Abstract
Background
Homeodomain-leucine zipper (HD-Zip) transcription factors are plant-specific and play important roles in plant defense against environmental stresses. Identification and functional studies have been carried out in model plants such as rice, Arabidopsis thaliana, and poplar, but comprehensive analysis on the HD-Zip family of Salix suchowensis have not been reported.
Results
A total of 55 HD-Zip genes were identified in the willow genome, unevenly distributed on 18 chromosomes except for chromosome 19. And segmental duplication events containing SsHD-Zip were detected on all chromosomes except chromosomes 13 and 19. The SsHD-Zip were classified into 4 subfamilies subfamilies (I-IV) according to the evolutionary analysis, and members of each subfamily shared similar domain structure and gene structure. The combination of GO annotation and promoter analysis showed that SsHD-Zip genes responded to multiple abiotic stresses. Furthermore, the results of qPCR analysis showed that the SsHD-Zip I gene exhibited different degrees of expression under salt stress, PEG treatment and heat treatment. Moreover, there was a synergistic effect between SsHD-Zip I genes under stress conditions based on coregulatory networks analysis.
Conclusions
In this study, HD-Zip transcription factors were systematically identified and analyzed at the whole genome level. These results preliminarily clarified the structural characteristics and related functions of willow HD-Zip family members, and it was found that SsHox34, SsHox36 and SsHox51 genes were significantly involved in the response to various stresses. Together, these findings laid the foundation for further research on the resistance functions of willow HD-Zip genes.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-024-10067-x.
Keywords: Salix suchowensis, HD-Zip gene, Comprehensive analysis, Abiotic stress, Expression pattern
Introduction
The homeodomain leucine zipper (HD-Zip) transcription factor is one of the larger families of transcription factors in plants, and it contains the highly conserved homeodomain (HD) and leucine zipper (LZ) [1]. The HD consists of 60 conserved amino acids and binds specifically to DNA, and the LZ is a dimeric motif that is critical for the recognition of DNA binding sites [2]. Based on structure and function, the HD-Zip gene family can be divided into four distinct subfamilies, namely HD-Zip I to HD-Zip IV. The HD-Zip I subfamily has the simplest gene structure, containing only the HD and LZ domains, and HD-Zip II contains an additional conserved region at the N-terminus [3]. In addition to the HD and LZ domains, HD-Zip III and IV have a START domain associated with sterol binding. However, a C-terminal MEKHLA domain has also been found in the HD-Zip III subfamily [4, 5].
The four subfamily members differ in their respective functions because of the differences in their structure. The HD-Zip I gene plays an important role in helping plants adapt to different environmental conditions, including drought, salt stress and temperature extremes [6, 7]. For example, Zmhdz10 and Oshox22, members of the HD-Zip I subfamily in maize and rice, regulated drought and salt tolerance stress in plants through ABA-dependent signaling pathways [8, 9]. TaHDZ5-6 A, a member of the HD-Zip I subfamily in wheat, increased drought tolerance when overexpressed in Arabidopsis [10]. Under salt stress, overexpression of MdHB7-like (an apple HD-Zip I gene) increased photosynthetic efficiency and reduced ROS and Na+ accumulation [11]. The results of RNA-Seq and RT-qPCR showed that radish HDZ17 from HD-Zip I was highly expressed under high temperature and salt stress. Further studies showed that overexpression of RsHDZ17 improved the heat tolerance of transgenic Arabidopsis [12]. Members of HD-ZIP II subfamily mainly play an important roles in light avoidance or light signalling and plant development [13, 14]. Five of the ten genes of the subfamily in Arabidopsis were response to changes in light quality [15]. The expression of ATHB2 and HAT4 was negatively correlated with the R/FR (red light/far-red light) ratio. Moreover, HAT1 and ATHB4 had been implicated in the regulation of the shade avoidance response of plants [16, 17]. And ATHB2 inhibits the germination of seeds of Arabidopsis [18]. It has been reported that the HD-Zip III subfamily is mainly involved in meristem formation, vascular bundle development and leaf polarity development [19]. In soybean, both GmREV-L-1 and GmHB14-L-2 were highly expressed in vascular cambium cells and maintained consistent high expression levels throughout all xylem maturation stages. GmREV-L-1 and GmHB14-L-2 are key components in xylem differentiation [20]. HD-Zip IV subfamily genes had been shown to be specifically expressed in epidermal cells and play an important role in epidermal cell development and trichosome formation [21, 22]. For instance, SlHDZIV8, a member of the HD-Zip IV subfamily in tomato, controlled the morphology of multicellular trichomes by regulating the expression of Hairless-2 [23].
Genome-wide investigations of the HD-Zip gene family have been carried out in several species, including Arabidopsis [19], Dendrobium officinale [24], Ginseng [25], and peach [26]. However, such work has not been carried out in willow. Salix suchowensis is a woody plant of the genus Salix, its branches are strong and can be used to weave wickerwork, baskets and agricultural tools, but it can also be used as a windbreak, sand fixing tree species with high ecological and economic value [27, 28]. The completion of whole genome sequencing allowed us to identify and analyse the expression of abiotic stress genes at the whole genome level [29]. However, little is known about the expression, structure and function of the willow HD-Zip genes.
In this study, 55 HD-Zip genes were identified in the willow genome, and their evolutionary relationships, conserved domains, gene replication, and cis-acting elements were comprehensively analyzed. At the same time, we also investigated the expression patterns of the HD-Zip I gene in willow under salt, PEG and heat stress. These results provide a basis for further studies on the expression and tolerance function of the HD-Zip genes in willow.
Results
A total of 55 HD-Zip genes were identified in S. suchowensis
A total of 55 HD-Zip genes were identified in S. suchowensis by whole-genome retrieval and validation. All putative HD-Zip genes contained a homeobox domain (PF00046). The 55 HD-Zip genes were named from SsHox1 to SsHox55 based on their physical location and conserved domain. The length of these proteins ranged from 153 ~ 877 amino acids (aa), with the coding region sequences corresponding 462 bp ~ 2643 bp. Moreover, the theoretical isoelectric point (pI) of the SsHox genes varied from 4.56 to 9.49. Table 1 provided additional information on the characterizatio of HD-Zip genes. Additionally, according to the predicted results of subcellular localization, the majority of the willow HD-Zip proteins were located in the nucleus, SsHox26 and SsHox49 were located in chloroplasts, and SsHox29 was located in the cytoplasm (Table S1).
Table 1.
Details of the identified HD-Zip genes in S. suchowensis
| Name | Gene Identifier | Location | ORF length (bp) | Protein | |||
|---|---|---|---|---|---|---|---|
| Length (a.a.) | PI | Mol.Wt. (Da) | Exons | ||||
| SsHox1 | willow_GLEAN_10002342 | chr13:5209149.5210205 | 954 | 317 | 5.05 | 36086.77 | 2 |
| SsHox2 | willow_GLEAN_10003886 | chr05:2501516.2504989 | 1428 | 475 | 6.56 | 53823.25 | 4 |
| SsHox3 | willow_GLEAN_10004942 | chr17:487110.487631 | 522 | 173 | 6.29 | 19451.87 | 1 |
| SsHox4 | willow_GLEAN_10005026 | chr05:7848777.7849826 | 789 | 262 | 5.71 | 30538.8 | 3 |
| SsHox5 | willow_GLEAN_10005056 | chr11:7473266.7473769 | 504 | 167 | 9.37 | 19337.64 | 1 |
| SsHox6 | willow_GLEAN_10005194 | chr12:274045.274900 | 774 | 257 | 5.32 | 29284.43 | 2 |
| SsHox7 | willow_GLEAN_10005705 | chr06:6228916.6230045 | 654 | 217 | 8.85 | 25318.29 | 3 |
| SsHox8 | willow_GLEAN_10005832 | chr01:12229293.12230670 | 963 | 320 | 8.69 | 35689.61 | 4 |
| SsHox9 | willow_GLEAN_10006690 | chr07:7817079.7817615 | 537 | 178 | 9.17 | 20350.79 | 1 |
| SsHox10 | willow_GLEAN_10007184 | chr18:2092915.2099179 | 2571 | 856 | 5.85 | 94346.81 | 18 |
| SsHox11 | willow_GLEAN_10007942 | chr18:7540908.7545445 | 948 | 315 | 6.95 | 35491.58 | 6 |
| SsHox12 | willow_GLEAN_10008337 | chr17:8082883.8084041 | 462 | 153 | 7.74 | 17504.55 | 3 |
| SsHox13 | willow_GLEAN_10008909 | chr10:5275904.5277549 | 870 | 289 | 6.01 | 32,954 | 3 |
| SsHox14 | willow_GLEAN_10009247 | chr02:8801327.8803781 | 885 | 294 | 9.08 | 33006.88 | 4 |
| SsHox15 | willow_GLEAN_10009523 | chr12:3864207.3865121 | 795 | 264 | 4.56 | 30333.3 | 2 |
| SsHox16 | willow_GLEAN_10009658 | chr16:11668377.11673053 | 2274 | 757 | 5.96 | 84363.34 | 11 |
| SsHox17 | willow_GLEAN_10009743 | chr15:8411038.8415237 | 2223 | 740 | 5.54 | 81513.69 | 8 |
| SsHox18 | willow_GLEAN_10009808 | chr15:1853449.1856582 | 2127 | 708 | 6.53 | 77753.34 | 10 |
| SsHox19 | willow_GLEAN_10010028 | chr04:720621.724207 | 2040 | 679 | 5.51 | 74826.89 | 10 |
| SsHox20 | willow_GLEAN_10010090 | chr05:5707352.5709223 | 726 | 241 | 9.25 | 27999.9 | 4 |
| SsHox21 | willow_GLEAN_10010591 | chr08:7569496.7570759 | 708 | 235 | 5.91 | 26951.08 | 2 |
| SsHox22 | willow_GLEAN_10011112 | chr15:3371825.3373099 | 930 | 309 | 4.83 | 34765.26 | 3 |
| SsHox23 | willow_GLEAN_10011227 | chr11:5749314.5755253 | 2529 | 842 | 5.86 | 92313.32 | 18 |
| SsHox24 | willow_GLEAN_10011792 | chr02:9983842.9990080 | 2490 | 829 | 5.94 | 90567.88 | 10 |
| SsHox25 | willow_GLEAN_10012615 | chr07:440589.441742 | 846 | 281 | 8.57 | 31623.73 | 3 |
| SsHox26 | willow_GLEAN_10013067 | chr04:11864612.11872078 | 2613 | 870 | 5.91 | 95497.31 | 18 |
| SsHox27 | willow_GLEAN_10013903 | chr07:5920647.5922183 | 1014 | 337 | 4.83 | 37882.69 | 3 |
| SsHox28 | willow_GLEAN_10014069 | chr01:4791570.4792971 | 1038 | 345 | 8.69 | 38266.97 | 4 |
| SsHox29 | willow_GLEAN_10014688 | chr16:11311833.11318838 | 2556 | 851 | 6.06 | 93492.87 | 18 |
| SsHox30 | willow_GLEAN_10014867 | chr16:7124426.7129402 | 2412 | 803 | 5.52 | 89332.98 | 11 |
| SsHox31 | willow_GLEAN_10015140 | chr17:4534451.4535861 | 732 | 243 | 5.91 | 27663.95 | 2 |
| SsHox32 | willow_GLEAN_10015458 | chr08:6462504.6465411 | 699 | 232 | 8.54 | 25539.89 | 4 |
| SsHox33 | willow_GLEAN_10016083 | chr14:3617075.3622033 | 2499 | 832 | 5.76 | 90238.59 | 9 |
| SsHox34 | willow_GLEAN_10016271 | chr14:5066221.5067038 | 702 | 233 | 5.26 | 26708.69 | 2 |
| SsHox35 | willow_GLEAN_10016696 | chr14:1979359.1983173 | 1017 | 338 | 8.38 | 37672.41 | 5 |
| SsHox36 | willow_GLEAN_10016966 | chr02:11557986.11558804 | 711 | 236 | 4.88 | 27134.03 | 2 |
| SsHox37 | willow_GLEAN_10017080 | chr01:20984214.20990008 | 2634 | 877 | 6.03 | 96673.33 | 17 |
| SsHox38 | willow_GLEAN_10017524 | chr01:22569925.22570446 | 522 | 173 | 8.42 | 20055.49 | 1 |
| SsHox39 | willow_GLEAN_10017797 | chr16:8705805.8707465 | 906 | 301 | 8.84 | 33448.78 | 4 |
| SsHox40 | willow_GLEAN_10019718 | chr12:1153935.1157059 | 2127 | 708 | 6.19 | 77667.02 | 10 |
| SsHox41 | willow_GLEAN_10020038 | chr02:14549645.14553648 | 2283 | 760 | 5.49 | 83273.13 | 11 |
| SsHox42 | willow_GLEAN_10020281 | chr14:7939892.7944022 | 2280 | 759 | 5.57 | 82998.92 | 11 |
| SsHox43 | willow_GLEAN_10020955 | chr02:6588033.6589139 | 939 | 312 | 6.28 | 35742.99 | 3 |
| SsHox44 | willow_GLEAN_10021056 | chr02:7575862.7576910 | 807 | 268 | 8.02 | 30076.02 | 3 |
| SsHox45 | willow_GLEAN_10021385 | chr10:6571681.6573779 | 678 | 225 | 8.28 | 25419.91 | 4 |
| SsHox46 | willow_GLEAN_10021964 | chr03:2109069.2113988 | 1998 | 665 | 6.88 | 73596.48 | 10 |
| SsHox47 | willow_GLEAN_10021975 | chr03:2292482.2298568 | 2418 | 805 | 6.18 | 88596.3 | 17 |
| SsHox48 | willow_GLEAN_10022161 | chr03:4404415.4406099 | 909 | 302 | 8.2 | 33782.95 | 4 |
| SsHox49 | willow_GLEAN_10022766 | chr09:1442600.1449942 | 2688 | 895 | 5.97 | 98426.55 | 18 |
| SsHox50 | willow_GLEAN_10022837 | chr09:2382283.2383617 | 1047 | 348 | 6.33 | 38307.18 | 4 |
| SsHox51 | willow_GLEAN_10023610 | chr16:3863371.3864068 | 582 | 193 | 9.26 | 22556.03 | 2 |
| SsHox52 | willow_GLEAN_10025698 | chr03:5546441.5550860 | 2427 | 808 | 5.53 | 89816.92 | 11 |
| SsHox53 | willow_GLEAN_10025934 | chr06:11760197.11761563 | 1020 | 339 | 9.49 | 37438.11 | 4 |
| SsHox54 | willow_GLEAN_10026270 | chr06:14407519.14413810 | 2436 | 811 | 5.8 | 89369.02 | 18 |
| SsHox55 | willow_GLEAN_10026836 | chr06:3548298.3549461 | 648 | 215 | 8.79 | 24778.76 | 3 |
Phylogenetic analysis and chromosomal distribution of the HD-Zip gene family
To further understand the evolutionary relationships among HD-Zip family members, a phylogenetic tree was constructed using the 44 Arabidopsis HD-Zip proteins, 55 maize HD-Zip proteins, 63 poplar HD-Zip proteins and 55 willow HD-Zip proteins identified in this study. Using MEGA 11.0, the phylogenetic tree was built using the Maximum Likelihood (ML) and Neighbor-Joining (NJ) methods, respectively. With the exception of a few modest adjustments at internal branches, the tree topologies generated by the two algorithms were essentially similar. The classification results were the same for both methods, the NJ phylogenetic tree is shown in Fig. 1, and the ML phylogenetic tree is shown in Fig. S1. The result showed that the willow HD-Zip gene family can be divided into four subfamilies (I-IV), each containing 20, 14, 8, and 13 members of the willow HD-Zip gene family, respectively (Fig. 1A). According to the classification of maize, Arabidopsis and poplar, subfamily I was further subdivided into eight clades, designated α, β1, β2, γ, δ, ε, ζ and φ. And the clade β1 and ζ did not contain any willow or poplar HD-Zip genes (Fig. S2). The results were consistent with those of Arabidopsis and poplar, with subfamily I among them being the largest and subfamily III the smallest (Fig. 1B). However, the HD-Zip gene members in maize were the largest in the subfamily II. In addition, the phylogenetic tree showed that PtHox and SsHox genes were evenly clustered together, indicating that poplar and willow are closely related.
Fig. 1.
Phylogenetic tree of HD-Zip genes from willow, Arabidopsis, poplar and maize. (A) Classification of HD-Zip gene family based on phylogenetic tree. 55 SsHD-Zip genes, 44 AtHD-Zip genes, 63 PtHD-Zip genes and 55 ZmHD-Zip genes are clustered into four subfamilies (I-IV). HD-Zip genes from S. suchowensis, Arabidopsis, poplar and maize are denoted by red, blue, green and yellow shape, respectively. The tree was generated with the Clustal X 2.0 software using the neighbor-joining (N-J) method. (B) The number distribution of HD-Zip gene family in four species
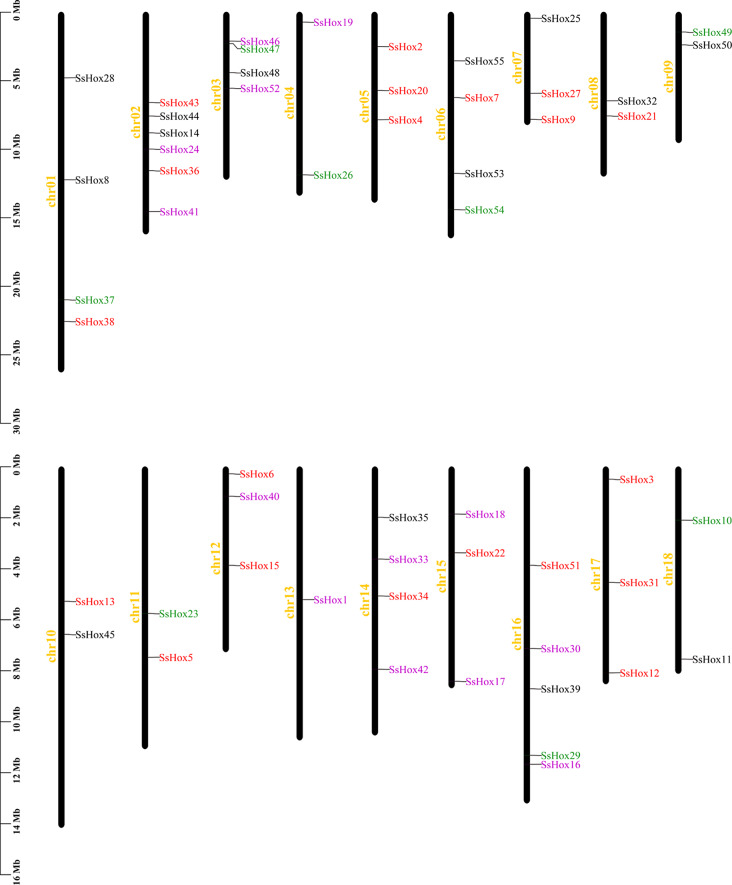
According to the SsHoxs genome annotation file obtained in the willow database, the locations of the 55 willow HD-Zips on the chromosomes were mapped (Fig. 2). The result demonstrated that, with the exception of chromosome 19, the 55 SsHox genes were randomly and unevenly distributed on 18 of the 19 chromosomes. Furthermore, Chr02 contains the maximum number of SsHox genes with a total of six, while the chr13 had only one gene. Specifically, both chr05 and chr17 had three HD-Zip genes, all belonging to subfamily I.
Fig. 2.
Chromosomal location of HD-Zip genes in willow. The 55 SsHD-Zip genes are widely mapped to 18 chromosomes of S. suchowensis. Different colors represent different subfamilies
Conserved motifs, domains, and gene structural analysis
Motif Elicitation tool was used to predict the conserved motif of the SsHox protein, and 20 different motifs were identified. Each motif sequence identified from the MEME was annotated using the Pfam and Smart websites, and it was found that Motif1 and Motif2 encoded the HD domain, Motif3, Motif4 and Motif6 encoded the START domain, Motif5 encoded the Zip domain, and Motif7 encoded the MEKHLA domain (Table S2). Figure 3A showed that Motif1 and Motif2 were common to all members of the willow HD-Zip family, while HD-Zip genes of the same subfamily contained similar number and type of motifs. Moreover, HD-Zip I and HD-Zip II subfamily members have similar domains and contain fewer simple motifs, while HD-Zip III and HD-Zip IV subfamily members had more motifs and more complex domain. Domain pattern analysis also revealed that HD domain was highly conserved in all SsHox proteins (Fig. S3). Subfamily I and II possessed HD and LZ domains, while certain members of subfamily II also contained the N-terminal domain. The START domain were present in HD-Zip genes of the subfamilies III and IV, whereas the MEKHLA domain was specific to subfamily III.
Fig. 3.
Gene structure and Conserved motifs of HD-Zip genes in willow. (A) Conserved motifs of HD-Zip genes in S. suchowensis. Distribution of the 20 conserved motifs in the SsHD-Zip genes following analysis by MEME tool. The different-colored boxes represent different motifs and their position in each protein sequence of SsHD-Zip. (B) Gene structure of HD-Zip genes in S. suchowensis. Exons are indicated by green rectangles. Gray lines connecting two exons represent introns
Exon/intron structure analysis was performed on the HD-Zip gene of S. suchowensis to understand its structural diversity (Fig. 3B). The number of introns of the 55 SsHox genes ranged from 1 to 18, with no significant differences in the number of introns in the same subfamilies with similar structures. The subfamilies I and II had a simple gene structure with a small number of introns (1–6), while the SsHoxs in subfamily III contained 17 or 18. In addition, subfamily IV genes ranged in intron number from 2 to 11, with the majority of genes having 10 or 11 introns.
Gene replication and collinearity analysis of HD-Zip gene family
To explore the evolutionary mechanism of the HD-Zip gene family in S. suchowensis, replication events in the willow genome were analyzed. A total of 36 pairs of homologous genes were obtained by the MCScanX method (Fig. 4A). These results suggested that segmental duplication might play a key role in the amplification of the SsHD-Zip gene family. Meanwhile, collinearity analysis was performed on the willow and three other plants (Fig. 4B). The results showed that there were 83 collinearity pairs of HD-Zip genes were identifed in Arabidopsis and willow, and the HD-Zip genes involved accounted for more than 72% of each genome. Moreover, collinearity analyses revealed that about 135 ccollinearity pairs were found between willow and poplar, and only SsHox1 and SsHox12 were not in the collinear regions.In addition, only 27 collinearity pairs were detected in the willow and maize, and none of the HD-Zip III members in willow were in the collinear regions. According to the results, poplar and willow had a closer genetic relationship and dicotyledons plants had a higher homology of the HD-Zip gene.
Fig. 4.
Collinearity analysis. (A) Collinearity analysis of HD-Zip gene in S. suchowensis. (B) HD-Zip gene collinearity between willow and other species genomes
In order to analyze the influence of selective pressure on the evolution of the HD-Zip gene family, we analyzed the Ka/Ks ratio of paralogs and orthologs in four species (Table S3). The Ka/Ks ratio for all paralogues ranged from 0.07 to 0.53, while the Ka/Ks ratio of all orthologues was less than 1, indicating that the HD-Zip genes in willow had undergone purifying selection pressure and and had high conservation.
GO annotation and promoter element analysis
The GO annotation using a cut-off value of P < 0.05 showed that a total of 149 GO items were enriched (Table S4). The results were divided into three categories: molecular function, biological process, and cellular component. In Fig. 5A, more than 90% of the terms were categorized into biological process. Analysis of the cell component annotation revealed that HD-Zip proteins were mainly located in the nucleus, which was consistent with the prediction of subcellular localization. It was also observed that some HD-Zip genes were assigned to categories associated with development, hormones, and stress response (Fig. 5B). For example, 28 genes were classified in the “response to hormone” category, while 37, 42 and 44 genes were classified in the “response to osmotic stress”, “response to water deprivation” and “response to salt stress” categories, respectively.
Fig. 5.
GO enrichment analysis of 55 HD-Zip genes in willow. (A) the GO annotation using a cut-off value of P ≤ 0.05 showed that a total of 149 GO items, including molecular function, biological process, and cellular component. (B) Some SsHD-Zip genes assigned to the categories associated with development, hormone, and stress response. The color gradient represents the size of the Pvalue and the size of circular represents number of SsHD-Zip genes. The X-axis shows the ratio of the number of the SsHD-Zip genes to the total gene number in certain categories
Promoter cis-acting elements are functional elements that regulate gene expression. Figure 6 showed the categorization of cis-acting elements based on their functions is illustrated, which include hormones, various stresses, and plant growth and development response elements. Many cis-elements of hormones had been found, focusing on 10 hormone response components. Of these, more than 80% and 70% of HD-Zip genes contained ABA-responsive elements and MeJA-responsive elements, respectively. Moreover, 274 cis-elements related to abiotic and biotic stresses were identified, including anaerobic induction response element (49.6%), low temperature response element (12.8%), wound-responsive element (10.9%), drought induced response element (10.2%), and defense stress response (14.2%). In addition, 72 elements related to plant growth and development were identified in the promoter region of the HD-Zip gene in S. suchowensis, including CAT-box, GCN4-motif, O2 site, HD-Zip 1, circadian, MSA-like, RY-element and MBSI element.
Fig. 6.
Cis-acting elements analysis of HD-Zip genes in promoter region of willow. Number of each cis-acting element in the promoter region (2000 bp) of SsHD-Zip genes
Expression pattern of the HD-Zip I genes in S. suchowensis following various stresses
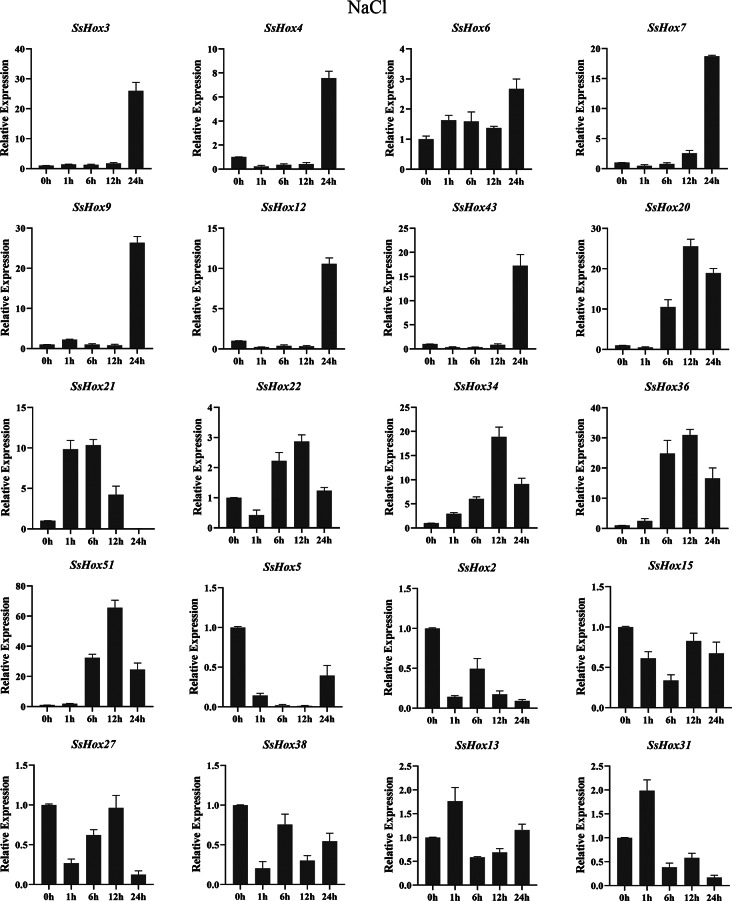
The HD-Zip I genes has been reported to play an important role in the plant’s response to abiotic stres [30, 31]. Therefore, we investigated the expression patterns of willow HD-Zip I genes under PEG, heat and NaCl treatments. During the NaCl stress, the seven genes were significantly upregulated at 24 h of treatment, including SsHox3, 4, 6, 7, 9, 12 and 43 (Fig. 7). However, five SsHoxs were down-regulated apparently. Additionally, SsHox34/-36, SsHox34/-51 and SsHox51/-36 showed the same expression pattern after NaCl treatment. For example, the expression of SsHox34 was up-regulated and reached a maximum at 12 h, but then gradually decreased. As for the heat stress (Fig. 8), SsHox5, SsHox15, SsHox27, and SsHox38 were downregulated by heat treatment across all time points. The expression level of 5 SsHox genes exhibited a rapid rapid strong up-regulation and peaked at 12 h after exposure to high temperature, but 9 SsHox genes were significantly up-regulated at 1 h. Interestingly, the expression of SsHox31 differed from other SsHox genes, being slightly down-regulated at the first time point, drastically up-regulated sixfold at 6 h, but then gradually decreased at subsequent time points. Additionally, qRT-PCR was performed to investigate the response to PEG treatment (Fig. 9). Of the 20 HD-Zip genes, 19 were up-regulated or down-regulated at some time points, while only SsHox6 was not expressed at all time points. Notably, SsHox20 was up-regulated more than 40-fold of drought stress. Furthermore, five paralogs (SsHox2/-27, SsHox3/-9, SsHox34/-36, SsHox34/-51 and SsHox51/-36) exhibited similar expression patterns in response to PEG treatment. For example, the expression level of SsHox3/-9 was significantly up-regulated and peaked at 24 h. However, the expression patterns of two paralogous genes (SsHox2/-43 and SsHox27/-43) were opposite. Like SsHox2/-43, SsHox2 was continuously down-regulated, whereas SsHox43 remained up-regulated at its maximum value.
Fig. 7.
Expression analysis of HD-Zip I genes following NaCl treatments by qRT-PCR. The Y-axis and X-axis indicates relative expression levels and the time courses of stress treatments, respectively. Mean values and standard deviations (SDs) were obtained from three biological and three technical replicates. The error bars indicate standard deviation
Fig. 8.
Expression analysis of HD-Zip I genes following heat treatments by qRT-PCR. The Y-axis and X-axis indicates relative expression levels and the time courses of stress treatments, respectively. Mean values and standard deviations (SDs) were obtained from three biological and three technical replicates. The error bars indicate standard deviation
Fig. 9.
Expression analysis of HD-Zip I genes following drought treatments by qRT-PCR. The Y-axis and X-axis indicates relative expression levels and the time courses of stress treatments, respectively. Mean values and standard deviations (SDs) were obtained from three biological and three technical replicates. The error bars indicate standard deviation
Correlations and coregulatory networks of SsHD-Zip I genes under various stresses
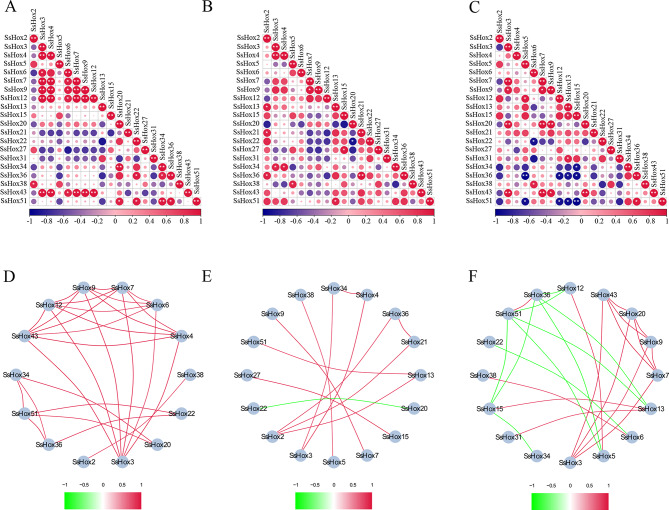
Based on the PCCs of their relative expression levels of HD-Zip I genes, correlation and coregulatory networks were constructed to examine the connections between genes in response to PEG, NaCl, and heat treatment. Under salt treatment, positive correlations and coregulatory network were observed between HD-Zip I genes. Under salt treatment, positive correlations (Pvalue ≤ 0.05 and 0.8 < PCC) were observed between SsHoxs, such as SsHox51, SsHox36, and SsHox22 (Fig. 10A and D). Among them, SsHox3, SsHox9, SsHox7, SsHox20, and SsHox43 also showed positive correlations with each other under the PEG treatments (Fig. 10C and F). Moreover, 9 gene pairs showed negative correlations (Pvalue ≤ 0.05 and − 1.0 < PCC< -0.8) in response to the PEG treatments. In addition, there was a significant positive correlation between 14 SsHoxs under heat stress (Fig. 10B and E), and only SsHox22/SsHox20 showed a negative correlation.
Fig. 10.
Correlations among HD-Zip I genes under NaCl, PEG and heat treatment. Correlation analysis of HD-Zip I genes under NaCl (A), heat (B) and PEG (C) treatment was performed based on the PCCs of qRT-PCR data. Correlations are indicated by the size and colour of circles. The lower bar represents the PCC values. * and ** represent correlations with P-value ≤ 0.05 and P-value ≤ 0.01, respectively. The coregulatory network of HD-Zip I genes under NaCl (D), heat (E) and PEG (F) treatment was illustrated by Cytoscape. The significant PCCs of gene pairs (P-value ≤ 0.05) are included, and the different correlation levels of the gene pairs are marked by edge lines with different colors, as shown below the coregulatory networks
Discussion
Plant transcription factors often interact with DNA and other proteins or transcription factors to promote or inhibit gene expression, thus participating in plant growth and development and response to stress [32]. With the development of molecular bioinformatics, the HD-Zip gene family has been identified in more and more plants. In this study, we used bioinformatics and qRT-PCR analyses to perform genome-wide analysis of willow HD-Zip genes to investigate their regulatory roles in stress response.
A total of 55 HD-Zip genes were identified in willow, distributed unevenly over 18 chromosomes (Figs. 1 and 2). There are 15 members of the HD-Zip I subfamily in Arabidopsis, 21 in poplar and 17 in maize [33, 34]. The genome sizes vary greatly between species, but the number of genes in the same subfamily is similar, indicating that the evolution of genes in this subfamily is relatively conservative. Previous studies have shown that the HD-Zip I genes in Arabidopsis and maize can be further subdivided into eight subclasses (α, β1, β2, γ, δ, ε, ζ and φ) (Fig. S2), and these subclasses may have a common origin in early organisms [35]. However, subclasses β1 and ζ did not contain HD-Zip I genes in poplar and willow. One of the ζ subclass contained only the maize HD-Zip I genes, suggesting that maize acquired an additional branch. According to the structural similarity, evolutionary relationship analysis and motif distribution of the HD-Zip gene family, the willow HD-Zip gene family could be divided into 4 subfamilies. Among these 4 subfamilies, the HD-Zip III subfamily had the least number of members. This was consistent with the findings in poplar, watermelon and Zanthoxylum armatum [33, 36, 37]. It was found that members of different subfamilies differ greatly in sequence length, exon number and conserved domain (Fig. 3). For example, SsHox37 (HD-Zip III) had 17 exons and the longest protein sequence, whereas SsHox12 (HD-Zip I) had 3 exons and the shortest protein sequence. In addition, the gene structure and conserved motifs of most genes in the same subfamily are similar, which may be related to the function and phylogenetic clustering of the family.
Collinearity analysis revealed that segmental duplications containing SsHoxs could be detected on all chromosomes except chromosome 13 and 19, indicating that segmental duplications were the main cause of the expansion of the SsHD-Zip gene family (Fig. 4). Similar results of collinearity analysis had been found in other species such as potato and peach [26, 38]. The collinear pairs of HD-Zip family genes between willow and poplar genomes were more than those between other genomes, indicating that willow and poplar were closely related. Moreover, genes with collinear relationships could also be grouped into the same class in the phylogenetic tree, indicating that these genes are relatively conserved in the process of genome evolution. These results may also be related to the conserved domains of these genes. Combined with the promoter analysis and GO annotation (Figs. 5 and 6), it was found that there were many cis-acting elements related to growth and development, stress response and hormone response in the promoters of many SsHD-Zip genes, and most of the genes were annotated to the enriched categories related to salt, osmotic stress and water deprivation response. It is speculated that the willow HD-Zip family may be transcriptionally regulated in adverse growth environments.
Many HD-Zip I subfamily genes have been reported to be involved in the regulation of abiotic stresses such as drought, salinity and temperature stress [6, 39]. For example, the HaHB4 gene of sunflower HD-Zip I subfamily regulated drought resistance through ethylene-mediated senescence, and the PeHDZ genes of Moso bamboo HD-Zip I subfamily were significantly induced by PEG and NaCl [40, 41]. In contrast to subfamilies II, III, and IV, the HD-Zip I subfamily plays an important role in response to abiotic stresses [1]. Therefore, we investigated the expression patterns of willow HD-Zip I genes in response to PEG, salt and heat treatment by qRT-PCR analysis, and the expression of most genes increased under stress (Figs. 7, 8 and 9). The expression levels of SsHox5, SsHox28 and SsHox23 were suppressed under all three stresses. On the contrary, SsHox3, SsHox7, SsHox9, SsHox36 and SsHox51 were strongly expressed under the different treatments. These five genes may play an important role in the response of willow to abiotic stress. SsHox36 and SsHox51 were homologous to AtHB7 and AtHB12, while two Arabidopsis genes were induced by ABA, drought, and salt stress, and improve plant drought resistance by influencing stomatal closure [42]. Moreover, SsHox36 and SsHox51 were homologous to PsnHDZ63 (Potri.002G176300.1), and overexpression of PsnHDZ63 confers salt tolerance in transgenic plants [43]. These results suggested that SsHox36 and SsHox51 may play essential roles in responses to PEG, heat and NaCl treatment. At the same time, some genes were discovered to be paralogous pairs whose expression levels differed significantly under a given stress treatment, in particular the expression patterns of SsHox3/-9, SsHox34/-36, SsHox34/-51 and SsHox51/-36 differed under all three stresses. It is possible that some paralogous pairs have functionally diverged during the evolutionary process. Under 37 °C high temperature treatment, the expression levels of SsHox36, SsHox3, SsHox4, SsHox7, SsHox9 and SsHox31 were strongly up-regulated (more than 4-fold) at some points. Similarly, 6 HD-Zip I genes of potato were differentially expressed in different tissues at high temperature (37 °C) [38]. Other HD-Zip genes such as HaHB1, AtHB13 and TaHDZIPI-5 had been implicated d in plant cold tolerance [44, 45].
Conclusion
In this study, 55 HD-Zip genes of S. suchowensis were identified, which were unevenly distributed on 18 out of 19 chromosomes. The willow HD-Zip genes were classified into four subfamilies using phylogenetic analysis and conserved domain analysis. Results from GO annotation and promoter analysis showed that the SsHox gene was controlled by a complex regulatory network. Furthermore, combining with the results of HD-Zip I gene expression analysis in willow, it was speculated that HD-Zip I gene played an important role in the resistance to abiotic stress, but the specific functions of each gene needed to be further studied. In this study, the whole genome of the willow HD-Zip gene family members was identified and analyzed, providing a theoretical basis for the subsequent functional verification of this gene family.
Materials and methods
Identification of putative HD-Zip genes in Salix suchowensis
The protein sequence of S.suchowensis were downloaded from the website (https://figshare.com/articles/dataset/Willow_gene_family/9878582/1?file=17720912, accessed 20 December 2022). The Hidden Markov model of the HD-Zip gene family domain (PF00046) from the Pfam database (http://pfam.xfam.org/, accessed 22 December 2022) were used to search the HD-Zip genes of S.suchowensis. And a local blast search of the S.suchowensis protein database was performed using the BlastP tool (E value-5) in TBtools, with 63 protein sequences of poplar HD-Zip as query sequences. After manual de-duplication, the 64 putative HD-Zip genes in S.suchowensis were obtained. Meanwhile, the NCBI Conserved Domain Database and the SMART database were used to verify 64 putative genes, leaving candidate genes that included the known conserved domains (HD domain). ExPASy (http://www.expasy.ch/tools/pi_tool.html, accessed 26 December 2022) and WoLP PSORT (https://wolfpsort.hgc.jp/, accessed 30 December 2022) were used to determine the molecular weight, isoelectric point and localization for each HD-Zip genes from S.suchowensis.
Multiple alignment and phylogenetic analysis
Multiple sequence alignments of the full-length HD-Zip protein sequences from Populus, Arabidopsis, maize and S.suchowensis were performed using ClustalW in MEGA 11.0.10 with default parameters. With default settings and a bootstrap value of 1000, we created a phylogenetic tree using the neighbor-joining method (NJ) and Maximum Likelihood (ML) in the MEGA 11.0.10 program [46].
Chromosomal distribution, collinearity and Ka/Ks analysis
The chromosomal locations of the HD-Zip genes in S.suchowensis were extracted from the GFF3 annotation file downloaded from the website (https://figshare.com/articles/dataset/Willow_gene_family/9878582/1?file=17720912, accessed 20 December 2022) and were displayed using TBtools-II v2.003 software [47]. Genome annotation file of Arabidopsis, maize and poplar were obtained from Phytozome database. One Step MCScanX-Super Fast in TBtools-II v2.003 was used to analyze the genome-wide collinearity between willow and three other species, and the collinear results were mapped using TBtools-II v2.003 with default settings [48]. The ratio of non-synonymous to synonymous substitutions (Ka/Ks) of orthologues and paralogues was calculated by TBtools-II v2.003.
Prediction of gene structure, conserved motifs, and cis-regulatory elements
The exon and intron location information of SsHD-Zip genes were extracted from the GFF3 annotation file, and the results were uploaded to the Gene Structure Display Server 2.0 website. To predict and analyze conserved protein motifs, all candidate SsHD-Zip protein sequences were uploaded to the MEME online tool [49]. A maximum of 20 motifs were set, while all other parameters were kept as default. The 2 kb sequences upstream of each SsHD-Zip gene were retrieved from the Genome annotation file. These sequences were then submitted to PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) for identification and prediction of Cis-elements.
Gene ontology annotation analysis
Gene ontology (GO) analysis was conducted for the SsHox genes using the agriGO database (http://systemsbiology.cau.edu.cn/agriGOv2/index.php, accessed 15 May 2023). The reference set consisted of all 26,599 genes of S.suchowensis, while the test set included 55 SsHox genes. The GO analysis diagram was generated using ChiPlot (chiplot.online).
Plant materials, growth conditions, and stress treatments
S. suchowensis were cultured by hydroponics (Hoagland’s nutrient solution) for six weeks in plant climate incubator (16 h light/8 h dark and 25 /22°C day/night). In order to investigate the expression profiles of SsHD-Zip I genes under abiotic stresses, the seedlings were treated under 200 mM NaCl, 20% (w/v) polyethylene glycol (PEG 6000), 37 °C, respectively. All leaves were harvested at 0, 1, 6, 12 and 24 h after each treatment, and then rapidly frozen in liquid nitrogen and stored at -80 °C for total RNA extraction.
RNA extraction and qRT-PCR analysis
Total RNA was extracted from the samples using the Aidlab plant RNA kit (Aidlab Biotech, Beijing, China) and the first-strand cDNA was synthesized using the UnionScript First-strand cDNA Synthesis Mix (Gensand, Bejing, China). The OTU (OTU-like cysteineprotease familyprotein) gene was used as the reference gene for salt stress, and UBC (Ubiquitin-conjugating enzyme E2) both for heat and drought treatment [50, 51]. All primers for qRT-PCR experiments were designed with Primer 5.0 and checked for primer specificity with TBtools (Table S5). Real-time PCR was performed on a CFX96TM Real-Time System (BIO-RAD, CA, USA) with TB Green Premix Ex Taq II (Tli RNaseH Plus; TaKaRa Biotechnology) in a sample volume of 10 µL. For each sample, we conducted three biological and three technical replicates. In addition, the relative expression level of each gene was calculated by standard 2−∆∆CT method was calculated [52].
Statistical and Pearson correlation analysis
The mean values and standard deviations (SDs) were calculated from three biological and three technical replicates. Pearson correlation coefficients (PCCs) and p-values of stress-induced SsHox gene pairs were obtained from the qRT-PCR results and plotted using the R package. The coexpression network was constructed in Cytoscape v3.9.1 [53] by including gene pairings with PCC values greater than 0.8 and significant at the 0.05 significance level (P-value). Significant differences are indicated at **P < 0.01 and *P < 0.05.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Not applicable.
Author contributions
Conceived and designed the experiments: YJW, JBZ and BLJ. Performed the experiments: YJW, CY and HJW. Analyzed the data: YJW, HJW, XMY and JSC. Wrote the paper: YJW. Participated in the design of this study and revised manuscript: YJW, JBZ and BLJ. The authors read and approved the final manuscript.
Funding
This work was supported by a grant from the research on key technology of standardised production and industrialisation of willow along Huaihe (202003b06020026).
Data availability
The genome sequences of Salix suchowensis were downloaded from the website (https://figshare.com/articles/dataset/Willow_gene_family/9878582/1?file=17720912, accessed 20 December 2022). The genome sequences of A. thaliana were downloaded from Phytozome database (https://phytozome-next.jgi.doe.gov/info/Athaliana_TAIR10, accessed 22 December 2022). The genome sequences of rice were downloaded from Phytozome database (https://phytozome-next.jgi.doe.gov/info/Osativa_v7_0, accessed 22 December 2022). The genome sequences of maize were downloaded from Phytozome database (https://phytozome-next.jgi.doe.gov/info/Zmays_RefGen_V4, accessed 22 December 2022). The genome sequences of poplar were downloaded from Phytozome database https://phytozome-next.jgi.doe.gov/info/Ptrichocarpa_v4_1, accessed 20 December 2022). The datasets supporting the results of this article are included in the article and Additional files.
Declarations
Ethics approval and consent to participate
Experimental research and field studies on plants including the collection of plant material are comply with relevant guidelines and regulation.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yujiao Wang and Hongjuan Wang contributed equally to this work.
Contributor Information
Benli Jiang, Email: 543806899@qq.com.
Jiabao Zhu, Email: 13955611798@139.com.
References
- 1.Ariel FD, Manavella Pa Fau - Dezar CA, Dezar Ca Fau -, Chan RL, Chan RL. The true story of the HD-Zip family. Trends Plant Sci 2007, 12(9):419–426. [DOI] [PubMed]
- 2.Viola IL, Gonzalez DH. Structure and evolution of Plant Homeobox genes. Plant Transcription Factors; 2016.
- 3.Sharif R, Raza A, Chen P, Li Y, El-Ballat EM, Rauf A, Hano C, El-Esawi MAJG. HD-ZIP Gene Family: potential roles in improving Plant Growth and regulating stress-responsive mechanisms in plants. Genes (Basel) 2021;12(8):1256. doi: 10.3390/genes12081256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Côté CL, Boileau F, Fau - Roy V, Roy V, Fau - Ouellet M, Ouellet M, Fau - Levasseur C, Levasseur C, Fau - Morency M-J. Morency Mj Fau - Cooke JEK, Cooke Je Fau - Séguin A, Séguin A Fau - MacKay JJ, MacKay JJ: Gene family structure, expression and functional analysis of HD-Zip III genes in angiosperm and gymnosperm forest trees. BMC Plant Biol. 2010;10:1471–2229. doi: 10.1186/1471-2229-10-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pandey A, Misra P, Alok A, Kaur N, Sharma S, Lakhwani D, Asif MH, Tiwari S, Trivedi PK. Genome-wide identification and expression analysis of homeodomain leucine Zipper Subfamily IV (HDZ IV) Gene Family from Musa Accuminata. Front Plant Sci. 2016;7:20. doi: 10.3389/fpls.2016.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gong S, Ding YA-O, Hu S, Ding L, Chen Z, Zhu C. The role of HD-Zip class I transcription factors in plant response to abiotic stresses. Physiol Plant. 2019;167(4):516–25. doi: 10.1111/ppl.12965. [DOI] [PubMed] [Google Scholar]
- 7.Li Y, Yang Z, Zhang Y, Guo J, Liu L, Wang C, Wang B, Han G. The roles of HD-ZIP proteins in plant abiotic stress tolerance. Front Plant Sci. 2022;13:1027071. doi: 10.3389/fpls.2022.1027071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang S, Haider I, Fau - Kohlen W, Kohlen W, Fau - Jiang L, Jiang L, Fau - Bouwmeester H, Bouwmeester H, Fau - Meijer AH, Meijer Ah Fau . Schluepmann H, Schluepmann H. Fau - Liu C-M, Liu Cm Fau - Ouwerkerk PBF, Ouwerkerk PB: function of the HD-Zip I gene Oshox22 in ABA-mediated drought and salt tolerances in rice. Plant Mol Biol. 2012;80(6):571–85. doi: 10.1007/s11103-012-9967-1. [DOI] [PubMed] [Google Scholar]
- 9.Zhao Y, Ma Q, Jin X, Peng X, Liu J, Deng L, Yan H, Sheng L, Jiang H, Cheng B. A novel maize homeodomain-leucine zipper (HD-Zip) I gene, Zmhdz10, positively regulates drought and salt tolerance in both rice and Arabidopsis. Plant Cell Physiol. 2014;55(6):1142–56. doi: 10.1093/pcp/pcu054. [DOI] [PubMed] [Google Scholar]
- 10.Li S, Chen N, Li F, Mei F, Wang Z, Cheng X, Kang Z, Mao HA-O. Characterization of wheat homeodomain-leucine zipper family genes and functional analysis of TaHDZ5-6A in drought tolerance in transgenic Arabidopsis. BMC Plant Biol. 2020;20(1):50. doi: 10.1186/s12870-020-2252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao S, Wang H, Jia X, Gao H, Mao K, Ma FA-O. The HD-Zip I transcription factor MdHB7-like confers tolerance to salinity in transgenic apple (Malus domestica) Physiol Plant. 2021;173(3):1452–64. doi: 10.1111/ppl.13330. [DOI] [PubMed] [Google Scholar]
- 12.Wang K, Xu L, Wang Y, Ying J, Li J, Dong J, Li C, Zhang X, Liu LA-O. Genome-wide characterization of homeodomain-leucine zipper genes reveals RsHDZ17 enhances the heat tolerance in radish (Raphanus sativus L) Physiol Plant. 2022;174(5):e13789. doi: 10.1111/ppl.13789. [DOI] [PubMed] [Google Scholar]
- 13.Elhiti M, Stasolla C. Structure and function of homodomain-leucine zipper (HD-Zip) proteins. Plant Signal Behav. 2009;4(2):86–8. doi: 10.4161/psb.4.2.7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruberti I, Sessa G, Fau - Ciolfi A, Ciolfi A, Fau - Possenti M, Possenti M, Fau - Carabelli M, Carabelli M, Fau - Morelli G, Morelli G. Plant adaptation to dynamically changing environment: the shade avoidance response. Biotechnol Adv. 2012;30(5):1047–58. doi: 10.1016/j.biotechadv.2011.08.014. [DOI] [PubMed] [Google Scholar]
- 15.Ciarbelli AR, Ciolfi A, Fau - Salvucci S, Salvucci S, Fau - Ruzza V, Ruzza V, Fau - Possenti M, Possenti M, Fau - Carabelli M, Carabelli M, Fau - Fruscalzo A, Fruscalzo A, Fau - Sessa G, Sessa G. Fau - Morelli G, Morelli G Fau - Ruberti I, Ruberti I: the Arabidopsis homeodomain-leucine zipper II gene family: diversity and redundancy. Plant Mol Biol. 2008;68(4):465–78. doi: 10.1007/s11103-008-9383-8. [DOI] [PubMed] [Google Scholar]
- 16.Carabelli M, Possenti M, Fau - Sessa G, Sessa G, Fau - Ciolfi A, Ciolfi A, Fau - Sassi M, Sassi M, Fau - Morelli G, Morelli G. Fau - Ruberti I, Ruberti I: canopy shade causes a rapid and transient arrest in leaf development through auxin-induced cytokinin oxidase activity. Genes Dev. 2007;21(15):1863–8. doi: 10.1101/gad.432607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sorin C, Salla-Martret M, Fau - Bou-Torrent J, Bou-Torrent JF, Roig-Villanova I, Roig-Villanova I. Fau - Martínez-García JF, Martínez-García JF: ATHB4, a regulator of shade avoidance, modulates hormone response in Arabidopsis seedlings. Plant J. 2009;59(2):266–77. doi: 10.1111/j.1365-313X.2009.03866.x. [DOI] [PubMed] [Google Scholar]
- 18.Tognacca RS, Carabelli M, Morelli G, Ruberti I, Botto JF. ATHB2 is a negative regulator of germination in Arabidopsis thaliana seeds. Sci Rep. 2021;11(1):9688. doi: 10.1038/s41598-021-88874-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prigge MJ, Otsuga D, Fau - Alonso JM, Alonso Jm Fau . Ecker JR, Ecker Fau - Drews GN, Drews Gn Fau - Clark SE, Clark SE: class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell. 2005;17(1):61–76. doi: 10.1105/tpc.104.026161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao J, Chen J, Feng L, Wang Q, Li S, Tan X, Yang F, Yang W. HD-Zip III Gene Family: identification and expression profiles during Leaf Vein Development in soybean. Plants (Basel) 2022;11(13):1728. doi: 10.3390/plants11131728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Javelle M, Vernoud V, Fau - Depège-Fargeix N, Depège-Fargeix N, Fau - Arnould C, Arnould C, Fau - Oursel D, Oursel D, Fau - Domergue F, Domergue F, Fau - Sarda X, Sarda X, Fau - Rogowsky PM, Rogowsky PM. Overexpression of the epidermis-specific homeodomain-leucine zipper IV transcription factor outer cell Layer1 in maize identifies target genes involved in lipid metabolism and cuticle biosynthesis. Plant Physiol. 2010;154(1):273–86. doi: 10.1104/pp.109.150540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schrick K, Ahmad B, Nguyen HV. HD-Zip IV transcription factors: drivers of epidermal cell fate integrate metabolic signals. Curr Opin Plant Biol. 2023;75:102417. doi: 10.1016/j.pbi.2023.102417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie Q, Gao Y, Li J, Yang Q, Qu X, Li H, Zhang J, Wang T, Ye Z, Yang C. The HD-Zip IV transcription factor SlHDZIV8 controls multicellular trichome morphology by regulating the expression of Hairless-2. J Exp Bot. 2020;71(22):7132–45. doi: 10.1093/jxb/eraa428. [DOI] [PubMed] [Google Scholar]
- 24.Yang Q, Xiang W, Li Z, Nian Y, Fu X, Zhou G, Li L, Zhang J, Huang G, Han X, et al. Genome-wide characterization and expression analysis of HD-ZIP Gene Family in Dendrobium officinale. Front Genet. 2022;13:797014. doi: 10.3389/fgene.2022.797014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li L, Lv B, Zang K, Jiang Y, Wang C, Wang Y, Wang K, Zhao M, Chen P, Lei J, et al. Genome-wide identification and systematic analysis of the HD-Zip gene family and its roles in response to pH in Panax ginseng Meyer. BMC Plant Biol. 2023;23(1):30. doi: 10.1186/s12870-023-04038-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Z, Wu X, Zhang B, Xiao Y, Guo J, Liu J, Chen Q, Peng F. Genome-wide identification, bioinformatics and expression analysis of HD-Zip gene family in peach. BMC Plant Biol. 2023;23(1):122. doi: 10.1186/s12870-023-04061-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuzovkina YA, Quigley MF. Willows beyond wetlands: uses of Salix L. Species for Environmental projects. Water Air Soil Pollution. 2005;162(1/4):183–204. doi: 10.1007/s11270-005-6272-5. [DOI] [Google Scholar]
- 28.Qu Y, Bi C, He B, Ye N, Yin T, Xu LA. Genome-wide identification and characterization of the MADS-box gene family in Salix suchowensis. PeerJ. 2019;7:e8019. doi: 10.7717/peerj.8019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dai X, Hu Q, Cai Q, Feng K, Ye N, Tuskan GA, Milne R, Chen Y, Wan Z, Wang Z, et al. The willow genome and divergent evolution from poplar after the common genome duplication. Cell Res. 2014;24(10):1274–7. doi: 10.1038/cr.2014.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harris JC, Hrmova M, Lopato S, Langridge P. Modulation of plant growth by HD-Zip class I and II transcription factors in response to environmental stimuli. New Phytol. 2011;190(4):823–37. doi: 10.1111/j.1469-8137.2011.03733.x. [DOI] [PubMed] [Google Scholar]
- 31.Li Y, Bai B, Wen F, Zhao M, Xia Q, Yang DH, Wang G. Genome-wide identification and expression analysis of HD-ZIP I gene subfamily in Nicotiana tabacum. Genes (Basel) 2019;10(8):575. doi: 10.3390/genes10080575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakashima K, Ito Y, Fau - Yamaguchi-Shinozaki K, Yamaguchi-Shinozaki K. Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 2009;149(1):88–95. doi: 10.1104/pp.108.129791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu R, Chi X, Fau - Chai G, Chai G, Fau - Kong Y, Kong Y, Fau - He G, He G, Fau - Wang X, Wang X, Fau - Shi D, Shi D, Fau - Zhang D, Zhang D, Fau - Zhou G, Zhou G. Genome-wide identification, evolutionary expansion, and expression profile of homeodomain-leucine zipper gene family in poplar (Populus trichocarpa) PLoS ONE. 2012;7(2):e31149. doi: 10.1371/journal.pone.0031149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao Y, Zhou Y, Fau - Jiang H, Jiang H, Fau - Li X, Li X, Fau - Gan D, Gan D, Fau - Peng X, Peng X, Fau - Zhu S, Zhu S, Fau - Cheng B, Cheng B. Systematic analysis of sequences and expression patterns of drought-responsive members of the HD-Zip gene family in maize. PLoS ONE. 2011;6(12):e28488. doi: 10.1371/journal.pone.0028488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henriksson E, Olsson ASB, Johannesson H, Johansson H, Hanson J, Engström P, Söderman E. Homeodomain leucine Zipper Class I genes in Arabidopsis. Expression patterns and phylogenetic relationships. Plant Physiol. 2005;139(1):509–18. doi: 10.1104/pp.105.063461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan X, Yue Z, Pan X, Si F, Li J, Chen X, Li X, Luan F, Yang J, Zhang X et al. The HD-ZIP Gene Family in Watermelon: Genome-Wide Identification and Expression Analysis under Abiotic Stresses. Genes (Basel) 2022, 13(12). [DOI] [PMC free article] [PubMed]
- 37.Zhang X, Chen Z, Wang C, Zhou X, Tang N, Zhang WA-O, Xu F, Yang Z, Luo C, Liao Y, et al. Genome-wide identification of HD-ZIP gene family and screening of genes related to prickle development in Zanthoxylum Armatum. Plant Genome. 2023;16(1):e20295. doi: 10.1002/tpg2.20295. [DOI] [PubMed] [Google Scholar]
- 38.Li W, Dong J, Cao M, Gao X, Wang D, Liu B, Chen Q. Genome-wide identification and characterization of HD-ZIP genes in potato. Gene. 2019;697:103–17. doi: 10.1016/j.gene.2019.02.024. [DOI] [PubMed] [Google Scholar]
- 39.Perotti MF, Ribone PA, Chan RA-O. Plant transcription factors from the homeodomain-leucine zipper family I. Role in development and stress responses. IUBMB Life. 2017;69(5):280–9. doi: 10.1002/iub.1619. [DOI] [PubMed] [Google Scholar]
- 40.Manavella PA, Dezar Ca Fau . Bonaventure G, Bonaventure G, Fau - Baldwin IT, Baldwin It Fau . Chan RL, Chan RL. HAHB4, a sunflower HD-Zip protein, integrates signals from the jasmonic acid and ethylene pathways during wounding and biotic stress responses. Plant J. 2008;56(3):376–88. doi: 10.1111/j.1365-313X.2008.03604.x. [DOI] [PubMed] [Google Scholar]
- 41.Chen D, Chen Z, Wu M, Wang Y, Wang Y, Yan H, Xiang Y. Genome-wide identification and expression analysis of the HD-Zip Gene Family in Moso Bamboo (Phyllostachys edulis) J Plant Growth Regul. 2017;36(2):323–37. doi: 10.1007/s00344-016-9642-x. [DOI] [Google Scholar]
- 42.Ré DA, Capella M, Bonaventure G, Chan RL. Arabidopsis AtHB7 and AtHB12evolved divergently to fine tune processes associated with growth and responses to water stress. BMC Plant Biol. 2014;14(1):150. doi: 10.1186/1471-2229-14-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo Q, Jiang J, Yao W, Li L, Zhao K, Cheng Z, Han L, Wei R, Zhou B, Jiang T. Genome-wide analysis of poplar HD-Zip family and over-expression of PsnHDZ63 confers salt tolerance in transgenic Populus simonii × P.nigra. Plant Sci. 2021;311:111021. doi: 10.1016/j.plantsci.2021.111021. [DOI] [PubMed] [Google Scholar]
- 44.Yang Y, Luang S, Harris J, Riboni M, Li Y, Bazanova N, Hrmova MA-O, Haefele S, Kovalchuk N, Lopato S. Overexpression of the class I homeodomain transcription factor TaHDZipI-5 increases drought and frost tolerance in transgenic wheat. Plant Biotechnol J. 2018;16(6):1227–40. doi: 10.1111/pbi.12865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cabello JV, Arce Al Fau . Chan RL, Chan RL. The homologous HD-Zip I transcription factors HaHB1 and AtHB13 confer cold tolerance via the induction of pathogenesis-related and glucanase proteins. Plant J. 2012;69(1):141–53. doi: 10.1111/j.1365-313X.2011.04778.x. [DOI] [PubMed] [Google Scholar]
- 46.Tamura K, Stecher G, Kumar S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol Biol Evol. 2021;38(7):3022–7. doi: 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen C, Chen H, Zhang Y, Thomas HR, Xia R. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020;13(8):1194–202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 48.Wang Y, Tang H, Fau - Debarry JD, Debarry Jd Fau - Tan X, Tan X, Fau - Li J, Li J, Fau - Wang X, Wang X Fau - Lee T-h, Lee Th Fau - Jin, Jin H, Fau H, Marler B, Marler B, Fau - Guo H, Guo H et al. Fau - Kissinger JC: MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res 2012, 40(7):e49. [DOI] [PMC free article] [PubMed]
- 49.Bailey TL, Boden M, Fau - Buske FA, Buske Fa Fau - Frith M, Frith M, Fau - Grant CE, Grant Ce Fau -, Clementi L, Clementi L, Fau - Ren J, Ren J, Fau - Li WW, Li Ww Fau - Noble WS, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 2009, 37:W202-W208. [DOI] [PMC free article] [PubMed]
- 50.Li J, Jia H, Han X, Zhang J, Sun P, Lu M, Hu J. Selection of Reliable reference genes for Gene Expression Analysis under Abiotic stresses in the Desert Biomass Willow, Salix psammophila. Front Plant Sci. 2016;7:1505. doi: 10.3389/fpls.2016.01505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li J, Zhang JA-O, Jia H, Yue Z, Lu M, Xin X, Hu J. Genome-wide characterization of the sHsp Gene Family in Salix suchowensis reveals its functions under different abiotic stresses. Int J Mol Sci. 2018;19(10):3246. doi: 10.3390/ijms19103246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 53.Doncheva NA-O, Morris JA-O, Gorodkin J, Jensen LA-OX. Cytoscape StringApp: Network Analysis and Visualization of Proteomics Data. J Proteome Res. 2019;18(2):623–32. doi: 10.1021/acs.jproteome.8b00702. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome sequences of Salix suchowensis were downloaded from the website (https://figshare.com/articles/dataset/Willow_gene_family/9878582/1?file=17720912, accessed 20 December 2022). The genome sequences of A. thaliana were downloaded from Phytozome database (https://phytozome-next.jgi.doe.gov/info/Athaliana_TAIR10, accessed 22 December 2022). The genome sequences of rice were downloaded from Phytozome database (https://phytozome-next.jgi.doe.gov/info/Osativa_v7_0, accessed 22 December 2022). The genome sequences of maize were downloaded from Phytozome database (https://phytozome-next.jgi.doe.gov/info/Zmays_RefGen_V4, accessed 22 December 2022). The genome sequences of poplar were downloaded from Phytozome database https://phytozome-next.jgi.doe.gov/info/Ptrichocarpa_v4_1, accessed 20 December 2022). The datasets supporting the results of this article are included in the article and Additional files.