Abstract
The M1inv+ subclone of M1 group A streptococci that spread globally in the late 1980s and early 1990s was previously identified by restriction fragment length polymorphism (RFLP), M protein, and SpeA exotoxin sequence analyses. Strains representing this subclone were characterized with regard to carriage of bacteriophage and capacity to invade cultured human epithelial cells. The M1inv+ subclone was found to harbor two entirely different prophages, phage T13 and phage T14, which together supplement its genome with nearly 70 kb of DNA. Phage T14 encodes the SpeA exotoxin and is closely related to the classic converting phage T12. Plaque-forming characteristics and RFLP analyses of phages T13 and T14 were compared to each other and to phage T12. Other subclones of M1, isolated in the 1970s to the early 1980s, lacked both prophages. The M1inv+ subclone was previously reported to be efficiently internalized by human epithelial cells. This potential was confirmed and expanded by comparing a variety of clinical isolates. The capacity for high-frequency invasion of epithelial cells was not transmitted to a laboratory strain of group A streptococci by the above-mentioned bacteriophages.
Since 1985, numerous geographically and temporally restricted outbreaks of systemic infections caused by serotype M1 group A streptococci have been reported (1, 5, 16, 23, 24, 26–29, 32, 34, 36, 37). These outbreaks occurred on at least five continents of the world. Based on genomic restriction fragment length polymorphism (RFLP), we suggested that a specific subclone or clonal variant of this serotype, designated M1inv+, emerged in the mid-1980s to become the most-common M1 strain isolated from cases of uncomplicated pharyngitis (5). This same variant was also associated with the increased incidence of toxic shock in North America (5, 28), Scandinavia (24, 26, 34), and New Zealand (23). Other M1 subclones, based on distinctive RFLP, are designated M1inv− for ease of discussion. Although outbreaks of toxic shock associated with M1 infection continued to occur in the 1990s, health authorities began to recognize clusters of toxic shock and necrotizing fasciitis associated with other serotypes (2, 7, 8, 10). This observation is puzzling but could be explained by increased awareness of public health authorities, changes in the health status of susceptible populations, or the horizontal dissemination of an unidentified virulence factor for other serotypes.
The acquisition of plasmids, prophages, or transposons could account for differences in RFLP and virulence that distinguish M1 subclones. This streptococcal species seldom harbors plasmids or transposons, but most strains carry one or more prophages (18, 19, 35, 38, 40). To investigate this possibility, phages were isolated from an M1inv+ strain and compared to the bacteriophage T12, a known and well-studied vector of the speA toxin gene (19, 38, 40, 41). Preliminary data suggested that M1inv+ strains invade cultured human epithelial cells at a significantly higher frequency than M1inv− strains. This finding was confirmed, and the relationship between intracellular invasion and prophages carried by M1inv+ strains was investigated.
Streptococcal cultures were grown and stored as previously described (6). Fifty-eight group A streptococcal M1 serotype strains that were isolated from different areas of the United States and the world were used in this study. Among them, 27 strains were isolated from individuals with systematic streptococcal disease (toxic shock, sepsis, or necrotizing fasciitis). Thirty-one strains were isolated from the throats of patients with uncomplicated pharyngitis. Most strains were obtained from the World Health Organization Collaborating Center for Reference and Research on Streptococci at the University of Minnesota. Strains 89, 80, 68, and 62 are blood isolates that represent the predominant M1 subclone in Finland and Norway (24, 26, 34). The New Zealand strains, 88-711, 89-385, 85-1532, and 84-221, represent the RFLP Ia subclone and were obtained from D. Martin, Communicable Disease Center, Porirua, New Zealand (23, 28). The T253c strain, which was cured of endogenous plaque-forming bacteriophages (18, 41), was used as the recipient strain for phage propagation and the construction of lysogens. The T253c(phage T12) lysogen served as the donor strain for the preparation of phage T12 DNA. The serotype M12 strain CS24 was used as a noninvasive control in invasion assays (35). Mitomycin-induced phage lysates from M1inv+ strain 90-226 (9) were prepared as previously described (35, 40). Lysogens were obtained by streaking bacteria from turbid plaques onto Todd-Hewitt agar medium.
Chromosomal DNAs, used for Southern blots, were extracted from streptococci as previously described (6). Template chromosomal DNA for PCR was extracted by a rapid, microwave extraction method (3). DNA probes for Southern blots were labelled with digoxigenin. Hybridization conditions and the methods for detection of hybrids are described in the Genius System user’s guide (Boehringer Mannheim Biochemicals). Phage particles, extraction, and purification of phage DNA were described previously by Yu and Ferretti (40). The probe used to identify speA sequence was produced by PCR. The speA primers were speaFor (5′TTTAAATCTAGAGGAGA ACCCAGATATAAAATGGAGG3′) and speaRev (5′GACGATAAAA TAGTTGCTAAGCTACAAGCTCCTG3′) (6). The product from these primers extends beyond the 3′ end of the speA gene by 271 nucleotides. A gentamicin-penicillin resistance intracellular invasion assay was used to determine the frequency of internalization of streptococci by cultured A549 cells (22).
The M1inv+ subclone carries a T12-like prophage.
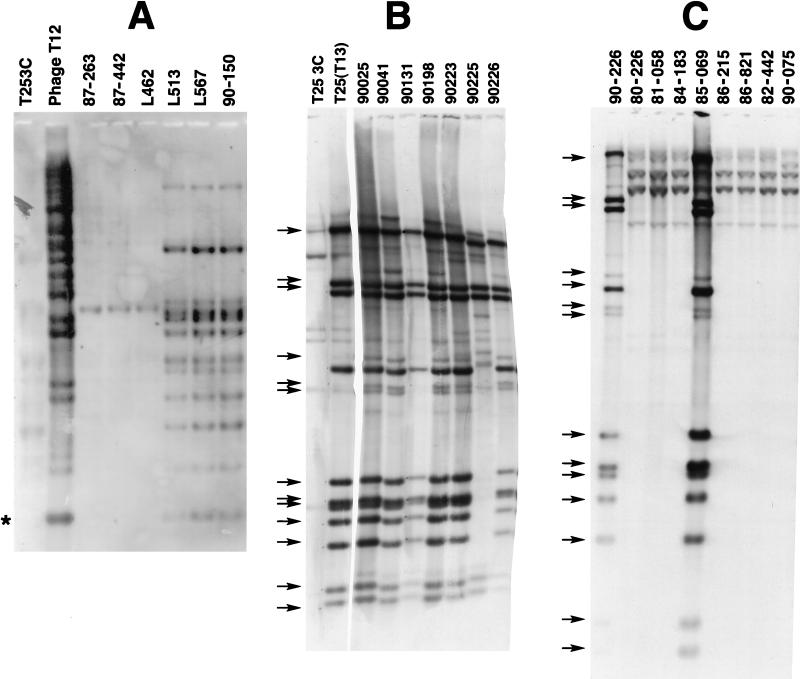
Earlier studies showed that the M1inv+ subclone harbored the speA gene (5, 27–29, 36); therefore, M1inv+ cultures were presumed to harbor a T12-like prophage that is not present in the M1inv− strains. To explore this possibility, Southern blots of genomic DNA from 58 M1 clinical isolates, with and without the invasive restriction profile (5), were probed with digoxigenin-labelled phage T12 DNA. Figure 1 shows representative results. Multiple HaeIII restriction fragments from 47 strains with the invasive restriction profile, whether isolated from systemic disease or uncomplicated infections, hybridized to the probe (Fig. 1A). Phage T12 DNA was included for comparison. The T12 and T14 bacteriophages are highly related but not identical (Fig. 1A and 2). Estimations of the size of the T14 genome obtained by summing the sizes of hybridizing fragments approached 36 kb. An asterisk marks a fragment of speA. At least two fragments are chromosomal junction fragments, so 36 kb exceeds the actual size of the phage T14 genome. DNAs from M1inv+ strains from Finland and New Zealand also harbored the same prophage. The hybridizing fragments were identical to those from North America, confirming that the M1inv+ subclone associated with serious disease was globally dispersed (data not shown). A recent analysis of Scandinavian strains showed that this strain waxed and waned in dominance and in being the cause of systemic infections between 1988 and 1995 (26). As predicted, DNA from 11 M1inv− strains lacked a T12-like prophage (Fig. 1B). Strain T253c DNA was included as a negative control. The phage T12 probe weakly hybridized to DNA fragments in T253c and M1inv− DNAs. These weaker signals were due to the fact that T253c, although cured of plaque-forming phage, retains a defective prophage that contaminates all phage lysates prepared in this strain (40). Yu et al. suggested that this or a related defective phage was present in most clinical isolates of group A streptococci (40).
FIG. 1.
Southern hybridization of restricted DNAs from M1inv+ and M1inv− clinical isolates to dioxigenin-labelled phage DNAs. Strain T253c has been cured of phage T12 and other plaque-forming phages. (A) HaeIII-digested DNAs were probed with digoxigenin-labelled phage T12 DNA. Strains 87-263, 87-442, and L462 are M1inv− isolates. The remaining strains are M1inv+. The asterisk marks an speA HaeIII fragment. (B) ClaI-digested DNAs from M1inv+ clinical isolates were probed with labelled phage T13 DNA. Strain T253c(T13) is a T13 lysogen that was purified from a single plaque. (C) ClaI-digested DNA from M1inv− strains probed with labelled phage T13 DNA. DNA from the M1inv+ strain 90-226 was included as a positive control. Strain 85-069 was designated M1inv− on the basis of RFLP (data not shown) and because it lacks phage T14.
FIG. 2.
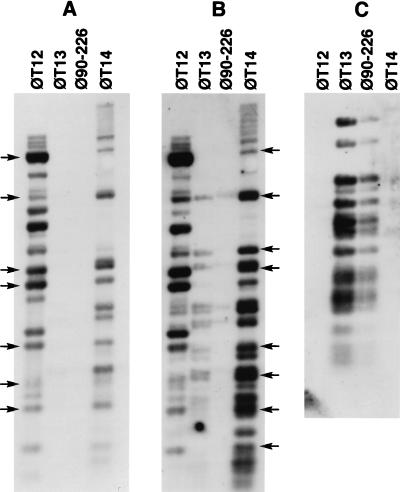
Southern hybridization of HaeIII-digested DNA purified from T12, T13, and T14 phage (φ) particles to labelled phage DNAs. DNA was probed with digoxigenin-labelled phage T12 DNA (A), labelled phage T14 DNA (B), and phage T13 DNA (C). Arrows mark phage T12-specific (A) and phage T14-specific (B) fragments.
M1inv+ strains contain a second plaque-forming bacteriophage.
The prophage carried by two M1inv+ cultures, strains 90-131 and 90-226, were induced by mitomycin and plaqued onto strain T253c. Both large and small turbid plaques were observed, and these plaques were easily distinguished from phage T12 plaques. Individual plaques from a mitomycin-induced culture of strain 90-226 were resuspended and replated onto strain T253c soft agar lawns. The turbid areas of single plaques were streaked onto agar media to obtain T253c lysogens, T253c(T13) from large, highly turbid plaques, and T253c(T14) from small, lightly turbid plaques. These lysogens were used for the production of high-titer lysates and phage DNA. Phage T12, T13, and T14 lysogens exhibited different immunities to superinfection (Table 1). Phage T12 formed plaques on M1inv+ cultures and on T253c(T13) and T253c(T14) lysogens. These phages also have somewhat different temperature optima for plaque formation. Phages T13 and T12 form plaques at 30 and 37°C, whereas phage T14 plaqued more efficiently at 30°C. Yu and coworkers described other T12-related phage with unique immunities and restriction fragment profiles, indicating a high level of genetic diversity among T12-like bacteriophages (40).
TABLE 1.
Abilityc of bacteriophages T12, T13, and T14 to form plaques on lysogenic host strains
| Phage | Plaque-forming ability on indicated host strain
|
||||||
|---|---|---|---|---|---|---|---|
| T25b | T25 | 90-226 | 90-131 | T25(T12) | T25(T13) | T25(T14) | |
| 90-226a | + | + | − | − | + | + | + |
| T12 | + | + | + | + | − | + | + |
| T13 | + | + | − | − | + | − | + |
| T14 | + | ± | − | − | + | + | − |
Lysates contained both T13 and T14.
These plates and those containing phage T14 were incubated at 30°C. Plates for all other strains were incubated at 37°C.
+, plaques formed; −, no plaques formed; ±, barely discernible.
Preparations of all three phages were contaminated with the defective phage harbored by T253c. Phages T12 and T13 produced very high-titered lysates, whereas phage T14 produced low-titered lysates that were heavily contaminated by the defective phage. Therefore, phage T14 DNA probes contained nearly equal amounts of DNA from the defective phage, complicating hybridization experiments. Genomic DNAs from M1inv+ and M1inv− clinical isolates were probed with labelled phage T13 DNA to determine whether both carried this prophage. Phage T13 DNA hybridized extensively to 13 ClaI fragments from 9 of 10 M1inv+ DNAs (Fig. 1B). Strain 90-225 was the exception; its T13 prophage lacked several fragments common to the other strains. As expected, T253c lacked the T13 prophage. The weakly hybridizing bands in T253c DNA are derived from the defective prophage. ClaI-digested genomic DNAs from M1inv− strains were also probed with phage T13 DNA (Fig. 1C). Seven of eight of these strains lacked T13 prophage DNA. M1inv− strain 85-069 contained a T13 prophage but did not harbor the T14 prophage, suggesting that phage T13 has been transmitted between different M1 subclones. The phage T13 probe hybridized weakly to four large ClaI fragments, suggesting that M1inv− strains carry a prophage that is related to phage T13 or to the T253c-defective phage. Since phage T14 DNA hybridizes to the same fragments, the latter explanation is likely to be correct. Genomic DNA from an M1inv+ strain, 90-226, was included as a positive control for T13 prophage.
Phages T12 and T14 are highly similar and unrelated to phage T13.
DNAs extracted from phage particles were probed in order to investigate genomic similarities between T12, T13, and T14 bacteriophages (Fig. 2). As expected, the T12 probe hybridized extensively to HaeIII digests of itself and phage T14 DNA but did not hybridize to phage T13 DNA (Fig. 2A). The T14 probe hybridized extensively to phage T12 DNA and to itself (Fig. 2B) but did not hybridize to phage DNA from 90-226 cultures because the ratio of phage T13 to T14 in induced 90-226 cultures approached 2 × 105:1. The few phage T14 HaeIII fragments that weakly hybridized to phage T13 DNA correspond to the defective-phage DNA that contaminates both DNA preparations. The phage T13 probe hybridized to itself and to a mixture of phage DNAs obtained from a mitomycin-induced culture of 90-226 (Fig. 2C). Phage T12 and T14 DNAs did not hybridize to the T13 DNA probe, indicating that these phages are unrelated to phage T13. Estimations of the genomic sizes of T14 and T13 genomes from hybridizing fragments on agarose gels were 36 and 34.5 kb, respectively.
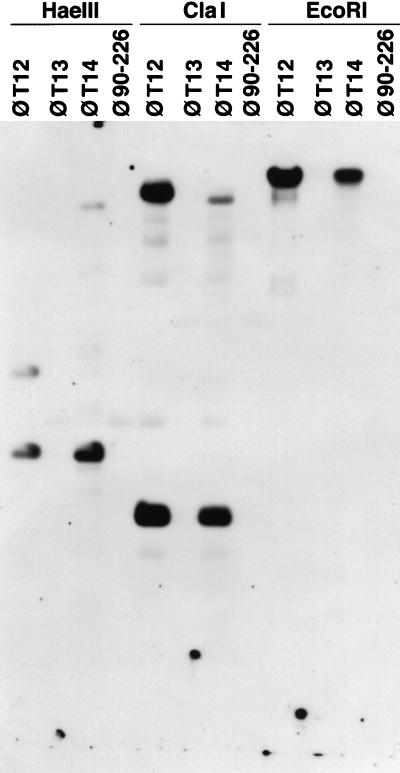
Phage DNAs were probed with a labelled fragment of the speA gene that was produced by PCR to determine which phage encoded the speA gene. HaeIII-, ClaI-, and EcoRI-digested phage T12 and T14 DNAs hybridized to the speA probe (Fig. 3). The speA gene contains one HaeIII site, one ClaI site, and no EcoRI sites (18, 38). As expected, two HaeIII fragments, two ClaI fragments, and one EcoRI fragment from the T12 and T14 DNAs produced signals. The smaller speA HaeIII and ClaI fragments were the same size in phages T12 and T14. The larger speA fragments differed in size and hybridization intensity, suggesting that sequence adjacent to one side of speA was not the same in both phages. Restricted phage T13 DNA did not hybridize to the speA probe under the same conditions. With longer exposures, a 3.1-kb ClaI fragment of T13 DNA hybridized weakly to the speA and phage T12 and T14 probes. This fragment was cloned and sequenced to determine its relationship to speA. The sequence revealed that cross-hybridization between phage T13 DNA and the speA probe was due to the presence of a 152-bp DNA segment that was 72% similar to a region immediately 3′ to speA (data not shown). While this result indicated that phage T13 did not possess an speA gene, it suggested that phage T13 may have acquired a host gene(s) that was closely linked to the ancestral speA gene acquired by phage T12.
FIG. 3.
Southern hybridization of restricted phage DNAs to a labelled PCR fragment that corresponds to a segment of the speA gene. Enzymes used for restriction digests are indicated at the top. Phage 90-226 (φ90-226), phage DNA isolated from mitomycin-induced cultures of strain 90-226. Although this strain harbors both T13 and T14 phages, induced lysates contain predominately T13 particles.
The discovery that M1inv+ strains carry two different prophages, contributing 70.5 kb of DNA sequence that is not present in M1inv− strains, further supports the claim that the former represents a phylogenetically distinct subclone. Numerous reports have documented reassortment and recombination between genetic loci in group A streptococci (4, 15, 31, 39). Although lateral movement of exotoxin genes speA (18, 38, 41) and speC (12) was clearly demonstrated to be mediated by converting temperate bacteriophages, it is not known which mechanisms are responsible for horizontal transfer of other chromosomal genes. Transposons and plasmids are seldom identified in group A streptococci. Since most, if not all, strains of group A streptococci contain temperate phage (40), it is reasonable to assume that they are the most-common vehicles of genetic exchange. Acquisition of one or more bacteriophages greatly increases the genetic potential of an organism and could substantially increase the fitness of a subclone in humans.
The M1inv+ subclone invades epithelial cells more efficiently than other M1 subclones.
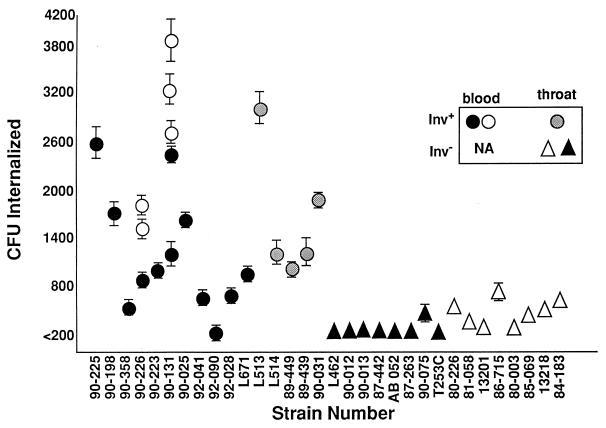
Group A streptococci have been shown to invade epithelial cells at a high frequency (9, 13, 14, 22, 33). Expression of M protein or a fibronectin binding protein was demonstrated to be sufficient for efficient ingestion of these streptococci (9, 14, 25). Cultured lung epithelial cells were shown to much more efficiently internalize an M1inv+ strain than an M1inv− culture (22), suggesting that prophage carried by the former may be responsible for the more-invasive phenotype. The capacities of a variety of M1inv+ and M1inv− clinical isolates to invade A549 cells were compared in order to confirm this initial observation (Fig. 4). Although the invasion frequency varied considerably from day to day, M1inv+ cultures invaded these epithelial cells at significantly higher frequencies than did M1inv− cultures. This variability prompted us to perform an experiment in which the genotype of the strains was concealed (Fig. 4, open symbols). The variability associated with strain 90-131 is the result of genetic instability in expression of the Vir regulon (6). High- and low-frequency internalization correlated with the genotype of the strain and did not reflect the source of the culture.
FIG. 4.
Internalization of M1 clinical isolates by lung epithelial A549 cells. Monolayers were infected with approximately 105 CFU of each culture. Internalized streptococci were determined by viable counts following 2 h of exposure to gentamicin and penicillin (22). Circles indicate M1inv+ strains. Stippled circles, M1inv+ strains isolated from patients with uncomplicated disease; triangles, M1inv− strains isolated from patients with uncomplicated pharyngitis. Strain 85-069 is M1inv− but harbors phage T13. Open circles and triangles represent strains whose genotypes were unknown to the investigator at the time of the experiment. NA, not available.
Serial passage of an M1inv+ strain through cultured epithelial cells enriched for streptococci that invaded cells at a higher frequency, whereas passage of an M1inv− strain had no effect on its invasive phenotype (6), suggesting that M1inv− strains lacked the genetic potential to revert to a highly invasive phenotype and prompting us to test whether the T13 or T14 prophage could increase the low frequency of invasion of a laboratory strain. This was explored by comparing the invasion efficiencies of T253c lysogens. No differences were observed between the frequency of the parent strain, T253c, and those of T253c(T13), T253c(T12), and T253c(T14), single lysogens, or that of T253c(T13, T14), a double lysogen.
Lysogeny by temperate bacteriophages can enhance bacterial virulence by a number of different mechanisms. Bacteriophages can encode proteins that contribute to adherence, serum resistance, and antigenic variation of bacterial hosts. The production of SpeA has been associated with severe streptococcal infection. Whether this toxin acts alone or in concert with other superantigens is debatable. Assuming that SpeA contributes to the pathogenesis of toxic shock, the dissemination of phage T14 or other T12-like phage has the potential to significantly influence the epidemiology of streptococcal infections. Whether this phage encodes additional factors that contribute to the propensity of streptococci to cause serious disease is unclear. The retention of phage T13 by M1inv+ strains suggests that this phage may also confer a selective advantage on streptococci.
Cockerill et al. discovered that 30% of schoolchildren in a southern Minnesota community carried the same subclone of M3 group A streptococci that was associated with an outbreak of invasive disease and several deaths among adults in the same region (7). Others have noted that the most-common subclone of a given serotype isolated from cases of uncomplicated pharyngitis is also the subclone that is most-often associated with invasive disease (17). The carriage of streptococci by healthy individuals and the frequent persistence of these bacteria in the throat after vigorous antibiotic therapy is well documented but not understood (11). No clinical isolate of group A streptococci has been documented to be genetically resistant to penicillin (20), yet 10 to 30% of individuals retain this organism in their throats after a 10-day course of penicillin. Our observation that M1inv+ strains are internalized into a penicillin-impervious compartment of cultured epithelial cells at a significantly higher frequency than other strains could account for the persistence and widespread dissemination of this subclone. A strain with greater potential to be internalized by mucosal or tonsillar tissue may better resist many antibiotics and continue to be spread to others in the community. Data reported by Österlund et al. are consistent with this explanation (30). These researchers demonstrated that 13 of 14 tonsils that were removed from children plagued by recurrent pharyngitis contained intracellular streptococci. Tonsils removed from healthy adults for other unrelated reasons did not contain intracellular streptococci. In a recent comparison of streptococcal isolates from a variety of patients, Molinari and Chhatwal observed that isolates from carriers invaded Hep2 cells at a higher frequency than blood isolates (25). We did not observe a correlation between the frequency with which epithelial cells are invaded and the origin of the culture. High-frequency invasion correlated only with genotype. Our collection, however, did not contain strains from carriers. Based on Molinari and Chhatwal’s study, we predict that carriers may shed the M1inv+ subclone more frequently than strains that are poorly internalized by epithelial cells.
The globally disseminated M1inv+ subclone, also termed Ia (28) and d3B (26), can be distinguished from other serotype M1 strains by its emm1, speB, speA, ska, and scpA alleles (21, 28). Moreover, this subclone also efficiently invades human epithelial cells and harbors two prophages, or nearly 70 kb of DNA sequence—potentially 50 to 60 genes. Both characteristics could affect the propensity of this subclone to cause serious infections and its dissemination in humans.
Acknowledgments
This study was financed by a grant from the Public Health Service (AI34503), and D.C. was supported by Public Health Service training grant AI07421.
We thank Patrick Schlievert, Edward Kapland, H. Seppälä, and Diane Martin for supplying us with M1 clinical isolates and Tim Leonard for his assistance with the presentation of our data.
REFERENCES
- 1.Abolnik I Z, Sexton D J. Necrotizing fasciitis and myositis caused by group A streptococci. Epidemiology, diagnosis, and treatment of “flesh-eating bacteria.”. NC Med J. 1994;55:464–466. [PubMed] [Google Scholar]
- 2.Beall B, Facklam R, Hoenes T, Schwartz B. Survey of emm gene sequences and T-antigen types from systemic Streptococcus pyogenes infection isolates collected in San Francisco, California; Atlanta, Georgia; and Connecticut in 1994 and 1995. J Clin Microbiol. 1997;35:1231–1235. doi: 10.1128/jcm.35.5.1231-1235.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bollet C, Gevaudan M J, de Lamballerie X, Zandotti C, de Micco P. A simple method for the isolation of chromosomal DNA from Gram+ or acid-fast bacteria. Nucleic Acids Res. 1991;19:1955. doi: 10.1093/nar/19.8.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cleary P P, Johnson D, Wannamaker L W. Genetic variation in the M antigen of group A streptococci: reassortment of type-specific markers and possible antigenic drift. J Infect Dis. 1979;140:747–757. doi: 10.1093/infdis/140.5.747. [DOI] [PubMed] [Google Scholar]
- 5.Cleary P P, Kaplan E L, Handley J P, et al. Clonal basis for resurgence of serious streptococcal disease in the 1980’s. Lancet. 1992;321:518–521. doi: 10.1016/0140-6736(92)90339-5. [DOI] [PubMed] [Google Scholar]
- 6.Cleary P P, McLandsborough L, Ikeda L, Cue D, Krawezak J, Lam H. High frequency intracellular infection and erythrogenic toxin A expression undergo phase variation in M1 group A streptococci. Mol Microbiol. 1998;28:157–167. doi: 10.1046/j.1365-2958.1998.00786.x. [DOI] [PubMed] [Google Scholar]
- 7.Cockerill F R, Kristine L, MacDonald M D, et al. An outbreak of invasive group A streptococcal disease associated with high carriage rates of the invasive clone among school-aged children. JAMA. 1997;277:38–43. [PubMed] [Google Scholar]
- 8.Coleman G, Tanna A, Efstratiou A, Gaworzewska E. The serotypes of Streptococcus pyogenes presented in Britain during 1980–1990 and their association with disease. J Med Microbiol. 1993;39:165–178. doi: 10.1099/00222615-39-3-165. [DOI] [PubMed] [Google Scholar]
- 9.Cue D, Dombek P E, Lam H, Cleary P P. Streptococcus pyogenes serotype M1 encodes multiple pathways for entry into human epithelial cells. Infect Immun. 1998;66:4593–4601. doi: 10.1128/iai.66.10.4593-4601.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gamba M A, Martinelli M, Schaad H J, et al. Familial transmission of a serious disease-producing group A streptococcal clone: case reports and reviews. Clin Infect Dis. 1997;24:1118–1121. doi: 10.1086/513636. [DOI] [PubMed] [Google Scholar]
- 11.Gerber M. Treatment failures and carriers: perception of problems? Pediatr Infect Dis J. 1994;13:576–579. doi: 10.1097/00006454-199406000-00036. [DOI] [PubMed] [Google Scholar]
- 12.Goshorn S C, Schlievert P M. Bacteriophage association of streptococcal pyrogenic exotoxin C. Infect Immun. 1989;171:3068–3073. doi: 10.1128/jb.171.6.3068-3073.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greco R, De Martino L, Donnarumma G, Conte M P, Seganti L, Valenti P. Invasion of cultured human cells by Streptococcus pyogenes. Res Microbiol. 1995;146:5551–5560. doi: 10.1016/0923-2508(96)80561-4. [DOI] [PubMed] [Google Scholar]
- 14.Fluckiger U, Jones K F, Fischetti V A. Immunoglobulins to group A streptococcal surface molecules decrease adherence to and invasion of human pharyngeal cells. Infect Immun. 1998;66:974–979. doi: 10.1128/iai.66.3.974-979.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harbaugh M P, Podbielski A, Hugl S, Cleary P P. Nucleotide substitutions and small-scale insertion produce size and antigenic variation in group A streptococcal M1 protein. Mol Microbiol. 1993;8:981–991. doi: 10.1111/j.1365-2958.1993.tb01642.x. [DOI] [PubMed] [Google Scholar]
- 16.Johnson D R, Stevens D L, Kaplan E L. Epidemiologic analysis of group A streptococcal serotypes associated with severe systemic infections, rheumatic fever, or uncomplicated pharyngitis. J Infect Dis. 1992;166:374–382. doi: 10.1093/infdis/166.2.374. [DOI] [PubMed] [Google Scholar]
- 17.Johnson D R, Romana C L, Rehder C D, Dehnbostel J, Kaplan E L. Restriction enzyme analysis (REA) of group A streptococcal (GAS) M-serotypes 1, 3, and 28. In: Horaud T, editor. Streptococci and the host. New York, N.Y: Plenum Press; 1997. pp. 221–223. [PubMed] [Google Scholar]
- 18.Johnson L P, Schlievert P M. A physical map of the group A streptococcal pyrogenic exotoxin bacteriophage genome. Mol Gen Genet. 1983;189:251–255. doi: 10.1007/BF00337813. [DOI] [PubMed] [Google Scholar]
- 19.Johnson L P, Schlievert P M. Group A streptococcal phage T12 carries the structural gene for pyrogenic exotoxin type A. Mol Gen Genet. 1984;194:52–56. doi: 10.1007/BF00383496. [DOI] [PubMed] [Google Scholar]
- 20.Kaplan E L. Recent evaluation of antimicrobial resistance in β-hemolytic streptococci. Clin Infect Dis. 1997;24:S89–S92. doi: 10.1093/clinids/24.supplement_1.s89. [DOI] [PubMed] [Google Scholar]
- 21.Kapur V, Kanjilal S, Hamrick M R, et al. Molecular population analysis of the streptokinase gene of Streptococcus pyogenes: mosaic alleles generated by recombination. Mol Microbiol. 1995;16:509–519. doi: 10.1111/j.1365-2958.1995.tb02415.x. [DOI] [PubMed] [Google Scholar]
- 22.LaPenta D, Rubens C, Chi E, Cleary P P. Group A streptococci efficiently invade human respiratory epithelial cells. Proc Natl Acad Sci USA. 1994;91:12115–12119. doi: 10.1073/pnas.91.25.12115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin D R, Single L A. Molecular epidemiology of group A streptococcus M type 1 infections. J Infect Dis. 1993;167:1112–1117. doi: 10.1093/infdis/167.5.1112. [DOI] [PubMed] [Google Scholar]
- 24.Martin P R, Hoiby E A. Streptococcal serogroup A epidemic in Norway 1987–1988. Scand J Infect Dis. 1990;22:421–429. doi: 10.3109/00365549009027073. [DOI] [PubMed] [Google Scholar]
- 25.Molinari G, Chhatwal G. Invasion and survival of Streptococcus pyogenes in eukaryotic cells correlates with the source of the clinical isolates. J Infect Dis. 1998;177:1600–1607. doi: 10.1086/515310. [DOI] [PubMed] [Google Scholar]
- 26.Muotiala H, Seppälä H, Huovinen P, Vuopio-Varkila J. Molecular comparison of group A streptococci of T1M1 serotype from invasive and noninvasive infections in Finland. J Infect Dis. 1997;175:392–399. doi: 10.1093/infdis/175.2.392. [DOI] [PubMed] [Google Scholar]
- 27.Musser J M, Hauser A R, Kim M H, Schlievert P M, Nelson K, Selander R K. Streptococcus pyogenes causing toxic shock-like syndrome and other invasive diseases: clonal diversity and pyrogenic exotoxin expression. Proc Natl Acad Sci USA. 1991;88:2668–2672. doi: 10.1073/pnas.88.7.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Musser J M, Kapur V, Szeto J, Pan X, Swanson D S, Martin D R. Genetic diversity and relationships among Streptococcus pyogenes strains expressing serotype M1 protein: recent intercontinental spread of a subclone causing episodes of invasive disease. Infect Immun. 1995;63:994–1003. doi: 10.1128/iai.63.3.994-1003.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nelson K, Schlievert P M, Selander R K, Musser J M. Characterization and clonal distribution of four alleles of the speA gene encoding pyrogenic exotoxin A (scarlet fever toxin) in Streptococcus pyogenes. J Exp Med. 1991;174:1271–1274. doi: 10.1084/jem.174.5.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Österlund A, Popa R, Nikkila T, Scheynius A, Engstrand L. Intracellular reservoir of Streptococcus pyogenes in vivo: a possible explanation for recurrent pharyngotonsillitis. Laryngoscope. 1997;107:640–647. doi: 10.1097/00005537-199705000-00016. [DOI] [PubMed] [Google Scholar]
- 31.Podbielski A, Hawlitzky J, Pack T D, Flosdorff A. A group A streptococcal enn protein potentially resulting from intergenomic recombination exhibits atypical immunoglobulin-binding characteristics. Mol Microbiol. 1994;12:725–736. doi: 10.1111/j.1365-2958.1994.tb01060.x. [DOI] [PubMed] [Google Scholar]
- 32.Schlievert P M, Assimacopoulos A P, Cleary P P. Severe invasive group A streptococcal disease: clinical description and mechanisms of pathogenesis. J Lab Clin Med. 1996;127:13–22. doi: 10.1016/s0022-2143(96)90161-4. [DOI] [PubMed] [Google Scholar]
- 33.Schrager H M, Wessels M R. Hyaluronic acid capsule modulates interactions of group A streptococci with human epidermal keratinocytes. Adv Exp Med Biol. 1997;418:517–523. doi: 10.1007/978-1-4899-1825-3_122. [DOI] [PubMed] [Google Scholar]
- 34.Seppälä H, Vuopio-Varkila J, Osterblad M, et al. Evaluation of methods for epidemiologic typing of group A streptococci. J Infect Dis. 1994;169:519–525. doi: 10.1093/infdis/169.3.519. [DOI] [PubMed] [Google Scholar]
- 35.Spanier J, Cleary P P. A restriction map and analysis of the terminal redundancy in the group A streptococcal bacteriophage SP24. Virology. 1983;130:502–513. doi: 10.1016/0042-6822(83)90103-4. [DOI] [PubMed] [Google Scholar]
- 36.Stevens D L, Tanner M H, Winship J, Swarts R, Ries K, Schlievert P, Kapland E. Severe group A streptococcal infections associated with a toxic shock-like syndrome and scarlet fever toxin A. N Engl J Med. 1989;321:1–7. doi: 10.1056/NEJM198907063210101. [DOI] [PubMed] [Google Scholar]
- 37.Talkington D F, Schwartz B, Black C M, Todd J K, Elliott J, Breiman R F, Facklam R R. Association of phenotypic and genotypic characteristics of invasive Streptococcus pyogenes isolates with clinical components of streptococcal toxic shock syndrome. Infect Immun. 1993;61:3369–3374. doi: 10.1128/iai.61.8.3369-3374.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weeks C R, Ferretti J J. The gene for type A streptococcal exotoxin (erythrogenic toxin) is located in bacteriophage T12. Infect Immun. 1984;46:531–536. doi: 10.1128/iai.46.2.531-536.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whatmore A, Kapur V, Sullivan D J, Musser J M, Kehoe M A. Non-congruent relationships between variation in gene sequences and the population genetic structures of group A streptococci. Mol Microbiol. 1994;14:619–631. doi: 10.1111/j.1365-2958.1994.tb01301.x. [DOI] [PubMed] [Google Scholar]
- 40.Yu C, Ferretti J J. Molecular characterization of new group A streptococcal bacteriophages containing the gene for streptococcal erythrogenic toxin (speA) Mol Gen Genet. 1991;231:161–168. doi: 10.1007/BF00293833. [DOI] [PubMed] [Google Scholar]
- 41.Zabriskie J. The role of temperate bacteriophage in the production of erythrogenic toxin by group A streptococci. J Exp Med. 1964;119:761–780. doi: 10.1084/jem.119.5.761. [DOI] [PMC free article] [PubMed] [Google Scholar]