Abstract
Objective
Carbapenemase production and biofilm formation in K. pneumoniae are crucial factors influencing the pathogenicity and antibiotic resistance of this bacterium. This study investigated the interplay between carbapenemase production and biofilm formation in K. pneumoniae clinical isolates.
Results
The distribution of biofilm-forming ability significantly differed between carbapenemase-producing (CP-Kp) (n = 52) isolates and carbapenemase-nonproducing (CN-Kp) isolates (n = 37), suggesting a potential link between carbapenemase production and biofilm formation. All the blaNDM-1-harbouring isolates demonstrated biofilm formation, with varying levels classified as strong (33.33%), moderate (22.22%), or weak (44.45%). blaNDM-1 and blaKPC-coharbouring isolates did not exhibit strong or moderate biofilm formation. blaNDM-1 and blaOXA-48-coharbouring isolates were predominantly moderate (48.65%), followed by weak (32.43%), with none showing strong biofilm production. These findings suggest a correlation between the presence of carbapenemases and biofilm-forming ability; however, the heterogeneity in biofilm-forming abilities associated with different carbapenemase types and the absence of strong biofilm producers in the detected carbapenemase combinations prompt a closer look at the complex regulatory mechanisms governing biofilm formation in CP-Kp isolates.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13104-024-06708-9.
Keywords: Klebsiella pneumoniae, Carbapenem resistance, NDM-1, OXA-48, Biofilm
Introduction
K. pneumoniae is a clinically significant gram-negative pathogen known to cause various infections in hospital and community settings [1]. K. pneumoniae develops carbapenem resistance, mainly through the production of carbapenemases along with other mechanisms, such as outer membrane impermeability and efflux pumps [2]. The most clinically significant carbapenemases in K. pneumoniae are classified into Ambler class A β-lactamases (encoded by blaKPC), class B metallo-β-lactamases (MBLs) (encoded by blaNDM−1 and blaVIM), and class D β-lactamases (encoded by blaOXA−48) [3, 4]. The emergence and spread of carbapenemase-producing K. pneumoniae (CP-Kp) strains have posed a serious threat to public health and continue to occur at alarming rates [5] due to their extensive antibiotic resistance [6], including resistance to carbapenem antibiotics, which are considered the last resort for the treatment of infections caused by multidrug-resistant (MDR) K. pneumoniae [7].
K. pneumoniae has the ability to produce virulence factors that play a role in its pathogenesis, including the crucial virulence trait of biofilm formation. The biofilm is a complex matrix that frequently consists of a dense matrix of proteins, polysaccharides, and DNA [8, 9], within which bacteria are highly resistant to antibiotics and host immune responses [10], making it challenging to eradicate. Moreover, biofilms of K. pneumoniae that develop on medical devices such as catheters and endotracheal tubes pose a substantial risk of infection for patients who are catheterized [11].
The development of antibiotic resistance is often intertwined with infection, highlighting its close association with virulence. This connection becomes especially apparent in the cases of microorganisms capable of producing biofilms [12]. Consequently, both the formation of biofilms and the production of carbapenemases contribute to heightened levels of antibiotic resistance. The aim of this study was to investigate the correlation between carbapenemase production and biofilm formation in K. pneumoniae clinical isolates.
Methods
Bacterial strains
Eighty-nine K. pneumoniae clinical isolates were included in this study (52 CP-Kp and 37 CN-Kp) from different clinical sources: urine (n = 38), blood (n = 19), wound (n = 9), sputum (n = 6), pus (n = 5) and other (n = 12), collected between March 2021 and January 2022 from Egypt Air Hospital in Cairo, Egypt. Identification of the isolates was carried out using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDITOF/MS, SAI, UK) with score values ranging from 0.82 to 0.87, as per the manufacturer’s recommendations.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was employed by the disk diffusion technique using Mueller-Hinton agar (Oxoid, Thermo Fisher Scientific), following the guidelines established by the Clinical and Laboratory Standards Institute (CLSI) [13]. Imipenem (10 µg), meropenem (10 µg), and ertapenem (10 µg) (Oxoid, Thermo Fisher Scientific) were tested. E. coli ATCC 25922 was used as the control strain. Interpretation of zones of inhibition was performed according to CLSI guidelines [14]. The inclusion criterion for the CN-Kp group was based on susceptibility, which was defined as having phenotypes sensitive to all tested carbapenems.
Phenotypic detection of carbapenemases
Carbapenemase-positive isolates were detected using the modified carbapenem inactivation method (mCIM) in accordance with the CLSI guidelines [14].
DNA extraction
To obtain crude DNA, 2 colonies were lysed in 500 µL of sterile distilled water at 100 °C for 10 min, followed by centrifugation. The supernatant was stored at − 80 °C for subsequent PCR assays. The concentration and purity of the DNA extract were detected using a NanoDrop spectrophotometer at wavelengths of 260 and 280 nm.
Detection of resistance genes by PCR
The carbapenemase-encoding genes blaIMP, blaVIM, blaKPC, blaNDM-1, blaSPM, and blaOXA-48 were amplified as follows: One multiplex PCR for the detection of blaKPC, blaNDM-1, and blaOXA-48 and three uniplex PCRs for the detection of blaIMP, blaVIM, and blaSPM were carried out in a 25 µL volume using COSMO PCR RED master mix (Willowfort, Birmingham, England). Primers (Invitrogen®, Thermo Fisher Scientific Inc., MA, USA) sequences and sizes are listed in Table 1, and the PCR conditions were as follows: For the multiplex PCR, 10 min at 94 °C and 30 cycles of amplification consisting of 30 s at 94 °C, 40 s at 52 °C, and 50 s at 72 °C were used, with 5 min at 72 °C for the final extension. For the three uniplex PCRs, 10 min at 94 °C and 30 cycles of amplification consisting of 30 s at 94 °C, 40 s at 55 °C, and 50 s at 72 °C, with 5 min at 72 °C for the final extension. A control without a template was included. The amplicons were separated by electrophoresis on a 2% (w/v) agarose gel containing ethidium bromide (0.5 µg/ml) using Thermo Scientific™ GeneRuler™ 100 bp DNA Ladder (Thermo Fisher Scientific Baltics UAB, Lithuania). Prior to their use in the multiplex PCR assay, the primer pairs were individually tested to confirm their functionality and specificity.
Table 1.
Primers used for detection of carbapenemase genes
| Gene | Primer DNA sequence | Amplicon size (bp) | Reference |
|---|---|---|---|
| bla NDM−1 |
F-5’GGTTTGGCGATCTGGTTTTC 3’ R-5’CGGAATGGCTCATCACGATC 3’ |
621 bp | [15] |
| bla KPC |
F-5’CGTCTAGTTCTGCTGTCTTG 3’ R-5’CTTGTCATCCTTGTTAGGCG3’ |
798 bp | [15] |
| bla OXA−48 |
F-5’GCGTGGTTAAGGATGAACA3’ R-5’CATCAAGTTCAACCCAACCG3’ |
438 bp | [15] |
| bla IMP |
F-5’GGAATAGAGTGGCTTAAYTCTC3’ R-5’GGTTTAAYAAAACAACCACC3’ |
232 bp | [16] |
| bla SPM |
F-5’AAAATCTGGGTACGCAAACG3’ R-5’ACATTATCCGCTGGAACAGG3’ |
271 bp | [16] |
| bla VIM |
F-5’GATGGTGTTTGGTCGCATA3’ R-5’CGAATGCGCAGCACCAG3’ |
390 bp | [16] |
Biofilm assay
The microtiter plate (96-well plate) assay was used to study biofilm formation, as described elsewhere [17]. The bacterial isolates were grown overnight at 37 ℃, and the bacterial suspension turbidity was adjusted to match the OD of a 0.5 McFarland standard in saline using a spectrophotometer (GeneQuant, Biochrom Ltd., England). Twenty µL aliquots of each suspension were added to the wells of polystyrene microtiter plates containing 180 µL TSB supplemented with 1% glucose, with three wells per bacterial isolate. The plate was incubated for 48 h under static conditions, and the broth was gently aspirated. Each well was washed thrice with 300 µL of PBS at pH 7.2, and the adherent biofilm layer in each well was stained with 150 µL of 0.5% (w/v) crystal violet solution for 14 min at room temperature. Crystal violet was removed using sterile distilled water, and the plates were air-dried. The biofilm was solubilized by adding 150 µL of 95% ethanol. The OD of each well was measured at 570 nm using a microtiter plate reader (Synergy 2, BioTek, WI, USA). Sterile broth was used as a negative control. Each assay was performed in triplicate on three occasions. The results were interpreted according to the following criteria: no biofilm production (ODsample<ODcontrol), weak biofilm production (ODcontrol<ODsample<2xODcontrol), moderate biofilm production (2xODcontrol < ODsample<4xODcontrol), and strong biofilm production (4xODcontrol < ODsample) [17].
Data analysis
Chi-square tests (Fisher’s exact test where appropriate) were performed using GraphPad Prism version 5.01 for Windows and GraphPad InStat version 3.05 (GraphPad Software, San Diego, California, USA) to assess the relationship between carbapenemase production and biofilm formation. In this context, any correlation analyses that resulted in p values less than 0.05 were statistically significant.
Results
Characterization of isolates
Among the CP-Kp isolates, 37 exhibited simultaneous resistance to all three tested carbapenems. Specifically, all CP-Kp isolates were resistant to ertapenem, with the exception of two isolates that showed intermediate resistance. Out of 52 CP-Kp isolates, 46 were resistant to meropenem and 37 to imipenem, with intermediate resistance in four and five isolates, respectively. In contrast, 10 isolates were found to be sensitive to imipenem, and two isolates were sensitive to meropenem.
Prevalence and distribution of carbapenemase genes
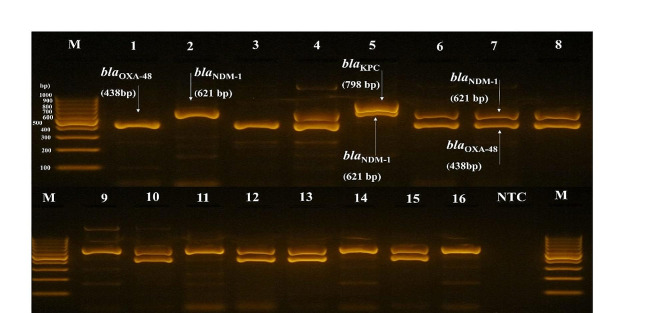
All CR-Kp isolates that phenotypically expressed carbapenemase, as determined by the mCIM, were positive for one or more carbapenemase genes, according to PCR results (Fig. 1). Out of the 52 CP-Kp isolates, the blaNDM-1 gene was detected in 49 (94.23%), and blaOXA-48 was detected in 40 (76.92%) isolates. None of the carbapenemase genes tested (blaVIM,blaIMP, and blaSPM) were detected. Out of the 52 carbapenemase gene-carrying isolates, 37 (71.15%) coharboured blaNDM-1 and blaOXA-48, while 3 (5.77%) isolates carried both blaNDM-1 and blaKPC. None of the isolates were found to harbour blaKPC only.
Fig. 1.
Carbapenemase gene amplification profile in K. pneumoniae using multiplex PCR. The PCR products were separated on a 2% agarose gel. A molecular size marker (in bp, measurements on the left, middle, and right) was used, featuring a reference band at 500 bp. No template control (NTC) was included. The specific lanes correspond to the following amplified genes: Lane 1 and 3 for blaOXA-48 (438 bp); Lane 2, 9, 11, 14 and 16 for blaNDM-1 (621 bp); Lane 5 for blaNDM-1 and blaKPC (621 and 798 bp); and Lane 4, 6, 7, 8, 10, 12, 13 and 15 for blaNDM-1 and blaOXA-48 (621 bp and 438 bp)
Evaluation of biofilm formation
In total, 63 out of 89 (70.79%) isolates were identified as biofilm producers, and 26 isolates (29.21%) were biofilm non-producers from different clinical sources. The distribution of biofilm-forming ability varied significantly (p = 0.0037; chi-square test) between the CP-Kp and CN-Kp isolates. For the CP-Kp isolates, 42.31% exhibited moderate biofilm production, 34.62% were weak producers, 5.77% were strong producers, and 17.31% were non-producers. Among the CN-Kp isolates, 37.84% were weak producers, 10.81% were moderate producers, 5.41% were strong producers, and 45.95% were non-producers (see Table 2 and Supplementary Fig. 1). The biofilm-forming abilities of K. pneumoniae isolates were assessed based on various carbapenemase types. Among the blaNDM-1-harbouring isolates, 33.33% exhibited strong biofilm formation, 22.22% had moderate biofilm formation, and 44.45% had weak biofilm formation. None of the isolates were classified as biofilm non-producers. In blaNDM-1 and blaKPC-coharbouring isolates, 66.67% were weak biofilm producers, and 33.33% were biofilm non-producers. In blaNDM-1 and blaOXA-48-coharbouring isolates, no strong biofilm producers were observed. Instead, 48.65% were moderate producers, 32.43% were weak producers, and 18.92% were non-biofilm producers. In the blaOXA-48 harbouring isolates, 66.67% were classified as moderate biofilm producers, and 33.33% were biofilm non-producers. (see Supplementary Table 1).
Table 2.
Comparison of biofilm-forming ability between carbapenemase-producing (CP-Kp) and nonproducing Klebsiella pneumoniae (CN-Kp) isolates (Fisher’s exact test)
| Biofilm formation | CP-Kp, n = 52 (%) | CN-Kp, n = 37 (%) | Total, n = 89 (%) | p value |
|---|---|---|---|---|
| No biofilm | 9 (17.31) | 17 (45.95) | 26 (29.21) | 0.0046 |
| Weak | 18 (34.62) | 14 (37.84) | 32 (35.96) | 0.8243 |
| Moderate | 22 (42.31) | 4 (10.81) | 26 (29.21) | 0.0018 |
| Strong | 3 (5.77) | 2 (5.41) | 5 (5.62) | 1.0000 |
Discussion
Over the past few years, there has been a notable increase in the occurrence of CP-Kp in hospitals worldwide. In this study, 52 CP-Kp isolates were included. blaNDM-1 was the most predominant (94.23%) carbapenemase-encoding gene detected, followed by blaOXA-48 (76.92%). This finding is in line with previously reported data that found that 35 out of 37 (94.59%) of their carbapenemase-producing isolates harboured blaNDM-1 and 26 (70.27%) harboured blaOXA-48 [18]. In contrast to our findings, another study reported that blaOXA-48 (25/62, 40.32%) was the most predominant and that blaNDM-1 had only a minor incidence (6/62, 9.68%) [19]. When a carbapenemase-producing isolate carries multiple carbapenemases, it becomes highly resistant to treatment as it increases the range of hydrolytic activity, making it challenging to target with antibiotics [20]. Here, we found that the coexistence of blaNDM-1 and blaOXA-48 accounted for 71.15% of the CP-Kp isolates. This high occurrence is consistent with previous work, which reported a prevalence of 64.86% (24 out of 37 isolates) of tested CP-Kp isolates [18]. In contrast, a previous study conducted in the ICUs of Zagazig University Hospitals reported that only 5.71% (6 out of 105 isolates) of tested CP-Kp isolates coharboured blaNDM-1 and blaOXA-48 [21]. In this study, we found a low incidence (5.77%) of CP-Kp isolates that coharbored blaNDM-1 and blaKPC. These findings are in accordance with previous studies conducted in Egypt, which reported a low incidence (11.29%) (7 out of 62 isolates) [19] or absence [18] of the blaKPC gene in their CP-Kp isolates. This suggests that KPC is less prevalent in our geographic area. However, opposite findings were reported by others who found a higher prevalence of blaKPC at 17.14% (18 out of 105) in the tested CP-Kp [21]. Here, we could not detect the presence of blaIMP or blaVIM in any of our CP-Kp isolates. These findings are in line with other studies conducted in Egypt and other countries that reported the absence of these genes in their CP-Kp isolates [18, 21]. However, in contrast to our findings, some studies reported a higher prevalence [19]. Here, we could not detect the presence of blaSPM in any isolate.
One of the important virulence factors of K. pneumoniae is its ability to form biofilms [22]. The current study revealed that the majority of the K. pneumoniae isolates were biofilm producers (70.79%), regardless of their clinical source. This finding is consistent with that of a study conducted in 2023 [23]. However, a previous study reported a greater propensity for biofilm formation, with 91.2% of the isolates forming biofilms [24]. In this study, there was a significant difference in biofilm-forming ability between CP-Kp and CN-Kp (p = 0.0037; chi-square test). These findings corroborate those of the 2022 study on reference strains of K. pneumoniae [25]. Notably, a smaller percentage of CP-Kp isolates (17.31%) were biofilm non-producers compared to CN-Kp isolates (45.95%) (p = 0.0046; Fisher’s exact test). Therefore, the presence of carbapenem resistance, indicated by the presence of carbapenemases, suggested a greater overall propensity for biofilm formation in K. pneumoniae. This could be attributed to several factors, such as the interplay of carbapenemases with regulatory pathways governing biofilm formation, which may lead to the upregulation of key biofilm-associated genes in CP-Kp isolates [25]. Additionally, the environmental stress induced by the presence of carbapenemases might support the formation of biofilms as a survival strategy in CP-Kp isolates. Despite the suggested association, it is noteworthy that there is a subset of CP-Kp isolates that demonstrated an inability to produce biofilms, suggesting that carbapenem resistance in K. pneumoniae alone does not guarantee biofilm formation and is likely multifactorial.
CP-Kp isolates, despite being primarily biofilm producers, display varied biofilm-forming capabilities. The strong biofilm producers of the CP-Kp group were exclusively found among the isolates harbouring only blaNDM-1, consistent with the findings of a previous study [25]. Notably, the blaNDM-1 and blaKPC-coharbouring isolates did not exhibit strong or moderate biofilm formation, and one such isolate was a biofilm non-producer. This finding aligns with that of a prior study [23]. In contrast, the majority of blaNDM-1 and blaOXA-48coharbouring isolates exhibited a moderate biofilm phenotype (48.65%), followed by a weak biofilm phenotype (32.43%), while 18.92% of the isolates exhibited no biofilm formation. No strong biofilm producers were identified.
Most of the literature indicated a higher prevalence of strong biofilm producers among β-lactamase producers [26, 27]. In the present study, unexpectedly, no correlation was observed between the formation of strong biofilms and the presence of detected carbapenemases. Additionally, none of the isolates detected with more than one resistance gene exhibited strong biofilm formation, contrary to the findings of a recent study [23]. Conversely, another study revealed a notable association between the prevalence of the blaVIM1 and blaIMP1 genes and the formation of strong biofilms [24]. These findings suggest that certain carbapenemase subtypes or combinations may enhance biofilm formation, while others may have a more modest impact or even a negative influence. However, other factors, such as quorum sensing [25], the HMV phenotype [28], high adhesion capacity, and cell death, have been reported to have significant impacts on the formation of strong biofilms on K. pneumoniae isolates [29].
In conclusion, this study suggested a link between carbapenemases and biofilm formation, but the diverse biofilm-forming capabilities of various carbapenemase types and the lack of strong producers in the detected combinations emphasise the need for a deeper understanding of the intricate regulatory mechanisms governing biofilm formation in CP-Kp isolates.
Limitations
One of the limitations of this study is the lack of further carbapenemase subtyping and correlation with biofilm phenotypes, which could provide insights into the varying effects of specific carbapenemases or combinations on biofilm formation. In addition, performing functional assays, such as gene knockout experiments or overexpression studies, can validate the role of specific genetic elements in biofilm formation.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Material 1: Supplementary Figure 1. Distribution of biofilm-forming ability among carbapenemase-producing (CP-Kp) and nonproducing Klebsiella pneumoniae (CN-Kp) isolates. Supplementary Table 1.. Distribution of the biofilm-forming ability across different carbapenemase types in carbapenemase-producing Klebsiella pneumoniae (CP-Kp) isolates
Acknowledgements
Not applicable.
Abbreviations
- ATCC
American Type Culture Collection
- bla
Beta-lactamase genes
- CP-Kp
Carbapenemase-producing Klebsiella pneumoniae
- CN-Kp
Carbapenemase-nonproducing Klebsiella pneumoniae
- CR-Kp
Carbapenem-resistant Klebsiella pneumoniae
- CLSI
Clinical and Laboratory Standards Institute
- dNTP
Deoxynucleoside triphosphate
- DNA
Deoxyribonucleic acid
- eDNA
Extracellular DNA
- EPS
Exopolysaccharides
- E. coli
Escherichia coli
- HMV phenotype
Hypermucoviscous phenotype
- ICU
Intensive Care Unit
- IMP
Imipenemase
- K. pneumoniae
Klebsiella pneumoniae
- KPC
Klebsiella pneumoniae carbapenemase
- MALDITOF/MS
Matrix-Assisted Laser Desorption-Ionization-Time of Flight Mass Spectrometry
- mCIM
Modified Carbapenem Inactivation Method
- MDR
Multidrug-resistant
- MBLs
Metallo-β-lactamases
- NDM-1
New Delhi metallo-β-lactamase type 1
- OD
Optical density
- OXA-48
Oxacillinase-48
- PBS
Phosphate Buffered Saline
- PCR
Polymerase Chain Reaction
- SPM
Sao Paulo metallo-β-lactamase
- Taq
Thermus aquaticus
- TSA
Tryptone Soy Agar
- TSB
Tryptic Soy Broth
- VIM
Verona integron-encoded metallo-β-lactamase
Author contributions
N.M.E Material preparation, isolate collection, analysis of results and writing the manuscript. R.M.S Revision of the manuscript. F.M.E.S study conception and design and revision of the manuscript. M.E., Study conception and design, analysis of results and writing, revision and approval of the manuscript.
Funding
No funding was received to assist with the preparation of this manuscript.
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB).
Data availability
All data generated or analysed during this study are included in this article and its supplementary information files.
Declarations
Ethics approval and consent to participate
The study was ethically approved by the Ethics Committee of the Faculty of Pharmacy, Helwan University (registration number: 07H2023). Permission was received from the hospital administration to utilize the samples for research purposes. Other patient identifier information was kept confidential. Clinical samples were collected as part of routine laboratory analysis to check for potential infections in referred and admitted patients and not as a part of this study. Therefore, written informed consent was waived by the Ethics Committee of the Faculty of Pharmacy, Helwan University.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: Epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev. 1998;11:589–603. doi: 10.1128/CMR.11.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Han YL, Wen XH, Zhao W, Cao XS, Wen JX, Wang JR, et al. Epidemiological characteristics and molecular evolution mechanisms of carbapenem-resistant hypervirulent Klebsiella pneumoniae. Front Microbiol. 2022;13:1003783. doi: 10.3389/fmicb.2022.1003783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hansen GT. Continuous evolution: perspective on the epidemiology of Carbapenemase Resistance among enterobacterales and other Gram-negative Bacteria. Infect Dis Ther. 2021;10:75–92. doi: 10.1007/s40121-020-00395-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elemary NM, Emara MM, Tahoun AAE, Eloomany RA. Correlation between antimicrobial resistance and virulence genes in Klebsiella pneumoniae isolates from Egypt. J Pak Med Assoc. 2023;73:274–81. doi: 10.47391/JPMA.EGY-S4-54. [DOI] [PubMed] [Google Scholar]
- 5.Solgi H, Nematzadeh S, Giske CG, Badmasti F, Westerlund F, Lin YL, et al. Molecular epidemiology of OXA-48 and NDM-1 producing enterobacterales species at a University Hospital in Tehran, Iran, between 2015 and 2016. Front Microbiol. 2020;11:529632. doi: 10.3389/fmicb.2020.00936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonomo RA, Burd EM, Conly J, Limbago BM, Poirel L, Segre JA, et al. Carbapenemase-producing organisms: A Global Scourge. Clin Infect Dis. 2018;66:1290–7. doi: 10.1093/cid/cix893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, et al. Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2001;45:1151–61. doi: 10.1128/AAC.45.4.1151-1161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rabin N, Zheng Y, Opoku-Temeng C, Du Y, Bonsu E, Sintim HO. Biofilm formation mechanisms and targets for developing antibiofilm agents. Future Med Chem. 2015;7:493–512. doi: 10.4155/fmc.15.6. [DOI] [PubMed] [Google Scholar]
- 9.Wahman S, Emara M, Shawky RM. In-vitro assessment of staphylococci biofilms formed under biologically-relevant conditions and correlation to the biofilm genotype. Res J Pharm Technol. 2023;16:2273–9. doi: 10.52711/0974-360X.2023.00373. [DOI] [Google Scholar]
- 10.Jagnow J, Clegg S. Klebsiella pneumoniae MrkD-mediated biofilm formation on extracellular matrix- and collagen-coated surfaces. Microbiology. 2003;149:2397–405. doi: 10.1099/mic.0.26434-0. [DOI] [PubMed] [Google Scholar]
- 11.Schroll C, Barken KB, Krogfelt KA, Struve C. Role of type 1 and type 3 fimbriae in Klebsiella pneumoniae biofilm formation. BMC Microbiol. 2010;10:1–10. doi: 10.1186/1471-2180-10-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patel R. Biofilms and antimicrobial resistance. Clin Orthop Relat Res. 2005;437:41–7. doi: 10.1097/01.blo.0000175714.68624.74. [DOI] [PubMed] [Google Scholar]
- 13.CLSI. Performance standards for Antimicrobial Disk susceptibility tests; approved standard—Tenth Edition. CLSI document M02-A10. Wayne, PA: Clinical and Laboratory Standards Institute; 2009.
- 14.CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 31st ed. CLSI supplement M100. Clinical and Laboratory Standards Institute; 2021. [DOI] [PMC free article] [PubMed]
- 15.Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70:119–23. doi: 10.1016/j.diagmicrobio.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 16.Ellington MJ, Kistler J, Livermore DM, Woodford N. Multiplex PCR for rapid detection of genes encoding acquired metallo-β-lactamases. J Antimicrob Chemother. 2007;59:321–2. doi: 10.1093/jac/dkl481. [DOI] [PubMed] [Google Scholar]
- 17.Stepanović S, Vuković D, Hola V, Di Bonaventura G, Djukić S, Ćirković I, et al. Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. Apmis. 2007;115:891–9. doi: 10.1111/j.1600-0463.2007.apm_630.x. [DOI] [PubMed] [Google Scholar]
- 18.El-Domany RA, El-Banna T, Sonbol F, Abu-Sayedahmed SH. Co-existence of NDM-1 and OXA-48 genes in Carbapenem resistant Klebsiella pneumoniae clinical isolates in Kafrelsheikh, Egypt. Afr Health Sci. 2021;21:489–96. doi: 10.4314/ahs.v21i2.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taha MS, Hagras MM, Shalaby MM, Zamzam YA, Elkolaly RM, Abdelwahab MA et al. Genotypic Characterization of Carbapenem-Resistant Klebsiella pneumoniae Isolated from an Egyptian University Hospital. Pathog 2023, Vol 12, Page 121. 2023;12:121. [DOI] [PMC free article] [PubMed]
- 20.Meletis G, Chatzidimitriou D, Malisiovas N. Double- and multi-carbapenemase-producers: the excessively armored bacilli of the current decade. Eur J Clin Microbiol Infect Dis. 2015;34:1487–93. doi: 10.1007/s10096-015-2379-9. [DOI] [PubMed] [Google Scholar]
- 21.Gandor NHM, Amr GES, Eldin Algammal SMS, Ahmed AA. Characterization of Carbapenem-Resistant K. Pneumoniae isolated from Intensive Care units of Zagazig University Hospitals. Antibiotics. 2022;11. [DOI] [PMC free article] [PubMed]
- 22.Wang G, Zhao G, Chao X, Xie L, Wang H. The characteristic of virulence, Biofilm and Antibiotic Resistance of Klebsiella pneumoniae. Int J Environ Res Public Heal 2020. 2020;17(6278):6278. doi: 10.3390/ijerph17176278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sabença C, Costa E, Sousa S, Barros L, Oliveira A, Ramos S et al. Evaluation of the Ability to Form Biofilms in KPC-Producing and ESBL-Producing Klebsiella pneumoniae Isolated from Clinical Samples. Antibiot 2023, Vol 12, Page 1143. 2023;12:1143. [DOI] [PMC free article] [PubMed]
- 24.Khodadadian R, Rahdar HA, Javadi A, Safari M, Khorshidi A. Detection of VIM-1 and IMP-1 genes in Klebsiella pneumoniae and relationship with biofilm formation. Microb Pathog. 2018;115:25–30. doi: 10.1016/j.micpath.2017.12.036. [DOI] [PubMed] [Google Scholar]
- 25.Al-Bayati M, Samarasinghe S. Biofilm and Gene Expression Characteristics of the Carbapenem-Resistant Enterobacterales, Escherichia coli IMP, and Klebsiella pneumoniae NDM-1 Associated with Common Bacterial Infections. Int J Environ Res Public Heal. 2022, Vol 19, Page 4788. 2022;19:4788. [DOI] [PMC free article] [PubMed]
- 26.Rahdar HA, Malekabad ES, Dadashi AR, Takei E, Keikha M, Kazemian H, et al. Correlation between biofilm formation and carbapenem resistance among clinical isolates of Klebsiella pneumoniae. Ethiop J Health Sci. 2019;29:745. doi: 10.4314/ejhs.v29i6.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanchez CJ, Mende K, Beckius ML, Akers KS, Romano DR, Wenke JC, et al. Biofilm formation by clinical isolates and the implications in chronic infections. BMC Infect Dis. 2013;13:1–12. doi: 10.1186/1471-2334-13-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Di Domenico EG, Cavallo I, Sivori F, Marchesi F, Prignano G, Pimpinelli F, et al. Biofilm production by Carbapenem-Resistant Klebsiella pneumoniae significantly increases the risk of death in oncological patients. Front Cell Infect Microbiol. 2020;10:561741. doi: 10.3389/fcimb.2020.561741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Desai S, Sanghrajka K, Gajjar D. High Adhesion and Increased Cell Death Contribute to Strong Biofilm Formation in Klebsiella pneumoniae. Pathog 2019, Vol 8, Page 277. 2019;8:277. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1: Supplementary Figure 1. Distribution of biofilm-forming ability among carbapenemase-producing (CP-Kp) and nonproducing Klebsiella pneumoniae (CN-Kp) isolates. Supplementary Table 1.. Distribution of the biofilm-forming ability across different carbapenemase types in carbapenemase-producing Klebsiella pneumoniae (CP-Kp) isolates
Data Availability Statement
All data generated or analysed during this study are included in this article and its supplementary information files.