Abstract
Peanut (Arachis hypogaea L.) is one of the most important oil crops in the world due to its lipid-rich seeds. Lipid accumulation and degradation play crucial roles in peanut seed maturation and seedling establishment, respectively. Here, we utilized lipidomics and transcriptomics to comprehensively identify lipids and the associated functional genes that are important in the development and germination processes of a large-seed peanut variety. A total of 332 lipids were identified; triacylglycerols (TAGs) and diacylglycerols were the most abundant during seed maturation, constituting 70.43 and 16.11%, respectively, of the total lipids. Significant alterations in lipid profiles were observed throughout seed maturation and germination. Notably, TAG (18:1/18:1/18:2) and (18:1/18:2/18:2) peaked at 23386.63 and 23392.43 nmol/g, respectively, at the final stage of seed development. Levels of hydroxylated TAGs (HO-TAGs) increased significantly during the initial stage of germination. Accumulation patterns revealed an inverse relationship between free fatty acids and TAGs. Lipid degradation was determined to be regulated by diacylglycerol acyltransferase, triacylglycerol lipase, and associated transcription factors, predominantly yielding oleic acid, linoleic acid, and linolenic acid. Collectively, the results of this study provide valuable insights into lipid dynamics during the development and germination of large-seed peanuts, gene resources, and guiding future research into lipid accumulation in an economically important crop.
Keywords: peanut, lipidomics, HO-TAGs, development, germination
Introduction
Peanut (Arachis hypogaea L.) is one of the most important oilseed crops globally, providing both edible oil and protein-rich seeds for human consumption.1 The seeds of peanuts are the main components of this crop consumed by humans. The composition of lipid species in peanuts is critical for both human health and nutrition. Therefore, understanding lipid metabolism in peanut seeds is crucial for human health and nutritional value.
Lipids are a vast group of naturally occurring molecules that are insoluble in water but soluble in nonpolar solvents. They store energy, act as structural components of cell membranes, and signal biological processes. Lipids are also involved in many biological processes in plants, including seed germination, organ differentiation, leaf senescence, and pollination.2 With advances in genomics, transcriptomics, and proteomics, researchers have adopted metabolomics, a qualitative and quantitative analysis of all metabolites, to facilitate functional genomics research. Lipidomics, first defined by Han and Gross, is a discipline concerned with studying the characterization of all lipid molecules in organisms and their roles in protein expression and gene regulation.3,4 Recent years have seen much attention being paid to lipid research, and lipidomics research methods have made significant breakthroughs.
The application of advanced mass spectrometry has enabled the quantitative analysis of eight lipid types, including fatty acids (FAs), glycerolipids (GLs), glycerophospholipids (GPs), sphingolipids (SPs), sterol lipids (STs), prenol lipids, saccharolipids, and polyketides, in various crops, including peanut, hickory (Carya cathayensis Sarg.), rape (Brassica napus L.), and hazelnut (Corylus heterophylla Fisch. ex Trautv.).5−8 The local concentrations of lipid species are essential at different plant growth stages.6 Our earlier study revealed that the initiation of FA accumulation in high-oleic peanut seeds occurred in the earliest stages after flowering.9
Seed size is a critical agronomic trait for crops, and peanut has over 300 varieties grown in tropical and subtropical regions.10 Usually, large seeds contain more nutrients and energy stored and thus have a higher potential for germination and stronger environmental stress tolerance during seedling establishment.11 Peanut seeds contain protein, starch, lipids, flavonoids, and other chemical constituents.12 Seed development quality directly affects seed germination, seedling morphogenesis, and even crop yield. The size of the seeds is closely related to the characteristics of the seedlings, especially if their growth primarily depends on substances stored in the seeds. Additionally, the amount of lipid accumulation in peanut seeds determines their yield and quality.
Peanut is an allotetraploid, with its diploid ancestors being Arachis duranensis (AA) and Arachis ipaensis (BB). The genome sequences of cultivated tetraploid peanut species, including A. hypogaea cv. Tifrunner, Shitouqi, and Fuhuasheng, the wild allotetraploid species Arachis monticola, and the diploid wild species A. duranensis and A. ipaensis, have been completed.7,13−17 Advances in transcriptomics and lipidomics offer insights into transcriptional regulation and metabolic flux. Screening lipid metabolites and their associated genes provides valuable information regarding the lipid biosynthetic and metabolic pathways, as well as the associated regulatory networks.18 In recent studies, researchers have investigated the lipid composition and changes in embryogenesis, maturation, and roasting of peanut seeds, especially high-oleic peanut seeds.7,19 To date, however, no study has been conducted on the lipidomics profile during seed development and germination stages in large-seeded peanut varieties, which can reveal the dynamic changes of key nutrient lipid molecules at the different seed mature stages as well as the germinated stages.
Here, the objectives of this research were (1) to study the lipidomics of the peanut seeds of the large-seed peanut cultivar A. hypogaea cv. ZP06 at the four seed developmental stages (D1–D4) and three germination stages (G1–G3), and (2) to conduct the transcriptomics of the seeds at different seed development and germination stages to sequence 21 RNA samples. The identification of individual lipid species and their dynamic changes in their quantities in peanuts at different stages could provide deeper insights into their biological and nutritional functions. Furthermore, the transcriptomics would reveal the regulatory mechanisms of the lipid synthesis pathway in peanut seeds during maturation and germination. This research could also contribute to the development of healthier and more sustainable peanut-based products. Our research could provide a comprehensive overview of the relationship between peanut lipids and genes and offer new insights into the lipid nutrition of large peanut seeds.
Materials and Methods
Plant Material and Lipid Standards
A large seeded peanut cultivar (A. hypogaea cv. ZP06) was developed in this laboratory and used as an experimental material in the present study. The ZP06 peanut is a Virginia-type variety with large pods and seeds. The mature seeds of ZP06 (hundred-seed weight: 130.1 g) are approximately 2.11 cm in length and 1.11 cm in width.20 The growth period of ZP06 is approximately 130 days. The plants were grown in the scientific and educational park of Henan Agricultural University, Zhengzhou City, China (113.63E, 37.75N). Seeds for germination were placed in a growth chamber at 22 °C with 250–300 μmol/m2/s photosynthetically available radiation and a 16 h light/8 h dark cycle. Seed samples were collected at different specific developmental stages (15, 30, 45, and 60 days after gynophore entered the soil, named D1, D2, D3, and D4, respectively) and different germination stages (1, 5, and 10 days after germination, named G1, G2, and G3, respectively). The seed samples (1–5 g) were frozen in liquid nitrogen immediately and stored at −80 °C for lipid extraction and subsequent transcriptomics and lipidomics analysis. Three replications were performed for these experiments. All lipid standards, including phosphatidylethanolamine (d7-PE-15:0/18:1), phosphatidylserine (d7-PS-15:0/18:1), phosphatidylglycerol (d7-PG-15:0/18:1), phosphatidylinositol (d7-PI-15:0/18:1), glycerophosphatidic acid (d7-PA-15:0/18:1), lysophosphatidylethanolamine (Lyso PE 18:1(d7)), phosphatidylcholine (d7-PC-15:0/18:1), lysophosphatidylcholine (Lyso PC 18:1(d7)), sphingomyelin (SM-d18:1–18:1(d9)), cholesterol ester (CE-18:1(d7)), triacylglycerol (TAG 15:0–18:1(d7)-15:0), diacylglycerol (DAG-15:0–18:1(d7)-15:0), monacylglycerol (MAG-18:1(d7)), ceramide (Cer-d18:1-d7/15:0), and fatty acid (d4-FA-16:0) were purchased from Avanti Polar Lipids (Alabaster, AL, U.S.A.). A mixed solution of all internal standards was prepared in DCM/MeOH (3:1, v/v), and the mixture was stored in −20 °C until further use. The concentration of 15 internal standards was 10 μg/mL.
Lipid Extraction
A total of 21 seed samples were divided into seven groups for the lipidomics study, including D1, D2, D3, D4, G1, G2, and G3. Each group had three biological replicates. All lipids from the seed were extracted by a modified method.6 Briefly, about 15 mg of a sample freeze-dried and ground to powder was combined with 80 μL of internal standard (10 μg/mL), and 2 mL of methanol was added to precipitate the protein overnight at −20 °C. Next, 4 mL of dichloromethane was added, followed by vortexing for 1 h. The sample was then mixed with 1.6 mL of ultrapure water, vortexed for 30 s, centrifuged, and the supernatant was removed. This process was repeated twice with the addition of 4 mL of dichloromethane to extract any remaining supernatant. The merged supernatant was dried using nitrogen blowing, redissolved with 1 mL of CHCl3/MeOH (v/v, 1:1), filtered through a 0.22 μm organic membrane, and injected into the machine.
LC–MS Analysis
Large-seed peanut lipidomics analysis using the previous method with partially modified.21 In brief, liquid chromatography mass spectrometry consisted of the Shimadzu UPLC 30A system (Shimadzu) coupled to a Sciex TripleTOF 6600 system (AB Scienx). The lipid extract sample was separated by the nonpolar C18 column (Kinetex C18 column, 100 × 2.1 mm, 2.6 μm, Phenonex) with a SecurityGuard precolumn (C18 guard column, Phenomenex). The injection volume was 1 μL, the column temperature was maintained at 60 °C, and the flow rate was 400 μL/min. Initially, gradient elution started with 20% solvent B (IPA/ACN = 5:1, v/v; 5 mM ammonium acetate) and 80% solvent A (water/MeOH/ACN = 1:1:1, v/v/v; 5 mM ammonium acetate) for 0.5 min. Solvent B increased from 40% at 0.5 min to 60% at 1.5 to 3 min and then increased to 98% between 3 and 13 min, subsequently decreasing to 20% at 13 to 13.1 min, and finally 20% from 13.1 to 17 min; the total running time is 17 min. The MS parameter settings are as follows: the ion injection voltage was configured to be 5500 V (+) and −4500 V (−) within the mass range of m/z 100 to 1200. The curtain gas was at 35 psi, and the two ion source gases (Gas1 and Gas2) were set at 50 psi, while the temperature of the interface heater was 600 °C.
Data Analysis and Lipid Identification
Freely available MS-DIAL, version 4.92 (http://prime.psc.riken.jp), and commercially available software packages PeakView, MasterView, and MultiQuant were applied to lipid profile analysis studies.22 Before dealing with MS-DIAL data processing, the original mass spectrometry data files were converted from the original SCIEX format (.wiff) to the analysis base file format (.abf). Each of the MS/MS spectra features was matched with the integrated LipidBlast database by MS-DIAL software to identify lipidomics. All lipid species were identified by mass accuracy precursor ions (<0.01 Da), fragment ions (<0.05 Da), isotope patterns (differences, <10%), MS accurate mass tolerance (0.01 Da), MS/MS accurate mass tolerance (0.05 Da), and identification score cutoff (80%). The lipid quantification method was conducted by MasterView (version 2.0) and MultiQuant (version 3.0.3), including lipid name, retention time, m/z. Identified lipid species was quantified via internal standards (IS) and the relative peak area (analyte area/IS area).
Quality Assurance
In order to monitor the stability and evaluate the data quality produced by the AB Sciex Triple TOF 6600 system in real time, equal amounts of each sample were injected as quality control (QC) samples. One QC sample was injected after every ten analytical samples. Unsupervised principal component analysis (PCA) was conducted by the statistical function prcomp within R (www.R-project.org). PCA showed that the QC samples of LC–MS lipid profile data were closely clustered throughout the entire analysis process, indicating that the measurement had good instrument reproducibility and stability.21
mRNA Library Construction and Sequencing
Total RNA was isolated from 21 peanut seed samples (three biological replication/each seed stage) and extracted using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s procedure. The RNA amount and purity of each sample were quantified using NanoDrop ND-1000 (NanoDrop, Wilmington, DE, USA). The sequencing library preparation and high-throughput sequencing were carried out by Illumina Novaseq 6000 (LC-Bio Technology CO., Ltd., Hangzhou, China). Only clean reads with a perfect match or one mismatch to the reference genome sequence were further analyzed and annotated based on the reference genome. All subsequent analyses were based on clean, high-quality data.
Statistical Analysis
Differential gene/lipid screening, GO functional annotation, and KEGG pathway analysis were performed for all other periods using the data on D-60 as a control, and the following are the screening criteria. Significantly regulated metabolites between groups were determined by variable importance in projection (VIP) ≥ 1 and absolute Log2 fold change (FC) ≥ 1. VIP values were extracted from the OPLS-DA result, which also contains score plots and permutation plots and was generated using the R package MetaboAnalystR. The data was log-transformed (log2) and mean-centered before OPLS-DA. In order to avoid overfitting, a permutation test (200 permutations) was performed. Differentially expressed genes are evaluated from both fold change and significance level, and here, the fold change FC ≥ 2 or FC ≤ 0.5 and p-value <0.05 were taken as the criteria for significance, and the genes screened were considered differentially expressed and displayed as yes in the signal column of the result file. The correlation of gene expression with lipid accumulation was detected using the WGCNA R software package.22
Results
Lipidomic Profiling of the Large-Seed Peanut Variety ZP06
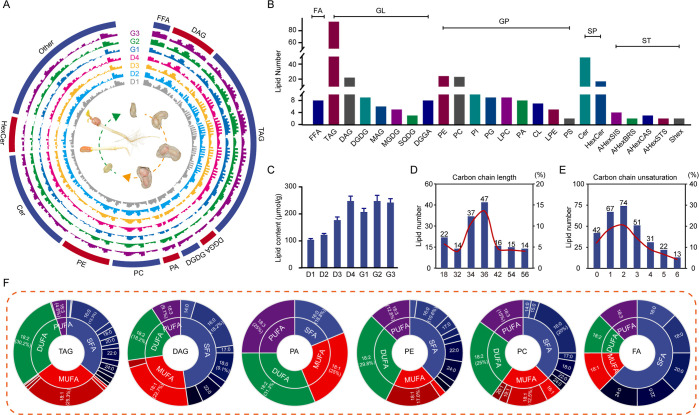
ZP06 samples were collected at four developmental stages (D1, D2, D3, and D4) and three germination stages (G1, G2, and G3) for lipidomic analysis with ultraperformance liquid chromatography (UPLC) quadrupole time-of-flight (Q-TOF) mass spectrometry (MS). Chromatograms (Figure S1A) and PCA (Figure S1B) demonstrated consistency between biological replicates, indicating that the lipid data were accurate and reliable. Diagnosis and identification of fatty acid chains were based on matching MS/MS spectra ion fragment information characteristics with the MS-DIAL database (Figure S1C). In total, 332 types of lipids were identified (Table S1), including DAGs, TAGs, free fatty acids (FFAs), PCs, PEs, and others (Figure S2), and their content was displayed in the Circos map at different seed developmental and germination stages (Figure 1A,B). They were classified into five main categories (class I): GLs (148 types), GPs (97 types), FAs (8 types), SPs (66 types), and STs (13 types), with proportions of 44.58, 29.22, 2.41, 19.88, and 3.92%, respectively (Table S1). Lipids were further classified into 24 subclasses, including TAGs (95 types), DAGs (22 types), PEs (24 types), FFAs (8 types), and PCs (23 types) (Figure 1B; Table S2). Notably, there were more different types of TAGs than any other subclass. The total lipid contents gradually increased across both the seed development and germination stages (Figure 1C), peaking at D4 (247.83 μmol/g) and G3 (207.11 μmol/g), respectively (Figure S3).
Figure 1.
Lipidomic analysis of large-seed peanuts during the seed development and germination processes. (A) Circos plot showing contents of all lipid types at each of the analyzed different stages. (B) Abundance of lipids in each category. (C) Total lipid contents at each of the analyzed different stages. (D) Distribution of lipid carbon chain lengths. (E) Distribution of lipid carbon chain unsaturation numbers. (F) Proportions of fatty acid types among TAG, DAG, PC, PE, PA, and FA lipid molecules.
Because the lipid carbon chain length and saturation are closely related to lipid function, we analyzed these characteristics among the identified lipids (Table S3). The lipid chain lengths ranged from 16 to 78 carbons, with most falling between 18 and 36; lipids with 36 carbons were the most abundant (Figure 1D). Carbon chain unsaturation numbers varied from 0 to 12, with the most common unsaturation numbers being 2 (22.2%), 1 (20.1%), 3 (15.3%), and 0 (12.6%) (Figure 1E). Notably, the hydroxylated-triacylglycerols (HO-TAGs), monogalactosyldiacylglycerols (MGDGs), diacylglyceryl glucuronides (DGGAs), PAs, cardiolipin (CLs), PSs, and glucosylceramides (HexCers) did not contain any saturated fatty acids (SFAs) (Table S3).
Figure 1F shows the FA compositions of specific lipid classes. In TAGs, lipid molecules were composed of SFAs, monounsaturated fatty acids (MUFAs), diunsaturated fatty acids (DUFAs), and polyunsaturated fatty acids (PUFAs) with their respective proportions of 37.3, 28.1, 28.9, and 5.7%, where palmitic acid (16:0, 12.7%), oleic acid (18:1, 24.7%), linoleic acid (18:2, 28.5%), and linolenic acid (18:3, 5.7%) were found to be their major FAs, respectively (Figure 1F). DAGs were predominantly composed of SFAs (47.7%), MUFAs (25.0%), DUFAs (18.2%), and PUFAs (9.1%), with the primary FAs being palmitic acid (16:0, 18.2%), oleic acid (18:1, 22.7%), linoleic acid (18:2, 18.2%), and linolenic acid (18:3, 9.1%), respectively (Figure 1F). The primary FA molecules were nearly identical in the PA, PE, and PC groups. Overall, these results showed that large-seed peanut is rich in long-chain fatty acids and that the most abundant FA molecules in all lipid classes were UFAs, particularly oleic acid, linoleic acid, and linolenic acid, similar to most grains.
Dynamic Changes in Large-Seed Peanut Lipid Profiles during Seed Development and Germination
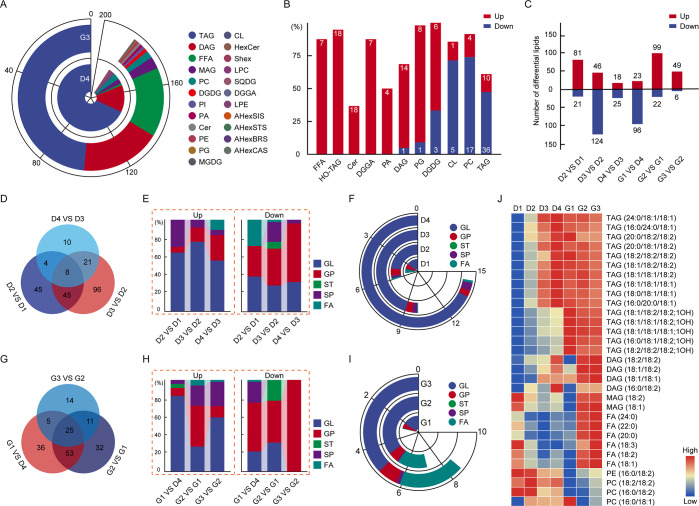
In mature seeds (D4), the content of TAGs and DAGs in GLs is as high as 173.99 and 39.93 μmol/g, accounting for 77.90 and 17.88% of total lipids, respectively (Figures 2A and S2; Table S4). Notably, 18:1/18:2/18:2, 18:1/18:1/18:2, and 18:1/18:1/18:1 were the most abundant TAG species, accounting for 13.96, 13.96, and 11.91%, respectively, of the total lipids (Table S1; Figure S4). Among DAGs, 18:1/18:2 and 18:2/18:2 were the most abundant (8.71 and 6.97%, respectively). The most abundant TAGs and DAGs were consistent between the G3 and D4 samples. Of the GPs, PCs (16:0/18:2, 18:1/18:2, and 18:2/18:2) were present at the highest levels in both D4 and G3 samples. Significantly, most PCs (17 of 21) were reduced in germinated peanut (G3) compared with the mature seeds (D4), as were TAGs and CLs. In contrast, most FFAs, DAGs, HO-TAGs, digalactosyldiacylglycerols (DGDGs), PGs, and Cers were accumulated at higher levels in germinated peanuts than in mature seeds (Figure 2B). Except for these lipid species, overall, the lipid composition is similar in mature seeds and sprouted peanuts, but the lipid content is different, which is due to the dynamic changes in the lipid profile during seed development and germination processes, respectively.
Figure 2.
Differentially accumulated lipids (DALs) in large-seed peanuts during seed development and germination processes. (A) Lipid composition in mature seeds (D4) and peanut sprouts (G3). (B) Distribution of up-regulated and down-regulated lipid species at the G3 stage compared to the D4 stage. Lipids were classified as differentially accumulated at thresholds of fold change ≥1.2 or ≤0.80 and p < 0.05. (C) Total number of up-regulated and down-regulated lipid species between each pair of consecutive time points. Lipids were classified as differentially accumulated at thresholds of fold change ≥2 or ≤0.50 and p < 0.05. (D) Unique and shared DALs throughout seed development. (E) Relative abundance of up-regulated DALs (up-DALs) and down-regulated DALs (down-DALs) in several lipid classes for each pair of consecutive time points during seed development. (F) Levels of up-DALs and down-DALs at each stage of seed development. (G) Unique and shared DALs throughout germination. (H) Relative abundance of up-DALs and down-DALs in several lipid classes for each pair of consecutive time points during germination. (I) Levels of up-DALs and down-DALs at each stage of germination. (J) Levels of representative DALs during seed development and germination.
The lipid content of different lipid species varies with stages, and thus the differently accumulated lipids (DALs) between each adjacent growth stage were compared. A total of 282 DALs (Table S5) were obtained (Figures 2C and S5). During seed development, the largest number of DALs (Tables S6–S8) were present between D2 and D3 (Figure 2C,D), indicating that the metabolism of life activities is more vigorous in these stages, which are the rapid filling period for the seed. The number of up-regulated DALs (up-DALs) decreased gradually across the comparisons, suggesting that levels of specific lipids peaked at different periods. The majority of up-DALs were GLs, whereas the majority of down-regulated DALs (down-DALs) were GPs (Figure 2E). The total GL and GP contents steadily increased during these periods (Figure 2F). Some of the GLs and GPs (including 49 TAGs, 12 HO-TAGs, and 24 Cers) showed a continuous increase during the developmental stages; the three most abundant TAGs were 18:1/18:2/18:2, 18:1/18:1/18:2, and 18:1/18:1/18:1. A majority of continuously decreasing lipids were GPs, including PIs, PAs, PGs, and PCs (reflecting the necessity for seeds to store energy while also continuing to grow and divide). Moreover, we observed a consistent reduction in PAs (especially 16:0/18:2, 16:0/18:3, and 18:2/18:3) during all developmental stages, possibly as a result of membrane reorganization during seed enlargement.
In the germination stage, there were more DALs in the comparisons of G1 to D4 (Tables S9–S11) and of G2 to G1 than in G3 compared to G2 (Figure 2C,G), indicating that changes in the lipid composition occurred primarily during the first two germination stages. At the first germination stage, the majority of DALs were down-regulated GLs and GPs (Figure 2H). Although the total GL and GP contents increased as germination progressed (Figure 2I), there were distinct changes in the levels of the individual lipid components. Among the FFAs, levels of palmitic acid (16:0), stearic acid (18:0), oleic acid (18:1), linoleic acid (18:2), linolenic acid (18:3), and docosanoic acid (22:0) significantly increased during germination, but to varying degrees (Figure 2J). Differences in the abundances of these compounds may have been due to alterations in the cell membrane resulting from germination; in this stage, energy is supplied to the plant and the seeds absorb water, causing cells to swell.23
Notably, there was a dramatic increase (1679%) in the levels of HO-TAGs, a potentially beneficial class of lipids, at the first germination stage. Total HO-TAG contents were 79.39, 195.69, 362.55, 539.42, 9056.00, 5604.07, and 4986.31 nmol/g in stages D1–D4 and G1–G3, respectively. There were also significant changes in FFA contents during the development and germination stages; total FFA contents were 15003.63, 4560.22, 3038.97, 4063.77, 6278.78, 25976.99, and 36529.32 nmol/g at the D1–D4 and G1–G3 stages, respectively. In contrast to the sharp decreases and increases observed for FFAs throughout seed development and germination, total HexCer contents gradually decreased throughout development, with total contents of 1044.46, 836.60, 246.61, 163.30, 87.46, 132.17, and 185.58 nmol/g at the D1–D4 and G1–G3 stages, respectively. Levels of β-galactosidase (GLB1) were highest in the early stage of seed development, suggesting that GLB1-mediated cell wall remodeling was involved in plant growth, development, and regulation of HexCer metabolism.
Transcriptomic Analysis of Large-Seed Peanuts during Seed Development and Germination
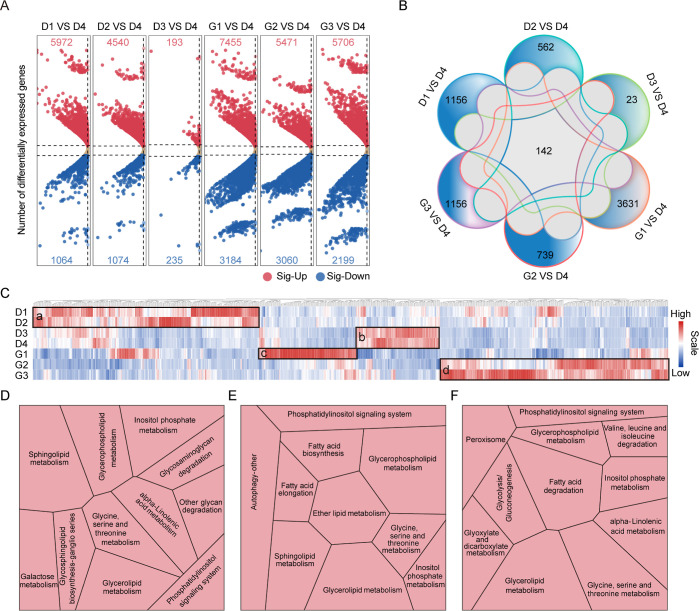
According to the sequencing analysis of the first generation full-length transcriptome of peanut and the original reference genome information, we obtained a valid data file size of 8.08–9.50 G for each sample (Table S12). This corresponded to 84,714 transcripts, including 67,124 peanut genomes. The transcripts were analyzed with the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases for functional annotation, yielding a total of 54,702 GO terms and 22,536 KEGG biochemical pathways (Table S13). Pearson correlation showed that the correlation coefficient between samples was high, and the sample clustering was significant (Figure S6). Three independent biological replicates for each sample were consistent with each other, demonstrating the reliability of the data (Table S13). Correlation analysis (Figure S6) showed that samples from the same period can be clustered together and each growth period can be distinguished, indicating that there are differences in the profiles of transcriptomes in each period.
The differences in the whole transcriptome map changed with seed development and germination. Taking D-60d as a control, in the developmental stages, there are 7036, 5614, and 428 differentially expressed genes (DEGs) with the criteria of |FC| ≥ 2 and P < 0.05, respectively, in the comparison group D1 vs D4 (Table S14), D2 vs D4 (Table S15), and D3 vs D4 (Table S16). Among them, most DEGs were upregulated (Figure 3A). The number of DEGs decreased as the development stage approached seed maturity, indicating that seed accelerated development significantly at the early stage of development, and the development process slowed down after 45 days of flowering and approached maturity. In the germination stages, 10,639, 8531, and 7905 genes were up- or down-regulated in the comparison group G1 vs D4 (Table S17), G2 vs D4 (Table S18), and G3 vs D4 (Table S19), respectively (Figure 3A). These results show that genes are activated as a part of the transformation mechanism of development and germination, and the transcriptional regulation of genes in the early germination stage is significant. The Venn diagram shows the number in common among all comparison groups (Figure 3B), and 142 DEGs were found to be commonly differentially expressed in all stages. Among them, we found the expression of gene-encoded vicilin 47 kDa protein (6TFR0H) increased during seed maturation, was highly expressed at maturity, and decreased during germination. Moreover, unique DEGs were in the majority in G1 from D4 (3631 DEGs), with less in D3 to D4 (23 DEGs). The results reported above indicated there were biological differences in the development and germination stages of seeds, and there were also certain similarities in each stage.
Figure 3.
Transcriptomics sequencing analysis of large-seed peanuts during seed development and germination. (A) Expression levels of differentially expressed genes (DEGs) in each comparison group (seed development stages D1–D3 and germination stages G1–G3 compared to D4). (B) Unique and shared DEGs between comparison groups. (C) Expression levels of DEGs involved in lipid metabolism pathways. DEGs with high expression in early seed development, late seed development, early germination, and late germination are indicated with the letters a, b, c, and d. (D) KEGG pathways involved by DEGs in early development. (E) KEGG pathways involved by DEGs in early germination. (F) KEGG pathways involved by DEGs in late germination. Modules represent metabolic pathways, and the size of the modules represents the proportion of DEGs.
The KEGG pathway enrichment analysis showed that DEGs were significantly enriched in biological pathways across seven different stages. In the pathway of the top 20 KEGG-annotated genes, “Metabolism” contained the most target genes (Figure S7). Thus, lipid metabolism-related DEGs were selected (Table S20), and their expressions were shown in the heatmap (Figure 3C). The numbers of genes with higher expression in early and late development and germination were 305, 113, 131, and 308, indicating block a, b, c, and d in Figure 3C, respectively. Further analysis by KEGG showed that highly expressed DEGs in early development stages were mostly involved in the pathways of sphingolipid metabolism, inositol phosphate metabolism, and fatty acid degradation (Figure 3D, Table S21). The higher-expressed DEGs in the initial stage of germination were mostly involved in the pathways of glycerophospholipid metabolism, ether lipid metabolism, and glycerolipid metabolism (Figure 3E, Table S21); the high-expressed DEGs in the late period of seed germinating were mostly involved in the pathways of fatty acid degradation, glycerolipid metabolism, glycine, serine, and threomine metabolism (Figure 3F, Table S21).
Correlations between Lipid Contents and Gene Expression in Large-Seed Peanuts during Seed Development and Germination
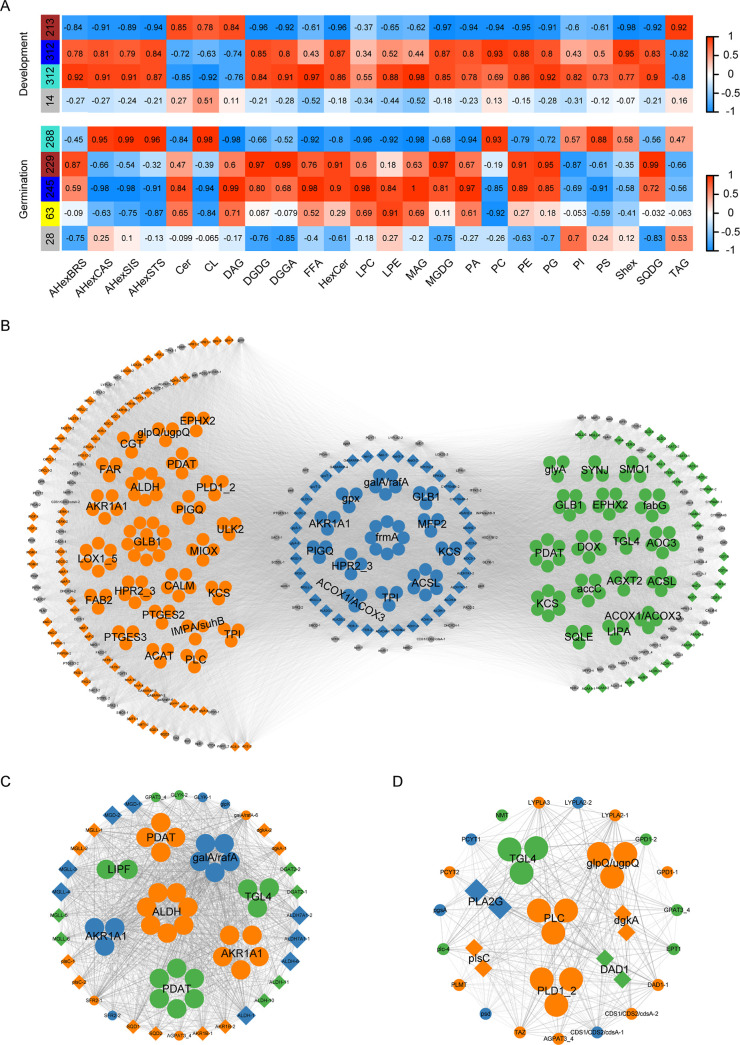
To comprehensively analyze lipid and gene expression dynamics during the peanut seed development and germination processes, weighted gene coexpression network analysis (WGCNA) was conducted using the lipidomic and transcriptomic data. The above 857 DEGs (Table S20) were used for joint analysis with all the subclass lipids by comparing the GO and KEGG analysis results of RNA-seq data. Quality control analysis was performed on developmental and germination gene data, suitable for in-depth analysis in the next step (Figures S8 and S9). The hierarchical cluster tree shows that the lipid-related genes can be classified into three modules indicated by different colors, i.e., turquoise (312 genes) (Table S22, Figure S10), brown (213 genes), blue (312 genes), and gray (14 genes) for development, and gray (28 genes), turquoise (288 genes), brown (229 genes), blue (245 genes), and yellow (63 genes) (Figure S11, Table S23) for germination (Figure 4A). The turquoise and blue modules have a higher correlation with subclass lipid compounds during development and germination, respectively (Figure 4A). The high correlations and low p values indicated that these two modules could be used to identify hub genes associated with the corresponding lipids. Thus, these two module genes were further analyzed.
Figure 4.
Weighted gene coexpression network analysis (WGCNA) from transcriptomic and lipidomic data. (A) Significant coexpression modules during seed development and germination. (B) Network showing correlations of genes in the turquoise and blue expression modules during seed development and germination. Blue node genes are shared between the two processes. Gray and orange (left) are genes unique to seed development; gray and green (right) are genes unique to germination. (C) Correlation of glycerolipid metabolism WGCNA genes. (D) Correlation of glycerophospholipid metabolism WGCNA genes.
For the turquoise and blue module genes, KEGG pathway enrichment analysis showed that they were commonly involved in the pathways of fatty acid biosynthesis, fatty acid degradation, glycosphingolipid biosynthesis, and glycerolipid metabolism (Figure S12). To investigate the gene relationships in the modules, the above-related genes were subjected to network visualization by Cytoscape (Figure 4B; Tables S24 and S25). The 446 nodes are grouped into different clusters, indicated by colors. In common genes involved in the metabolism pathways of development and germination (blue nodes), most of them share a high degree with others (see Tables S26 and S27 for gene name abbreviations). Glycerolipid metabolism genes encoding phospholipid: diacylglycerol acyltransferase (PDAT; ZE5M4X), alcohol dehydrogenase (AKR1A1; CP5KSN, VZ85GY, 3M1BNC, and 7ND1W), aldehyde dehydrogenase (ALDH; 6QU7U5), α-galactosidase (KWHJ4Z), sulfoquinovosyl transferase (SQD2; L6AUEE, YIK7D0), and galactolipid galactosyltransferase (SFR2; FT2DSH) in these two modules coexisted and were involved in glycerolipid metabolism (Figure 4C). The expression of almost all these above-indicated genes decreased with development and then gradually increased with germination, similar to V-shaped, except for PDAT. In addition, TAG lipase (TGL4-1, E4P4QB; TGL4-2, K52S6H; TGL4-3, and 92S4AT), glycerol-3-phosphate O-acyltransferase 3/4 (GPAT3_4; 0G9KRE), and lysophosphatidic acid acyltransferase (AGPAT3_4; S2 V0TW) were both involved in the pathway of glycerolipid metabolism and glycerophospholipid metabolism (Figure 4C,D). The expression of TGLs was higher in germination stages than in development stages. Moreover, related transcription factors were also found to be differentially expressed, including the APETALA2 transcription factor WRINKLED1 (WRI1; QI820R and UVRI66), ABSCISIC ACID INSENSITIVE3 (ABI3; 6PZ1IJ, PN7UB6, Z6Z4XE, and 4K8YXT), FUSCA3 (FUS3; D9C186 and HB4W2S), and transcription factor MYB118 (VP8GIF and 57FMZL), whose expression was mainly high during development but germination stage, except for MYB118, involving controlling seed oil accumulation.
Integrated Analysis of DALs and DEGs in the Assembly Pathway of Large-Seed Peanut Seeds during the Development and Germination Processes
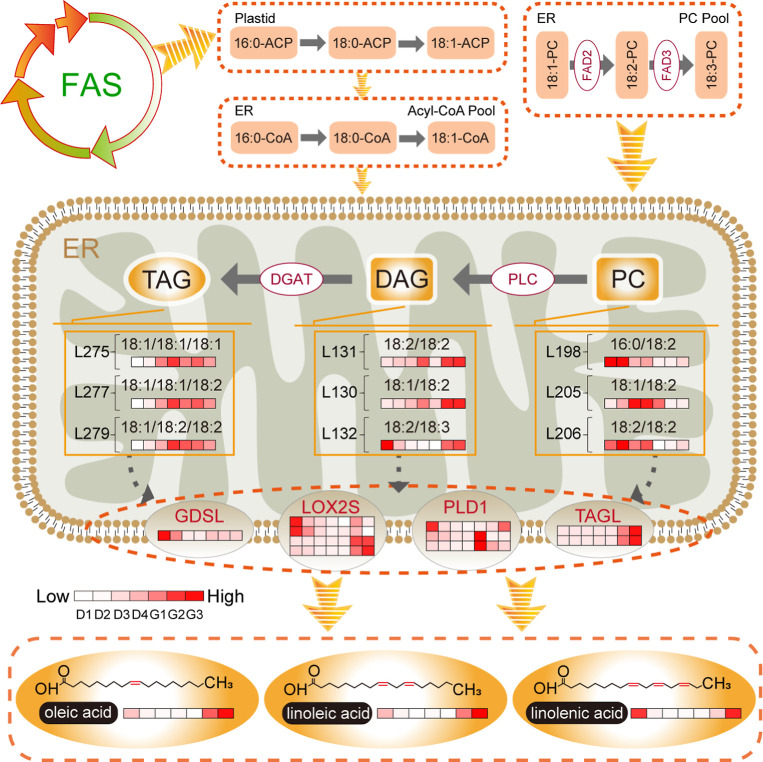
Seed development is accompanied by lipid accumulation. The molecular pathways include carbohydrate metabolism, FA synthesis, TAG accumulation/packaging pathways, and oil body or lipid droplet formation (Figure 5). In de novo FA synthesis in the plastid, acetyl-CoA (derived from sucrose) is the original substrate from which malonyl-CoA is formed by acetyl-CoA carboxylase. Malonyl-CoA is then catalyzed by FA synthases in sequential condensation reactions, ultimately producing FAs. Fatty acid desaturase 2 (FAD2) and FAD3 on the endoplasmic reticulum catalyze oleic acid (18:1) to form linoleic acid (18:2) and linolenic acid (18:3) sequentially. We here identified eight FAD3 genes; expression levels of three (40PHQK, BC0JZ1, and ZDHF3I) were consistent with linolenic acid contents during the seven seed and sprout growth stages, showing decreases in the seed development and early germination stages but increases during late germination. The FAD2 genes identified, 8TPQ4A and BY45PL, were highly expressed, and their expression trend was similar to the trend in the change in the content of linoleic acid.
Figure 5.
Key changes in lipid metabolism pathways during seed development and germination, as identified with an integrated lipidomic and transcriptomic analysis.
The FAs formed in seeds generally exist in the form of TAG, which is synthesized through two main pathways. In the Kennedy pathway, FAs are activated by long-chain acyl-CoA synthetase to form CoA esters, which are then esterified to glycerol-3-phosphate and form LPA. LPA is sequentially acylated by 1-acylglycerol-3-phosphate acyltransferase (LPAAT), phosphatidic acid phosphatase (PAP), and diacylglycerol acyltransferase (DGAT) to produce PA, DAG, and TAG, respectively. The transcript levels for LPAAT (SD0DCP and T5GNGC) were higher at D2 than at other developmental stages. DGAT (Q8CNLJ and U0HZSX) was highly expressed in both the late seed development and late germination stages. In the acyl-CoA-independent pathway, DAG is acylated using PC as the acyl donor, then catalyzed by PDAT to produce TAG.24PDAT (10ET88 and F78IM4) was most highly expressed at the early development and late germination stages, whereas PDAT (91WX57) was more highly expressed during germination than seed development stages. After synthesis, TAGs are packaged to form oil bodies attached to various oil body proteins. Oleosin is the main lipid droplet protein. The transcriptome data showed that the expression of oleosin (N1YYK3 and YSQ5WX) was higher at D3 and D4 than at D1 and D2.
The molecular pathways involved in lipid degradation include lipid decomposition, β oxidation, and the glyoxylate cycle, among others. TAGs are degraded to free FAs by TGLs. FAs then undergo β-oxidation in the mitochondria and glyoxysomes to produce acetyl-CoA, which is subsequently used in the tricarboxylic acid (TCA) cycle and in carbohydrate synthesis. We found significant oil body disintegration and decreased oleosin expression during germination compared to seed development. Moreover, TGLs were gradually up-regulated throughout germination; this was particularly notable for TAGL (DY4FLS), which was expressed 94.08- and 54.65-fold higher at G3 and G2, respectively, than at D4 (Table S28). GDSL lipases are one type of lipolytic enzyme. In the early germination stage, the expression of GDSL lipase (VMW7K4, SLB9HH, and X1KXE8) was higher at G1 than at D4. At the same time, phospholipase D 1_2 (PLD1_2; 79BS07 and PYQ6LW), which cleaves PC to produce PA, had higher transcript levels at G1 than at D4. Lipoxygenase (LOX2S; 6TJ5BP and S6TVHJ) was similarly up-regulated during early germination, which may have contributed to FFA oxidation (Table S28).
Discussion
Lipids in peanuts have direct effects on the sensory and nutritional properties of the peanut products. In recent years, lipidomics has emerged as a powerful tool for investigating the lipid composition and metabolism of various plant species. Seeds contain critical resources for plant survival.24 An important metabolic function of mature seeds is the deposition of storage compounds, which are later mobilized to promote seed germination and seedling growth. Seeds undergo complex dynamic and physiological changes during their development, which are associated with a significant change in the levels of organic acids, sugars, and amino acids, suggesting that they are effectively incorporated into storage reserves.25 During seed maturation and germination, changes in metabolism are associated with distinct expression patterns of the metabolic genes. Thus, here, we conducted lipidomics and transcriptomics during seed development and germination processes, aiming to aid understanding of the related regulation mechanisms for large-seeded peanut.
Storage lipids are known to accumulate in peanut seed. Our present study shows that various types of lipids already exist at the early stage of seed development, and lipid content changes with time, indicating that the lipid network map has been established at the early stage of development. Comparing peanut ZP06 seed lipids with other species, which showed the presence of 141 GLs, 89 GPs, 22 SPs, 18 STs, and 13 PRs in green tea and the presence of 2 FAs, 98 GLs, 383 GPs, 10 SPs, and 51 SLs in mature hickory nuts, the results showed that FAs are more abundant in peanut than other species, which conformed to the characteristics of oil crop of peanut.6,26 In high-oleic peanut H176 seeds, there were 547 lipids identified, including 18 FAs, 253 GLs, 182 GPs, 93 SPs, and 1 STs.5 Thus, the comparison of large seed variety ZP06 and high-oleic peanut seeds indicated that the species of GLs, FAs, and SPs are more abundant in the high-oleic peanut H176, and the species of STs are more abundant in ZP06,5 indicating diverse lipid molecules and complex metabolic mechanisms existing in the peanut variety with specific characteristics. These also could result from detection methodologies and/or technological disparities. In H176, 60 days after the emergence of the gynophores, the TAG contents were lower than found at the 50-day time point. This was attributed to some unknown TAG degradation or synthesis termination pathways, such as FAs β-oxidation.5 In this study, the total content of TAGs and DAGs was continuously accumulated in ZP06 through development processes. The largest accumulation of TAG occurred in the middle development stage, when seeds rapidly expanded.
DGATs, including DGAT1, DGAT2, soluble DGAT3, and wax ester synthase/DGAT, are rate-limiting enzymes in the final step of TAG biosynthesis.27 Among them, soluble DGAT3, at present, has only been identified in peanut and Arabidopsis thaliana.28,29 A previous study showed that AhDGAT1-1, AhDGAT1-2, and AhDGAT3-3 expression levels peak at the late seed development stage.30 We found here that DGAT was highly expressed during both late seed development and late germination. Moreover, DGAT was more highly expressed than PDAT. These results indicated that the Kennedy pathway was the primary mechanism by which acyl groups were transferred from fatty acyl-CoA to the DAG backbone for TAG synthesis and lipid storage.
In the complex transcriptional network that controls seed oil accumulation, MYB transcription factors regulate seed oil biosynthesis by controlling DGAT1 and PDAT1 expression.18,31 By repressing FA biosynthesis-associated genes, MYB118 functions as a repressor of FA biosynthesis, and its expression was very low in this study.32 Other transcription factors, including WRI1, ABI3, and FUS3, appeared to play positive roles in the processes of seed development, maturation, and oil biosynthesis. Expression of these genes enabled the high accumulation of storage lipids in large-seed peanuts at the seed development stage, preparing the seeds to provide various metabolites and energy for later seedling growth at the germination stage.
TAG is a significant energy storage molecule in plant biosynthesis, which is essential for seed development and germination. Our results showed that TAG was the most abundant lipid molecule in peanut seeds and therefore played a crucial role in germination and growth. Here, a novel type of TAG, HO-TAG, consisting of 19 subtypes, was found in peanut seeds. HO-TAG is highly expressed during the peanut germination stage, with its content spiking at early germination. HO-TAGs have a structure similar to that of TAGs, except for the hydroxylation of one fatty acid. The biological functions of HO-TAGs during seed development remain unclear. Research has shown that hydroxylated fatty acids can improve glucose tolerance and insulin sensitivity and reduce inflammation in obesity, diabetes, and immune-mediated diseases.33 Therefore, germinated peanut seeds with high levels of HO-TAGs may have unique health benefits and are worthy of further in-depth investigation. Sprouted peanuts are also rich in many other nutrients and have been shown to possess antioxidant and antidiabetic properties, making them an ideal candidate for functional food development.34 The potential beneficial properties of germinated peanut seeds should be further investigated in future studies.
FFAs are differentially accumulated during seed development and germination. FAs, which are FFAs, are a major family of structurally complex lipids. Oleic acid (18:1), linoleic acid (18:2), and linolenic acid (18:3) were found to be the primary FA molecules of most major lipid types, such as TAG, DAG, PC, PE, and PA, also in high-oleic and normal peanuts.5 In large-seed peanuts, UFAs showed a higher relative abundance than SFAs, comparable to findings in mature hickory nuts.6 Oleic acid (18:1), linoleic acid (18:2), and linolenic acid (18:3) have high nutritional value and are necessary for human health; importantly, linoleic acid and linolenic acid cannot be synthesized by humans and must therefore be supplied by foods. Furthermore, humans convert α-linolenic acid to eicosapentaenoic acid (C20:5) and docosahexaenoic acid (C22:6), which enhance immunity.35 We found levels of FFAs, particularly linolenic acid, decreased overall during peanut seed development (Figure S2). Linolenic acid contents were high during the early stages of seed development (D1 and D2) but decreased over time, consistent with results in high-oleic and normal peanut seeds.5 Levels of α-linolenic acid are generally low in other dietary oil plants, including rapeseed (7.90%), soybean (6.10%), palm (0.30%), olive (0.60%), corn germ (0.10%), and sunflower (0.20%).36 These results indicate that FFAs may be involved in the assembly process of other lipids in seed development. In these lipid assembly pathways, BnDGAT1 preferentially incorporates oleic acid (18:1) into TAG.37 However, further studies will be required to identify functionally related genes and enzymes in peanuts.
TAGs are the primary energy storage substance in the seed development and germination stages; TAG degradation can produce FFAs and provide energy and raw materials for seed germination and sprout establishment. Similarly, phospholipases such as PLA, PLC, and PLD are involved in lipid turnover through phospholipid hydrolysis and FFA release.38 We here found that levels of FFAs (16:0, 18:0, 18:1, 18:2, 18:3, and 22:0) were low during the initial stages of seed germination but increased with time, suggesting the mobilization of various lipid types during germination. Changes in the peanut seed FFA composition may have significant implications for the nutritional value of the derivative products. Oleic acid has beneficial effects on blood cholesterol levels and cardiovascular health; increases in oleic acid concentrations during seed germination suggest that the nutritional value of peanut as a cholesterol-lowering food may increase throughout seed maturation and could be particularly high in sprouted peanut.39 Linoleic and alpha-linolenic acids, which are essential omega-6 and omega-3 FAs, respectively, also increased during peanut seed germination, further suggesting potential health benefits of sprouted peanuts. Therefore, sprouted large-seed peanuts could be used as a food material for α-linolenic acid intake. Overall, large seeds of peanut ZP06 could be used as materials to explore health-influencing factors and also for developing healthcare peanut products.
In summary, we successfully established and analyzed the dynamic lipidomic and transcriptomic profiles of the large-seed peanut variety ZP06 throughout the seed development and germination stages. The seed lipid metabolism profile has been established at early development, especially for TAGs and DAGs that are highly accumulated during embryogenesis. Storage lipids and phospholipids were subsequently mobilized to provide energy and raw materials for seed germination and seedling establishment. Notably, we found a significant increase in HO-TAGs, potential healthy lipid species, during the early germination stage. The integrated lipidomic and transcriptomic analyses allowed us to develop a comprehensive understanding of the coordinated changes in primary lipid metabolism that drive seed development and germination in large-seed peanut. These findings provide valuable information on the health-promoting benefits of large-seed peanut lipids based on the dynamic changes in comprehensive lipid species at different stages of seed development and germination. This knowledge may be particularly useful for the development of healthful peanut products using large-seed peanuts.
Acknowledgments
We would like to thank Shangde Sun from Henan University of Technology for expert assistance with the analysis and critical proofreading of this manuscript.
Glossary
Abbreviations
- FAs
fatty acids
- GLs
glycerolipids
- STs
sterol lipids
- SPs
sphingolipids
- FFA
free fatty acid
- TAG
triacylglycerol
- DAG
diacylglycerol
- HO-TAG
hydroxylated-triacylglycerol
- DGDG
digalactosyldiacylglycerol
- MAG
monoacylglycerol
- MGDG
monogalactosyldiacylglycerol
- SQDG
sulfoquinovosyldiacylglycerol
- DGGA
diacylglyceryl glucuronide
- PE
phosphatidylethanolamine
- PC
phosphatidylcholine
- PI
phosphatidylinositol
- PG
phosphatidylglycerol
- LPC
lysophophatidylcholine
- PA
phosphatidic acid
- PS
phosphatidylserine
- CL
cardiolipin
- LPE
lysophosphatidylethanolamine
- Cer
ceramide
- HexCer
hexosylceramide
- AHexSIS
acylhexosyl sitosterol
- AHexBRS
acylhexosyl brassicasterol
- AHexCAS
acylhexosyl campesterol
- AHexSTS
acylhexosyl stigmasterol
- Shex
stigmasterol hexoside
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jafc.3c06697.
Qualitative and quantitative analysis of lipidomic analysis of peanut large-seed; content analysis of 24 subclasses of lipids in different stages; content analysis of 24 subclasses of lipids in the same stage; lipid content in TAG, HO-TAG, DGA, PA, PC, and PE with 16:0, 18:1, 18:2, and 18:3; analysis of the main characteristics of DALs; correlation analysis of transcriptome data; expression analysis of DEGs; quality control analysis for WGCNA of DEGs and DALs in development; quality control analysis for WGCNA of DEGs and DALs in germination; turquoise module gene analysis for development; blue module gene analysis for germination; and development and germination modular gene contrasts (PDF)
Comprehensive information on all identified lipids in peanut large-seed; number of subclass lipids in seed development and germination; number of lipids with different saturations; content of subclass lipids; information on all differently accumulated lipids; differently accumulated lipids in D2 vs D1; differently accumulated lipids in D3 vs D2; differently accumulated lipids in D4 vs D3; differently accumulated lipids in G1 vs D4; differently accumulated lipids in G2 vs G1; differently accumulated lipids in G3 vs G2; overview of sequencing data quality control; transcriptome information on peanut large seeds at development and germination; differentially expressed genes in D1 vs D4; differentially expressed genes in D2 vs D4; differentially expressed genes in D3 vs D4; differentially expressed genes in G1 vs D4; differentially expressed genes in G2 vs D4; differentially expressed genes in G3 vs D4; differentially expressed genes involved in the lipid metabolism pathway; differentially expressed gene KEGG analysis; turquoise module gene at development; blue module gene at germination; weighting relationship of WGCNA at the development stage; weighting relationship of WGCNA at the germination stage; gene annotation of turquoise module genes in development; gene annotation of blue module genes in germination; and gene heatmap information (XLSX)
Author Contributions
Dongmei Yin: conceptualization, project administration, and funding acquisition. Di Cao, Yongzhe Ma, Zenghui Cao, and Jinzhi Wang: methodology. Sasa Hu, Zhan Li, Yanzhe Li, and Di Cao: formal analysis. Fangping Gong, Mun Yhung Jung, and Dongmei Yin: writing—original draft. Kai Zhao, Kunkun Zhao, Kuopeng Wang, Xiaoxuan Wang, Ren Rui, Xingli Ma, Ding Qiu, Zhongfeng Li, and Xingguo Zhang: validation. All authors read and approved the manuscript.
This work was supported by grants from the Key Program of the National Natural Science Foundation of China (NSFC)-Henan United Fund (no. U22A20475), the Key Scientific and Technological Project of Henan Province (no. 221111110500; 161100111000; and HARS-22-05-G1), the Natural Science Foundation of Henan Province (no. 222300420178), the Key Scientific Research Project of Henan Higher Education Institutions (24A210007; 21A210018), and the Science and Technology Research Project of Henan Province (no. 222102110128).
The authors declare no competing financial interest.
Supplementary Material
References
- Mingrou L.; Guo S.; Ho C. T.; Bai N. S. Review on chemical compositions and biological activities of peanut (Arachis hypogeae L.). J. Food Biochem. 2022, 46, e14119 10.1111/jfbc.14119. [DOI] [PubMed] [Google Scholar]
- Wang X.; Devaiah S. P.; Zhang W.; Welti R. Signaling functions of phosphatidic acid. Prog. Lipid Res. 2006, 45, 250–278. 10.1016/j.plipres.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Han X.; Gross R. W. Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry: a bridge to lipidomics. J. Lipid Res. 2003, 44, 1071–1079. 10.1194/jlr.R300004-JLR200. [DOI] [PubMed] [Google Scholar]
- Lagarde M.; Géloën A.; Record M.; Vance D.; Spener F. Lipidomics is emerging. Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 2003, 1634, 61. 10.1016/j.bbalip.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Liu H.; Hong Y. B.; Lu Q.; Li H. F.; Gu J. Z.; Ren L.; Deng L.; Zhou B.; Chen X.; Liang X. Integrated analysis of comparative lipidomics and proteomics reveals the dynamic changes of lipid molecular species in high-oleic acid peanut seed. J. Agric. Food Chem. 2020, 68, 426–438. 10.1021/acs.jafc.9b04179. [DOI] [PubMed] [Google Scholar]
- Huang C.; Li Y.; Wang K.; Xi J.; Xu Y.; Si X.; Pei D.; Lyu S.; Xia G.; Wang J.; et al. Analysis of lipidomics profile of Carya cathayensis nuts and lipid dynamic changes during embryonic development. Food Chem. 2022, 370, 130975. 10.1016/j.foodchem.2021.130975. [DOI] [PubMed] [Google Scholar]
- Lu Q.; Li H.; Hong Y.; Zhang G.; Wen S.; Li X.; Zhou G.; Li S.; Liu H.; Liu H.; et al. Genome sequencing and analysis of the peanut B-genome progenitor (Arachis ipaensis). Front. Plant Sci. 2018, 9, 604. 10.3389/fpls.2018.00604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J.; Feng X.; Lyu C.; Zhou S.; Liu Z. Effects of different processing methods on the lipid composition of hazelnut oil: A lipidomics analysis. Food Sci. Hum. Wellness 2022, 11, 427–435. 10.1016/j.fshw.2021.11.024. [DOI] [Google Scholar]
- Wang Y.; Ma X. L.; Zhang X. G.; He X. Y.; Li H. M.; Cui D.; Yin D. ITRAQ-based proteomic analysis of the metabolic mechanisms behind lipid accumulation and degradation during peanut seed development and postgermination. J. Proteome Res. 2016, 15, 4277–4289. 10.1021/acs.jproteome.6b00345. [DOI] [PubMed] [Google Scholar]
- Bodoira R.; Cecilia Cittadini M.; Velez A.; Rossi Y.; Montenegro M.; Martínez M.; Maestri D. An overview on extraction, composition, bioactivity and food applications of peanut phenolics. Food Chem. 2022, 381, 132250. 10.1016/j.foodchem.2022.132250. [DOI] [PubMed] [Google Scholar]
- Zhang B.; Li C.; Li Y.; Yu H. Mobile TERMINAL FLOWER1 determines seed size in Arabidopsis. Nat. Plants 2020, 6, 1146–1157. 10.1038/s41477-020-0749-5. [DOI] [PubMed] [Google Scholar]
- King J. C.; Blumberg J.; Ingwersen L.; Jenab M.; Tucker K. L. Tree nuts and peanuts as components of a healthy diet. J. Nutr. 2008, 138, 1736S–1740S. 10.1093/jn/138.9.1736S. [DOI] [PubMed] [Google Scholar]
- Bertioli D. J.; Jenkins J.; Clevenger J.; Dudchenko O.; Gao D.; Seijo G.; Leal-Bertioli S. C. M.; Ren L.; Farmer A. D.; Pandey M. K.; et al. The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat. Genet. 2019, 51, 877–884. 10.1038/s41588-019-0405-z. [DOI] [PubMed] [Google Scholar]
- Zhuang W.; Chen H.; Yang M.; Wang J.; Pandey M. K.; Zhang C.; Chang W. C.; Zhang L.; Zhang X.; Tang R.; et al. The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nat. Genet. 2019, 51, 865–876. 10.1038/s41588-019-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.; Lu Q.; Liu H.; Zhang J.; Hong Y.; Lan H.; Li H.; Wang J.; Liu H.; Li S.; et al. Sequencing of cultivated peanut, Arachis hypogaea, yields insights into genome evolution and oil improvement. Mol. Plant 2019, 12, 920–934. 10.1016/j.molp.2019.03.005. [DOI] [PubMed] [Google Scholar]
- Yin D. M.; Ji C. M.; Ma X. L.; Li H.; Zhang W. K.; Li S.; Liu F.; Zhao K.; Li F.; Li K.; et al. Genome of an allotetraploid wild peanut Arachis monticola: a de novo assembly. GigaScience 2018, 7, giy066. 10.1093/gigascience/giy066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.; Li H.; Pandey M. K.; Yang Q.; Wang X.; Garg V.; Li H.; Chi X.; Doddamani D.; Hong Y.; et al. Draft genome of the peanut A-genome progenitor (Arachis duranensis) provides insights into geocarpy, oil biosynthesis, and allergens. Proc. Natl. Acad. Sci. U.S.A. 2016, 113, 6785–6790. 10.1073/pnas.1600899113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.; Kong Q.; Lim A. R. Q.; Lu S.; Zhao H.; Guo L.; Yuan L.; Ma W. Transcriptional regulation of oil biosynthesis in seed plants: current understanding, applications and perspectives. Plant Commun. 2022, 3, 100328. 10.1016/j.xplc.2022.100328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D.; Guo X.; Wang Q.; Zhao L.; Sun Q.; Duan X.; Cao Y.; Sun H. Investigation on lipid profile of peanut oil and changes during roasting by lipidomic approach. Food Sci. Technol. 2022, 154, 112594. 10.1016/j.lwt.2021.112594. [DOI] [Google Scholar]
- Yin D. M.; Ji C. M.; Song Q. X.; Zhang W. K.; Zhang X. G.; Zhao K.; Chen C. Y.; Wang C.; He G.; Liang Z.; et al. Comparison of Arachis monticola with diploid and cultivated tetraploid genomes reveals asymmetric subgenome evolution and improvement of peanut. Adv. Sci. 2020, 7, 1901672. 10.1002/advs.201901672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu A.; Wei F.; Huang F.; Xie Y.; Wu B.; Lv X.; Chen H. Comprehensive and high-coverage lipidomic analysis of oilseeds based on ultrahigh-performance liquid chromatography coupled with electrospray ionization quadrupole time-of-flight mass spectrometry. J. Agric. Food Chem. 2021, 69, 8964–8980. 10.1021/acs.jafc.0c07343. [DOI] [PubMed] [Google Scholar]
- Han Z.; Ahsan M.; Adil M. F.; Chen X.; Nazir M. M.; Shamsi I. H.; Zeng F.; Zhang G. Identification of the gene network modules highly associated with the synthesis of phenolics compounds in barley by transcriptome and metabolome analysis. Food Chem. 2020, 323, 126862. 10.1016/j.foodchem.2020.126862. [DOI] [PubMed] [Google Scholar]
- Wang H.; Zhang S. H.; Fu Q. Q.; Wang Z. D.; Liu X. J.; Sun l. l.; Zhao Z. Y. Transcriptomic and metabolomic analysis reveals a protein module involved in preharvest apple peel browning. Plant Physiol. 2023, 192, 2102–2122. 10.1093/plphys/kiad064. [DOI] [PubMed] [Google Scholar]
- Bareke T. Biology of seed development and germination physiology. Adv. Plants Agric. Res. 2018, 8, 336–346. 10.15406/apar.2018.08.00335. [DOI] [Google Scholar]
- Fait A.; Angelovici R.; Less H.; Ohad I.; Urbanczyk-Wochniak E.; Fernie A. R.; Galili G. Arabidopsis seed development and germination is associated with temporally distinct metabolic switches. Plant Physiol. 2006, 142, 839–854. 10.1104/pp.106.086694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.; Hua J.; Yuan H.; Deng Y.; Zhou Q.; Yang Y.; Dong C.; Zeng J.; Jiang Y. Investigation on green tea lipids and their metabolic variations during manufacturing by nontargeted lipidomics. Food Chem. 2021, 339, 128114. 10.1016/j.foodchem.2020.128114. [DOI] [PubMed] [Google Scholar]
- Liu Q.; Siloto R. M.; Lehner R.; Stone S. J.; Weselake R. J. Acyl-CoA: diacylglycerol acyltransferase: molecular biology, biochemistry and biotechnology. Prog. Lipid Res. 2012, 51, 350–377. 10.1016/j.plipres.2012.06.001. [DOI] [PubMed] [Google Scholar]
- Saha S.; Enugutti B.; Rajakumari S.; Rajasekharan R. Cytosolic triacylglycerol biosynthetic pathway in oilseeds. Molecular cloning and expression of peanut cytosolic diacylglycerol acyltransferase. Plant Physiol. 2006, 141, 1533–1543. 10.1104/pp.106.082198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández M. L.; Whitehead L.; He Z.; Gazda V.; Gilday A.; Kozhevnikova E.; Vaistij F. E.; Larson T. R.; Graham I. A. A cytosolic acyltransferase contributes to triacylglycerol synthesis in sucrose-rescued Arabidopsis seed oil catabolism mutants. Plant Physiol. 2012, 160, 215–225. 10.1104/pp.112.201541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X.; Hu R.; Zhang X.; Chen M.; Chen N.; Pan L.; Wang T.; Wang M.; Yang Z.; Wang Q.; et al. Cloning and functional analysis of three diacylglycerol acyltransferase genes from peanut (Arachis hypogaea L.). PLoS One 2014, 9, e105834 10.1371/journal.pone.0105834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. G.; Kim H.; Suh M. C.; Kim H. U.; Seo P. J. The MYB96 transcription factor regulates triacylglycerol accumulation by activating DGAT1 and PDAT1 expression in Arabidopsis seeds. Plant Cell Physiol. 2018, 59, 1432–1442. 10.1093/pcp/pcy073. [DOI] [PubMed] [Google Scholar]
- Barthole G.; To A.; Marchive C.; Brunaud V.; Soubigou-Taconnat L.; Berger N.; Dubreucq B.; Lepiniec L.; Baud S. MYB118 represses endosperm maturation in seeds of Arabidopsis. Plant Cell 2014, 26, 3519–3537. 10.1105/tpc.114.130021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel R.; Santoro A.; Hofer P.; Tan D.; Oberer M.; Nelson A. T.; Konduri S.; Siegel D.; Zechner R.; Saghatelian A.; Kahn B. B. ATGL is a biosynthetic enzyme for fatty acid esters of hydroxy fatty acids. Nature 2022, 606, 968–975. 10.1038/s41586-022-04787-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Márton M.; Mandoki Z.; Csapó-Kiss Z.; Csapó J. The role of sprouts in human nutrition. A review. Acta Univ. Sapientiae Alimentaria 2010, 3, 81–117. [Google Scholar]
- Baker E. J.; Miles E. A.; Burdge G. C.; Yaqoob P.; Calder P. C. Metabolism and functional effects of plant-derived omega-3 fatty acids in humans. Prog. Lipid Res. 2016, 64, 30–56. 10.1016/j.plipres.2016.07.002. [DOI] [PubMed] [Google Scholar]
- Ramos M. J.; Fernández C. M.; Casas A.; Rodríguez L.; Pérez Á. Influence of fatty acid composition of raw materials on biodiesel properties. Bioresour. Technol. 2009, 100 (1), 261–268. 10.1016/j.biortech.2008.06.039. [DOI] [PubMed] [Google Scholar]
- Aznar-Moreno J.; Denolf P.; Van Audenhove K.; De Bodt S.; Engelen S.; Fahy D.; Wallis J. G.; Browse J. Type 1 diacylglycerol acyltransferases of Brassica napus preferentially incorporate oleic acid into triacylglycerol. J. Exp. Bot. 2015, 66, 6497–6506. 10.1093/jxb/erv363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong W.; Lv H.; Xia G.; Wang M. Does diacylglycerol serve as a signaling molecule in plants?. Plant Signaling Behav. 2012, 7, 472–475. 10.4161/psb.19644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikehara S.; Iso H.; Kokubo Y.; Yamagishi K.; Saito I.; Yatsuya H.; Kimura T.; Sawada N.; Iwasaki M.; Tsugane S. Peanut consumption and risk of stroke and ischemic heart disease in japanese men and women: the JPHC study. Stroke 2021, 52 (11), 3543–3550. 10.1161/strokeaha.120.031212. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.