Abstract
Pasteurella haemolytica serotype 1 is the bacterium most commonly associated with bovine shipping fever. The presence of antibodies against P. haemolytica outer membrane proteins (OMPs) correlates statistically with resistance to experimental P. haemolytica challenge in cattle. Until now, specific P. haemolytica OMPs which elicit antibodies that function in host defense mechanisms have not been identified. In this study, we have cloned and sequenced the gene encoding one such protein, PlpE. Analysis of the deduced amino acid sequence revealed that PlpE is a lipoprotein and that it is similar to an Actinobacillus pleuropneumoniae lipoprotein, OmlA. Affinity-purified, anti-PlpE antibodies recognize a protein in all serotypes of P. haemolytica except serotype 11. We found that intact P. haemolytica and recombinant E. coli expressing PlpE are capable of absorbing anti-PlpE antibodies from bovine immune serum, indicating that PlpE is surface exposed in P. haemolytica and assumes a similar surface-exposed conformation in E. coli. In complement-mediated killing assays, we observed a significant reduction in killing of P. haemolytica when bovine immune serum that was depleted of anti-PlpE antibodies was used as the source of antibody. Our data suggest that PlpE is surface exposed and immunogenic in cattle and that antibodies against PlpE contribute to host defense against P. haemolytica.
Pasteurella haemolytica serotype 1 (S1) is the organism most commonly associated with shipping fever, a disease of beef cattle characterized by fibrinous pleuropneumonia (reviewed in references 9 and 15). The disease is of significant economic importance to the beef industry in the United States, accounting for annual losses approaching 1 billion dollars (18). Shipping fever pneumonia is precipitated by stress-inducing conditions such as shipping, viral infections, inhalation of diesel fumes, overcrowding, and weaning (9, 15). P. haemolytica S1 resides in small numbers in the upper respiratory tracts of cattle, and the tonsil has been shown to be a reservoir (16, 46). Cells proliferate under stressful conditions and are aerosolized in large numbers into lung alveoli, where they cause the disease (16).
Numerous surface and secreted molecules of P. haemolytica S1 have been studied to evaluate their roles in immunity to P. haemolytica infection (reviewed in reference 6). A secreted cytolytic toxin, leukotoxin (Lkt) (44), is a significant P. haemolytica virulence factor. In one study, a vaccine consisting of recombinant Lkt (rLkt) did not provide protection against experimental P. haemolytica challenge (11). However, inclusion of that rLkt in a commercial vaccine resulted in enhanced resistance to challenge (11). Those results are in agreement with data from a prior study (45), which suggested that antibodies against Lkt and surface antigens are necessary for protective immunity to P. haemolytica.
The P. haemolytica surface antigens likely to be most important in contributing to protective immunity are outer membrane proteins (OMPs). Vaccination of cattle with an OMP-enriched fraction of P. haemolytica cell envelopes significantly reduces lung damage following experimental challenge with a P. haemolytica strain of the homologous serotype (29). Bovine antibody responses to proteins present in P. haemolytica surface extracts correlate statistically with resistance to pneumonia (10, 47). Our group and others have analyzed the bovine antibody response to PomA, a protein belonging to the OmpA family (28), to a 94-kDa P. haemolytica OMP (34), and to several membrane lipoproteins (12–14, 37). These studies suggest a role for outer membrane antigens in eliciting protective immunity. However, the capacity for P. haemolytica OMP-specific antibodies to function in host defense mechanisms remains uncharacterized. For the development of more-effective vaccines, it will be important to characterize individual OMPs and identify those that elicit host antibodies which enhance resistance to P. haemolytica infection.
Complement-mediated lysis is an important host defense mechanism against microbial infection and is believed to play a role in controlling P. haemolytica pneumonia. Serum complement concentrations were found to be lower in stressed cattle after transport to a feedlot (40). Lower complement concentrations were associated with higher morbidity in the feedlot, and morbid calves had significantly lower complement levels than did healthy calves in the same feedlots (40). These data suggest that a decrease in serum complement levels might facilitate P. haemolytica infection. However, complement-mediated killing of P. haemolytica requires sensitization with antibodies (27). Antibodies against surface-exposed epitopes of OMPs are likely to play an important role in complement-mediated lysis of P. haemolytica.
Cattle that are resistant to P. haemolytica-induced pneumonia develop antibodies to a surface-exposed, ∼45-kDa OMP (36). The purpose of this study was to determine, through genetic cloning and DNA sequencing, the specific identity of the immunogenic 45-kDa protein and to evaluate the contribution of antibodies against this protein to complement-mediated killing of P. haemolytica. We found that the 45-kDa protein is a lipoprotein, designated PlpE, and that antibodies against PlpE, present in bovine immune sera, contribute to complement-mediated killing of P. haemolytica.
MATERIALS AND METHODS
Bacteria, bacteriophage, culture media, and genomic library.
P. haemolytica (89010807N) S1 was grown in BHI broth or on BHI agar (Difco Laboratories, Detroit, Mich.) as previously described (32). Escherichia coli BB4 and XL1-Blue and bacteriophages λZAPII and R408 were supplied with a P. haemolytica genomic DNA library (Clontech Laboratories, Palo Alto, Calif.) (37) and were grown according to the manufacturer’s instructions. Recombinant E. coli strains were grown in the presence of ampicillin (50 μg/ml).
Bovine immune sera and purification of antibodies.
Two bovine immune sera were used, one from a calf hyperimmunized with live P. haemolytica (25) and one from a calf that was vaccinated with P. haemolytica OMPs and was resistant to experimental P. haemolytica challenge (7). Briefly, the OMP-vaccinated calf was vaccinated subcutaneously on day 0 and day 21 with P. haemolytica S1 OMPs (2 mg in 1 ml of phosphate buffered saline [PBS] and 1 ml of an aluminum hydroxide-DDA-bromide adjuvant which has been described elsewhere in more detail [8]). On day 36, the calf was experimentally challenged transthoracically with 5 ml of a mixture containing 109 CFU of P. haemolytica S1/ml in each caudal lung lobe. Lung damage was evaluated upon necropsy 4 days after challenge, by using a previously described lung lesion score system (35). The serum used in this experiment was collected on the day of experimental challenge.
Bovine sera usually contain antibodies that are immunoreactive to E. coli antigens. This may be because cattle are frequently exposed to numerous bacteria, including E. coli. Alternatively, antibodies against P. haemolytica proteins may be cross-reactive with E. coli antigens. Our studies with P. haemolytica PomA have shown that antibodies against PomA are cross-reactive with E. coli OmpA and vice versa (reference 28 and data not shown). As shown in Fig. 1, antibodies in the serum used in this study recognized E. coli antigens of approximately 28, 32, and 40 kDa. The immunoreactive bands at 32 and 40 kDa likely correspond to the two different forms of the heat-modifiable E. coli OmpA. The nature of the immunoreactive E. coli antigen of 28 kDa is unknown.
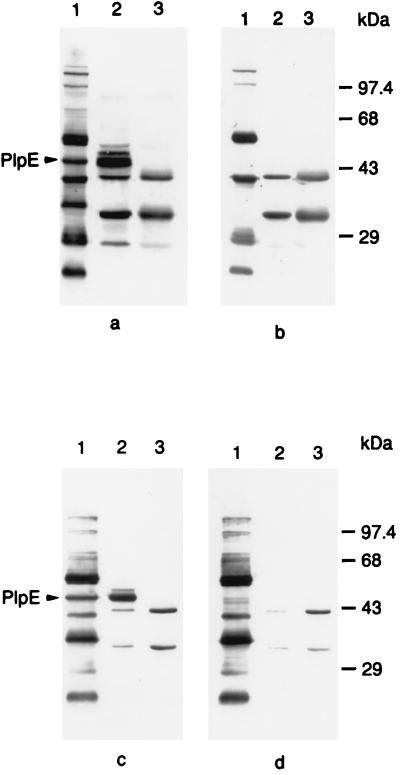
FIG. 1.
Western immunoblots demonstrating expression and surface exposure of PlpE in P. haemolytica and recombinant E. coli. Lanes contain whole-cell lysates of P. haemolytica (lanes 1), E. coli(pB4522) (lanes 2), and nonrecombinant E. coli(pBluescript SK−) (lanes 3). Blots were probed with various forms of bovine immune serum from a calf vaccinated with P. haemolytica OMPs: bovine immune serum (a), immune serum absorbed with P. haemolytica (b), immune serum absorbed with E. coli(pBluescript SK−) (c), and immune serum absorbed with E. coli(pB4522) (d). The arrows designate the 45-kDa band corresponding to PlpE. The immunoreactive E. coli antigens are discussed in the description of bovine immune sera in Materials and Methods.
Antibodies against P. haemolytica ∼45-kDa OMPs were purified from the bovine hyperimmune serum by immunoaffinity (20). Briefly, P. haemolytica outer membranes were purified as described previously (37), separated by preparative sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (14), and transferred to nitrocellulose membranes (37). The region of the nitrocellulose membrane containing ∼45-kDa proteins was identified by comparison with prestained molecular weight standards (Bio-Rad Laboratories, Richmond, Calif.) on the membrane. The 45-kDa region of the nitrocellulose was excised and incubated for 1 h with the bovine hyperimmune serum (1:25) in 0.01 M Tris-HCl (pH 7.4)–0.2 M NaCl (TS), containing 0.05% gelatin and 0.05% Tween (TSGT). Nitrocellulose strips were washed with TS, three times for 5 min each time. Anti-45-kDa-protein antibodies bound to antigen on the nitrocellulose strips were eluted as described elsewhere (20). The eluted antibodies were concentrated by using a Centriprep 10 concentrator (Amicon Inc., Beverly, Mass.). These antibodies are referred to throughout this paper as anti-45-kDa antibodies. Antibodies against at least three OMPs that migrate at ∼45 kDa are present in this preparation. However, the predominant antibodies in the preparation are those against PlpE. When antibodies are affinity purified by the method we used, contaminating antibodies are frequently present. We found antibodies against P. haemolytica PomA, a heat-modifiable protein belonging to the OmpA family (28), to be contaminants when the anti-45-kDa antibodies were purified by this procedure. PomA migrates at ∼30 and 38 kDa, and immunoreactive bands are apparent in Western immunoblots that used the anti-45-kDa antibodies purified in this study.
Gene cloning, DNA sequencing, site-directed mutagenesis, and sequence analysis.
The plpE gene was cloned by screening a P. haemolytica expression library (described above), according to the manufacturer’s instructions, with affinity-purified anti-45-kDa antibodies. Additional DNA cloning was performed as described previously (37). Plasmid inserts were progressively deleted from both ends by using the Erase A Base kit (Promega Corp., Madison, Wis.), and progressively smaller plasmid inserts were sequenced by using the universal and reverse primers. Both DNA strands were sequenced. DNA sequencing was performed at the Oklahoma State University Recombinant DNA/Protein Resource Facility, on an Applied Biosystems (Foster City, Calif.) model 373A automated DNA sequencer. Site-directed mutagenesis was performed by using the Gene Editor in vitro Mutagenesis System (Promega Corp.). Mutations were confirmed by DNA sequence analysis. Sequences were analyzed with MacVector/Assemblylign software (Oxford Molecular Group, Inc., Campbell, Calif.). The deduced amino acid sequence of PlpE was compared with other sequences in GenBank by using BLAST 2.0 (1), and alignments were generated with CLUSTALW 1.7 at the Baylor College of Medicine Search Launcher.
Antigen preparation and Western immunoblots.
Whole-cell lysates were prepared and Western immunoblots were performed as described previously (14, 37). Primary antibodies used for each Western immunoblot experiment are described in Results. Alkaline phosphatase-conjugated mouse monoclonal, anti-bovine immunoglobulin G antibody (Sigma Immunochemicals, St. Louis, Mo.) (1:20,000 in TSGT) was used as the secondary antibody in Western immunoblots.
For Western immunoblots, bovine immune serum was absorbed with intact P. haemolytica by using a modification of a previously described method (19). Logarithmic-phase P. haemolytica cells, from 1 liter of culture (A600 of 0.5), were pelleted by centrifugation, washed once in PBS, and resuspended in immune serum diluted 1:100 in Tris-saline-nonfat dry milk (TSM) (10 mM Tris [pH 7.4], 0.9% [wt/vol] NaCl, 1% nonfat dried milk). Cells resuspended in serum were incubated at 4°C for 3 h on a rocking platform. Following incubation, cells were pelleted by centrifugation at 11,000 × g. The supernatant was carefully removed and stored at −20°C after the addition of sodium azide at 0.02%. Unabsorbed immune serum was used as a control and was diluted 1:100 in TSM before use. Serum absorptions with recombinant E. coli(pB4522) and nonrecombinant E. coli(pBluescript SK−) were performed similarly except that stationary-phase organisms were used.
[3H]palmitic acid labeling of bacterial lipoproteins.
Labeling and analysis of P. haemolytica, recombinant E. coli expressing PlpE, and nonrecombinant E. coli were performed by using [9,10-3H]palmitic acid (Dupont, NEN, Boston, Mass.) as described previously (14).
Complement-mediated killing assay.
Serum from a calf with a low antibody titer against P. haemolytica as determined by an enzyme-linked immunosorbent assay was used as a complement source. The complement source serum was depleted of any existing antibodies against P. haemolytica by incubation with excess stationary-phase P. haemolytica at 4°C on a rocking platform for 1 h. Before incubation with complement source serum, P. haemolytica cells were washed once with cold PBS (4°C).
Serum from an OMP-vaccinated calf (described above) was used as the source of antibodies. The antibody source serum was heat inactivated at 56°C for 30 min and was used in two different forms. For the first form, anti-PlpE antibodies were removed from the antibody source serum by absorption of the serum with recombinant E. coli expressing PlpE. This process would also remove any anti-E. coli antibodies that were present in the serum. For the second form, control serum from which only anti-E. coli antibodies were removed was prepared by absorption of the antibody source serum with nonrecombinant E. coli(pBluescript SK−). For absorbing sera to be used in complement killing assays, recombinant or nonrecombinant E. coli cells were grown overnight in 100 ml of Luria broth (Life Technologies Inc., Grand Island, N.Y.) and harvested by centrifugation (11,000 × g) at 4°C. Cells were washed once with PBS and resuspended in 2 ml of antibody source serum that had been diluted 1:1 with PBS. Cells and serum were incubated on a rocking platform for 3 h at 4°C. After incubation, cells were removed from serum by centrifugation. The process was repeated until the serum absorbed with recombinant E. coli no longer recognized PlpE in a Western immunoblot (data not shown).
The complement-mediated killing assay was developed by modifying the techniques described by Chae et al. (5) and Murphy et al. (33). To ensure that the assay was capable of detecting a change in the amount of bactericidal antibody, numbers of bacteria and the concentration of complement were evaluated in preliminary experiments (33). For complement-mediated killing assays, bacteria were grown in BHI broth for 18 h at 37°C, on a rotary shaker (200 rpm). Cells were washed once with PBS and resuspended in PBS to an A600 of 0.5. Complement source serum (50 μl) and antibody source serum (form 1 or 2 described above) (50 μl) were added to 150 μl of PBS. P. haemolytica cells (∼9,000 to 18,000 CFU in 40 μl of PBS) were then added. Immediately after the addition of P. haemolytica (t = 0) and after incubation for 30 min in a 37°C water bath (t = 30), 100-μl samples were removed and diluted 1:100 in PBS. Dilutions were prepared and plated on BHI agar plates for determination of CFU. To monitor the killing activity of the complement source serum alone, a control with only complement source serum and no antibody source serum was evaluated in an identical manner. We also determined that the heat-killed antibody source serum alone had no killing activity (data not shown). Percent killing was calculated by the following formula: {(CFUt=0 − CFUt=30)/CFUt=0} × 100. Percent survival was calculated as 100 − (percent killing).
Statistical analysis.
Within each experiment, complement-mediated killing assays were done in triplicate for each antibody source serum and the complement source serum control. Additionally, three separate experiments were done on different days. Statistically significant differences between percent killing by different sera within experiments were determined by Student’s t test (2).
Nucleotide sequence accession number.
The nucleotide sequence of P. haemolytica plpE has been deposited in GenBank under accession no. AF059036.
RESULTS
Cloning of plpE.
To isolate a clone expressing the immunogenic 45-kDa protein, we screened a genomic library of P. haemolytica S1 with anti-45-kDa antibodies that were affinity purified from bovine immune serum. We isolated recombinant λZAPII phage that reacted with the affinity-purified antibodies. A recombinant plasmid containing a 4.5-kbp insert was excised from one phage clone and transformed into E. coli XL1-Blue. We subcloned a 2.2-kbp fragment from this insert into pBluescript SK(−) and named this plasmid pB4522. E. coli(pB4522) expressed a 45-kDa protein that was recognized by the affinity-purified antibodies and bovine immune serum (Fig. 1a). We named this protein PlpE.
Surface exposure of PlpE.
As mentioned earlier, we previously demonstrated the presence of an immunogenic 45-kDa P. haemolytica protein that is surface exposed (36). To determine if PlpE expressed by the recombinant E. coli strain corresponds to the strongly immunogenic P. haemolytica surface protein, we examined surface exposure of the protein on intact P. haemolytica. Absorption of bovine immune serum with intact P. haemolytica resulted in a loss of antibody reactivity on Western immunoblots with rPlpE and a protein of the same Mr in P. haemolytica whole-cell lysates (Fig. 1a and b), suggesting that PlpE is surface exposed in P. haemolytica. Similarly, absorption of the same bovine immune sera with intact recombinant E. coli(pB4522) expressing PlpE resulted in a loss of reactivity to rPlpE and to a 45-kDa protein in P. haemolytica (Fig. 1c and d). These data suggest that PlpE is also exposed on the surface of recombinant E. coli and that PlpE is the primary 45-kDa surface-exposed immunogen of P. haemolytica.
DNA sequence analysis of PlpE.
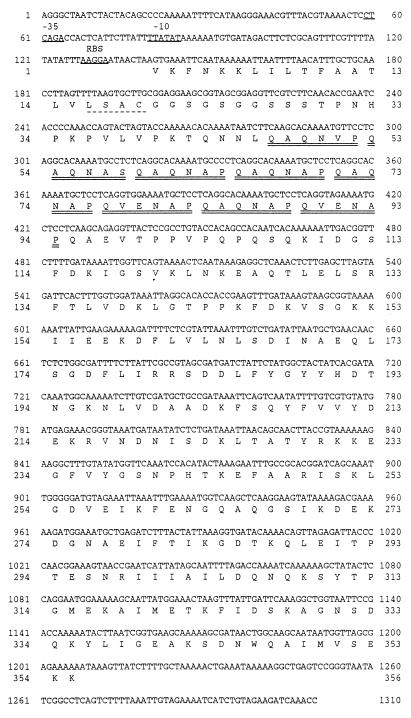
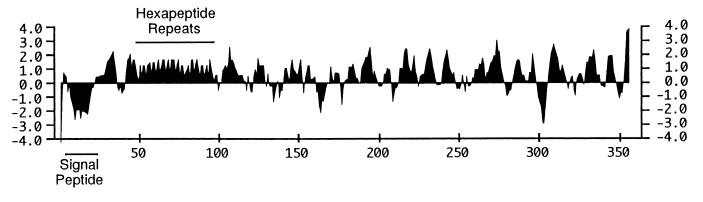
DNA sequencing of the cloned insert in pB4522 revealed an open reading frame of 1,068 nucleotides that begins with a GTG codon and encodes a protein with a calculated molecular mass of 39.1 kDa (Fig. 2). The deduced amino acid sequence contains a putative hydrophobic signal peptide followed by a consensus lipoprotein processing site (LSAC) (Fig. 2). The calculated molecular mass of the putative mature form of PlpE is 37.03 kDa. The N-terminal region of the mature PlpE contains eight imperfect copies of a repeated hexapeptide (Fig. 2) that are encoded by 18-nucleotide repeats (Fig. 3). The hexapeptide repeat is predicted to form a large hydrophilic domain in PlpE (Fig. 4). PlpE also has numerous other hydrophilic domains that correspond to regions with a high probability of being surface exposed (Fig. 4). A search of GenBank sequences and subsequent sequence alignments revealed that the deduced amino acid sequence of PlpE has 18% identity and 32% similarity to an outer membrane lipoprotein (OmlA) produced by Actinobacillus pleuropneumoniae serotype 1 and 20% identity and 35% similarity to the OmlA protein from A. pleuropneumoniae serotype 5 (data not shown). Although both of those proteins lack the hexapeptide repeat mentioned above, they contain regularly spaced PK and PQ repeats in the same region (4, 17, 23).
FIG. 2.
Nucleotide sequence of plpE and deduced amino acid sequence of PlpE. Bases 5′ of the gene and corresponding to E. coli consensus −35 and −10 sequences and ribosome binding sites (RBS) are underlined. A consensus lipoprotein processing site is underlined with a dashed line. Cleavage of the signal peptide (residues 1 to 18) would occur after the alanine residue at position 18. The eight hydrophilic hexapeptide repeats in the amino-terminal region of PlpE are doubly underlined.
FIG. 3.
Repeated nucleotide sequences encoding the eight hexapeptide repeats within the N-terminal region of PlpE. The consensus nucleotide and peptide repeats are shown. Nucleotides within repeats that match the consensus are indicated with a dash, and those that differ from the consensus are indicated with lowercase letters. Numbers shown correspond to amino acid residues in Fig. 2 and 4.
FIG. 4.
Hydrophilicity plot of the deduced amino acid sequence of PlpE. The signal peptide and the hydrophilic hexapeptide repeats are indicated. Positive values represent hydrophilic regions. The plot was generated in MacVector (Oxford Molecular Group, Inc.) by using the Kyte-Doolittle algorithm with a window size of 7.
Site-directed mutagenesis of the GTG start codon.
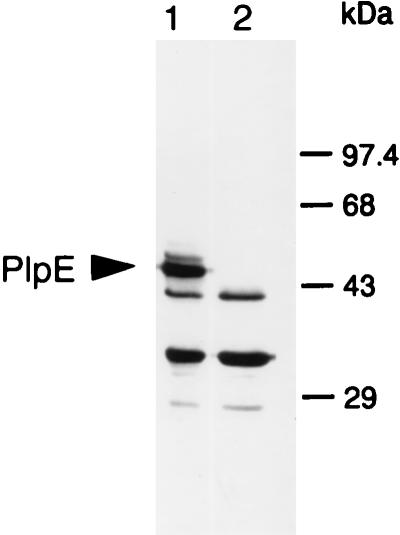
To verify that GTG functions as a translational start codon for PlpE, we performed site-directed mutagenesis and converted the codon to GGG. Western immunoblots of whole-cell lysates from the wild-type and mutant E. coli(pB4522) strains, probed with anti-45-kDa antibodies, revealed that the mutant no longer produced a 45-kDa immunoreactive protein, suggesting that GTG functions as the translational start codon (Fig. 5).
FIG. 5.
Western immunoblot demonstrating the effect of mutagenesis of the GTG codon on production of PlpE. The blot was probed with anti-45-kDa antibodies. Lanes: 1, E. coli(pB4522) with GTG and expressing PlpE; 2, E. coli(pB4522) in which GTG was changed to GGG by site-directed mutagenesis.
Lipid modification of PlpE.
Because the deduced amino acid sequence contained a consensus lipoprotein processing site, we examined P. haemolytica and E. coli(pB4522) for the presence of 45-kDa lipid-modified proteins. P. haemolytica, E. coli(pB4522), and nonrecombinant E. coli(pBluescript SK−) were grown in the presence of [3H]palmitic acid. A 45-kDa, 3H-labeled lipoprotein is present in whole-cell lysates of P. haemolytica and E. coli(pB4522) but absent from the nonrecombinant E. coli strain (Fig. 6).
FIG. 6.
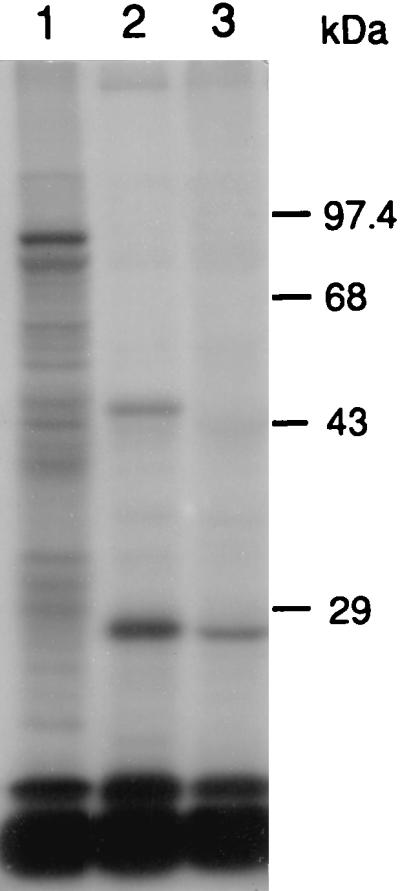
Autoradiograph of an SDS-polyacrylamide gel of [3H]palmitate-labeled total cellular proteins. Lanes: 1, P. haemolytica; 2, E. coli(pB4522); 3, E. coli(pBluescript SK−).
Conservation of PlpE among serotypes of P. haemolytica.
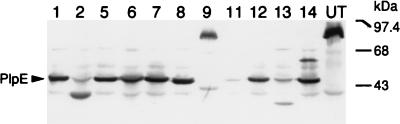
To determine if PlpE is expressed by other P. haemolytica serotypes, we examined whole-cell lysates of P. haemolytica serotypes 1, 2, 5, 6, 7, 8, 9, 11, 12, 13, and 14 and an untypeable strain of P. haemolytica, by Western immunoblot analysis, for reactivity with the anti-45-kDa antibodies (Fig. 7). The antibodies reacted strongly with a 45-kDa protein in serotypes 1, 5, 6, 7, 8, 12, and 14; a 38-kDa protein in serotype 2; a 36-kDa protein in serotype 13; and an ∼80-kDa protein in serotype 9 and the untypeable strain. The antibodies reacted weakly with a unique band at 43 kDa in serotype 9. The antibodies did not react strongly with a protein in the serotype 11 strain.
FIG. 7.
Western immunoblot of whole-cell lysates of P. haemolytica serotypes 1, 2, 5, 6, 7, 8, 9, 11, 12, 13, and 14 and an untypeable strain, probed with anti-45-kDa antibodies. The lane numbers represent the serotypes, with UT representing the untypeable strain. Weakly reactive bands that are common in all lanes and weakly reactive bands at 45 kDa in lanes 2, 11, and 13 are likely due to nonspecific antibodies that were eluted with the anti-PlpE antibodies during the affinity purification process (see Materials and Methods). The immunoblot was performed twice with different whole-cell lysate preparations for each strain and yielded similar results each time.
Role of anti-PlpE antibodies in complement-mediated killing of P. haemolytica.
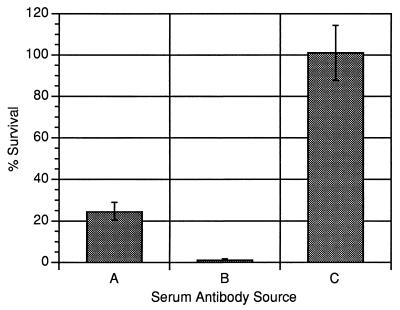
We sought to determine if anti-PlpE antibodies contribute to complement-mediated killing of P. haemolytica. Since we had observed that E. coli(pB4522) has the capacity to remove anti-PlpE antibodies from bovine serum (Fig. 1d), we used the recombinant strain to remove those antibodies from immune serum of a calf that was vaccinated with P. haemolytica OMPs. We then compared rates of complement-mediated killing of P. haemolytica using, as an antibody source, bovine immune serum or immune serum depleted of anti-PlpE antibodies. As shown in Fig. 8, immune serum that was depleted of anti-PlpE antibodies caused less killing of P. haemolytica than did immune serum that was not depleted of those antibodies. For each of three separate experiments, the difference in killing activity between the two sera was statistically significant (P < 0.003).
FIG. 8.
Complement-mediated killing activity of anti-OMP bovine immune serum with and without anti-PlpE antibodies. Sera used as the source of antibodies were bovine immune serum absorbed with E. coli(pB4522) (A), bovine immune serum absorbed with E. coli(pBluescript SK−) (B), and complement source alone (C). Values greater than 100% represent growth. Data shown are the means and standard deviations (error bars) of three replicates from a single experiment and are representative of three separate experiments. The differences between the means for the experiments represented by bars A and B are statistically significant (P < 0.003).
DISCUSSION
As mentioned earlier, numerous studies have indicated that P. haemolytica OMPs are important in eliciting protective immunity in cattle and that antibody responses of cattle to P. haemolytica OMPs correlate with resistance to experimental P. haemolytica challenge. However, not all antibody responses to individual antigens contribute significantly to host defense, and some may actually be detrimental to certain defense mechanisms (30, 38, 49). Therefore, one of our goals is to identify and characterize individual P. haemolytica OMPs that elicit antibodies which function in host immune mechanisms.
In our previous work, we observed an immunoreactive band at 45 kDa on Western immunoblots of P. haemolytica OMPs that were probed with three different sera from cattle resistant to P. haemolytica infection (36). Those studies also revealed the 45-kDa antigen to be a surface-exposed protein. Here, we present genetic and immunologic characterization of a P. haemolytica 45-kDa surface-exposed lipoprotein (PlpE) and demonstrate that bovine antibodies against PlpE contribute to complement-mediated killing of P. haemolytica. The results of our study demonstrate that PlpE is the major P. haemolytica antigen responsible for the significant, immunoreactive band at 45 kDa in Western immunoblots probed with immune sera from cattle.
Our analysis of the deduced amino sequence of PlpE revealed several interesting features. Although PlpE migrates at an Mr of 45 kDa on SDS-polyacrylamide gels, the calculated molecular mass of the putative mature form of PlpE is ∼37 kDa. This discrepancy may possibly be a result of the high proline content of PlpE (∼6%). Proline is a turn-inducing amino acid residue that often causes proteins to migrate slower on SDS-PAGE (39). The deduced amino acid sequence of PlpE also contains a typical signal peptide, followed by a consensus lipoprotein processing site, and in this study we demonstrated lipid modification of rPlpE. Another interesting feature of plpE is that GUG functions as the translational start codon. GUG, AUG, and UUG are in the group of class I initiation codons that support efficient translation (48). In E. coli, the intrinsic activity of GUG is 12 to 15% that of AUG (41). About 8% of known genes from E. coli and other bacteria use GUG as the start codon (43). The aroA gene of Pasteurella multocida appears to use GUG as an initiation codon (21). However, to our knowledge plpE is the first example of a P. haemolytica gene with GUG as a start codon.
Protein sequence similarities are present between the QAQNAP repeats in PlpE and a newly identified peptide repeat (NAP) in some forms of the polymorphic merozoite surface protein 2 (MSP2) from Plasmodium falciparum (22). The NAP repeats, like the QAQNAP repeats in PlpE, occur near the amino terminus of MSP2, and similarities between PlpE and MSP2 include glycines and serines on the amino-terminal side of the repeat regions. Hydrophilicity plots of PlpE and this form of MSP2 are also similar from the amino terminus, extending across the repeated region (data not shown). DNA sequence identity exists as well between the consensus nucleotide sequence encoding the NAP repeat unit of MSP2 (AATGCTCCA) and the consensus nucleotides encoding the corresponding region in the PlpE hexapeptide repeat (AATGCTCCT). In mouse immunization experiments with synthetic peptides and a rMSP2, an immunodominant T-cell determinant was mapped to a region spanning the NAP sequence (42). More detailed immunological studies are required to determine if such a role may exist for the hexapeptide repeat in PlpE.
Within the hexapeptide repeat of PlpE, codon sequences are generally conserved for each amino acid position, and unusual codons for P. haemolytica are also conserved in this region. Seven of the eight codons for the first glutamine of each repeat are CAG, whereas for the second glutamine, six of six codons are CAA. In P. haemolytica, CAA is the preferred codon for glutamine (26) and is reflective of the high moles percent A+T content (∼60%) of P. haemolytica chromosomal DNA. For the first alanine in the repeat, six of six codons are GCA, whereas for the second alanine, five of seven codons are GCT. Similarly, all asparagines are encoded by AAT, and all prolines are encoded by CCT. The relative synonymous codon usage in P. haemolytica for CCT was calculated by Lo et al. (26) to be slightly lower than that for CCA. Our recent analyses with a larger number of P. haemolytica genes revealed similar results (31). These data suggest that expansion of the hexapeptide repeat may have occurred by duplication of one or more of the 18 nucleotide repeats.
Additional amino acid sequence alignments revealed that the A. pleuropneumoniae OmlA lipoproteins are similar to PlpE over its entire sequence. A. pleuropneumoniae is a pathogen of pigs that causes a fibrinous pleuropneumonia very similar to that caused by P. haemolytica in cattle. Vaccination of pigs with protein aggregates containing rOmlA, cloned from A. pleuropneumoniae serotype 1, significantly reduced lung damage and death of pigs upon subsequent experimental challenge with a homologous A. pleuropneumoniae serotype (17). Similarly, vaccination of pigs with gel-purified rOmlA, cloned from A. pleuropneumoniae serotype 5a, significantly lowered mortality upon challenge with a serotype 5a strain (4).
The OmlA proteins from different A. pleuropneumoniae serotypes may be more antigenically heterogeneous than are the PlpE proteins from the different P. haemolytica serotypes. A. pleuropneumoniae serotypes 1, 5, and 7 are the most common in North America. Sera from pigs vaccinated with rOmlA (serotype 1) failed to recognize proteins in 6 of 13 A. pleuropneumoniae serotypes and only weakly recognized a protein in 3 of those serotypes (17). Similarly, rabbit antisera against rOmlA from serotype 5a recognized a protein only in serotypes 5a, 5b, and 10 (4, 23). These data suggest that OmlA proteins from a single serotype are unlikely to be cross-protective for heterologous serotypes. Indeed, one A. pleuropneumoniae vaccine currently under evaluation includes, among other antigens, rOmlA proteins from both serotypes 1 and 5 (24).
In contrast, our Western immunoblots revealed that anti-45-kDa antibodies, affinity purified from bovine immune sera, recognize a protein in all but one P. haemolytica serotype (serotype 11). P. haemolytica serotypes 1 and 6 are most frequently isolated from the lungs of pneumonic cattle in the United States. Thus, PlpE may have potential for being a significant cross-protective antigen for those serotypes. Several P. haemolytica serotypes possess immunoreactive antigens with Mrs different from that of PlpE (Fig. 7), including serotypes 2 (∼38 kDa) and 9 (∼80 kDa) and the untypeable strain (∼80 kDa). Although purely speculative at this time, it is possible that variation in the number of hexapeptide repeats could account for these differences in Mrs. Alternatively, as discussed above, different percentages of proline residues in the proteins from different serotypes could alter the mobility of these proteins. Significant differences in P. haemolytica serotype 1 and serotype 9 OMPs were also suggested by the results of a recent vaccine trial. Vaccination of calves with OMPs purified from a serotype 9 strain did not provide significant protection from experimental challenge with a serotype 1 strain (29). Future studies will be necessary to evaluate the capacity of PlpE to enhance protection of cattle against experimental challenge with homologous and heterologous serotypes of P. haemolytica.
Results of the complement-mediated killing assays demonstrate that anti-PlpE antibodies contribute to this mechanism of bovine defense, one that is believed to be important in protection against P. haemolytica pneumonia. Immune serum that was depleted of anti-PlpE antibodies also caused significant amounts of killing, ∼75% under these assay conditions. These results suggest that antibodies against other surface antigens also contribute to complement-mediated killing. Absorption of bovine immune serum with intact P. haemolytica results in the loss or reduction of immunoreactivity in Western blots to numerous antigens (Fig. 1a and b), any of which may bind antibodies that effect complement-mediated killing. Identification of these antigens and elucidation of the role of antibodies directed against them in bovine immune mechanisms are currently major focuses of our research.
Control complement-mediated killing assays with only complement source and no antibody source indicated that complement alone failed to kill P. haemolytica, suggesting that complement activation strictly through the alternative pathway does not play a significant role in the killing of the organism. These data are in agreement with those of previous studies which demonstrated that only activation of the complement cascade through the classical pathway is important in killing P. haemolytica (3, 27).
Studies in our laboratory to evaluate the role of anti-PlpE antibodies in other mechanisms of host defense, such as neutrophil phagocytosis and killing, and to identify additional OMPs that may be important in protective immunity against P. haemolytica are under way. The addition of PlpE and other OMPs which elicit protective antibodies to vaccines containing leukotoxin may provide significant protection against pneumonic pasteurellosis.
ACKNOWLEDGMENTS
This work was supported by grant 95-37204-1999 from the National Research Initiative Competitive Grants Program of the USDA, the Oklahoma Agricultural Experiment Station (project OKL02179), and the Oklahoma State University College of Veterinary Medicine. K.P. was supported by an OSU-CVM graduate research assistantship award.
REFERENCES
- 1.Altschul S F, Schaffer T L, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey N T J. Statistical methods in biology. New York, N.Y: John Wiley & Sons; 1981. pp. 43–47. [Google Scholar]
- 3.Blau K A, Ward A C S, Prieur D J, Corbeil L B. Serum susceptibility of bovine pasteurellas. Can J Vet Res. 1987;51:157–161. [PMC free article] [PubMed] [Google Scholar]
- 4.Bunka S, Christensen C, Potter A A, Willson P J, Gerlach G-F. Cloning and characterization of a protective outer membrane lipoprotein of Actinobacillus pleuropneumoniae serotype 5. Infect Immun. 1995;63:2797–2800. doi: 10.1128/iai.63.7.2797-2800.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chae C H, Gentry M J, Confer A W, Anderson G A. Resistance to host immune defense mechanisms afforded by capsular material of Pasteurella haemolytica, serotype 1. Vet Microbiol. 1990;25:241–251. doi: 10.1016/0378-1135(90)90081-6. [DOI] [PubMed] [Google Scholar]
- 6.Confer A W. Immunogens of Pasteurella. Vet Microbiol. 1993;37:353–368. doi: 10.1016/0378-1135(93)90034-5. [DOI] [PubMed] [Google Scholar]
- 7.Confer, A. W. 1998. Unpublished data.
- 8.Confer A W, Clinkenbeard K D, Gatewood D M, Driskel B A, Montelongo M. Serum antibody responses of cattle vaccinated with partially purified native Pasteurella haemolytica leukotoxin. Vaccine. 1997;15:1423–1429. doi: 10.1016/s0264-410x(97)84247-8. [DOI] [PubMed] [Google Scholar]
- 9.Confer A W, Panciera R J, Mosier D A. Bovine pneumonic pasteurellosis: immunity to Pasteurella haemolytica. J Am Vet Med Assoc. 1988;193:1308–1316. [PubMed] [Google Scholar]
- 10.Confer A W, Simons K R, Panciera R J, Mort A J, Mosier D A. Serum antibody response to carbohydrate antigens of Pasteurella haemolytica serotype 1: relation to experimentally induced bovine pneumonic pasteurellosis. Am J Vet Res. 1989;50:98–105. [PubMed] [Google Scholar]
- 11.Conlon J A, Shewen P E, Lo R Y C. Efficacy of recombinant leukotoxin in protection against pneumonic challenge with live Pasteurella haemolytica A1. Infect Immun. 1991;59:587–591. doi: 10.1128/iai.59.2.587-591.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooney B J, Lo R Y C. Three contiguous lipoprotein genes in Pasteurella haemolytica A1 which are homologous to a lipoprotein gene in Haemophilus influenzae type b. Infect Immun. 1993;61:4682–4688. doi: 10.1128/iai.61.11.4682-4688.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Craven R C, Confer A W, Gentry M J. Cloning and expression of a 30 kDa surface antigen of Pasteurella haemolytica. Vet Microbiol. 1991;27:63–78. doi: 10.1016/0378-1135(91)90063-l. [DOI] [PubMed] [Google Scholar]
- 14.Dabo S M, Styre D, Confer A W, Murphy G L. Expression, purification, and immunologic analysis of Pasteurella haemolytica A1 28–30 kDa lipoproteins. Microb Pathog. 1994;17:149–158. doi: 10.1006/mpat.1994.1061. [DOI] [PubMed] [Google Scholar]
- 15.Frank G H. Pasteurellosis of cattle. In: Adlam C, Rutter J M, editors. Pasteurella and pasteurellosis. London, United Kingdom: Academic Press Limited; 1989. pp. 197–222. [Google Scholar]
- 16.Frank G H, Briggs R E. Colonization of the tonsils of calves with Pasteurella haemolytica. Am J Vet Res. 1992;53:481–484. [PubMed] [Google Scholar]
- 17.Gerlach G-F, Anderson C, Klashinsky S, Rossi-Campos A, Potter A A, Willson P J. Molecular characterization of a protective outer membrane lipoprotein (OmlA) from Actinobacillus pleuropneumoniae serotype 1. Infect Immun. 1993;61:565–572. doi: 10.1128/iai.61.2.565-572.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griffin D. Economic impact associated with respiratory disease in beef cattle. In: Vestweber J, St. Jean G, editors. Veterinary clinics of North America: food animal practice. W. B. Philadelphia, Pa: Saunders Co.; 1997. pp. 367–377. [DOI] [PubMed] [Google Scholar]
- 19.Hansen E J, Hart D A, McGehee J L, Toews G B. Immune enhancement of pulmonary clearance of nontypable Haemophilus influenzae. Infect Immun. 1988;56:182–190. doi: 10.1128/iai.56.1.182-190.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 21.Homchampa P, Strugnell R A, Adler B. Molecular analysis of the aroA gene of Pasteurella multocida and vaccine potential of a constructed aroA mutant. Mol Microbiol. 1992;6:3585–3593. doi: 10.1111/j.1365-2958.1992.tb01794.x. [DOI] [PubMed] [Google Scholar]
- 22.Irion A, Beck H-P, Felger I. New repeat unit and hot spot for recombination in FC27-type alleles of the gene coding for Plasmodium falciparum merozoite surface protein 2. Mol Biochem Parasitol. 1997;90:367–370. doi: 10.1016/s0166-6851(97)00165-5. [DOI] [PubMed] [Google Scholar]
- 23.Ito H, Uchida I, Sekizaki T, Ooishi E, Kawai T, Okabe T, Taneno A, Terakado N. Molecular cloning of an Actinobacillus pleuropneumoniae outer membrane lipoprotein (OmlA) from serotype 5a. Microb Pathog. 1995;18:29–36. [PubMed] [Google Scholar]
- 24.Keich R L, Baarsch M J, Rossi-Campos A, Yancey R J. Conference of Research Workers in Animal Disease. Proceedings of the 78th Annual Meeting. 1997. The evaluation of a 4-way recombinant Actinobacillus pleuropneumoniae vaccine, abstr. P140. [Google Scholar]
- 25.Lessley B A, Confer A W, Mosier D A, Gentry M J, Durham J A, Rummage J A. Saline-extracted antigens of Pasteurella haemolytica: separation by chromatofocusing, preliminary characterization, and evaluation of immunogenicity. Vet Immunol Immunopathol. 1985;10:279–296. doi: 10.1016/0165-2427(85)90053-4. [DOI] [PubMed] [Google Scholar]
- 26.Lo R Y C. An analysis of the codon usage of Pasteurella haemolytica A1. FEMS Microbiol Lett. 1992;100:125–132. doi: 10.1111/j.1574-6968.1992.tb14030.x. [DOI] [PubMed] [Google Scholar]
- 27.MacDonald J T, Maheswaran S K, Opuda-Asibo J, Townsend E L, Theis E S. Susceptibility of Pasteurella haemolytica to the bactericidal effects of serum, nasal secretions, and bronchoalveolar washings from cattle. Vet Microbiol. 1983;8:585–599. doi: 10.1016/0378-1135(83)90007-x. [DOI] [PubMed] [Google Scholar]
- 28.Mahasreshti P J, Murphy G L, Wyckoff III J H, Farmer S, Hancock R E W, Confer A W. Purification and partial characterization of the OmpA family of proteins of Pasteurella haemolytica. Infect Immun. 1997;65:211–218. doi: 10.1128/iai.65.1.211-218.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morton R J, Panciera R J, Fulton R W, Frank G H, Ewing S A, Homer J T, Confer A W. Vaccination of cattle with outer membrane protein-enriched fractions of Pasteurella haemolytica and resistance against experimental challenge exposure. Am J Vet Res. 1995;56:875–879. [PubMed] [Google Scholar]
- 30.Munkley A, Tinsley C R, Virji M, Heckels J E. Blocking of bactericidal killing of Neisseria meningitidis by antibodies directed against class 4 outer membrane protein. Microb Pathog. 1991;11:447–452. doi: 10.1016/0882-4010(91)90041-8. [DOI] [PubMed] [Google Scholar]
- 31.Murphy, G. L. 1998. Unpublished observations.
- 32.Murphy G L, Whitworth L C. Construction of isogenic mutants of Pasteurella haemolytica by allelic replacement. Gene. 1994;148:101–105. doi: 10.1016/0378-1119(94)90241-0. [DOI] [PubMed] [Google Scholar]
- 33.Murphy T F, Bartos L C, Rice P A, Nelson B M, Dudas K C, Apicella M A. Identification of a 16,600-dalton outer membrane protein on nontypeable Haemophilus influenzae as a target for human serum bactericidal antibody. J Clin Invest. 1986;78:1020–1027. doi: 10.1172/JCI112656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nelson S L, Frank G H. Purification and characterization of a 94-kDa Pasteurella haemolytica antigen. Vet Microbiol. 1989;21:57–66. doi: 10.1016/0378-1135(89)90018-7. [DOI] [PubMed] [Google Scholar]
- 35.Panciera R J, Corstvet R E, Confer A W, Gresham C N. Bovine pneumonic pasteurellosis: effect of vaccination with live Pasteurella species. Am J Vet Res. 1984;45:2538–2542. [PubMed] [Google Scholar]
- 36.Pandher, K., A. W. Confer, and G. L. Murphy. Submitted for publication.
- 37.Pandher K, Murphy G L. Genetic and immunologic analyses of a 38 kDa surface-exposed lipoprotein of Pasteurella haemolytica A1. Vet Microbiol. 1996;51:331–341. doi: 10.1016/0378-1135(96)00029-6. [DOI] [PubMed] [Google Scholar]
- 38.Plummer F A, Chubb H, Simonsen J N, Bosire M, Slaney L, Maclean I, Ndinya-Achola J O, Waiyaki P, Brunham R C. Antibody to Rmp (outer membrane protein 3) increases susceptibility to gonococcal infection. J Clin Invest. 1993;91:339–343. doi: 10.1172/JCI116190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Postle K. TonB and the Gram-negative dilemma. Mol Microbiol. 1990;4:2019–2025. doi: 10.1111/j.1365-2958.1990.tb00561.x. [DOI] [PubMed] [Google Scholar]
- 40.Purdy C W, Richards A B, Foster G S. Market stress-associated changes in serum complement activity in feeder calves. Am J Vet Res. 1991;52:1842–1847. [PubMed] [Google Scholar]
- 41.Rinquist S, Shinedling S, Barrick D, Green L, Binkley J, Stormo G, Gold L. Translation initiation in Escherichia coli: sequences within the ribosome-binding site. Mol Microbiol. 1992;6:1219–1229. doi: 10.1111/j.1365-2958.1992.tb01561.x. [DOI] [PubMed] [Google Scholar]
- 42.Rzepczyk C M, Csurhes P A, Saul A J, Jones G L, Dyer S, Chee D, Goss N, Irving D O. Comparative study of the T cell response to two allelic forms of a malarial vaccine candidate protein. J Immunol. 1992;148:1197–1204. [PubMed] [Google Scholar]
- 43.Schneider T D, Stormo G D, Gold L. Information content of binding sites on nucleotide sequences. J Mol Biol. 1986;188:415–431. doi: 10.1016/0022-2836(86)90165-8. [DOI] [PubMed] [Google Scholar]
- 44.Shewen P E, Wilkie B N. Cytotoxin of Pasteurella haemolytica acting on bovine leukocytes. Infect Immun. 1982;35:91–94. doi: 10.1128/iai.35.1.91-94.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shewen P E, Wilkie B N. Vaccination of calves with leukotoxic culture supernatant from Pasteurella haemolytica. Can J Vet Res. 1988;52:30–36. [PMC free article] [PubMed] [Google Scholar]
- 46.Shoo M K, Wiseman A, Allan E M, Dalgleish R G, Gibbs H A, Al-Hendi A B, Selman I E. Distribution of Pasteurella haemolytica in the respiratory tracts of carrier calves and those subsequently infected experimentally with Dictyocaulus viviparus. Res Vet Sci. 1990;48:383–385. [PubMed] [Google Scholar]
- 47.Sreevatsan S, Ames T R, Werdin R E, Yoo H S, Maheswaran S K. Evaluation of three experimental subunit vaccines against pneumonic pasteurellosis in cattle. Vaccine. 1996;14:147–154. doi: 10.1016/0264-410x(95)00138-q. [DOI] [PubMed] [Google Scholar]
- 48.Sussman J K, Simons E L, Simons R W. Escherichia coli translation initiation factor 3 discriminates the initiation codon in vivo. Mol Microbiol. 1996;21:347–360. doi: 10.1046/j.1365-2958.1996.6371354.x. [DOI] [PubMed] [Google Scholar]
- 49.Virji M, Zak K, Heckels J E. Outer membrane protein III of Neisseria gonorrhoeae: variations in biological properties of antibodies directed against different epitopes. J Gen Microbiol. 1987;133:3393–3401. doi: 10.1099/00221287-133-12-3393. [DOI] [PubMed] [Google Scholar]