imaging capability.10–13 When incorporated with a series of probes, Raman imaging illustrates metabolic dynamics of a variety of biomolecules including lipids, proteins, and DNA in living cells and organisms in situ.14–18 We developed an imaging platform that integrates heavy water (D2O) probing with stimulated Raman scattering (SRS) (DO-SRS) microscopy for in situ imaging of metabolic dynamics of different types of biomolecules simultaneously in living animals.17 Like water, D2O can diffuse freely into the cell, and the deuterium (D) from D2O is incorporated into newly synthesized lipids through de novo lipogenesis. This enables us to visualize the spatiotemporal metabolic dynamics in organisms.
In this study, we first demonstrated label free SRS imaging in aging D. melanogaster fat bodies and the necessity of labeled SRS imaging to uncover the metabolic dynamics in flies. We then present our new approach using DO-SRS microscopy to visualize lipid (and protein) metabolic activities in the fat body of aging Drosophila. By qualitatively and quantitatively examining the lipid turnover rate in the lipid droplets (LDs) of the fat body, we can achieve a better understanding of the roles lipid metabolism play in aging.
Results
Label free SRS imaging for detecting proteins, retinoids, and unsaturated FAs in aging Drosophila fat bodies
SRS was initially developed as a label free chemical imaging modality and demonstrates chemical selectivity when visualizing different types of molecules in cells and tissues, including lipids and proteins. Using label free SRS imaging and Raman spectroscopy we detected and tracked endogenous molecules in aging Drosophila. We observed changes in multiple peaks in the Raman spectra of aging Drosophila, particularly, peaks at 1580, 2850, 2926, and 3005 cm−1 (Fig. 1A). The peaks at 1580 cm−1 and 3005 cm−1 resemble retinoids and unsaturated fatty acids (FAs),19 and the peaks at 2850 and 2926 cm−1 are due to CH2 stretching of lipids and proteins, respectively. We visualized and quantified their contents and distributions in the fat bodies of flies during aging using SRS imaging, and further compared the ratiometric images of 1580 cm−1, 2926 cm−1, and unsaturated FAs (3005 cm−1) by dividing them by the CH2 stretching of lipids at 2850 cm−1 (see Fig. S1† for quantification results of all age groups).
Fig. 1.
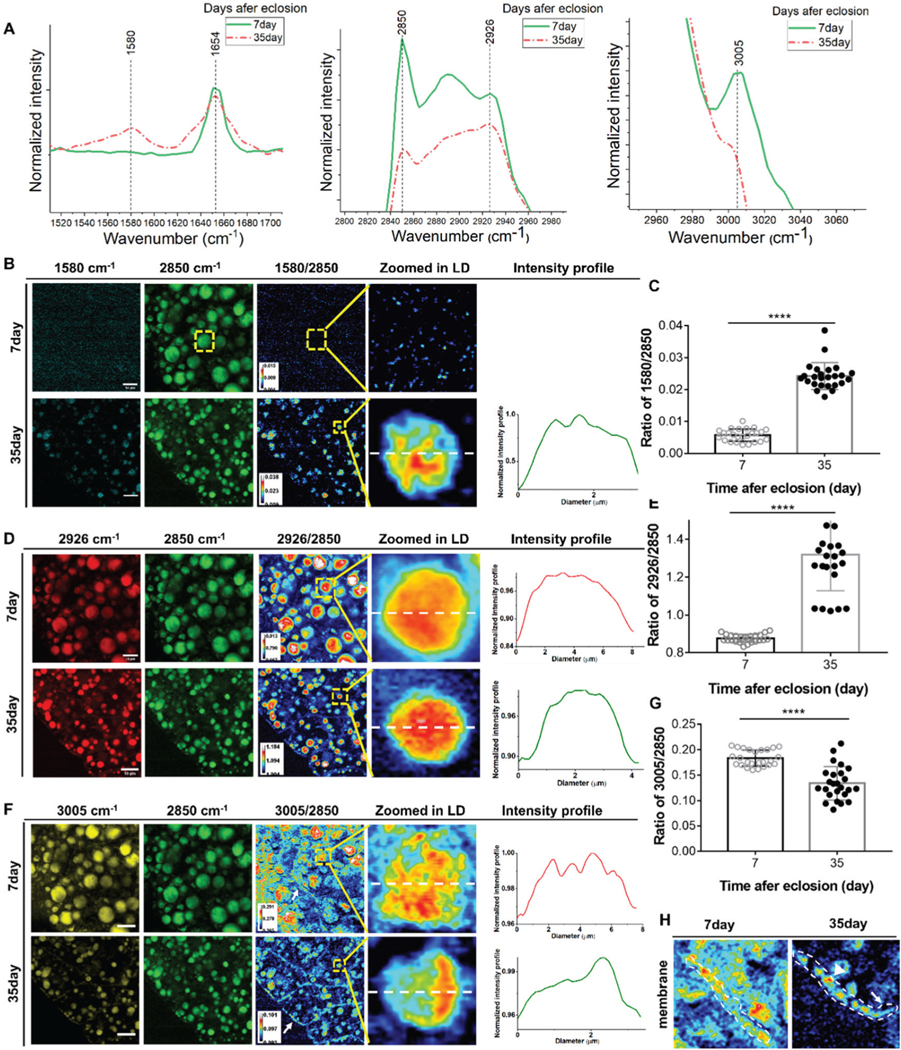
SRS imaging detection of biomolecular changes in LDs. SRS images demonstrate more and larger LDs in young flies and smaller LDs were observed in old flies. (A) Raman spectra of the fat body in 7-day-old flies and 35-day-old flies at 1580 cm−1 (retinoid C=C), 2850 cm−1 (CH2 stretching of lipids), 2926 cm−1, and 3005 cm−1 (unsaturated fatty acids). (B) SRS images of retinoids in the fat bodies of 7- and 35-day-old flies. Ratiometric images of 1580 cm−1 to 2850 cm−1 show a significant increase of retinoids in 35-day-old flies. Retinoids were not detectable in 7-day-old flies. The distribution profile of the retinoids inside single LD is also shown. (C) Quantification of changes in retinoids relative to lipids in 7- and 35-day-old flies. There was a significant increase in retinoids in old flies. (D) SRS images of 2926 cm−1 in the fat bodies of 7- and 35-day flies. Ratiometric images of 2926 cm−1 to 2850 cm−1 demonstrate significantly stronger intensity in old flies, indicating more protein synthesis. Curves also showed that proteins were evenly distributed in the LDs in both young and old flies. (E) Quantification of protein changes relative to lipids in 7- and 35-day-old flies. The relative content of proteins significantly increased in old flies. (F) SRS images of unsaturated FAs (at 3005 cm−1) in young and old flies. Ratiometric images of unsaturated FAs to lipids (at 2850 cm−1) show weaker signals in old flies, indicating the reduction of unsaturated FAs. Plots of the intensity profile show unsaturated FAs were more located in certain regions inside the LD in old flies. (G) Quantification of changes in unsaturated FAs in 7- and 35-day-old flies. In old flies, unsaturated FAs decreased significantly. (H) Ratiometric images of unsaturated FAs near/on the fat body cell membrane. There were more unsaturated FAs on the membrane in young flies. Results are presented as mean ± SD (n = 25 in each group). Statistical significance was determined by using unpaired Student’s t-test. ****, p < 0.0001.
Retinoids play crucial roles in lipid metabolism and development. A study19 showed that retinoids were positively correlated with fat storage. In response to high glucose stress, they significantly increased FA desaturation and incorporation into LDs, which reduced lipotoxicity by high glucose uptake and prolonged the lifespan of C. elegans.
Excessive glucose uptake provokes de novo synthesis of saturated FAs, which can lead to lipotoxicity when lacking sufficient desaturation and subsequent storage in lipid droplets. Our SRS images of retinoids at 1580 cm−1 show that the signals in 7-day-old flies could hardly be recognized but are readily apparent in 35-day-old flies (Fig. 1B). Ratiometric images highlight a significant increase of retinoids in 35-day-old flies, indicating the accumulation of retinoids within the fat bodies of aged flies. We quantified the ratios of retinoids at 1580 cm−1 to the CH2 stretching of lipids at 2850 cm−1 and found a 4.6-fold increase in 35-day-old flies (Fig. 1C), showing dramatic enhancement of the retinoids level in old flies.
SRS images of 2926 cm−1 display strong signals in both young and old flies, but ratiometric images (Fig. 1D) and quantification results (Fig. 1E) revealed significant differences. The ratio of 2926 cm−1 to lipid stretching (2850 cm−1) was 0.9 in 7-day-old flies, but was dramatically enhanced to 1.34 in 35-day-old flies, indicating more protein contents than lipids in the old flies’ fat body, which is most likely due to declined lipid storage in old flies (as shown by the reduction in lipids turnover rates measured, as shown in Fig. 3D). The distribution of proteins inside individual LDs could also be mapped using SRS imaging, which was uniform in both young and old flies (Fig. 1D).
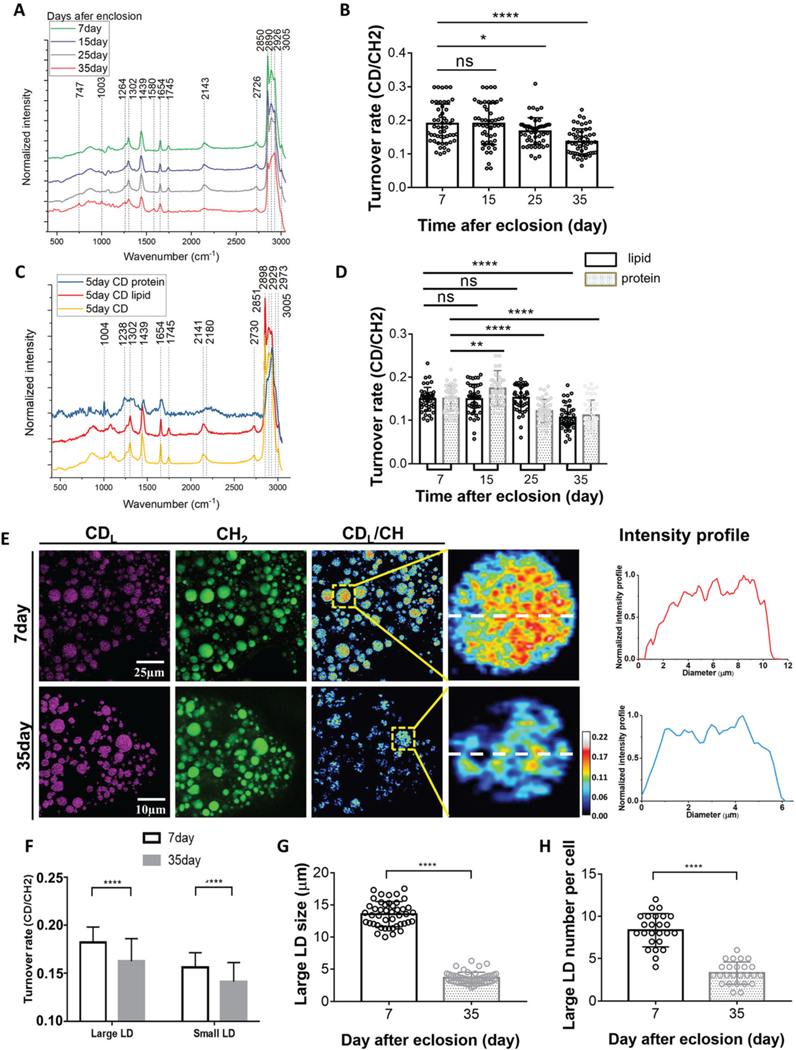
Fig. 3.
The metabolic changes in the fat bodies of Drosophila during aging. (A) Averaged Raman spectra (n = 50) were collected from flies of different ages (7, 15, 25, 35-day posteclosion). The flies were labeled by 20% D2O label for 5 days and then imaged. (B) Quantification of the C–D turnover by dividing the intensity of 2143 cm−1 to the peak at 2850 cm−1. (C) Unmixing of the C–D spectra into protein and lipid spectra. The peak at 2141 cm−1 is corresponding to the newly synthesized D-labeled lipids, and the peak at 2180 cm−1 is the newly synthesized D-labeled protein. (D) Quantification of lipids and protein turnover rates by 2141 cm−1 to 2850 cm−1 and 2180 cm−1 to 2929 cm−1, respectively. (E) SRS imaging of lipid metabolic activities in 7-day (young) and 35-day-old flies. Old (35-day) flies showed reduced lipid metabolism compared with young flies. SRS imaging could visualize the lipids turnover inside single LDs at the subcellular scale. (F) Quantification of the turnover rates of large (diameter ≥4 μm) and small (diameter <4 μm) LDs in the fat bodies of 7-day and 35-day-old flies. Lipid turnover rates inside both large and small LDs declined significantly in old flies. (G) Comparison of large LD size in young (7-day) and old (35-day) flies. (H) Comparison of large LD numbers per cell in young and old flies. Both the size and number per cell reduced drastically in old flies. Results are presented as mean ± SD (n = 50, from 3 individuals). Statistical significance was determined by using unpaired Student’s t-test. *, p < 0.05; **, p < 0.01; ****, p < 0.0001; ns, non-significant difference.
We then examined alterations in the unsaturated FA content and distribution in flies during aging using SRS imaging at 3005 cm−1 (Fig. 1F). Unlike the increased retinoid presence observed in 35-day-old flies (Fig. 1B), we observed a significant decrease in the level of unsaturated FAs in 35-day-old flies (Fig. 1F). This contrasts the previous study in C. elegans in which unsaturated FA was positively correlated with retinoids.19 Quantitative results further showed the ratio of unsaturated FAs (at 3005 cm−1) to CH2 lipid stretching (at 2850 cm−1) was reduced to 0.13 from 0.18 in 7-day-old flies (Fig. 1G). Enlarged ratiometric images (Fig. 1F) demonstrate that in young flies unsaturated FAs were scattered in various regions inside single LDs, while in old flies they were more concentrated in one area. Different distributions of unsaturated FAs in young and old flies may be due to interactions between LDs and other organelles such as mitochondria and endoplasmic reticulum (ER). For example, unsaturated FAs that are in close proximity to mitochondria may facilitate the transfer of FAs for beta oxidation. Another possibility of the uneven distribution may be associated with proteins at the monolayer surface of LDs, such as enzymes that control LD growth and degradation. The relocalization of several enzymes from the ER to the LD surface leads to the localized synthesis of triacylglycerols.20
We also observed a significant reduction of unsaturated lipids on the fat body cell membrane in 35-day-old flies compared with young flies in the ratiometric images (Fig. 1H). This may be due to the direct reduction of unsaturated lipids in the membrane or a decrease of small LDs (which contain a high level of unsaturated lipids) attached to the cell membrane in aged flies. Future study is needed for validation.
Although we visualized different endogenous molecules in Drosophila fat bodies and quantified their relative content changes during the aging processes, using label free SRS we could only measure the total contents of lipids and protein in the fat body without knowing the quantities of newly-synthesized lipids and proteins. To resolve this, we integrated D2O labeling with SRS imaging (DO-SRS) and applied it to the Drosophila model, allowing us to visualize newly-synthesized biomolecules (namely lipids and proteins) that can be distinguished from existing endogenous molecules and to quantify their turnover rates in situ.
DO-SRS enables the detection of lipid turnover in the aging Drosophila fat bodies
The fat body is a major energy storage organ in Drosophila. Excess lipids are converted into neutral lipids, such as triglycerides (TGs) or cholesteryl esters (CEs) and stored in the LDs of the fat body. In this study we examined the lipid turnover in the LDs of the fat body. To use D2O for in vivo labeling in Drosophila, we first determined the concentration of D2O that would generate a detectable C–D signal without disturbing the physiological environment in flies. We then determined the duration of D2O treatment that would cause optimal deuteium incorporation in the fat body.
We fed 3-day-old adult flies a standard yeast-sugar food mixed with various concentrations (0–100%) of D2O for 24 h. The deuterium (D) from D2O would form C–D bonds in newly-synthesized molecules with a characteristic vibrational frequency in the Raman silent cellular region (wavenumber 1800–2800 cm−1).17 By averaging the Raman spectra of 20–30 randomly selected fat body regions from 5 individual flies in each group, we observed a distinct signal peak at 2143 cm−1 (Fig. 2B and S2A†), corresponding to the vibration of C–D bonds in lipids (and some protein). The C–D signal was detected as early as 24 h post-cultivation in flies fed 10% D2O-containing food.
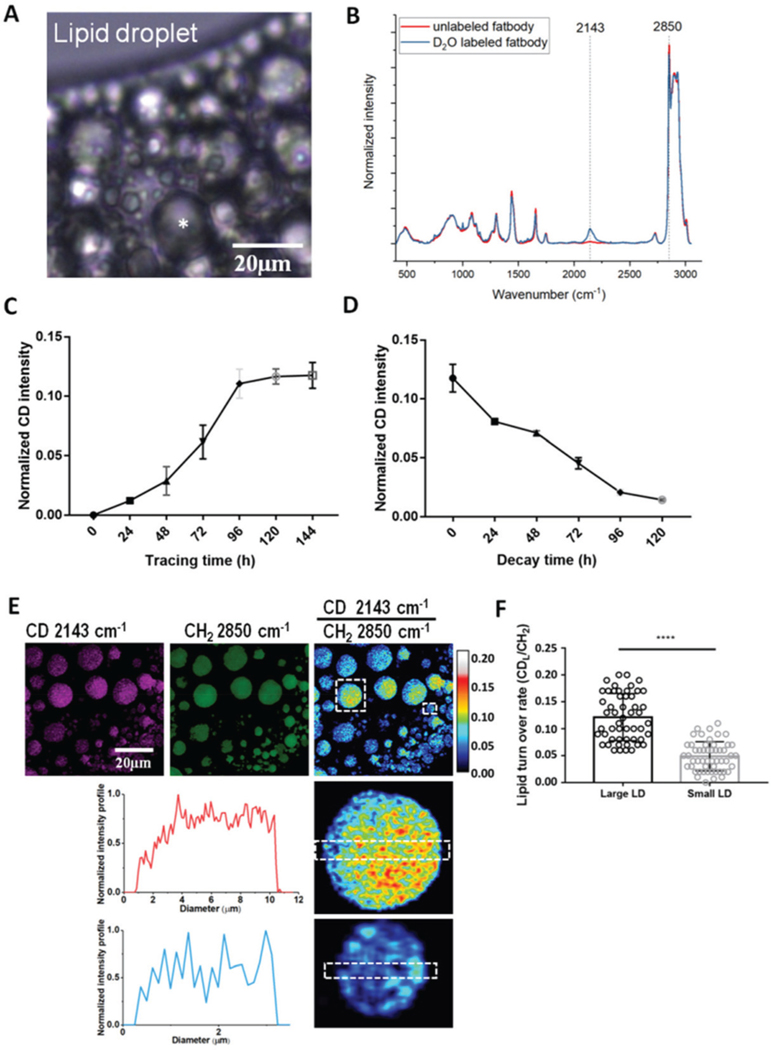
Fig. 2.
Incorporation of deuterium (D) in Drosophila fat bodies from heavy water detected by Raman microspectroscopy and SRS. (A) The fat body cells of Drosophila adults were filled with lipid droplets of different sizes. Scale bar, 10 μm. (B) The averaged Raman spectrum from the fat body of 3-day-old adult flies treated with 20% D2O for 5 days. A distinct C–D signal peak at 2143 cm−1 is observed in the cell silent region (ranging from 1800 cm−1 to 2800 cm−1 in the Raman spectra, no signal found from natural biological samples) compared to the spectrum from unlabeled flies. (C) 3-day-old adult flies were fed with 20% D2O-labeled food for 6 days and the deuterium incorporation at different time points was quantified. The deuterium incorporation level reached a plateau on day 5, which was around 12% of the intensity peaked at 2850 cm−1 (CH2 stretching of lipids). (D) After 6-day D2O labeling, flies were transferred to unlabeled standard food to monitor the utilization of the deuterium labeled metabolites at different time points. The level of deuterium signal decreased gradually with time and was hardly detectable on the 5th day after chasing. (E) SRS imaging of fat body cells in flies treated with 20% D2O labeling for 5 days. Ratiometric image of the CD signal (at 2143 cm−1) to the peak at 2850 (CH2 stretching of lipids) displays the metabolic activity inside individual LDs. Zoomed-in images show more active metabolism inside large LDs than in small LDs. The intensity profiles display spatial information of C–D turnover, which was more scattered in small LDs than in large LDs. Scale bar, 20 μm. (F) Quantification of the metabolic activity of large (dia. ≥4 μm) and small LDs from Raman spectra from 7-day-old adults. The results were shown as mean ± SD, n = 50 from 3 individuals. Statistical significance was determined by unpaired Student’s t-test. ****, p < 0.0001.
We evaluated the effect of D2O concentration on deuterium incorporation into LD by calculating the turnover,17 the ratio of the C–D signal at 2143 cm−1 to the C–H signal at 2850 cm−1. We found a significant increase in the turnover when D2O was increased from 10% to 30% in their feed (Fig. S2B†), indicating active metabolism in Drosophila in the culture environment. However, when the D2O concentration was above 30% deuterium incorporation started to decline, suggesting the possibility of saturation with deuterium incorporation at 30% D2O and the potential inhibitory effect of higher D2O on Drosophila metabolism. The latter is verified by the shortened lifespan of both female and male flies fed with high D2O-containing food. Male flies proved more sensitive to high D2O concentrations (≥30%) than females (Fig. S2C and D†). Compared with flies on standard food, the median lifespan of male flies fed 30% D2O-containing food was reduced from 35 ± 1 (days) to 30 ± 1 (days), but there was no significant change in female flies. Female and male flies respond differently to higher D2O concentrations, this may suggest different metabolic activities or metabolic pathways are involved in different genders. This paper will focus on DO-SRS metabolic imaging in Drosophila and we won’t further discuss this gender difference. The median lifespans of both male and female flies showed no significant difference when treated with 10% or 20% D2O, or standard food. A lower concentration (10% D2O) might lead to quicker exhaustion of the deuterium source in the body or a longer time to reach a stable condition. Therefore we selected 20% D2O for in vivo labeling of flies.
To examine when D2O incorporation would reach a stable state, we traced the flies fed 20% D2O labeled food for 6 days and quantified the temporal deuterium incorporation. We found that deuterium incorporation reached a plateau on day 4 with a maximum of 12% occurring on day 5 (Fig. 2C). Hence, we treated flies with D2O for 5 days to obtain detectable and stable incorporation of deuterium for Raman imaging. We also tracked the utilization of D-labeled metabolites by transferring the 6-day D2O-labeled flies to unlabeled standard food. We observed a gradual decay of the deuterium signal until it became barely detectable on the 5th day after chasing (Fig. 2D), indicating that these D-labeled metabolites were completely utilized.
After determining the D2O concentration (20%) and treatment duration (5-day), we employed DO-SRS imaging to visualize and quantify metabolism in LDs in Drosophila. After digestion and absorption in the gut, lipids in the food will enter the hemolymph, which is analogous to blood in vertebrates, and are then transported to the fat body for storage in LDs. Under specific conditions such as development, metamorphosis and starvation, the neutral lipids stored in LDs will be mobilized into fatty acids and provide energy through β-oxidation. The whole process is analogous to lipid metabolism in mammals.21 The size and number of LDs in the fat body may act as an indicator of the metabolic level of the fat body cells; however, this has not been examined, nor has the metabolic dynamics in LDs been visualized in vivo. For the first time, metabolic activities have been visualized in LDs at the subcellular scale in situ (Fig. 2E). Like in different species or different cell types, different sizes of LDs exist in Drosophila the fat body cells (Fig. 2E) range from 0.1 to ∼15 μm in diameter. The ratiometric images from CD (at 2143 cm−1) to CH (at 2850 cm−1) further illustrate higher metabolism in large LDs and non-uniform distribution of lipid metabolism inside LDs (Fig. 2E). DO-SRS images also allowed us to quantify the turnover rate in large LDs (defined as diameter ≥4 μm) and small LDs (diameter <4 μm), as shown in Fig. 2F. A significantly higher C–D turnover rate was measured in large LDs, indicating more active metabolism in large LDs, which suggests that large and small LDs play different roles in metabolism to maintain cellular homeostasis. For comparison, we quantified the turnover rates using both DO-SRS imaging (Fig. 2F) and Raman spectroscopy (Fig. S2E†) and obtained consistent results across the two platforms.
Lipid and protein metabolism changes in the Drosophila fat bodies during aging
The lifespan of an organism is strongly influenced by environmental factors including diet and internal factors, notably reproductive status. Lipid metabolism is critical for adaptation to external conditions or reproduction.22 Interestingly, specific lipid profiles are associated with longevity and increased uptake of certain lipids extends longevity in C. elegans23 and ameliorates disease phenotypes in humans.24,25 How lipids impact longevity and how lipid metabolism is regulated during aging are just beginning to be unraveled. Studies have demonstrated that lipid profiles change with age in worms, fruit flies, mice, and humans.22,23,26,27 Consistent with the idea that lipids are important for the regulation of lifespan, lipid profiles are altered in many long-lived C. elegans and Drosophila mutants.28,29 But all these studies were based on mass spectrometry, in which tissues are destructed and the spatial information cannot be preserved during sample preparation. Raman microscopy can elegantly overcome these limitations and allow us to visualize and quantitatively examine lipid metabolic changes in situ.
To examine metabolic activity changes in the fat body cells during Drosophila aging, we synchronized the larvae and allowed them to grow on standard food until eclosion. Female flies at day-2, 10, 20, and 30 posteclosion were fed with 20% D2O-labeled food for 5 days, respectively, prior to subsequent Raman spectroscopy and SRS imaging to assess their turnover rates in the fat body (hence, at 7-, 15, 25, and 35-day posteclosion). The spectra of 7, 15, and 25-day-old flies showed no obvious difference in spectral peaks, but 35-day-old flies displayed a less distinct spectral peak at 2143 cm−1 (Fig. 3A), indicating lower turnover in aged flies based on the lower signal. The quantification of the turnover rate (the ratio of C–D signal at 2143 cm−1 to CH2 stretching of lipids at 2850 cm−1) showed significantly decreased metabolism in flies at 35 and 25 days after eclosion (Fig. 3B). In addition to the peak in the C–D region, 35-day-old flies also displayed evident differences in the peaks at 747, 1580, and 3005 cm−1, as well as the spectral patterns between 2850 and 2926 cm−1.
The C–D peak at 2143 cm−1 is a mixture of signals from newly-synthesized lipids and proteins. To distinguish them, we unmixed the C–D spectra and separated them into protein and lipid specific spectra, respectively (Fig. 3C). By removing the protein using proteinase K treatment in fat body tissues, we obtained a peak at 2141 cm−1, corresponding to newly-synthesized D-labeled lipids; and by removing lipids using methanol we obtained a peak at 2180 cm−1, corresponding to a newly synthesized D-labeled protein.17 In addition to the C–D region, the fingerprint and C–H stretching regions from Raman measurements also provide valuable information on biochemical compositions of the fat body. For example, after dissolving lipids, a spectrum enriched in protein information appeared (at 2929 cm−1), allowing us to detect the subtle changes of endogenous protein components that were usually masked by the strong lipid signal (Fig. 3C). It is worth noting that by comparing Fig. 3A and C the spectral pattern from younger flies is more lipid-like while more protein-like in old flies, which suggests alterations of lipids and protein contents, in particular, loss of lipid storage in flies during aging.
To examine how lipid and protein turnover changed during aging, we next quantified lipids and protein turnover rates by calculating the ratios of the signals from 2141 cm−1 to 2850 cm−1 and 2180 cm−1 to 2929 cm−1, respectively (Fig. 3D). A significant decrease in the lipid turnover was observed only between 7 day-old and 35 day-old flies. Protein metabolic activity showed more fluctuations with an increase from 15-day-old flies but then a decrease from 25- and 35-day-old flies. Decreases in the protein turnover rate occurred earlier than lipids (25-day vs. 35-day), suggesting that protein metabolism may act as a prerequisite for lipid metabolism during aging. Studies have shown that triglyceride (TG) absorption can be impaired by as much as 50% in aging rodents, either due to a defect in lipoprotein assembly or a decline in the abundance of fatty acid-binding proteins.30 At the same time, there are many proteins localized on the cell and lipid droplet membrane, which regulate lipid absorption, transportation, lipogenesis and lipolysis.31–33 The alteration of maintenance for this protein turnover could also disrupt lipid metabolism. However, more experiments are needed to determine which protein components are closely related to the loss of fat storage in aged flies.
Using DO-SRS microscopy, we were able to visualize metabolic activities in fat body cells in situ and more importantly, we were able to distinguish individual large and small LDs and quantitatively map lipid turnover in single LDs (Fig. 3E). This is unattainable by mass spectrometry. SRS images showed dramatically reduced lipid turnover in 35-day-old flies compared with 7-day-old flies (Fig. 3E). Quantification results further verified these significant decreases in both large and small LDs in aged flies (Fig. 3F). In addition, SRS images showed more large LDs in young (7-day) flies while fewer large LDs but much more small LDs in old (35-day) flies. In Fig. 3G and H, we observed reductions in both the number and size of LDs for storage in 35-day-old flies, which may lead to lipotoxicity, but the enhanced level of retinoids observed in old flies may compensate for the loss of unsaturated FAs. The average size of large LDs decreased from 13.8 μm in young flies to 3.8 μm in old flies (Fig. 3G), and the quantity of large LDs also reduced from 8.5 per cell to 3.4 LDs per cell (Fig. 3H). The significant reductions of both LD size and quantity in old Drosophila fat bodies indicate the dramatic decline of lipid storage capability in Drosophila during aging. A key function of the fat bodies in Drosophila is to maintain the balance between energy storage and utilization under different circumstances, the reduced metabolic activity in old flies may be due to the lower rate of synthesis or higher rate of mobilization.
Discussion
In this study, we establish for the first time a platform that employs DO-SRS microscopy imaging in a Drosophila model to directly visualize spatiotemporal alterations in lipid metabolism in situ during the aging process. SRS is a rapidly evolving optical imaging technique. Conventional mass spectrometry for metabolic studies cannot provide spatial information of biomolecules inside cells/tissues, while optical imaging offers a direct means to observe this activity. SRS microscopy further allows for subcellular and quantitative imaging. Using D2O as the probe, we for the first time tracked and quantified lipid turnover rates in the fat body cells of fruit flies during the aging process. The experimental results from this study showed a tremendous decrease in lipid turnover in 35-day-old flies by quantitative DO-SRS imaging of CD labeled newly synthesized lipids. In contrast, the decrease of the protein turnover rate occurred earlier than lipid (25-day vs. 35-day), moreover, we observed many proteins localized on the cell and lipid droplet membrane, suggesting protein metabolism may play an important role as a prerequisite for the changing lipid metabolism during the aging process. We hypothesize that the disrupted lipid metabolism during the aging process might be the consequence of altered protein maintenance, and our planned future follow-up work will show consolidated imaging evidence. For the quantitative results of a significantly higher lipid/protein turnover rate in large LDs, we take it as an indication of more active metabolism in large LDs, while large and small LDs play different roles in metabolism to maintain cellular homeostasis. Lipid metabolism has been associated with aging and age-related diseases, as more evidence emerges, showing that rather than a consequence of aging and age-related diseases, lipid metabolism and its metabolites play active roles in regulating aging.4,34 Our established DO-SRS and Raman spectroscopy imaging modalities for Drosophila enable us to directly visualize and track spatiotemporal dynamics of lipid metabolism, to further explore the interplay of lipid synthesis and turnover with aging and aging-related diseases.
In conclusion, our study establishes the first optical imaging platform for the study of metabolic dynamics in live Drosophila in situ. This imaging platform is widely applicable to a broad range of biological studies in metabolism, aging, and aging-related diseases.
Materials and methods
Fly stocks
Wild type (w1118 stock #5905) flies were originally obtained from the Bloomington Stock Center and have been maintained in the lab for several generations.
Drosophila lifespan analysis
The w1118 parents were raised in vials containing standard food. To standardize the effects of parental age on offspring fitness, parents of experimental flies were of the same age (4–5 days and reared at a constant density for at least two generations). To synchronize larval development, we allowed flies to lay eggs on yeast apple juice plates for 1 h, discarded the first batch of embryos, and then collected for another 1.5 h. Groups of 20–25 embryos were put into vials containing standard food and allowed to develop until pupal eclosion. The groups of adult progenies that emerged from the puparium within 24 h were transferred to fresh bottles and allowed to mate for 2 days.
For the lifespan experiments, 2 days aged w1118 females were separated under CO2 anesthesia and randomly allocated to different media (approximately 20–30 flies per vial). All enclosures were maintained at 25 °C in a controlled light (12/12 h light/dark cycle) and humidity (>70%) environment. Flies were scored for survival daily and provided with a fresh medium every 2 days. To minimize any density effects on mortality, two vials with cohorts were merged when the density of flies reached five or fewer individuals. Enclosures were placed randomly in the incubator, and positions were rotated after each transfer to minimize the effects of the microclimate.
D2O-labeling experiments
The metabolic activity changes of wild type flies at different ages were labeled by transferring the 2-day and 30-day-old female adult flies to the corresponding 20% D2O labeled food conditions for 5 days, then the 7-day and 35-day old flies were sacrificed, and fat bodies were dissected and subjected to Raman measurements and SRS imaging.
To quantify the LD size, the diameters of 50 LDs (larger than 4 μm that can be accurately measured) in 20 fat cells from 3 different individuals were measured using ImageJ software. When we calibrated the lipid turnover rate in large and small lipid droplets separately, where the threshold setup is 4 microns, droplets bigger than 4 microns are considered large and smaller than 4 microns are considered small.
Drosophila sample preparation for Raman and SRS microscopy
The fat body tissues were dissected in PBS and fixed in 4% formaldehyde for 25 min at room temperature (RT). After fixation, the tissues were washed four times with PBS in glass wells. The tissues were then sandwiched between a cover slide and the bottom slide with PBS solution. To prevent tissue drying, nail polish was used to seal the surrounding of cover slides.
Resonance Raman spectroscopy
Raman spectra of all tissue samples were recorded using a Raman spectrometer connected to a confocal Raman microscope (XploRA PLUS, Horiba). A 532 nm diode line focus laser (∼40 mW at the sample) was focused on the cells with the help of a 100× objective (MPLN100X, Olympus). The laser power on the sample was optimized so as to avoid any damage to the cells. A cooled charge coupled device (CCD) detector fitted to 2400 grooves per mm grating spectrometer was used to detect the signal. We measured the Raman spectra with 60s acquisition time. The background spectra were recorded for each tissue point at the same focus plane and were subtracted from the original spectrum immediately. All ratio calculations were carried out on the raw data before any normalization or baseline correction steps. Peaks were normalized to the phenylalanine peak at 1003 cm−1. The instrumental calibration was verified using the silicon line at 520 cm−1. The observed data were processed and analyzed using Prism software (Origin Lab Corporation, Northampton, MA).
Stimulated Raman scattering microscopy
An upright laser-scanning microscope (DIY multiphoton, Olympus) with a 25× water objective (XLPLN, WMP2, 1.05 NA, Olympus) was applied for near-IR throughput. A synchronized pulsed pump beam (tunable 720–990 nm wavelength, 5–6 ps pulse width, and 80 MHz repetition rate) and Stokes beam (wavelength at 1032 nm, 6 ps pulse width, and 80 MHz repetition rate) were provided by a picoEmerald system (Applied Physics & Electronics) coupled to a microscope. The pump and Stokes beams were collected in transmission using a high NA oil condenser (1.4 NA). A high O.D. shortpass filter (950 nm, Thorlabs) was used that would completely block the Stokes beam and transmit the pump beam only onto a Si photodiode for detecting the stimulated Raman loss signal. The output current from the photodiode was terminated, filtered, and demodulated using a lock-in amplifier at 20 MHz. The demodulated signal was fed into the FV3000 software module FV-OSR (Olympus) to form images during laser scanning. All images obtained were 512 × 512 pixels, with a dwell time of 80 μs and the imaging speed of ∼23 s per image. A background image was acquired at 1900 cm−1 and subtracted from all SRS images using Fiji (ImageJ).
Statistical analysis
The Kaplan–Meier Log-rank test was performed for survival data in MATLAB. Others were tested by Student’s t-test, ANOVA with a post hoc Tukey’s multiple comparison or ANOVA with a post hoc Dunnett’s comparison test using GraphPad Prism software. Ratio data were analyzed from raw intensity values without baseline correction and normalization.
Supplementary Material
Acknowledgements
We thank Dr Jane Wu, Christian Metallo, and Wei Min, for helpful discussion. This work is partially supported by UCSD Startup funds, NIH U54 pilot grant 2U54CA132378-11A1, and the Hellman Fellow Award.
Footnotes
Electronic supplementary information (ESI) available. See DOI: 10.1039/d1an01638e
Conflicts of interest
The authors declare no competing interests.
References
- 1.Yazıcı D. and Sezer H, Insulin Resistance, Obesity and Lipotoxicity, Adv. Exp. Med. Biol, 2017, 960, 277–304. [DOI] [PubMed] [Google Scholar]
- 2.Young SG and Zechner R, Biochemistry and pathophysiology of intravascular and intracellular lipolysis, Genes Dev, 2013, 27(5), 459–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Meer G, Voelker DR and Feigenson GW, Membrane lipids: where they are and how they behave, Nat. Rev. Mol. Cell Biol, 2008, 9(2), 112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson AA and Stolzing A, The role of lipid metabolism in aging, lifespan regulation, and age-related disease, Aging Cell, 2019, 18(6), e13048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mahmoudi S, Xu L. and Brunet A, Turning back time with emerging rejuvenation strategies, Nat. Cell Biol, 2019, 21(1), 32–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schneiter R, Brugger B, Sandhoff R, Zellnig G, Leber A, Lampl M, et al. , Electrospray ionization tandem mass spectrometry (ESI-MS/MS) analysis of the lipid molecular species composition of yeast subcellular membranes reveals acyl chain-based sorting/remodeling of distinct molecular species en route to the plasma membrane, J. Cell Biol, 1999, 146(4), 741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lubes G. and Goodarzi M, GC–MS based metabolomics used for the identification of cancer volatile organic compounds as biomarkers, J. Pharm. Biomed. Anal, 2018, 147, 313–322. [DOI] [PubMed] [Google Scholar]
- 8.Bowman AP, Bogie JFJ, Hendriks JJA, Haidar M, Belov M, Heeren RMA, et al. , Evaluation of lipid coverage and high spatial resolution MALDI-imaging capabilities of oversampling combined with laser post-ionisation, Anal. Bioanal. Chem, 2020, 412(10), 2277–2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murphy RC, Hankin JA and Barkley RM, Imaging of lipid species by MALDI mass spectrometry, J. Lipid Res, 2009, 50(Suppl), S317–S322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ember KJI, Hoeve MA, McAughtrie SL, Bergholt MS, Dwyer BJ, Stevens MM, et al. , Raman spectroscopy and regenerative medicine: a review, npj Regener. Med., 2017, 2, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghita A, Pascut FC, Mather M, Sottile V. and Notingher I, Cytoplasmic RNA in undifferentiated neural stem cells: a potential label-free Raman spectral marker for assessing the undifferentiated status, Anal. Chem, 2012, 84(7), 3155–3162. [DOI] [PubMed] [Google Scholar]
- 12.Daudon M. and Bazin D, Vibrational spectroscopies to investigate concretions and ectopic calcifications for medical diagnosis, C. R. Chim, 2016, 19(11–12), 1416–1423. [Google Scholar]
- 13.Kong K, Rowlands CJ, Varma S, Perkins W, Leach IH, Koloydenko AA, et al. , Diagnosis of tumors during tissue-conserving surgery with integrated autofluorescence and Raman scattering microscopy, Proc. Natl. Acad. Sci. U. S. A, 2013, 110(38), 15189–15194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berry D, Mader E, Lee TK, Woebken D, Wang Y, Zhu D, et al. , Tracking heavy water (D2O) incorporation for identifying and sorting active microbial cells, Proc. Natl. Acad. Sci. U. S. A, 2015, 112(2), E194–E203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Manen AL H-J and Otto C, Noninvasive Imaging of Protein Metabolic Labeling in Single Human Cells Using Stable Isotopes and Raman Microscopy, Anal. Chem, 2008, 80(24), 9576–9582. [DOI] [PubMed] [Google Scholar]
- 16.Zhang L, Shi L, Shen Y, Miao Y, Wei M, Qian N, et al. , Spectral tracing of deuterium for imaging glucose metabolism, Nat. Biomed. Eng., 2019, 3(5), 402–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi L, Zheng C, Shen Y, Chen Z, Silveira ES, Zhang L, et al. , Optical imaging of metabolic dynamics in animals, Nat. Commun, 2018, 9(1), 2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, Xu J, Kong L, Liu T, Yi L, Wang H, et al. , Raman-deuterium isotope probing to study metabolic activities of single bacterial cells in human intestinal microbiota, Microbiol. Biotechnol, 2020, 13(2), 572–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen AJ, Li J, Jannasch A, Mutlu AS, Wang MC and Cheng JX, Fingerprint Stimulated Raman Scattering Imaging Reveals Retinoid Coupling Lipid Metabolism and Survival, ChemPhysChem, 2018, 19(19), 2500–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilfling F, Wang H, Haas JT, Krahmer N, Gould TJ, Uchida A, et al. , Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets, Dev. Cell, 2013, 24(4), 384–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gibbons GF, Islam K. and Pease RJ, Mobilisation of triacylglycerol stores, Biochim. Biophys. Acta, 2000, 1483(1), 37–57. [DOI] [PubMed] [Google Scholar]
- 22.Hahn O, Gronke S, Stubbs TM, Ficz G, Hendrich O, Krueger F, et al. , Dietary restriction protects from age-associated DNA methylation and induces epigenetic reprogramming of lipid metabolism, Genome Biol, 2017, 18(1), 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao AW, Chatzispyrou IA, Kamble R, Liu YJ, Herzog K, Smith RL, et al. , A sensitive mass spectrometry platform identifies metabolic changes of life history traits in C. elegans, Sci. Rep, 2017, 7(1), 2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martino A, Pezzi L, Magnano R, Salustri E, Penco M. and Calo L, Omega 3 and atrial fibrillation: Where are we?, World J. Cardiol., 2016, 8(2), 114–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robinson VM and Kowey PR, The Use of Omega-3 Polyunsaturated Fatty Acids (n-3 PUFAs) in Atrial Fibrillation, in Handbook of Lipids in Human Function, 2016, pp. 181–206. [Google Scholar]
- 26.Hansen M, Flatt T. and Aguilaniu H, Reproduction, fat metabolism, and life span: what is the connection?, Cell Metab., 2013, 17(1), 10–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Houtkooper RH, Argmann C, Houten SM, Canto C, Jeninga EH, Andreux PA, et al. , The metabolic footprint of aging in mice, Sci. Rep, 2011, 1, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Papsdorf K. and Brunet A, Linking Lipid Metabolism to Chromatin Regulation in Aging, Trends Cell Biol, 2019, 29(2), 97–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moghadam NN, Holmstrup M, Pertoldi C. and Loeschcke V, Age-induced perturbation in cell membrane phospholipid fatty acid profile of longevity-selected Drosophila melanogaster and corresponding control lines, Exp. Gerontol, 2013, 48(11), 1362–1368. [DOI] [PubMed] [Google Scholar]
- 30.Christian P, Sacco J. and Adeli K, Autophagy: Emerging roles in lipid homeostasis and metabolic control, Biochim. Biophys. Acta, 2013, 1831(4), 819–824. [DOI] [PubMed] [Google Scholar]
- 31.Thiele C. and Spandl J, Cell biology of lipid droplets, Curr. Opin. Cell Biol, 2008, 20(4), 378–385. [DOI] [PubMed] [Google Scholar]
- 32.Walther TC and Farese RV, The life of lipid droplets, Biochim. Biophys. Acta, 2009, 1791(6), 459–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walther TC and Farese RV, Lipid droplets and cellular lipid metabolism, Annu. Rev. Biochem, 2012, 81, 687–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chung KW, Advances in Understanding of the Role of Lipid Metabolism in Aging, Cells, 2021, 10(4), 880. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.