Abstract
Lynch Syndrome (LS) markedly increases risks of colorectal (CRC) and endometrial (EC) cancers. Early detection biomarkers for LS cancers could reduce the needs for invasive screening and surgical prophylaxis.
To validate a panel of methylated DNA markers (MDMs) previously identified in sporadic CRC and EC for discrimination of these cancers in LS.
In a case-control design, previously identified MDMs for the detection of CRC and EC were assayed by qMSP on tissue-extracted DNA. Results were normalized to ACTB values within each sample. LASSO models to classify CRC and EC were trained on sporadic cases and controls and then applied to classify CRC and EC, in those with LS, and cross-validated.
We identified CRC cases (23 with LS, 48 sporadic), colorectal controls (32 LS, 48 sporadic), EC cases (30 LS, 48 sporadic), and endometrial controls (29 LS, 37 sporadic). A 3-MDM panel (LASS4, LRRC4, and PPP2R5C) classified LS-CRC from LS controls with an AUC of 0.92 (0.84–0.99); results were similar for sporadic CRC. A 6-MDM panel (SFMBT2, MPZ, CYTH2, DIDO1, chr10.4479, and EMX2OS) discriminated LS-EC from LS controls with an AUC of 0.92 (0.83–1.0); the AUC for sporadic EC vs sporadic controls was nominally higher, 0.99 (0.96–1.0).
MDMs previously identified in sporadic EC and CRC discriminate between EC and benign endometrium and CRC and benign colorectum in LS. This supports the inclusion of patients with LS within future prospective clinical trials evaluating EC and CRC MDMs and may provide a new avenue for cancer screening or surveillance in this high-risk population.
INTRODUCTION:
Lynch Syndrome (LS), is an autosomal dominant hereditary cancer syndrome affecting 1 in 600 to 1 in 3,000 individuals and is due to a mutation in one of five mismatch repair genes, including MLH1, MSH2, MSH6, PMS2 or EPCAM.(1,2) LS poses the greatest risk for endometrial (EC) and colorectal cancers (CRC), and also elevates risk for development of small bowel, ureteral, gastric, hepatobiliary, and ovarian cancers. Those with LS have a markedly increased lifetime risk of developing CRC and EC, estimated to reach 80% and 60%, respectively. In the United States population, LS is identified in about 3% of CRC and 2% of EC case (3).
Among other tests, cancer prevention surveillance in LS patients involves annual colonoscopy and endometrial biopsy. Once childbearing is completed, risk-reducing hysterectomy and bilateral salpingo-oophorectomy upon completion of childbearing is recommended by several expert consensus guidelines (3–7). Cancer screening and risk-reducing surgery as part of the above protocols have several limitations including, observational/expert consensus evidence base, inaccuracy, invasiveness, and surgical morbidity. Thus, highly accurate and less invasive approaches for screening and early detection are clearly needed.
In persons at average risk for CRC, the multitarget stool DNA test (mt-sDNA), which assays methylated DNA markers (MDMs) BMP3 and NDRG4, in addition to fecal hemoglobin, has been FDA-approved and is an option for colorectal cancer screening (8–10). We and others have shown that MDMs and other biomarkers exfoliated by EC into vaginal fluid can be detected in self-collected tampon samples (11–13). These approaches to early detection of CRC and EC have not been widely studied in the LS patient population.
To begin to address the applicability of MDMs as biomarkers of disease in LS, we initially verified that BMP3 and NDRG4 are insufficiently representative in CRC and advanced precursors in patients with LS(14,15), further discouraging use of the current generation mt-sDNA test in this population. However, in the interim, our group has conducted extensive MDM biomarker discovery in both CRC and EC, using next-generation DNA sequencing. These novel MDMs have not only been shown to have higher discrimination than older candidates in sporadic tissues but have been successfully piloted by assay of DNA extracted from archival, average-risk, case-control stool and tampon specimens, respectively (13). These MDMs are currently undergoing phase 2–4 validation in multi-center clinical studies in average risk populations for CRC and EC (ClinicalTrials.gov Identifiers: NCT04144738 and NCT05051722, respectively).
To be applied to early detection testing in LS, we hypothesized that these novel candidate biomarkers would have to be strongly represented in LS-associated primary tumors but not otherwise in normal carrier tissues. The aim of this study was to validate a panel of MDMs previously identified in sporadic CRC and EC that could also discriminate cancer from benign epithelium using DNA extracted from primary tissues exclusively from patients with LS.
METHODS:
Overview:
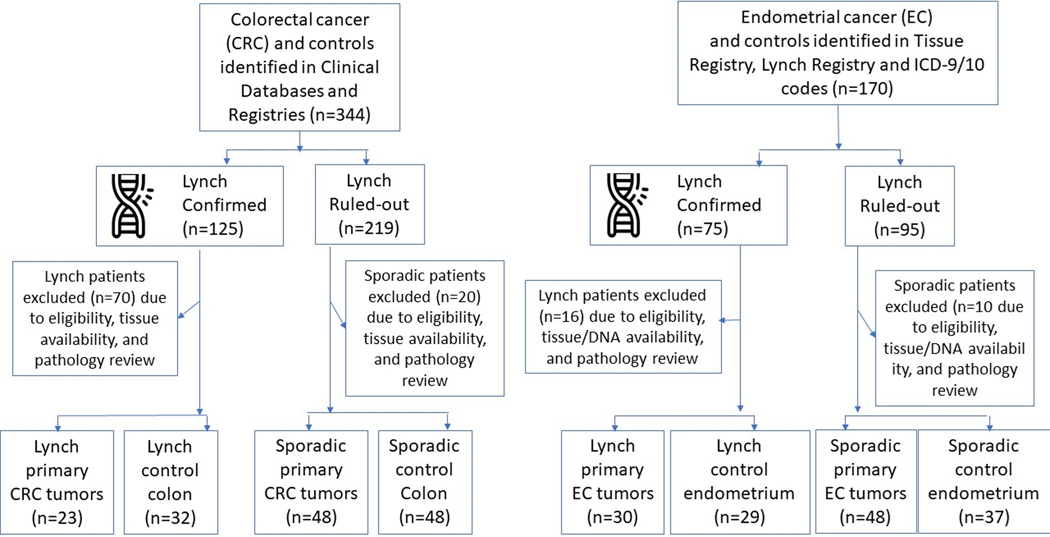
In two parallel case-control studies, panels of previously identified, highly discriminant MDMs for the detection of CRC and EC were assayed on tissue-extracted DNA to measure discrimination between benign and malignant lesions in individuals with LS (LS-CRC study and LS-EC study, respectively). In both studies, the MDMs were also assayed in DNA extracted from tissues of CRC and EC case and control patients without a known hereditary predisposition to cancer. The study schema is shown in Figure 1. This study was approved by the Mayo Clinic Institutional Review Board and performed in accordance with the ethical guidelines set for by the U.S. Common Rule.
Figure 1.
Study flow diagram. Summary of initial populations, sample dropout and final numbers in each group.
Study population and sample sources:
For this single center multi-site study, we used archival tissue samples obtained from the Mayo Clinic Tissue Registry, an archive of surplus clinical tissue specimens maintained for reserach by the Mayo Clinic Department of Anatomic Pathology. All frozen and Formalin-Fixed Paraffin-Embeded (FFPE) tissues underwent research histopathology review by one expert pathologist (LC) before macrodissection and DNA extraction.
LS-CRC study:
LS patients were identified from institutional clinical databases and registries of patients undergoing hereditary cancer germline testing. LS case patients with CRC (LS-CRC-CASE) and LS carrier control (LS-CRC-CTRL) patients were all required to have a germline pathogenic variant in the MMR genes (MLH1, MSH2, MSH6, PMS2) or the gene EPCAM. Comparison sporadic CRC cases (SPOR-CRC-CASE) and non-LS controls (SPOR-CRC-CTRL) were identified using an institutional tissue registry. Colorectal tumor tissue was sampled at the time of surgical resection from both patients with and without LS, prior to exposure to local/regional therapy or systemic chemotherapy. LS-CRC-CRTL tissues were colorectal tissue from LS patients without CRC, frequency matched, based on age and sex to SPOR-CRC-CTRL tissues (benign colorectal mucosae samples without LS); an additional comparison was made to sporadic CRC cases (SPOR-CRC-CASE tissues from individuals without LS, balanced on age and sex to LS-CRC-CASEs).
LS-EC study:
For inclusion, subjects had to have a history of a hysterectomy with histopathologically confirmed EC or benign endometrium. Females with LS, confirmed by germline testing, were identified using a combination of any of the following: a LS clinical registry, an institutional surgical database, and the electronic medical record utilizing International Classification of Diseases codes. This generated the LS EC case and control groups (LS-EC-CASE and LS-EC-CRTL, respectively). EC with only absent expression of MLH1 and normal germline MLH1 sequencing were excluded. Comparison sporadic EC cases (SPOR-EC-CASE) and non-LS controls (SPOR-EC-CTRL) were identified using an institutional tissue registry. Each SPOR-EC-CASE had either proficient mismatch repair (MMR) enzyme expression alone or were both MMR proficient and microsatellite stable. All SPOR-EC-CASE and SPOR-EC-CTRL subjects had negative hereditary germline genetic testing or a pedigree that did not meet Amsterdam criteria based on review of their medical records. Exclusion criteria for all groups included a history of pelvic radiation prior to hysterectomy. Demographic and clinical data was abstracted from the medical record.
Assay procedures:
As previously described (16), DNA was extracted from FFPE tissues using the QIAamp FFPE DNA mini kits (Qiagen, Valencia, CA) and quantified by Quant-iT Picogreen fluorescence (Thermo Fisher Scientific, Waltham MA). DNA was then bisulfate treated (EZ DNA Methylation Kit; Zymo Research, Orange, CA) and eluted in buffer. Annealing temperatures and designs of PCR primers for candidate MDMs were tested and optimized for quantitative methylation-specific PCR on universally methylated and unmethylated genomic DNA controls. Each sample was tested by quantitative PCR to ensure sufficient genomic equivalents of DNA prior to candidate marker assays. MDMs were assayed (10 ng/marker) with SYBR Green detection using the LightCycler 480 instrument and reagents (Roche Diagnostics, IN, USA).
For the LS-CRC study, candidate MDMs included ADCY4, ADM, AKLBH5, ANTXR1, ARHGEF4, BMP3, CBLN2, CNTFR, ELMO1, LASS4, LRRC4, NDRG4, OPLAH, PITX1, PPP2R5C, SFMBT2, STK32B, USP44, and VAV3. Results were normalized by the products of a CpG agnostic region in the control gene, ACTB, as previously described (17).
For the LS-EC study, candidate MDMs were ones identified in a prior study, chr8.3829, chr10.4479, CYTH2, DIDO1, EMX2OS, JSRP1, LRRC8D, MPZ, NBPF8, OBSCN, SFMBT2, ZNF506, and ZNF90. Results were normalized by the products of a CpG agnostic region in the control gene, ACTB, from each sample.
Statistical methods:
Sample size considerations were based on minimizing the distance of the lower bound of a 95% confidence interval (CI) for an assumed specificity of 90% and sensitivity of 85%. With a target sample size of 25 samples in the benign control groups and 36 samples in the cancer case groups, the lower bound of the 95% CI should not be below 80% and 75% respectively.
Areas under the receiver operating characteristics curve (AUCs) and the corresponding 95% CI’s were calculated for individual markers and were used to assess discrimination between cancer and benign tissues within LS and non-LS patients. AUCs were compared using a Z-test. The distribution of marker levels between subgroups was summarized as a median with corresponding 25th and 75th percentiles and depicted using boxplots. Wilcoxon rank-sum tests were used to evaluate differences in clinical covariates. Marker intensity above a specified cutoff determined using the benign CTRL groups, respectively, were used to generate a colored scale yellow to red based on deciles above the threshold. Least absolute shrinkage and selection operator (LASSO) regression was used to identify a reduced marker set based on SPOR-CRC and SPOR-EC samples. Cross-validation was used to find the optimal lambda value and markers with a non-zero coefficient were included in the reduced marker set. These models were then used to calculate the predicted probability of being a CASE within the LS-CRC and LS-EC samples.
To assess applicability of a prototype 2nd generation mt-sDNA test (mt-sDNA 2.0) a sub-analysis of the LS-CRC study was performed using a multi-marker (LASS4, LRRC4, PPP2R5C) logistic regression model, trained in the SPOR-CRC-CASEs and SPOR-CRC-CTRLs and applied to the LS-CRC samples.
Data Availability:
Data that support the findings of this study are available from the corresponding authors upon reasonable request.
RESULTS:
LS-CRC study
A total of 151 subjects met LS-CRC study inclusion criteria generating 23 LS-CRC-CASE, 48 SPOR-CRC-CASE, 32 LS-CRC-CTRL, and 48 SPOR-CRC-CTRL. To reach the final LS-CRC cohort of 151, the following were excluded: 83 participants judged ineligible and 44 based on pathology review or unavailability of tissues. Of 153 tissue blocks requested, 2 were not available and 2 had low tissue-derived DNA quality and were excluded (Figure 1). Demographic characteristics of the study population are summarized in Table 1. The 19 candidate MDMs were tested by qMSP on DNA extracted from these colorectal tissues.
Table 1:
Demographics and characteristics of cases and controls in the Lynch syndrome colorectal cancer study
| Lynch | Sporadic | |||
|---|---|---|---|---|
|
|
||||
| Colorectal cancers | Benign colorectum mucosa | Colorectal cancers | Benign colorectum mucosa | |
|
|
||||
| N | 23 | 32 | 48 | 48 |
| Germline LS gene mutation, N (%)* | 23 (100%) | 32 (100%) | - | - |
| MLH1 | 6 (25%) | 7 (21%) | - | - |
| MSH2 | 9 (38%) | 15 (44%) | - | - |
| MSH6 | 6 (25%) | 8 (24%) | - | - |
| PMS2 | 2 (8%) | 4 (12%) | - | - |
| EPCAM | 1 (4%) | 0 (0%) | - | - |
| Age, median [IQR] | 48 [42–57] | 47 [42–60] | 49 [44–59] | 48 [38–56] |
| Caucasian (%) | 21 (91%) | 28 (88%) | 40 (83%) | 47 (98%) |
| Females (%) | 8 (35%) | 20 (62%) | 15 (31%) | 22 (46%) |
| Tobacco use, current (%) | 6 (26%) | 3 (9%) | 7 (15%) | 6 (12%) |
| Site, proximal (%) | 13 (57%) | - | 17 (35%) | - |
| Stage (%) | ||||
| I | 10 (43%) | 7 (15%) | ||
| II | 6 (26%) | 8 (17%) | ||
| III | 7 (30%) | 21 (44%) | ||
| IV | 0 (0%) | 12 (25%) | ||
IQR, interquartile range; LS, Lynch Syndrome
Some patients had more than one mutation: One cancer and one benign had MSH2 and MSH6 and one benign had MLH1 and PMS2
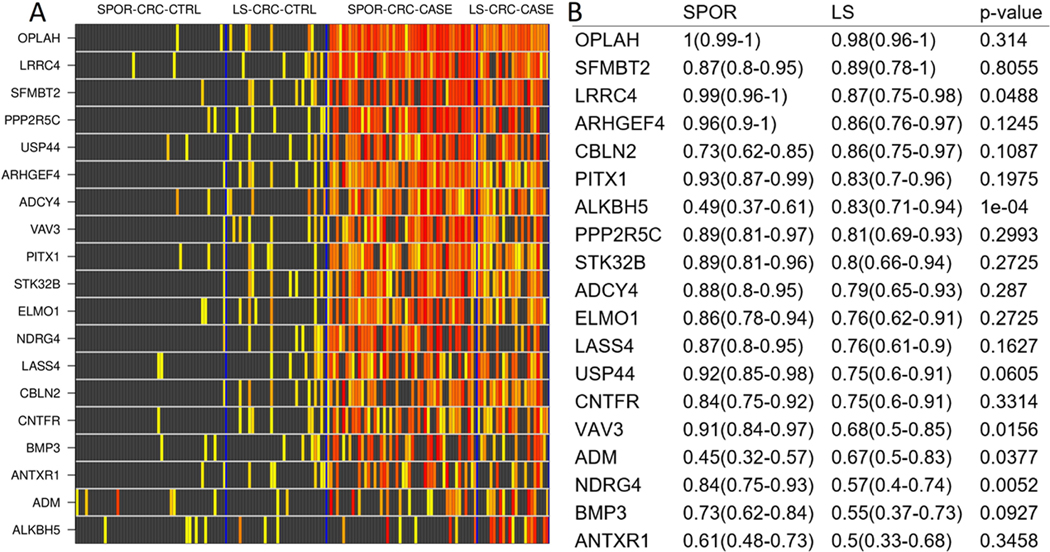
Several MDM candidates from the selected panel showed excellent separation between normal colorectal epithelium and CRC from subjects with and without LS. Boxplots demonstrating higher methylation in CRC tissue compared to benign colorectal epithelium are presented in Supplemental Figure 1. For these markers, tissue methylation levels of MDMs were several- fold higher in LS-CRC-CASE and SPOR-CRC-CASE tissues versus benign colorectal epithelium, which in some cases yielded low or negligible levels.
MDMs discriminated similarly between CRC and benign colorectal epithelium in both LS and non-LS groups as demonstrated in the heat matrix in Figure 2, which shows individual MDM signal intensity for each subject in case and control tissue. Overall, signal intensity is highest for each candidate MDM in both LS-CRC-CASE and SPOR-CRC-CASE tissues. Discrimination between benign colorectum and CRC as assessed by AUCs ranged from 0.5 to 0.98 for those with LS and did not differ significantly between those with and without LS except for two candidate MDMs, ALKBH5 (AUC 0.49 non-LS vs. AUC 0.83 LS, p=0.0004) and NDRG4 (AUC 0.84 non-LS vs. AUC 0.57, p = 0.0052) (Figure 2). The candidate MDMs with the highest discrimination between CRC and benign colorectum were OPLAH, SFMBT2, LRRC4, ARHGEF4, CBLN2, all with AUCs >0.85, and not significantly different between LS and non-LS groups.
Figure 2:
A.) Heat matrix showing methylation intensity of methylated DNA marker candidates for each colorectal cancer case and control tissue sample. Each row is an MDM quantitative PCR product, normalized to ACTB, and each row is an individual patient DNA sample. Black boxes represent normalized MDM values below the 90%-ile cut off in control tissues (LS-CRC-CTRL and SPOR-CRC-CTRL benign colorectal mucosa, combined); increasing color intensity on the yellow-red spectrum corresponds to increasing deciles of MDM levels exceeding the cut-off. LS-CRC-CTRL, Lynch-associated benign colorectal epithelium; SPOR-CRC-CTRL, sporadic benign colorectal epithelium; SPOR-CRC-CASE, sporadic colorectal cancer; LS-CRC-CASE, Lynch-associated colorectal cancer. B.) Corresponding area under the receiver operating characteristics curve values with 95% confidence intervals for each normalized MDM stratified by LS or sporadic status
LS-EC study
A total of 144 subjects met LS-EC study inclusion criteria generating 30 LS-EC-CASE, 48 SPOR-EC-CASE, 29 LS-EC-CRTL, and 37 SPOR-EC-CRTL. To reach the final LS-EC cohort of 144, the following were excluded: 9 did not meet clinical eligibility, 15 were excluded after pathology review or tissue was not available, and 2 had low tissue-derived DNA quality and were excluded (Figure 1). Patient demographics and tumor characteristics of the 144 LS-EC study subjects included in the final analysis are shown in Table 2. Median age was higher for SPOR-EC-CASEs at 61 (IQR 55, 64) compared to LS-EC-CASEs at 53 (IQR 51, 58) years (p= 0.01) and median BMI was also higher among SPOR-EC-CASEs at 33 (IQR 26–39) compared to LS-EC-CASEs at 28 [23–32] Kg/m2 (p= 0.08).
Table 2:
Demographics and characteristics of cases and controls in the Lynch syndrome endometrial cancer study
| Lynch | Sporadic | |||
|---|---|---|---|---|
|
|
||||
| Endometrial Cancer | Benign Endometrium | Endometrial Cancer | Benign Endometrium | |
|
|
||||
| N | 30 | 29 | 48 | 37 |
| Germline LS gene mutation, N (%)* | 30 (100%) | 29 (100%) | - | - |
| MLH1 | 0 (0%) | 5 (17%) | - | - |
| MSH2 | 10 (33.3%) | 14 (46%) | - | - |
| MSH6 | 16 (53.3%) | 3 (10%) | - | - |
| PMS2 | 3 (10%) | 6 (20%) | - | - |
| EPCAM | 1 (3.3%) | 2 (7%) | - | - |
| Age, median [IQR] | 53 [51–58] | 46 [41–53] | 61 [55–64] | 48 [44–53] |
| Caucasian (%) | 29 (97%) | 29 (100%) | 42 (88%) | 35 (95%) |
| Tobacco use, Current, N (%) | 5 (17%) | 2 (7%) | 7 (15%) | 0 (0%) |
| BMI, median [IQR] | 28 [23–32] | 25 [22–28] | 33 [26–39] | 29 [25–36] |
| Post-menopause, N (%) | 15 (50%) | 7 (24%) | 32 (67%) | 7 (19%) |
| Cancer Histology, N (%) | ||||
| Endometrioid | 28 (93%) | - | 39 (81%) | - |
| Clear Cell | 2 (7%) | - | 3 (6%) | - |
| Serous | 0 (0%) | - | 6 (13%) | - |
IQR, interquartile range; LS, Lynch Syndrome
Some patients had more than one mutation: one benign had MSH2 and EPCAM
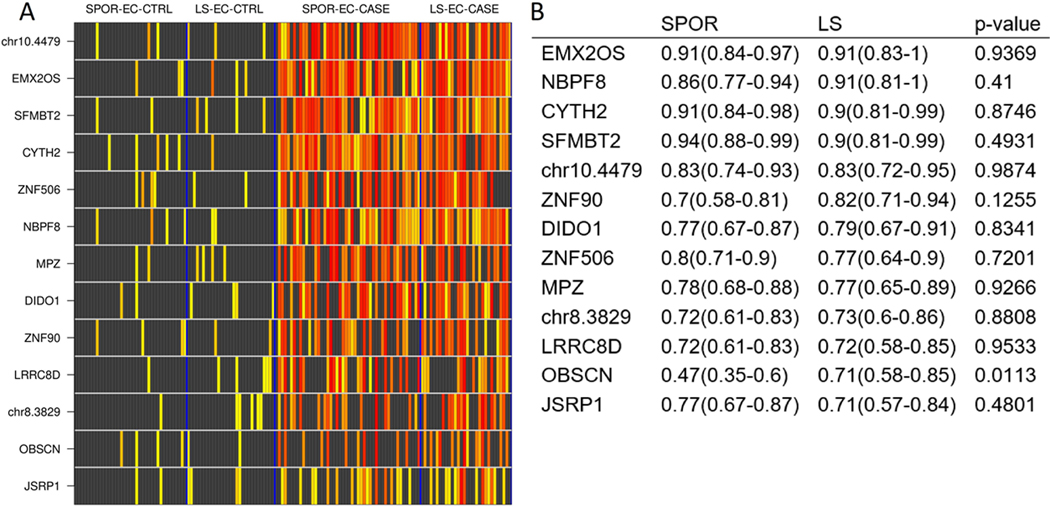
Several EC MDM candidates demonstrated discrimination between benign endometrium and EC in both LS groups (LS-EC-CASE and LS-EC-CRTL) and non-LS groups (SPOR-EC-CASE and SPOR-EC-CRTL). Boxplots demonstrating higher relative methylation of MDMs in EC tissue compared to benign endometrium are shown in Supplemental Figure 2. Specifically, methylation was low or negligible in benign endometrium from both LS and non-LS groups with several fold increases in methylation observed in both LS-EC-CASE and SPOR-EC-CASE tissues.
EC MDMs discriminated similarly between EC and benign endometrium in both LS and non-LS groups as demonstrated in the heat matrix in Figure 3, which shows individual MDM signal intensity for each patient in case and control tissue. Overall, signal intensity is higher for each candidate MDM in both LS-EC-CASE and SPOR-EC-CASE tissues compared to LS-EC-CRTL and SPOR-EC-CRTL, respectively. Areas under the receiver operator curve (AUCs) ranged from 0.71 to 0.91 in discriminating between EC and benign endometrium in the LS groups. Per MDM, AUCs did not differ significantly between LS groups’ comparisons and non-LS groups’ comparisons with the exception of OBSCN (LS AUC 0.71, non-LS AUC 0.47; p=0.01) (Figure 3B). The candidate MDMs with the greatest discrimination between EC and benign endometrium were SFMBT2, CYTH2, NBPF8, and EMXOS, all with AUCs ≥ 0.90.
Figure 3:
A.) Heat matrix showing methylation intensity of methylated DNA marker candidates for each endometrial cancer case and control tissue sample. Each row is an MDM quantitative PCR product, normalized to ACTB, and each row is an individual patient DNA sample. Black boxes represent normalized MDM values below the 90%-ile cut off in control tissues (LS-EC-CTRL and SPOR-EC-CTRL benign endometrium, combined); increasing color intensity on the yellow-red spectrum corresponds to increasing deciles of MDM levels exceeding the cut-off. LS-EC-CTRL, Lynch-associated benign endometrium; SPOR-EC-CTRL, sporadic benign endometrium; SPOR-EC-CASE, sporadic endometrial cancer; LS-EC-CASE, Lynch associated endometrial cancer. B.) Corresponding area under the receiver operating characteristics curve values with 95% confidence intervals for each normalized MDM stratified by LS or sporadic status
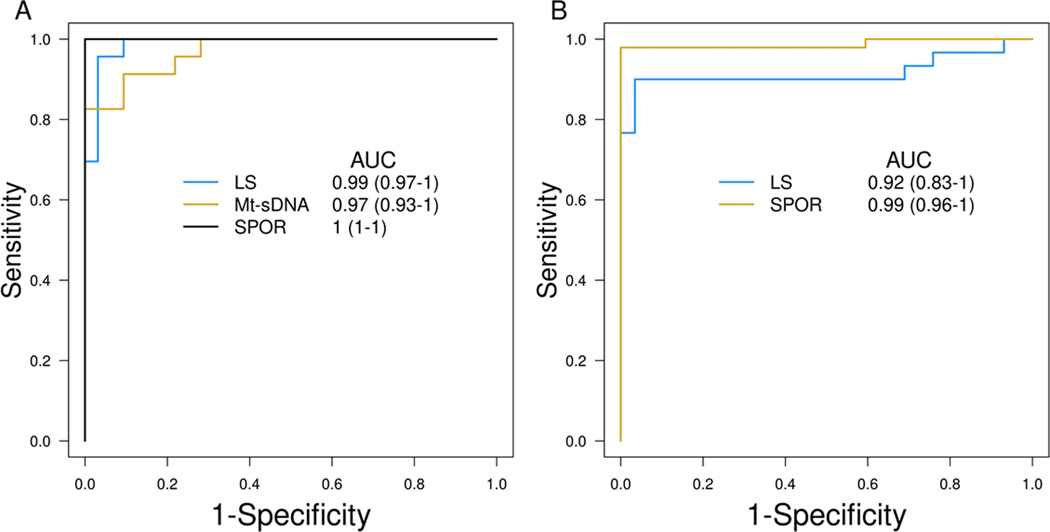
Multi-marker Panel
In a sub-analysis of the LS-CRC study performed using a multi-marker (LASS4, LRRC4, PPP2R5C) logistic regression model based on the 2nd generation mt-sDNA test, the AUC was 0.92 (Figure 4A). For EC, AUCs constructed from a 6-MDM panel (SFMBT2, MPZ, CYTH2, DIDO1, chr10.4479, and EMX2OS) normalized to ACTB, in both LS and non-LS groups exceeded 0.90 and had overlapping 95% CI values (Figure 4B).
Figure 4:
Receiver operating characteristics curves for A.) normalized MDM panel in LS-CRC and SPOR-CRC, based on NDRG4, LRRC4, PPP2R5C, and OPLAH, as well as mt-sDNA 2.0 panel (LASS4, LRRC4, PPP2R5C); and B.) normalized MDM panel in LS-EC and SPOR-EC, based on SFMBT2, MPZ, CYTH2, DIDO1, chr10.4479, and EMX2OS.
DISCUSSION:
In this case-control study using DNA from archival tissues, we demonstrate that previously-identified candidate MDMs that discriminate between cancer (CRC or EC) and benign control tissue in patients without LS perform similarly in discriminating between cancer and benign in those with LS. Several candidate markers in both CRC and EC exhibited excellent discrimination between cancer and benign tissue with high AUCs as well as several fold increases in tissue levels in those with CRC or EC. This suggests that MDMs that could identify the presence of CRC or EC in biospecimens from average risk people may also have similar performances in people with LS.
Lynch syndrome is the prototypical example of a cancer predisposition syndrome where early diagnosis through genetic testing and then implementation of rigorous cancer screening of the colorectum and risk-reducing gynecologic surgery has led to significant improvement in cancer free survival (18–20). However, the search for novel biomarkers has not yet identified candidates that can either out-perform or supplement current invasive screening for CRC, truly screen for EC, or provide an alternative to risk-reducing gynecologic surgery.
Aberrant DNA methylation is an early event in carcinogenesis(21) and appears to be more broadly informative than most protein and gene mutation biomarkers for early detection of cancer (22,23). Furthermore, recent advances in technology have resulted in improved analytic sensitivity for MDMs when assayed in stool and plasma(24–27). Overall, the role of MDMs in LS is at a nascent stage and continues to evolve.
While this technology is not new, this is the first report of leveraging unique MDMs for EC identification in the setting of LS and only the second report for CRC detection in LS. The multitarget stool DNA test (mt-sDNA, Cologuard® Exact Sciences, WI, USA) is a commercially available, FDA-approved, non-invasive screening test for the detection of CRC and high risk precancers in the average risk screening setting. Since the development of mt-sDNA, additional highly discriminant MDMs for the detection of sporadic CRC have been described and validated, showing excellent performance in both tissues and stool samples of patients with early-stage disease as well as advanced precursor lesions (15). As such the 2nd generation mt-sDNA test is currently under study in a >20,000 patient cohort trial, intended for FDA review [NCT04144738](13,15). In our prior work, we discovered MDMs for the detection of CRC in those with LS and found that the MDMs in the current, first-generation multi-target stool DNA test (Cologuard) perform better for sporadic CRC than LS-associated CRC, which dampened enthusiasm for further study of these markers in the LS population.(16) In the current analysis, a broader panel of novel MDMs discovered and validated in sporadic CRC were applied to LS-associated CRC. The cross-validated AUC of 0.92 for the classification of LS-CRC using MDMs under development for a second-generation stool DNA test (28) portends the possible expanded the use of such a product in patients with LS.
This study also demonstrated that EC-unique MDMs appear to discriminate between benign endometrium and EC in females with LS similar to females without LS. We previously observed that three candidate MDMs, EMX2OS, NBPF8, and SFMBT2, included in this study of females with LS were highly discriminant between sporadic EC and control tissues exhibiting 97% specificity, 97% sensitivity, and an AUC of 0.98. MDMs exhibiting a high signal strength in tissue-derived DNA are anticipated to be detectable in alternative biospecimens, such as lower genital tract samples, lending to non-invasive or minimally invasive means of collection. In fact, leveraging molecular markers within lower genital tract biospecimens has been the focus of several approaches aiming to identify EC through less invasive means. This has ranged from microscopic assessment of cytologic abnormalities in both cervicovaginal scrapings (29) and direct endometrial brushings (30) to determining the presence of EC-associated DNA mutations, methylation, and/or aneuploidy in liquid-based Papanicolaou smears (31–34). Perhaps the least invasive is that of leveraging self-collected vaginal fluid as a biospecimen. We and others have demonstrated that EC-associated methylated DNA, mutations, and/or aneuploidy can be detected in tampon-collected vaginal fluid. Given the high sensitivity and specificity of MDMs to detect EC in vaginal fluid from people without known LS(35), the findings of this present study suggest the potential for similar promise in people with LS.
Therefore, highly discriminant candidate MDMs noted in this study should be further evaluated to confirm detection with minimally invasive sample collection methods and may serve as a means of CRC and/or EC surveillance or early detection in people with LS.
One challenge is that the relatively low prevalence of LS has thwarted past efforts to cost-effectively develop non-invasive screening from a single medium based on a rationale that leverages their cumulative prevalence. Though it is possible to identify and validate MDMs specific for the detection of CRC/EC in patients with LS, there are several potential advantages to validating sporadic EC/CRC markers in patients with LS. First, it is logistically difficult to enroll prospective clinical samples from LS case patients who may undergo treatment for cancer before confirmation of the LS diagnosis. Second, those with LS who might benefit from early detection of CRC or EC might not yet have a known LS diagnosis. Third, the regulatory requirements for an CRC or EC detection test are anticipated to be equally as stringent for LS patients as they are for non-LS populations.
Longitudinal surveillance efforts with colonoscopy and endometrial biopsy can detect CRC and potentially EC at early stages and historical observational studies have reported improvement in survival compared to no surveillance in CRC (19,20). However, endometrial sampling as a screening test has not been shown to impact EC outcomes in LS (36). Additionally, colonoscopy and endometrial biopsy come with several disadvantages including: a) small risks of bleeding and organ perforation during the procedures; b) time off work or school for bowel preparation and endoscopic procedure or the provider-collected endometrial sampling. For females with LS, the current highest impact defense against EC is that of risk-reducing hysterectomy which poses the additional risks of surgical morbidity(36). A biomarker-based test that can detect early CRC or EC could transform the screening paradigm in LS by refining case selection for those most likely to benefit from invasive procedures or risk-reducing surgery and optimize the utilization of LS-associated healthcare resources.
There are several potential limitations to the current study. First, the retrospective design required us to rely on data from the medical record and could have introduced selection biases for identification of cases and controls. In the LS-EC study, age and menopausal status imbalances likely reflect inherent clinical selection biases associated with a younger average age of EC onset in women with LS compared to sporadic EC. Inclusion criteria for LS-EC required hysterectomy to ensure adequate tissue for DNA extraction and we included all LS subjects identified that met inclusion criteria. Women with LS tend to undergo risk-reducing hysterectomy in their mid-to-late 40s and once risk-reducing hysterectomy has been performed, all endometrium has been eliminated from that person (36). Archival formalin fixed specimens may have been susceptible to DNA degradation; however, all samples underwent DNA quantification and had adequate ACTB amplification prior to methylation specific PCR. In addition, the discovery effort for the panel utilized in this study was completed with frozen tissue and subsequently validated in FFPE tissue samples. There was limited racial and ethnic diversity in the samples utilized in this study which could impact the validity and performance of the MDM markers in more diverse populations and should be investigated in future studies. Although colorectal case and control tissues were balanced on age and LS control tissues came from a broad representation of colon segments, we were not able to match the segmental locations of case primary tumors and control mucosae. Further, the majority of non-LS control patients underwent resections for diverticulitis. This limitation is mitigated, in part, by recent observations that the markers we studied here also show very high specificity across a broad range of patient ages and benign conditions, when assayed from stool samples, representative of whole colon mucosa, in a recent study of over 20,000 patients(37). Our exclusion of microsatellite instability-high sporadic CRC tumors could also be viewed as a limitation; these cancers are frequently hypermethylated (38) and have been shown to arise from sessile serrated precursors (39), which are readily detected by both 1st and 2nd generation mt-sDNA tests (15,23). A strength of this study was the genetic confirmation of LS in patients with the disease, and inclusion of mostly early-stage LS cancer tissues. Lastly, while we did not meet our pre-specified sample size in all groups, this was mitigated in part by a larger than assumed effect size.
In conclusion, this study provides evidence that previously identified MDMs found to discriminate between CRC and benign colorectal epithelium and EC and benign endometrium in general population risk-based studies perform similarly well among people with LS. While this study was done in tissue samples, several markers performed with AUCs >0.85 and demonstrated several fold higher tissue-derived DNA methylation of candidate MDMs in cancer compared to benign tissue. These findings support the feasibility of cancer detection in stool and lower gynecologic genital tract samples in the setting of LS and warrant further testing of the highest performing candidates. An attractive finding was that a panel of markers included in the second-generation stool DNA test performed extremely well in discriminating CRC in LS. This has now led to a prospective multi-center clinical trial of patients with LS for evaluation of MDMs for CRC detection (CORAL, NCT05410977) and would justify a similar trial for EC detection. This would provide a new avenue for cancer surveillance and screening in this high-risk population.
Supplementary Material
Prevention Relevance Statement.
Lynch Syndrome (LS) markedly increases risks of colorectal (CRC) and endometrial (EC) cancers. Early detection biomarkers for LS cancers could reduce the needs for invasive screening and surgery. Methylated DNA markers previously identified in sporadic EC and CRC discriminate between benign and cancer tissue in LS.
Acknowledgements
Funding:
Funding for this study was from the Mayo Clinic Center for Individualized Medicine and Gerstner Foundation Career Development Award (N.J. Samadder). Additional support was provided by a sponsored research agreement between Mayo Clinic and Exact Sciences (Madison WI) as well as CA214679 to J.B. Kisiel and funding from the V Foundation (T2016–001-03) to J.N. Bakkum-Gamez. This work utilized Pathology Research Core, Biospecimens Accessioning & Processing, and Pathology Research Core shared resources of the Mayo Clinic Cancer Center Support Grant (P30 CA015083). Its contents are solely the responsibility of the authors and do not necessarily represent the official view of the National Institutes of Health.
Mark E. Sherman has received collaborative research funding supported by Exact Sciences. Portions of this research were presented in abstract form at the Society of Gynecologic Oncology Annual Meeting in March 2022 in Phoenix, AZ.
Abbreviations:
- AUC
area under (receiver operating characteristics) curve
- CRC
colorectal cancer
- CpG
cytosine-phosphate-guanine
- DNA
deoxyribonucleic acid
- EC
endometrial cancer
- FFPE
formalin fixed paraffin embedded
- LS
Lynch syndrome
- LS-CRC-CASE
Lynch syndrome colorectal cancer case patient
- LS-CRC-CTRL
Lynch syndrome colorectal cancer control patient
- LS-EC-CASE
Lynch syndrome endometrial cancer case patient
- LS-EC-CTRL
Lynch syndrome endometrial cancer control patient
- MDMs
methylated DNA markers
- Mt-sDNA
(multitarget stool DNA test)
- PCR
polymerase chain reaction
Footnotes
Conflict of Interest Statement: Jamie N. Bakkum-Gamez, William R. Taylor, Doulas W. Mahoney, and John B. Kisiel contributed to Mayo Clinic intellectual property which is licensed to Exact Sciences (Madison WI) and may receive royalties, paid to Mayo Clinic.
REFERENCES
- 1.de la Chapelle A. The incidence of Lynch syndrome. Fam Cancer 2005;4(3):233–7 doi 10.1007/s10689-004-5811-3. [DOI] [PubMed] [Google Scholar]
- 2.de la Chapelle A, Palomaki G, Hampel H. Identifying Lynch syndrome. Int J Cancer 2009;125(6):1492–3 doi 10.1002/ijc.24491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stoffel EM, Mangu PB, Gruber SB, Hamilton SR, Kalady MF, Lau MW, et al. Hereditary colorectal cancer syndromes: American Society of Clinical Oncology Clinical Practice Guideline endorsement of the familial risk-colorectal cancer: European Society for Medical Oncology Clinical Practice Guidelines. J Clin Oncol 2015;33(2):209–17 doi 10.1200/JCO.2014.58.1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weiss JM, Gupta S, Burke CA, Axell L, Chen LM, Chung DC, et al. NCCN Guidelines(R) Insights: Genetic/Familial High-Risk Assessment: Colorectal, Version 1.2021. J Natl Compr Canc Netw 2021;19(10):1122–32 doi 10.1164/jnccn.2021.0048. [DOI] [PubMed] [Google Scholar]
- 5.Giardiello FM, Allen JI, Axilbund JE, Boland CR, Burke CA, Burt RW, et al. Guidelines on genetic evaluation and management of Lynch syndrome: a consensus statement by the US Multi-society Task Force on colorectal cancer. Am J Gastroenterol 2014;109(8):1159–79 doi 10.1038/ajg.2014.186. [DOI] [PubMed] [Google Scholar]
- 6.Stjepanovic N, Moreira L, Carneiro F, Balaguer F, Cervantes A, Balmana J, et al. Hereditary gastrointestinal cancers: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-updagger. Ann Oncol 2019;30(10):1558–71 doi 10.1093/annonc/mdz233. [DOI] [PubMed] [Google Scholar]
- 7.Braun MM, Overbeek-Wager EA, Grumbo RJ. Diagnosis and Management of Endometrial Cancer. Am Fam Physician 2016;93(6):468–74. [PubMed] [Google Scholar]
- 8.Shaukat A, Kahi CJ, Burke CA, Rabeneck L, Sauer BG, Rex DK. ACG Clinical Guidelines: Colorectal Cancer Screening 2021. Am J Gastroenterol 2021;116(3):458–79 doi 10.14309/ajg.0000000000001122. [DOI] [PubMed] [Google Scholar]
- 9.Force USPST Davidson KW, Barry MJ Mangione CM, Cabana M Caughey AB, et al. Screening for Colorectal Cancer: US Preventive Services Task Force Recommendation Statement. JAMA 2021;325(19):1965–77 doi 10.1001/jama.2021.6238. [DOI] [PubMed] [Google Scholar]
- 10.Wolf AMD, Fontham ETH, Church TR, Flowers CR, Guerra CE, LaMonte SJ, et al. Colorectal cancer screening for average-risk adults: 2018 guideline update from the American Cancer Society. CA Cancer J Clin 2018;68(4):250–81 doi 10.3322/caac.21457. [DOI] [PubMed] [Google Scholar]
- 11.Bakkum-Gamez JN, Wentzensen N, Maurer MJ, Hawthorne KM, Voss JS, Kroneman TN, et al. Detection of endometrial cancer via molecular analysis of DNA collected with vaginal tampons. Gynecol Oncol 2015;137(1):14–22 doi 10.1016/j.ygyno.2015.01.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fiegl H, Gattringer C, Widschwendter A, Schneitter A, Ramoni A, Sarlay D, et al. Methylated DNA collected by tampons--a new tool to detect endometrial cancer. Cancer Epidemiol Biomarkers Prev 2004;13(5):882–8. [PubMed] [Google Scholar]
- 13.Bakkum-Gamez JN, Sherman ME, Slettedahl SW, Mahoney DW, Lemens MA, Laughlin-Tommaso SK, et al. Detection of endometrial cancer using tampon-based collection and methylated DNA markers. Gynecol Oncol 2023;174:11–20 doi 10.1016/j.ygyno.2023.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Veroushka Ballester MG, Tracy C. Yab, Taylor William R., Foote Patrick, Devens Mary E., Mahoney Douglas W., Boardman Lisa A., Kisiel John B., Allawi Hatim T., Lidgard Graham P., Marcia R. Cruz-Correa, David A. Ahlquist. Neoplasia: Do Markers That Target Acquired DNA Alterations in Sporadic Cases Also Discriminate Lynch Syndrome Cases? Gastroenterology 2015;148(4):S–355. [Google Scholar]
- 15.Bosch LJW, Melotte V, Mongera S, Daenen KLJ, Coupe VMH, van Turenhout ST, et al. Multitarget Stool DNA Test Performance in an Average-Risk Colorectal Cancer Screening Population. Am J Gastroenterol 2019;114(12):1909–18 doi 10.14309/ajg.0000000000000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ballester V, Taylor WR, Slettedahl SW, Mahoney DW, Yab TC, Sinicrope FA, et al. Novel methylated DNA markers accurately discriminate Lynch syndrome associated colorectal neoplasia. Epigenomics 2020;12(24):2173–87 doi 10.2217/epi-2020-0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kisiel JB, Klepp P, Allawi HT, Taylor WR, Giakoumopoulos M, Sander T, et al. Analysis of DNA Methylation at Specific Loci in Stool Samples Detects Colorectal Cancer and High-Grade Dysplasia in Patients With Inflammatory Bowel Disease. Clin Gastroenterol Hepatol 2019;17(5):914–21 e5 doi 10.1016/j.cgh.2018.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jarvinen HJ, Aarnio M, Mustonen H, Aktan-Collan K, Aaltonen LA, Peltomaki P, et al. Controlled 15-year trial on screening for colorectal cancer in families with hereditary nonpolyposis colorectal cancer. Gastroenterology 2000;118(5):829–34. [DOI] [PubMed] [Google Scholar]
- 19.Dove-Edwin I, Sasieni P, Adams J, Thomas HJ. Prevention of colorectal cancer by colonoscopic surveillance in individuals with a family history of colorectal cancer: 16 year, prospective, follow-up study. BMJ 2005;331(7524):1047 doi 10.1136/bmj.38606.794560.EB. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moller P, Seppala T, Bernstein I, Holinski-Feder E, Sala P, Evans DG, et al. Cancer incidence and survival in Lynch syndrome patients receiving colonoscopic and gynaecological surveillance: first report from the prospective Lynch syndrome database. Gut 2017;66(3):464–72 doi 10.1136/gutjnl-2015-309675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suzuki K, Suzuki I, Leodolter A, Alonso S, Horiuchi S, Yamashita K, et al. Global DNA demethylation in gastrointestinal cancer is age dependent and precedes genomic damage. Cancer Cell 2006;9(3):199–207 doi 10.1016/j.ccr.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 22.Levin TR, Corley DA, Jensen CD, Marks AR, Zhao WK, Zebrowski AM, et al. Genetic Biomarker Prevalence Is Similar in Fecal Immunochemical Test Positive and Negative Colorectal Cancer Tissue. Dig Dis Sci 2017;62(3):678–88 doi 10.1007/s10620-016-4433-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heigh RI, Yab TC, Taylor WR, Hussain FT, Smyrk TC, Mahoney DW, et al. Detection of colorectal serrated polyps by stool DNA testing: comparison with fecal immunochemical testing for occult blood (FIT). PLoS One 2014;9(1):e85659 doi 10.1371/journal.pone.0085659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berger BM, Ahlquist DA. Stool DNA screening for colorectal neoplasia: biological and technical basis for high detection rates. Pathology 2012;44(2):80–8 doi 10.1097/PAT.0b013e3283502fdf. [DOI] [PubMed] [Google Scholar]
- 25.Zou H, Allawi H, Cao X, Domanico M, Harrington J, Taylor WR, et al. Quantification of methylated markers with a multiplex methylation-specific technology. Clin Chem 2012;58(2):375–83 doi 10.1373/clinchem.2011.171264. [DOI] [PubMed] [Google Scholar]
- 26.Kisiel JB, Dukek BA, R VSRK, Ghoz HM, Yab TC, Berger CK, et al. Hepatocellular Carcinoma Detection by Plasma Methylated DNA: Discovery, Phase I Pilot, and Phase II Clinical Validation. Hepatology 2019;69(3):1180–92 doi 10.1002/hep.30244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marinelli LM, Kisiel JB, Slettedahl SW, Mahoney DW, Lemens MA, Shridhar V, et al. Methylated DNA markers for plasma detection of ovarian cancer: Discovery, validation, and clinical feasibility. Gynecol Oncol 2022;165(3):568–76 doi 10.1016/j.ygyno.2022.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kisiel JB GZ, Krockenberger M, Bhattacharya A, Bourne BL, Leduc CM, Matter MB, Fourrier KD, Edwards D, Limburg PJ, Domanico MJ. Can second-generation multitarget stool DNA panels reliably detect colorectal cancer and advanced precancerous lesions?. Journal of Clinical Oncology 2022;40:63.34793256 [Google Scholar]
- 29.Frias-Gomez J, Benavente Y, Ponce J, Brunet J, Ibanez R, Peremiquel-Trillas P, et al. Sensitivity of cervico-vaginal cytology in endometrial carcinoma: A systematic review and meta-analysis. Cancer Cytopathol 2020;128(11):792–802 doi 10.1002/cncy.22266. [DOI] [PubMed] [Google Scholar]
- 30.DeJong SR, Bakkum-Gamez JN, Clayton AC, Henry MR, Keeney GL, Zhang J, et al. Tao brush endometrial cytology is a sensitive diagnostic tool for cancer and hyperplasia among women presenting to clinic with abnormal uterine bleeding. Cancer Med 2021;10(20):7040–7 doi 10.1002/cam4.4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kinde I, Bettegowda C, Wang Y, Wu J, Agrawal N, Shih Ie M, et al. Evaluation of DNA from the Papanicolaou test to detect ovarian and endometrial cancers. Sci Transl Med 2013;5(167):167ra4 doi 10.1126/scitranslmed.3004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y, Li L, Douville C, Cohen JD, Yen TT, Kinde I, et al. Evaluation of liquid from the Papanicolaou test and other liquid biopsies for the detection of endometrial and ovarian cancers. Sci Transl Med 2018;10(433) doi 10.1126/scitranslmed.aap8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schreiberhuber L, Herzog C, Vavourakis CD, Redl E, Kastner C, Jones A, et al. The WID-qEC test: Performance in a hospital-based cohort and feasibility to detect endometrial and cervical cancers. Int J Cancer 2023;152(6):1269–74 doi 10.1002/ijc.34275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herzog C, Marin F, Jones A, Evans I, Reisel D, Redl E, et al. A Simple Cervicovaginal Epigenetic Test for Screening and Rapid Triage of Women With Suspected Endometrial Cancer: Validation in Several Cohort and Case/Control Sets. J Clin Oncol 2022;40(33):3828–38 doi 10.1200/JCO.22.00266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bakkum-Gamez JN. Repurposing the vaginal tampon for endometrial cancer detection. Biomark Med 2015;9(8):715–7 doi 10.2217/BMM.15.44. [DOI] [PubMed] [Google Scholar]
- 36.Lim N, Hickey M, Young GP, Macrae FA, Kelly C. Screening and risk reducing surgery for endometrial or ovarian cancers in Lynch syndrome: a systematic review. Int J Gynecol Cancer 2022;32(5):646–55 doi 10.1136/ijgc-2021-003132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dickinson L. 2023. Next generation cologuard test demonstrates 94 percent sensitivity for colorectal cancer at 91 percent specificity, raising the bar in non-invasive screening. [cited 2023 Aug 30] <https://www.prnewswire.com/news-releases/next-generation-cologuard-test-demonstrates-94-percent-sensitivity-for-colorectal-cancer-at-91-percent-specificity-raising-the-bar-in-non-invasive-screening-301855721.html#:~:text=Next%2Dgeneration%20Cologuard%20Test%20Demonstrates,Bar%20in%20Non%2Dinvasive%20Screening>. [Google Scholar]
- 38.Guinney J, Dienstmann R, Wang X, de Reynies A, Schlicker A, Soneson C, et al. The consensus molecular subtypes of colorectal cancer. Nat Med 2015;21(11):1350–6 doi 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bettington M, Walker N, Clouston A, Brown I, Leggett B, Whitehall V. The serrated pathway to colorectal carcinoma: current concepts and challenges. Histopathology 2013;62(3):367–86 doi 10.1111/his.12055. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data that support the findings of this study are available from the corresponding authors upon reasonable request.